Abstract
Polycomb group protein enhancer of zeste homolog 2 (EZH2) is a methyltransferase that correlates with the regulation of invasion and metastasis and is overexpressed in human cancers such as colorectal cancer. MicroRNA-31 (miR-31) plays an oncogenic role and is associated with BRAF mutation and poor prognosis in colorectal cancer. EZH2 is functionally considered to suppress miR-31 expression in human cancers; however, no study has reported its relationship with colon cancer. We therefore evaluated EZH2 expression using immunohistochemistry and assessed miR-31 and epigenetic alterations using 301 colorectal carcinomas and 207 premalignant lesions. Functional analysis was performed to identify the association between EZH2 and miR-31 using cancer cell lines. In the current study, negative, weak, moderate, and strong EZH2 expressions were observed in 15%, 19%, 25%, and 41% of colorectal cancers, respectively. EZH2 was inversely associated with miR-31 (P < 0.0001), independent of clinicopathological and molecular features. In a multivariate stage-stratified analysis, high EZH2 expression was related to favorable prognosis (P = 0.0022). Regarding premalignant lesions, negative EZH2 expression was frequently detected in sessile serrated adenomas/polyps (SSA/Ps) (76%; P < 0.0001) compared with hyperplastic polyps, traditional serrated adenomas, and non-serrated adenomas (25–36%). Functional analysis demonstrated that the knockdown of EZH2 increased miR-31 expression. In conclusion, an inverse association was identified between EZH2 and miR-31 in colorectal cancers. Our data also showed that upregulation of EZH2 expression may be rare in SSA/Ps. These results suggest that EZH2 suppresses miR-31 in colorectal cancer and may correlate with differentiation and evolution of serrated pathway.
Keywords: EZH2, CIMP, EGFR, ChIP, H3K27me3
INTRODUCTION
A polycomb group protein, enhancer of zeste homolog 2 (EZH2), is a methyltransferase and the core catalytic element of polycomb repressive complex 2 (PRC2), which plays a critical role in the regulation of cancer initiation, progression, invasion, metastasis, and drug resistance [1-19]. Various oncogenic transcription factors and cancer-associated non-coding RNAs regulate EZH2 expression [1-4, 6, 9, 10, 16, 17, 20, 21]. Increased EZH2 activity induces the genomewide histone H3 lysine 27 trimethylation (H3K27me3) and may act as an oncogene via the repression of tumor suppressor genes in human cancers [1-16, 20, 22-24]. In gastrointestinal cancers, EZH2 overexpression has been shown in colorectal [8, 11-14], esophageal (squamous cell carcinoma) [1, 15], gastric [10, 17, 20, 25], pancreatic [5, 26], and bile duct cancers [4]. Associations have also been reported between EZH2 overexpression and poor prognosis in esophageal [15], gastric [25], pancreatic [26], and bile duct cancers [4]. In contrast, previous studies on colorectal cancer have reported associations between EZH2 overexpression and better prognosis [11-13].
MicroRNAs constitute a class of small non-coding RNA molecules that function as post-transcriptional gene regulators and have been increasingly recognized as useful biomarkers in various human cancers [1, 2, 6, 17, 20, 27-34]. Recent evidence has shown that microRNAs can act as both oncogenes and tumor suppressors, depending on the genes they regulate [6, 17]; for example, microRNA-31 (miR-31) is reportedly deregulated in human cancers [1, 2, 6, 16, 27, 28, 30, 35], and provides oncogenic potential in colorectal cancer [27-29, 31, 36]. Using microRNA array analysis, we recently identified that miR-31 expression was significantly upregulated in BRAF-mutated colorectal cancer compared with wild-type colorectal cancer [28]. Moreover, associations were identified between miR-31 expression and poor prognosis in colorectal cancer [28]. We also reported that high miR-31 expression may correlate with evolution of the serrated pathway [31, 37].
A recent study has reported that EZH2 suppresses miR-31 expression by inducing H3K27me3 on the miR-31 promoter and that the inhibition of EZH2 increased miR-31 expression in prostate cancer [2]. Furthermore, EZH2-mediated histone methylation has been shown to suppress miR-31 expression in adult T-cell leukemia [16]. With regard to melanoma, genetic and epigenetic loss of miR-31 produced a feed-forward EZH2 expression [6]. Thus, accumulating evidence indicates that EZH2 may downregulate miR-31 expression in human cancers; however, no study has reported the relationship between EZH2 and miR-31 in colorectal cancer.
We conducted this study to clarify the association of EZH2 expression with miR-31 and epigenetic alterations using a database comprising more than 500 colorectal tumors. Furthermore, we performed functional analyses to identify whether EZH2 suppressed miR-31 expression in colorectal cancers.
RESULTS
EZH2 expression in colorectal cancer tissue and matched normal mucosa
Using immunohistochemistry, we assessed 310 formalin-fixed paraffin-embedded (FFPE) specimens of colorectal cancer tissues in the EZH2 expression assay (Figure 1) and successfully obtained 301 (97%) valid results. We also examined matched samples of normal colorectal mucosa (controls). EZH2 expression scores of 0 (negative), 1 (weak), 2 (moderate), and 3 (strong) were observed in 15%, 19%, 25%, and 41% of the colorectal cancer tissues, respectively (Table 1). EZH2 expression in colorectal cancer tissues was significantly higher than its expression in normal mucosa tissues (P < 0.0001) (Supplementary Figure 1).
Figure 1. Immunohistochemical findings related to EZH2 expression in colorectal cancers.
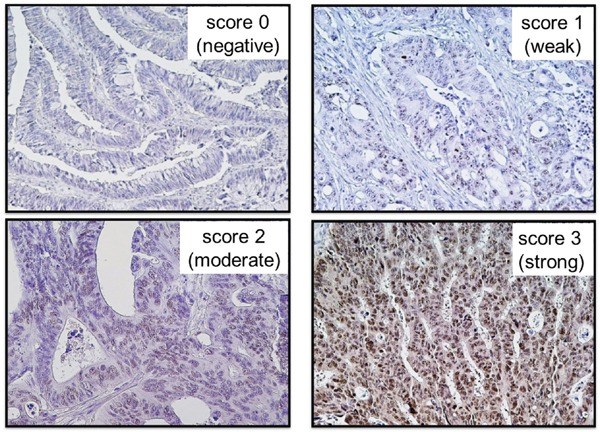
Score 0 (negative), score 1 (weak), score 2 (moderate), and score 3 (strong) EZH2 expressions were observed in 15%, 19%, 25%, and 41% of the 301 colorectal cancer tissues, respectively.
Table 1. Clinicopathological and molecular features of 301 colorectal cancers according to EZH2 expression.
| Clinicopathological or molecular feature | Total N | EZH2 expression | p | |||
|---|---|---|---|---|---|---|
| score 0 (negative) | score 1 (weak) | score 2 (moderate) | score 3 (strong) | |||
| All cases | 301 | 44 | 58 | 76 | 123 | |
| Gender | ||||||
| Male | 166 (55%) | 27 (61%) | 39 (67%) | 41 (54%) | 59 (48%) | 0.078 |
| Female | 135 (45%) | 17 (39%) | 19 (33%) | 35 (46%) | 64 (52%) | |
| Age (mean ± SD) | 65.5 ± 12.3 | 67.3 ± 11.3 | 65.3 ± 11.8 | 65.7 ± 11.4 | 64.8 ± 13.4 | 0.71 |
| Tumor size (mm) (mean ± SD) | 52.6 ± 25.0 | 58.5 ± 31.7 | 53.3 ± 24.9 | 51.5 ± 22.4 | 50.9 ± 24.0 | 0.37 |
| Year of diagnosis | ||||||
| Prior to 2003 | 155 (52%) | 29 (66%) | 32 (55%) | 39 (51%) | 55 (45%) | 0.16 |
| 2004-2006 | 79 (26%) | 6 (14%) | 17 (29%) | 21 (28%) | 35 (28%) | |
| Posterior to 2007 | 67 (22%) | 9 (20%) | 9 (16%) | 16 (21%) | 33 (27%) | |
| Tumor location | ||||||
| Rectum and Distal colon (splenic flexure to sigmoid) | 213 (71%) | 30 (68%) | 36 (62%) | 53 (70%) | 94 (76%) | 0.24 |
| Proximal colon (cecum to transverse) | 88 (29%) | 14 (32%) | 22 (38%) | 23 (30%) | 29 (24%) | |
| Tumor differentiation | ||||||
| Well to Moderate | 270 (90%) | 36 (82%) | 52 (90%) | 72 (95%) | 110 (89%) | 0.17 |
| Poor | 31 (10%) | 8 (18%) | 6 (10%) | 4 (5.3%) | 13 (11%) | |
| Disease stage | ||||||
| I | 8 (2.7%) | 0 (0%) | 1 (1.7%) | 2 (2.6%) | 5 (4.1%) | 0.30 |
| IIA | 34 (11%) | 7 (16%) | 5 (8.6%) | 9 (12%) | 13 (11%) | |
| IIB | 22 (7.3%) | 6 (14%) | 3 (5.2%) | 5 (6.6%) | 8 (6.5%) | |
| IIIA | 28 (9.3%) | 2 (4.6%) | 6 (10%) | 3 (4.0%) | 17 (14%) | |
| IIIB | 103 (34%) | 13 (30%) | 21 (36%) | 24 (32%) | 45 (37%) | |
| IIIC | 56 (19%) | 7 (16%) | 15 (26%) | 16 (21%) | 18 (15%) | |
| IV | 50 (17%) | 9 (20%) | 7 (12%) | 17 (22%) | 17 (14%) | |
| BRAF mutation | ||||||
| Wild-type | 288 (96%) | 42 (95%) | 52 (90%) | 74 (97%) | 120 (98%) | 0.13 |
| Mutant | 13 (4.3%) | 2 (4.6%) | 6 (10%) | 2 (2.6%) | 3 (2.4%) | |
| KRAS mutation | ||||||
| Wild-type | 204 (68%) | 23 (52%) | 42 (72%) | 49 (64%) | 90 (73%) | 0.066 |
| Mutant | 97 (32%) | 21 (48%) | 16 (28%) | 27 (36%) | 33 (27%) | |
| PIK3CA mutation | ||||||
| Wild-type | 267 (89%) | 40 (91%) | 49 (84%) | 67 (88%) | 111 (90%) | 0.68 |
| Mutant | 34 (11%) | 4 (9.1%) | 9 (16%) | 9 (12%) | 12 (9.8%) | |
| CIMP status | ||||||
| CIMP-low/zero | 283 (94%) | 39 (89%) | 54 (93%) | 75 (99%) | 115 (94%) | 0.10 |
| CIMP-high | 18 (6.0%) | 5 (11%) | 4 (6.9%) | 1 (1.3%) | 8 (6.5%) | |
| MSI status | ||||||
| MSS/MSI-low | 279 (93%) | 40 (91%) | 55 (95%) | 72 (95%) | 112 (91%) | 0.66 |
| MSI-high | 22 (7.3%) | 4 (9.1%) | 3 (5.2%) | 4 (5.3%) | 11 (8.9%) | |
| miR-31 expression | ||||||
| Low expression (Q1, Q2 and Q3) | 226 (75%) | 20 (45%) | 36 (62%) | 60 (79%) | 110 (89%) | < 0.0001 |
| High expression (Q4) | 75 (25%) | 24 (55%) | 22 (38%) | 16 (21%) | 13 (11%) | |
Percentage (%) indicates the proportion of cases with a specific clinicopathological or molecular feature within a given category (score 0, score 1, score 2, score 3) of EZH2 expression by immunohistochemistry. P-values were calculated by analysis of variance for age and tumor size and by a chi-square test or Fisher's exact test for all other variables. To account for multiple hypothesis testing in associations between EZH2 expression and other 13 covariates, the P-value for significance was adjusted by Bonferroni correction to P = 0.0038 (= 0.05/13).
CIMP, CpG island methylator phenotype; MSI, microsatellite instability; MSS, microsatellite stable; miR-31, microRNA-31; SD, standard deviation.
MicoroRNA-31 expression in colorectal cancer
The distributions of miR-31 expression in the 301 colorectal cancers were as follows: mean 55.4; median 10.7; standard deviation (SD) 211; range 0.11–2108; interquartile range 3.9–32.2 (Supplementary Figure 2). Cases with miR-31 expression were divided into quartiles Q1 (<3.9), Q2 (3.9–10.6), Q3 (10.7–32.1), and Q4 (≥32.2) for further analysis.
The association between EZH2 expression and clinical, pathological and molecular characteristics in colorectal cancer
Table 1 summarises the clinical features of all 301 patients with colorectal cancer according to EZH2 expression. No significant association existed between EZH2 expression and the clinical or pathological characteristics such as gender, age, tumor size, year of diagnosis, tumor location, tumor differentiation and disease stage; BRAF, KRAS, and PIK3CA mutations; and the MSI (microsatellite instability) and CpG island methylator phenotype (CIMP) status (Table 1). CIMP-specific promoter methylation (CACNA1G, IGF2, MLH1, or RUNX3) was not associated with EZH2 expression (data not shown). However, EZH2 expression was inversely associated with miR-31 expression (P < 0.0001) (Table 1 and Figure 2).
Figure 2. The association between EZH2 expression and microRNA-31 expression in 301 colorectal cancers.
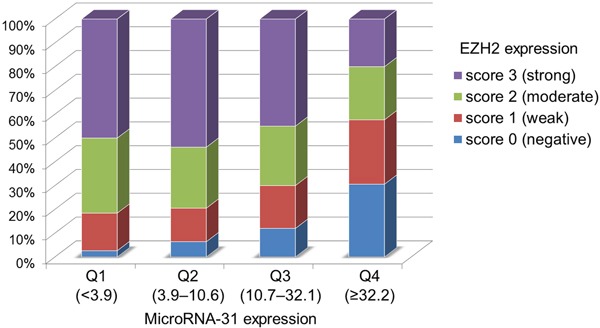
EZH2 expression levels were inversely associated with microRNA-31 expressions in colorectal cancers (P < 0.0001).
EZH2 expression and patient survival
The influence of EZH2 expression on clinical outcomes was assessed in 299 patients with colorectal cancer (stages I–IV). During follow-up among eligible patients, 100 patients died, of which 81 deaths were attributed to colorectal cancer. The median follow-up time for censored patients was 4.4 years. Kaplan–Meier analysis was performed using categorical variables (score 0, score 1, score 2, or score 3). In terms of cancer-specific survival, significantly lower mortality was observed in patients with high EZH2 expression (log-rank test: P = 0.010) than in those with low EZH2 expression (Figure 3A).
Figure 3. Kaplan–Meier survival curves for colorectal cancer (stages I-IV) according to the EZH2 expression level.
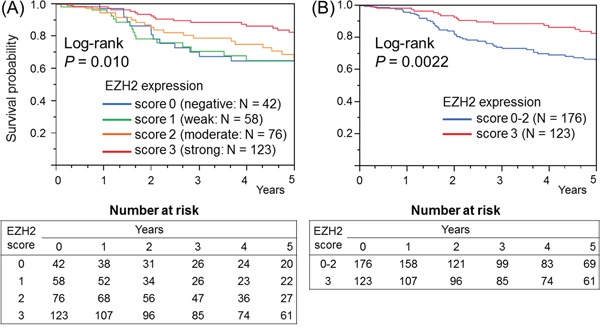
A. In terms of cancer-specific survival, significantly lower mortality was observed in patients with high EZH2 expression than in those with low EZH2 expression (log-rank test: P = 0.010). B. In terms of cancer-specific survival, significantly lower mortality was observed in patients with high expression (score 3) than in those with low expression (score 0–2) (log-rank test: P = 0.0022).
We made a dichotomous expression variable for EZH2, defining a score of 3 as high expression and scores of 0–2 as low expression. In terms of cancer-specific survival, significantly lower mortality was observed in patients with high expression (log-rank test: P = 0.0022) than in those with low expression (Figure 3B). However, no significant differences were observed between the high and low expression groups when we defined scores of 1–3 as high expression and score of 0 as low expression (log-rank test: P = 0.69) or when we defined scores of 2–3 as high expression and scores of 0–1 as low expression (P = 0.075).
In univariate Cox regression analysis for cancer-specific survival, significantly lower mortality was observed in the high expression group (score 3) compared with the low expression group (score 0–2) [hazard ratio (HR): 0.48; 95% confidence interval (CI): 0.29–0.77; P = 0.0018] (Supplementary Table 1). Similarly, in comparison with the low expression group, an independent association with a favorable prognosis was observed in the high expression group in both stage-stratified (HR: 0.51; 95% CI: 0.29−0.77; P = 0.0022) and multivariate analyses (HR: 0.46; 95% CI: 0.27−0.76; P = 0.0022) (Supplementary Table 1).
Multivariate logistic regression analysis in high EZH2 expression group
Considering potential confounders and cause-and-effect sequences, we performed multivariate logistic regression analyses to assess the relationships with EZH2 expression. In the final model, high EZH2 expression (score 3) was inversely associated with high miR-31 expression (Q4) [odds ratio (OR): 0.22; 95% CI: 0.11–0.42; P < 0.0001] (Table 2).
Table 2. Multivariate logistic regression analysis of EZH2 expression in colorectal cancers.
| Variables in the final model for EZH2 expression (as an outcome variable) [High expression group (score 3) vs. Low expression group (score 0-2)] |
Adjusted odds ratio (95% CI) | P |
|---|---|---|
| High microRNA-31 expression (vs. Low expression) | 0.22 (0.11-0.42) | < 0.0001 |
| Female gender (vs. Male) | 1.78 (1.08-2.98) | 0.025 |
A multivariate logistic regression analysis assessing the relationships with EZH2 expression status initially included gender, age, tumor size, year of diagnosis, tumor location, tumor differentiation, disease stage, CIMP status, MSI status, mutations of BRAF, KRAS and PIK3CA, and microRNA-31 expression, considering potential confounding and causal relationships. For multiple hypothesis testing, the P-value for significance was adjusted by Bonferroni correction to 0.0038 (= 0.05/13).
CI, confidence interval; CIMP, CpG island methylator phenotype; MSI, microsatellite instability.
Association of EZH2 expression with clinicopathological and molecular features in premalignant colorectal lesions
We assessed 215 FFPE tissue specimens from premalignant colorectal lesions using immunohistochemistry (Supplementary Figure 3) and successfully obtained 207 (96%) valid results. Table 3 shows the clinicopathological and molecular features, including EZH2 expression in serrated lesions and non-serrated adenomas. Negative EZH2 expression (score 0) was frequently detected in sessile serrated adenomas/polyps (SSA/Ps) (76%) compared with hyperplastic polyps (HPs) (36%), traditional serrated adenomas (TSAs) (25%), and non-serrated adenomas (36%). EZH2 expression [score 2 (moderate) or score 3 (strong)] was not detected in any SSA/Ps.
Table 3. Clinicopathological and molecular features of 152 serrated lesions and 55 non-serrated adenomas.
| Clinicopathological or molecular feature | Total N | Histological type | p | |||
|---|---|---|---|---|---|---|
| Hyperplastic polyp (HP) | Sessile serrated adenoma/polyp (SSA/P) | Traditional serrated adenoma (TSA) | Non-serrated adenoma | |||
| All cases | 207 | 50 | 51 | 51 | 55 | |
| Gender | ||||||
| Male | 124 (60%) | 36 (72%) | 23 (45%) | 26 (51%) | 39 (71%) | 0.0065 |
| Female | 83 (40%) | 14 (28%) | 28 (55%) | 25 (49%) | 16 (29%) | |
| Age (mean ± SD) | 61.7 ± 10.8 | 58.9 ± 11.6 | 57.4 ± 10.8 | 65.1 ± 11.5 | 65.3 ± 8.7 | < 0.0001 |
| Tumor size (mm) (mean ± SD) | 11.0 ± 5.3 | 9.0 ± 3.7 | 13.5 ± 6.7 | 9.9 ± 4.7 | 11.4 ± 5.4 | 0.0002 |
| Tumor location | ||||||
| Rectum and Distal colon | 98 (47%) | 24 (48%) | 5 (9.8%) | 38 (75%) | 31 (56%) | < 0.0001 |
| Proximal colon | 109 (53%) | 26 (52%) | 46 (90%) | 13 (25%) | 24 (44%) | |
| BRAF mutation | ||||||
| Wild-type | 107 (52%) | 27 (54%) | 7 (14%) | 19 (37%) | 54 (98%) | < 0.0001 |
| Mutant | 100 (48%) | 23 (46%) | 44 (86%) | 32 (63%) | 1 (1.8%) | |
| CIMP status | ||||||
| CIMP-low/zero | 174 (84%) | 46 (92%) | 32 (63%) | 41 (80%) | 55 (100%) | < 0.0001 |
| CIMP-high | 33 (16%) | 4 (8.0%) | 19 (37%) | 10 (20%) | 0 (0%) | |
| MicroRNA-31 | ||||||
| Low expression (Q1-3) | 155 (75%) | 39 (78%) | 40 (78%) | 27 (53%) | 49 (89%) | < 0.0001 |
| High expression (Q4) | 52 (25%) | 11 (22%) | 11 (22%) | 24 (47%) | 6 (11%) | |
| EZH2 expression | ||||||
| Score 0 (negative) | 90 (43%) | 18 (36%) | 39 (76%) | 13 (25%) | 20 (36%) | < 0.0001 |
| Score 1 (weak) | 83 (40%) | 20 (40%) | 12 (24%) | 23 (45%) | 28 (51%) | |
| Score 2 (moderate) | 34 (16%) | 12 (24%) | 0 (0%) | 15 (29%) | 7 (13%) | |
| Score 3 (strong) | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) | |
P-values were calculated by analysis of variance for age and tumor size, and by a chi-square test for all other variables. CIMP, CpG island methylator phenotype; HP, hyperplastic polyp; SSA/P, sessile serrated adenoma/polyp; SD, standard deviation; TSA, traditional serrated adenoma.
Knockdown of EZH2 increases miR-31 expression in colon cancer cell lines
To examine whether EZH2 suppressed miR-31 expression, we knocked down EZH2 mRNA by siRNAs in colon cancer cell lines and measured the resulting miR-31 expression. Figure 4A shows the expression level of EZH2 in 7 colon cancer cell lines using quantitative reverse transcription-PCR (qRT-PCR) (Figure 4A). Our data demonstrated that in RKO cells, EZH2 expression was successfully downregulated by approximately 30% and 56% when transfecting two different EZH2 siRNAs (siEZH2_7644 and siEZH2_7882, respectively) (Figure 4B). Moreover, we found that there was a considerable increase in miR-31 expression in RKO cells transfected with EZH2 siRNAs (Figure 4C). Similar results were observed in HT29 and SW480 cells (Supplementary Figure 4).
Figure 4. EZH2 knockdown caused microRNA-31 (miR-31) overexpression on quantitative RT-PCR.
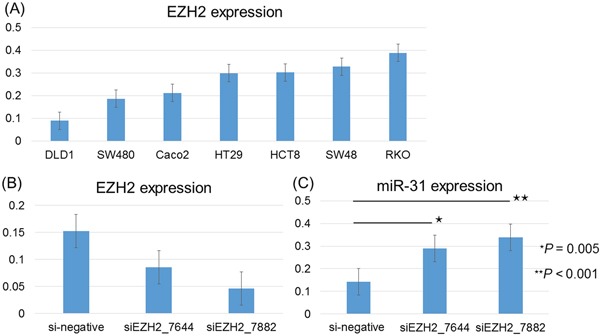
A. EZH2 expression is shown according to each of the 7 colorectal cancer cell lines studied. Error bars represent the standard deviations. B. The siRNA-mediated knockdown of EZH2 caused a significant reduction in EZH2 expression in RKO cells transfected with EZH2 siRNAs (siEZH2_7644 and siEZH2_7882). C. There was a considerable increase in miR-31 expression in RKO cells transfected with EZH2 siRNAs (siEZH2_7644 and siEZH2_7882). The P-value was analyzed using paired T-test.
Promoter region of miR-31 is marked by histone H3 lysine 27 trimethylation (H3K27me3)
To clarify the functional link between EZH2 and miR-31 expression, we examined H3K27me3 levels around the promoter region of miR-31 by performing chromatin immunoprecipitation (ChIP) assay. We observed that H3K27me3 was steadily enriched at the promoter region of miR-31 in RKO cells, and the H3K27me3 levels were decreased after knockdown of EZH2 (Supplementary Figure 5). Taken together with increased expression of miR-31 after EZH2 knockdown (Figure 4C), these results suggest the expression of miR-31 is suppressed by EZH2 through H3K27me3 in RKO cells.
DISCUSSION
We performed this study to identify the association of EZH2 expression with molecular alterations in colorectal tumors. In a database comprising 301 patients with colorectal cancer, high EZH2 expression was inversely associated with miR-31 expression, independent of clinicopathological and molecular features. Our data also showed that high EZH2 expression was a favorable prognostic factor in colorectal cancer. Regarding premalignant lesions, negative EZH2 expression was higher in SSA/Ps than in HPs, TSAs or non-serrated adenomas. In functional analysis, we showed that the siRNA knockdown of EZH2 increased miR-31 expression in colon cancer cell lines.
EZH2 reportedly downregulates miR-31 expression in human cancers such as prostate cancer [2] and adult T-cell leukemia [16]. However, no study has reported the potential role of EZH2 in the regulation of miR-31 expression in colorectal cancer. The present multivariate analysis showed that high EZH2 expression was inversely associated with miR-31 expression in colorectal cancer, whereas functional analysis showed that EZH2 knockdown increased miR-31 expression in colon cancer cell lines. Recent studies that used colon cancer cell lines have reported that EZH2 expression was inversely associated with microRNA-506 [38], microRNA-26a, and let-7b [39]; these microRNAs downregulated EZH2 expression by directly targeting 3′-UTR. Conversely, EZH2 has been reported to suppress miR-31 expression by inducing H3K27me3 on the miR-31 promoter in prostate cancer. Therefore, to determine if EZH2 is involved in the regulation of miR-31 expression through histone H3K27me3, we examined H3K27me3 levels around the transcription start site on the miR-31 promoter by performing ChIP assay and found that EZH2-mediated histone methylation downregulates miR-31 expression in colon cancer. To the best of our knowledge, this is the first report describing the downregulation of miR-31 by EZH2 in colorectal cancer. Recently, we also reported that high miR-31 expression was associated with shorter progression-free survival in patients with colorectal cancer treated using anti-epidermal growth factor receptor (EGFR) therapy [27]. Therefore, because of the relationship with miR-31, EZH2 may represent a new prognostic biomarker for molecular targeted therapies and can provide a promising therapeutic target in patients with colorectal cancer.
With regard to the association between EZH2 expression and outcomes in patients with colorectal cancer, previous studies have reported that EZH2 overexpression was associated with a favorable prognosis [11-13]. Our data also showed that high EZH2 expression was associated with favorable survival using multivariate stage-stratified Cox analysis. These results are reasonable because we recently reported that high miR-31 expression, which is inversely correlated with EZH2 expression, is an unfavorable prognostic factor in patients with colorectal cancer [28]. We summarized the association of EZH2 expression with miR-31 and cancer-specific survival in Supplementary Figure 6. Our current study had some important limitations, particularly because of its cross-sectional nature and unknown bias (i.e. selection bias). Nevertheless, our multivariate regression analysis was adjusted for potential confounders, including clinical and molecular features, and we were able to show that high EZH2 expression was inversely associated with miR-31 expression and associated with better prognosis.
CIMP is a distinct form of epigenomic instability that causes most cases of sporadic MSI-high colorectal cancers through the epigenetic inactivation of MLH1 [30, 31, 40-44]. RUNX3 belongs to the RUNX family of genes, which has been reported to be the single best marker for diagnosing CIMP-high status [41]. Previous studies have reported that EZH2 downregulates RUNX3 by inducing histone H3 methylation in various human cancers such as gastric [10], bile duct [4], and pancreatic cancers [5]. With regard to colorectal cancer, Kodach et al. reported, that despite higher levels of EZH2 and lower levels of RUNX3, no inverse correlation was present between EZH2 and RUNX3 [9]. Consistent with those results, when using 4 CIMP-specific promoters (CACNA1G, IGF2, MLH1, and RUNX3), we showed that neither RUNX3 methylation nor CIMP-high status was associated with EZH2 expression. These results suggest that EZH2 overexpression may not correlate with the epigenetic silencing of CIMP-specific promoters by histone H3 methylation in colorectal cancer.
Premalignant colorectal neoplasms appear to be important precursor lesions in the pathogenesis of colorectal cancer. In particular, the serrated neoplasia pathway has attracted considerable attention as an alternative pathway of colorectal cancer development, and serrated lesions exhibit unique clinicopathological and molecular features [45-48]. Both SSA/Ps and TSAs are recognized premalignant lesions, but SSA/Ps are the principle serrated precursors of colorectal cancer [47-50]. Because there are many clinicopathological and molecular similarities between SSA/Ps and CIMP-high colorectal cancers, including proximal tumor location, BRAF mutation, and MLH1 methylation, SSA/Ps are hypothesized to be precursor lesions that develop into CIMP-high colorectal cancers [31, 42, 46, 50, 51].
We recently reported that high miR-31 expression was more pronounced in SSA/Ps with cytological dysplasia than in other SSA/Ps, but we did not find a significant difference between TSAs with high-grade dysplasia and other TSAs [31]. These data imply that miR-31 correlate with the progression of SSA/Ps. However, our current data revealed that EZH2 expression (moderate or strong) was not detected in any SSA/Ps and that negative EZH2 expression was higher in SSA/Ps than in HPs, TSAs, and non-serrated adenomas. In addition, we demonstrated that the knockdown of EZH2 by siRNAs increased miR-31 expression in colon cancer cell lines, suggesting that negative EZH2 expression causes miR-31 upregulation in the progression of SSA/Ps. Therefore, EZH2 may be a key molecule in the differentiation and evolution of serrated lesions, and microvesicular HPs without EZH2 expression may progress to SSA/Ps. Future independent studies are required to clarify the role of EZH2 expression in the serrated pathway.
In conclusion, we identified an inverse association between the expressions of EZH2 and miR-31 in colorectal cancer and that the upregulation of EZH2 expression may be a rare event in SSA/Ps. Hence, we suggest that EZH2 suppresses miR-31 expression in colorectal cancer and may correlate with differentiation and evolution of the serrated pathway. These findings improve our understanding of the mechanism of colorectal tumorigenesis and have the potential to significantly affect clinical and translational research on colorectal cancer.
MATERIALS AND METHODS
Patients and tissue specimens
Formalin-fixed paraffin-embedded (FFPE) tissues of 310 colorectal cancers (stages I–IV), 158 serrated lesions and 57 non-serrated adenomas (i.e. tubular or tubulovillous adenomas) of patients who underwent endoscopic resection or other surgical treatment at Sapporo Medical University Hospital and Keiyukai Sapporo Hospital between 1999 and 2014 were collected. To avoid selection bias as much as possible, we consecutively collected FFPE tissue specimens of colorectal cancers, serrated lesions, and non-serrated adenomas.
The criterion for diagnosis of colorectal cancer was invasion of malignant cells beyond the muscularis mucosa. Intramucosal carcinoma and carcinoma in situ were classified as adenoma. Colorectal tumors were classified by location as follows: the proximal colon (cecum, ascending, and transverse colon), distal colon (splenic flexure, descending, and sigmoid colon) and rectum. The patients were followed until death or December 2014, whichever came first.
Serrated lesions [hyperplastic polyps (HPs) (N = 54), sessile serrated adenomas/polyps (SSA/Ps) (N = 53) and traditional serrated adenomas (TSAs) (N = 51)] were classified on the basis of the current World Health Organization (WHO) criteria [52]. All HPs were found to be microvesicular. Informed consent was obtained from all the patients before specimen collection. This study was approved by the respective institutional review boards of the participating institutions.
RNA extraction and qRT-PCR of microRNA-31
Total RNA was extracted from FFPE tissues using the miRNeasy FFPE Kit (Qiagen, Valencia, CA, USA) [28]. MicroRNA-31 (miR-31)-5p expression was analyzed by qRT-PCR using TaqMan MicroRNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA, USA) and TaqMan microRNA Assays (Applied Biosystems) as described previously [28]. U6 snRNA (RNU6B; Applied Biosystems) served as an endogenous control. miR-31 expression was calculated using the equation 2−ΔCT, where ΔCT = (CT miR-31 − CT U6). To calculate the relative expression of miR-31 in each colorectal cancer, 2−ΔCT of cancer tissue was divided by 2−ΔCT of normal tissue, as described previously [28].
DNA extraction and pyrosequencing of KRAS, BRAF, and PIK3CA mutations and MSI analysis
Genomic DNA was extracted from FFPE tissues of colorectal tumors using QIAamp DNA FFPE Tissue Kit (Qiagen) [28]. Using extracted genomic DNA, PCR and targeted pyrosequencing were performed for KRAS (codon 12 or 13) [27], BRAF (codon 600) [28], and PIK3CA (exon 9 or 20) [40]. MSI analysis was performed as described previously [28].
Sodium bisulfite treatment and real-time PCR (MethyLight) to measure promoter methylations of CACNA1G, IGF2, MLH1, and RUNX3
Bisulfite modification of genomic DNA was performed using a BisulFlash™ DNA Modification Kit (Epigentek, Brooklyn, NY, USA) [28]. We quantified DNA methylation in 4 CIMP-specific promoters (CACNA1G, IGF2, MLH1, and RUNX3) by real-time PCR (MethyLight), as described previously [30, 31, 41]. CIMP-high was defined as the presence of three/four or more methylated promoters and CIMP low/zero as zero/four to two/four methylated promoters [31].
Immunohistochemistry for EZH2 expression
Immunohistochemistry was performed on 5 μm FFPE sections. Sections were autoclave-pretreated in target retrieval solution (Dako Cytomation, Carpinteria, CA, USA). Endogenous peroxidase activity was blocked using 3% hydrogen peroxide, and the sections were incubated overnight at 4°C with anti-EZH2 antibody (#612667, BD Biosciences, San Jose, CA, USA) at a dilution of 1:100. A subsequent reaction was performed using a horseradish peroxidase enzyme-labeled polymer of the EnVision™ Plus detection system (Dako). When a positive reaction was visualized using a diaminobenzidine (DAB) solution, counterstaining was performed using Mayer's hematoxylin. Five random high-power fields were evaluated per lesion to determine the mean nuclear positivity, which was categorized as follows: score 0 (negative, <5%), score 1 (weak, 5%–39%), score 2 (moderate, 40%–79%), or score 3 (strong, ≥80%). EZH2 expression was visually interpreted by H.K., who was unaware of the other data. For the agreement study on EZH2 expression, 147 randomly selected cases were examined by a second pathologist (by K.N.), who was also unaware of the other data. The concordance between the two pathologists (P < 0.0001) was 0.87 (κ = 0.74), indicating substantial agreement.
Colon cancer cell line and small interfering RNA (siRNA) transfection
In this study, we used 7 colon cancer cell lines (Caco2, DLD1, HCT8, HT29, RKO, SW48, and SW480). Twenty-four hours after plating, the cells were transfected with either negative control siRNA (Sigma-Aldrich, St. Louis, MO, USA) or two different EZH2-targeting siRNAs (Sigma-Aldrich; siRNA ID SASI_Hs01_00147882, SASI_Hs02_00337644) using the Lipofectamine® RNAiMax (Invitrogen by Life Technologies, Carlsbad, CA, USA). Fourty-eight hours after transfection, the cells were harvested for qRT-PCR.
RNA isolation and quantitative RT-PCR
Total RNA was extracted from cell pellets using the TRIzol® reagent (Invitrogen by Life Technologies) and reverse transcribed to cDNA with the PrimeScript™ RT Reagent Kit (Takara Bio Inc., Kusatsu, Japan). Quantitative RT-PCR was performed using the TaqMan® universal master mix with specific primers and a probe set for EZH2 (Applied Biosystems; TaqMan Gene Expression Assay). Actin-beta (ACTB) expression was used to normalize for variance. All genes were tested in triplicates.
Chromatin immunoprecipitation (ChIP)-PCR
ChIP was performed as described previously [35] with minor modifications. Briefly, 1 × 106 cells were fixed in 1% formaldehyde for 10 min at room temperature, rinsed in glycine, then washed in cold phosphate buffered saline (PBS) twice. The cells were re-suspended in SDS lysis buffer and were sonicated using a Covaris S2 device (Covaris Inc., Woburn, MA, USA), following the manufacturers' instructions. Sheared chromatin was immunoprecipitated for more than eight hours at 4°C using 2 μg of anti-H3K27me3 antibody (#9733; Cell Signaling Technology, Danvers, MA, USA). Before adding antibodies, 10 μl of sheared chromatin was saved as input DNA sample. After washing, elution and reversal of the cross-links, DNA was purified using Agencourt AMPure XP® (Agencourt Biosciences, Beverly, MA, USA), according to the product manual. The purified DNA was amplified by real-time quantitative PCR with SYBR® Select Master Mix (Life Technologies) and 7500 fast real-time PCR system (Life Technologies) and was analysed for enrichment. The sequences of the ChIP primers are provided in Supplementary Table 2.
Statistical analysis
JMP (version 10) and SAS (version 9) software programs were used for statistical analyses (SAS Institute, Cary, NC, USA). All P-values were two-sided. Univariate analyses were performed to investigate clinicopathological and molecular characteristics according to the EZH2 expression level; a chi-square test or Fisher's exact test was used for categorical data, while analysis of variance was used to compare the mean patient age and tumor size. To account for multiple hypothesis testing in associations between EZH2 expression and other 13 covariates, the P-value for significance was adjusted by Bonferroni correction to P = 0.0038 (= 0.05/13).
In survival analysis, the Kaplan–Meier method and log-rank test were used to assess the survival time distribution. Cox proportional hazards regression models were used to compute mortality HRs according to the EZH2 expression status. Stratification by the tumor-node-metastasis (TNM) disease stage (I, IIA, IIB, IIIA, IIIB, IIIC and IV) was performed using the “strata” option in the SAS “proc phreg” command. The multivariate, stage-stratified Cox model included the EZH2 expression variable stratified by gender (male vs. female), age at diagnosis (continuous), tumor size (continuous), year of diagnosis (continuous), tumor location (proximal colon vs. distal colon and rectum), tumor differentiation (well to moderate vs. poor), MSI status (MSI-high vs. MSS/MSI-low), CIMP status (CIMP-high vs. CIMP-low/zero), mutations of BRAF, KRAS and PIK3CA (present vs. absent), and miR-31 (high expression vs. low expression). A backward elimination was performed with a threshold of P = 0.10, to avoid overfitting.
A multivariate logistic regression analysis assessing the relationships with EZH2 expression status initially included gender, age, tumor size, year of diagnosis, tumor location, tumor differentiation, disease stage, MSI, CIMP, mutations of BRAF, KRAS and PIK3CA, and miR-31, considering potential confounding and causal relationships. For multiple hypothesis testing, the P-value for significance was adjusted by Bonferroni correction to 0.0038 (= 0.05/13).
SUPPLEMENTARY FIGURES AND TABLE
Acknowledgments
We thank the pathology departments of Sapporo Medical University Hospital and Keiyukai Sapporo Hospital for providing the tissue specimens.
Abbreviations
- ChIP
chromatin immunoprecipitation
- CI
confidence interval
- CIMP
CpG island methylator phenotype
- EGFR
epidermal growth factor receptor
- EZH2
enhancer of zeste homolog 2
- FFPE
formalin-fixed paraffin-embedded
- HP
hyperplastic polyp
- HR
hazard ratio
- miR-31
microRNA-31
- MSI
microsatellite instability
- MSS
microsatellite stable
- PRC2
polycomb repressive complex 2
- qRT-PCR
quantitative reverse transcription-PCR
- SD
standard deviation
- siRNA
small interfering RNA
- SSA/P
sessile serrated adenoma/polyp
- TSA
traditional serrated adenoma
- H3K27me3
histone H3 lysine 27 trimethylation
- WHO
World Health Organization.
Footnotes
CONFLICTS OF INTEREST
The authors declare no conflicts of interest.
GRANT SUPPORT
This work was supported by The Japanese Society of Gastroenterology Research Foundation (to K.N.), Takeda Science Foundation (to K.N.), Daiwa Securities Health Foundation (to K.N.), Kobayashi Foundation for Cancer Research (to K.N.), Ono Cancer Research Foundation (to M.I.), Sapporo Jikeikai Tomoiki Foundation (to S.K.), and The Japan Society for the Promotion of Science (JSPS) Grant-in-Aid for Scientific Research (grant number 15K08954 to Y.S.).
Statement of author contributions
Study concept and design: H.K., K.N. Data acquisition: K.M., S.K., I.Y., K.I., M.I., H.I. Data analysis and interpretation: H.K., K.N., H.I., R.M., K.I. Drafting of the manuscript: H.K., R.M., K.N. Critical revision of the manuscript for important intellectual content: K.N., H.Y., Y.S. Statistical analysis: K.N., H.K., R.M., Y.S. Material support: T.T., K.O., T.H. Study supervision: K.N., H.K., K.I., Y.S., H.Y. Final approval of manuscript: all authors.
REFERENCES
- 1.Koumangoye RB, Andl T, Taubenslag KJ, Zilberman ST, Taylor CJ, Loomans HA, Andl CD. SOX4 interacts with EZH2 and HDAC3 to suppress microRNA-31 in invasive esophageal cancer cells. Mol Cancer. 2015;14:24. doi: 10.1186/s12943-014-0284-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang Q, Padi SK, Tindall DJ, Guo B. Polycomb protein EZH2 suppresses apoptosis by silencing the proapoptotic miR-31. Cell Death Dis. 2014;5:e1486. doi: 10.1038/cddis.2014.454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yamaguchi H, Hung MC. Regulation and Role of EZH2 in Cancer. Cancer Res Treat. 2014;46:209–222. doi: 10.4143/crt.2014.46.3.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tang B, Du J, Li Y, Tang F, Wang Z, He S. EZH2 elevates the proliferation of human cholangiocarcinoma cells through the downregulation of RUNX3. Med Oncol. 2014;31:271. doi: 10.1007/s12032-014-0271-6. [DOI] [PubMed] [Google Scholar]
- 5.Fujii S, Fukamachi K, Tsuda H, Ito K, Ito Y, Ochiai A. RAS oncogenic signal upregulates EZH2 in pancreatic cancer. Biochem Bioph Res Co. 2012;417:1074–1079. doi: 10.1016/j.bbrc.2011.12.099. [DOI] [PubMed] [Google Scholar]
- 6.Asangani IA, Harms PW, Dodson L, Pandhi M, Kunju LP, Maher CA, Fullen DR, Johnson TM, Giordano TJ, Palanisamy N, Chinnaiyan AM. Genetic and epigenetic loss of microRNA-31 leads to feed-forward expression of EZH2 in melanoma. Oncotarget. 2012;3:1011–1025. doi: 10.18632/oncotarget.622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fujii S, Tokita K, Wada N, Ito K, Yamauchi C, Ito Y, Ochiai A. MEK-ERK pathway regulates EZH2 overexpression in association with aggressive breast cancer subtypes. Oncogene. 2011;30:4118–4128. doi: 10.1038/onc.2011.118. [DOI] [PubMed] [Google Scholar]
- 8.Wang CG, Ye YJ, Yuan J, Liu FF, Zhang H, Wang S. EZH2 and STAT6 expression profiles are correlated with colorectal cancer stage and prognosis. World J Gastroenterol. 2010;16:2421–2427. doi: 10.3748/wjg.v16.i19.2421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kodach LL, Jacobs RJ, Heijmans J, van Noesel CJ, Langers AM, Verspaget HW, Hommes DW, Offerhaus GJ, van den Brink GR, Hardwick JC. The role of EZH2 and DNA methylation in the silencing of the tumour suppressor RUNX3 in colorectal cancer. Carcinogenesis. 2010;31:1567–1575. doi: 10.1093/carcin/bgq147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fujii S, Ito K, Ito Y, Ochiai A. Enhancer of zeste homologue 2 (EZH2) down-regulates RUNX3 by increasing histone H3 methylation. J Biol Chem. 2008;283:17324–17332. doi: 10.1074/jbc.M800224200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fluge O, Gravdal K, Carlsen E, Vonen B, Kjellevold K, Refsum S, Lilleng R, Eide TJ, Halvorsen TB, Tveit KM, Otte AP, Akslen LA, Dahl O. Expression of EZH2 and Ki-67 in colorectal cancer and associations with treatment response and prognosis. Br J Cancer. 2009;101:1282–1289. doi: 10.1038/sj.bjc.6605333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Benard A, Goossens-Beumer IJ, van Hoesel AQ, Horati H, Putter H, Zeestraten EC, van de Velde CJ, Kuppen PJ. Prognostic value of polycomb proteins EZH2, BMI1 and SUZ12 and histone modification H3K27me3 in colorectal cancer. PLoS One. 2014;9:e108265. doi: 10.1371/journal.pone.0108265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ishikawa S, Hayashi H, Kinoshita K, Abe M, Kuroki H, Tokunaga R, Tomiyasu S, Tanaka H, Sugita H, Arita T, Yagi Y, Watanabe M, Hirota M, Baba H. Statins inhibit tumor progression via an enhancer of zeste homolog 2-mediated epigenetic alteration in colorectal cancer. Int J Cancer. 2014;135:2528–2536. doi: 10.1002/ijc.28672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Meng X, Huang Z, Wang R, Jiao Y, Li H, Xu X, Feng R, Zhu K, Jiang S, Yan H, Yu J. The prognostic role of EZH2 expression in rectal cancer patients treated with neoadjuvant chemoradiotherapy. Radiat Oncol. 2014;9:188. doi: 10.1186/1748-717X-9-188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.He LR, Liu MZ, Li BK, Jia WH, Zhang Y, Liao YJ, Chen YC, Zhang LJ, Guan XY, Zeng YX, Kung HF, Xie D. High expression of EZH2 is associated with tumor aggressiveness and poor prognosis in patients with esophageal squamous cell carcinoma treated with definitive chemoradiotherapy. Int J Cancer. 2010;127:138–147. doi: 10.1002/ijc.25031. [DOI] [PubMed] [Google Scholar]
- 16.Yamagishi M, Nakano K, Miyake A, Yamochi T, Kagami Y, Tsutsumi A, Matsuda Y, Sato-Otsubo A, Muto S, Utsunomiya A, Yamaguchi K, Uchimaru K, Ogawa S, Watanabe T. Polycomb-mediated loss of miR-31 activates NIK-dependent NF-kappaB pathway in adult T cell leukemia and other cancers. Cancer cell. 2012;21:121–135. doi: 10.1016/j.ccr.2011.12.015. [DOI] [PubMed] [Google Scholar]
- 17.Chen DL, Zhang DS, Lu YX, Chen LZ, Zeng ZL, He MM, Wang FH, Li YH, Zhang HZ, Pelicano H, Zhang W, Xu RH. microRNA-217 inhibits tumor progression and metastasis by downregulating EZH2 and predicts favorable prognosis in gastric cancer. Oncotarget. 2015;6:10868–10879. doi: 10.18632/oncotarget.3451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Varambally S, Dhanasekaran SM, Zhou M, Barrette TR, Kumar-Sinha C, Sanda MG, Ghosh D, Pienta KJ, Sewalt RG, Otte AP, Rubin MA, Chinnaiyan AM. The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature. 2002;419:624–629. doi: 10.1038/nature01075. [DOI] [PubMed] [Google Scholar]
- 19.Vire E, Brenner C, Deplus R, Blanchon L, Fraga M, Didelot C, Morey L, Van Eynde A, Bernard D, Vanderwinden JM, Bollen M, Esteller M, Di Croce L, de Launoit Y, Fuks F. The Polycomb group protein EZH2 directly controls DNA methylation. Nature. 2006;439:871–874. doi: 10.1038/nature04431. [DOI] [PubMed] [Google Scholar]
- 20.Carvalho J, van Grieken NC, Pereira PM, Sousa S, Tijssen M, Buffart TE, Diosdado B, Grabsch H, Santos MA, Meijer G, Seruca R, Carvalho B, Oliveira C. Lack of microRNA-101 causes E-cadherin functional deregulation through EZH2 up-regulation in intestinal gastric cancer. J Pathol. 2012;228:31–44. doi: 10.1002/path.4032. [DOI] [PubMed] [Google Scholar]
- 21.Gu L, Frommel SC, Oakes CC, Simon R, Grupp K, Gerig CY, Bar D, Robinson MD, Baer C, Weiss M, Gu Z, Schapira M, Kuner R, Sultmann H, Provenzano M, Yaspo ML, et al. BAZ2A (TIP5) is involved in epigenetic alterations in prostate cancer and its overexpression predicts disease recurrence. Nature genetics. 2015;47:22–30. doi: 10.1038/ng.3165. [DOI] [PubMed] [Google Scholar]
- 22.Kondo Y. Targeting histone methyltransferase EZH2 as cancer treatment. J Biochem. 2014;156:249–257. doi: 10.1093/jb/mvu054. [DOI] [PubMed] [Google Scholar]
- 23.Fillmore CM, Xu C, Desai PT, Berry JM, Rowbotham SP, Lin YJ, Zhang H, Marquez VE, Hammerman PS, Wong KK, Kim CF. EZH2 inhibition sensitizes BRG1 and EGFR mutant lung tumours to TopoII inhibitors. Nature. 2015;520:239–242. doi: 10.1038/nature14122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McCabe MT, Ott HM, Ganji G, Korenchuk S, Thompson C, Van Aller GS, Liu Y, Graves AP, Della Pietra A, 3rd, Diaz E, LaFrance LV, Mellinger M, Duquenne C, Tian X, Kruger RG, McHugh CF, et al. EZH2 inhibition as a therapeutic strategy for lymphoma with EZH2-activating mutations. Nature. 2012;492:108–112. doi: 10.1038/nature11606. [DOI] [PubMed] [Google Scholar]
- 25.Matsukawa Y, Semba S, Kato H, Ito A, Yanagihara K, Yokozaki H. Expression of the enhancer of zeste homolog 2 is correlated with poor prognosis in human gastric cancer. Cancer Sci. 2006;97:484–491. doi: 10.1111/j.1349-7006.2006.00203.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen S, Chen JZ, Zhan Q, Zhu Y, Chen H, Deng XX, Hou ZY, Shen BY, Chen YL, Peng CH. H2AK119Ub1 and H3K27Me3 in molecular staging for survival prediction of patients with pancreatic ductal adenocarcinoma. Oncotarget. 2014;5:10421–10433. doi: 10.18632/oncotarget.2126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Igarashi H, Kurihara H, Mitsuhashi K, Ito M, Okuda H, Kanno S, Naito T, Yoshii S, Takahashi H, Kusumi T, Hasegawa T, Sukawa Y, Adachi Y, Okita K, Hirata K, Imamura Y, et al. Association of MicroRNA-31-5p with Clinical Efficacy of Anti-EGFR Therapy in Patients with Metastatic Colorectal Cancer. Ann Surg Oncol. 2015;22:2640–2648. doi: 10.1245/s10434-014-4264-7. [DOI] [PubMed] [Google Scholar]
- 28.Nosho K, Igarashi H, Nojima M, Ito M, Maruyama R, Yoshii S, Naito T, Sukawa Y, Mikami M, Sumioka W, Yamamoto E, Kurokawa S, Adachi Y, Takahashi H, Okuda H, Kusumi T, et al. Association of microRNA-31 with BRAF mutation, colorectal cancer survival and serrated pathway. Carcinogenesis. 2014;35:776–783. doi: 10.1093/carcin/bgt374. [DOI] [PubMed] [Google Scholar]
- 29.Cekaite L, Rantala JK, Bruun J, Guriby M, Agesen TH, Danielsen SA, Lind GE, Nesbakken A, Kallioniemi O, Lothe RA, Skotheim RI. MiR-9, -31, and -182 deregulation promote proliferation and tumor cell survival in colon cancer. Neoplasia. 2012;14:868–879. doi: 10.1593/neo.121094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mitsuhashi K, Nosho K, Sukawa Y, Matsunaga Y, Ito M, Kurihara H, Kanno S, Igarashi H, Naito T, Adachi Y, Tachibana M, Tanuma T, Maguchi H, Shinohara T, Hasegawa T, Imamura M, et al. Association of Fusobacterium species in pancreatic cancer tissues with molecular features and prognosis. Oncotarget. 2015;6:7209–7220. doi: 10.18632/oncotarget.3109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ito M, Mitsuhashi K, Igarashi H, Nosho K, Naito T, Yoshii S, Takahashi H, Fujita M, Sukawa Y, Yamamoto E, Takahashi T, Adachi Y, Nojima M, Sasaki Y, Tokino T, Baba Y, et al. MicroRNA-31 expression in relation to BRAF mutation, CpG island methylation and colorectal continuum in serrated lesions. Int J Cancer. 2014;135:2507–2515. doi: 10.1002/ijc.28920. [DOI] [PubMed] [Google Scholar]
- 32.Hur K, Toiyama Y, Schetter AJ, Okugawa Y, Harris CC, Boland CR, Goel A. Identification of a metastasis-specific MicroRNA signature in human colorectal cancer. J Natl Cancer Inst. 2015:107. doi: 10.1093/jnci/dju492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mitsuhashi K, Yamamoto I, Kurihara H, Kanno S, Ito M, Igarashi H, Ishigami K, Sukawa Y, Tachibana M, Takahashi H, Tokino T, Maruyama R, Suzuki H, Imai K, Shinomura Y, Yamamoto H, et al. Analysis of the molecular features of rectal carcinoid tumors to identify new biomarkers that predict biological malignancy. Oncotarget. 2015;6:22114–22125. doi: 10.18632/oncotarget.4294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Iino I, Kikuchi H, Miyazaki S, Hiramatsu Y, Ohta M, Kamiya K, Kusama Y, Baba S, Setou M, Konno H. Effect of miR-122 and its target gene cationic amino acid transporter 1 on colorectal liver metastasis. Cancer Sci. 2013;104:624–630. doi: 10.1111/cas.12122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lin PC, Chiu YL, Banerjee S, Park K, Mosquera JM, Giannopoulou E, Alves P, Tewari AK, Gerstein MB, Beltran H, Melnick AM, Elemento O, Demichelis F, Rubin MA. Epigenetic repression of miR-31 disrupts androgen receptor homeostasis and contributes to prostate cancer progression. Cancer Res. 2013;73:1232–1244. doi: 10.1158/0008-5472.CAN-12-2968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Slattery ML, Herrick JS, Mullany LE, Valeri N, Stevens J, Caan BJ, Samowitz W, Wolff RK. An evaluation and replication of miRNAs with disease stage and colorectal cancer-specific mortality. Int J Cancer. 2015;137:428–438. doi: 10.1002/ijc.29384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Aoki H, Nosho K, Igarashi H, Ito M, Mitsuhashi K, Naito T, Yamamoto E, Tanuma T, Nomura M, Maguchi H, Shinohara T, Suzuki H, Yamamoto H, Shinomura Y. MicroRNA-31 expression in colorectal serrated pathway progression. World J Gastroenterol. 2014;20:12346–12349. doi: 10.3748/wjg.v20.i34.12346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang Y, Lin C, Liao G, Liu S, Ding J, Tang F, Wang Z, Liang X, Li B, Wei Y, Huang Q, Li X, Tang B. MicroRNA-506 suppresses tumor proliferation and metastasis in colon cancer by directly targeting the oncogene EZH2. Oncotarget. 2015;6:32586–32601. doi: 10.18632/oncotarget.5309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vishnubalaji R, Hamam R, Abdulla MH, Mohammed MA, Kassem M, Al-Obeed O, Aldahmash A, Alajez NM. Genome-wide mRNA and miRNA expression profiling reveal multiple regulatory networks in colorectal cancer. Cell Death Dis. 2015;6:e1614. doi: 10.1038/cddis.2014.556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liao XY, Lochhead P, Nishihara R, Morikawa T, Kuchiba A, Yamauchi M, Imamura Y, Qian ZR, Baba Y, Shima K, Sun RF, Nosho K, Meyerhardt JA, Giovannucci E, Fuchs CS, Chan AT, et al. Aspirin Use, Tumor PIK3CA Mutation, and Colorectal-Cancer Survival. New Engl J Med. 2012;367:1596–1606. doi: 10.1056/NEJMoa1207756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nosho K, Irahara N, Shima K, Kure S, Kirkner GJ, Schernhammer ES, Hazra A, Hunter DJ, Quackenbush J, Spiegelman D, Giovannucci EL, Fuchs CS, Ogino S. Comprehensive Biostatistical Analysis of CpG Island Methylator Phenotype in Colorectal Cancer Using a Large Population-Based Sample. PLoS One. 2008:3. doi: 10.1371/journal.pone.0003698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nosho K, Kure S, Irahara N, Shima K, Baba Y, Spiegelman D, Meyerhardt JA, Giovannucci EL, Fuchs CS, Ogino S. A prospective cohort study shows unique epigenetic, genetic, and prognostic features of synchronous colorectal cancers. Gastroenterology. 2009;137:1609–1620. e1601–1603. doi: 10.1053/j.gastro.2009.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tahara T, Yamamoto E, Madireddi P, Suzuki H, Maruyama R, Chung W, Garriga J, Jelinek J, Yamano HO, Sugai T, Kondo Y, Toyota M, Issa JP, Estecio MR. Colorectal carcinomas with CpG island methylator phenotype 1 frequently contain mutations in chromatin regulators. Gastroenterology. 2014;146:530–538. e535. doi: 10.1053/j.gastro.2013.10.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ichimura N, Shinjo K, An B, Shimizu Y, Yamao K, Ohka F, Katsushima K, Hatanaka A, Tojo M, Yamamoto E, Suzuki H, Ueda M, Kondo Y. Aberrant TET1 Methylation Closely Associated with CpG Island Methylator Phenotype in Colorectal Cancer. Cancer Prev Res (Phila) 2015;8:702–711. doi: 10.1158/1940-6207.CAPR-14-0306. [DOI] [PubMed] [Google Scholar]
- 45.Torlakovic E, Skovlund E, Snover DC, Torlakovic G, Nesland JM. Morphologic reappraisal of serrated colorectal polyps. Am J Surg Pathol. 2003;27:65–81. doi: 10.1097/00000478-200301000-00008. [DOI] [PubMed] [Google Scholar]
- 46.Leggett B, Whitehall V. Role of the serrated pathway in colorectal cancer pathogenesis. Gastroenterology. 2010;138:2088–2100. doi: 10.1053/j.gastro.2009.12.066. [DOI] [PubMed] [Google Scholar]
- 47.Rosty C, Hewett DG, Brown IS, Leggett BA, Whitehall VL. Serrated polyps of the large intestine: current understanding of diagnosis, pathogenesis, and clinical management. J Gastroenterol. 2013;48:287–302. doi: 10.1007/s00535-012-0720-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bettington M, Walker N, Clouston A, Brown I, Leggett B, Whitehall V. The serrated pathway to colorectal carcinoma: current concepts and challenges. Histopathology. 2013;62:367–386. doi: 10.1111/his.12055. [DOI] [PubMed] [Google Scholar]
- 49.Edelstein DL, Axilbund JE, Hylind LM, Romans K, Griffin CA, Cruz-Correa M, Giardiello FM. Serrated polyposis: rapid and relentless development of colorectal neoplasia. Gut. 2013;62:404–408. doi: 10.1136/gutjnl-2011-300514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rex DK, Ahnen DJ, Baron JA, Batts KP, Burke CA, Burt RW, Goldblum JR, Guillem JG, Kahi CJ, Kalady MF, O'Brien MJ, Odze RD, Ogino S, Parry S, Snover DC, Torlakovic EE, et al. Serrated lesions of the colorectum: review and recommendations from an expert panel. Am J Gastroenterol. 2012;107:1315–1329. doi: 10.1038/ajg.2012.161. quiz 1314, 1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Naito T, Nosho K, Ito M, Igarashi H, Mitsuhashi K, Yoshii S, Aoki H, Nomura M, Sukawa Y, Yamamoto E, Adachi Y, Takahashi H, Hosokawa M, Fujita M, Takenouchi T, Maruyama R, et al. IGF2 differentially methylated region hypomethylation in relation to pathological and molecular features of serrated lesions. World J Gastroenterol. 2014;20:10050–10061. doi: 10.3748/wjg.v20.i29.10050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bosman FT, World Health Organization . WHO classification of tumours of the digestive system. Lyon: International Agency for Research on Cancer; 2010. and International Agency for Research on Cancer. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


