Abstract
Listeria monocytogenes FrvA (Lmo0641) is critical for virulence in the mouse model and is an ortholog of the Bacillus subtilis Fur- and PerR-regulated Fe(II) efflux P1B4-type ATPase PfeT. Previously, FrvA was suggested to protect against heme toxicity. Here, we demonstrate that an frvA mutant is sensitive to iron intoxication, but not to other metals. Expression of frvA is induced by high iron and this induction requires Fur. FrvA functions in vitro as a divalent cation specific ATPase most strongly activated by ferrous iron. When expressed in B. subtilis, FrvA increases resistance to iron both in wild-type and in a pfeT null strain. FrvA is a high affinity Fe(II) exporter and its induction imposes severe iron limitation in B. subtilis resulting in derepression of both Fur- and PerR-regulated genes. FrvA also recognizes Co(II) and Zn(II) as substrates and can complement B. subtilis strains defective in the endogenous export systems for these cations. Building on these results, we conclude that FrvA functions in the efflux of Fe(II), and not heme, during listerial infection.
ABBREVIATED SUMMARY
We demonstrate that the Listeria monocytogenes Fur-regulated virulence gene frvA encodes a P1B4-type Fe(II) efflux ATPase orthologous to Bacillus subtilis PfeT. FrvA is induced by Fur in response to iron excess and mutants lacking frvA are iron sensitive. Expression of FrvA in B. subtilis depletes cells of iron leading to derepression of both Fur- and PerR-regulated genes.
Introduction
Listeria monocytogenes is a model intracellular pathogen that invades mammalian cells by phagocytosis, ultimately escaping the phagocytic vacuole to proliferate in the host cytosol and spread from cell to cell (Cossart, 2011). Despite intensive study, the roles of many genes critical for virulence have remained unclear. Our attention was drawn to the Fur-regulated virulence determinant frvA (lmo0641) which encodes a protein homologous to the Bacillus subtilis P1B4-type ATPase PfeT (Guan et al., 2015). The FrvA protein is critical for virulence, although the nature of the virulence defect is not yet clear (McLaughlin et al., 2012). The ΔfrvA strain is highly attenuated, but is still able to invade and replicate in a macrophage cell line. Inoculation into mice induces cellular immunity suggesting that this strain could be useful for vaccine development (McLaughlin et al., 2013).
FrvA is a member of the P1B4 subfamily of metal transport ATPases as it shares their invariant transmembrane metal binding site (Argüello, 2003, McLaughlin et al., 2012). In general, P1B-ATPases drive cytoplasmic metal ion efflux, and members of the P1B4 subfamily are most closely associated with efflux of Co(II) (Argüello, 2003, Raimunda et al., 2014, Raimunda et al., 2012, Smith et al., 2014, Zielazinski et al., 2012). Previous studies have highlighted the role of Cu(I) and Zn(II) efflux P-type ATPases in resistance to metal ion toxicity and in virulence (Argüello et al., 2011, Djoko et al., 2015, German et al., 2013). Similarly, some P1B4-type ATPases, including FrvA, are known to be critical for virulence (McLaughlin et al., 2012, Raimunda et al., 2014). However, the nature of the substrate transported by the corresponding proteins during infection is not established, and it would be surprising for Co(II) efflux to be relevant to pathogenesis.
The B. subtilis PfeT protein (formerly ZosA) is a prototypic member of the P1B4-class of ATPases and was originally ascribed a function in Zn(II) homeostasis (Gaballa & Helmann, 2002). Recently, we demonstrated that the primary function of PfeT is efflux of Fe(II) to prevent iron overload (Guan et al., 2015). We therefore reasoned that FrvA (and perhaps other P1B4-type ATPases) might also function in Fe(II) efflux. If correct, this would provide evidence that the role of metal efflux ATPases in bacterial virulence includes the export of Fe(II). However, this model contrasts with a previous report that FrvA likely functions as a heme exporter as inferred from sensitivity to heme in growth assays and an increase in accumulated 59Fe when cells were exposed to 59Fe-labeled heme (McLaughlin et al., 2012).
The physiological role of proteins is often revealed by a careful analysis of their patterns of expression. The frvA (lmo0641) gene was first highlighted as a member of the PerR regulon (Rea et al., 2005), consistent with the PerR regulation of the B. subtilis homolog (Fuangthong & Helmann, 2003, Gaballa & Helmann, 2002, Helmann et al., 2003). The frvA gene is highly derepressed in a perR null mutant and the promoter region is associated with candidate Per box sequences (Rea et al., 2005). These results are consistent with PerR acting as a repressor of frvA and further suggest that frvA is induced by hydrogen peroxide. It is likely that FrvA is coordinately induced with other PerR-regulated genes including catalase and the hemA operon (Rea et al., 2005), encoding enzymes of heme biosynthesis, as also observed in B. subtilis (Chen et al., 1995, Fuangthong et al., 2002). The presumed rationale for derepression of the hemA operon is that high level expression of catalase, a heme-containing enzyme, increases the cellular demand for heme. Indeed, expression of catalase in a B. subtilis perR mutant contributes to iron deficiency due to the high demand imposed for its heme cofactor (Faulkner et al., 2012, Ma et al., 2012). The proposed role of FrvA in heme export (McLaughlin et al., 2012) would seem inconsistent with its apparent co-induction with heme biosynthesis functions.
In addition to PerR, FrvA is also known to be regulated by Fur, the ferric uptake regulator. FrvA was named as a Fur-regulated virulence determinant in studies that sought to identify additional Fur regulated genes by searching the L. monocytogenes genome for candidate Fur binding sites (McLaughlin et al., 2012). Fur generally functions as an Fe(II)-dependent repressor, which led to the expectation that FrvA would be derepressed under iron limitation conditions. Indeed, RT-PCR analyses revealed an increased level of transcripts for all 12 tested Fur regulon genes (including frvA) in a Δfur strain (McLaughlin et al., 2012). However, an opposite result was noted in a prior transcriptomic study in which it was observed that frvA was expressed at a 5.6-fold lower level in a fur mutant under iron replete conditions (Ledala et al., 2010). Thus, the role of Fur in regulation of frvA is presently unclear.
The current model, in which FrvA functions as a Fur-regulated heme exporter induced by iron deficiency (McLaughlin et al., 2012), is counterintuitive: why would cells efflux heme under conditions of iron starvation when iron uptake functions (including heme import) are derepressed (Lechowicz & Krawczyk-Balska, 2015, Sheldon & Heinrichs, 2015)? We instead favor a model in which FrvA functions, like PfeT, as an Fe(II) efflux pump that is induced by conditions of iron excess.
Here, we have re-investigated the role of FrvA and demonstrate that this gene is indeed induced by elevated levels of available iron, consistent with a role in iron detoxification, and this induction requires Fur. Moreover, physiological studies monitoring the effects of FrvA expression in B. subtilis demonstrate a striking ability of this protein to deplete the bacterial cell of iron as well as an ability to export other divalent cations. These results correlate with biochemical studies showing a strong Fe(II)-dependent activation of the purified FrvA ATPase. These findings are integrated with previously published studies in L. monocytogenes to support a role for iron efflux in the growth and dissemination of this model intracellular pathogen.
Results and Discussion
An L. monocytogenes frvA mutant is sensitive to iron intoxication
A B. subtilis pfeT mutant strain is sensitive to iron overload, as monitored by the zone of growth inhibition in a disk diffusion assay (Guan et al., 2015), and prior work suggests that an L. monocytogenes frvA mutant may have a similar phenotype (McLaughlin et al., 2012). We therefore compared the metal ion sensitivity of the L. monocytogenes EGDe wild-type strain (WT) and an isogenic frvA null mutant, with and without complementation by an ectopic copy of frvA. The frvA mutant strain was more sensitive to iron intoxication than WT as indicated by a >15% increase in the diameter of growth inhibition as well as a large zone of reduced cell density surrounding the iron-loaded filter disk (Fig. 1 and Fig S1). This increase in iron sensitivity is metal specific; there was no significant increase in sensitivity to other tested metals or to hemin (Fig. S1). These results are consistent with the hypothesis that FrvA functions as an Fe(II) efflux pump.
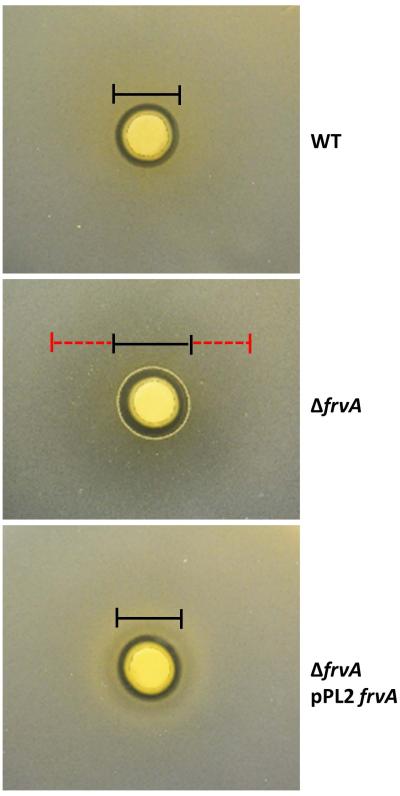
Fig 1. An frvA mutant is sensitive to iron intoxication in Listeria monocytogenes.
Representative photographs of a disk diffusion assay with wild-type EGDe (WT), an isogenic frvA null mutant (ΔfrvA), and an frvA complemented strain (ΔfrvA pPL2 frvA) on BHI plates with 10 μl of 1 M FeSO4 added onto the filter paper disk. Black lines indicate the clearance zone and red dashed lines indicate the sensitivity zone. ΔfrvA null mutant has a bigger clearance zone (2 ± 0.3 mm difference) and a large sensitivity zone with reduced cell density (28 ± 1 mm).
L. monocytogenes frvA is regulated by Fur in response to bioavailable iron
Prior studies have suggested the frvA promoter region contains a binding site for the iron-sensing transcription factor Fur. However, reports are conflicted as to the role of Fur with one study presenting results suggesting that frvA is repressed by Fur (McLaughlin et al., 2012, Rea et al., 2005) and another including data consistent with activation by Fur (Ledala et al., 2010). We therefore set out to determine if frvA is induced by iron and/or heme, and whether or not this induction is mediated by Fur acting as a repressor or an activator.
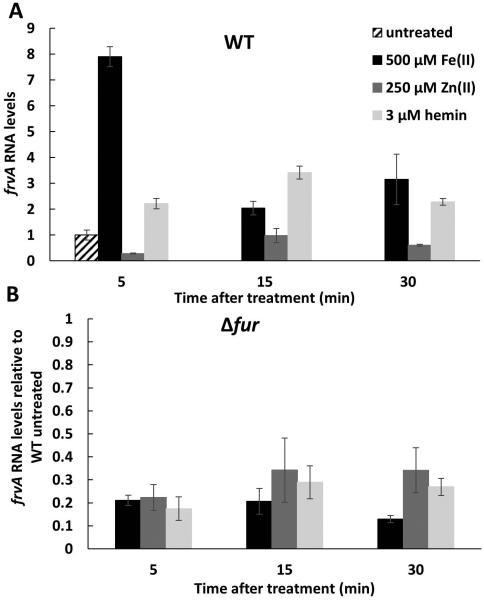
If FrvA functions in Fe(II) efflux, one would expect that expression would be induced by Fe(II). Indeed, frvA mRNA levels are elevated ~8-fold within 5 minutes of treatment with iron (Fig. 2A). Induction by iron was rapid with the highest mRNA levels at 5 min after treatment and lower levels at later times tested. In contrast, induction by heme (a preferred iron source for L. monocytogenes) was more modest with the highest level at 15 min after treatment, a delay consistent with the idea that heme is first imported into the cell and then iron is released intracellularly, ultimately leading to iron stress. This induction was specific for iron since treatment of cells with high levels of Zn(II), known to strongly induce Zn(II) efflux in B. subtilis (Ma et al., 2014), did not lead to induction of frvA (Fig. 2A).
Fig 2. Induction of Listeria monocytogenes frvA by Fe(II) and hemin.
L. monocytogenes wild-type EGDe (A) and an isogenic null mutant EGDe Δfur (B) were grown at 37°C in BHI medium overnight and subcultured at a 1:100 ratio into fresh BHI medium to an OD600 of 0.4. Cells were treated with 500 μM FeSO4, 250 μM ZnCl2, or 3 μM hemin, respectively. Aliquots of 5 ml of cells were harvested at different time points as indicated and total RNA from both treated and untreated samples was extracted and used for reverse transcription and subsequent quantitative real time-PCR to evaluate mRNA expression levels for frvA. The expression levels of frvA in WT untreated cells is set as 1 in both Fig 2A and 2B. L. monocytogenes 16S rRNA was used as a housekeeping control gene. The data are expressed as the mean ± SD (n=3).
To add ress the role of Fur in frvA gene regulation we repeated these measurements using a fur null mutant (Fig. 2B). The basal level of frvA expression was lower in the fur mutant strain (nearly 5-fold below the level in WT), and no further increase was noted within 5 min of treatment (and only modest changes were noted thereafter). These results indicate that Fur functions as a positive regulator required for the induction of frvA in response to elevated Fe(II) levels. The participation of Fur in the regulation of frvA is further evidence in support of an iron- rather than heme-related function since Fur senses Fe(II) directly. As noted previously, frvA is also repressed by PerR (Rea et al., 2005), suggesting that this gene is also likely to be induced in response to peroxide stress.
A direct role for Fur in positive regulation of frvA is also congruent with a prior transcriptomic study in which frvA was expressed at a 5.6-fold lower level in the fur mutant (almost identical to the effect noted here) under iron replete conditions (Ledala et al., 2010). In contrast, in this same study genes encoding predicted iron uptake proteins (based on homology to the characterized iron uptake functions of B. subtilis; Ollinger et al., 2006) were, as expected, derepressed between 3- and 15-fold in the fur mutant compared to the WT strain. Moreover, the predicted iron uptake functions were up-regulated in medium depleted of iron, whereas frvA was not. These results suggest that Fur binds DNA in response to elevated intracellular iron and serves to both repress genes for iron uptake and activate expression of frvA.
Expression of frvA complements the iron-sensitivity of a B. subtilis pfeT null mutant
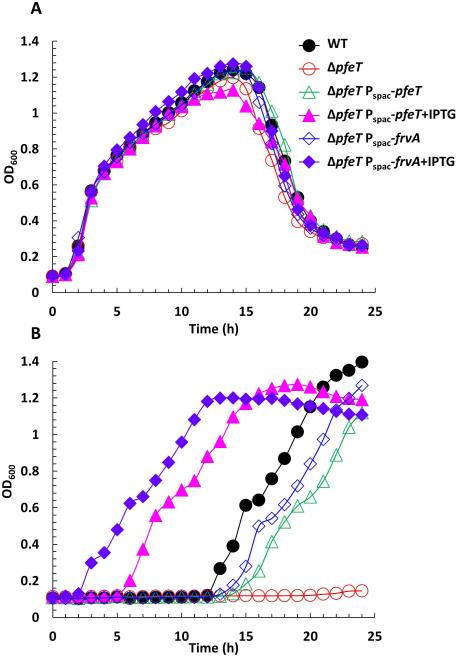
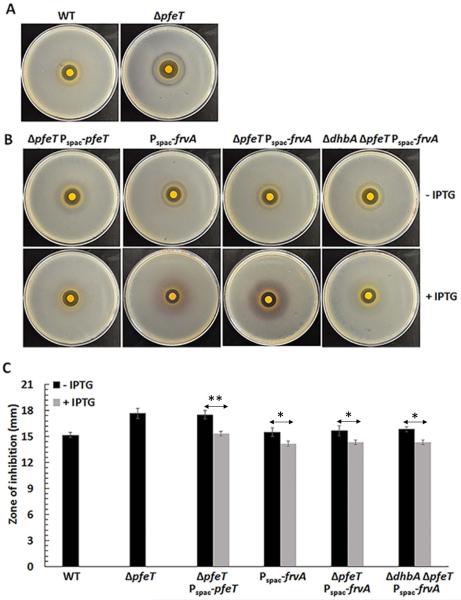
To further investigate the function of FrvA, we have used a heterologous expression strategy taking advantage of the detailed knowledge of the B. subtilis PerR, Fur and other metal ion responsive regulons (Faulkner & Helmann, 2011, Helmann, 2014). As shown previously, a mutant strain lacking PfeT (32% identical to FrvA) is sensitive to iron intoxication (Guan et al., 2015). We integrated an IPTG-inducible copy of the frvA gene into the amyE locus of the B. subtilis pfeT null mutant, and checked the effect of frvA induction on resistance to iron intoxication using a growth curve assay. In LBC medium (LB medium amended with 3.4 mM citrate trisodium dihydrate) amended with 4 mM Fe(II), the pfeT null mutant is severely compromised in growth whereas the WT strain grows well after an extended lag phase, as previously reported (Guan et al., 2015). The severe growth defect, which is dependent on added iron, is complemented by either PfeT or FrvA (Fig. 3). Significant complementation is observed in this assay even in the absence of added IPTG, indicating that even the low level of expression in uninduced cells is sufficient to provide protection against iron intoxication. Consistent with our prior results (Guan et al., 2015), the growth lag upon exposure to toxic levels of iron can be eliminated if PfeT is induced prior to sub-culturing, and similar results are noted for FrvA. Complementation of the pfeT null mutant is also apparent in a spot dilution assay to monitor cell growth under conditions of iron intoxication. In this assay, the pfeT mutant is unable to grow on LBC medium with 4 mM Fe(II) and grows as very small colonies on plates amended with 3 mM Fe(II). Mutant strains expressing either PfeT or FrvA from an IPTG-inducible promoter form colonies with high efficiency and grow at a rate comparable to WT under these conditions (Fig. S2). To further characterize the effects of FrvA on iron intoxication, a disk diffusion assay was used to evaluate the sensitivity of strains to high Fe(II). The pfeT null mutant displays an elevated sensitivity to Fe(II) compared to the WT cells (Fig. 4A). Induction of FrvA or PfeT by IPTG in the pfeT null mutant significantly increased iron resistance to a level at least as high as for WT (Fig. 4B and 4C). These results demonstrate that frvA complements the pfeT null mutation in B. subtilis.
Fig 3. Induction of L. monocytogenes FrvA complements a pfeT deletion of B. subtilis.
A. Representative growth curves of WT (CU1065), ΔpfeT, ΔpfeT Pspac-pfeT, and ΔpfeT Pspac-frvA grown in LBC medium with no added iron.
B. Representative growth curves of the same strains as panel (A) in LBC medium amended with 4 mM FeSO4. For IPTG treated cells, 1 mM IPTG was added to cell cultures 30 min before 2 μl of cell culture was inoculated to 200 μl of growth medium.
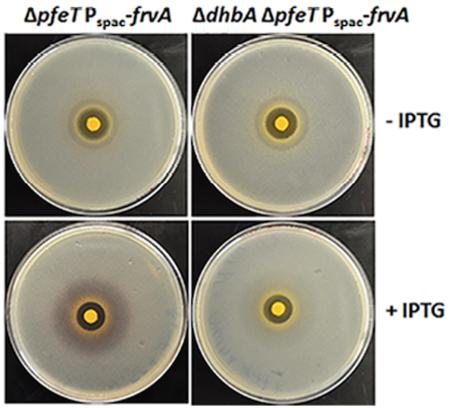
Fig 4. Induction of L. monocytogenes FrvA leads to iron starvation and induces the Fur regulon.
Representative photographs of a disk diffusion assay with (A) WT and pfeT null mutant as shown previously (Guan et al., 2015) and (B) ΔpfeT Pspac-pfeT, Pspac-frvA, ΔpfeT Pspac-frvA, and ΔdhbA ΔpfeT Pspac-frvA strains, each grown without (top) or with (bottom) IPTG induction. The plates contained LBC medium with 10 μl of 1 M FeSO4 added onto the filter paper disk.
C. The data are expressed as the diameter (mean ± SD; n=3) of the clearance zone (mm). *P<0.05 and **P<0.01 indicate statistically significant difference between the indicated groups.
Induction of FrvA imposes iron limitation and induces the Fur regulon
In the course of these complementation studies, we were surprised to note that induction of FrvA (but not PfeT) resulted in a readily visible purple halo around the inhibition zone (Fig. 4B). This color is reminiscent of the Fe(III)-complex formed by B. subtilis siderophores (Ito & Neilands, 1958, Peters & Warren, 1968), suggesting that FrvA may be imposing iron starvation on the cells despite the presence of high or even toxic levels of iron in the medium. Most laboratory strains of B. subtilis (strain 168 and its derivatives) encode the catecholate siderophore bacillibactin, but are unable to synthesize the mature siderophore due to a null mutation of sfp (sfp0), encoding 4'-phosphopantetheinyl transferase (May et al., 2001). Instead, B. subtilis 168 strains produce 2, 3-dihydroxybenzoate (DHBA) and its derivative, dihydroxybenzoylglycine (DHBG), precursors of bacillibactin which form purple colored ferric complexes (Ito & Neilands, 1958, Peters & Warren, 1968). As predicted, the purple color disappeared after introduction of a dhbA null mutation to the frvA complemented strain (Fig. 4B). These observations indicate that induction of FrvA imposes iron limitation and induces the Fur regulon. To further test this hypothesis, we measured the effect of FrvA induction on expression of a dhbA-lacZ reporter fusion (Fig. S3). In LB medium, which is iron sufficient, this reporter fusion is largely repressed by Fur. However, β-galactosidase levels increased ~25-fold upon induction of frvA. Together, these results indicate that induction of FrvA (but not PfeT) leads to iron limitation sufficient to derepress the Fur-regulated dhb operon (Baichoo et al., 2002, Bsat & Helmann, 1999). One possible explanation for this result is that FrvA may be a higher affinity Fe(II) efflux transporter compared to PfeT. Indeed, we find that induction of FrvA, but not PfeT, increases iron tolerance to a level slightly higher than WT cells in all three tested backgrounds (Fig. 4C).
FrvA is a divalent metal ion activated ATPase selective for Fe(II)
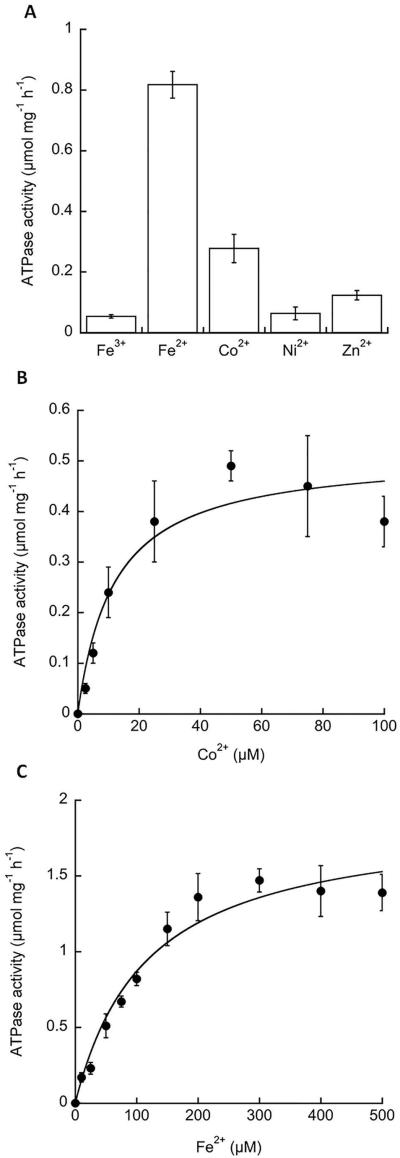
The metal ion selectivity of P1B-type ATPases can be inferred from biochemical studies that monitor the metal-dependency of ATPase activation (Argüello et al., 2011). We therefore purified FrvA after overexpression in E. coli and assayed metal ion-activated ATPase activity in detergent micelles as described previously for PfeT (Guan et al., 2015). Of the metal ions tested, the highest level of ATPase activity was observed with Fe(II), with lower levels of activity with Zn(II) and Co(II), which is similar to Fe(II) in its coordination chemistry (Fig 5A). The overall pattern of FrvA metal ion activation shows similarities with that reported for PfeT (Guan et al., 2015), although the latter protein was not significantly activated by Zn(II). In addition, the apparent K1/2 for activation of FrvA by Fe(II) (116±24 μM) was substantially lower than for PfeT (520±120 μM) (Fig. 5C). Similarly, the K1/2 for Co(II) (Fig. 5B) was also ~3-4-fold lower than measured for PfeT (Guan et al., 2015), but quite similar to that observed in Mycobacterium smegmatis CtpJ (Raimunda et al. 2012). These differences in relative metal selectivity and K1/2 are likely associated with differences in the second spheres of metal coordination present in the compared proteins. The physiological implications of these observations are not straightforward since alternative forms of Fe(II), rather than the hydrated metal, might be relevant in vivo. Nevertheless, this lower K1/2 for activation of the ATPase is consistent with the inference that FrvA may have a higher affinity for Fe(II) in vivo when compared to PfeT.
Fig 5. Activation of FrvA ATPase activity by metal ions.
A. FrvA was purified and its ATPase activity (μmol mg−1 h−1) was measured in vitro in the presence of 100 μM of metal ions as indicated.
B. Kinetic characterization of the FrvA ATPase activity in the presence of various concentrations of Co(II) reveals a Vmax = 0.51 ± 0.05 μmol mg−1 h−1 and K1/2 = 12 ± 4 μM.
C. Kinetic characterization of the FrvA ATPase activity in the presence of various concentration of Fe(II) reveals a Vmax = 1.88 ± 0.14 μmol mg−1 h−1 and K1/2 = 116 ± 24 μM.
Data are expressed as the mean ± SD (n=3).
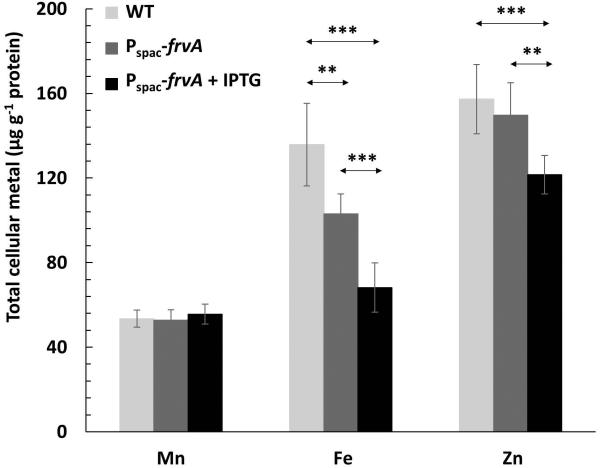
To monitor the effects of FrvA on intracellular metal ion levels we grew cells in LB medium and, using inductively coupled plasma mass spectrometry (ICP-MS), we monitored the intracellular levels of Mn(II), Fe(II), and Zn(II) in both WT and the WT strain carrying an IPTG-inducible copy of frvA. Even in the uninduced cells, the presence of frvA led to a reduction in intracellular Fe(II) levels, but did not affect Mn(II) or Zn(II) levels (Fig. 6). This is consistent with the finding that frvA can partially complement a pfeT mutant strain for growth under conditions of iron intoxication even when it is not induced (Figs. 3 and 4), presumably due to leaky expression from the Pspac promoter. When frvA is induced, there is a further reduction of Fe(II) levels and under these conditions Zn(II) levels also decline (Fig. 6). This suggests that, at least when overproduced, FrvA can export Zn(II) from cells, consistent with the weak but demonstrable activation of FrvA ATPase activity by added Zn(II) (Fig. 5A).
Fig 6. Measurements of intracellular metal ion concentrations by ICP-MS.
Cells were grown in LB medium to an OD600 of about 0.1 and supplemented with 1 mM IPTG where indicated, then harvested after two additional hours. Levels of intracellular Mn, Fe and Zn were analyzed by inductively coupled plasma mass spectrometry (ICP-MS). Strains included in the study were WT and Pspac-frvA (with or without IPTG induction). The total concentration of ions was expressed as μg ion per gram of protein content (mean ± SD; n=9). **P<0.01 and ***P<0.001 indicate statistically significant difference between the indicated groups.
FrvA can increase tolerance to Co(II) and Zn(II) in efflux-defective mutants
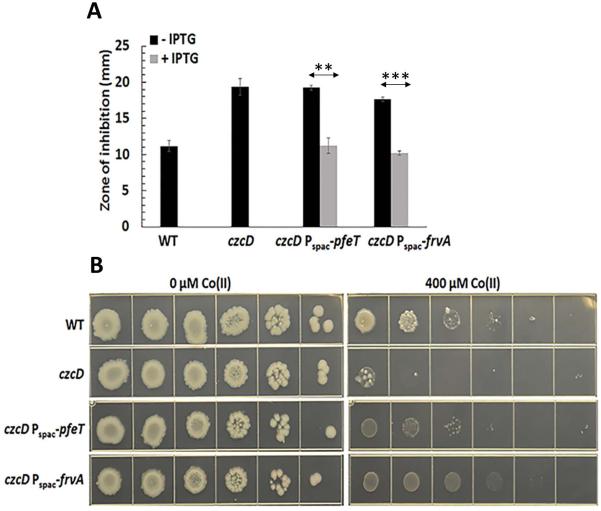
We previously demonstrated that artificial induction of PfeT could increase the Co(II) tolerance of a B. subtilis mutant lacking the endogenous Co(II) efflux system, CzcD (Guan et al., 2015). Here we have confirmed this effect, and also tested FrvA for the ability to increase Co(II) tolerance as measured in both a zone of inhibition assay (Fig. 7A) and a spot dilution assay (Fig. 7B). Indeed, a low level induction of FrvA (with 25 μM IPTG to avoid iron deprivation) does increase Co(II) resistance in a B. subtilis czcD mutant strain, suggesting that FrvA can also serve to efflux Co(II).
Fig 7. Induction of FrvA increases Co(II) resistance in a B. subtilis czcD mutant.
A. Disk diffusion assay of strains WT, czcD, czcD Pspac-pfeT and czcD Pspac-frvA. 10 μl of 100 mM CoCl2 was added onto the filter paper disk. The data are expressed as the mean ± SD (n=3) in mm.
B. Representative photographs of a spot dilution assay with the same strains as Fig 7A on LB agar plates. 25 μM and 1 mM IPTG were used to induce FrvA and PfeT, respectively. **P<0.01 and ***P<0.001 indicate a statistically significant difference between the indicated groups.
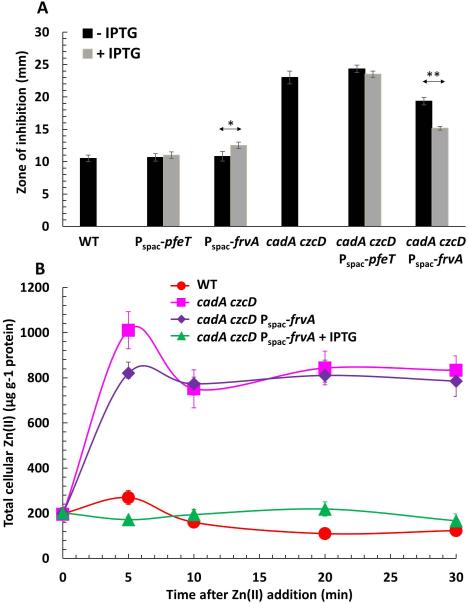
In contrast with PfeT, FrvA ATPase activity was activated (albeit weakly; Fig. 5A) by Zn(II). This motivated us to test whether or not FrvA might function in vivo in Zn(II) export, as suggested by the ICP-MS measurements of intracellular metal levels (Fig. 6). We therefore introduced an IPTG-inducible copy of frvA into a cadA czcD double mutant known to be highly sensitive to Zn(II) intoxication (Ma et al., 2014). In this background, induction of FrvA but not PfeT significantly increased Zn(II) tolerance (Fig. 8A). As expected, induction of FrvA or PfeT in the WT background (containing the Zn(II)-inducible CadA and CzcD efflux pumps) did not lead to any further increase in Zn(II) tolerance and may even lead to a slight increase in Zn(II) sensitivity. To further characterize the contribution of FrvA to Zn(II) tolerance, we treated cells with 250 μM ZnCl2 and monitored metal ion levels over time using ICP-MS. Within five min after Zn(II) addition, there is a significant increase in intracellular Zn(II) in the cadA czcD double mutant. Induction of FrvA can prevent this accumulation of intracellular Zn(II) upon shift to zinc excess (Fig. 8B).
Fig 8. Induction of FrvA increases Zn(II) resistance in a B. subtilis strain lacking Zn(II) efflux systems.
A. Disk diffusion assay of strains WT, Pspac-pfeT, Pspac-frvA, cadA czcD, cadA czcD Pspac-pfeT, and cadA czcD Pspac-frvA. For IPTG treated cells, 1 mM IPTG was added to both soft agar and LB agar plates. 10 μl of 100 mM ZnCl2 was added onto the filter paper disk. The data are expressed as the mean ± SD (n=3). *P<0.05 and **P<0.01 indicate a statistically significant difference between the indicated groups.
B. Levels of intracellular Zn monitored by ICP-MS for WT, cadA czcD, and cadA czcD Pspac-frvA (without and with 1 mM IPTG induction) before and at the time points indicated after addition of 250 μM ZnCl2 to LB medium. The data are expressed as the mean ± SD (n=3).
Collectively, these results indicate that FrvA can export Co(II) and Zn(II), in addition to Fe(II), when induced. However, as noted previously for PfeT (Guan et al., 2015), this does not imply that this is a normal in vivo activity of FrvA. L. monocytogenes encodes predicted Zn(II) and Co(II) efflux transporters that are likely orthologs of CzcD (Lmo2575; 56% identity) and CadA (Lmo1100; 34% identity) and these would likely obscure any role for FrvA in Zn(II) or Co(II) efflux in vivo (Jesse et al., 2014). Moreover, it is unclear whether FrvA would be expressed under conditions of Zn(II) or Co(II) toxicity. Indeed, PerR regulated genes may be repressed under conditions of excess Zn(II) or Co(II) (Chen et al., 1993, Moore et al., 2005), and excess Zn(II) failed to induce frvA in vivo (Fig. 1). However, a physiologically relevant ability of P1B4 subfamily ATPases to efflux Zn(II) is supported by the observation that derepression of the FrvA homolog PmtA in a Streptococcus pyogenes perR mutant leads to induction of genes regulated by the Zn(II)-sensing repressor AdcR (Brenot et al., 2007, Sanson et al., 2015). This can now be understood as likely resulting from depletion of cytosolic Zn(II) pools by the constitutively expressed PmtA.
Induction of FrvA leads to protoporphyrin IX accumulation
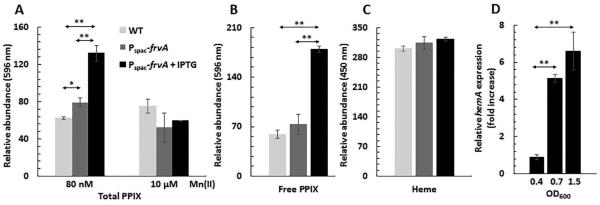
Although the preceding biochemical and physiological results strongly suggest that FrvA functions in Fe(II) efflux, we sought to further test the suggestion that FrvA might play a role in heme export (McLaughlin et al., 2012). As noted above, an L. monocytogenes frvA mutant was not more sensitive to heme than WT (Fig. S1). Next, we tested whether induction of FrvA would protect B. subtilis from heme toxicity. In these experiments we failed to note any increase in heme tolerance in the induced cells (data not shown), however B. subtilis (unlike L. monocytogenes) is not known to import heme and the mechanism of heme toxicity is not well understood in this system. We hypothesized that if FrvA did have a robust heme efflux activity it might lead to a measurable reduction in intracellular heme pools. Therefore, we measured intracellular heme levels using a fluorescence-based assay. In this assay, iron chelated by heme is removed by heating in 2 M oxalic acid and the resultant protoporphyrin IX (PPIX) is measured by fluorescence. To avoid contamination from heme in the medium, we grew cells using a metal-limiting minimal medium (MLMM) with 80 nM Mn(II) (Chen et al., 1993). Unexpectedly, induction of frvA led to a more than 2-fold increase in total PPIX compared with WT (Fig. 9A). To determine whether this increase was due to higher levels of heme or to higher levels of the heme precursor PPIX, we extracted the tetrapyrroles from cells and monitored PPIX and heme separately using fluorescence assays optimized for PPIX and heme detection (Fig. 9B, 9C). These results indicate that FrvA induction leads to elevated levels of PPIX, but not to a notable change in heme levels. Since FrvA did not reduce, under any tested condition, the level of heme in the cell we suggest that FrvA has no significant heme efflux activity under these conditions.
Fig 9. Effects of FrvA on B. subtilis heme biosynthesis.
A. Overnight cultures of WT and Pspac-frvA were subcultured (1:100 ratio) in MOPS metal-limiting minimal medium (MLMM; Chen et al., 1993) supplemented with 80 nM or 10 μM MnCl2. FrvA was induced with 1 mM IPTG at mid-log phase (OD600=0.25) for 3 h. Total protoporphyrin IX (PPIX) was extracted with 2 M oxalic acid. The fluorescence emission of PPIX was scanned from 560 to 680 nm after excitation at 400 nm and fluorescence intensity at 596 nm (peak) was normalized and plotted.
B. Measurements of PPIX in the same strains listed as Fig. 9A by acidic acetone extraction. The fluorescence emission of PPIX was scanned from 500 to 650 nm after excitation at 410 nm and fluorescence intensity at 596 nm (peak) was normalized and plotted.
C. Measurements of heme in the same strains listed as Fig. 9A by acidic acetone extraction. The fluorescence emission of heme was scanned from 400 to 500 nm after excitation at 380 nm and fluorescence intensity at 450 nm (peak) was normalized and plotted.
D. Strain CU1065 Pspac-frvA was grown in LB medium in the absence or presence of 1 mM IPTG. Total RNA was extracted from cells harvested at different growth points as indicated (n=3) and used for reverse transcription and subsequent quantitative real-time PCR to evaluate mRNA expression levels for hemA. The hemA mRNA levels in cells with 1 mM IPTG were presented as fold increase relatively to those of uninduced cells. The data are expressed as the mean ± SD (n=3). *P<0.05 and **P<0.01 indicate a statistically significant difference between the indicated groups.
Since induction of FrvA depletes cells of Fe(II) leading to induction of the Fur regulon, we speculated that Fe(II) depletion might also lead to induction of the PerR regulon, including the hemA biosynthesis operon, and this might account for the elevation of intracellular PPIX levels. To test this notion, we repeated the study using the same MLMM amended with 10 μM Mn(II), a concentration sufficient to activate the PerR repressor even under conditions of low Fe(II) availability (Chen et al., 1995, Fuangthong et al., 2002, Helmann, 2014). As predicted, in this Mn(II) amended medium PPIX overproduction was no longer observed (Fig. 9A). We confirmed these results using qRT-PCR which revealed that induction of FrvA leads to ~5-7-fold derepression of hemA (the first gene of the hemAXCDBL operon) in mid-logarithmic and stationary phase cells (Fig. 9D). Therefore, we conclude that FrvA depletes intracellular Fe(II) leading to derepression of the PerR-regulated heme biosynthesis operon. Despite upregulation of PPIX biosynthesis, there is no detectable increase in cellular heme levels. This may be due to a restriction of the activity of ferrochelatase imposed by the ability of FrvA to deplete the cytosol of iron. The ability of FrvA to derepress a PerR-repressed operon is notable since PerR binds Fe(II) with substantially higher affinity than Fur (Ka >108 M−1 for PerR vs. 1.2 × 106 M−1 for Fur) (Ma et al., 2011, Ma et al., 2012). This result further highlights the extent to which FrvA can impose iron limitation on B. subtilis.
Concluding Remarks
We propose, based on the evidence provided here, that the critical role of FrvA in virulence is resistance to Fe(II) intoxication. The inclusion of FrvA as a member of the PerR regulon suggests that this gene is upregulated in response to oxidative stress, which is likely to be adaptive since hydrogen peroxide toxicity results largely from its reaction with Fe(II) to generate toxic hydroxyl radicals via Fenton chemistry (Imlay, 2013).
The inclusion of FrvA as part of the Fur regulon has led to some confusion since published results are conflicting as to whether frvA transcription is increased or decreased in fur mutant strains (Ledala et al., 2010, McLaughlin et al., 2012). We demonstrate here that frvA is positively regulated by Fur, and that this provides a mechanism for the induction of frvA in response to high Fe(II) levels. The basis for the attenuated virulence of the frvA mutant strain is still unresolved (McLaughlin et al., 2013, McLaughlin et al., 2012). We favor a model in which this attenuation is based on a defect in Fe(II) efflux and subsequent sensitivity to iron intoxication in the host. Based on its inclusion in the PerR regulon, one might expect that FrvA could function in the activated macrophage. However, frvA does not appear to be upregulated (and is actually downregulated) in the macrophage phagosome (Chatterjee et al., 2006). This may reflect the fact that this is generally considered to be an Fe(II) restricted environment due to the action of host NRAMP1 (Wessling-Resnick, 2015). In contrast, the host cytosol is often considered an iron replete environment. However, the iron status of these compartments may change upon infection or depending on the nutritional status of the host cell. For example, internalization of L. monocytogenes in macrophages has been linked to upregulation of the major cellular Fe(II) exporter, ferroportin (Haschka et al., 2015). Moreover, supplementation of cultured phagocytes with iron salts led to an increase in intracellular proliferation of L. monocytogenes suggesting that the cytosol may be an iron-restricted environment, perhaps due to the induction of ferroportin. In contrast, iron supplementation led to increased killing of an L. monocytogenes llo mutant unable to escape from the phagocytic vacuole, consistent with a model in which cells employ iron intoxication as a killing mechanism (Haschka et al., 2015). It is therefore presently unclear at which stage(s) (invasion, escape from the vacuole, intercellular translocation, and dissemination) FrvA is likely to play the most important role, but we speculate that this role is correlated in general with the transition of cells from iron-limited to iron replete environments.
There are several reasons to question the previously proposed role of FrvA in heme efflux (McLaughlin et al., 2012). First, the evidence in support of this notion derives from the elevated accumulation of 59Fe from heme in frvA mutant cells, and this cannot distinguish between a role in Fe(II) efflux or heme efflux. Indeed, frvA mutants are sensitive to elevated Fe(II) (Fig. 1), consistent with prior findings (McLaughlin et al., 2012). Second, the coordination of Co(II) by the invariant transmembrane residues present in P1B4-ATPases, as verified by X-ray spectroscopy and site directed mutagenesis (Zielazinski et al., 2012), defines a divalent ion binding site poorly suited to accommodate the bulky heme group. Third, L. monocytogenes can efficiently acquire iron from heme through the combined actions of hemolysin and the HupDCG heme import system (Klebba et al., 2012, Lechowicz & Krawczyk-Balska, 2015). The importance of this pathway is evident from the 100-fold attenuation in virulence of hupDCG mutant cells in the mouse model. Once imported, heme is degraded to liberate Fe(II) (Lechowicz & Krawczyk-Balska, 2015). If iron-limited cells encounter a heme rich environment it can be imagined that the highly efficient import of heme could lead to transient iron overload leading to induction of FrvA (as noted in Fig. 2A) to help alleviate iron toxicity. While some imported heme may be directly shuttled into heme proteins, as shown for Staphylococcus aureus (Hammer & Skaar, 2011), this has not been demonstrated in L. monocytogenes, and the major fate of imported heme under iron limitation conditions is presumed to be degradation and Fe(II) release (Lechowicz & Krawczyk-Balska, 2015, Hammer & Skaar, 2011). It is possible that heme overload might occur under specific conditions in which heme degradation is inhibited or in which the rate of heme uptake exceeds that of degradation. In this situation L. monocytogenes may export excess heme using a system (Lmo2580, 48% identity to HrtA; Lmo2581, 30% identity to HrtB) orthologous to the S. aureus HrtAB heme exporter (Anzaldi & Skaar, 2010), although this remains to be tested.
The finding that B. subtilis PfeT functions physiologically as a Fe(II) exporter (Guan et al., 2015), and that the L. monocytogenes FrvA protein has a very similar activity, suggests that Fe(II) efflux may be important in other human pathogens. Notably, the Mycobacterium tuberculosis CtpD protein is also a member of the P1B4 subfamily of metal-activated ATPases, is known to be activated in vitro by Co(II) (which is structurally similar to Fe(II)) and is important for survival in the mouse lung (Raimunda et al., 2014). The results reported here provide evidence for the notion that Fe(II) efflux is an important, although only recently appreciated, contributor to bacterial iron homeostasis. Previous results have highlighted the role of cation diffusion facilitator proteins in efflux of Fe(II) in E. coli (FieF) (Grass et al., 2005) and, more recently, during Fe(III) respiration in Shewanella oneidensis (FeoE) (Bennett et al., 2015). Salmonella enterica encodes a major facilitator superfamily (MFS) protein, IceT, that exports iron-citrate complexes (Frawley et al., 2013), but the physiological role of this system is unclear. Indeed, IceT seems to be induced by nitric oxide (which may lead to elevated iron toxicity) rather than specifically by iron overload, and also effluxes citrate alone, suggesting a possible role in curtailing activity of the TCA cycle. As reported here, the characterization of L. monocytogenes FrvA as an Fe(II) efflux ATPase provides compelling evidence directly linking iron efflux to bacterial pathogenesis.
Experimental Procedures
Bacterial strains, plasmids, and growth conditions
All B. subtilis strains are derivatives of strain CU1065 (WT) and L. monocytogenes strains included EGDe (WT) and its mutant derivatives. Strains and plasmids used in the study are listed in Table S1. E. coli DH5α was used for standard cloning procedures. Bacteria were grown in LB medium (B. subtilis) or BHI medium (L. monocytogenes) with vigorous shaking or on solid LB (B. subtilis) or BHI (L. monocytogenes) agar with appropriate antibiotic selection at 37°C. LBC medium (LB medium supplemented with 3.4 mM of citrate trisodium dihydrate) was used to reduce iron precipitation in iron intoxication experiments (Guan et al., 2015). Antibiotics were used for selection at the following concentration: ampicillin (amp, 100 μg ml−1), spectinomycin (spec, 100 μg ml−1), chloramphenicol (cm, 10 μg ml−1), tetracycline (tet, 5μg ml−1), kanamycin (kan, 15 μg ml−1), neomycin (neo, 8 μg ml−1), and macrolide lincosoamide-streptogramin B (MLS, 1 μg ml−1 erythromycin and 25 μg ml−1 lincomycin).
Strain constructions
The frvA gene, amplified from L. monocytogenes 10403S chromosomal DNA, was cloned into plasmid pPL82 (Quisel et al., 2001) after digestion with HindIII and XbaI. The sequence of the insert was confirmed by DNA sequencing (Cornell DNA sequencing facility). Primer pairs used for PCR amplification, confirmation and sequencing are listed in Table S2. The recombinant plasmid was transformed into B. subtilis and integrated into the amyE locus. Chromosomal DNA transformation was conducted as described previously (Harwood & Cutting, 1990). SPβ606 lysate was prepared from strain HB606 by heating at 50°C for 15 min and transduced to strain HB19208 (amyE::Pspac-frvA) to make lacZ reporter strain HB19253 (HB19208 SPβ PdhbA-cat-lacZ).
RNA extraction and qRT-PCR
To monitor frvA induction, L. monocytogenes EGDe and EGDe Δfur was grown at 37°C in BHI medium overnight and subcultured with 1:100 ratio into fresh BHI medium to an OD600 of 0.4. Cells were treated with Fe(II), Zn(II), or hemin at the indicated concentrations. Aliquots of 5 ml of cells were harvested at different time points as indicated and total RNA from both treated and untreated samples was extracted using RNeasy Mini Kit following the manufacturer’s instructions (Qiagen Sciences, Germantown, MD).
To monitor hemA induction, strains were grown in LB medium overnight and subcultured at 1% into fresh LB medium in the presence or absence of 1mM IPTG where indicated. Aliquots of 2 mL of cells were harvested at different growth phases as indicated and total RNA from each sample was extracted using RNeasy Mini Kit according to the manufacturer’s instructions.
All RNA samples were treated with Turbo-DNA freeTM DNase (AmbionTM) and precipitated with ethanol and sodium acetate at -80°C overnight. RNA samples were washed with 70% ethanol and dissolved in RNase-free water then quantified by NanoDrop spectrophotometer. 200 ng of total RNA from each sample was used for cDNA synthesis with TaqMan reverse transcription reagent (Applied Biosystems, Foster City, CA). Primers used for cDNA synthesis are listed in Table S2. Quantitative PCR (qPCR) was then conducted using iQ SYBR green supermix in an Applied Biosystems 7300 Real Time PCR System. 16S rRNA (L. monocytogenes) or 23S rRNA (B. subtilis) was used as a housekeeping gene as an internal control.
Growth curves
Strains were grown overnight in LB medium and then subcultured at 1% into LB medium and grown to an OD600 of 0.4. For IPTG treated cells, 1mM IPTG was added to cell cultures 30 min before 2 μl of cell culture was inoculated to 200 μl of LBC medium or LBC medium supplemented with 4 mM FeSO4 (Guan et al., 2015). Cell Growth (OD600) was monitored every 15 min for 25 h using a Bioscreen growth analyzer (Growth Curves USA, Piscataway, NJ) at 37°C with continuous shaking. Data shown were representative growth curves and experiments were performed at least three times with at least three biological replicates.
Disk diffusion assays
All strains tested were grown overnight in LB medium (B. subtilis) or BHI medium (L. monocytogenes) then subcultured at 1% into fresh medium to an OD600 of 0.4. Aliquots of 100 μl (B. subtilis) or 150 μl (L. monocytogenes) of cell cultures were mixed with 4 ml of 0.75% LB (B. subtilis) or BHI (L. monocytogenes) soft agar and poured onto LB (B. subtilis) or BHI (L. monocytogenes) agar plates. The plates were dried for 10 to 15 min in a laminar flow hood at room temperature. Filter paper disks (6.5 mm in diameter) soaked with 10 μl of desired chemicals were placed on the top of the agar plates, and the plates were incubated at 37°C for 16 to 18 h. The diameter of the zone of clearance was measured. The data shown are the mean and standard deviation of three biological replicates. For IPTG-treated cells, the indicated concentration of IPTG was added to both the soft agar and the LB agar plates.
β-galactosidase assays
To check the effects of frvA induction on dhbA expression, the WT strain and HB19208 harboring a dhbA-cat-lacZ reporter fusion were grown overnight in LB medium and then subcultured at 1% into fresh LB medium to different growth phases as indicated in the presence or absence of 1 mM IPTG and assayed for β-galactosidase as described previously (Chen et al., 1993, Miller, 1972).
Measurements of intracellular metal ion concentrations by ICP-MS
For experiments performed in Fig. 6, cells were grown in LB medium to an OD600 of about 0.1 and 1 mM IPTG was added into the cell culture where indicated. Aliquots of 4 ml of cell culture were harvested after two additional hours of incubation. For experiments performed in Fig. 8B, cells were grown in LB medium to an OD600 of 0.4 in the presence or absence of 1 mM IPTG where indicated and then 250 μM ZnCl2 were added to cell cultures. Aliquots of 4 ml cell culture were harvested at different time points before and after addition of 250 μM ZnCl2.
All samples were prepared as described previously (Guan et al., 2015). Briefly, samples were washed once with buffer 1 (1X PBS buffer, 0.1 M EDTA) then twice with buffer 2 (1X chelex-treated PBS buffer). Cell pellets were resuspended in 400 μl buffer 3 (1X chelex-treated PBS buffer, 75 mM NaN3, 1% Triton X-100) and incubated at 37°C for 90 min for cell lysis. Lysed samples were centrifuged and subject to Bradford assay to quantify the total protein content. Then, samples were mixed with 600 μl buffer 4 (5% HNO3, 0.1% (v/v) Triton X-100) and heated in a 95°C sand bath for 30 min. Samples were centrifuged and supernatants were diluted in 1% HNO3. Levels of intracellular Zn, Fe and Mn were analyzed by Perkin-Elmer ELAN DRC II ICP-MS. Gallium was used as an internal standard. The total concentration of metal ions was calculated as the average of three biological replicates (± standard deviation) and expressed as μg ion per gram of protein.
Fluorescence-based PPIX and heme assays
For experiments performed in Fig. 9A, strains were grown overnight in LB medium and then subcultured with 1:100 ratio in MOPS minimum medium amended with 80 nM or 10 μM of MnCl2 (Fig. 9A). At OD600 ~0.3, 1 mM IPTG was added to induce FrvA expression. After three additional hours of incubation, OD600 was recorded for each sample and aliquots of 4 ml of cell culture were harvested. Wild type cells and uninduced HB19208 cells (amyE::Pspac-frvA) were included as controls. All samples were washed once with PBS buffer. Cell pellets were resuspended in 400 μl chelex-treated PBS buffer containing 75 mM NaN3 and 1% Triton X-100 and incubated at 37°C for 90 min to lyse the cells. Lysed cells were mixed with 500 μl of 2 M oxalic acid and heated in a 95°C sand bath for 30 min to remove iron from heme. The fluorescence emission of the resultant protoporphyrin IX (PPIX) was scanned from 560 nm to 680 nm with excitation at 400 nm as described (Sinclair et al., 2001). Fluorescence intensity at 596 nm (peak) was normalized and plotted. Data shown represent the mean and standard deviation of three biological replicates.
For experiments performed in Fig. 9B & C, strains were grown overnight in LB medium and then subcultured with 1:100 ratio in fresh LB medium with or without 1 mM IPTG induction to an OD600 of 0.5. OD600 was recorded and aliquots of 5 ml of cell culture were harvested. Cell pellets were resuspended in 1 ml 50 mM TrisHCl buffer (pH 7.4) containing 100 μg ml−1 of lysozyme and were incubated at 37°C for 30 min to lyse the cells. Cell debris was removed by centrifugation. PPIX and heme were extracted from 500 μl of the clear lysates using 500 μl acidic acetone (20% (v/v) 1.6 M HCl). Precipitate was removed by centrifugation and the supernatant was analyzed by fluorescence spectroscopy. The fluorescence emission of PPIX was scanned from 500 nm to 680 nm with excitation at 410 nm as described (Mielcarek et al., 2015). Fluorescence intensity at 596 nm (peak) was normalized and plotted. The fluorescence emission of heme was scanned from 400 nm to 500 nm with excitation at 380 nm as described (Mielcarek et al., 2015). Fluorescence intensity at 450 nm (peak) was normalized and plotted.
Spot dilution assays
All strains tested were grown overnight in LB medium and then inoculated at 1% into fresh LB medium to an OD600 of 0.4. A serial of 10-fold dilutions were prepared for each strain and aliquots of 5 μl cell dilutions were spotted on LB plates amended with different concentrations of metal ions as indicated. Data shown represent the efficiency of plating of cell culture from 1, 10−1, 10−2, 10−3, 10−4, and 10−5-fold serial dilutions (from left to right). Different concentrations of IPTG as indicated were amended in the LB agar plates to induce either PfeT or FrvA expression. The plates were incubated at 37°C for 16-18 hours. Data shown are representative photographs from at least three individual experiments.
FrvA expression and purification
L. monocytogenes frvA cDNA was amplified using genomic DNA as template and primers that introduced a Tobacco etch virus (TEV) protease site coding sequence at the amplicon 3′ ends. The PCR product was cloned into the pBAD-TOPO/His vector (Invitrogen) that introduces a C-terminal 6xHis tag. The plasmid was introduced into E. coli Top10 chemically competent cells. For FrvA-TEV-6xHis expression, cells were grown at 37°C in ZYP-505 media supplemented with 0.05% arabinose and 100 μg ml−1 ampicillin (Studier, 2005). Affinity purification of membrane proteins and removal of the 6xHis tag was performed as previously described (Raimunda et al., 2014, Raimunda et al., 2012). Solubilized lipid/detergent micellar forms of FrvA proteins were stored at −20°C in buffer C containing 25 mM Tris, pH 8.0, 50 mM NaCl, 0.01% n-dodecyl-β-d-maltopyranoside, 0.01% asolectin and 20% glycerol until use. The 6xHis tag was cleaved from the FrvA-6xHis fusion protein by treatment with 6xHis-tagged TEV protease (Rosadini et al., 2011) at 5:1 FrvA:TEV weight ratio for 1 h at 22°C in buffer C plus 1 mM Tris(2-carboxyethyl)phosphine (TCEP) and 0.5 mM EDTA. TEV-6xHis was removed by affinity purification with Ni-NTA resin. Protein determinations were performed in accordance to Bradford (Bradford, 1976). Protein purity was assessed by Coomassie brilliant blue staining of overloaded SDS-PAGE gels and by immunostaining of Western blots with rabbit anti-6xHis polyclonal primary antibody (GenScript) and goat anti-rabbit IgG secondary antibody coupled to horseradish peroxidase. Prior to ATPase activity determinations, proteins (1 mg ml−1) were treated with 0.5 mM EDTA and 0.5 mM tetrathiomolybdate for 45 min at room temperature. Chelators were removed using Ultra-30 Centricon (Millipore) filtration devices.
ATPase assays
The ATPase activity of isolated FrvA was measured as previously described (Guan et al., 2015). The assay was performed at 37°C in a medium containing 50 mM Tris (pH 7.4), 50 mM NaCl, 3 mM MgCl2, 3 mM ATP, 0.01% asolectin, 0.01% n-dodecyl-β-d-maltopyranoside, 2.5 mM TCEP, 20 μg ml−1 purified protein, and freshly prepared transition metal ions at the desired concentrations. Fe2+ and Zn2+ were included in the assay media as the sulfate salts, whereas Co2+ and Ni2+ were included as their chloride salts. Fe3+ was added as FeCl3, where TCEP was not included in the assay media. ATPase activity was stopped after 20 min incubation and released Pi determined (Lanzetta et al., 1979). ATPase activity measured in the absence of transition metals was subtracted from plotted values. Curves of ATPase activity versus metal concentrations were fit to v = Vmax[metal]/([metal] +K1/2). The reported standard errors for Vmax and K1/2 are asymptotic standard errors reported by the fitting software KaleidaGraph (Synergy).
Supplementary Material
Acknowledgements
We thank Phillip E. Klebba and Salete M. Newton for providing the Listeria strains used in this study. This work was supported by a grant from the NIH (GM059323; to JDH).
Footnotes
Author Contributions: HP and JDH were responsible for conception and design of the study, HP was responsible for acquisition of all experimental data exclusive of the ATPase assays which were done by SP. All authors participated in data analysis, interpretation, and writing of the manuscript.
REFERENCES CITED
- Anzaldi LL, Skaar EP. Overcoming the heme paradox: heme toxicity and tolerance in bacterial pathogens. Infection and immunity. 2010;78:4977–4989. doi: 10.1128/IAI.00613-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Argüello JM. Identification of ion-selectivity determinants in heavy-metal transport P1B-type ATPases. The Journal of membrane biology. 2003;195:93–108. doi: 10.1007/s00232-003-2048-2. [DOI] [PubMed] [Google Scholar]
- Argüello JM, Gonzalez-Guerrero M, Raimunda D. Bacterial transition metal P1B-ATPases: transport mechanism and roles in virulence. Biochemistry. 2011;50:9940–9949. doi: 10.1021/bi201418k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baichoo N, Wang T, Ye R, Helmann JD. Global analysis of the Bacillus subtilis Fur regulon and the iron starvation stimulon. Mol Microbiol. 2002;45:1613–1629. doi: 10.1046/j.1365-2958.2002.03113.x. [DOI] [PubMed] [Google Scholar]
- Bennett BD, Brutinel ED, Gralnick JA. A Ferrous Iron Exporter Mediates Iron Resistance in Shewanella oneidensis MR-1. Applied and environmental microbiology. 2015;81:7938–7944. doi: 10.1128/AEM.02835-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Analytical biochemistry. 1976;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- Brenot A, Weston BF, Caparon MG. A PerR-regulated metal transporter (PmtA) is an interface between oxidative stress and metal homeostasis in Streptococcus pyogenes. Mol Microbiol. 2007;63:1185–1196. doi: 10.1111/j.1365-2958.2006.05577.x. [DOI] [PubMed] [Google Scholar]
- Bsat N, Helmann JD. Interaction of Bacillus subtilis Fur (Ferric Uptake Repressor) with the dhb Operator In Vitro and In Vivo. J Bacteriol. 1999;181:4299–4307. doi: 10.1128/jb.181.14.4299-4307.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee SS, Hossain H, Otten S, Kuenne C, Kuchmina K, Machata S, Domann E, Chakraborty T, Hain T. Intracellular gene expression profile of Listeria monocytogenes. Infection and immunity. 2006;74:1323–1338. doi: 10.1128/IAI.74.2.1323-1338.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, James LP, Helmann JD. Metalloregulation in Bacillus subtilis: Isolation and Characterization of Two Genes Differentially Repressed by Metal Ions. Journal of Bacteriology. 1993;175:5428–5437. doi: 10.1128/jb.175.17.5428-5437.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Keramati L, Helmann JD. Coordinate regulation of Bacillus subtilis peroxide stress genes by hydrogen peroxide and metal ions. Proc Natl Acad Sci U S A. 1995;92:8190–8194. doi: 10.1073/pnas.92.18.8190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cossart P. Illuminating the landscape of host-pathogen interactions with the bacterium Listeria monocytogenes. Proc Natl Acad Sci U S A. 2011;108:19484–19491. doi: 10.1073/pnas.1112371108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djoko KY, Ong CL, Walker MJ, McEwan AG. The Role of Copper and Zinc Toxicity in Innate Immune Defense against Bacterial Pathogens. J Biol Chem. 2015;290:18954–18961. doi: 10.1074/jbc.R115.647099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faulkner MJ, Helmann JD. Peroxide stress elicits adaptive changes in bacterial metal ion homeostasis. Antioxidants & redox signaling. 2011;15:175–189. doi: 10.1089/ars.2010.3682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faulkner MJ, Ma Z, Fuangthong M, Helmann JD. Derepression of the Bacillus subtilis PerR peroxide stress response leads to iron deficiency. J Bacteriol. 2012;194:1226–1235. doi: 10.1128/JB.06566-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frawley ER, Crouch ML, Bingham-Ramos LK, Robbins HF, Wang W, Wright GD, Fang FC. Iron and citrate export by a major facilitator superfamily pump regulates metabolism and stress resistance in Salmonella Typhimurium. Proc Natl Acad Sci U S A. 2013;110:12054–12059. doi: 10.1073/pnas.1218274110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuangthong M, Helmann JD. Recognition of DNA by three ferric uptake regulator (Fur) homologs in Bacillus subtilis. J Bacteriol. 2003;185:6348–6357. doi: 10.1128/JB.185.21.6348-6357.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuangthong M, Herbig AF, Bsat N, Helmann JD. Regulation of the Bacillus subtilis fur and perR genes by PerR: not all members of the PerR regulon are peroxide inducible. J Bacteriol. 2002;184:3276–3286. doi: 10.1128/JB.184.12.3276-3286.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaballa A, Helmann JD. A peroxide-induced zinc uptake system plays an important role in protection against oxidative stress in Bacillus subtilis. Mol. Microbiol. 2002;45:997–1005. doi: 10.1046/j.1365-2958.2002.03068.x. [DOI] [PubMed] [Google Scholar]
- Gaballa A, Helmann JD. Bacillus subtilis CPx-type ATPases: characterization of Cd, Zn, Co and Cu efflux systems. Biometals. 2003;16:497–505. doi: 10.1023/a:1023425321617. [DOI] [PubMed] [Google Scholar]
- German N, Doyscher D, Rensing C. Bacterial killing in macrophages and amoeba: do they all use a brass dagger? Future Microbiol. 2013;8:1257–1264. doi: 10.2217/fmb.13.100. [DOI] [PubMed] [Google Scholar]
- Grass G, Otto M, Fricke B, Haney CJ, Rensing C, Nies DH, Munkelt D. FieF (YiiP) from Escherichia coli mediates decreased cellular accumulation of iron and relieves iron stress. Archives of microbiology. 2005;183:9–18. doi: 10.1007/s00203-004-0739-4. [DOI] [PubMed] [Google Scholar]
- Guan G, Pinochet-Barros A, Gaballa A, Patel SJ, Arguello JM, Helmann JD. PfeT, a P -type ATPase, effluxes ferrous iron and protects Bacillus subtilis against iron intoxication. Mol Microbiol. 2015;98:787–803. doi: 10.1111/mmi.13158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammer ND, Skaar EP. Molecular mechanisms of Staphylococcus aureus iron acquisition. Annual review of microbiology. 2011;65:129–147. doi: 10.1146/annurev-micro-090110-102851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harwood CR, Cutting SM. Molecular Biological Methods for Bacillus. John Wiley and Sons, Ltd.; 1990. [Google Scholar]
- Haschka D, Nairz M, Demetz E, Wienerroither S, Decker T, Weiss G. Contrasting regulation of macrophage iron homeostasis in response to infection with Listeria monocytogenes depending on localization of bacteria. Metallomics: integrated biometal science. 2015;7:1036–1045. doi: 10.1039/c4mt00328d. [DOI] [PubMed] [Google Scholar]
- Helmann JD. Specificity of Metal Sensing: Iron and Manganese Homeostasis in Bacillus subtilis. J Biol Chem. 2014;289:28112–28120. doi: 10.1074/jbc.R114.587071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmann JD, Wu MF, Gaballa A, Kobel PA, Morshedi MM, Fawcett P, Paddon C. The global transcriptional response of Bacillus subtilis to peroxide stress is coordinated by three transcription factors. J Bacteriol. 2003;185:243–253. doi: 10.1128/JB.185.1.243-253.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imlay JA. The molecular mechanisms and physiological consequences of oxidative stress: lessons from a model bacterium. Nature reviews. Microbiology. 2013;11:443–454. doi: 10.1038/nrmicro3032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T, Neilands JB. Products of "Low-iron Fermentation" with Bacillus subtilis: Isolation, Characterization and Synthesis of 2,3-Dihydroxybenzoylglycine. Journal of the American Chemical Society. 1958;80:4645–4647. [Google Scholar]
- Jesse HE, Roberts IS, Cavet JS. Metal ion homeostasis in Listeria monocytogenes and importance in host-pathogen interactions. Advances in microbial physiology. 2014;65:83–123. doi: 10.1016/bs.ampbs.2014.08.003. [DOI] [PubMed] [Google Scholar]
- Klebba PE, Charbit A, Xiao Q, Jiang X, Newton SM. Mechanisms of iron and haem transport by Listeria monocytogenes. Mol Membr Biol. 2012;29:69–86. doi: 10.3109/09687688.2012.694485. [DOI] [PubMed] [Google Scholar]
- Lanzetta PA, Alvarez LJ, Reinach PS, Candia OA. An improved assay for nanomole amounts of inorganic phosphate. Analytical biochemistry. 1979;100:95–97. doi: 10.1016/0003-2697(79)90115-5. [DOI] [PubMed] [Google Scholar]
- Lechowicz J, Krawczyk-Balska A. An update on the transport and metabolism of iron in Listeria monocytogenes: the role of proteins involved in pathogenicity. Biometals. 2015;28:587–603. doi: 10.1007/s10534-015-9849-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ledala N, Sengupta M, Muthaiyan A, Wilkinson BJ, Jayaswal RK. Transcriptomic response of Listeria monocytogenes to iron limitation and Fur mutation. Applied and environmental microbiology. 2010;76:406–416. doi: 10.1128/AEM.01389-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Z, Chandrangsu P, Helmann TC, Romsang A, Gaballa A, Helmann JD. Bacillithiol is a major buffer of the labile zinc pool in Bacillus subtilis. Mol Microbiol. 2014;94:756–770. doi: 10.1111/mmi.12794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Z, Faulkner MJ, Helmann JD. Origins of specificity and cross-talk in metal ion sensing by Bacillus subtilis Fur. Mol Microbiol. 2012;86:1144–1155. doi: 10.1111/mmi.12049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Z, Lee JW, Helmann JD. Identification of altered function alleles that affect Bacillus subtilis PerR metal ion selectivity. Nucleic acids research. 2011;39:5036–5044. doi: 10.1093/nar/gkr095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- May JJ, Wendrich TM, Marahiel MA. The dhb operon of Bacillus subtilis encodes the biosynthetic template for the catecholic siderophore 2,3-dihydroxybenzoate-glycine-threonine trimeric ester bacillibactin. J Biol Chem. 2001;276:7209–7217. doi: 10.1074/jbc.M009140200. [DOI] [PubMed] [Google Scholar]
- McLaughlin HP, Bahey-El-Din M, Casey PG, Hill C, Gahan CG. A mutant in the Listeria monocytogenes Fur-regulated virulence locus (frvA) induces cellular immunity and confers protection against listeriosis in mice. J Med Microbiol. 2013;62:185–190. doi: 10.1099/jmm.0.049114-0. [DOI] [PubMed] [Google Scholar]
- McLaughlin HP, Xiao Q, Rea RB, Pi H, Casey PG, Darby T, Charbit A, Sleator RD, Joyce SA, Cowart RE, Hill C, Klebba PE, Gahan CG. A putative P-type ATPase required for virulence and resistance to haem toxicity in Listeria monocytogenes. PloS one. 2012;7:e30928. doi: 10.1371/journal.pone.0030928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mielcarek A, Blauenburg B, Miethke M, Marahiel MA. Molecular Insights into Frataxin-Mediated Iron Supply for Heme Biosynthesis in Bacillus subtilis. PLoS ONE. 2015;10(3):e0122538. doi: 10.1371/journal.pone.0122538. doi:10.1371/journal.pone.0122538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore CM, Gaballa A, Hui M, Ye RW, Helmann JD. Genetic and physiological responses of Bacillus subtilis to metal ion stress. Mol Microbiol. 2005;57:27–40. doi: 10.1111/j.1365-2958.2005.04642.x. [DOI] [PubMed] [Google Scholar]
- Ollinger J, Song KB, Antelmann H, Hecker M, Helmann JD. Role of the Fur regulon in iron transport in Bacillus subtilis. J Bacteriol. 2006;188:3664–3673. doi: 10.1128/JB.188.10.3664-3673.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters WJ, Warren RA. Itoic acid synthesis in Bacillus subtilis. J Bacteriol. 1968;95:360–366. doi: 10.1128/jb.95.2.360-366.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quisel JD, Burkholder WF, Grossman AD. In vivo effects of sporulation kinases on mutant Spo0A proteins in Bacillus subtilis. J Bacteriol. 2001;183:6573–6578. doi: 10.1128/JB.183.22.6573-6578.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raimunda D, Long JE, Padilla-Benavides T, Sassetti CM, Argüello JM. Differential roles for the Co(2+) /Ni(2+) transporting ATPases, CtpD and CtpJ, in Mycobacterium tuberculosis virulence. Mol Microbiol. 2014;91:185–197. doi: 10.1111/mmi.12454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raimunda D, Long JE, Sassetti CM, Argüello JM. Role in metal homeostasis of CtpD, a Co2+ transporting P1B4-ATPase of Mycobacterium smegmatis. Mol Microbiol. 2012;84:1139–1149. doi: 10.1111/j.1365-2958.2012.08082.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rea R, Hill C, Gahan CG. Listeria monocytogenes PerR mutants display a small-colony phenotype, increased sensitivity to hydrogen peroxide, and significantly reduced murine virulence. Applied and environmental microbiology. 2005;71:8314–8322. doi: 10.1128/AEM.71.12.8314-8322.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosadini CV, Gawronski JD, Raimunda D, Argüello JM, Akerley BJ. A novel zinc binding system, ZevAB, is critical for survival of nontypeable Haemophilus influenzae in a murine lung infection model. Infection and immunity. 2011;79:3366–3376. doi: 10.1128/IAI.05135-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanson M, Makthal N, Flores AR, Olsen RJ, Musser JM, Kumaraswami M. Adhesin competence repressor (AdcR) from Streptococcus pyogenes controls adaptive responses to zinc limitation and contributes to virulence. Nucleic acids research. 2015;43:418–432. doi: 10.1093/nar/gku1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheldon JR, Heinrichs DE. Recent developments in understanding the iron acquisition strategies of gram positive pathogens. FEMS microbiology reviews. 2015;39:592–630. doi: 10.1093/femsre/fuv009. [DOI] [PubMed] [Google Scholar]
- Sinclair PR, Gorman N, Jacobs JM. Measurement of heme concentration. Curr Protoc Toxicol. 2001 doi: 10.1002/0471140856.tx0803s00. Chapter 8: Unit 8 3. [DOI] [PubMed] [Google Scholar]
- Smith AT, Smith KP, Rosenzweig AC. Diversity of the metal-transporting P1B-type ATPases. Journal of biological inorganic chemistry: JBIC: a publication of the Society of Biological Inorganic Chemistry. 2014;19:947–960. doi: 10.1007/s00775-014-1129-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studier FW. Protein production by auto-induction in high density shaking cultures. Protein Expr Purif. 2005;41:207–234. doi: 10.1016/j.pep.2005.01.016. [DOI] [PubMed] [Google Scholar]
- Wessling-Resnick M. Nramp1 and Other Transporters Involved in Metal Withholding during Infection. J Biol Chem. 2015;290:18984–18990. doi: 10.1074/jbc.R115.643973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zielazinski EL, Cutsail GE, 3rd, Hoffman BM, Stemmler TL, Rosenzweig AC. Characterization of a cobalt-specific P1B-ATPase. Biochemistry. 2012;51:7891–7900. doi: 10.1021/bi3006708. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.