Abstract
Bacillithiol is a low-molecular-weight thiol analogous to glutathione and is found in several Firmicutes, including Staphylococcus aureus. Since its discovery in 2009, bacillithiol has been a topic of interest because it has been found to contribute to resistance during oxidative stress and detoxification of electrophiles, such as the antibiotic fosfomycin, in S. aureus. The rapid increase in resistance of methicillin-resistant Staphylococcus aureus (MRSA) to available therapeutic agents is a great health concern, and many research efforts are focused on identifying new drugs and targets to combat this organism. This review describes the discovery of bacillithiol, studies that have elucidated the physiological roles of this molecule in S. aureus and other Bacilli, and the contribution of bacillithiol to S. aureus fitness during pathogenesis. Additionally, the bacillithiol biosynthesis pathway is evaluated as a novel drug target that can be utilized in combination with existing therapies to treat S. aureus infections.
Keywords: bacillithiol, bacillithiol conjugate amidase, drug resistance, fosfomycin, oxidative stress, pathogenesis, Staphylococcus aureus
Thiols in aerobic bacteria
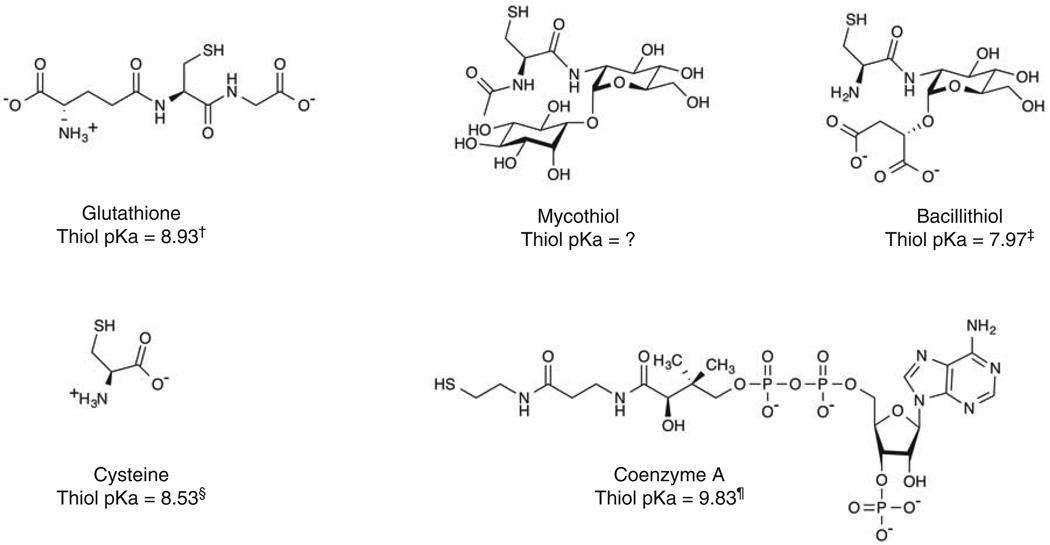
Aerobic bacteria live in an oxidizing environment and often require this environment for life, yet they maintain a reducing intracellular environment. Generating and maintaining this reducing environment requires not only energy but also specialized molecules in the cell that store this reducing potential and use it for various biochemical processes. These molecules include low-molecular-weight thiols such as glutathione (GSH), cysteine (Cys) and coenzyme A (CoA), each of which play different cellular roles [1]. Free cysteine levels are typically kept low because cysteine is prone to metal-catalyzed oxidation [2], which produces hydrogen peroxide, which can damage both DNA and proteins. CoA has a high thiol pKa compared to cysteine and glutathione (see “Biophysical Properties” below, [3–5]), such that the availability of the thiolate anion form of CoA required for reaction with electrophiles and thiol-disulfide exchange reactions is very limited. CoA is not a storage form of cysteine and cannot resupply the cell with Cys for protein synthesis during times of shortage, which can occur during oxidative stress. GSH is oxidized more slowly than is Cys, and it can be broken down to release Cys when necessary, so it plays a key role in redox reactions and redox homeostasis. GSH is present in Gram-negative bacteria and also in eukaryotic cells and was at one time assumed to be present in all living aerobic cells. However, many Gram-positive bacteria were subsequently shown to lack GSH and contain instead other low-molecular-weight thiols [1,6,7]. For example, the Actinobacteria, which include human pathogens such as Mycobacterium tuberculosis, use mycothiol (MSH), whereas several Firmicutes, including the human pathogens Bacillus anthracis and Staphylococcus aureus, use bacillithiol (BSH). Since its discovery, BSH has become the focus of considerable biochemical and physiological attention. In this review, we summarize this work and we evaluate the BSH system in Staphylococcus aureus as a potential target for the development of chemotherapeutic agents.
The discovery of mycothiol & bacillithiol
Once it became clear that GSH was not ubiquitous, the search was underway for alternative low-molecular-weight thiols. To study these thiols, an assay was developed utilizing the thiol-labeling compound monobromobimane in combination with reverse phase separation of the resulting fluorescent thiolbimane derivatives (RSmB) [8,9]. The bimanes (such as the widely used 9,10-dioxa-syn-dimethylbimanes [10]) are among the smallest sulfhydryl-reactive fluorophores known and allow the thiol moiety to dominate the physical chemical properties of the conjugate and its chromatographic behavior. Another advantage of the bimane label is that it is highly conjugated and has a simple proton nuclear magnetic resonance spectrum, which is beneficial in solving novel biological thiol structures [11–14]. Application of the bimane assay to a variety of bacteria revealed novel thiols, but most were present at low levels relative to GSH or CoA. An exception was the Actinomycetes where a major novel thiol, U17, was identified in Streptomyces clavuligerus and in other actinobacteria [15]. The structure of U17 was solved by three laboratories and renamed mycothiol (Figure 1, [11,12,16]). Mycothiol would later guide the determination of the structure and discovery of the biosynthetic pathway for bacillithiol.
Figure 1. Structures and pKa values of the major low-molecular-weight thiols found in bacteria.
Thiols shown as the major ionized form at pH = 7.7 and represent microscopic thiol pKa values.
†Rabenstein 1973 [5]; ‡Sharma et al. 2013 [20]; §Benesch and Benesch 1955 [4]; ¶Keire et al. 1992 [3].
Although the Firmicutes were recognized to lack glutathione [7], the biologically relevant low-molecular-weight thiol in these bacteria remained unclear for several years because the bimane derivative of bacillithiol (BSmB) coeluted with the cysteine bimane derivative (CySmB) [17] in the sodium acetate buffer that was used in the HPLC separation method #1 [9]. The addition of an online mass detector in parallel with the HPLC proved extremely productive because bimane-thiol derivatives ionize easily in electrospray mass spectrometry. During a systematic thiol survey of B. anthracis in 2006, an unknown thiol was found to coelute with CySmB on the HPLC analysis and had a different mass. After the pH of the buffer was adjusted from 3.4 to 4.0, this novel thiol was fully resolved from CySmB. In retrospect, this was due to the ionization of the malate carboxylates of bacillithiol at the higher pH. U12 had a mass of 398, which did not match any of the biological thiols identified in previous surveys [18]. Subsequently, it was reported that treatment of Bacillus subtilis with cumene hydroperoxide led to the formation of mixed disulfides between the organic hydroperoxide regulator OhrR and either cysteine, CoA or an unknown thiol of the same mass as U12 [19]. U12 was reported soon afterward in Deinococcus radiodurans [14]. Thus, the common occurrence of the novel thiol justified solving the structure.
The structure of U12-bimane from D. radiodurans was determined from a 2D NMR and analysis of acid hydrolysis products, the same strategy used to solve the bimane derivative of mycothiol [12]. U12 was found to be the α-anomeric glycoside of L-cysteinyl-D-glucosamine with L-malic acid (Figure 1) and was named BSH because of its widespread occurrence within the Bacilli [14]. Bacillithiol resembles mycothiol in that it has the same central cysteine moiety amide bonded to glucosamine, but it has L-malate in an α-glycosidic bond to glucosamine in place of the inositol moiety in mycothiol, and the cysteine of MSH is N-acetylated (Figure 1). Bacillithiol was found in seven species of Bacillus, in Geobacillus and in some but not all species of Staphylococcus and Streptococcus [14]. A survey of Firmicutes growing exponentially in trypticase soy broth with shaking at 37°C, including B. subtilis and S. aureus, revealed BSH levels at 0.2–0.7 µmol/g dry weight [0.1–0.2 mM], some 10-fold lower than the concentration of GSH in Gram-negative bacteria or of MSH in Actinomycetes, but similar to the levels of Cys and CoA in these organisms. Thus, the Firmicutes appear to have three major low-molecular-weight thiols (cysteine, coenzyme A and bacillithiol) that are maintained at similar, low levels during exponential growth.
Biophysical properties of bacillithiol
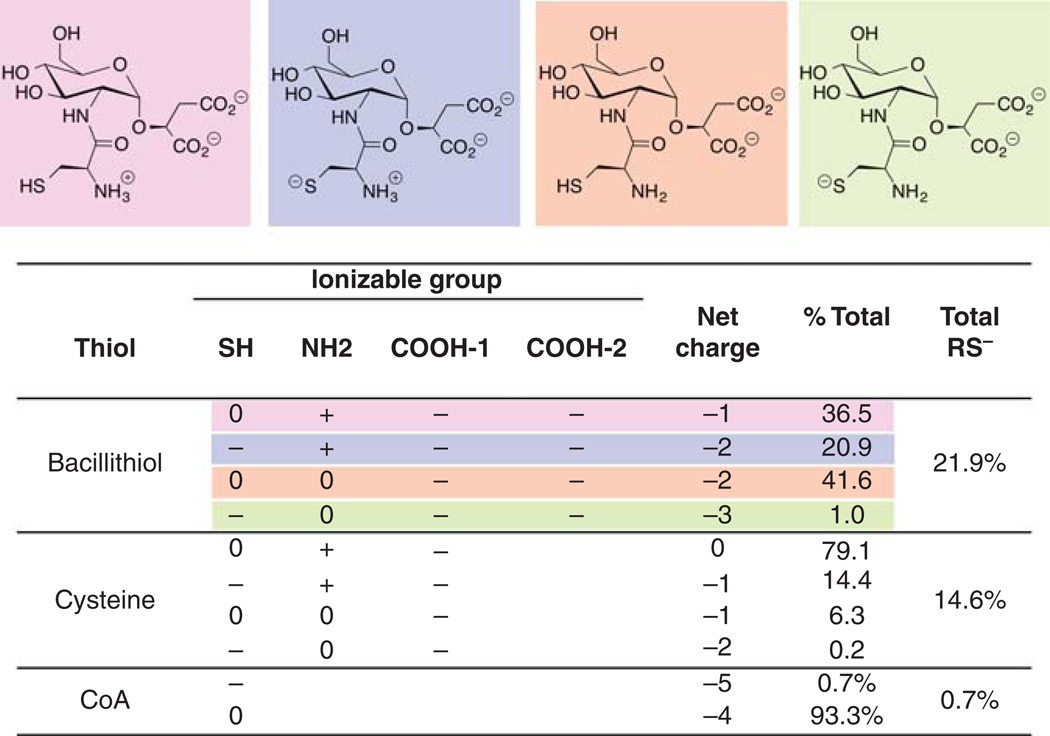
The solution structure of BSH is dominated by its zwitterionic character at physiological pH [20], which also contributes to its biological activities. The redox potential and the microscopic and macroscopic pKas for each ionizable group were determined by a combination of spectroscopic techniques. The redox potential for BSH is −221 mV, which is similar to that of most biological monothiols including Cys (−223 mV), CoA (−234 mV) and GSH (−240 mV). In contrast, all of the pKa values for BSH are lower than the corresponding values for Cys or GSH. The first two pKa values of BSH (3.1 and 4.4) correspond to those of the malate moiety, which are lower than the respective values for malic acid alone (3.4 and 5.1). This indicates that the malate moiety in BSH is a dianion at physiological pH (Figure 2). The most unusual aspect of BSH ionization is the microscopic amine pKn of 7.63, which is just ~0.3 pH units below that of the thiol pKs of 7.97. Hence when BSH is at the physiological pH of 7.7, there will be two major forms of BSH dianion, the − + − − and the 00− − forms ([20], Figure 2). The amount of BSH in the reactive thiolate form (RS−, 21.9%) is greater than that of Cys (14.6%) and much greater than that of CoA, the two other major thiols found in the B. subtilis and in S. aureus. Thus, the major nucleophilic thiol available for reaction with electrophiles or disulfides in these organisms is BSH.
Figure 2. Comparison of the major forms of bacillithiol, cysteine and CoA at pH 7.7.
Proportions of each form are based on microscopic pKa values. The four major forms of bacillithiol are depicted. The ionizable phosphate groups of CoA are not listed.
Figure from Sharma et al. 2013 [20].
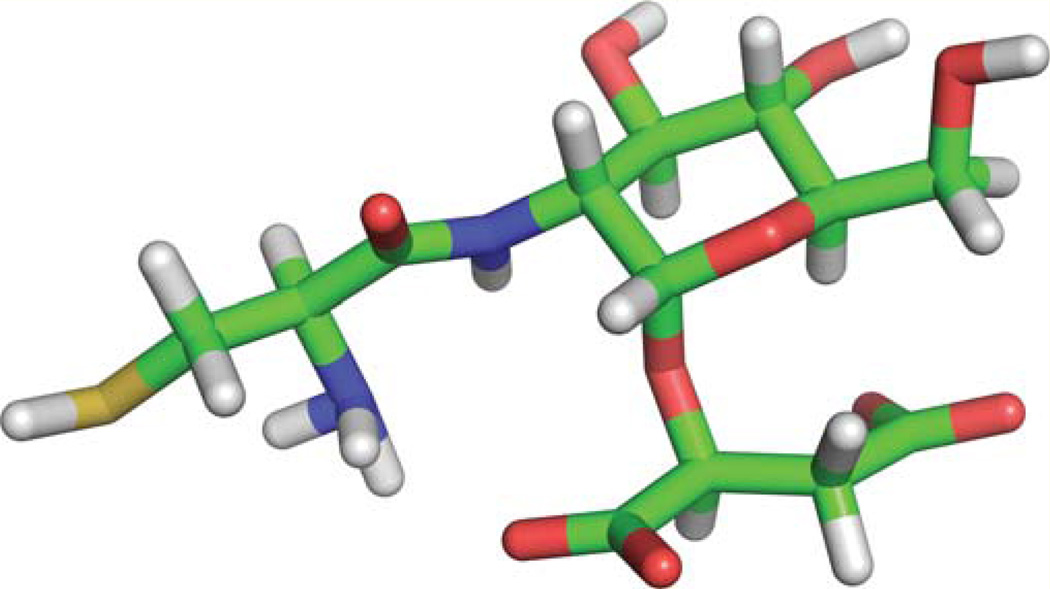
Many of the unique biophysical properties of BSH may be clarified by the solution structure of this thiol. A 3D structure for a major physiologically relevant form of BSH shows a close proximity of the carboxylate of malate to the amine of cysteine, which may contribute to the deprotonation of the amine and the low amine pKa (Figure 3). When the major 0+ − − ionized form of bacillithiol (Figure 2) was analyzed in ChemDraw 3D, the energy minimized structure rapidly converged to that shown in (Figure 3) The proximity of the ammonium, carboxylate and thiol groups in BSH have previously been postulated to contribute to metal chelation [14]. Importantly, in B. subtilis bacillithiol has recently been shown to chelate Zn2+ and to act as a Zn2+ buffer for metal homeostasis [21]. B. subtilis wildtype and a bacillithiol (bshC) mutant did not show a Zn2+ or Cd2+ stress phenotype in the wildtype background; however a double metal exporter mutant, cadA czcD, did show increased sensitivity to Zn2+ and Cd2+ in the absence of BSH [21]. Bacillithiol-dependent Zn2+ sensitivity has not been reported for S. aureus, but BSH mutants of S. aureus USA300 were reported to be sensitive to Cd2+ [22]. Gene expression studies revealed that metal-sensing regulators are more responsive to zinc perturbation in a mutant lacking bacillithiol, presumably because BSH buffers the zinc labile pool. This suggests that bacillithiol plays a role in zinc homeostasis by holding free zinc in a cytoplasmic pool, perhaps keeping the metal available for the estimated 5–10% cellular enzymes that require a Zn2+ cofactor [23]. It has been noted that during S. aureus infection, neutrophil proteins sequester and limit Zn2+ availability to the pathogen, reducing S. aureus growth and making it more vulnerable to oxidative stress [21,24].
Figure 3. 3-dimensional structure of bacillithiol in the ionized 0 + − − form (Figure 2).
The proximal location of thiol, amine and carboxylate moieties in the energy-minimized structure may support divalent metal ion chelation. Atom labels: C, green; N, blue; O, red; H, white, and S, yellow.
The dianion character of BSH drives the substrate specificity of the BSH biosynthesis enzymes BshB1 and BshB2 and of the fosfomycin detoxification enzyme FosB (see below) [25,26]. The catalytic efficiency (kcat/Km) of BshB1 and BshB2 was diminished by ~104–105 when GlcNAc-Mal was replaced with the des-malate analog GlcNAc. This specificity is likely determined by opposing arginines (Arg53 and Arg109) present in the active site near the malate moiety when GlcNAc-Mal is computationally docked in the crystal structure of B. cereus BshB (BC1534, [25],). Similarly, the catalytic efficiency of FosB was diminished by 103 when BSH (Cys-GlcN-Mal) was replaced with the des-malate analogs Cys-GlcN-O-methyl or Cys-GlcN-O-benzyl [26]. In the catalytic sites of the crystallized FosB proteins, Arg35 and Lys36 (B. cereus and B. anthracis), or Lys35 and Lys36 (S. aureus), are positioned in the active site ~10 Å apart in an orientation to bind the malate moiety of BSH [26]. It may be possible to find BSH-binding enzymes by identifying opposing positively charged residues with the appropriate geometry for malate binding near their active sites.
Bacillithiol biosynthesis enzymes
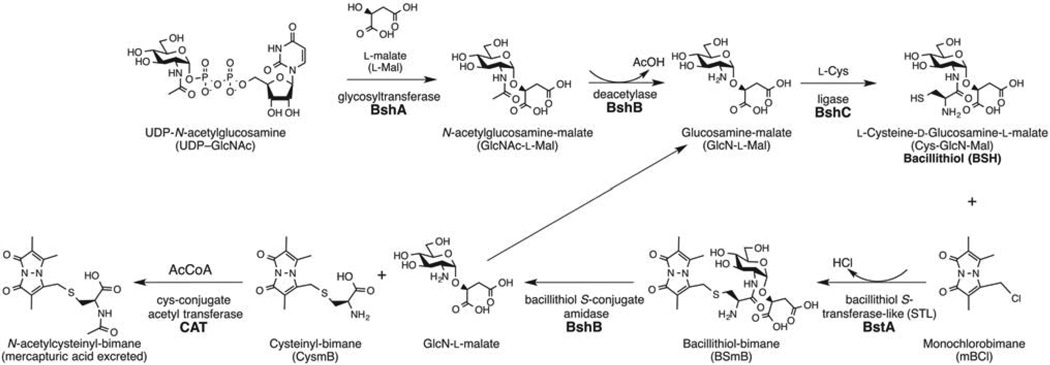
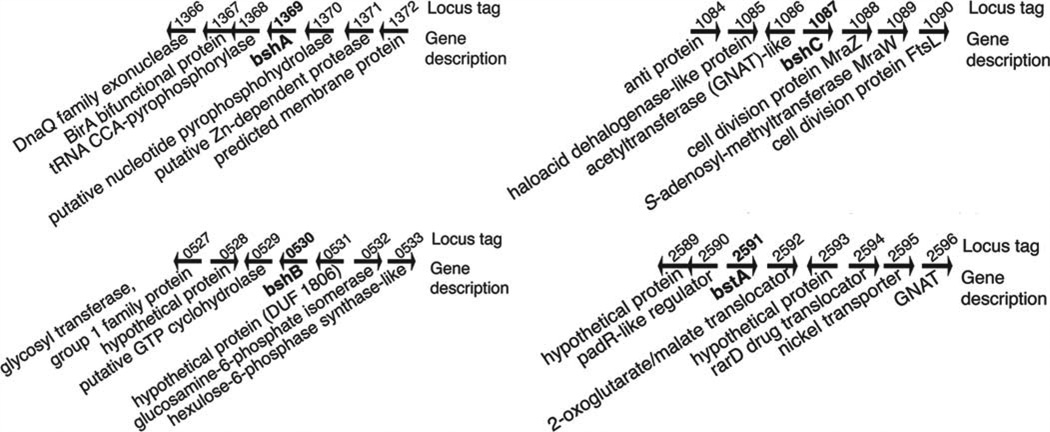
BshA
The structural similarity of MSH and BSH made identification of the first two BSH biosynthetic genes relatively straightforward since they encoded homologs of the previously identified MSH biosynthetic enzymes [27]. For example, the first step in mycothiol biosynthesis is the formation of the glycosidic linkage between 1-L-inositol-1-phosphate (Ins-P) and UDP-N-acetylglucosamine (UDP-GlcNAc), which is catalyzed by the glycosyltransferase (MshA) [28]. B. subtilis has five glycosyltransferases with low sequence homology to MshA, but the prime bshA candidate with ~25% identity over 40% of the sequence was ypjH, which is located immediately downstream from the mshB homolog ypjG. ypjH was shown to be essential for BSH biosynthesis and the purified YpjH protein catalyzed the production of N-acetylglucosamine-malate (GlcNAc-Mal). The BshA glycosyltransferase reaction was able to utilize UDP-GlcNAc and L-malate as substrates (Figure 4), but not D-malate. The B. anthracis homolog of B. subtilis BshA (BA_1558, 65% identity) had been crystallized without ligands prior to the discovery of BSH and was observed to have an active site that was similar to MshA (PDB entry 2JJM, [29]). This structure aided in the solution of a ternary complex of B. anthracis BshA, UDP and L-malate (PDB entry 3MBO, [30]). Kinetic studies of the B. anthracis BshA [30] and the S. aureus BshA [31], which share ~56% sequence homology, indicated that the proteins perform similar catalytic functions. Allosteric inhibition of B. subtilis BshA by BSH (IC50 ~ 0.7 mM) suggests that the enzyme is subject to feedback regulation to modulate the intracellular level of BSH by inhibiting the first biosynthetic enzyme in the pathway, as is the case for many pathways, including those involved in GSH and MSH biosynthesis [31]. Significantly, the heterologous expression of bshA in non-BSH-containing bacteria led to the production of significant quantities of GlcNAc-Mal, providing a source for this non-commercially available substrate for BshB.
Figure 4. Bacillithiol biosynthesis (top) and bacillithiol-dependent detoxification (bottom) in Staphylococcus aureus.
Data from Gaballa et al. 2010 [27], Newton et al. 2011 [56], Newton et al. 2012 [48], Fang et al. 2013 [25], and Perera et al. 2014 [52].
BshB
The second bacillithiol biosynthesis gene bshB is analogous to the corresponding MSH biosynthesis gene mshB, which encodes a deacetylase. bshB (ypjG) is immediately upstream of the bshA gene in some BSH-producing organisms, such as B. subtilis, and its product was proposed to catalyze the deacetylation of GlcNAc-Mal. The BSH levels in a B. subtilis bshB1 deletion strain were at about 40% of wildtype levels. A second unlinked homolog of bshB1, bshB2 (yojG), was then discovered in the B. subtilis genome, and deletion of both bshB1 and bshB2 yielded a mutant that failed to produce bacillithiol [27]. Thus, the B. subtilis BshB1 and BshB2 enzymes have redundant functions, but BshB1 can provide all of the necessary deacetylase activity since the bshB2 mutant has wildtype levels of bacillithiol [27]. The genome of B. anthracis appears to have one bshB1 and two bshB2 homologs. The B. anthracis bshB1 gene (BA_1557) was cloned into E. coli, and the purified protein showed high deacetylation activity with GlcNAc-Mal [27,30]. Unlike B. subtilis, the genome sequence of S. aureus USA300 indicated that there was only one deacetylase gene closely related to bshB2, leading to the prediction that a mutation in this gene would produce no BSH. Analysis of a S. aureus USA300 strain containing a transposon insertion in bshB demonstrated that this is the case [22].
A Bacillus cereus BC1534 (BshB1) crystal structure, published before the discovery of BSH, revealed a metalloprotein with structural similarity to the metalloproteins LpxC and MshB from M. tuberculosis [32]. Although the native substrate of the BC1534 was not known, the protein was reported to deactylate GlcNAc and chitobiose [33]. However, a detailed analysis of the substrate specificity of BshB1 and BshB2 type proteins from B. cereus and B. anthracis failed to confirm this activity [25], perhaps because the malate moiety of the native substrate GlcNAc-Mal is a major determinant for BshB substrate specificity. Deacetylation of GlcNAc by BshB proteins is found to be at least 105 fold lower (kcat/Km) than that of the native substrate GlcNAc-Mal [25,30]. Interestingly, the BshB enzymes have a second function in addition to their deacetylation activity, as they can also act as a bacillithiol conjugate amidase (BCA) and cleave bacillithiol S-conjugates during detoxification reactions (Figure 4). This activity is described below.
BshC
The enzyme utilized in the third step in mycothiol biosynthesis is the L-Cys, GlcN-Ins ligase MshC. This enzyme was purified from crude extracts of M. smegmatis by following activity, and the protein was identified by aminoterminal sequencing [34]. The corresponding open reading frame (ORF) in the M. tuberculosis genome, ORF Rv2130c, had been designated cysS2 and annotated as a second Cys tRNA synthase. However, neither the B. subtilis nor S. aureus genome encoded a MshC homolog or a second gene annotated as a putative Cys tRNA synthase, and no BshC activity was associated with the one Cys tRNA synthase in S. aureus [27]. Therefore, to identify candidate BshC proteins, EMBL STRING was queried with a BshA input sequence to identify genes that that co-occur with bshA in the genomes of bacteria that produce bacillithiol. This phylogenetic profiling identified yllA as a bshC candidate in B. subtilis. The product of this gene is a member of DUF2317, but it has no recognizable Pfam domains. The null mutant lacked BSH and produced excess GlcN-Mal, which would be the expected substrate for BshC [27]. Thus, the bshC gene is essential for BSH production, and in both B. subtilis and S. aureus bshC mutants are completely devoid of BSH [22,27]. However, when the Bacillus subtilis bshC gene was cloned and expressed in E. coli, activity was not detected when purified BshC was assayed under conditions that had been used to assay MshC activity [27]. The lack of activity may have been due to the assay conditions or to misfolding of the 539 amino acid BshC protein. Recently, B. subtilis BshC was crystallized as a dimer with an overall architecture of the Rossman fold active site similar to MshC and Cys t-RNA synthase except for the absence of the canonical adenylate and zinc-binding residues in MshC [35]. However, because BshC was enzymatically inactive, it is difficult to evaluate the biological significance of this structure. It remains possible that additional cofactors or protein partners are necessary for BshC enzymatic activity.
Bacillithiol as a redox buffer
Any thiol that serves as a redox buffer in aerobic organisms must have the ability to regenerate the reduced thiol after it is oxidized to the disulfide form. Glutathione disulfide reductase (Gor) was originally isolated from E. coli and was thought to be necessary for reduction of GSH during redox stress [36]. The first complete genome of Mycobacterium tuberculosis H37Rv [37] contained a gene that was annotated as glutathione disulfide reductase (gor). It seemed likely that the mycobacterial gor was instead a gene for a mycothiol disulfide reductase because M. tuberculosis contains millimolar levels of mycothiol but no glutathione. This was demonstrated when the M. tuberculosis gene product Mtr was found to reduce only disulfides of mycothiol and truncated versions containing at least cysteinyl-glucosamine, but not oxidized GSH (GSSG) or cystine [38].
Identifying a redox thiol in the Firmicutes was more difficult than in the Actinomycetes given the variety of low-molecular-weight thiols present at low levels in the cell and the absence of likely candidates for proteins involved in redox recycling of these molecules. CoA disulfide reductase (CoADR) was identified in Staphylococcus aureus and some Bacillaceae including Bacillus anthracis [39–41]. A paradox arose in the model organism Bacillus subtilis when a CoA disulfide reductase (CoADR) homolog was not identified in the complete genome sequence. Additionally, no cystine disulfide reductase from Firmicutes had been reported, so the search began for a bacillithiol disulfide reductase in Bacillus subtilis. Although candidate proteins for the bacillithiol disulfide reductase have been proposed, no flavin-containing, NADH- or NADPH-utilizing reductase has been identified in either B. subtilis or S. aureus [14,22]. It is possible that the highly reduced state of bacillithiol [14,20] is maintained by a thioredoxin-thioredoxin reductase type system as found for glutathione in gor mutants of E. coli K12 [42].
Interestingly, a fluorescent reporter system has recently been used to image real-time changes in the redox status of MSH during M. tuberculosis infection of macrophages [43]. This technology could in principle be applied in S. aureus to evaluate the BSH redox status during various stages of growth and mammalian cell infection.
The role of bacillithiol in Firmicutes: phenotypes of biosynthesis defective mutants
The first BSH mutants were produced in the genetically tractable B. subtilis [27]. Deletion of bshA, or of both bshB1 and bshB2, or of bshC gives rise to completely bacillithiol-deficient B. subtilis. Each of these deletion strains was readily constructed and appeared to grow normally, demonstrating that BSH is not essential for growth in nutrient-rich media. However, B. subtilis BSH mutants were reported to be sensitive to salt stress and to acid stress, and mildly sensitive to oxidants and electrophiles such as N-ethylmaleimide, iodoacetamide, monobromobimane, diamide and methyglyoxal [27].
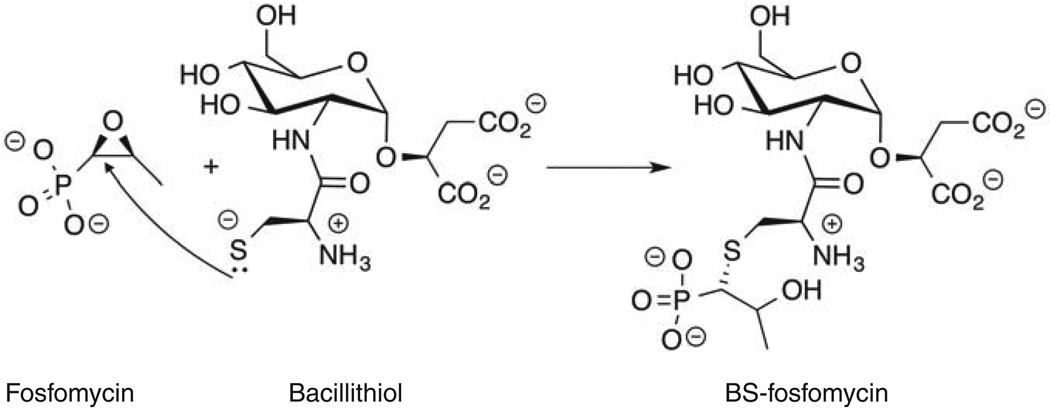
Bacillithiol also participates in the detoxification of fosfomycin by serving as the cosubstrate for the FosB enzyme [22,26,27,44]. FosB is a fosfomycin resistance protein found in Firmicutes that was initially thought to catalyze the reaction of cysteine with the epoxide moiety of fosfomycin, a cell wall inhibitor [45]. However, after the discovery of bacillithiol, several biochemical studies found that bacillithiol is the preferred FosB thiol cosubstrate for fosfomycin detoxification, making FosB the first bacillithiol transferase to be described (see “Bacillithiol Transferases” section below) [26,46,47]. The importance of bacillithiol in fosfomycin detoxification was confirmed in B. subtilis [22,27,44] and B. anthracis Sterne [30] as mutants lacking bacillithiol were as sensitive to fosfomycin as the fosB mutant. This has also been observed in S. aureus [22,26] where mutants lacking bshA are 15- to 60-fold more sensitive to fosfomycin than their corresponding wildtype methicillin-sensitive and methicillin-resistant (MRSA) S. aureus strains [44].
One sequenced S. aureus strain, NCTC 8325 (www.phe-culturecollections.org.uk), harbors an 8 basepair frameshift mutation in the bshC gene [44,48,49]. This naturally occurring mutation is also found in lineages derived from this strain, such as RN4220 an SH1000, and was the first BSH mutant of S. aureus to be studied. NCTC 8325 was found to lack BSH and contains elevated levels of GlcNAc-L-Mal and GlcN-L-Mal, the biosynthetic precursors of BSH [48]. NCTC 8325 and other S. aureus bacillithiol mutants do not exhibit growth defects in nutrient-rich media, but are severely hindered when challenged with fosfomycin [22,26,27].
Staphylococcus aureus bshA mutants are also sensitized to exogenous H2O2 and diamide [44]. As expected, a S. aureus strain with the bshC frameshift (SH1000) treated with monochlorobimane or the antibiotic rifamycin S did not show N-acetylcysteinyl S-conjugates (mercapturic acid products) of the toxins in the medium. The production of mercapturic acid products (see Figure 4 and bacillithiol S-conjugate amidase below) was dependent on the presence of bacillithiol even though SH1000 cells contained normal levels of cysteine, demonstrating that cysteine does not directly react with target substrates. Interestingly, S. aureus SH1000 converted all of the monochlorobimane to dimethylbimane [48], indicating the presence of a source of electrons not normally utilized in the wildtype strain. Dimethylbimane was also observed during the labeling of spinach chloroplasts and was thought to be produced by electron transfer from photosystem II [50].
In a recent metabolomics study, a S. aureus NCTC 8325 derived natural bshC mutant strain (HG001) was treated with fosfomycin and UDP-GlcNAc, GlcNAc-Mal, GlcN-Mal levels were observed by LCMS [51]. Interestingly, fosfomycin treatment significantly increased levels of UDP-GlcNAc (3.4-fold) and GlcNAc-Mal (the product of BshA, 2.2-fold), but levels of GlcN-Mal (product of BshB) were similar to treatment by the other drugs tested (ciprofloxacin, erythromycin, vancomycin and ampicillin). UDP-GlcNAc is a substrate for the first enzyme in BSH biosynthesis BshA, and the fosfomycin targets MurA and MurZ, which UDP-N-acetylglucosamine enolpyruvyl transferase activity. Thus, it is possible that the inhibition of MurA and MurZ by fosfomycin can increase flux through the BSH biosynthesis pathway, leading to an increase in the thiol cofactor utilized in FosB-dependent resistance to fosfomycin. This could limit the utility of BshA as a drug target during fosfomycin treatment of S. aureus (see “Expert commentary & five-year view”).
Identification of bacillithiol-dependent & bacillithiol-related enzymes
Bacillithiol transferases
Thiol transferases are enzymes that catalyze the addition of a low-molecular-weight thiol to target substrates forming a new thioether bond. The native substrates for two mycothiol-dependent enzymes from the S-transferase like (STL) superfamily [52], formerly known as the DinB/YfiT-like superfamily, have been characterized from Actinomycetes. Corynebacterium glutamicum was found to grow on aromatic hydrocarbons like gentisate as a sole carbon source in a mycothiol-dependent manner. The key step in the degradation of gentisate in C. glutamicum is the isomerization of malylpyruvate to fumarylpyruvate in which mycothiol serves as a cofactor for the STL enzyme Ncgl2918 [53]. In Streptomyces lincolnensis LmbE, a mycothiol conjugate amidase (MCA), and LmbV, a STL mycothiol transferase, have been shown to be involved in the biosynthesis of lincomycin A [54]. LmbV installs the sulfur moiety derived from mycothiol in a lincomycin A precursor. This is the first example of mycothiol, a MCA, and a STL mycothiol transferase being used in the biosynthesis rather than the detoxification of secondary metabolites.
The two bacillithiol transferases identified in the Firmicutes thus far are members of two distinct superfamilies. The first bacillithiol transferase to be identified in S. aureus is FosB (Figure 5), which confers resistance to the antibiotic fosfomycin. FosB is a member of the vicinal oxygen chelate (VOC) superfamily. VOCs are metalloenzymes that catalyze a diverse set of reactions, but all coordinate a divalent metal cation through vicinal oxygen atoms of a substrate, intermediate or transition state in the reaction [55]. FosB was identified before the discovery of bacillithiol, and cysteine was believed to be the thiol cofactor of FosB [45]. Early studies with the B. subtilis FosB reported a Km value for cysteine of 35 ± 3 mM, which is well above the calculated cysteine concentration for this organism (~230 µM, [14],). Since the discovery of bacillithiol, the thiol cofactor of both the B. subtilis and S. aureus FosB enzymes was reevaluated and determined to be bacillithiol [26,46,47]. The S. aureus FosB Km for bacillithiol was determined to be 4.2 ± 0.7 mM [26]. This value is also above the intracellular concentration of bacillithiol. However, due to the low thiol pKa of bacillithiol (see “Biophysical Properties” above), there is more of the reactive thiolate form of bacillithiol present than that of cysteine in S. aureus. Genetic data also support these findings: fosB and bacillithiol null mutants show similar sensitivity to fosfomycin [22,27,44]. Thus, the thiol cofactor of FosB is bacillithiol.
Figure 5. Bacillithiol-dependent detoxification of fosfomycin is catalyzed by S. aureus FosB.
Figure from Roberts et al. 2013 [26].
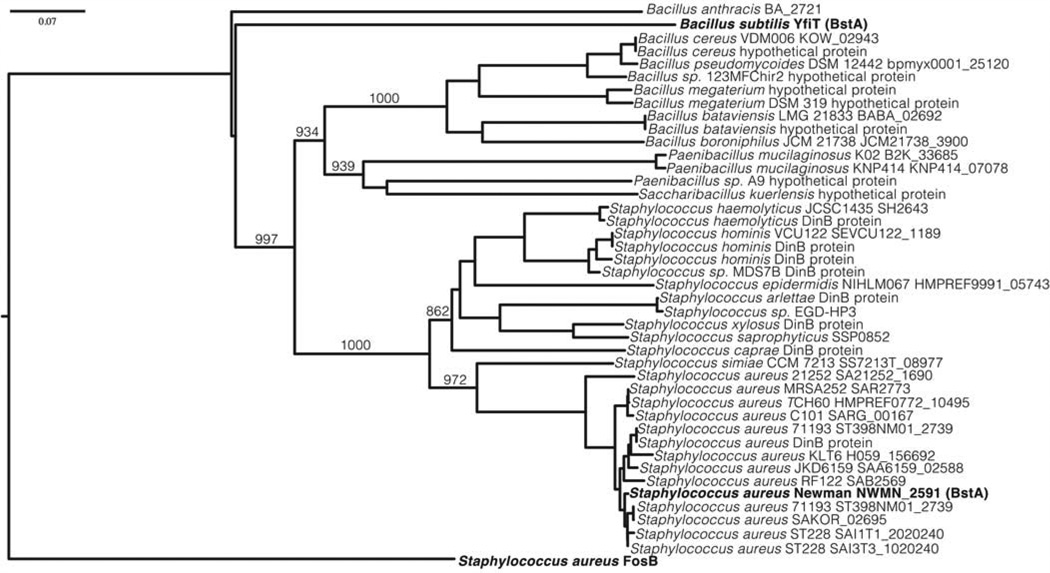
The first non-FosB bacillithiol transferase identified was B. subtilis YfiT (B. subtilis BstA), a member of the STL superfamily [56]. The single predicted S. aureus Newman STL enzyme (ORF ID NWMN_2591) was identified in a Superfamily search (http://supfam.org/SUPERFAMILY/) using B. subtilis BstA as a query sequence against the S. aureus Newman genome. The S. aureus enzyme was confirmed to be a bacillithiol transferase on the basis of biochemical studies of cell free extracts [22] and the purified protein and was named S. aureus BstA [52]. Kinetic studies with S. aureus BstA demonstrated that the Km for BSH is 16 ± 4 µM, which is ~10-fold lower than the intracellular concentration of bacillithiol in S. aureus, indicating that the enzyme is saturated with bacillithiol in vivo [52]. S. aureus BstA was found to catalyze the addition of bacillithiol to cerulenin [22] at a low rate in vitro [52]. This newly identified STL superfamily is a diverse protein family that contains ~30,000 proteins with structural similarity but very little sequence identity. Phylogenetic studies have demonstrated that B. subtilis BstA and S. aureus BstA are distantly related, but are more related to each other than they are to the non-STL bacillithiol transferase S. aureus FosB (Figure 6). The number of predicted STL transferases varies between species, with only one predicted structural homolog of YfiT in S. aureus Newman (BstA) and eight in B. subtilis [56].
Figure 6. Close relatives of S. aureus Newman BstA in the S-transferase like (STL, formerly DinB/YfiT-like) superfamily.
Staphylococcus aureus BstA is distantly related to B. subtilis YfiT (BstA) and a B. anthracis STL superfamily member (BA_2721), and unrelated to the bacillithiol transferase protein FosB. Bootstrap values are shown for the major branches.
Figure from Perera et al. 2014 [52].
Although additional substrates of S. aureus BstA have not been identified, the genomic context of bstA may give insight into its function (Figure 7). Five genes downstream and in the same orientation as bstA is a gene encoding an N-acetyltransferase, an enzyme that could N-acetylate the Cys-adduct product of the bacillithol conjugate amidase, and three genes downstream is a gene encoding RarD, an efflux transporter that might export acetylated Cysadducts (mercapturic acids, Figure 4). Products of these types of enzymatic reactions have been previously observed in the detoxification of electrophiles by S. aureus Newman [48]. A 2-oxoglutarate/malate translocator is adjacent to the bstA gene, which is significant because L-malate is a BshA substrate. Upstream of S. aureus bstA is a gene encoding PadR, a component of the phenolic acid stress response in B. subtilis that functions as a negative regulator of padC, a gene encoding a phenolic acid decarboxylase that converts toxic phenolic acids to their vinyl phenol derivatives [57]. The ortholog of padC in Lactobacillus plantarum, padA, is transcribed divergently from its negative regulator padR [58], just as S. aureus padR is transcribed divergently from bstA. This organization of genes can indicate a negative regulation relationship. In Lactococcus lactis, the padR analog lmrR encodes a negative regulator of lmrCD, which encodes an ATP-binding-cassette transporter, and is in the operon with these genes [59]. The gene upstream of padR, Newman_2589, encodes a hypothetical protein that shares structural motifs with efflux pumps. If PadR regulates expression of bstA, then the natural BstA substrate(s) might bind PadR and relieve transcriptional repression of bstA and could provide a method to identify the BstA natural substrate(s).
Figure 7. Genomic context of S. aureus bshA, bshB, bshC and bstA genes in S. aureus Newman.
The bacillithiol biosynthesis genes bshA, bshB and bshC and the bstA gene are unlinked (in bold). Overlapping arrows represent overlapping ORFs.
BshB has bacillithiol conjugate amidase activity in addition to GlcNAc-Mal deacetylase activity
The first mycothiol-dependent enzyme identified was mycothiol conjugate amidase (MCA) from M. smegmatis (MSMEG_5261) and M. tuberculosis (Rv1082) [60]. This enzyme cleaves a mycothiol S-adduct at the amide linkage between the cysteinyl and glucosamine moieties to generate a N-acetylcysteinyl S-adduct. The adduct is excreted and the residual glucosaminyl-inositol pseudo-disaccharide is retained in the cell and cycles back to MSH biosynthesis [60,61]. This is a commonly observed detoxification reaction among mycothiol-containing actinomycetes. Mycothiol conjugate amidase is a zinc-containing hydrolase and a close homolog of the mycothiol biosynthetic amidase, MshB [62]. These enzymes have both mycothiol conjugate amide hydrolase and GlcNAc-Ins deacetylase activities [63].
It appears there are specialized enzymes that control the degradation of the major low-molecular-weight thiols such as GSH, MSH and BSH, which can ultimately supply the cell with cysteine. The unique γ-glutamyl peptide bond in GSH (γ-Glu-Cys-Gly) prevents the unwanted breakdown of GSH by peptidases. For example, γ-glutamyltranspeptidase is required to hydrolyze the γ-glutamyl-cysteine peptide bond, which releases Cys-Gly, a substrate for carboxypeptidase. γ-glutamyltranspeptidase is found in eukaryotes and GSH-producing bacteria. The obligate human pathogen M. tuberculosis, which does not produce GSH, contains a glutathione ABC transport protein and γ-glutamyltranspeptidase that can supply Cys to M. tuberculosis from host GSH [64]. MSH-containing organisms possess MCA, which has been shown to hydrolyze the unusual Cys-GlcN amide bond of MSH to provide AcCys and ultimately Cys for protein synthesis [60,61,65]. It was predicted that the same holds in true in BSH-containing organisms: hydrolysis of the Cys-GlcN amide bond of BSH by bacillithiol conjugate amidase should release of Cys in the cell.
A similar pathway was observed in the BSH containing organisms B. cereus and B. anthracis, which encode two BshB enzymes (BshB1 and BshB2) that have dual GlcNAc-Mal deacetylase and bacillithiol S-conjugate amidase activities [25]. A sensitive assay that monitors hydrolysis of the fluorescent bimane derivative of bacillithiol (BSmB) to the corresponding derivative of cysteine (CySmB) was developed to measure bacillithiol conjugate amidase activity. Assays of purified B. anthracis BshB1 (BA_1557) showed low amidase activity, about 10−5 in comparison with the deacetylation rate with GlcNAc-Mal [25,30]. B. anthracis BshB2 (BA_3888) showed considerably more BSmB deacetylase activity, only 100-fold less than the GlcNAc-Mal deactylase activity. Both B. anthracis enzymes show low amidase activity toward unmodified BSH that is about 10−3– 10−4 of the GlcNAc-Mal deacetylase rate. The authors indicated that this low level of BSH amide hydrolysis activity was unlikely to be sufficient to supply cysteine to the cell. However, the MCA activity in M. smegmatis was also about 103 below that of the MSmB amidase reaction and was shown to be responsible for MSH degradation [60,65]. It remains unclear if this low level of BCA activity with BSH is capable of supplying Cys in a rapidly growing organism such as S. aureus.
S. aureus contains only one BshB2-type enzyme and the bshB NARSA transposon mutant (SAUSA300_0552) was determined by thiol analysis to be a complete BSH knockout [22,52]. The same bshB transposon mutant also lacks 90% of the wildtype bacillithiol S-conjugate amidase activity, indicating that this protein is responsible for all of the BshB activity and the majority of bacillithiol conjugate amidase activity in S. aureus [52]. An inhibitor of S. aureus BshB in combination with fosfomycin might provide a novel cotherapy method against S. aureus. Such a method would eliminate both BSH synthesis and BCA detoxification and would prevent fosfomycin detoxification.
Glyoxylases GlxA & GlxB
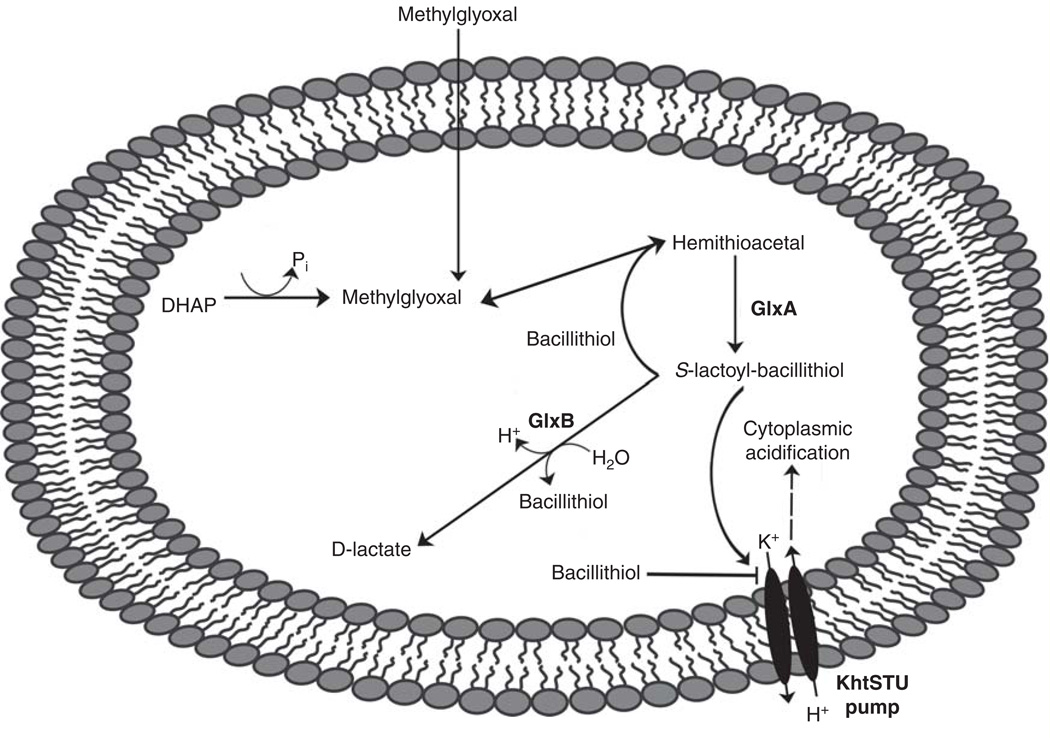
Methylglyoxal is a toxic, electrophilic byproduct formed during many metabolic pathways such as glycolysis (e.g., the conversion of dihydroxyacetone phosphate (DHAP) to methylgloxal) [66] and can also diffuse into the cell from the environment (Figure 8) [67]. Methylglyoxal detoxification in E. coli and can be separated into glutathione-dependent and -independent pathways. In the glutathione-dependent pathway, methyglyoxal reacts spontaneously with glutathione to form GS-hemithioacetal. The glyoxylase enzyme GlxI then converts GS-hemithioacetal to S-lactoyl-glutathione, which GlxII then converts to D-lactate and glutathione [68]. Production of S-lactoyl-glutathione activates the KefGB and KefKC potassium efflux pumps, leading to cytoplasmic acidification and protection from methylglyoxal presumably by antagonizing its reaction with cellular targets [69].
Figure 8. Bacillithiol-dependent methylglyoxal detoxification pathway in Bacillus subtilis.
Methylglyoxal can enter the cell via diffusion or can be formed intracellularly (i.e., from dihydroxyacetone phosphate, abbreviated DHAP). Bacillithiol and both glyoxalase A (GlxA) and glyoxylase B (GlxB) are utilized to convert methylglyoxal to D-lactate. Cytoplasmic acidification is mediated by the interplay of the KhtSTU pump with bacillithiol and S-lactoyl-bacillithiol.
Model from Chandrangsu et al. 2013 [67].
Recently bacillithiol-independent and dependent pathways for methylglyoxal detoxification in B. subtilis have been characterized [67]. Unlike E. coli, the primary resistance pathway in B. subtilis is thiol-independent. The BSH-dependent pathway is similar to glutathione-dependent methylglyoxal detoxification. Methylglyoxal reacts spontaneously with bacillithiol to form BS-hemithioacetal. The glyoxylase GlxA then converts BS-hemithioacetal to S-lactoyl-bacillithiol, which GlxB converts to D-lactate and bacillithiol. This pathway involves the acidification of the cytoplasm, which is both necessary and sufficient for bacillithiol-dependent methylglyoxal resistance. Acidification depends on both bacillithiol and a K+ efflux pump KhtU, which is in the same family of transporters as the E. coli KefGB and KefKC, and the ancillary proteins KhtS and KhtT (collectively called KhtSTU). The authors propose that the S-lactoyl-BSH conjugate gates the KhtU efflux pump in the manner similar to the gating of the E. coli efflux pump by S-lactoyl-glutathione, although direct evidence is still lacking. Thus, bacillithiol is proposed to mediate methylglyoxal resistance through cytoplasmic acidification rather than methyglyoxal detoxification (Figure 8).
The closest B. subtilis KhtU analog in S. aureus Newman is NWMN_0880, a Na+/H+ transporter (23% sequence identity with 69% coverage) in the same CPA2 family as the E. coli transporter. Interestingly, glxA shares 25–30% sequence identity with S. aureus FosB, depending on the subspecies. Structural and biochemical studies could elucidate whether these enzymes also bind bacillithiol.
Bacilliredoxins debacillithiolate proteins
Upon exposure of cells to oxidants such as cumene hydroperoxide or sodium hypochlorite (NaOCl), bacillithiol has been shown to form mixed disulfides with the cysteine residues of certain proteins. This process, termed S-bacillithiolation, protects protein cysteine residues from overoxidation and irreversible damage of the protein [19,70,71]. Transcriptomic and proteomic studies revealed that several proteins are bacillithiolated, including the redox-sensing regulator OhrR, the methionine synthase MetE and two proteins of unknown function YtxJ and YphP. Inactivation of OhrR results in the upregulation of ohrA, which encodes a peroxiredoxin that contributes to NaOCl resistance. Bacillithiol biosynthetic gene mutants show the same sensitivity to NaOCl as do ohrA knockouts, suggesting that bacillithiolation of OhrR is essential for ohrA expression [70]. S-bacillithiolation of MetE decreased the expression of methionine biosynthesis genes, resulting in transient methionine autotrophy [71], which also occurs during S-glutathionylation of E. coli MetE [72].
Debacillithiolation of protein mixed disulfides was hypothesized to be analogous to deglutathionylation, which is a process catalyzed by glutaredoxins. The first evidence that Firmicutes contained glutaredoxin-like proteins emerged from a bioinformatics search for the bacillithiol biosynthesis gene BshC. An EMBL STRING analysis remarkably identified four putative thiol-disulfide oxidoreductase proteins that had yet to be characterized, including YtxJ, a putative monothiol (TCPIS) active site glutaredoxin-like protein, and YpdA, a thioredoxin reductase-like protein and putative bacilliredoxin reductase. The other two proteins, YphP and YqiW, are paralogs of the DUF1094 family (53% identity) and contain a CxC motif that is typical of glutaredoxins [27]. Both proteins were recently demonstrated to have bacilliredoxin activity and were renamed BrxA (YphP) and BrxB (YqiW) [73]. These proteins were found to debacillithiolate active site cysteine residues of OhrR and MetE, two bacillithiolated redox-sensing proteins [19,70]. The missing part of the bacilliredoxin story is the disulfide reductase needed to recycle the bacillithiolated forms of BrxA and BrxB. One candidate for this enzyme is the thioredoxin reductase like protein YpdA found in early bioinformatics studies of BSH [27], which was observed to be bacillithiolated in its putative active site [70,71].
The B. subtilis BrxA and BrxB proteins share a high level of sequence identity with their S. aureus Newman homologs NWMN_1339 (57% with 97% sequence coverage) and NWMN_1420 (69% with 99% sequence coverage), respectively. Thus, bacilliredoxins are likely to participate in the oxidative stress response in S. aureus.
Response of Firmicutes to electrophilic & oxidative stress
Data on cellular responses to electrophiles and oxidants predates the discovery of bacillithiol, and reexamination of these transcriptomic and proteomic studies provides significant evidence for bacillithiol-dependent stress responses. Catabolism of phenolic acids like salicylic acid occurs during the decay of plant material in the soil and the response of Bacillus subtilis to salicylic acid indicates a role for bacillithiol transferases in this process. One of the eight putative STL enzymes from B. subtilis, yuaE, is upregulated in response to salicylic acid along with bshB2, which encodes an enzyme with both BshB deacetylase and bacillithiol conjugate amidase activity (Tables 1 & 2, [25,74]). Exposure of B. subtilis to the fungal-related antimicrobial compounds methylhydroquinone and 6-brom-2-vinyl-chroman-4-on elicited a response similar to that of salicylic acid: two bacillithiol transferases (yjoA, yuaE), the bshB2 gene yojG and the bacillithiol biosynthesis gene bshC were upregulated (Tables 1 & 2, [75]). Bacilliredoxin (brxB) was also upregulated, indicating that excessive protein thiolation might be occurring.
Table 1.
Bacillithiol-related genes in S. aureus and Bacillus strains.
| Bacillithiol-related gene | S. aureus Newman |
S. aureus USA300_ FPR3757 |
B. anthracis Ames |
B. subtilis 168 |
|---|---|---|---|---|
| bshA | NWMN_1369 | SAUSA300_1349 | BA_1558 | BSU22460 (ypjH) |
| bshB1 | – | – | BA_1557 | BSU22470 (ypjG) |
| bshB2 | NWMN_0530 | SAUSA300_0552 | BA_3524 BA_3888 |
BSU19460 (yojG) |
| bshC | NWMN_1087 | SAUSA300_1071 | BA_4058 | BSU15120 (yllA) |
| brxA | NWMN_1339 | SAUSA300_1321 | BA_2173 | BSU21860 (yphP) |
| brxB | NWMN_1420 | SAUSA300_1463 | BA_4378 | BSU23990 (yqiW) |
| Thioredoxin-reductase like enzyme | NWMN_1388 | SAUSA300_1369 | BA_1515 | BSU22950 (ypdA) |
| Thioredoxin family enzyme | NWMN_0710 | SAUSA300_0725 | BA_4931 | BSU29760 (ytxJ) |
| glxA | NWMN_2420 | SAUSA300_2461 | BA_3208 | BSU38370 (ywbC) |
| glxB | NWMN_1416 | SAUSA300_1458 | BA_5452 BA_2111 |
BSU32660 (yurT) |
| fosB | NWMN_2234 | SAUSA300_2280 | BA_4109 | BSU17840 (yndN) |
| S-transferase-like (STL) superfamily genes | NWMN_2591 | SAUSA300_2626 | ||
| BA_2700 | BSU08390 (yfiT/bstA) | |||
| BA_1354 | BSU31030 (yuaE) | |||
| BA_2379 | BSU05630 (dinB)† | |||
| BA_2379 | BSU10860 (yisT)† | |||
| BA_2937 | BSU13070 (ykkA) | |||
| BA_2558 | BSU10800 (yizA) | |||
| BA_4768 | BSU26780 (yrdA) | |||
| BSU12410 (yjoA) | ||||
| BA_2721 | ||||
| BA_3104 | ||||
| BA_2777 | ||||
| BA_3538 | ||||
| BA_2007 | ||||
| BA_3539 | ||||
| BA_2065 | ||||
| BA_2078 | ||||
| BA_2990 | ||||
B. subtilis 168 ORF names are in parenthesis. STL proteins listed on the same line share ≥30% amino acid identity with ≥90% sequence coverage. ORF names in grey share homology with more than one B. subtilis protein.
B. subtilis genes that share >30% amino acid sequence identity with ≥90% sequence coverage.
B. anthracis: Bacillus anthracis; B. subtilis: Bacillus subtilis; S. aureus: Staphylococcus aureus; ORF: Open reading frame.
Table 2.
Transcriptomic and proteomic analyses of bacillithiol-related enzymes during treatment with stressors.
| Compound | Structure | Organism | Gene | Transcriptome | Proteome |
|---|---|---|---|---|---|
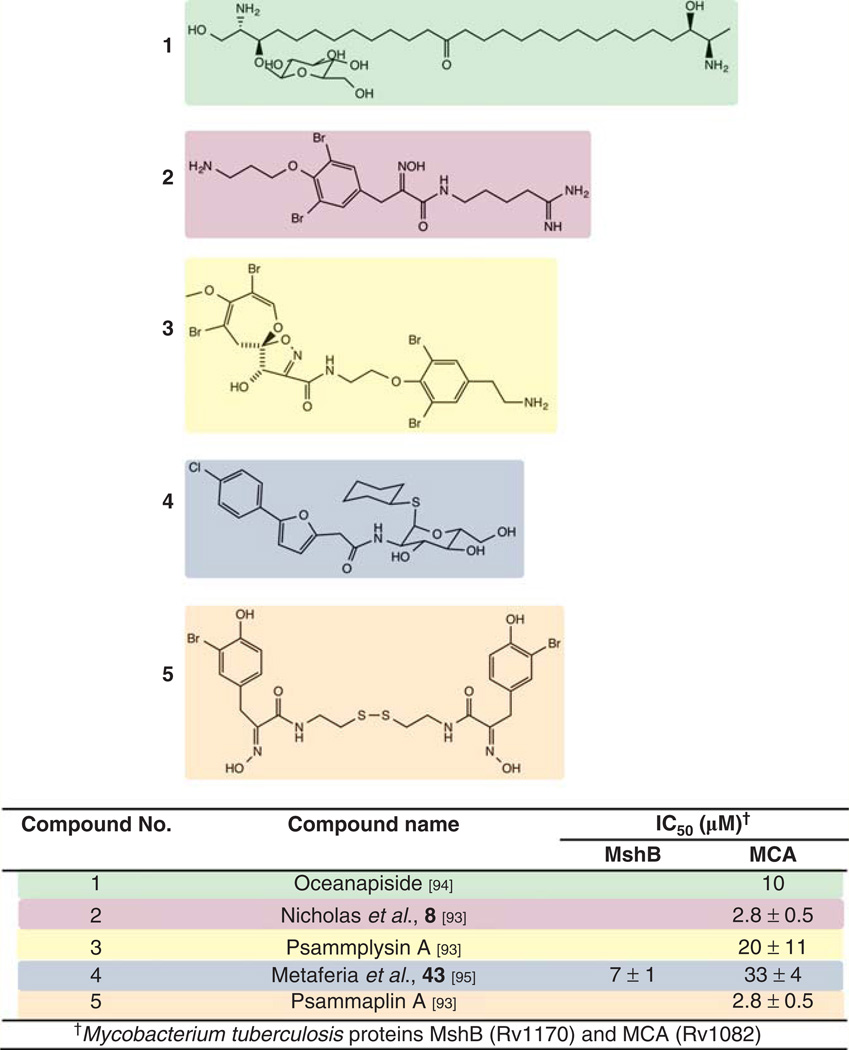
| Salicylic acid | 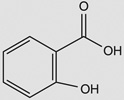 |
Bacillus subtilis 168† | yuaE (STL) | 6–8 | 5–6 |
| yojG (BshB2) | 3–4 | – | |||
| yhdN (FosB) | 3–4 | 3–4 | |||
| 2-methylhydroquinone | 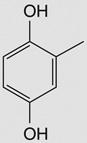 |
Bacillus subtilis 168‡ | yjoA (STL) | 6 | – |
| yojG (BshB2) | 6–7 | – | |||
| yuaE (STL) | 10–14 | 4–10 | |||
| yqiW (BrxB) | 4–6 | – | |||
| 6-brom-2-vinyl-chroman-4-on |  |
Bacillus subtilis 168‡ | yjoA (STL) | 4–5 | – |
| yojG (BshB2) | 9–10 | – | |||
| yuaE (STL) | 7–10 | 3–8 | |||
| yllA (BshC) | 4 | – | |||
| yqiW (BrxB) | 4–6 | – | |||
| Hydrogen peroxide | H2O2 | Staphylococcus aureus MW2§ | MW1348 (BshA) | 0.5 | ND |
| MW2611 (BstA/STL) | 3 | ND | |||
| MW2442¶¶ (GlxA) | 2 | ND | |||
| MW1463¶¶ (GlxB) | 3 | ND | |||
| MW1467¶¶ (BrxB) | 6 | ND | |||
| Bacillus anthracis Sterne¶ | BA_1558 (BshA) | 2 | ND | ||
| BA_1557 (BshB1) | 2 | ND | |||
| BA_3524 (BshB2) | 2 | ND | |||
| BA_4058 (BshC) | 3 | ND | |||
| BA_1515¶¶ (YpdA) | 2 | ND | |||
| Diamide |  |
Staphylococcus aureus COL# | SACOL1498 (BshA) | 2 | ND |
| SACOL1190 (BshC) | 5 | ND | |||
| SACOL2717 (BstA/ STL) | 0.3 | ND | |||
| SACOL1553¶¶ (GlxB) | 2 | ND | |||
| SACOL1520¶¶ (YpdA) | 4 | ND | |||
| Nitric oxide | NO | Bacillus subtilis 168†† | yuaE (STL) | ND | 2–3 |
| Staphylococcus aureus COL†† | SACOL0614 (BshB2) | ND | 3 | ||
| SACOL1520¶¶ (YpdA) | ND | 1–3 | |||
| Nitrite | NO2− | Staphylococcus aureus N315‡‡ | SA1291 (BshA) | 3 | ND |
| SA0525 (BshB2) | 3 | ND | |||
| SA1020 (BshC) | 5 | ND | |||
| SA2310¶¶ (GlxA) | 8 | ND | |||
| SA1311¶¶ (YpdA) | 3 | ND | |||
| Azurophilic granule proteins | - | Staphylococcus aureus MW2§ | MW1348 (BshA) | 0.6 | ND |
| MW0552 (BshB2) | 0.5 | ND | |||
| MW2611 (BstA/STL) | 2 | ND | |||
| MW1463¶¶ (GlxB) | 0.7 | ND | |||
| MW1318¶¶ (BrxA) | 0.6 | ND | |||
| MW1467¶¶ (BrxB) | 2 | ND | |||
| Hypochlorous acid | HOCl | Staphylococcus aureus MW2§ | MW1348 (BshA) | 0.5 | ND |
| MW2611 (BstA/STL) | 2 | ND | |||
| MW2442¶¶ (GlxA) | 3 | ND | |||
| MW1318¶¶ (BrxA) | 0.6 | ND | |||
| MW1467¶¶ (BrxB) | 2 | ND | |||
| Mammalian peptidoglycan recognition protein (PGLYRP4) | – | Bacillus subtilis 168§§ | yuaE (STL) | 8 | ND |
Numbers < 1 show down-regulated genes, where 1 is no change.
Van Duy et al. 2007; treatment with 4 mM salicylic acid for 10 min, range shown is from duplicate experiments.
Van Duy et al. 2007; 100 µg/ml 6-brom-2-vinyl-chroman-4-on or 63 µg/ml of 2-methylhydroquinone (treatment for 10 min for both conditions), range shown is from duplicate experiments.
Palazzolo-Balance et al. 2008; treatment with 100 µg/ml azurophilic granule proteins, 5 mM H2O2, or 25 µM HOCl for 15 min for all conditions.
Pohl et al. 2011; treatment with 1.0 mM H2O2 for 10 min.
Posada et al. 2014; treatment with 1 mM diamide for 20 min.
Hochgrafe et al. 2008; treatment with 500 µM MAHMA NONOate (nitric oxide donor) 10 min, range shown is from duplicate experiments.
Schlag et al. 2007; treatment with 5 mM sodium nitrite for 6 h at 37°C.
Kashyap et al. 2014; treatment with 100 µg/ml PGLYRP4 for 30 min at 37°C.
Closest homolog to B. subtilis 168 gene.
ND: Not determined.
Firmicutes are exposed to hydrogen peroxide during infection, where bacillithiol-dependent defenses function as an adjunct to catalase activity. In Bacillus anthracis exposed to H2O2, all three bacillithiol biosynthesis genes as well as bshB2 and the bacilliredoxin reductase candidate ypdA are upregulated (Tables 1 & 2, [76]). Exposure of S. aureus MW2 to H2O2 resulted in a twofold downregulation of bshA, but upregulation of bstA, the glyoxylase genes glxA and glxB, and the two putative bacilliredoxins brxA and brxB (Tables 1 & 2).
Furthermore, mammalian peptidoglycan recognition proteins (PGRP) are a component of the host response and have been shown to kill B. subtilis by a combination of oxidative stress, thiol depletion and metal stress from release of Zn2+ and Cu2+ ions [77]. Treatment with one isoform of PGRP (PGLYRP4) increased expression of the bacillithiol transferase yuaE ~8 fold [77]. Diamide leads to disulfide stress by oxidizing cellular thiols to disulfides. S. aureus COL responds with upregulation of the bacillithiol biosynthesis genes bshA and bshC and downregulation of bstA [78]. The putative bacilliredoxin reductase gene ypdA is also upregulated, which is consistent with the observation that diamide increases protein thiolation [79]. Oxidizing agents also induce bacillithiol-related genes in S. aureus. Nitrite upregulates all three bacillithiol biosynthesis genes, glxA and ypdA in S. aureus N315 [80]. Nitric oxide leads to the upregulation of genes encoding the BSH detoxification proteins bshB2 and ypdA [81]. Oxidizing agents produced in neutrophils, hypochlorous acid, azurophilic granule proteins and H2O2 downregulate bshA and brxA ~2-fold, but induce bstA, glxA and brxB ~2-fold [78].
From these studies, we see that electrophilic compounds tend to induce specific bacillithiol transferases in the Firmicutes, whereas the major bacillithiol conjugate amidase protein BshB2 is upregulated in response to both electrophiles and oxidants. The bacillithiol biosynthesis genes are most often upregulated in response to oxidants such as H2O2, diamide and nitrite. Thus, bacillithiol is likely to play a key role in responding to these toxic molecules for both B. subtilis in soil and S. aureus infections.
Spontaneous enzyme-independent reactions with bacillithiol
Thiols can react spontaneously with electrophilic substrates such as monobromobimane and methylglyoxal. Preliminary evidence indicates that bacillithiol participates in a wider range of spontaneous reactions than other major low-molecular-weight thiols, presumably because its thiol group is more acidic and thus more reactive at cytoplasmic pH (Figures 2 and 3). This property would help ensure rapid detoxification of certain harmful substances. The uncatalyzed reaction of H2O2 with bacillithiol was the first such reaction to be described, and it was found to proceed at a rate of ~80 nmol min−1 mg−1, 40-fold higher than the rate with glutathione or mycothiol [56]. Bacillithiol has been found to react in an enzyme-independent manner with a number of substrates, such as the antibiotic rifamycin S and the thiol-reactive molecules etacrynic acid, 4-hydroxynonenal and sulforaphane [52]. The antibiotic cerulenin also reacts with bacillithiol spontaneously [52], but the S. aureus bacillithiol transferase BstA enhances this rate of reaction 2 fold [20,52,56]. These results suggest that bacillithiol may be more reactive than glutathione or mycothiol in uncatalyzed reactions with certain electrophiles.
Contribution of bacillithiol to pathogenesis
The model for bacillithiol-dependent detoxification
The models for GSH- and MSH-dependent detoxification pathways involve similar enzymatic components and both produce a mercapturic acid, an N-acetylated cysteine-toxin conjugate that is excreted from the organism and is usually non-toxic [60,61,82,83]. In S. aureus, bacillithiol conjugate amidase and N-acetyltransferase activity were both detected using cell-free extracts [48]. Studies with whole cells demonstrated that BSH was added to the toxins prior to the appearance of the cysteine-toxin conjugate, demonstrating that the cysteine-toxin conjugate was likely the product of cleavage of the bacillithol-conjugate. The cysteine-toxin conjugate is then N-acetylated and excreted from the cells. In support of this interpretation, the levels of GlcN-L-malate increased with the appearance of the cysteine-toxin conjugate [48].
Bacillithiol protects S. aureus against mammalian cells involved in immune responses
Bacteria that have been engulfed by macrophages and neutrophils are exposed to a number of reactive oxygen and nitrogen species, and bacillithiol likely serves as a cellular redox buffer that can neutralize these highly damaging molecules. Indeed, the BSH-deficient Staphylococcus aureus strain NCTC 8325 showed diminished survival in murine macrophages in comparison to strains that produce BSH [49]. A similar deficiency was observed in whole blood with the BSH deficient S. aureus strain SH1000 as compared with the BSH containing methicillin-resistant S. aureus strain COL [44]. Taken together with the increased sensitivity of bacillithiol-deficient mutants to H2O2 [44] and with the rapid rate of reaction of bacillithiol with H2O2 [56], the decreased survival of the bacillithiol mutants in macrophages might be explained by their increased susceptibility to oxidative stress.
Bacillithiol levels have been reported to be relatively low in exponentially growing S. aureus [14,26,44] compared with the glutathione levels in Gram-negative bacteria, but comparable to the bacillithiol levels in B. subtilis. Bacillithiol levels in B. subtilis continue to increase to ~5.2 mM during late stationary phase, but it is unclear whether the same occurs in S. aureus. Cysteine and bacillithiol levels did rise rapidly in S. aureus in response to treatment with rifamycin S, however [48].
Interestingly, S. aureus has been found to import glutathione from the growth medium during stationary phase [49]. Although S. aureus does not encode any known enzymes that utilize this thiol as a cofactor, glutathione can react spontaneously with thiol-reactive molecules and may still serve as a protective redox buffer for the bacterium. Other Gram-positive bacteria such as Streptococcus mutans [84] and Streptococcus pneumoniae have also been reported to import glutathione, which conferred oxidative stress resistance and increased survival in the mouse model of infection [85]. Interestingly, the S. aureus genome encodes a protein (NWMN_0147) that shares 27% sequence identity with 91% coverage to the M. tuberculosis γ-glutamyltranspeptidase, which is required to hydrolyze the γ-glutamyl-cysteine peptide bond of GSH to release cysteine in glutathione producing organisms [86]. M. tuberculosis γ-glutamyltranspeptidase provides this intracellular pathogen the critical activity to utilize macrophage GSH as a source of cysteine for protein synthesis during infection [64]. The S. aureus γ-glutamyltranspeptidase analog has not been studied to the same extent as the M. tuberculosis γ-glutamyltranspeptidase, and it is unclear whether this enzyme is expressed during uptake of GSH. It remains an open question whether glutathione is imported by S. aureus during eukaryotic infection and whether it contributes to the fitness of the pathogen.
Expert commentary & five-year view
The rapid rate of the development of resistance of methicillin-resistant S. aureus to the current therapeutic options is a great health concern, and many research efforts are focused on identifying new antibiotics and new antibiotic targets to combat this organism [87,88]. The cell wall synthesis inhibitor fosfomycin is currently used in the treatment of urinary tract and gastrointestinal S. aureus infections. Fosfomycin can enter the cell via the L-α-glycerophosphate transport system and/or the hexose-monophosphate transport system (UhpT) [89]. Mutations in these transporters contribute to fosfomycin resistance and are presumably a major mechanism of fosfomycin resistance [90]. The plasmid borne fosB gene has been identified in 34% of fosfomycin-resistant clinical Staphylococci isolates [91]. Recently, deletion of the chromosomally encoded fosB or bshA in clinically relevant MRSA isolates was determined to be the major source of inherent fosfomycin susceptibility [44]. The use of fosfomycin as a monotherapy has resulted in rapid development of resistance [92]. The FDA recently recommended that one antimicrobial therapy that should be considered for streamlined development are those incorporating an agent that inhibits drug resistance. We predict that the bacillithiol biosynthetic enzymes would be good targets for neutralizing resistance, because mutants lacking bacillithiol show increased sensitivity both to oxidative stress and to drugs such as fosfomycin that are detoxified by a FosB/bacillithiol-dependent process [22,27,44,49]. Thus, inhibition of BSH biosynthesis should sensitize S. aureus to the innate immune response and to antibiotics such as fosfomycin.
The glycosyltransferase BshA may be a problematic drug target when used in combination therapy with fosfomycin due to increases in UDP-GlcNAc levels. Treatment of S. aureus with fosfomycin, which targets peptidoglycan biosynthesis by inhibiting MurA and MurZ, has been shown to increase levels of UDP-GlcNAc, the substrate of BshA [51]. This accumulation of a BshA substrate could hinder the ability of a potential competitive inhibitor to effectively block BshA activity and decrease bacillithiol levels. Thus, bacillithiol would still be synthesized at some level and would confer resistance to fosfomycin. BshC, the final enzyme in the BSH biosynthesis pathway, has yet to be characterized, so significant development would be required before a BshC inhibitor screen could be devised.
S. aureus BshB may be a good candidate drug target, as this enzyme is involved in bacillithiol biosynthesis and also provides most of the bacillithiol conjugate amidase detoxification activity [22,25,52]. Unlike the Bacillus species discussed in this review, there are no compensating activities for S. aureus BshB, so the appropriate drug could completely inhibit BSH biosynthesis. S. aureus BshB is ~25% identical with M. tuberculosis MshB (Rv1170) and MCA (Rv1082), and is easily expressed in E. coli. A screen for natural product inhibitors yielded µM inhibitors of M. tuberculosis MCA and MshB (Figure 9, [93–95]), and these would constitute a logical starting point for S. aureus BshB2 inhibition studies. Inhibition of S. aureus BshB2 activity would diminish the production of bacillithiol and thereby render the organism more susceptible to being killed by the host defenses such as the oxidizing environment of macrophages, neutrophils and blood [44,49]. Depletion of bacillithiol would also diminish the BCA detoxification activity of BshB2 and inhibit excretion of mercapturic acids. A decrease in bacillithiol levels could result in a 15- 60-fold decrease in resistance to fosfomycin [44], suggesting a combination therapy of fosfomycin and a BshB/BCA inhibitor could be effective against multidrug resistant S. aureus infections. Identifying an inhibitor of this enzyme should be feasible within the next 5 years, especially with the numerous inhibitor chemotypes identified for the homologous M. tuberculosis MCA (Figure 9).
Figure 9. Natural product and synthetic inhibitors of M. tuberculosis metalloproteins MshB and MCA.
Oceanapiside (compound 1) is a non-competitive inhibitor of M. tuberculosis MCA; a brominated derivative of tyrosine (compound 2) and psammplysin A (compound 3) are competitive inhibitors of MCA. Psammaplin A (compound 5) and (compound 2) contain oxime moieties, known to chelate metal ions. Compound 4 is a synthetic substrate analog of GlcNAc-Ins (substrate of MshB), and one of the few compounds that inhibits both M. tuberculosis MshB and MCA.
Key issues.
Bacillithiol is a recently discovered low-molecular-weight thiol found in the Firmicutes, which include the pathogens Staphylococcus aureus and Bacillus anthracis.
The low-molecular-weight thiols bacillithiol and mycothiol are related by their Cys-glucosamine core moiety.
In S. aureus, three unlinked genes encode the bacillithiol biosynthesis enzymes, which are each essential for bacillithiol biosynthesis.
The BshB enzyme has dual activities: it is a deacetylase utilized in the second step of bacillithiol biosynthesis and a bacillithiol conjugate amidase that can cleave S-conjugates.
Compared to other low-molecular-weight thiols found in bacteria, bacillithiol has structural differences that make it more reactive with hydrogen peroxide, xenobiotics and drugs, particularly fosfomycin.
Bacillithiol is the thiol cofactor of the enzyme FosB. Collectively, bacillithiol and FosB-derived fosfomycin resistance is the major detoxification mechanism of multi-drug resistant Staphylococcus aureus to fosfomycin.
S. aureus mutants deficient in bacillithiol are more susceptible to killing by macrophages, neutrophils and whole blood.
A possible new strategy for treatment of multidrug resistant Staphylococcus aureus is the combination therapy of a bacillithiol biosynthesis inhibitor with fosfomycin.
Acknowledgments
We thank Dr Alan Derman and Eammon Riley for their insightful comments on the review. We also thank Dr James LaClair for ChemDraw3D structures of bacillithiol.
This research was supported by the National Institutes of Health (R01-AI095125).
Footnotes
Financial & competing interests disclosure
Kit Pogliano is a founder of Linnaeus Bioscience Incorporated, holds equity interest in the company and receives consulting income. This arrangement has been reviewed and approved by the University of California, San Diego in accordance with its conflict of interest policies. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
References
Papers of special note have been highlighted as:
• of interest
•• of considerable interest
- 1. Fahey RC. Glutathione analogs in prokaryotes. Biochim Biophys Acta. 2013;1830(5):3182–3198. doi: 10.1016/j.bbagen.2012.10.006. • A comprehensive review of bacterial low molecular weight thiols.
- 2.Park S, Imlay JA. High levels of intracellular cysteine promote oxidative DNA damage by driving the fenton reaction. J Bacteriol. 2003;185(6):1942–1950. doi: 10.1128/JB.185.6.1942-1950.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Keire DA, Robert JM, Rabenstein DL. Microscopic Protonation equilibria and solution conformations of coenzyme-a and coenzyme-a disulfides. J Org Chem. 1992;57(16):4427–4431. [Google Scholar]
- 4.Benesch RE, Benesch R. The acid strength of the -Sh group in cysteine and related compounds. J Am Chem Soc. 1955;77(22):5877–5881. [Google Scholar]
- 5.Rabenstein DL. Nuclear magnetic resonance studies of the acid-base chemistry of amino acids and peptides. I. Microscopic ionization constants of glutathione and methylmercury-complexed glutathione. J Am Chem Soc. 1973;95(9):2797–2803. [Google Scholar]
- 6.Setlow B, Setlow P. Levels of acetyl coenzyme A, reduced and oxidized coenzyme A, and coenzyme A in disulfide linkage to protein in dormant and germinated spores and growing and sporulating cells of Bacillus megaterium. J Bacteriol. 1977;132(2):444–452. doi: 10.1128/jb.132.2.444-452.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fahey RC, Brown WC, Adams WB, Worsham MB. Occurrence of glutathione in bacteria. J Bacteriol. 1978;133(3):1126–1129. doi: 10.1128/jb.133.3.1126-1129.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Newton GL, Dorian R, Fahey RC. Analysis of biological thiols: derivatization with monobromobimane and separation by reverse-phase high-performance liquid chromatography. Anal Biochem. 1981;114(2):383–387. doi: 10.1016/0003-2697(81)90498-x. [DOI] [PubMed] [Google Scholar]
- 9.Fahey RC, Newton GL. Determination of low-molecular-weight thiols using monobromobimane fluorescent labeling and high-performance liquid chromatography. Methods Enzymol. 1987;143:85–96. doi: 10.1016/0076-6879(87)43016-4. [DOI] [PubMed] [Google Scholar]
- 10.Radkowsky AE, Kosower EM. Bimanes. 17. (Haloalky1)- 1,5-diazabicyclo[3.3.0loctadienediones (Halo-9,10-dioxabimanes): Reactivity toward the Tripeptide Thiol. Glutathione. J Am Chem Soc. 1986;108(15):4527–4531. [Google Scholar]
- 11.Spies HS, Steenkamp DJ. Thiols of intracellular pathogens. Identification of ovothiol A in Leishmania donovani and structural analysis of a novel thiol from Mycobacterium bovis. Eur J Biochem. 1994;224(1):203–213. doi: 10.1111/j.1432-1033.1994.tb20013.x. [DOI] [PubMed] [Google Scholar]
- 12.Newton GL, Bewley CA, Dwyer TJ, et al. The structure of U17 isolated from Streptomyces clavuligerus and its properties as an antioxidant thiol. Eur J Biochem. 1995;230(2):821–825. doi: 10.1111/j.1432-1033.1995.0821h.x. [DOI] [PubMed] [Google Scholar]
- 13.Newton GL, Buchmeier N, Fahey RC. Biosynthesis and functions of mycothiol, the unique protective thiol of Actinobacteria. Microbiol Mol Biol Rev. 2008;72(3):471–494. doi: 10.1128/MMBR.00008-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Newton GL, Rawat M, La Clair JJ, et al. Bacillithiol is an antioxidant thiol produced in Bacilli. Nat Chem Biol. 2009;5(9):625–627. doi: 10.1038/nchembio.189. • The first report of the structure and distribution of bacillithiol in bacteria.
- 15.Newton GL, Fahey RC, Cohen G, Aharonowitz Y. Low-molecular-weight thiols in streptomycetes and their potential role as antioxidants. J Bacteriol. 1993;175(9):2734–2742. doi: 10.1128/jb.175.9.2734-2742.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sakuda S, Zhou ZY, Yamada Y. Structure of a novel disulfide of 2-(N-acetylcysteinyl) amido-2-deoxy-alpha-D-glucopyranosylmyo-inositol produced by Streptomyces sp. Biosci Biotechnol Biochem. 1994;58(7):1347–1348. doi: 10.1271/bbb.58.1347. [DOI] [PubMed] [Google Scholar]
- 17.Fahey RC, Newton GL. Occurrence of low molecular weight thiols in biological systems. In: Larsson A, Orrenius S, Holmgren A, Mannervik B, editors. Functions of glutathione: biochemical, physiological, toxicological and clinical aspects. New York: Raven Press; 1983. [Google Scholar]
- 18.Nicely NI, Parsonage D, Paige C, et al. Structure of the type III pantothenate kinase from Bacillus anthracis at 2.0 A resolution: implications for coenzyme A-dependent redox biology. Biochem. 2007;46(11):3234–3245. doi: 10.1021/bi062299p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee JW, Soonsanga S, Helmann JD. A complex thiolate switch regulates the Bacillus subtilis organic peroxide sensor OhrR. Proc Natl Acad Sci USA. 2007;104(21):8743–8748. doi: 10.1073/pnas.0702081104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sharma SV, Arbach M, Roberts AA, et al. Biophysical Features of Bacillithiol, the Glutathione Surrogate of Bacillus subtilis and other Firmicutes. ChemBioChem. 2013;14(16):2160–2168. doi: 10.1002/cbic.201300404. • A physical chemistry study of bacillithiol suggesting unusual reactivity of the cysteine derived thiol and ionization of the moieties involved in chelation of metals.
- 21.Ma Z, Chandrangsu P, Helmann TC, et al. Bacillithiol is a major buffer of the labile zinc pool in Bacillus subtilis. Mol Microbiol. 2014;94(4):756–770. doi: 10.1111/mmi.12794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rajkarnikar A, Strankman A, Duran S, et al. Analysis of mutants disrupted in bacillithiol metabolism in Staphylococcus aureus. Biochem Biophys Res Commun. 2013;436(2):128–133. doi: 10.1016/j.bbrc.2013.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Andreini C, Banci L, Bertini I, Rosato A. Zinc through the three domains of life. J Proteome Res. 2006;5(11):3173–3178. doi: 10.1021/pr0603699. [DOI] [PubMed] [Google Scholar]
- 24.Kehl-Fie TE, Chitayat S, Hood MI, et al. Nutrient metal sequestration by calprotectin inhibits bacterial superoxide defense, enhancing neutrophil killing of Staphylococcus aureus. Cell Host Microbe. 2011;10(2):158–164. doi: 10.1016/j.chom.2011.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Fang Z, Roberts AA, Weidman K, et al. Cross-functionalities of Bacillus deacetylases involved in bacillithiol biosynthesis and bacillithiol-S-conjugate detoxification pathways. Biochem J. 2013;454(2):239–247. doi: 10.1042/BJ20130415. • An in depth kinetic study of the role of BshB in BSH biosynthesis and detoxification reactions in Bacillus.
- 26. Roberts AA, Sharma SV, Strankman AW, et al. Mechanistic studies of FosB: a divalent-metal-dependent bacillithiol-S-transferase that mediates fosfomycin resistance in Staphylococcus aureus. Biochem J. 2013;451(1):69–79. doi: 10.1042/BJ20121541. •• A signifcant study establishing bacillithiol as cofactor for FosB mediated fosfomycin resistance in S. aureus.
- 27. Gaballa A, Newton GL, Antelmann H, et al. Biosynthesis and functions of bacillithiol, a major low-molecular-weight thiol in Bacilli. Proc Natl Acad Sci USA. 2010;107(14):6482–6486. doi: 10.1073/pnas.1000928107. • The initial study elucidating the bacillithiol biosynthesis pathway in Bacillus subtilis.
- 28.Newton GL, Ta P, Bzymek KP, Fahey RC. Biochemistry of the initial steps of mycothiol biosynthesis. J Biol Chem. 2006;281(45):33910–33920. doi: 10.1074/jbc.M604724200. [DOI] [PubMed] [Google Scholar]
- 29.Ruane KM, Davies GJ, Martinez-Fleites C. Crystal structure of a family GT4 glycosyltransferase from Bacillus anthracis ORF BA1558. Proteins. 2008;73(3):784–787. doi: 10.1002/prot.22171. [DOI] [PubMed] [Google Scholar]
- 30.Parsonage D, Newton GL, Holder RC, et al. Characterization of the N-acetyl-alpha-D-glucosaminyl l-malate synthase and deacetylase functions for bacillithiol biosynthesis in Bacillus anthracis. Biochemistry. 2010;49(38):8398–8414. doi: 10.1021/bi100698n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Upton H, Newton GL, Gushiken M, et al. Characterization of BshA, bacillithiol glycosyltransferase from Staphylococcus aureus and Bacillus subtilis. FEBS Lett. 2012;586(7):1004–1008. doi: 10.1016/j.febslet.2012.02.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fadouloglou VE, Deli A, Glykos NM, et al. Crystal structure of the BcZBP, a zinc-binding protein from Bacillus cereus. FEBS J. 2007;274(12):3044–3054. doi: 10.1111/j.1742-4658.2007.05834.x. [DOI] [PubMed] [Google Scholar]
- 33.Deli A, Koutsioulis D, Fadouloglou VE, et al. LmbE proteins from Bacillus cereus are de-N-acetylases with broad substrate specificity and are highly similar to proteins in Bacillus anthracis. FEBS J. 2010;277(13):2740–2753. doi: 10.1111/j.1742-4658.2010.07691.x. [DOI] [PubMed] [Google Scholar]
- 34.Sareen D, Steffek M, Newton GL, Fahey RC. ATP-dependent L-cysteine: 1D-myo-inosityl 2-amino-2-deoxy-alpha-D-glucopyranoside ligase, mycothiol biosynthesis enzyme MshC, is related to class I cysteinyl-tRNA synthetases. Biochemistry. 2002;41(22):6885–6890. doi: 10.1021/bi012212u. [DOI] [PubMed] [Google Scholar]
- 35.VanDuinen AJ, Winchell KR, Keithly ME, Cook PD. The X-ray Crystallographic Structure of BshC, a Unique Enzyme Involved in Bacillithiol Biosynthesis. Biochemistry. 2014;54(2):100–103. doi: 10.1021/bi501394q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Asnis RE. A glutathione reductase from Escherichia coli. J Biol Chem. 1955;213(1):77–85. [PubMed] [Google Scholar]
- 37.Cole ST, Brosch R, Parkhill J, et al. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393(6685):537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- 38.Patel MP, Blanchard JS. Expression, purification, and characterization of Mycobacterium tuberculosis mycothione reductase. Biochemistry. 1999;38(36):11827–11833. doi: 10.1021/bi991025h. [DOI] [PubMed] [Google Scholar]
- 39.delCardayre SB, Davies JE. Staphylococcus aureus coenzyme A disulfide reductase, a new subfamily of pyridine nucleotide-disulfide oxidoreductase. Sequence, expression, and analysis of cdr. J Biol Chem. 1998;273(10):5752–5757. doi: 10.1074/jbc.273.10.5752. [DOI] [PubMed] [Google Scholar]
- 40.delCardayre SB, Stock KP, Newton GL, et al. Coenzyme A disulfide reductase, the primary low molecular weight disulfide reductase from Staphylococcus aureus. Purification and characterization of the native enzyme. The J Biol Chem. 1998;273(10):5744–5751. doi: 10.1074/jbc.273.10.5744. [DOI] [PubMed] [Google Scholar]
- 41.Wallen JR, Mallett TC, Boles W, et al. Crystal structure and catalytic properties of Bacillus anthracis CoADR-RHD: implications for flavin-linked sulfur trafficking. Biochemistry. 2009;48(40):9650–9667. doi: 10.1021/bi900887k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tuggle CK, Fuchs JA. Glutathione reductase is not required for maintenance of reduced glutathione in Escherichia coli K-12. J Bacteriol. 1985;162(1):448–450. doi: 10.1128/jb.162.1.448-450.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bhaskar A, Chawla M, Mehta M, et al. Reengineering redox sensitive GFP to measure mycothiol redox potential of Mycobacterium tuberculosis during infection. PLoS Pathog. 2014;10(1):e1003902. doi: 10.1371/journal.ppat.1003902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Posada AC, Kolar SL, Dusi RG, et al. Importance of bacillithiol in the oxidative stress response of Staphylococcus aureus. Infect Immun. 2014;82(1):316–332. doi: 10.1128/IAI.01074-13. • A detailed study on bacillithiol dependent fosfomycin sensitivity in multidrug resistant S. aureus bacillithiol mutants.
- 45.Cao M, Bernat BA, Wang Z, et al. FosB, a cysteine-dependent fosfomycin resistance protein under the control of sigma(W), an extracytoplasmic-function sigma factor in Bacillus subtilis. J Bacteriol. 2001;183(7):2380–2383. doi: 10.1128/JB.183.7.2380-2383.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lamers AP, Keithly ME, Kim K, et al. Synthesis of bacillithiol and the catalytic selectivity of FosB-type fosfomycin resistance proteins. Org Lett. 2012;14(20):5207–5209. doi: 10.1021/ol302327t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Thompson MK, Keithly ME, Harp J, et al. Structural and chemical aspects of resistance to the antibiotic fosfomycin conferred by FosB from Bacillus cereus. Biochemistry. 2013;52(41):7350–7362. doi: 10.1021/bi4009648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Newton GL, Fahey RC, Rawat M. Detoxification of toxins by bacillithiol in Staphylococcus aureus. Microbiology. 2012;158(Pt 4):1117–1126. doi: 10.1099/mic.0.055715-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pother DC, Gierok P, Harms M, et al. Distribution and infection-related functions of bacillithiol in Staphylococcus aureus. Int J Med Microbiol. 2013;303(3):114–123. doi: 10.1016/j.ijmm.2013.01.003. [DOI] [PubMed] [Google Scholar]
- 50.Melis A, Kosower NS, Crawford NA, et al. Bimanes—26. An Electron Transfer Reaction Between Photosystem II and Monobromobimane Induces Static Chlorophyll a Quenching in Spinach Chloroplasts. Photochem Photobio. 1986;43(5):583–589. [Google Scholar]
- 51.Dorries K, Schlueter R, Lalk M. The impact of antibiotics with various target sides on the metabolome of Staphylococcus aureus. Antimicrob Agents Chemother. 2014;58(12):7151–7163. doi: 10.1128/AAC.03104-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Perera VR, Newton GL, Parnell JM, et al. Purification and characterization of the Staphylococcus aureus bacillithiol transferase BstA. Biochimi Biophys Acta. 2014;1840(9):2851–2861. doi: 10.1016/j.bbagen.2014.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Feng J, Che Y, Milse J, et al. The gene ncgl2918 encodes a novel maleylpyruvate isomerase that needs mycothiol as cofactor and links mycothiol biosynthesis and gentisate assimilation in Corynebacterium glutamicum. J Biol Chem. 2006;281(16):10778–10785. doi: 10.1074/jbc.M513192200. [DOI] [PubMed] [Google Scholar]
- 54.Zhao Q, Wang M, Xu D, et al. Metabolic coupling of two small-molecule thiols programs the biosynthesis of lincomycin A. Nature. 2015;518(7537):115–119. doi: 10.1038/nature14137. [DOI] [PubMed] [Google Scholar]
- 55.He P, Moran GR. Structural and mechanistic comparisons of the metal-binding members of the vicinal oxygen chelate (VOC) superfamily. J Inorg Biochem. 2011;105(10):1259–1272. doi: 10.1016/j.jinorgbio.2011.06.006. [DOI] [PubMed] [Google Scholar]
- 56.Newton GL, Leung SS, Wakabayashi JI, et al. The DinB superfamily includes novel mycothiol, bacillithiol, and glutathione S-transferases. Biochemistry. 2011;50(49):10751–10760. doi: 10.1021/bi201460j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tran NP, Gury J, Dartois V, et al. Phenolic acid-mediated regulation of the padC gene, encoding the phenolic acid decarboxylase of Bacillus subtilis. J Bacteriol. 2008;190(9):3213–3224. doi: 10.1128/JB.01936-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gury J, Barthelmebs L, Tran NP, et al. Cloning, deletion, and characterization of PadR, the transcriptional repressor of the phenolic acid decarboxylase-encoding padA gene of Lactobacillus plantarum. Appl Environ Microbiol. 2004;70(4):2146–2153. doi: 10.1128/AEM.70.4.2146-2153.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lubelski J, de Jong A, van Merkerk R, et al. LmrCD is a major multidrug resistance transporter in Lactococcus lactis. Mol Microbiol. 2006;61(3):771–781. doi: 10.1111/j.1365-2958.2006.05267.x. [DOI] [PubMed] [Google Scholar]
- 60.Newton GL, Av-Gay Y, Fahey RC. A novel mycothiol-dependent detoxification pathway in mycobacteria involving mycothiol S-conjugate amidase. Biochemistry. 2000;39(35):10739–10746. doi: 10.1021/bi000356n. [DOI] [PubMed] [Google Scholar]
- 61.Steffek M, Newton GL, Av-Gay Y, Fahey RC. Characterization of Mycobacterium tuberculosis mycothiol S-conjugate amidase. Biochemistry. 2003;42(41):12067–12076. doi: 10.1021/bi030080u. [DOI] [PubMed] [Google Scholar]
- 62.Newton GL, Av-Gay Y, Fahey RC. N-Acetyl-1-D-myo-inosityl-2-amino-2-deoxy-alpha-D-glucopyranoside deacetylase (MshB) is a key enzyme in mycothiol biosynthesis. J Bacteriol. 2000;182(24):6958–6963. doi: 10.1128/jb.182.24.6958-6963.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Newton GL, Ko M, Ta P, et al. Purification and characterization of Mycobacterium tuberculosis 1D-myo-inosityl-2-acetamido-2-deoxy-alpha-D-glucopyranoside deacetylase, MshB, a mycothiol biosynthetic enzyme. Protein Expr Purif. 2006;47(2):542–550. doi: 10.1016/j.pep.2006.03.003. [DOI] [PubMed] [Google Scholar]
- 64.Dayaram YK, Talaue MT, Connell ND, Venketaraman V. Characterization of a glutathione metabolic mutant of Mycobacterium tuberculosis and its resistance to glutathione and nitrosoglutathione. J Bacteriol. 2006;188(4):1364–1372. doi: 10.1128/JB.188.4.1364-1372.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bzymek KP, Newton GL, Ta P, Fahey RC. Mycothiol import by Mycobacterium smegmatis and function as a resource for metabolic precursors and energy production. J Bacteriol. 2007;189(19):6796–6805. doi: 10.1128/JB.00644-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ozyamak E, de Almeida C, de Moura AP, et al. Integrated stress response of Escherichia coli to methylglyoxal: transcriptional read through from the nemRA operon enhances protection through increased expression of glyoxalase I. Mol Microbiol. 2013;88(5):936–950. doi: 10.1111/mmi.12234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chandrangsu P, Dusi R, Hamilton CJ, Helmann JD. Methylglyoxal resistance in Bacillus subtilis: contributions of bacillithiol-dependent and independent pathways. Mol Microbiol. 2014;91(4):706–715. doi: 10.1111/mmi.12489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ferguson GP, Booth IR. Importance of glutathione for growth and survival of Escherichia coli cells: detoxification of methylglyoxal and maintenance of intracellular K+ J Bacteriol. 1998;180(16):4314–4318. doi: 10.1128/jb.180.16.4314-4318.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Krymkiewicz N. Reactions of methylglyoxal with nucleic acids. FEBS Lett. 1973;29(1):51–54. doi: 10.1016/0014-5793(73)80013-4. [DOI] [PubMed] [Google Scholar]
- 70.Chi BK, Gronau K, Mader U, et al. S-bacillithiolation protects against hypochlorite stress in Bacillus subtilis as revealed by transcriptomics and redox proteomics. Mol Cell Proteomics. 2011;10(11):M111. doi: 10.1074/mcp.M111.009506. 009506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chi BK, Roberts AA, Huyen TT, et al. S-bacillithiolation protects conserved and essential proteins against hypochlorite stress in firmicutes bacteria. Antioxid Sedox Signal. 2013;18(11):1273–1295. doi: 10.1089/ars.2012.4686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hondorp ER, Matthews RG. Oxidative stress inactivates cobalamin-independent methionine synthase (MetE) in Escherichia coli. PLoS Biol. 2004;2(11):e336. doi: 10.1371/journal.pbio.0020336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gaballa A, Chi BK, Roberts AA, et al. Redox regulation in Bacillus subtilis: the bacilliredoxins BrxA (YphP) and BrxB (YqiW) function in de-bacillithiolation of S-bacillithiolated OhrR and MetE. Antioxid Redox Signal. 2013;21(3):357–367. doi: 10.1089/ars.2013.5327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Duy NV, Mader U, Tran NP, et al. The proteome and transcriptome analysis of Bacillus subtilis in response to salicylic acid. Proteomics. 2007;7(5):698–710. doi: 10.1002/pmic.200600706. [DOI] [PubMed] [Google Scholar]
- 75.Nguyen VD, Wolf C, Mader U, et al. Transcriptome and proteome analyses in response to 2-methylhydroquinone and 6-brom-2-vinyl-chroman-4-on reveal different degradation systems involved in the catabolism of aromatic compounds in Bacillus subtilis. Proteomics. 2007;7(9):1391–1408. doi: 10.1002/pmic.200700008. [DOI] [PubMed] [Google Scholar]
- 76.Pohl S, Tu WY, Aldridge PD, et al. Combined proteomic and transcriptomic analysis of the response of Bacillus anthracis to oxidative stress. Proteomics. 2011;11(15):3036–3055. doi: 10.1002/pmic.201100085. [DOI] [PubMed] [Google Scholar]
- 77.Kashyap DR, Rompca A, Gaballa A, et al. Peptidoglycan recognition proteins kill bacteria by inducing oxidative, thiol, and metal stress. PLoS Pathog. 2014;10(7):e1004280. doi: 10.1371/journal.ppat.1004280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Palazzolo-Ballance AM, Reniere ML, Braughton KR, et al. Neutrophil microbicides induce a pathogen survival response in community-associated methicillin-resistant Staphylococcus aureus. J Immunol. 2008;180(1):500–509. doi: 10.4049/jimmunol.180.1.500. [DOI] [PubMed] [Google Scholar]
- 79.Pother DC, Liebeke M, Hochgrafe F, et al. Diamide triggers mainly S Thiolations in the cytoplasmic proteomes of Bacillus subtilis and Staphylococcus aureus. J Bacteriol. 2009;191(24):7520–7530. doi: 10.1128/JB.00937-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Schlag S, Nerz C, Birkenstock TA, et al. Inhibition of staphylococcal biofilm formation by nitrite. J Bacteriol. 2007;189(21):7911–7919. doi: 10.1128/JB.00598-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hochgrafe F, Wolf C, Fuchs S, et al. Nitric oxide stress induces different responses but mediates comparable protein thiol protection in Bacillus subtilis and Staphylococcus aureus. J Bacteriol. 2008;190(14):4997–5008. doi: 10.1128/JB.01846-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hinchman CA, Ballatori N. Glutathione conjugation and conversion to mercapturic acids can occur as an intrahepatic process. J Toxicol Environ Health A. 1994;41(4):387–409. doi: 10.1080/15287399409531852. [DOI] [PubMed] [Google Scholar]
- 83.Rawat M, Uppal M, Newton G, et al. Targeted mutagenesis of the Mycobacterium smegmatis mca gene, encoding a mycothiol-dependent detoxification protein. J Bacteriol. 2004;186(18):6050–6058. doi: 10.1128/JB.186.18.6050-6058.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Sherrill C, Fahey RC. Import and metabolism of glutathione by Streptococcus mutans. J Bacteriol. 1998;180(6):1454–1459. doi: 10.1128/jb.180.6.1454-1459.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Potter AJ, Trappetti C, Paton JC. Streptococcus pneumoniae uses glutathione to defend against oxidative stress and metal ion toxicity. J Bacteriol. 2012;194(22):6248–6254. doi: 10.1128/JB.01393-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Suzuki H, Kumagai H, Tochikura T. gamma-Glutamyltranspeptidase from Escherichia coli K-12: formation and localization. J Bacteriol. 1986;168(3):1332–1335. doi: 10.1128/jb.168.3.1332-1335.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.U.S. Department of Health and Human Services, Food and Drug Administration. Guidance for Industry: Antibacterial therapies Guidance for industry: antibacterial therapies for patients with unmet medical need for the treatment of serious bacterial diseases. 2014 [Google Scholar]
- 88.Burke SL, Rose WE. New pharmacological treatments for methicillin-resistant Staphylococcus aureus infections. Expert Opin Pharmacother. 2014;15(4):483–491. doi: 10.1517/14656566.2014.876991. [DOI] [PubMed] [Google Scholar]
- 89.Park JY, Kim JW, Moon BY, et al. Characterization of a novel two-component regulatory system, HptRS, the regulator for the hexose phosphate transport system in Staphylococcus aureus. Infect Immun. 2015;83(4):1620–1628. doi: 10.1128/IAI.03109-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Popovic M, Steinort D, Pillai S, Joukhadar C. Fosfomycin: an old, new friend? Eur J Clin Microbiol Infect Dis. 2010;29(2):127–142. doi: 10.1007/s10096-009-0833-2. [DOI] [PubMed] [Google Scholar]
- 91.Etienne J, Gerbaud G, Fleurette J, Courvalin P. Characterization of staphylococcal plasmids hybridizing with the fosfomycin resistance gene fosB. FEMS Microbiol Lett. 1991;68(1):119–122. doi: 10.1016/0378-1097(91)90406-z. [DOI] [PubMed] [Google Scholar]
- 92.Falagas ME, Giannopoulou KP, Kokolakis GN, Rafailidis PI. Fosfomycin: use beyond urinary tract and gastrointestinal infections. Clin Infect Dis. 2008;46(7):1069–1077. doi: 10.1086/527442. [DOI] [PubMed] [Google Scholar]
- 93.Nicholas GM, Eckman LL, Ray S, et al. Bromotyrosine-derived natural and synthetic products as inhibitors of mycothiol-S-conjugate amidase. Bioorg Med Chem Lett. 2002;12(17):2487–2490. doi: 10.1016/s0960-894x(02)00385-2. [DOI] [PubMed] [Google Scholar]
- 94.Nicholas GM, Eckman LL, Newton GL, et al. Inhibition and kinetics of mycobacterium tuberculosis and mycobacterium smegmatis mycothiol-S-conjugate amidase by natural product inhibitors. Bioorg Med Chem Lett. 2003;11(4):601–608. doi: 10.1016/s0968-0896(02)00345-0. [DOI] [PubMed] [Google Scholar]
- 95.Metaferia BB, Fetterolf BJ, Shazad-Ul-Hussan S, et al. Synthesis of natural product-inspired inhibitors of Mycobacterium tuberculosis mycothiol-associated enzymes: the first inhibitors of GlcNAc-Ins deacetylase. J Med Chem. 2007;50(25):6326–6336. doi: 10.1021/jm070669h. [DOI] [PubMed] [Google Scholar]