Abstract
Chromosome replication, gene expression and chromatin assembly all occur on the same template, necessitating a tight spatial and temporal coordination to maintain genomic stability. The distribution of replication initiation events is responsive to local and global changes in chromatin structure and is affected by transcriptional activity. Concomitantly, replication origin sequences, which determine the locations of replication initiation events, can affect chromatin structure and modulate transcriptional efficiency. The flexibility observed in the replication initiation landscape might help achieve complete and accurate genome duplication while coordinating the DNA replication program with transcription and other nuclear processes in a cell-type specific manner. This review discusses the relationships among replication origin distribution, local and global chromatin structures and concomitant nuclear metabolic processes.
Introduction
Genome duplication involves creating two identical copies of all DNA sequences along with exact replicas of their chromatin modifications to insure proper nuclear packaging in the next generation. The locations of replication initiation sites as well as local and global chromatin interaction domains, determined at the onset of interphase, inscribe the spatial and temporal replication program. Replication initiation sites are established by a process called replication licensing, during which pre-replication complexes (pre-RCs) are recruited to distinct chromatin sites that can potentially initiate replication (replication origins - box 1 and figure 1). As cells prepare to synthesize DNA, additional components are added to the pre-RCs to form pre-initiation complexes (pre-ICs). Those structures, in turn, can be activated by a series of signaling events to initiate replication. As many of these events were recently summarized in several excellent reviews [1–6], the discussion below will focus on the relationships among replication origin distribution, local and global chromatin structures and concomitant nuclear metabolic processes.
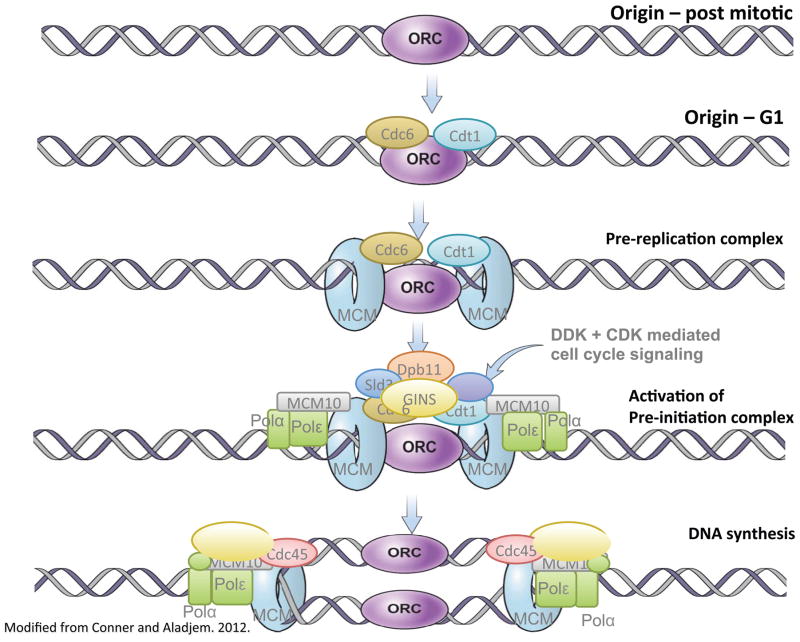
BOX 1. Initiation of DNA replication at replication origins.
Origin recognition complexes (ORC) bind to potential initiation sites, which in turn allows licensing factors Cdc6 and Cdt1 that facilitate recruitment of the inactive form of the replicative helicase (MCM2-7) to create the pre-replication complex (pre-RC). Following phosphorylation by Cdc7/Dbf4-dependent kinase (DDK) and cyclin dependent kinases (CDK), pre-RCs form pre initiation complexes (Pre-IC s) containing the active helicase composed of Cdc45, MCMs, and GINS (CMG) [13,87]. CDKs interact with pre-ICs to initiate DNA replication concomitant with degradation of released pre-RC components [5,87]. As replication forks progress, pre-replication complexes from passively replicated origins dissociate and degrade. The locations of pre-RC formation are determined during G1 largely based on replicator sequences, and the timing of initiation largely reflect chromatin structure.
CDK and DDK levels fluctuate throughout the cell cycle to activate and deactivate replication machinery. Low CDK activity during the late M-early G1-phase is required for pre-RC formation [87,88]. CDK and DDK levels increase during G1 because members of the pre-IC require phosphorylation to load onto the replication machinery. Phosphorylation of residues on the MCM2-7 complex activates helicase activity [5,89,90], which allows DNA polymerases and CMG complex to bind and activate origins [5,6,91]. CDKs prevent reformation of pre-RC and re-replication at already used origins by phosphorylating MCM2-7, ORC1, Cdt1, and Cdc6 [5]. Re-replication is also prevented by Cdt1 inhibition by Geminin and ubiquitin-directed proteolysis of Cdt1, ORC1, and Cdc6 [5]. Beyond interacting directly with proteins belonging to the Pre-RC and Pre-IC components, CDKs and DDKs interact with transacting factors that regulate recruitment of replication machinery [2].
Figure 1.
Cellular signaling cascades interact with pre-replication complexes to initiate DNA replication when cells have accumulated sufficient resources and the extracellular environment is favorable to growth [1]. Within each cell type, the order of origin activation reflects gene expression patterns and provides coping mechanisms to address replication stress [7]. The spatial and temporal distribution of replication initiation events respond to changes in chromatin structure to coordinate transcription and chromosome condensation with DNA synthesis [5,8]. Conversely, replication origin sequences can affect chromatin structure and modulate transcriptional efficiency [9]. Irregularities in the replication timing programs are associated with aberrant gene expression and structural chromosomal variations, underlying the importance of distribution and timing of replication origins to genomic integrity [10].
Eukaryotic genomes exhibit an excess of potential replication origins; only 10–20% of potential origins initiate replication at any given somatic cell cycle [11]. Combined data from single fiber and whole-genome analyses of DNA replication [5] indicate that in many loci, replication initiates alternately within clusters of adjacent origins so that each cell in a population uses slightly different combinations of replication origins at any given cell cycle. The use of alternate locations for replication initiation (also known as “origin choice”) might help coordinate the DNA replication program with transcription and other nuclear processes in a cell-type specific manner. Flexible initiation of DNA replication during development and differentiation might affect the transcription program by altering chromatin condensation, and thus transcription factor accessibility, of select genomic regions. Furthermore, excess origins and pre-RCs are necessary for genomic integrity, as they can be utilized to complete replication when approaching replication forks collapse or stall, leaving un-replicated chromatin [12–14].
Although the basic biochemical events that lead to initiation of DNA replication have been described, outstanding questions remain about origin location, the genomic distribution of replication origins, the determinants that activate origins at specific times, and the impact of origin distribution on DNA stability and integrity. Origin flexibility and the apparent excess of potential replication origins suggest that the role of origins in maintaining genome integrity does not reflect a mechanistic requirement for genome duplication; rather, the particular distribution of replication origins and their sequential activation might help coordinate replication and transcription on their shared chromatin template. Origin activation dynamics, therefore, might affect or and be affected by local and global chromatin structure.
Distribution and sequence determinants of replication origins
Large-scale topological domains, which are several hundred kilobases to megabases long sequences characterized by a common structural feature, exhibit high concordance with replication timing domains (contiguous regions exhibiting similar replication time during S-phase – Fig. 2) [3,15–20]. High-resolution whole-genome analyses reveal that replication timing domains often reflect chromatin modifications and the topological organization of chromatin [21,22] and that cis-acting genetic elements determine, at least in part, the location of megabase-scale replication timing domains [23]. Most replication timing domains exhibit multiple initiation sites [24], reflecting concomitant replication from several replication origins [5,25].
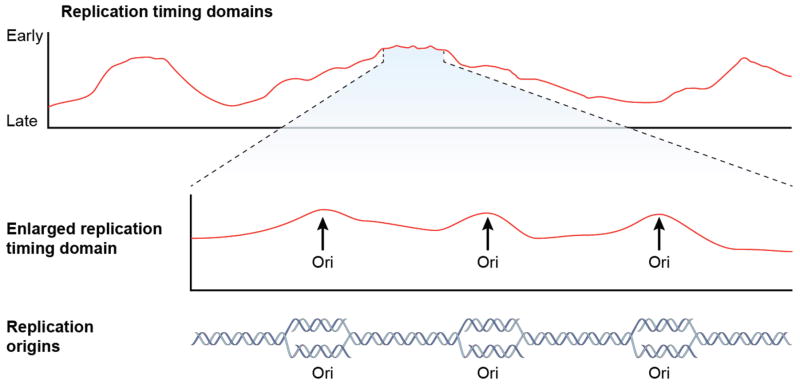
Figure 2.
Replication timing domains are structurally determined regions that contain origins that initiate replication at similar times. Mapping of replication timing events in a population of cells reveals large, megabase-size domains with similar time of replication (top panel). High-resolution mapping of replication time reveals that the replication timing domain peaks contain short, low-amplitude subpeaks (“ripples” seem in the middle panel)[24]. Nascent strand abundance analyses reveal that those subpeaks correspond to replication origins (bottom panel) [24].
Genetic and structural features associate with replication initiation sites on mammalian chromosomes [2,5,7,26]. Although mammalian replication origins do not share a single “consensus” sequence, a large fraction of origins is located adjacent to transcriptional start sites (TSSs), regions of DNase hypersensitivity [11,24,27–30] and G-rich sequences, including CpG islands and sequences that can potentially form G-quadruplex structures [11,24,27,30]. It is unclear whether these colocalizations represent causal relationships. For example, placement of origins near transcription start sites might affect transcription levels and prevent transcription-replication collisions or might reflect a consequence of the enhanced chromatin accessibility near transcriptionally active regions. Similarly, G-quadruplex forming sequences often associate with promoters, euchromatin, and CpG islands [31] and their locations near replication origins might either imply an effect on replication or reflect negative selection against both G-quadruplexes and origins in gene bodies [32]. G-quadruplexes’ ability to interfere with replication fork progression [32] and the requirement for specialized helicases for their unwinding [33,34] might provide an additional selective factor favoring origin-proximal G-quadruplexes. An analysis of allele-specific origins suggests that the genetic determinants of origin activity are base composition asymmetry and high GC content rather than the ability to form quadruplexes [25,35].
Transcriptional regulatory sequences at replication origins
Replication frequently initiates near transcribed genes, but high levels of transcription can be inhibitory to replication initiation [28,29]. This relationship might reflect a competition between replication initiation complexes and transcription initiation complexes on the same template. Conversely, replication initiation events occur more frequently at unmethylated CpGs than at methylated CpG tracks, suggesting that heterochromatin packaging also lowers the frequency of replication initiation events and consistent with the preferential association of replication origins with active chromatin modifications [11,28,29,36,37].
Distal DNA sequences, which often affect transcriptional activity, influence replication origin activity by establishing long-distance interactions or responding to developmental cues [7,38,39]. Such interactions could be mediated by chromatin remodeling factors and transcriptional activators that bind enhancers and locus control regions [5,7]. Long-distance interactions can also be mediated by long non-coding RNAs (lncRNAs) such as Xist and HOTAIR, which guide histone and chromatin remodeling proteins to specific DNA sequences and facilitate chromatin interactions [5,40]. lncRNAs can stabilize ORC to origins in viruses by creating G-quadruplexes [5,40] and can regulate pre-RC components and DNA polymerase to promote cell proliferation [41].
DNA is packaged into the nucleus in a cell-type specific manner that underlies the plasticity required for multiple differentiation states to arise from the same genomic information. Replicating chromatin accurately, therefore, needs to address two challenges. First, replication should duplicate DNA sequence, but also create an exact copy of the chromatin states associated with each locus each cell cycle [42]. Second, chromatin must decondense ahead of the replication fork to allow the double helix to unwind; compact chromatin packaging delineates cell-type specific chromatin that might form a barrier to elongating replication forks. A tight coordination of the DNA replication machinery, chromatin remodeling complexes and chromatin modifiers can address these two challenges. In addition, some chromatin-associated proteins establish and maintain replication timing domains and nuclear structure [8].
Origin selection and replication timing
As originally proposed by the replicon model [43], DNA sequences (replicators), including those found at replication origins, can dictate initiation of DNA replication. Replicators can affect particular histone modifications when moved to ectopic sites [9,44–46], providing one avenue for re-establishing chromatin structure following duplication. For example, Ubiquitous Chromatin Opening Elements (UCOEs) [47] exhibit a high prevalence of replication origins [9]. UCOEs maintain an open chromatin structure that protects transcriptional activity despite local repressive chromatin modifications [48]. By serving to recruit DNA sequence-specific transcription factors that can in turn recruit chromatin-modifying complexes, a group of replication origins might create a context permissive for both transcriptional activity and replication initiation [7,49,50].
The replication timing program is determined anew each cell cycle [3], suggesting that early chromatin packaging dictates a structural and genetic landscape that coordinates transcription and DNA replication, allowing for tight control between the transcription and DNA synthesis. Conversely, DNA sequences that dictate replication initiation rates can influence chromatin structure, which in turn regulates transcriptional efficiency [9,46]. The timing of initiation during the S-phase of the cell cycle correlates with gene expression levels and chromatin structure, with transcribed “open” chromatin replicating early and heterochromatin replicating later. This association could reflect a tightly regulated replication initiation program whereby replication of gene-encoding euchromatin occurs early in S-phase to allow essential proteins to be synthesized without interference from pre-RCs and the replication machinery. This association could also be a passive consequence of the higher accessibility of transcribed regions to DNA-protein interactions, including those that activate replication origins [16,17]. Although the severe effects of changes in replication timing support a critical role for replication timing regulation in maintaining genomic stability [3,4,5,7], a recent mathematical model of replication kinetics accurately predicts origin firing with only two factors: an “Initiation probability landscape” corresponding to replication origin activity, and the availability of a single rate-limiting activator [18]. This study implies that replication initiation sites and replication timing might not be regulated independently of each other and instead both might reflect a single set of determinants.
Histone modifications that impact origin licensing and activation
Certain histone modifications correlate with degrees of chromatin compaction and facilitate recruitment of transcription factors and pre-RC complexes [5]. Early replicating origins associate with euchromatic histone modifications (H3K4me1/2/3, H3K9ac, H3K18ac, H3K36me3, and H3K29ac,) whereas late replication is associated with repressive chromatin modifications like H3 and H4 hypoacetylation, H3K9me1/3, and H3K27me3 [2,36,51]. Specific histone modifications have distinct impacts on origin activity; histone acetyl transferase HBO1 facilitates initiation of DNA replication [52]. In plants, methylation of histone H3 on lysine 27 is required to induce re-replication [53] while in mammals, DOT1L catalyzes the methylation of H3K79, which prevents re-replication [37].
HBO1, ORCA, and PR-Set7 modify histones near replication origins, thus controlling the activity of local replication domains as elaborated below. Histone acetyltransferase HBO1, which is enhanced at H3K4me3, interacts with ORC1 and Cdt1 during G1 to acetylate H4K5 and H4K12 near origin replications [32,54]. Acetylation decondenses the chromatin, influencing origin activity and promoting early replication ([54] and references therein). H4K20me1/2/3 reduces acetylation by HBO1 and pre-RC formation [52].
PR-Set7, the only known 1mono-methylator of H4K20, regulates origin licensing and genome stability by mono-methylating H4K20 throughout G1 and S [32]. Thus, PR-Set7 plays an essential role in regulating replication in addition to mitosis, DNA damage responses, transcription, and formation of heterochromatin [32]. H4K20me1 serves as a binding domain for other methyltransferases (ie Suv4) [32,55]. H4K20me1/2/3 has also been shown to reduce pre-RC formation by decreasing H4 acetylation levels [52]. PR-Set7 may also influence pre-RC assembly by depleting binding domain for ORCA. H4K20me1 by PR-Set7 is needed to form H4K20me2/3, and cells with depleted H4K20me2/3 have depleted ORCA and ORC1 binding [32].
ORCA/LRWD1 regulates late replication by promoting chromatin compaction, stabilizing ORC in regions with low expression [56,57]. Depletion of ORCA causes disorganization of chromatin in post-G1 cells and a reduction of replicating origins [57]. ORCA typically associates with ORC1 and stabilizes ORC binding to heterochromatin with repressive modifications H3K9me3, H3K27me3, and H4K20me3 [54–59]. Like ORCA, HP1 associates with both H3K9 methylations and stabilizes ORC by binding ORC2 and ORC3 [56].
It is uncertain if these two proteins act in concert or separately [56]. Beyond stabilizing ORC in heterochromatin, ORCA encourages further chromatin condensation by recruiting methyltransferases at already repressed regions [57]. For example, ORCA recruits lysine methyltransferases (KMTs) (ie Suv2-30H1/2) to regions that already have repressive lysine modifications, and promotes further repressive histone modifications and cohesion recruitment [57,60].
Nuclear structures and trans-acting factors regulate origin activity
Rif1 (Rap1-interacting-factor-1) is a telomere-binding protein that promotes mid to late-S-Phase replication by regulating recruitment of pre-IC components to telomeric and sub-telomeric regions [61–64]. Rif1 acts downstream of Taz1, which is another telomere-binding protein found to control half of the chromosomal late origins by preventing activation by DDK and promotes mid to late-S-phase replication [64]. Both the Taz1-dependent and Taz1-independent Rif1 pathways structurally interfere with DDK’s ability to phosphorylate chromatin-bound Mcm2-7, which is necessary for loading of Cdc7, Cdc45, Sld3 [60,64,65]. Rif1 levels increase throughout G1 and regulate initiation at mid-S phase origins by facilitating chromatin loop formation [18,62,63,66], which delays early S-phase firing by physically restricting DDK access to phosphorylate MCM [63]. Additionally, Rif1 recruits protein phosphatase 1 (PP1) to chromatin to counteract MCM phosphorylation by DDK [66,67].
Trans-acting modifiers can also promote early replication by helping to recruit replication factors. Fkh1/2 recruits early replication factors to chromatin by encouraging inter-chromosomal interactions in budding yeast [2,68]. Fkh1/2 activates some origins, while it represses others [4]. Similarly, in Drosophila, HP1 can modulate replication initiation patterns. On one hand, HP1 facilitates inactivation of late origins by contributing to chromatin compaction near repressive H3K9me3 modifications [47]. On the other hand, like Fkh1/2, HP1 can promote early replication when bound to euchromatic regions [5].
Replication origin activation also involves interactions with structural features intermediate filaments (in particular, lamins), matrix and scaffold attachment sites (MARs and SARs, respectively) and Stabilizing Anti Repressor elements (STARs) [2,8]. MARs are known regulators of CCTC-binding factor (CTCF), a transcriptional repressor that acts in concert with the ring-shaped cohesins to anchor DNA to the nuclear matrix [8] and creating chromatin loops. Cohesin interacts with Pre-RC components (including ORC and Mcm2-7) and DDKs to control the number of active origins and inter-origin distance [8]. These data, suggesting that structural nuclear components might act as determinants of replication initiation events, support the hypothesis that spatial organization plays a regulatory role in replication.
The effects of transcription factors and modulators of the DNA damage response
The frequency and location of replication initiation events can be affected by changes in the cellular gene expression program. Cellular differentiation, associated with massive changes in gene expression, is often accompanied by altered replication initiation patterns [38,39] and reprogramming of replication timing [21]. Activation by distinct transcription factors can also facilitate pronounced changes in the replication program. For example, the c-Myc proto-oncogene (Myc) is a well-characterized transcription factor that also has non-transcriptional roles in cell-cycle progression [69,70]. In addition to transcribing genes encoding cyclin-D2 and CDK4 [71,72], Myc activates Cdt1, a protein directly involved in origin licensing [73]. Dysregulation of Myc, and the resulting aberrant Cdt1 levels, stimulate re-replication [74]. Overexpression of Myc in Xenopus extract induces premature origin firing, causes asymmetrical fork progression, and induces DNA damage [70,75]. It has been suggested that Myc interacts with the pre-RC prior to Cdc45-MCM2-7-GINS (CMG) loading at the G1-S phase transition [75].
In addition to regulating the response to DNA damage, Checkpoint kinase 1 (Chk1) plays a role in origin licensing through repression of replication initiation and fork progression, and by preventing fork collapse [4,76]. In the absence of genotoxic stress, Chk1 negatively regulates DNA synthesis by inhibiting replication at forks adjacent to activated origins of replication by binding to and phosphorylating Treslin (homologous to yeast Sld3), thus inhibiting Cdc45 loading [76–80]. However, under low stress, Chk1 stimulates origins neighboring the stalled forks to initiate [12]. It is thought that during periods of stress, Chk1 inhibits distant replication factories and redirects replication machinery to regions that are already replicating [4]. Chk1 degrades Cdc25 that decreases the amount of systematic Cdk2. This, in turn, reduces the amount of active Cdc45 available to form the CMG-helicase complex required to activate new origin [76,81].
An increased frequency of replication initiation events (also known as activation of “dormant origins”) is also observed in response to events that affect the rate of replication fork progression. For example, nucleotide pool levels [82] and exposure to various agents ranging from ultraviolet radiation [83] to histone deacetylase inhibitors [84] increase replication initiation levels in cancer cells (reviewed in [12]). In the absence of exogeneous DNA damage, deficiencies in enzymes involved in homologous recombination (HR) [85] and in Mus81 endonuclease activities [86] also slow down replication and increase initiation. The observed increased initiation under conditions that slow DNA replication might reflect a global compensatory mechanism that couples DNA polymerase progression with the activation of nascent replication forks. Alternatively, the slower overall replication rate observed in population-based studies might reflect severe replication stalling in a small group of loci (e.g. fragile sites) that are particularly prone to agents that perturb replication and might require interactions with the DNA repair machinery to facilitate replication fork progression.
Concluding remarks
The remarkable flexibility in spatial organization of replication origins is an important chromatin feature. Since DNA synthesis proceeds on a chromatin template while the cell continues its normal maintenance, replication requires coordination with other nuclear processes, particularly transcription. Furthermore, the density of replication initiation sites may influence DNA structural integrity and a cell’s ability to respond to stress. While chromatin structure and organization are strong determinants of the replication landscape (for a summary, see Table 1), structural changes associated with origin activation and the factors that help establish the replication program might also play a role in establishing global and local nuclear architecture. Consequently, understanding the mechanisms controlling the spatial and temporal programs of replication as well as genetic and epigenetic factors associated with origin licensing and activation will help connect seemingly distinct cellular pathways.
Table 1.
Chromatin modulators that affect spatial and temporal replication profiles.
| Early Replication | Late Replication | |
|---|---|---|
| Sequence Association | TSS High GC-content CpG islands Base composition asymmetry* |
Base composition asymmetry* |
| Structural Association | G4 UCOEs |
MARs, SARs, STARs cohesin |
| Protein Association | HBO1 Fkh1/2* Taz1* |
ORCA, HP1 Rif1 Fkh1/2* Taz1* |
| Histone Modifications | H3K4me1/2/3* H3K9ac H3K18ac H3K36me3 H3K27ac |
H3 and H4 - hypoacetylation H3K9me1/2/3 H3K27me3 H3K4me3 |
implied in both early and late replication
Acknowledgments
We thank Drs. Haiqing Fu and Christophe Redon for critical reading of the review. We thank many colleagues at the NCI Developmental Therapeutics Branch for helpful comments and apologize to our colleagues whose primary work could not be cited due to lack of space. This work was funded by the intramural program of the CCR, National Cancer Institute, National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Masai H, Matsumoto S, You Z, Yoshizawa-Sugata N, Oda M. Eukaryotic chromosome DNA replication: where, when, and how? Annu Rev Biochem. 2010;79:89–130. doi: 10.1146/annurev.biochem.052308.103205. [DOI] [PubMed] [Google Scholar]
- 2.Mechali M, Yoshida K, Coulombe P, Pasero P. Genetic and epigenetic determinants of DNA replication origins, position and activation. Curr Opin Genet Dev. 2013;23:124–131. doi: 10.1016/j.gde.2013.02.010. [DOI] [PubMed] [Google Scholar]
- 3.Rhind N, Gilbert DM. DNA Replication Timing. Cold Spring Harb Perspect Med. 2013;3:1–26. doi: 10.1101/cshperspect.a010132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Musialek MW, Rybaczek D. Behavior of replication origins in Eukaryota -spatio-temporal dynamics of licensing and firing. Cell Cycle. 2015;14:2251–2264. doi: 10.1080/15384101.2015.1056421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fragkos M, Ganier O, Coulombe P, Mechali M. DNA replication origin activation in space and time. Nat Rev Mol Cell Biol. 2015;16:360–374. doi: 10.1038/nrm4002. [DOI] [PubMed] [Google Scholar]
- 6.Remus D, Diffley JF. Eukaryotic DNA replication control: lock and load, then fire. Curr Opin Cell Biol. 2009;21:771–777. doi: 10.1016/j.ceb.2009.08.002. [DOI] [PubMed] [Google Scholar]
- 7.Aladjem MI. Replication in context: dynamic regulation of DNA replication patterns in metazoans. Nat Rev Genet. 2007;8:588–600. doi: 10.1038/nrg2143. [DOI] [PubMed] [Google Scholar]
- 8.Smith OK, Aladjem MI. Chromatin structure and replication origins: determinants of chromosome replication and nuclear organization. J Mol Biol. 2014;426:3330–3341. doi: 10.1016/j.jmb.2014.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Conner AL, Aladjem MI. The chromatin backdrop of DNA replication: lessons from genetics and genome-scale analyses. Biochim Biophys Acta. 2012;1819:794–801. doi: 10.1016/j.bbagrm.2012.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Donley N, Thayer MJ. DNA replication timing, genome stability and cancer: late and/or delayed DNA replication timing is associated with increased genomic instability. Semin Cancer Biol. 2013;23:80–89. doi: 10.1016/j.semcancer.2013.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cayrou C, Coulombe P, Vigneron A, Stanojcic S, Ganier O, Peiffer I, Rivals E, Puy A, Laurent-Chabalier S, Desprat R, et al. Genome-scale analysis of metazoan replication origins reveals their organization in specific but flexible sites defined by conserved features. Genome Res. 2011;21:1438–1449. doi: 10.1101/gr.121830.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Blow JJ, Ge XQ, Jackson DA. How dormant origins promote complete genome replication. Trends Biochem Sci. 2011;36:405–414. doi: 10.1016/j.tibs.2011.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.DePamphilis ML, Blow JJ, Ghosh S, Saha T, Noguchi K, Vassilev A. Regulating the licensing of DNA replication origins in metazoa. Curr Opin Cell Biol. 2006;18:231–239. doi: 10.1016/j.ceb.2006.04.001. [DOI] [PubMed] [Google Scholar]
- 14.Kawabata T, Luebben SW, Yamaguchi S, Ilves I, Matise I, Buske T, Botchan MR, Shima N. Stalled fork rescue via dormant replication origins in unchallenged S phase promotes proper chromosome segregation and tumor suppression. Mol Cell. 2011;41:543–553. doi: 10.1016/j.molcel.2011.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hiratani I, Ryba T, Itoh M, Rathjen J, Kulik M, Papp B, Fussner E, Bazett-Jones DP, Plath K, Dalton S, et al. Genome-wide dynamics of replication timing revealed by in vitro models of mouse embryogenesis. Genome Res. 2010;20:155–169. doi: 10.1101/gr.099796.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16*.Moindrot B, Audit B, Klous P, Baker A, Thermes C, de Laat W, Bouvet P, Mongelard F, Arneodo A. 3D chromatin conformation correlates with replication timing and is conserved in resting cells. Nucleic Acids Res. 2012;40:9470–9481. doi: 10.1093/nar/gks736. This paper shows that chromatin structure correlates with replication timing and identifies specific chromatin contacts that affect the shape of the replication domain. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yaffe E, Farkash-Amar S, Polten A, Yakhini Z, Tanay A, Simon I. Comparative analysis of DNA replication timing reveals conserved large-scale chromosomal architecture. PLoS Genet. 2010;6:e1001011. doi: 10.1371/journal.pgen.1001011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gindin Y, Valenzuela MS, Aladjem MI, Meltzer PS, Bilke S. A chromatin structure-based model accurately predicts DNA replication timing in human cells. Mol Syst Biol. 2014;10:722. doi: 10.1002/msb.134859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, Amit I, Lajoie BR, Sabo PJ, Dorschner MO, et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326:289–293. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rao SS, Huntley MH, Durand NC, Stamenova EK, Bochkov ID, Robinson JT, Sanborn AL, Machol I, Omer AD, Lander ES, et al. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell. 2014;159:1665–1680. doi: 10.1016/j.cell.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dileep V, Ay F, Sima J, Vera DL, Noble WS, Gilbert DM. Topologically associating domains and their long-range contacts are established during early G1 coincident with the establishment of the replication-timing program. Genome Res. 2015;25:1104–1113. doi: 10.1101/gr.183699.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22**.Pope BD, Ryba T, Dileep V, Yue F, Wu W, Denas O, Vera DL, Wang Y, Hansen RS, Canfield TK, et al. Topologically associating domains are stable units of replication-timing regulation. Nature. 2014;515:402–405. doi: 10.1038/nature13986. This paper reports a strong correlation between the boundaries of replication timing domains and the boundaries of chromatin domains in which intra-domain chromatin interactions are stronger than inter-domain interactions (topologically associating domains). This strong correlation was observed across cell types and differentiation stages, implying that topologically associated domains represent stable nuclear organization units that play an important role in establishing replication timing. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23**.Koren A, Handsaker RE, Kamitaki N, Karlic R, Ghosh S, Polak P, Eggan K, McCarroll SA. Genetic variation in human DNA replication timing. Cell. 2014;159:1015–1026. doi: 10.1016/j.cell.2014.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24**.Mukhopadhyay R, Lajugie J, Fourel N, Selzer A, Schizas M, Bartholdy B, Mar J, Lin CM, Martin MM, Ryan M, et al. Allele-specific genome-wide profiling in human primary erythroblasts reveal replication program organization. PLoS Genet. 2014;10:e1004319. doi: 10.1371/journal.pgen.1004319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25**.Bartholdy B, Mukhopadhyay R, Lajugie J, Aladjem MI, Bouhassira EE. Allele-specific analysis of DNA replication origins in mammalian cells. Nat Commun. 2015;6:7051. doi: 10.1038/ncomms8051. These three papers identify allele-specific changes in spatial and temporal replication program and identified genetic variants that affect replication timing and replication origin activity. Replication asynchrony reflected structural variants and imprinting rather than changes in individual replication initiation sites. Allele-specific replication origins in phased genomes, annotated for maternal and paternal sequences, allowed the identification of distinct genetic requirements for replication origin activity. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Urban JM, Foulk MS, Casella C, Gerbi SA. The hunt for origins of DNA replication in multicellular eukaryotes. F1000Prime Rep. 2015;7:30. doi: 10.12703/P7-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Besnard E, Babled A, Lapasset L, Milhavet O, Parrinello H, Dantec C, Marin JM, Lemaitre JM. Unraveling cell type-specific and reprogrammable human replication origin signatures associated with G-quadruplex consensus motifs. Nat Struct Mol Biol. 2012;19:837–844. doi: 10.1038/nsmb.2339. [DOI] [PubMed] [Google Scholar]
- 28.Martin MM, Ryan M, Kim R, Zakas AL, Fu H, Lin CM, Reinhold WC, Davis SR, Bilke S, Liu H, et al. Genome-wide depletion of replication initiation events in highly transcribed regions. Genome Res. 2011;21:1822–1832. doi: 10.1101/gr.124644.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sequeira-Mendes J, Diaz-Uriarte R, Apedaile A, Huntley D, Brockdorff N, Gomez M. Transcription initiation activity sets replication origin efficiency in mammalian cells. PLoS Genet. 2009;5:e1000446. doi: 10.1371/journal.pgen.1000446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Foulk MS, Urban JM, Casella C, Gerbi SA. Characterizing and controlling intrinsic biases of lambda exonuclease in nascent strand sequencing reveals phasing between nucleosomes and G-quadruplex motifs around a subset of human replication origins. Genome Res. 2015;25:725–735. doi: 10.1101/gr.183848.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huppert JL, Balasubramanian S. Prevalence of quadruplexes in the human genome. Nucleic Acids Res. 2005;33:2908–2916. doi: 10.1093/nar/gki609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sherstyuk VV, Shevchenko AI, Zakian SM. Epigenetic landscape for initiation of DNA replication. Chromosoma. 2014;123:183–199. doi: 10.1007/s00412-013-0448-3. [DOI] [PubMed] [Google Scholar]
- 33.Wickramasinghe CM, Arzouk H, Frey A, Maiter A, Sale JE. Contributions of the specialised DNA polymerases to replication of structured DNA. DNA Repair (Amst) 2015;29:83–90. doi: 10.1016/j.dnarep.2015.01.004. [DOI] [PubMed] [Google Scholar]
- 34.Sabouri N, Capra JA, Zakian VA. The essential Schizosaccharomyces pombe Pfh1 DNA helicase promotes fork movement past G-quadruplex motifs to prevent DNA damage. BMC Biol. 2014;12:101. doi: 10.1186/s12915-014-0101-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hyrien O, Rappailles A, Guilbaud G, Baker A, Chen CL, Goldar A, Petryk N, Kahli M, Ma E, d’Aubenton-Carafa Y, et al. From simple bacterial and archaeal replicons to replication N/U-domains. J Mol Biol. 2013;425:4673–4689. doi: 10.1016/j.jmb.2013.09.021. [DOI] [PubMed] [Google Scholar]
- 36.Picard F, Cadoret JC, Audit B, Arneodo A, Alberti A, Battail C, Duret L, Prioleau MN. The spatiotemporal program of DNA replication is associated with specific combinations of chromatin marks in human cells. PLoS Genet. 2014;10:e1004282. doi: 10.1371/journal.pgen.1004282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37*.Fu H, Maunakea AK, Martin MM, Huang L, Zhang Y, Ryan M, Kim R, Lin CM, Zhao K, Aladjem MI. Methylation of histone H3 on lysine 79 associates with a group of replication origins and helps limit DNA replication once per cell cycle. PLoS Genet. 2013;9:e1003542. doi: 10.1371/journal.pgen.1003542. This paper reports that dimethylation of H3K79 is highly associated with replication origins and possibly plays a role in preventing origin re-replication during S-phase. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38**.Gerhardt J, Tomishima MJ, Zaninovic N, Colak D, Yan Z, Zhan Q, Rosenwaks Z, Jaffrey SR, Schildkraut CL. The DNA replication program is altered at the FMR1 locus in fragile X embryonic stem cells. Mol Cell. 2014;53:19–31. doi: 10.1016/j.molcel.2013.10.029. This paper reports that fragile X syndrome, characterized by CGG repeats in the FMR1 locus, is accompanied by changes in the replication program in embryonic cells. The replication origin upstream of the FMR1 locus does not initiate replication in fragile X cells, possibly contributing to further repeat expansion and to the disease phenotype. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Norio P, Kosiyatrakul S, Yang Q, Guan Z, Brown NM, Thomas S, Riblet R, Schildkraut CL. Progressive activation of DNA replication initiation in large domains of the immunoglobulin heavy chain locus during B cell development. Mol Cell. 2005;20:575–587. doi: 10.1016/j.molcel.2005.10.029. [DOI] [PubMed] [Google Scholar]
- 40.Nagano T, Fraser P. No-nonsense functions for long noncoding RNAs. Cell. 2011;145:178–181. doi: 10.1016/j.cell.2011.03.014. [DOI] [PubMed] [Google Scholar]
- 41.Huang L, Damle SS, Booten S, Singh P, Sabripour M, Hsu J, Jo M, Katz M, Watt A, Hart CE, et al. Partial Hepatectomy Induced Long Noncoding RNA Inhibits Hepatocyte Proliferation during Liver Regeneration. PLoS One. 2015;10:e0132798. doi: 10.1371/journal.pone.0132798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Annunziato AT. The Fork in the Road: Histone Partitioning During DNA Replication. Genes (Basel) 2015;6:353–371. doi: 10.3390/genes6020353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jacob F, Brenner S. On the regulation of DNA synthesis in bacteria: the hypothesis of the replicon. C R Hebd Seances Acad Sci. 1963;256:298–300. [PubMed] [Google Scholar]
- 44*.Chen X, Liu G, Leffak M. Activation of a human chromosomal replication origin by protein tethering. Nucleic Acids Res. 2013;41:6460–6474. doi: 10.1093/nar/gkt368. This paper reports that tethering the pre-RC components Orc2 and Cdt1 to the c-Myc replication origin facilitates recruitment of downstream pre-RC members when the chromatin around is in an active (acetylated histone H3) conformation. This indicates that the both chromatin state and the replicon sequences and histone acetylation cooperate in origin activation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu G, Malott M, Leffak M. Multiple functional elements comprise a mammalian chromosomal replicator. Mol Cell Biol. 2003;23:1832–1842. doi: 10.1128/MCB.23.5.1832-1842.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fu H, Wang L, Lin CM, Singhania S, Bouhassira EE, Aladjem MI. Preventing gene silencing with human replicators. Nat Biotechnol. 2006;24:572–576. doi: 10.1038/nbt1202. [DOI] [PubMed] [Google Scholar]
- 47.Majocchi S, Aritonovska E, Mermod N. Epigenetic regulatory elements associate with specific histone modifications to prevent silencing of telomeric genes. Nucleic Acids Res. 2014;42:193–204. doi: 10.1093/nar/gkt880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Flickinger RA. Possible role of H1 histone in replication timing. Dev Growth Differ. 2015;57:1–9. doi: 10.1111/dgd.12190. [DOI] [PubMed] [Google Scholar]
- 49.Hassan-Zadeh V, Chilaka S, Cadoret JC, Ma MK, Boggetto N, West AG, Prioleau MN. USF binding sequences from the HS4 insulator element impose early replication timing on a vertebrate replicator. PLoS Biol. 2012;10:e1001277. doi: 10.1371/journal.pbio.1001277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Huang L, Fu H, Lin CM, Conner AL, Zhang Y, Aladjem MI. Prevention of transcriptional silencing by a replicator-binding complex consisting of SWI/SNF, MeCP1, and hnRNP C1/C2. Mol Cell Biol. 2011;31:3472–3484. doi: 10.1128/MCB.05587-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nakayama T, Takami Y. Participation of histones and histone-modifying enzymes in cell functions through alterations in chromatin structure. J Biochem. 2001;129:491–499. doi: 10.1093/oxfordjournals.jbchem.a002882. [DOI] [PubMed] [Google Scholar]
- 52.Iizuka M, Matsui T, Takisawa H, Smith MM. Regulation of replication licensing by acetyltransferase Hbo1. Mol Cell Biol. 2006;26:1098–1108. doi: 10.1128/MCB.26.3.1098-1108.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stroud H, Hale CJ, Feng S, Caro E, Jacob Y, Michaels SD, Jacobsen SE. DNA methyltransferases are required to induce heterochromatic re-replication in Arabidopsis. PLoS Genet. 2012;8:e1002808. doi: 10.1371/journal.pgen.1002808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.McConnell KH, Dixon M, Calvi BR. The histone acetyltransferases CBP and Chameau integrate developmental and DNA replication programs in Drosophila ovarian follicle cells. Development. 2012;139:3880–3890. doi: 10.1242/dev.083576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tardat M, Brustel J, Kirsh O, Lefevbre C, Callanan M, Sardet C, Julien E. The histone H4 Lys 20 methyltransferase PR-Set7 regulates replication origins in mammalian cells. Nat Cell Biol. 2010;12:1086–1093. doi: 10.1038/ncb2113. [DOI] [PubMed] [Google Scholar]
- 56.Chakraborty A, Shen Z, Prasanth SG. “ORCanization” on heterochromatin: linking DNA replication initiation to chromatin organization. Epigenetics. 2011;6:665–670. doi: 10.4161/epi.6.6.16179. [DOI] [PubMed] [Google Scholar]
- 57**.Giri S, Aggarwal V, Pontis J, Shen Z, Chakraborty A, Khan A, Mizzen C, Prasanth KV, Ait-Si-Ali S, Ha T, et al. The preRC protein ORCA organizes heterochromatin by assembling histone H3 lysine 9 methyltransferases on chromatin. Elife. 2015;4 doi: 10.7554/eLife.06496. This paper reports that the ORC associated protein ORCA recruits lysine methyltransferases to mono-methylated H3K9 to alter chromatin structure. ORCA’s effect on the replication landscape links chromatin packaging and replication timing. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bartke T, Vermeulen M, Xhemalce B, Robson SC, Mann M, Kouzarides T. Nucleosome-interacting proteins regulated by DNA and histone methylation. Cell. 2010;143:470–484. doi: 10.1016/j.cell.2010.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yokochi T, Poduch K, Ryba T, Lu J, Hiratani I, Tachibana M, Shinkai Y, Gilbert DM. G9a selectively represses a class of late-replicating genes at the nuclear periphery. Proc Natl Acad Sci U S A. 2009;106:19363–19368. doi: 10.1073/pnas.0906142106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hahn M, Dambacher S, Dulev S, Kuznetsova AY, Eck S, Worz S, Sadic D, Schulte M, Mallm JP, Maiser A, et al. Suv4-20h2 mediates chromatin compaction and is important for cohesin recruitment to heterochromatin. Genes Dev. 2013;27:859–872. doi: 10.1101/gad.210377.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61**.Cornacchia D, Dileep V, Quivy JP, Foti R, Tili F, Santarella-Mellwig R, Antony C, Almouzni G, Gilbert DM, Buonomo SB. Mouse Rif1 is a key regulator of the replication-timing programme in mammalian cells. EMBO J. 2012;31:3678–3690. doi: 10.1038/emboj.2012.214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62**.Hayano M, Kanoh Y, Matsumoto S, Renard-Guillet C, Shirahige K, Masai H. Rif1 is a global regulator of timing of replication origin firing in fission yeast. Genes Dev. 2012;26:137–150. doi: 10.1101/gad.178491.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63**.Yamazaki S, Ishii A, Kanoh Y, Oda M, Nishito Y, Masai H. Rif1 regulates the replication timing domains on the human genome. EMBO J. 2012;31:3667–3677. doi: 10.1038/emboj.2012.180. These three papers (61–63) report that the Rif1 protein plays an essential role in establishing the temporal replication program by stabilizing mid- to late-S-phase replication timing domains. Depletion of Rif1 leads to changes in G1/S transition, replication timing and chromatin structure in the next cell generation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tazumi A, Fukuura M, Nakato R, Kishimoto A, Takenaka T, Ogawa S, Song JH, Takahashi TS, Nakagawa T, Shirahige K, et al. Telomere-binding protein Taz1 controls global replication timing through its localization near late replication origins in fission yeast. Genes Dev. 2012;26:2050–2062. doi: 10.1101/gad.194282.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Francis LI, Randell JC, Takara TJ, Uchima L, Bell SP. Incorporation into the prereplicative complex activates the Mcm2-7 helicase for Cdc7-Dbf4 phosphorylation. Genes Dev. 2009;23:643–654. doi: 10.1101/gad.1759609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66**.Dave A, Cooley C, Garg M, Bianchi A. Protein phosphatase 1 recruitment by Rif1 regulates DNA replication origin firing by counteracting DDK activity. Cell Rep. 2014;7:53–61. doi: 10.1016/j.celrep.2014.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67**.Hiraga S, Alvino GM, Chang F, Lian HY, Sridhar A, Kubota T, Brewer BJ, Weinreich M, Raghuraman MK, Donaldson AD. Rif1 controls DNA replication by directing Protein Phosphatase 1 to reverse Cdc7-mediated phosphorylation of the MCM complex. Genes Dev. 2014;28:372–383. doi: 10.1101/gad.231258.113. These two papers report that Protein Phosphatase 1, recruited by Rif1, counteracts DDK-mediated phosphorylation of the MCM replicative helicase. These studies reveal a feedback loop involving Rif1, itself a DDK target, and suggest a biochemical mechanism underlying the effects of Rif1 on replication timing (papers 61–63) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68**.Knott SR, Peace JM, Ostrow AZ, Gan Y, Rex AE, Viggiani CJ, Tavare S, Aparicio OM. Forkhead transcription factors establish origin timing and long-range clustering in S. cerevisiae. Cell. 2012;148:99–111. doi: 10.1016/j.cell.2011.12.012. This paper reports that interactions with trans-acting factor Fkh1/2 control replication timing independent of the local environment by clustering early replicating origins and recruiting the Pre-RC interacting protein Cdc45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Fernandez PC, Frank SR, Wang L, Schroeder M, Liu S, Greene J, Cocito A, Amati B. Genomic targets of the human c-Myc protein. Genes Dev. 2003;17:1115–1129. doi: 10.1101/gad.1067003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Dominguez-Sola D, Ying CY, Grandori C, Ruggiero L, Chen B, Li M, Galloway DA, Gu W, Gautier J, Dalla-Favera R. Non-transcriptional control of DNA replication by c-Myc. Nature. 2007;448:445–451. doi: 10.1038/nature05953. [DOI] [PubMed] [Google Scholar]
- 71.Hermeking H, Rago C, Schuhmacher M, Li Q, Barrett JF, Obaya AJ, O’Connell BC, Mateyak MK, Tam W, Kohlhuber F, et al. Identification of CDK4 as a target of c-MYC. Proc Natl Acad Sci U S A. 2000;97:2229–2234. doi: 10.1073/pnas.050586197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Pelengaris S, Khan M, Evan G. c-MYC: more than just a matter of life and death. Nat Rev Cancer. 2002;2:764–776. doi: 10.1038/nrc904. [DOI] [PubMed] [Google Scholar]
- 73.Valovka T, Schonfeld M, Raffeiner P, Breuker K, Dunzendorfer-Matt T, Hartl M, Bister K. Transcriptional control of DNA replication licensing by Myc. Sci Rep. 2013;3:3444. doi: 10.1038/srep03444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Seo J, Chung YS, Sharma GG, Moon E, Burack WR, Pandita TK, Choi K. Cdt1 transgenic mice develop lymphoblastic lymphoma in the absence of p53. Oncogene. 2005;24:8176–8186. doi: 10.1038/sj.onc.1208881. [DOI] [PubMed] [Google Scholar]
- 75.Srinivasan SV, Dominguez-Sola D, Wang LC, Hyrien O, Gautier J. Cdc45 is a critical effector of myc-dependent DNA replication stress. Cell Rep. 2013;3:1629–1639. doi: 10.1016/j.celrep.2013.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gonzalez Besteiro MA, Gottifredi V. The fork and the kinase: a DNA replication tale from a CHK1 perspective. Mutat Res Rev Mutat Res. 2015;763:168–180. doi: 10.1016/j.mrrev.2014.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Feijoo C, Hall-Jackson C, Wu R, Jenkins D, Leitch J, Gilbert DM, Smythe C. Activation of mammalian Chk1 during DNA replication arrest: a role for Chk1 in the intra-S phase checkpoint monitoring replication origin firing. J Cell Biol. 2001;154:913–923. doi: 10.1083/jcb.200104099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zegerman P, Diffley JF. DNA replication as a target of the DNA damage checkpoint. DNA Repair (Amst) 2009;8:1077–1088. doi: 10.1016/j.dnarep.2009.04.023. [DOI] [PubMed] [Google Scholar]
- 79**.Guo C, Kumagai A, Schlacher K, Shevchenko A, Shevchenko A, Dunphy WG. Interaction of Chk1 with Treslin negatively regulates the initiation of chromosomal DNA replication. Mol Cell. 2015;57:492–505. doi: 10.1016/j.molcel.2014.12.003. This paper reports that activation of Treslin by CDK positively regulates DNA replication by promoting the binding of Cdc45 to Mcm2-7. Conversely, phosphorylation of Treslin by Chk1 negatively regulates DNA replication by limiting the Cdc45 binding the helicase complex. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Tanaka S, Umemori T, Hirai K, Muramatsu S, Kamimura Y, Araki H. CDK-dependent phosphorylation of Sld2 and Sld3 initiates DNA replication in budding yeast. Nature. 2007;445:328–332. doi: 10.1038/nature05465. [DOI] [PubMed] [Google Scholar]
- 81.Liu P, Barkley LR, Day T, Bi X, Slater DM, Alexandrow MG, Nasheuer HP, Vaziri C. The Chk1-mediated S-phase checkpoint targets initiation factor Cdc45 via a Cdc25A/Cdk2-independent mechanism. J Biol Chem. 2006;281:30631–30644. doi: 10.1074/jbc.M602982200. [DOI] [PubMed] [Google Scholar]
- 82.Anglana M, Apiou F, Bensimon A, Debatisse M. Dynamics of DNA replication in mammalian somatic cells: nucleotide pool modulates origin choice and interorigin spacing. Cell. 2003;114:385–394. doi: 10.1016/s0092-8674(03)00569-5. [DOI] [PubMed] [Google Scholar]
- 83.Kunnev D, Rusiniak ME, Kudla A, Freeland A, Cady GK, Pruitt SC. DNA damage response and tumorigenesis in Mcm2-deficient mice. Oncogene. 2010;29:3630–3638. doi: 10.1038/onc.2010.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Conti C, Leo E, Eichler GS, Sordet O, Martin MM, Fan A, Aladjem MI, Pommier Y. Inhibition of histone deacetylase in cancer cells slows down replication forks, activates dormant origins, and induces DNA damage. Cancer Res. 2010;70:4470–4480. doi: 10.1158/0008-5472.CAN-09-3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Daboussi F, Courbet S, Benhamou S, Kannouche P, Zdzienicka MZ, Debatisse M, Lopez BS. A homologous recombination defect affects replication-fork progression in mammalian cells. J Cell Sci. 2008;121:162–166. doi: 10.1242/jcs.010330. [DOI] [PubMed] [Google Scholar]
- 86*.Fu H, Martin MM, Regairaz M, Huang L, You Y, Lin CM, Ryan M, Kim R, Shimura T, Pommier Y, et al. The DNA repair endonuclease Mus81 facilitates fast DNA replication in the absence of exogenous damage. Nat Commun. 2015;6:6746. doi: 10.1038/ncomms7746. This paper reports that Mus81, an endonuclease active in the replication-stress response pathway, promotes replication fork progression in normally growing cells. When replication forks slow in the absence of Mus81, cells increase the frequency of initiation from flexible replication origins. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Depamphilis ML, de Renty CM, Ullah Z, Lee CY. “The Octet”: Eight Protein Kinases that Control Mammalian DNA Replication. Front Physiol. 2012;3:368. doi: 10.3389/fphys.2012.00368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hayashi MT, Masukata H. Regulation of DNA replication by chromatin structures: accessibility and recruitment. Chromosoma. 2011;120:39–46. doi: 10.1007/s00412-010-0287-4. [DOI] [PubMed] [Google Scholar]
- 89.Sheu YJ, Stillman B. Cdc7-Dbf4 phosphorylates MCM proteins via a docking site-mediated mechanism to promote S phase progression. Mol Cell. 2006;24:101–113. doi: 10.1016/j.molcel.2006.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Sheu YJ, Stillman B. The Dbf4-Cdc7 kinase promotes S phase by alleviating an inhibitory activity in Mcm4. Nature. 2010;463:113–117. doi: 10.1038/nature08647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ilves I, Petojevic T, Pesavento JJ, Botchan MR. Activation of the MCM2-7 helicase by association with Cdc45 and GINS proteins. Mol Cell. 2010;37:247–258. doi: 10.1016/j.molcel.2009.12.030. [DOI] [PMC free article] [PubMed] [Google Scholar]