SUMMARY
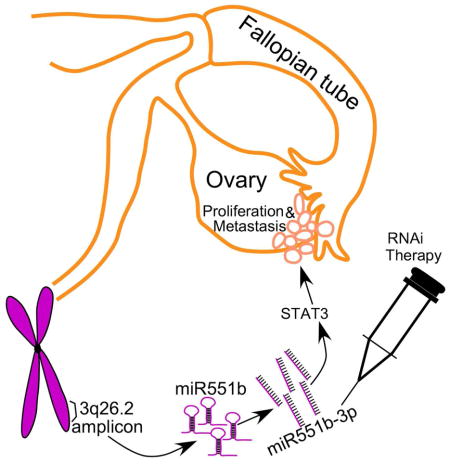
3q26.2 amplification in high-grade serous ovarian cancer leads to increased expression of mature microRNA miR551b-3p, which is associated with poor clinical outcome. Importantly, miR551b-3p contributes to resistance to apoptosis and increased survival and proliferation of cancer cells in vitro and in vivo. miR551b-3p up-regulates STAT3 protein levels with STAT3 being required for the effects of miR551b-3p on cell proliferation. Rather than decreasing levels of target mRNA as expected, we demonstrate that miR551b-3p binds a complementary sequence on the STAT3 promoter recruiting RNA Polymerase II and the TWIST1 transcription factor to activate STAT3 transcription and thus directly upregulates STAT3 expression. Furthermore, anti-miR551b reduced STAT3 expression in ovarian cancer cells in vitro and in vivo and reduced ovarian cancer growth in vivo. Together our data demonstrates a role for miR551b-3p in transcriptional activation. Thus miR551b-3p represents a promising candidate biomarker and therapeutic target in ovarian cancer.
Graphical abstract
INTRODUCTION
DNA copy number aberrations (CNA) occur commonly in cancer. 3q26 amplification is a frequent event in High Grade Serous Epithelial Ovarian Cancer (HGSEOC) (Bell et al., 2011; Chaluvally-Raghavan et al., 2014; Shayesteh et al., 1999), which is the most common and aggressive form of ovarian cancer. We have demonstrated that the 3q26 CNA is complex, resulting in amplification and aberrant function of several oncogenes including PIK3CA (Shayesteh et al., 1999), PKCι (Eder et al., 2005), Mecom (EVI1 and MDS1/EVI1) (Nanjundan et al., 2007) and a non-coding RNA, miR-569 (Chaluvally-Raghavan et al., 2014). While the coding genes in the 3q26.2 amplicon are well-studied, the potential roles of microRNAs (miRNA or miR) located at the 3q26 locus remain poorly characterized. Thus, it appears likely that the 3q26.2 amplicon contributes to tumor pathophysiology through coordinate dysregulation of multiple cellular functional processes as a consequence of altered expression of coding and non-coding RNAs.
miRNAs are thought to primarily regulate cellular functions through decreasing the stability or translation of multiple mRNAs based on binding to “seed” sequences located at the 3′ end of the mRNA (Bartel, 2004). While a small set of studies suggest that miRNA can increase gene transcription (Janowski et al., 2007; Place et al., 2008), the underlying mechanisms remain to be elucidated. Thus, miRNAs can directly increase or decrease the expression of key regulatory proteins as well as have secondary effects on multiple targets.
The signal transducer and activator 3 (STAT3) transcription factor (TF) is constitutively activated in cancer cells and high levels of STAT3 are associated with poor outcome in ovarian cancer patients (Rosen et al., 2006). STAT3 not only increases proliferation of ovarian cancer cells but also engenders resistance of stem cell-like cells to chemotherapy (Abubaker et al., 2014). STAT3 has been explored as a potential target in ovarian cancer models and has demonstrated utility both in monotherapy and combination therapy (Ma et al., 2015; Wen et al., 2015).
In this study, we demonstrate that miR551b, which is located near the peak of the 3q26.2 amplicon, is increased and functionally relevant in ovarian cancer, consistent with non-coding RNAs and miRNAs located in the 3q26.2 locus acting as unexpected drivers in ovarian cancer. Importantly, our studies identified complementarity between miR551b-3p and sequences in the promoter of STAT3. Indeed, miR551b-3p leads to recruitment of RNA Polymerase II (RNA-Pol-II or RPOL2) and the Twist1 transcription factor to the STAT3 promoter and increases STAT3 mRNA levels. Consistent with a key role of miR551b-3p and STAT3 in ovarian cancer pathophysiology, anti-miR551b incorporated into neutral nano-liposomes (Chaluvally-Raghavan et al., 2014; Landen et al., 2005) markedly decreased ovarian cancer growth in vivo.
RESULTS
Increased expression of miR551b-3p is associated with worsened outcome in HGSEOC
Our analysis using high-resolution SNP array datasets from The Cancer Genome Atlas (TCGA) demonstrated that miR-551b is located at the consensus peak of the 3q26.2 locus and is amplified in approximately 1/3 of patients with HGSEOC (Fig. 1A). Since two mature miRNAs, miR551b-5p and miR551b-3p, are derived from precursor miR551b (pre-miR551b), we employed TCGA ovarian cancer data to assess expression of miR551b mature forms. Our analysis demonstrated miR551b-3p to be highly expressed in tumors whereas the mature miR551b-5p was barely detectable (Supplementary Figure S1A–S1B). Consistent with the expression of miR551b-3p and miR551b-5p in TCGA data, our qPCR results demonstrate that miR551b-3p is highly expressed in ovarian cancer cell lines compared to miR551b-5p (Supplementary Figure S1C). Thus we focused on the functional roles and mechanism of action of miR551b-3p in the current study.
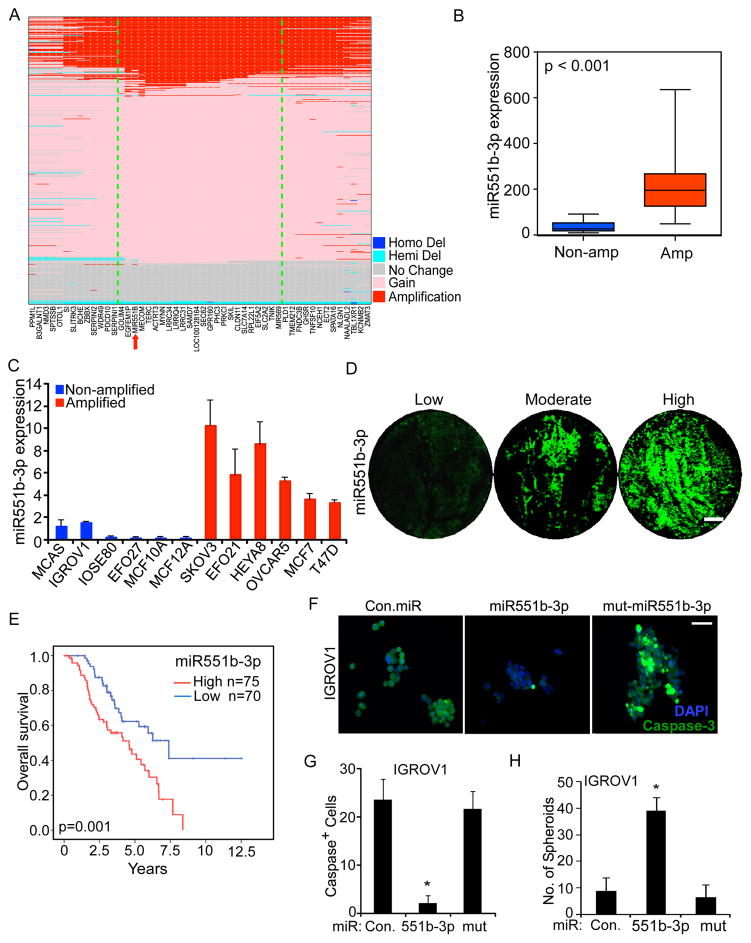
Figure 1. 3q26.2 amplification correlates with miR551b-3p expression and miR551b-3p increases proliferation of ovarian cancer cells.
A, Copy number status of genes located in 3q26.2 locus in 489 ovarian cancer patients in the TCGA SNP array data set. Genes were arranged based on the order in RefSeq (GRCh37/hg19, 2009). The core 3q26.2 amplicon is bounded by the green lines. B, miR551b-3p expression was quantified in 24 ovarian cancer samples by quantitative real time PCR (qPCR). Blue (n=8) and red (n=16) boxes indicate 3q26.2 amplification status from SNP data. C, miRNA was isolated from the indicated cell lines in triplicate. miR551b-3p expression was assessed by qRT-PCR and normalized to U6 RNA in breast and ovarian cell lines. Bars represent s.d. of triplicate determinations. D, miR551b-3p in situ hybridization was performed in 145 HGSEOC tumor samples. Representative samples from each group are displayed. Scale bar represents 200 μm. E, Kaplan-Meier plot of overall survival of 145 ovarian cancer patients stratified by high (red combination of moderate and strong staining) vs low (blue) miR551b-3p expression determined by RNA in situ hybridization. Log-rank test was used to compare differences between the two groups. F, IGROV1 cells were grown on low attachment culture plates and cells transfected with control (Con.) miR, miR551b-3p or mutated miR551b. Spheroids formed were fixed and stained with DAPI or anti-cleaved caspase 3. Scale bar represents 50 μm. G–H, Cleaved caspase-3 positive cells were counted from three different fields from three independent experiments as described in (F). Numbers of spheroids formed were quantified. Bars represent s.d. of three different experiments. * indicates p<0.001 compared to control or mutated miR transfected group. p values were determined using Student’s t test.
Analysis of two different ovarian cancer patient cohorts demonstrated that copy number gain (>2.8 copies) or amplification (>4 copies) of 3q26.2 is associated with increased miR551b-3p expression (Fig. 1B, Supplementary Fig. S2A). Furthermore, miR551b-3p is highly expressed in tumor samples compared to normal ovarian tissue (Supplementary Fig. S2B). Consistent with the patient data, miR551b amplified cell lines exhibit high expression of miR551b-3p (Fig. 1C). We further assessed miR-551b-3p expression using RNA in situ hybridization in 145 HGSEOC samples (Fig. 1D). Our analysis demonstrates that miR551b-3p expression is associated with poor outcome in HGSEOC (Fig. 1E and Table-S1) with approximately 40% of HGSEOC patients with low miR551b-3p demonstrating long-term survival and all patients with high miR551b-3p levels succumbing to their disease. Thus amplification and overexpression of miR551b-3p is associated with aggressive HGSEOC and predicts patient outcome. Together our results suggest that miR551b-3p, the key mature form derived from pre-miR551b, has an important role in the pathophysiology of HGSEOC.
miR551b-3p regulates ovarian cancer cell growth and survival in vitro
Consistent with a role of miR551b-3p in the pathophysiology of ovarian cancer, enforced expression of miR551b-3p in IGROV1 and HEYA8 ovarian cancer cells and immortalized ovarian epithelial cells IOSE-80 reduced apoptosis as indicated by low expression of cleaved caspase-3 and increased the number and size of spheroids formed in non-adherent culture conditions (Fig. 1F–1H and Supplementary Fig. S2C–S2E). miR551b-3p also increased proliferation of IGROV1, IOSE80, HEYA8, and MDAMB231 cells in 2D adherent culture conditions (Supplementary Fig. S2F). Thus, the effects of miR551b-3p on proliferation are generalizable across multiple cell lines encompassing both breast and ovarian lineages. In contrast, miR551b-5p or mutated miR551b-3p did not alter proliferation of ovarian cancer cells or immortalized ovarian epithelial cells (Supplementary Fig. S2F), again supporting miR551b-3p as the functionally relevant product of the miR551b precursor.
miR551b-3p increases tumor growth of ovarian cancer cells in vivo
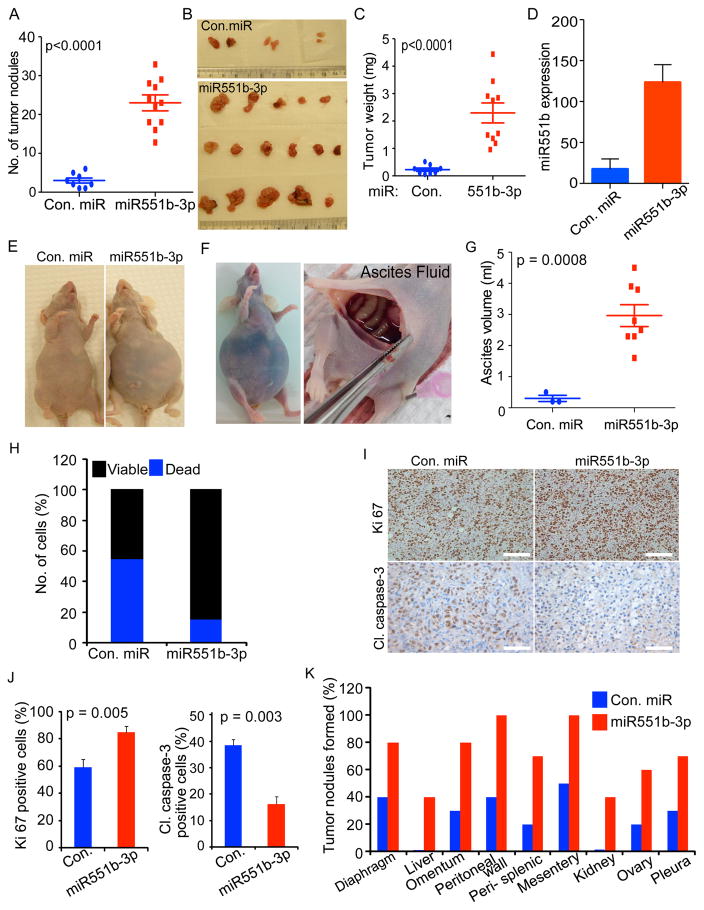
To determine whether miR551b-3p altered tumor growth in vivo, nude mice bearing IGROV1 tumors were treated with intraperitoneal injection of miR551b-3p incorporated in neutral 1,2-dioleoyl-sn-glycero-3-phosphotidylcholine (DOPC) nano-liposomes (Chaluvally-Raghavan et al., 2014; Landen et al., 2005). Intraperitoneal injection of miR551b-3p into nude mice markedly increased the number of tumor nodules and tumor weight (Fig. 2A–2D). Ascites formation is a common event in ovarian carcinoma, particularly HGSEOC, potentially contributing to peritoneal seeding and poor patient outcomes (Mills et al., 1990; Mills et al., 1988). Of note miR551b-3p increased ascites formation in mice bearing IGROV1 cancer cells (Fig. 2E–2G). Subsequently, we assessed the viability of epithelial cancer cells isolated from peritoneal fluid from IGROV1 tumor bearing mice. Strikingly miR551b-3p expression markedly reduced the number of apoptotic cells in ascites fluid (Fig. 2H). As expected, immunohistochemical analysis of tumor tissues from miR551b-3p-treated mice demonstrated an increase in the number of Ki67 positive cells and loss of cleaved caspase-3 expression, consistent with proliferative and anti-apoptotic effects of miR551b-3p in vivo (Fig. 2I–2J). miR551b-3p also increased the number of peritoneal implants in mice bearing IGROV1 tumors (Fig. 2K). Consistent with our results that miR55b-3p increased ascites formation in animal models (Fig. 2E–2G), there is a significant association of miR551b-3p expression with ascites formation in clinical samples of HGSEOC patients (Table S2).
Figure 2. miR551b-3p increases proliferation of ovarian cancer cells in vivo.
A–C, IGROV1 cells (1 × 106 cells/animal) were injected intraperitoneally. Mice were treated with control (Con.) miR or miR551b-3p (n=10 per group) twice weekly for five weeks. Number of abdominal tumors and tumor weight were quantified at the end of the experiment. Representative images are displayed. D, miR551b-3p expression in tumor tissues described in 1C were quantified by qPCR. miR551b-3p expression was normalized to U6 small RNA and p values were determined using Student’s t test. E, Representative images of mice described in A. F–G, Mice bearing ascites fluid were dissected and photographed. Ascites fluid volume was quantified. p values were determined using Student’s t-test. H, Epithelial cells from ascites fluid from three mice in each group were isolated and cell viability was assessed by propidium iodide staining. I–J, Immunohistochemistry (IHC) was performed on tumor tissues from A using Ki67 and cleaved caspase-3 antibody. Positive cells were quantified from tumor tissues isolated from three mice from each group. p values were determined using Student’s t-test. Scale bar represents 100 μm. K, Sites of tumor growth from mice in A.
Taken together, our results suggest that amplification and subsequent high expression of miR551b-3p is a key event contributing to survival and proliferation of ovarian cancer cells in vitro and in vivo.
miR551b-3p targets STAT3 expression with subsequent increases in self renewal capacity of spheroids
We used four different prediction tools (RNA22, miRanda, Targetscan and miRwalk) to identify potential targets of miR551b-3p. To test if miR551b-3p alters levels of the thirty-three genes predicted to be miR551b-3p targets (Supplementary Fig. S3A), we overexpressed miR551b-3p in IGROV1 cells and performed qPCR to determine levels of each potential target. Of the thirty-three potential targets, eight genes (ATXN7, GPT, NDEL1, NTRK2, RPP15, SLC7A1, TCHH and WDFY4) were modestly downregulated (between 30% to 60%) upon miR551b-3p expression (Supplementary Fig. S3B). To test whether the loss of expression of the eight potential miR551b-3p targets (Supplementary Fig. S3C) could explain the biological effects of miR551b-3p, we assessed the effect of knockdown of these genes on proliferation and spheroid formation, which is markedly altered by miR551b-3p in IGROV1 cells (see Fig. 1 and Supplementary Fig. S1). In contrast with the effects of miR551b-3p, knockdown of each of the eight genes did not substantially alter either proliferation or spheroid formation of IGROV1 cells (Supplementary Fig. S3D–S3G). These results suggest that miR551b-3p could act through a mechanism independent of binding to consensus sites in the 3′ untranslated regions of target genes.
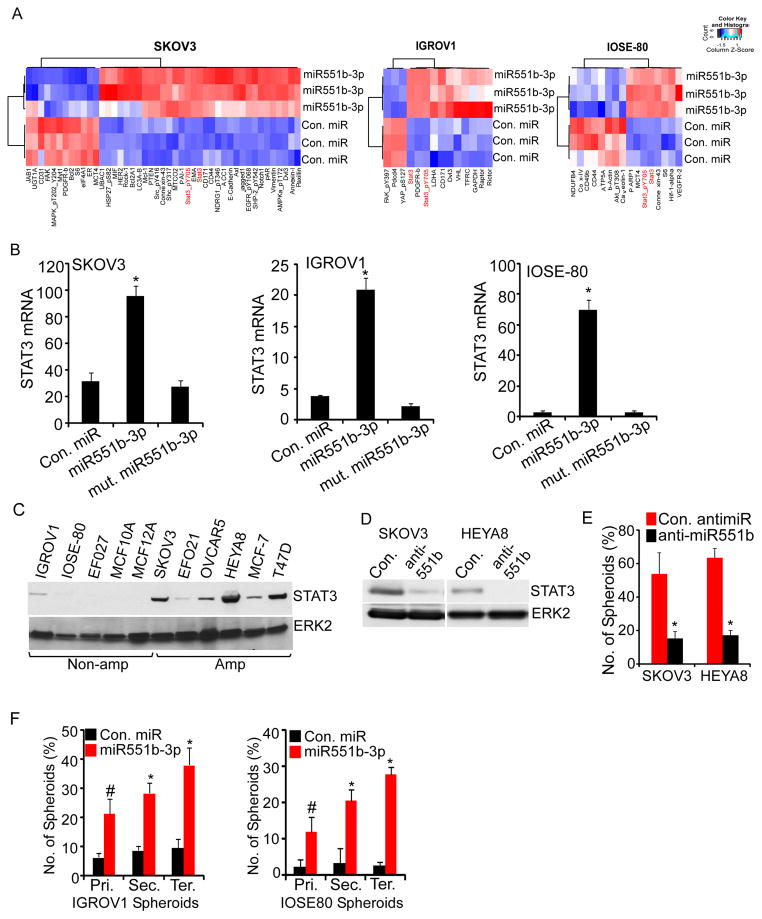
To identify potential functional consequences of miR551b-3p, we employed Reverse Phase Protein Arrays (RPPA) (Hennessy et al., 2010), where we expressed miR551b-3p in IGROV1 and SKOV3 ovarian cancer cells and IOSE-80 immortalized ovarian epithelial cells and assessed effects on the levels of 280 proteins involved in cell signaling. Our analysis demonstrated that total and phosphorylated (Y705) STAT3 are upregulated in all three cell lines (Fig. 3A). Consistent with our RPPA results, qPCR demonstrated that miR551b-3p upregulated STAT3 mRNA in SKOV3, IOSE-80 and IGROV1 cells (Fig. 3B). Furthermore, immunoblotting showed that miR551b amplified cancer cell lines (Fig. 1C) exhibit increased STAT3 protein levels (Fig. 3C). Conversely, anti-miR551b decreases STAT3 levels in HEYA8 and SKOV3 cells (Fig. 3D), which express high levels of miR551b-3p (see Fig. 1C) and concurrently reduces the number of spheroids under matrix-deprived conditions (Fig. 3E).
Figure 3. miR551b-3p regulates STAT3 expression.
A, SKOV3, IGROV1 and IOSE-80 cells were transfected with control miR or miR551b-3p in three independent experiments and effects on protein levels determined by RPPA 48h later. Proteins with greater than 20% change in expression are represented in the heatmap. B, RNA was extracted from the samples described in (A). Relative expression of STAT3 was analyzed by qRT-PCR and normalized to β-Actin. Bars represent s.d. of three independent experiments. C, Immunoblot analysis of STAT3 expression and 3q26.2 copy number status of indicated cell lines. D–E, Cells transfected with control antimiR or anti-miR551b-3p were lysed 72h after transfection and immunoblotting performed with the indicated antibodies. Number of spheroids formed on matrix-deprived conditions was quantified 5 days after antimiR transfection. Bars represent s.d. of three independent experiments. * indicates p<0.001 compared to the control as assessed by Student’s t test. F, Cells were grown under matrix-deprived conditions and then transfected with con. miR or miR551b-3p. Number of spheroids were quantified and then re-plated serially at five days intervals after miR transfection. Spheroids were re-transfected with control miR or miR551b-3p during serial passages. Bars represent s.d. of three independent experiments. # indicates p<0.002 and * indicates p<0.001 compared to respective controls as assessed by Student’s t test.
The ability of miR551b-3p to increase growth in non-adherent cultures suggested that miR551b-3p could promote tumor-initiating cell (TIC) or stem cell-like activity (Burgos-Ojeda et al., 2012; Medema, 2013). Consistent with the effects of miR551b-3p on STAT3 levels (Fig. 3A–3B), miR551b-3p increased the number of IOSE80 and IGROV1 spheroids formed in serial passages, which provides an in vitro surrogate for TIC and stem cell-like activity (Fig. 3F). Taken together, our data demonstrate that the miR551b-STAT3 axis regulates multiple oncogenic processes in ovarian cancer.
STAT3 is required for miR551b-induced proliferation and survival of ovarian cancer cells
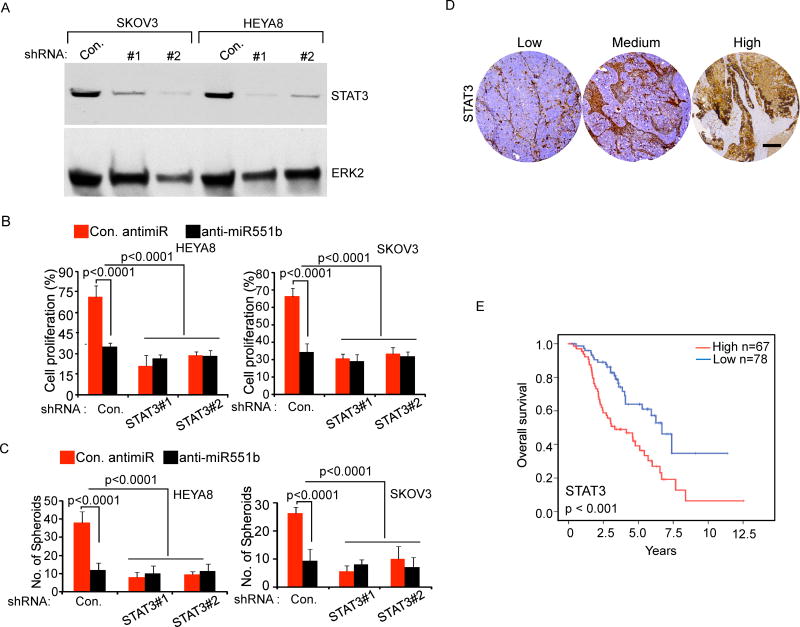
To determine whether the decrease in STAT3 induced by anti-miR551b was responsible for the inhibition of proliferation and spheroid formation, we stably reduced STAT3 expression using two different shRNAs (Fig. 4A). Consistent with our results that anti-miR551b reduced STAT3 levels and spheroid formation (Fig. 3D–3E), STAT3 shRNA markedly decreased proliferation and spheroid formation in both HEY8 and SKOV3 cells (Fig. 4B–4C). In cells with knockdown of STAT3, anti-miR551b did not further decrease either proliferation or spheroid formation (Fig. 4B–4C). In conclusion, our results demonstrate that the effects of miR551b-3p on cell survival and proliferation are dependent on STAT3 as an intermediary factor.
Figure 4. Knock down of STAT3 abolishes miR551b-3p mediated cell growth.
A, SKOV3 or HEYA8 cells stably expressing control or two different shRNA targeting STAT3 were lysed and immunoblotted using indicated antibodies. B–C, Two different clones of STAT3 knock down cells were transfected with control anti-miR or anti-miR551b-3p and cell proliferation assessed on day 3 using MTT and number of spheroids formed on low attachment plates assessed on day 5. Bars represent s.d. of three independent experiments. D, STAT3 protein level was quantified in 145 HGSEOC patient samples using immunohistochemistry (IHC). Scale bar represents 200 μm. Representative samples from each group are displayed. E, Kaplan-Meier plot of overall survival of 145 ovarian cancer patients stratified by high (red combination of moderate and strong staining) vs low (blue) STAT3 expression. Log-rank test was used to compare differences between the two groups.
To assess whether any of the eight genes identified as potential targets of miR551b-3p (Supplementary Fig. S3C) could explain the effect of miR551b-3p on STAT3, we knocked down each of the eight genes (ATXN7, GPT, NDEL1, NTRK2, RPP15, SLC7A1, TCHH and WDFY4) modestly downregulated by miR551b-3p (Supplementary Fig. S3C) and showed that this did not change STAT3 expression, again suggesting that the effect of miR551b-3p on STAT3 is likely independent of binding to 3′UTR of target genes (Supplemental Fig. S4A–S4B).
STAT3 levels correlate with outcomes in ovarian cancer
Consistent with the poor prognosis associated with high miR551b-3p expression (Fig. 1E), STAT3 expression by immunohistochemisty in 145 HGSEOC patient samples demonstrated that high STAT3 expression was associated with a decreased overall survival of women with HGSEOC (Fig. 4D–4E and Table S1). Similarly, our analysis demonstrated that increased STAT3 mRNA levels were associated with decreased progression free survival (Supplemental Fig. S4C) in an independent cohort of ovarian cancer patients (Tothill et al., 2008). Altogether our data support the contention that miR55b-3p induces STAT3 expression, which contributes to aggressiveness of HGSEOC.
miR551b-3p interacts with the STAT3 promoter to increase STAT3 transcription
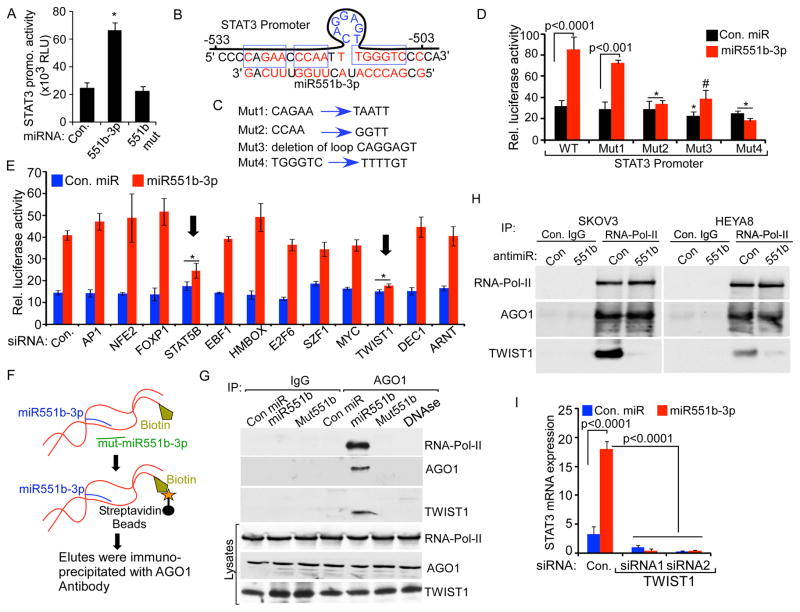
miRNAs primarily elicit their effects by suppressing translation or degrading mRNA through RNA interference (RNAi) (Ambros, 2001; Farh et al., 2005). Although miRNAs can also target promoter regions and increase transcription (Huang et al., 2012; Janowski et al., 2007; Place et al., 2008), the mechanism(s) by which miRNAs activate transcription is not well understood. Importantly, miR551b-3p increased STAT3 promoter driven luciferase activity while control miRs or mutant miR551b-3p did not increase luciferase activity (Fig. 5A) consistent with the contention that miR551b-3p activates the STAT3 promoter. Surprisingly, miR551b-3p demonstrated sequence complementarity with a loop structure in the STAT3 promoter (Fig. 5B). To determine if the miR551b-3p complementary sequences are critical for miR551b-3p induced STAT3 transcription, we mutated four sites on the STAT3 promoter that could play a role in the action of miR551b-3p (sequences in the blue boxes in Fig. 5B are mutated as indicated in Fig. 5C). Of note, deletion of the loop structure or sequences on either side of the loop structure abolished the effects of miR551b-3p on STAT3 promoter induced luciferase activity (Fig. 5C–5D). However mutations distant to the loop structure (Mut1 in Fig. 5C and Fig. 5D) did not block the effects of miR551b-3p expression on luciferase activity. Thus the core complementary sequences between miR551b-3p and the STAT3 promoter are critical for the ability of miR551b-3p to active the STAT3 promoter.
Figure 5. miR551b-3p promotes the interaction of TWIST1-AGO1 complex in the STAT3 promoter.
A, The STAT3 promoter including 962 bp upstream to transcription start site (TSS) was cloned in the pLight luciferase reporter. The construct was co-transfected with control miR or miR551b-3p into IGROV1 cells and luciferase activity assessed (bottom). Bars represent sd of quadruplicates. p values were determined using Student’s t test. B, miR551b-3p sequence complementarity with the STAT3 promoter sequence (−530bp to −503bp upstream to TSS). C, Four different mutant STAT3 promoters were constructed as indicated in blue boxes or loop in (B). D, Mutant STAT3 promoter constructs were co-transfected with control miR or miR551b-3p into IGROV1 cells and luciferase activity assessed. Bars represent s.d. of triplicates. p values were determined by Student’s t test. *p<0.001 and # p<0.005 compared to the WT group transfected with miR551b-3p. E, IGROV1 cells were transfected with the indicated siRNAs. STAT3 promoter cloned in the pLight luciferase reporter was co-transfected with control miR or miR551b-3p into IGROV1 cells 24h after siRNA transfection. Luciferase activity was assessed 48h after transfection of the reporter vector. Bars represent s.d. of triplicates. p values were determined by Student’s t test. *p<0.005 compared to the WT group transfected with miR551b-3p. F–G, Scheme of assay used to hybridize and pull down the biotinylated STAT3 promoter fragment incubated with con. miR, miR551b-3p or mutant miR551b. Promoter-miRNA complex were incubated with IGROV1 cell lysates and immunoprecipitated 72h later with control IgG or anti-AGO1. DNAase was used to degrade the indicated samples. Total lysate or AGO1 immunoprecipitates were western blotted with the indicated antibodies. Total lysate represents 1/10 the amount in the immunoprecipitates. H, HEYA8 or SKOV3 cells were transfected with either control antimiR or anti-miR551b-3p. Nuclear extract was prepared and immunoprecipitation was performed using RNA-Pol-II specific antibodies and immunoblotted using antibodies indicated. I, IGROV1 cells were transfected with the indicated siRNAs. RNA was extracted 48h after transfection. Relative expression of STAT3 was analyzed by qPCR and normalized to β-Actin. Bars represent s.d. of three independent experiments. p values were determined by Student’s t test.
To identify transcription factors (TFs) that bind in the region of the STAT3 promoter with complementarity to miR551b-3p, we employed Genomatix Matinspector software. We then knocked down the expression of each candidate TF, demonstrating that knock down of TWIST1 but not other candidates with the exception of STAT5B, abrogated the effects of miR551b-3p on the STAT3 promoter (Fig. 5E). Importantly, neither TWIST1 nor STAT5B knockdown altered basal (i.e. non-miR551b-3p mediated) promoter activity.
miR551b-3p recruits RNA-Pol-II to the STAT3 promoter
miRNAs that increase transcription have recently been shown to be part of a complex with Argonaute proteins and the RNA-Pol-II transcription machinery (Zhang et al., 2014). To determine whether miR551b-3p associates with RNA-Pol-II on the STAT3 promoter, we biotinylated the 3′end of a 328bp STAT3 promoter construct that includes the miR551b-3p complementary site and the TWIST1 site as well as other TF sites near the miR551b-3p complementary site (Figure 5F). The biotinylated STAT3 promoter fragment was incubated with control miR, miR551b-3p or mutant miR551b-3p. The promoter-miRNA complexes were then incubated with IGROV1, HEYA8 or SKOV3 cell lysates. Streptavidin beads were used to purify the complexes, which were then immunoprecipitated using an AGO1 antibody. Western blotting of the biotinylated DNA-miRNA complex, demonstrated that miR551b-3p, but not control miR or mutated miR551b-3p, associated with the STAT3 promoter fragment formed a complex including Argonaute, RNA-Pol-II and TWIST1 (Fig. 5G and Supplementary Fig. S5A). Although knockdown of STAT5B inhibited miR551b-3p mediated promoter activity (Fig. 5E), we were not able to detect STAT5B in the DNA-miRNA complex. We next determined whether miR551b-3p was required for the formation of the AGO1, RNA-Pol-II and TWIST1 complex. Intriguingly immunoprecipitation of RNA-Pol-II protein from nuclear extracts of SKOV3 or HEYA8 cells treated with anti-miR551b, demonstrated reduced association of TWIST1 with the AGO1/RNA-Pol-II complex (Fig. 5H). Together our results support the notion that miR551b-3p expression regulates the interaction of TWIST1 with the STAT3 transcription activation complex. Importantly, two different siRNAs targeting TWIST1 disrupted miR551b-3p induced increases in STAT3 expression (Fig. 5I), demonstrating TWIST1 to be required for miR551b-3p induced STAT3 expression. Of note, knockdown of either STAT3 or STAT5B decreased TWIST1 mRNA consistent with a regulatory loop wherein STAT3 and STAT5B regulate TWIST1 levels and TWIST1 in turn activates STAT3 transcription (Supplemental Fig. S5B–S5C).
Furthermore, biotinylated miR551b-3p, but not mutant miR551b-3p mixed with sonicated IGROV1 DNA extracts pulled down the STAT3 promoter (Supplemental Fig. S5D–S5E), again suggesting a direct interaction of miR551b-3p with the STAT3 promoter. Thus our results demonstrate that a miR551b-3p-AGO1 complex enhances the recruitment of RNA-Pol-II and TWIST1 transcription factor to the STAT3 promoter.
In an independent approach, we cloned 172 bp of the STAT3 promoter containing the miR551b-3p complementary site and Twist1 binding site into a thymidine kinase minimal promoter in the pGLuc reporter vector. miR551b-3p, but not mutant miR551b-3p, increased activity of the STAT3 promoter (Supplemental Fig. S6). Strikingly, mutation of the miR551b-3p complementary site or the Twist1 binding site abolished miR551b-3p mediated STAT3 promoter activity (Supplemental Fig. S6).
Altogether our results demonstrate an unexpected role for miR551b-3p, wherein miR551b-3p directly binds to the STAT3 promoter, which in turn facilitates the recruitment of RNA-Pol-II and TWIST1 to the promoter contributing to increased STAT3 transcription (Fig-6J).
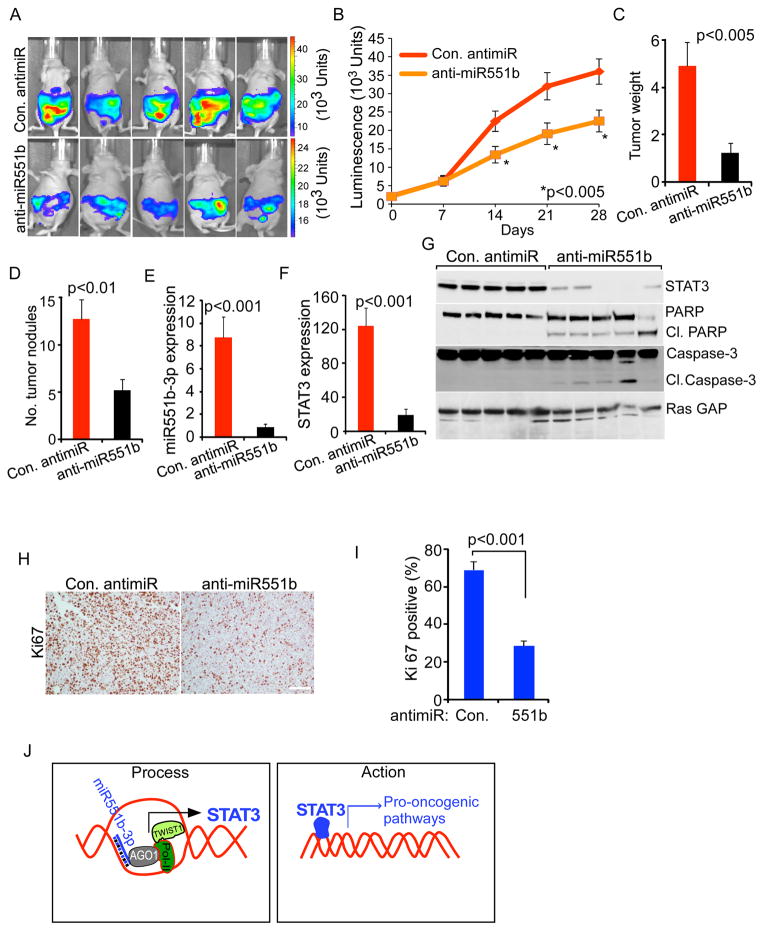
Silencing of miR551b-3p inhibits ovarian cancer tumor growth in nude mice
To explore the potential utility of miR551b-3p as a therapeutic target in ovarian cancers with miR551b amplification and overexpression, we established luciferase labeled HEYA8 cells in the peritoneal cavity of nude mice. Mice were treated with anti-miR551b-3p incorporated in neutral nano-liposomes (1,2-dioleoyl-sn-glycero-3-phosphotidylcholine-DOPC) (Chaluvally-Raghavan et al., 2014; Landen et al., 2005) twice a week for four weeks. As expected from our in vitro studies, treatment with anti-miR551b markedly decreased tumor growth (Fig. 6A–6B), tumor weight (Fig. 6C) and number of tumor nodules (Fig. 6D). Importantly, anti-miR551b, but not control antimiR decreased levels of both miR551b-3p and STAT3 transcripts (Fig. 6E–6F). Consistent with the effects of antimiR-551b on the growth of ovarian cancer cells, our results demonstrate that anti-miR551b increased levels of apoptotic cell death markers: cleaved PARP and cleaved Caspase-3 (Fig. 6G). Of note, anti-miR551b also reduced expression of the Ki67 proliferation marker in tumor tissues (Fig. 6H–6I). Together, our results demonstrate therapeutic potential of antimiR-551b in treating ovarian cancers with high levels of miR551b-3p. Future studies will need to examine the activity of combination therapy of anti-miR551b with other therapeutic interventions.
Figure 6. Liposomal delivery of anti-miR551b-3p reduces tumor burden and downregulates STAT3 in vivo.
A, HEYA8 cells (2.5 × 105 cells/animal) stably express luciferase reporter genes were injected into the peritoneal cavity of nude mice (n=10). Mice were treated with con. antimiR or anti-miR551b-3p encapsulated in DOPC nanoliposomes. Bioluminescence imaging was performed on day 28 and the images of five representative mice from each group is displayed. The color scale bars depict photon fluxes emitted from the tumor cells. B, Luminescence intensity was quantified (n=10) on the indicated days. Bars represent standard error (s.e.). Significance was determined by two-way Anova test. C–D, Tumor nodules were isolated on day 28. Numbers of abdominal tumors and total tumor weight were calculated. p values were determined by Student’s t-test. E–F, RNA was extracted from three representative tumor samples from each group. Relative expression of miR551b-3p and STAT3 was analyzed by qRT-PCR and normalized to U6 RNA or β-Actin respectively. p values were determined by Student’s t test. G, Tumor tissues from 5 separate tumors (A) were lysed and immunoblotting performed using the indicated antibodies. H–I, In vivo effects of anti-miR551b-3p treatment on tumor cell proliferation were determined by Ki67 immunohistochemistry (IHC). Ki67 positive cells were quantified in three different fields from the tumor tissues isolated from three mice from each group. p values were determined by Student’s t test. Scale bar represents 100 μm. J, Two-step model illustrates the process how STAT3 is regulated by miR551b-3p and the action of STAT3.
DISCUSSION
We have previously demonstrated that the 3q26.2 amplicon contains several oncogenes including PIK3CA, PKCι and Mecom (Eder et al., 2005; Nanjundan et al., 2007; Shayesteh et al., 1999). In a recent study, we demonstrated that miR569 located in the 3q26.2 amplicon, down-regulates the tumor suppressor TP53INP1 and increases growth and survival of ovarian cancer cells (Chaluvally-Raghavan et al., 2014). In this study, we demonstrate that the 3q26.2 amplicon also leads to increased miR551b-3p expression with subsequent STAT3 induction that contributes to cell proliferation and survival.
RNA activation (RNAa) wherein miRNAs increase transcription of target genes (Li et al., 2006), has been reported for E-cadherin, p21, VEGF and progesterone receptor (PR) (Huang et al., 2010; Place et al., 2008; Portnoy et al., 2011). Further studies of miRNA immunoprecipitation followed by deep sequencing revealed that AGO complex bound small RNAs are enriched in promoter regions of many genes (Cernilogar et al., 2011). Intriguingly, a recent report suggests that AGO1 interacts with RNA-Pol-II at active promoters throughout the genome (Huang et al., 2013). In this study we have identified an unexpected action of miR551b-3p wherein it activates STAT3 transcription by direct interaction with the STAT3 promoter increasing the recruitment of the transcription factor TWIST1 and RNA-Polymerase-II. This process appears to represents a key mechanism regulating STAT3 transcription in ovarian cancer. Further studies are required to determine whether miR551b-3p alters transcriptional activation of other genes that along with STAT3 combine to mediate the effects of miR551b-3p on ovarian cancer pathophysiology. Databases such as microPIR (Piriyapongsa et al., 2012) can identify predicted miRNA target sites located within promoter regions of human genes facilitating this effort. However these database searches will need to be confirmed experimentally as the effects of miRNA on different genes could be dependent on complementarity in the promoter region as well as organization and occupancy of transcription factor binding sites located near the potential miRNA binding sites (Portnoy et al., 2011).
We explored the potential effects of miR551b-3p on downregulation of RNA function through binding to consensus sequences in 3′ untranslated regions (3′UTR) of mRNAs. However, we failed to demonstrate marked effects of miR551b-3p on potential mRNA targets identified as having consensus miR551b-3p binding sites in their 3′UTR through database searches. Furthermore, knockdown of the mRNA modulated by miR551b-3p failed to recapitulate the functional effects of miR551b-3p on cells. However, we have not excluded the possibility that miR551b-3p regulates mRNA stability or translation through canonical mechanisms involving binding to 3′ UTR of mRNAs. These effects of miR551b-3p could, in turn, indirectly regulate the action of miR551b-3p on the STAT3 promoter or alternatively contribute to the functional effects of miR551b-3p by altering the activity of other pathways.
The ability of miRNAs to concurrently down-regulate stability or transcription of mRNAs as well as to upregulate expression of target genes, supports the concept that miRNA have the ability to fine tune the effects of transcription factors in controlling protein levels. The ability to alter protein levels through multiple mechanisms has the potential to allow miRNA to coordinately regulate critical phenotypic outcomes.
Our results support the concept that targeting miR551b expression, using antimiR, could block STAT3 activity and provide a useful therapeutic avenue for treating human ovarian cancers harboring the 3q26.2 amplification. STAT3 plays a broad role in physiological responses including differentiation, cell survival and metastasis supporting a therapeutic opportunity (Levy and Darnell, 2002). Furthermore metabolic rewiring and shift in glutamine levels upregulate STAT3 with a concomitant change in growth and metastatic capacity of ovarian cancer cells (Yang et al., 2014). Based on our results, anti-miR nanoliposome based therapeutic approaches warrant evaluation as an approach to improve outcomes for ovarian cancer patients. Importantly, such clinical trials are progressing at the MD Anderson Cancer Center.
Experimental Procedures
Patient Samples
After informed consent, serous epithelial ovarian cancer tissues were collected according to an Institutional Review Board approved protocol at the MD Anderson Cancer Center.
RNA Isolation and miR detection
Total RNA from cultured cells, with efficient recovery of small RNAs, was isolated using the mirVana miR Isolation Kit (Ambion, Austin, TX). Detection of mature miR was performed using cDNA generated with Qiagen miScript kit and subsequent qRT-PCR analysis with SYBR Green I (Qiagen, Valencia, CA) according to manufacturers’ instructions. Primers specific to mature miRNA-3p and miRNA-5p sequences were purchased from Qiagen. Primers to the U6 small nuclear RNA (RNU6B) from Qiagen were used as an internal normalization control.
miRNA Transfection
The miRIDIAN miRNA mimics from Dharmacon (Lafayette, CO) and antimiR from Ambion (Waltham, MA USA) were transfected using Lipofectamine RNAi Max from Invitrogen (Waltham, MA USA). antimiR-551b indicates anti-miRNAs targeting mature miRNA miR551b-3p in this study unless indicated otherwise. Mature Sequences of the miRNAs used in this study are follows:
miR551b-3p:GCGACCCAUACUUGGUUUCAG;
miR551b-5p:GAAAUCAAGCGUGGGUGAGACC;
control miR: UCACAACCUCCUAGAAAGAGUAGA; and
Mutated miR551b-3p: GCGUGGGAUACAACCUUUCAG
Analysis of public gene expression datasets
TCGA ovarian cancer SNP, mRNA data and TCGA breast cancer RPPA, and clinical data were downloaded from TCGA Data portal (http://tcga-data.nci.nih.gov/tcga/findArchives.htm). CNTools package from Bioconductor (http://www.bioconductor.org/) was used to process SNP data. Cox proportional hazard regression model was used for univariate survival analysis. Overall survival or Recurrence Free Survival (RFS) were used as endpoints. The cut-offs of high-expression and low-expression were optimized to achieve the lowest p value. Statistical analysis was performed using R Software.
Copy Number Analysis
The gene lists for human chromosome 3q26.1 to 3q26.3 were downloaded from the UCSC Genome Browser website (http://genome.ucsc.edu/cgi-bin/hgTables; assembly: GRCh37/hg19). DNA copy number alteration (CNA) data generated using the Agilent SurePrint G3 Human CGH Microarray Kit 1×1M platform, were downloaded from the cBio Portal (http://www.cbioportal.org/). CNA calls were made using GISTIC 2.0 as −2 for homozygous deletion; −1 for hemizygous deletion; 0 for no change; 1 for gain of copy number; and 2 for amplification (Mermel et al., 2011).
Promoter Hybridization and Pull-Down Assay
Promoter fragments were biotinylated using Purebiotech Kit (Middlesex, NJ), denatured, then hybridized with miR551b-3p or mutant miR551b-3p at 60°C for 2h and then incubated with 500 μg of nuclear lysate IGROV1 cell lysates for 6h. Samples were fixed in formaldehyde (1%) for 10 min, quenched with glycine (125 mM). Biotinylated promoter fragments were enriched using streptavidin-conjugated beads. Beads were washed with ice-cold NT2 buffer five times and incubated at 90°C for 5 min and elutes were immunoprecipitated using AGO1 antibody (WAKO, Richmond, VA). The beads were washed with ice-cold NT2 buffer five times and then boiled with 2x Laemmli loading buffer.
In situ hybridization and immunofluorescence staining
In situ hybridization was performed as described previously (Rupaimoole et al., 2016). Formalin-fixed, paraffin-embedded tissue sections were deparrafinized and rehydrated. Tissues were then digested with 15 μg/ml proteinase K for 20 min at room temperature and loaded onto Ventana Discovery Ultra for in situ hybridization. Tissue slides were incubated with double-DIG labeled mercury LNA miRNA probe (Exiqon, Woburn, MA, USA) for 2 h at 55 °C. H2O2 was used to inactivate endogenous peroxidases. Polyclonal anti-DIG antibody and horseradish peroxidase-conjugated secondary antibody (Ventana, Tucson, AZ) were incubated with the slide. Tyramine-conjugated fluorochrome (TSA) reaction was performed for 12 min. Slides were mounted with antifading ProLong Gold Solution (Life Technologies, Austin, TX).
Statistical analyses
Microsoft Excel (San Francisco, CA, USA) or GraphPad Prism (La Jolla, CA, USA) was used to analyze data. Statistical analysis was performed using Student’s t-test, two-way Anova or log-rank test as indicated in the figure legend. p<0.05 was considered statistically significant. Data are presented as mean±s.d. or mean±s.e. as indicated in figures. Cox proportional hazard regression model was used for univariate survival analysis. Statistical analysis was performed using R Software.
Supplementary Material
Highlights.
3q26.2 copy number alterations associate with miR551b-3p expression.
miR551b-3p interacts with the STAT3 promoter and activates STAT3 transcription.
miR551b-3p is an oncogenic microRNA that enhances tumor growth in vitro and in vivo.
Therapeutic delivery of anti-miR551b reduces tumor growth in vivo.
Acknowledgments
GBM is supported by NCI (2P50CA083639-11, 5P50CA058183-17, and 5R01CA123219-01), Stand up to Cancer/American Association of Cancer Research (SU2C-AACR-DT0209), Adelson Medical Research Foundation and Komen Promise Grant (KG081694). PCR is supported by Ovarian Cancer Research Fund and Marsha Rivkin Center for Ovarian Cancer Research. AKS is supported by NCI (P50CA083639, P50CA098258, U54CA151668, and UH2 TR000943). GAC is supported in part by the NIH/NCI grants 1UH2TR00943-01 and 1 R01 CA182905-01.
Footnotes
The authors disclose no potential conflicts of interest
AUTHORS CONTRIBUTIONS: GBM and PCR conceived the study, generated hypotheses and designed experiments. PCR designed and performed most of the experiments including tissue culture, animal experiments, immunofluorescence, and 3D morphogenesis. WL performed bioinformatics analysis. PCR, SP, AKS, and GBM designed the in vivo antimiR experiments. SP performed IHC experiments, assisted in animal experiments and in vivo delivery of antimiR. SY assisted in cloning and mutagenesis. YL, SY and DZ performed RPPA analysis. C-RA, GL-B and AKS developed antimiR in nanoliposomes for in vivo experiments. TM and PR performed computational analysis. GBM provided scientific direction, established collaborations, prepared the manuscript with PCR and allocated funding for the work.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abubaker K, Luwor RB, Escalona R, McNally O, Quinn MA, Thompson EW, Findlay JK, Ahmed N. Targeted Disruption of the JAK2/STAT3 Pathway in Combination with Systemic Administration of Paclitaxel Inhibits the Priming of Ovarian Cancer Stem Cells Leading to a Reduced Tumor Burden. Front Oncol. 2014;4:75. doi: 10.3389/fonc.2014.00075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambros V. microRNAs: tiny regulators with great potential. Cell. 2001;107:823–826. doi: 10.1016/s0092-8674(01)00616-x. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Bell D, Berchuck A, Birrer M, Chien J, Cramer DW, Dao F, Dhir R, DiSaia P, Gabra H, Glenn P, et al. Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474:609–615. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgos-Ojeda D, Rueda BR, Buckanovich RJ. Ovarian cancer stem cell markers: prognostic and therapeutic implications. Cancer Lett. 2012;322:1–7. doi: 10.1016/j.canlet.2012.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cernilogar FM, Onorati MC, Kothe GO, Burroughs AM, Parsi KM, Breiling A, Lo Sardo F, Saxena A, Miyoshi K, Siomi H, et al. Chromatin-associated RNA interference components contribute to transcriptional regulation in Drosophila. Nature. 2011;480:391–395. doi: 10.1038/nature10492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaluvally-Raghavan P, Zhang F, Pradeep S, Hamilton MP, Zhao X, Rupaimoole R, Moss T, Lu Y, Yu S, Pecot CV, et al. Copy Number Gain of hsa-miR-569 at 3q26.2 Leads to Loss of TP53INP1 and Aggressiveness of Epithelial Cancers. Cancer Cell. 2014;26:863–879. doi: 10.1016/j.ccell.2014.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eder AM, Sui X, Rosen DG, Nolden LK, Cheng KW, Lahad JP, Kango-Singh M, Lu KH, Warneke CL, Atkinson EN, et al. Atypical PKCiota contributes to poor prognosis through loss of apical-basal polarity and cyclin E overexpression in ovarian cancer. Proc Natl Acad Sci U S A. 2005;102:12519–12524. doi: 10.1073/pnas.0505641102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farh KK, Grimson A, Jan C, Lewis BP, Johnston WK, Lim LP, Burge CB, Bartel DP. The widespread impact of mammalian MicroRNAs on mRNA repression and evolution. Science. 2005;310:1817–1821. doi: 10.1126/science.1121158. [DOI] [PubMed] [Google Scholar]
- Hennessy BT, Lu Y, Gonzalez-Angulo AM, Carey MS, Myhre S, Ju Z, Davies MA, Liu W, Coombes K, Meric-Bernstam F, et al. A Technical Assessment of the Utility of Reverse Phase Protein Arrays for the Study of the Functional Proteome in Non-microdissected Human Breast Cancers. Clin Proteomics. 2010;6:129–151. doi: 10.1007/s12014-010-9055-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang V, Place RF, Portnoy V, Wang J, Qi Z, Jia Z, Yu A, Shuman M, Yu J, Li LC. Upregulation of Cyclin B1 by miRNA and its implications in cancer. Nucleic Acids Res. 2012;40:1695–1707. doi: 10.1093/nar/gkr934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang V, Qin Y, Wang J, Wang X, Place RF, Lin G, Lue TF, Li LC. RNAa is conserved in mammalian cells. PLoS One. 2010;5:e8848. doi: 10.1371/journal.pone.0008848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang V, Zheng J, Qi Z, Wang J, Place RF, Yu J, Li H, Li LC. Ago1 Interacts with RNA polymerase II and binds to the promoters of actively transcribed genes in human cancer cells. PLoS Genet. 2013;9:e1003821. doi: 10.1371/journal.pgen.1003821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janowski BA, Younger ST, Hardy DB, Ram R, Huffman KE, Corey DR. Activating gene expression in mammalian cells with promoter-targeted duplex RNAs. Nature chemical biology. 2007;3:166–173. doi: 10.1038/nchembio860. [DOI] [PubMed] [Google Scholar]
- Landen CN, Jr, Chavez-Reyes A, Bucana C, Schmandt R, Deavers MT, Lopez-Berestein G, Sood AK. Therapeutic EphA2 gene targeting in vivo using neutral liposomal small interfering RNA delivery. Cancer Res. 2005;65:6910–6918. doi: 10.1158/0008-5472.CAN-05-0530. [DOI] [PubMed] [Google Scholar]
- Levy DE, Darnell JE., Jr Stats: transcriptional control and biological impact. Nat Rev Mol Cell Biol. 2002;3:651–662. doi: 10.1038/nrm909. [DOI] [PubMed] [Google Scholar]
- Li LC, Okino ST, Zhao H, Pookot D, Place RF, Urakami S, Enokida H, Dahiya R. Small dsRNAs induce transcriptional activation in human cells. Proc Natl Acad Sci U S A. 2006;103:17337–17342. doi: 10.1073/pnas.0607015103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y, Zhang X, Xu X, Shen L, Yao Y, Yang Z, Liu P. STAT3 Decoy Oligodeoxynucleotides-Loaded Solid Lipid Nanoparticles Induce Cell Death and Inhibit Invasion in Ovarian Cancer Cells. PLoS One. 2015;10:e0124924. doi: 10.1371/journal.pone.0124924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medema JP. Cancer stem cells: the challenges ahead. Nat Cell Biol. 2013;15:338–344. doi: 10.1038/ncb2717. [DOI] [PubMed] [Google Scholar]
- Mermel CH, Schumacher SE, Hill B, Meyerson ML, Beroukhim R, Getz G. GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome Biol. 2011;12:R41. doi: 10.1186/gb-2011-12-4-r41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills GB, May C, Hill M, Campbell S, Shaw P, Marks A. Ascitic fluid from human ovarian cancer patients contains growth factors necessary for intraperitoneal growth of human ovarian adenocarcinoma cells. J Clin Invest. 1990;86:851–855. doi: 10.1172/JCI114784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills GB, May C, McGill M, Roifman CM, Mellors A. A putative new growth factor in ascitic fluid from ovarian cancer patients: identification, characterization, and mechanism of action. Cancer Res. 1988;48:1066–1071. [PubMed] [Google Scholar]
- Nanjundan M, Nakayama Y, Cheng KW, Lahad J, Liu J, Lu K, Kuo WL, Smith-McCune K, Fishman D, Gray JW, et al. Amplification of MDS1/EVI1 and EVI1, located in the 3q26.2 amplicon, is associated with favorable patient prognosis in ovarian cancer. Cancer Res. 2007;67:3074–3084. doi: 10.1158/0008-5472.CAN-06-2366. [DOI] [PubMed] [Google Scholar]
- Piriyapongsa J, Bootchai C, Ngamphiw C, Tongsima S. microPIR: an integrated database of microRNA target sites within human promoter sequences. PLoS One. 2012;7:e33888. doi: 10.1371/journal.pone.0033888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Place RF, Li LC, Pookot D, Noonan EJ, Dahiya R. MicroRNA-373 induces expression of genes with complementary promoter sequences. Proc Natl Acad Sci U S A. 2008;105:1608–1613. doi: 10.1073/pnas.0707594105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portnoy V, Huang V, Place RF, Li LC. Small RNA and transcriptional upregulation. Wiley Interdiscip Rev RNA. 2011;2:748–760. doi: 10.1002/wrna.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosen DG, Mercado-Uribe I, Yang G, Bast RC, Jr, Amin HM, Lai R, Liu J. The role of constitutively active signal transducer and activator of transcription 3 in ovarian tumorigenesis and prognosis. Cancer. 2006;107:2730–2740. doi: 10.1002/cncr.22293. [DOI] [PubMed] [Google Scholar]
- Rupaimoole R, Ivan C, Yang D, Gharpure KM, Wu SY, Pecot CV, Previs RA, Nagaraja AS, Armaiz-Pena GN, McGuire M, et al. Hypoxia-upregulated microRNA-630 targets Dicer, leading to increased tumor progression. Oncogene. 2016 doi: 10.1038/onc.2015.492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shayesteh L, Lu Y, Kuo WL, Baldocchi R, Godfrey T, Collins C, Pinkel D, Powell B, Mills GB, Gray JW. PIK3CA is implicated as an oncogene in ovarian cancer. Nat Genet. 1999;21:99–102. doi: 10.1038/5042. [DOI] [PubMed] [Google Scholar]
- Tothill RW, Tinker AV, George J, Brown R, Fox SB, Lade S, Johnson DS, Trivett MK, Etemadmoghadam D, Locandro B, et al. Novel molecular subtypes of serous and endometrioid ovarian cancer linked to clinical outcome. Clin Cancer Res. 2008;14:5198–5208. doi: 10.1158/1078-0432.CCR-08-0196. [DOI] [PubMed] [Google Scholar]
- Wen W, Wu J, Liu L, Tian Y, Buettner R, Hsieh MY, Horne D, Dellinger TH, Han ES, Jove R, et al. Synergistic anti-tumor effect of combined inhibition of EGFR and JAK/STAT3 pathways in human ovarian cancer. Mol Cancer. 2015;14:100. doi: 10.1186/s12943-015-0366-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Moss T, Mangala LS, Marini J, Zhao H, Wahlig S, Armaiz-Pena G, Jiang D, Achreja A, Win J, et al. Metabolic shifts toward glutamine regulate tumor growth, invasion and bioenergetics in ovarian cancer. Mol Syst Biol. 2014;10:728. doi: 10.1002/msb.20134892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Fan M, Zhang X, Huang F, Wu K, Zhang J, Liu J, Huang Z, Luo H, Tao L, et al. Cellular microRNAs up-regulate transcription via interaction with promoter TATA-box motifs. RNA. 2014;20:1878–1889. doi: 10.1261/rna.045633.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.