Summary
The kinetochore is a multi-protein complex that mediates the attachment of a eukaryotic chromosome to the mitotic spindle. The protein composition of kinetochores is similar across species as divergent as yeast and human. However, recent findings have revealed an unexpected degree of compositional diversity in kinetochores. For example, kinetochore proteins that are essential in some species have been lost in others, whereas new kinetochore proteins have emerged in other lineages. Even in lineages with similar kinetochore composition, individual kinetochore proteins have functionally diverged to acquire either essential or redundant roles. Thus, despite functional conservation, the repertoire of kinetochore proteins has undergone recurrent evolutionary turnover.
Keywords: Kinetochore, evolutionary dynamics, functional conservation, CenH3, CENP-B, CENP-T
The ship wherein Theseus and the youth of Athens returned from Crete had thirty oars, and was preserved by the Athenians………. they took away the old planks as they decayed, putting in new and stronger timber in their places, in so much that this ship became a standing example among the philosophers, for the logical question of things that grow; one side holding that the ship remained the same, and the other contending that it was not the same.
— Plutarch, Theseus
Kinetochores mediate chromosome segregation in eukaryotes
Chromosome segregation is an essential, conserved process that ensures faithful transmission of genetic information to daughter cells during cell division in all eukaryotes. Specialized chromosomal regions called centromeres coordinate this process of chromosome segregation. Centromeres were first cytologically described as primary constrictions on chromosomes where spindle microtubules attach to pull the sister chromatids apart during the mitotic and meiotic divisions [1]. Two different types of centromere arrangements are found in eukaryotes: monocentric or holocentric (Box 1). In both types of centromeres, the physical connection between the spindle microtubules and the centromeric DNA is provided by a large proteinaceous structure termed the kinetochore [2]. Micrographic images of the kinetochore reveal a bipartite structure consisting of an inner complex that connects to the centromeric DNA and an outer complex that connects to the spindle microtubules. Detailed biochemical studies resolved this bipartite structure into many protein subunits that function in a coordinated manner to enable dynamic assembly on centromeric DNA [3]. Perhaps the best elucidation of the kinetochore structure and biophysical mechanism comes from budding yeast, where in vivo isolation of intact kinetochores has led to unprecedented insights into its structure and regulation [4, 5].
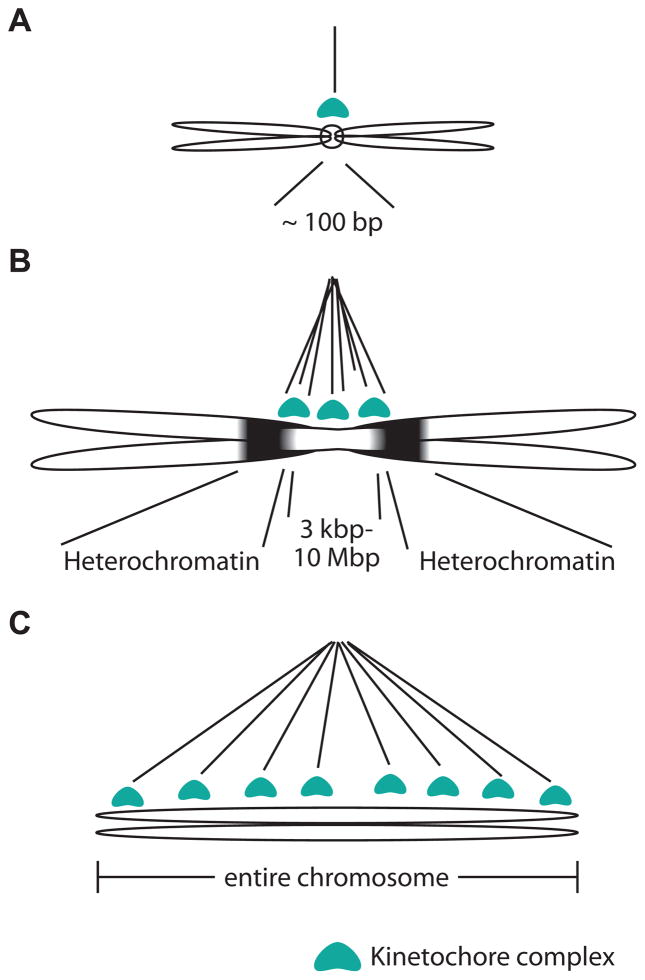
Box 1. Dramatic variation of number and arrangement of attached spindle microtubules.
Most eukaryotic chromosomes are monocentric i.e., the centromere is restricted to one region of the chromosome. Although monocentromeres are commonly visible as primary constrictions of chromosomes, the actual length of the centromeric region can vary over several orders of magnitude across different eukaryotes [14]. The simplest monocentromere is the budding yeast point centromere, which measures about 100 bp in length and captures one spindle microtubule [88] (Figure IA). In contrast, ‘regional’ centromeres found in most other eukaryotes can span up to a few megabases [89] (Figure IB) and capture several microtubules [90].
Several lineages have evolved holocentromeres, in which microtubule attachment sites extend along large portions or even the entire length of the chromosome [91] (Figure IC). Based on the phylogenetically dispersed pattern of holocentric species among eukaryotes, it appears likely that holocentricity repeatedly evolved from monocentric ancestors [11, 91]. Holocentromeres have been best characterized in C. elegans. Profiling of inner kinetochore components revealed both discrete sites along the C. elegans chromosomes that recruit kinetochore components, as well as enrichment over transcriptionally silent regions [92, 93]. Although this arrangement is likely to be conserved in other holocentric nematodes, it is unclear whether such a “polycentric” architecture also applies to other independently derived holocentric species. Indeed, a recent study in holocentric plants revealed that centromeric sites are composed of dispersed satellite DNA along the chromosomes [94]. Despite these dramatic differences in the number and spatial arrangement of attached spindle microtubules, none of those changes have been found to correlate with changes in kinetochore composition with the exception of holocentric insects (Figure 1D, Key Figure) [11]. In fact, most monocentric and holocentric species use the histone variant CenH3 as the chromatin determinant enabling kinetochore assembly.
Figure I. Dramatic variation in kinetochore architectures.
We highlight three types of kinetochore arrangements to illustrate their range, from the simplest ‘point’ monocentromere in budding yeast (A), to regional monocentromeres (B), and to holocentromeres seen in many lineages including nematodes C).
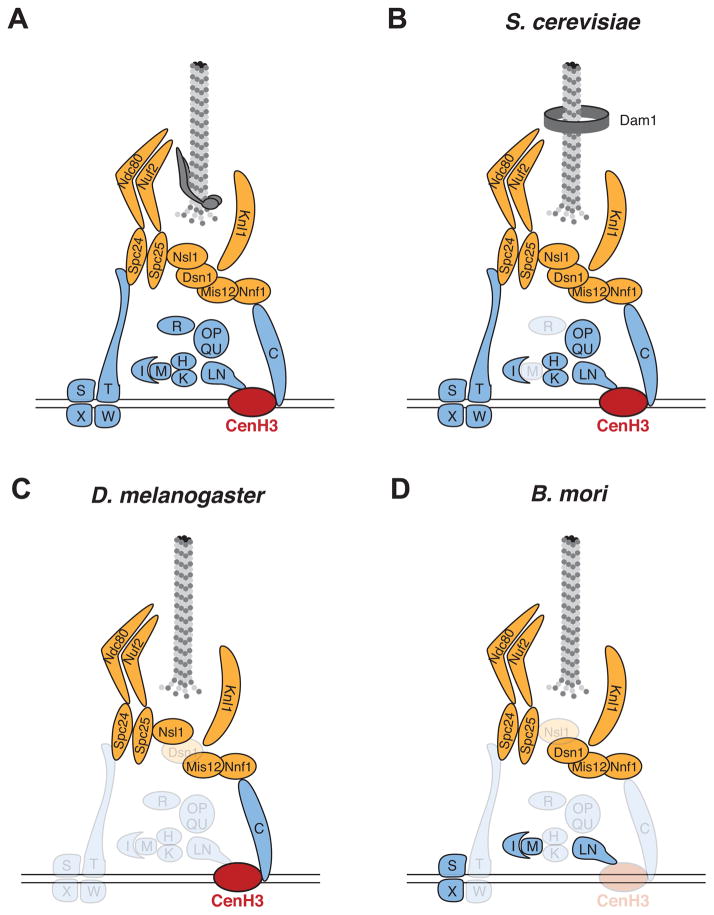
The kinetochore’s basic organization is highly conserved in all eukaryotes. Early studies of kinetochore function and composition were mainly performed in yeast and vertebrate cell lines as model systems. Not only did these studies reveal fundamental principles of kinetochore function, but they also uncovered a high degree of similarity between yeast and vertebrate kinetochores, especially of the inner complex, despite millions of years of divergence between the two lineages [6] (Figure 1A, B, Key Figure). However, analysis of kinetochore composition in other eukaryotes revealed an unexpected degree of variation. For instance, the D. melanogaster kinetochore, which has also been well dissected via biochemical and genetic means [7–10], is missing many of the inner kinetochore components found in vertebrate and budding yeast cells (Figure 1C, Key Figure). Similarly, the kinetochores of the holocentric Bombyx mori are distinct from those in vertebrate and budding yeast [11] (Figure 1D, Key Figure). Finally, a recent proteomic study of the kinetochore in trypanosomes, an early branching lineage of eukaryotes, revealed a completely different set of kinetochore proteins, with no detectable homology to fungal or vertebrate kinetochores [12, 13]. In addition to their different repertoires, animal and plant kinetochore proteins have undergone rapid protein evolution despite conservation of kinetochore function [14, 15].
Figure 1. Key Figure: Variation in kinetochore composition in four eukaryotic taxa.
Kinetochores have been best-dissected biochemically in vertebrates (A) and budding yeast (B). These two kinetochores are remarkably similar in composition, with the most notable difference being the absence of CENP-M in yeast (grayed out) and the fungal-specific origin of the Dam1 complex, the functional counterpart of the Ska1 complex. However, the kinetochores in D. melanogaster (C) and holocentric Bombyx mori (D) are quite distinct from those in vertebrate and budding yeast. The conclusions that certain CCAN components are missing in both (C) and (D) are tempered by the possibility that rapid evolution might have impaired homology searches; these conclusions await further confirmation via proteomic or other experimental means.
The evolutionary dynamics of the kinetochore complex is reminiscent of a thought experiment called the “Ship of Theseus” recorded by the Greek philosopher Plutarch. This paradox raises the question: does a ship that has all its wooden parts replaced one-by-one still remain the same? During the evolutionary process, the kinetochore “ship” has maintained its conserved function in chromosome segregation, despite apparently replacing many or all of its parts. We summarize three signatures of evolutionary turnover, focusing on inner kinetochore composition–loss of kinetochore components that are essential in some species, recurrent innovation of novel kinetochore components and differential importance of certain kinetochore components for the important function of chromosome segregation.
Loss of an ‘essential’ chromatin determinant of kinetochore assembly
In most eukaryotes, a specialized centromeric histone H3 variant called CenH3 [16] is found predominantly and ubiquitously at centromeres. In these eukaryotes, CenH3 is the major chromatin determinant enabling kinetochore assembly on centromeric regions. The first discovery of CenH3 (called CENP-A in mammals) hinged serendipitously on an observation made in 1985; antibodies present in the serum of patients with an autoimmune disease were found to react with kinetochore proteins [17]. Subsequent studies in other species of animals, fungi, plants and protists showed that the incorporation of CenH3 homologs is a common feature of their centromeres [18–21], thereby establishing CenH3 as the chromatin marker associated with all active centromeres, and likely responsible for their inheritance (see Box 2).
Box 2. Genetic versus epigenetic centromeres.
Although the meaning of “epigenetic” has been widely debated [95], there is general agreement that epigenetic inheritance is characterized by a lack of DNA sequence determinants. Epigenetic centromeres would then be defined as centromeres that lack specific DNA sequence requirements. The first characterized centromeres were the genetically defined ‘point’ centromeres of the budding yeast Saccharomyces cerevisiae, which are specified by a tripartite ~120-bp sequence that wraps a single CenH3 (called Cse4 in budding yeast) nucleosome [88]. In contrast, regional centromeres in fission yeast and Candida are not associated with specific DNA sequences, supporting the model that they are epigenetically defined.
In most eukaryotes, centromeres are embedded in megabases of tandemly repeated DNA “satellite” sequences, which strongly suggests that satellite DNA specifies centromeres. However, the discovery that a human neo-centromere lacks alpha-satellite sequences suggested that human centromeres could be entirely epigenetic [96]. Despite this evidence for very rare epigenetic centromeres, human artificial centromeres require alpha satellite arrays with a binding site for CENP-B protein (CENP-B box) in order to be propagated in culture [97]. Moreover, most human CENP-A was found to be enriched over two classes of highly homogenous CENP-B box-containing alpha-satellite dimers [98], suggesting that stable centromeres are consistently associated with particular alpha-satellite repeats. The stable inheritance of the centromere-defining chromatin determinant over sequence-defined alpha-satellite repeat dimers supports the distinction between epigenetically defined neo-centromeres and genetically defined alpha-satellite centromeres. Such a genetic definition might apply to other regional centromeres and even to holocentromeres that are composed of particular satellite DNAs [94].
Despite functional conservation across all eukaryotes, the evolutionary history of CenH3 remains unresolved. Due to its conserved presence at the eukaryotic centromere, we favor the more parsimonious model of a single origin for CenH3 in an early eukaryotic ancestor [22]. However, CenH3 homologs in different species are far less conserved compared to H3 and thus we cannot rule out the possibility of multiple independent origins of CenH3 in the H3 family [23, 24]. Even less conserved is the machinery that mediates the deposition of CenH3 at centromeres (Box 3).
Box 3. No designated driver for CenH3 delivery.
The deposition of CenH3 into chromatin is under tight control. Failure to deposit adequate amounts of CenH3 could lead to functionally acentric chromosomes whereas ectopic incorporation of CenH3 can cause dicentric chromosomes [99]; both outcomes are detrimental to accurate chromosome segregation. The deposition of CenH3 occurs during a defined window during the cell cycle, although this window varies among species [100]. Indeed, constitutive incorporation of CenH3 throughout the entire cell cycle results in mitotic defects [101]. Based on the requirement for tight temporal and spatial control of CenH3 incorporation, one might assume that the deposition machinery of CenH3 would be conserved across eukaryotes. This is not the case.
In vertebrates and budding yeast, homologs of the specialized histone chaperone HJURP/Scm3 mediate the deposition of CenH3 into centromeric chromatin [102–105]. HJURP/Scm3 probably arose in a common ancestor of fungi and animals. However, it has been apparently lost in several animal lineages including nematodes and flies, raising the question of how these lineages compensated for the loss of this essential gene [106]. In flies, the loss of HJURP/Scm3 was perhaps facilitated by the evolution of the non-orthologous chaperone CAL1 that deposits D. melanogaster CenH3/CID onto centromeric chromatin [107–109]. Intriguingly, the CenH3 interacting domains of CAL1 and Scm3 show similarity in both sequence and structure, which could be a result of convergent evolution [110]. The similar emergence of new chaperones in other lineages may have led to the loss of HJURP/Scm3. Alternatively, an existing histone chaperone might have acquired the additional function of CenH3 incorporation. In that context, it is worth noting that the histone chaperone DAXX was recently found to be involved ectopic incorporation in human cells overexpressing CENP-A [111]. Thus, CenH3 incorporation can also be mediated by chaperones that also deliver other histone variants.
Although it is clear that HJURP/Scm3 and CAL1 represent bona fide CenH3 assembly factors, other proteins also are needed to coordinate CenH3 deposition. For instance, proteins of the Mis18 complex (Mis18α, Mis18β, and Mis18BP1/KNL2) and the Mis16 complex are also required for CenH3 deposition in HJURP-containing (e.g. human, fission yeast) and HJURP-lacking (e.g. C. elegans, Arabidopsis) lineages [112–118]. Outside animals and fungi, even less is known about CenH3 incorporation in plants or protists, where no designated CenH3 chaperones have been identified.
Not only is CenH3 a marker of centromeres, but it is also a key player in the kinetochore assembly pathway. In species as diverse as fungi, worms, plants and humans, CenH3 is required to anchor the kinetochore to centromeric chromatin [2]. These findings led to the expectation that CenH3 would be found at all centromeres. However, this expectation was challenged by multiple discoveries of CenH3-independent kinetochore assembly pathways in eukaryotes. The first completely CenH3-independent chromosome segregation pathway was discovered in kinetoplastids [12]. Not only do these species lack CenH3, but detailed biochemical studies revealed a kinetoplastid kinetochore complex, whose components shared no detectable homology to other eukaryotic kinetochores [12]. This may reflect the fact that kinetoplastids branched early during eukaryotic evolution from the lineages that gave rise to plants, fungi and animals [25], which compromises the power of remote homology detection programs. It is formally possible that the CenH3-independent kinetoplastid kinetochore represents a system ancestral to the origin of the more well-studied kinetochore complex. However, the discovery of CenH3 in other early branching eukaryotes like Giardia lamblia [19] and Trichomonas vaginalis [26], together with recently revised models of eukaryotic evolution [27], suggest instead that CenH3-independent kinetochores in kinetoplastids represent a derived rather than ancestral state. Nevertheless, the discovery and characterization of the unconventional kinetochores in kinetoplastids provides unique insights into the origin of chromosome segregation machineries and the general design principles crucial for kinetochore function.
A second CenH3-independent chromosome segregation pathway was recently described in several orders of insects using genomic and transcriptomic surveys [11]. Unlike in the case of kinetoplastids, it is unambiguous that CenH3 absence in insects is due to at least four independent loss events, some of which occurred as long as 300 million years ago. In insects, CenH3 losses occurred in lineages that underwent dramatic changes in their centromeric architecture, from monocentricity to holocentricity (Box 1). This perfect correlation in insects suggests a causal relationship between the two events i.e., transition to holocentric chromosomes in insects might have facilitated the loss of CenH3, or vice versa. Notably, other kinetochore components are still present in CenH3-deficient insects (see Figure 1D for example), which implies that those species utilize CenH3-independent ways of initiating kinetochore assembly using at least some of the same components used in CenH3-encoding insects. Notably, outside of insects, other holocentric species with sequenced genomes such as C. elegans do encode CenH3, where it carries out essential functions in mitotic chromosome segregation [11, 18, 28]. This suggests that the association of CenH3 loss with holocentricity might be unique to the insect lineage.
Despite its essential mitotic role in C. elegans, CenH3 is not required for meiotic divisions, thus representing a third case of a CenH3-independent chromosome segregation pathway [29]. Although the outer kinetochore complex is required for proper chromosome alignment during early meiotic metaphase, even this complex disassembles before chromosomes separate during C. elegans meiosis. In this instance, lateral attachments to microtubule bundles along the sides of chromosomes and microtubule motor proteins may provide the major driving force [30, 31]. Intriguingly, neocentromeres formed on maize knob chromosomal elements during meiosis also do not require CenH3 for mediating attachments to the meiotic spindle [32].
The three cases of CenH3 loss we have highlighted here have different evolutionary origins; these species likely accomplish chromosome segregation by distinct means. Together these examples emphasize the fact that even kinetochore proteins that are otherwise considered most fundamental to centromere function and kinetochore assembly can be lost.
A variable requirement of CENP-T for kinetochore function
There are two primary modes of connecting the centromeric DNA and the outer kinetochore [2, 33, 34] (Figure 1, Key Figure). The first of these is mediated by CENP-C, which is a large, unstructured protein [35] that directly interacts with CenH3 to connect the inner kinetochore to the Mis12 outer kinetochore complex [36–38]. The interaction of CENP-C with CenH3 is mediated by the carboxy-terminus of CenH3 and a conserved carboxy-terminal motif in the CENP-C protein, termed the CENP-C motif [39, 40]. In mammals, CENP-C additionally binds to CenH3 via a central region; however this central region is only conserved in mammals [40]. Despite the essentiality of the CENP-C protein and the conservation of the CENP-C motif, the cognate interaction sites at the C-termini of CenH3 proteins are surprisingly not well conserved. Instead, CenH3 C-termini share the common feature of being more hydrophobic than those of the canonical H3. This has led to the suggestion that the hydrophobicity of CenH3’s C-terminal tail, rather than a specific amino acid sequence, is the key determinant for CENP-C recognition [40]. CENP-C and CenH3 are interdependent at centromeres: incorporation of CenH3 is necessary for CENP-C recruitment, and binding of CENP-C stabilizes CenH3 nucleosomes at centromeres, whereas CENP-C depletion leads to rapid loss of CenH3 from centromeres [41]. Consistent with the interdependence of CenH3 and CENP-C, CENP-C proteins are absent in holocentric insects that lack CenH3 [11].
The CENP-T protein provides a second mode of connecting the centromeric DNA and the outer kinetochore. Tethering CENP-T to ectopic sites is sufficient for kinetochore assembly in chicken cells [33]. The N-terminus of CENP-T interacts with the Ndc80 outer kinetochore complex [10, 33, 42]. CENP-T’s defining feature is a histone fold at the C-terminus followed by a conserved amino acid extension of unknown function [43, 44]. Its cognate histone fold interaction partner is CENP-W; a CENP-T/W dimer structurally resembles an H2A/H2B dimer [44]. The CENP-T/W dimer can be paired with a CENP-S/X dimer, completing a nucleosome-like complex that is found at centromeres [44]. This resemblance to nucleosomes had led to suggestions that CENP-T-W-S-X may also wrap centromeric DNA similar to centromeric nucleosomes [45]. Although this might suggest that all four proteins are equally important for kinetochore function, phenotypes of CENP-S and CENP-X deletions are rather mild compared to CENP-T and CENP-W deletions in chicken DT40 cells [43, 46]. Furthermore, though recruitment of CENP-S and CENP-X to kinetochores is dependent on CENP-T and CENP-W, some species containing CENP-S and CENP-X appear not to have CENP-S and CENP-W based on computational predictions [6, 11]. In those species, CENP-S and CENP-X might be retained due to their role in DNA repair [47], which is independent of their CENP-T/W-dependent function at the kinetochore.
In contrast to CENP-C, CENP-T is only sporadically conserved across eukaryotes. While CENP-T homologs are essential in vertebrates and fission yeast [43, 48, 49], no CENP-T homologs have been identified in D. melanogaster or C. elegans [6]. Thus, it appears that in some species, one axis of connecting the centromere to the outer kinetochore (via CenH3 & CENP-C) is sufficient for kinetochore function, whereas in others, additional recruitment of CENP-T is required for proper kinetochore formation. Yet, CENP-T homologs display extensive protein sequence divergence, which complicates homology-based detection of CENP-T homologs from genomic sequences even in well-sequenced taxa such as arthropods, nematodes or plants. Thus, it is hard to conclusively rule out the absence of CENP-T based purely on homology searches; such conclusions should be confirmed by additional experimental approaches when possible. Indeed, combining experimental approaches with remote homology predictions successfully enabled the identification of a budding yeast CENP-T homolog, which also binds to the Ndc80 complex as does in vertebrate CENP-T [10]. However, whereas CENP-T is essential in all other species tested, the budding yeast CENP-T is not essential for chromosome segregation [10]. This finding, among others, suggests that the functional composition of the fission yeast kinetochores more closely resemble vertebrate kinetochores, than those from budding yeast.
New genes for old functions: the enigma of recurrent domestications of CENP-B-like proteins
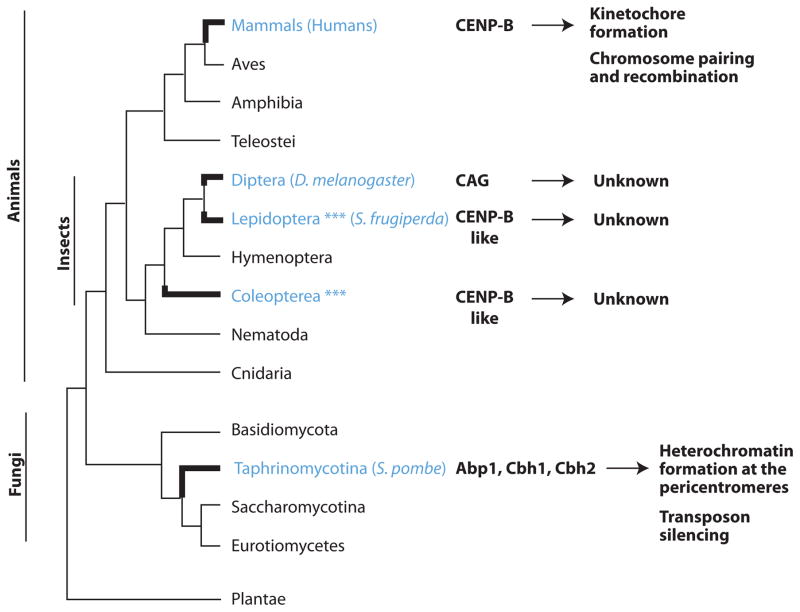
Another dramatic example of the rapid turnover of kinetochore proteins emerges is the recurrent domestication of CENP-B DNA-binding proteins for centromere functions (Figure 2). CENP-B proteins are derived from transposases of ancient pogo-like DNA transposons and have originated independently in mammals, fission yeast [50], Lepidoptera [51] and other metazoans [52]. In both mammals and fission yeast, CENP-B proteins localize to centromeres. CENP-B-like proteins in fission yeast are involved in heterochromatin formation surrounding the centromeric region [53, 54] but also serve additional roles in silencing of transposable elements [55, 56]. In contrast, the role of mammalian CENP-B proteins at centromeres is less understood. Thus, though all CENP-B proteins are homologous whether they perform the same function is unclear.
Figure 2. Recurrent domestication of pogo-like transposases to give rise to CENP-B DNA-binding proteins.
CENP-B proteins have been domesticated multiple independent times in animals and fungi. Three domesticated CENP-B-like proteins were found in fission yeast (highlighted in blue). Blue asterisks indicate potential domestications of additional CENP-B-like proteins in insects (often multiple instances per genome) predicted from bioinformatics analyses [52] that have yet to be confirmed experimentally.
Mammalian CENP-B proteins have DNA binding sites (called CENP-B boxes) in megabase-long centromeric repeat arrays [57] except on the Y chromosome [58]. Furthermore, CENP-B has been conserved through most of mammalian evolution. Despite this massive abundance of its recognition motif and its evolutionary conservation, CENP-B is not essential for centromere function and CENP-B boxes have not been strictly conserved during mammalian evolution [59]. This suggests that CENP-B conservation might be due to a non-centromeric function such as the silencing of transposable elements as described in fission yeast [55, 56]. The most direct demonstration of CENP-B’s role in kinetochore formation has come out of a recent study in which human CENP-B was shown to interact with the N-termini of CenH3 and CENP-C [60]. CENP-B mutations in human cells led to depletion of both CENP-C levels as well as defects in cell cycle-dependent loading of CenH3. This suggested a model in which CENP-B represents a parallel pathway for CENP-C recruitment and CenH3 stabilization at human centromeres [60].
Although CENP-B was lost in some mammalian lineages, it is difficult to reconcile this dispensability with the observation that it may mediate some important, albeit non-essential, function at the kinetochore. We speculate that this discrepancy could result from CENP-B’s specific DNA-binding properties and the unique battle waged between centromeres during female meiosis. Under the ‘centromere-drive’ hypothesis, chromosomes compete via their centromeric satellites in female meiosis for inclusion into the oocyte [14, 61]. Greater recruitment of centromeric proteins might directly translate to meiotic success [62]. In this regard, recruiting CENP-B proteins by virtue of CENP-B boxes may be one means by which certain centromeres could outcompete others ‘selfishly’ for meiotic dominance [63]. However, centromere drive can result in deleterious effects on male fertility [64, 65] and other functions [66]. To counter these deleterious effects, suppressor alleles are predicted to arise. Some of these suppressors could result in in centromere strength (and kinetochore function) being less dependent on CENP-B recruitment, thereby negating the ‘selfish’ advantage of some centromeric satellite repeats. Under this model, the loss of CENP-B could be a result of the transition between ‘centromere drive’ in which specific DNA sequences are selected by competition (a “genetic” state), and ‘suppression’ in which DNA specificity is ignored for centromere specification (an “epigenetic” state) [67]. This ‘centromere-drive’ hypothesis might especially apply to kinetochore proteins like CENP-B with DNA-binding specificity, but only in lineages that undergo female meiosis.
Centromere drive and its suppression can also explain the signatures of rapid evolution observed even in essential kinetochore components like CenH3 and CENP-C in multiple animal and plant lineages [14, 15, 35]. The relentless need to innovate at the centromere-kinetochore interface as a result of centromere drive has also been proposed to lead to the rapid acquisition of essential roles for centromere function on an evolutionary young gene in Drosophila [68]. It is currently unclear to what extent such lineage-specific, evolutionarily young genes have further diversified kinetochore repertoires across animal and plants.
Evolutionary turnover of the kinetochore complex
In the previous sections, we used the examples of three inner kinetochore proteins to highlight three facets of plasticity of kinetochore repertoires: loss of components essential in some lineages (CenH3), differential essentiality for kinetochore function (CENP-T) and invention of novel components (CENP-B). These facets can be extended to other kinetochore proteins of the inner kinetochore (collectively termed the constitutive centromere-associated network, or CCAN), the outer kinetochore and even to components of the mitotic checkpoint, which acts to safeguard against premature chromosome segregation.
The CCAN component CENP-M represents another example for the non-uniform presence of a kinetochore component. CENP-M homologs are easily recognizable as catalytically inactive members of the GTPase superfamily. At the kinetochore, CENP-M forms a tight complex with CENP-I in a way that is structurally reminiscent of a small GTPase bound to importin beta [6, 69]. The CENP-M/CENP-I complex associates with CENP-H and CENP-K and localization of the entire complex is interdependent with CENP-T in human cells [69]. Notably, though CENP-M is essential in vertebrates, it appears to be absent in fungi [10, 49, 70, 71]. It is still unclear whether this is because CENP-M is a vertebrate- or animal-specific invention or whether it was lost in fungi. In the latter case, it will be interesting to test if another GTPase-like protein performs the same function in fungi as CENP-M does in vertebrates [69], or whether they have adopted a completely non-homologous means to substitute for CENP-M.
The CENP-O complex is another example of a CCAN complex of proteins that is essential for kinetochore function in some species but not others [49, 70, 72, 73]. The CENP-O complex includes CENP-O, CENP-P, CENP-Q, CENP-U and CENP-R (CENP-R is not present in fungi). Two components of the CENP-O complex, CENP-Q and CENP-U are essential in budding yeast and have been shown to interact with the Mis12 complex along with CENP-C. Budding yeast centromeres consist of only one CenH3 and CENP-C, but several Mis12 complexes. The CENP-U/Q-mediated interaction with the Mis12 complex has therefore been proposed to provide additional bridges to the outer kinetochore [72]. In contrast to budding yeast, CENP-U is not required for viability in chicken DT40 and mouse embryonic fibroblasts. Yet, CENP-U mutant mice die during early embryonic development and CENP-U is essential in undifferentiated murine embryonic stem cells [74]. To explain this difference, it has been proposed that kinetochores are weaker in all cell types in the absence of CENP-U. This effect might lead to more detrimental consequences in rapidly dividing embryonic cells due to weaker mitotic checkpoints, which would otherwise allow cells to correct inappropriate kinetochore attachments [74].
With the exception of CENP-C, most CCAN components have only been found in vertebrates and fungi based on sequence homology [10, 75]. It is possible that CCAN components are present in other species but homology detection is challenged by a high degree of divergence and lack of pre-defined protein domains. If CCAN proteins are truly absent in some lineages, it will be interesting to determine whether their kinetochore function is replaced by other proteins, or whether kinetochore composition in those lineages is less complex. To explore these ideas, it would be useful to study the composition of kinetochores in additional lineages such as Dictyostelium, ciliates and plants that are accessible to genomics and proteomics.
Although we have focused in this review on inner kinetochore proteins, some outer kinetochore components also exhibit features of evolutionary lability. The function of the outer kinetochore KNL1 proteins in microtubule attachment and the mitotic checkpoint is highly conserved across eukaryotes. Yet, repeat arrays that recruit BUB1-BUB3 signaling proteins exhibit recurrent episodes of rapid evolution in both number and sequence [76]. It is unclear what drives these changes and whether these episodes affect the strength or fidelity of the signaling. Similar to the lineage-specific invention of some inner kinetochore proteins, computational predictions based on homology suggest that the Dam1 outer kinetochore complex arose in an early fungal ancestor. The Dam1 complex forms oligomers around spindle microtubules thereby contributing to the strength of microtubule attachment [77] (see Figure 1A, Key Figure). In humans and other species such as nematodes and plants, the Ska1 complex might be the functional equivalent of the Dam1 complex in fungi, in terms of coupling chromosome movements to microtubule dynamics [78–80] although both complexes do not share sequence or even structural similarity.
In addition to evolutionary turnover within the kinetochore complex, recent studies have found significant turnover within components of the mitotic checkpoint pathway [81]. For example, although the mitotic checkpoint kinase Mps1 is essential in fungi and several animal lineages [82, 83], it is lost in C. elegans [81]. Indeed, a recent study has confirmed that a different kinetochore-localized kinase, the Polo-like kinase 1, may have replaced Mps1 in nematodes [84]. Evolutionary turnover is even seen in protein complexes involved in the recruitment and removal of signaling proteins involved in the mitotic checkpoint. This turnover includes both evolutionary adaptation of existing proteins (RZZ complex components Rod and ZW10) [85] as well as invention of novel proteins (Spindly, RZZ complex component Zwilch) [81, 86, 87]. The fact that some of these components cannot be found outside the ophistokonts suggest that a more ancient mechanism for checkpoint regulation in eukaryotes may have gotten replaced by RZZ/Spindly in ophistokonts. Identification of these ancestral mechanisms will require studies beyond the traditional model organisms, which have been largely limited to within the ophistokonts thus far.
Concluding remarks
In this review, we have highlighted the evolutionary dynamics of the kinetochore complex. Despite accomplishing the same essential function of connecting centromeric DNA to spindle microtubules in all eukaryotes, kinetochore composition is unexpectedly evolutionarily labile. What is the effect of this lack of evolutionary conservation on core functions of the kinetochore, including kinetochore strength and regulation? While one kinetochore-microtubule attachment is sufficient to segregate budding yeast chromosomes, many kinetochore-microtubule attachments work in concert to segregate a human monocentric chromosome or a nematode holocentric chromosome. This suggests that the relative demands on kinetochore strength might be different among different species; whether this is a cause or consequence of changes in kinetochore composition remains unexplored. Similarly, it is unknown how number of kinetochore-microtubule interactions affects stringency of kinetochore regulation, which may explain the rapid evolution of kinetochore proteins like KNL1 [76].
Most importantly, our survey reveals the effects of a long, historical bias of studying kinetochore compositions in vertebrates, fungi and a handful of model invertebrate animals. Although genome sequencing and new proteomic approaches are helping to correct this bias, additional experimental approaches and remote homology predictions will be needed to determine kinetochore composition in additional major eukaryotic lineages including plants. Only then will a true picture of component fluidity and general design principles of kinetochores begin to emerge (see ‘Outstanding Questions’).
Outstanding questions.
Are the architectural principles of kinetochores and their regulation during cell division preserved in divergent organisms like trypanosomes despite lack of conservation of kinetochore components?
What were the intermediate evolutionary steps by which a previously essential centromeric protein was lost in certain lineages (e.g. CenH3 in holocentric insects) or rendered non-essential in others (e.g. CENP-T in budding yeast)?
What has replaced CenH3 function in trypanosomes and holocentric insects?
How do changes in kinetochore architecture (e.g. holocentric versus monocentric, chromosome fusions) or strength mediate such transitions?
What is the effect of evolutionary turnover of kinetochore proteins on kinetochore strength and fidelity of chromosome segregation?
Is the composition of meiotic kinetochores identical to mitotic kinetochores? If not, what are the meiosis-specific components, how do they function and how well are they conserved in evolution especially in taxa that do not undergo female or male meiosis?
What is the effect of evolutionary turnover on checkpoint proteins and even proteins that ‘moonlight’ at the kinetochore (e.g. some nuclear pore complex proteins) on kinetochore function?
In spite of the evolutionary turnover of kinetochore repertoires, we favor Aristotle’s resolution of the paradox of the Ship of Theseus: although the matter (composition) of the kinetochore varies over time, the formal cause - or design - remains the same.
Highlights.
CenH3 plays an essential role in chromosome segregation of most eukaryotes.
At least three CenH3-independent chromosome segregation systems have evolved: in trypanosomes, insects and during meiosis in nematodes.
The inner kinetochore protein CENP-T is essential in some organisms but dispensable in others.
CENP-B-like proteins have arisen multiple times in evolution.
Acknowledgments
We thank Kate Grey, Rini Kasinathan, Lisa Kursel, Matthew Miller, Paul Talbert, Jitendra Thakur and Janet Young for discussions and comments on this review. We are also grateful to the many helpful suggestions from the two anonymous reviewers. We are supported by a postdoctoral fellowship from the Jane Coffin Childs Foundation (IAD), grants from the Mathers Foundation (HSM), NIH R01-GM74108 (HSM) and the Howard Hughes Medical Institute (SH, HSM). SH and HSM are Investigators of the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Flemming W. Zellsubstanz, Kern und Zelltheilung. F.C.W. Vogel; 1882. [Google Scholar]
- 2.Cheeseman IM. The kinetochore. Cold Spring Harbor perspectives in biology. 2014;6:a015826. doi: 10.1101/cshperspect.a015826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rieder CL. The formation, structure, and composition of the mammalian kinetochore and kinetochore fiber. Int Rev Cytol. 1982;79:1–58. doi: 10.1016/s0074-7696(08)61672-1. [DOI] [PubMed] [Google Scholar]
- 4.Gonen S, et al. The structure of purified kinetochores reveals multiple microtubule-attachment sites. Nature structural & molecular biology. 2012;19:925–929. doi: 10.1038/nsmb.2358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Akiyoshi B, et al. Tension directly stabilizes reconstituted kinetochore-microtubule attachments. Nature. 2010;468:576–579. doi: 10.1038/nature09594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Westermann S, Schleiffer A. Family matters: structural and functional conservation of centromere-associated proteins from yeast to humans. Trends in cell biology. 2013;23:260–269. doi: 10.1016/j.tcb.2013.01.010. [DOI] [PubMed] [Google Scholar]
- 7.Barth TK, et al. Identification of novel Drosophila centromere-associated proteins. Proteomics. 2014;14:2167–2178. doi: 10.1002/pmic.201400052. [DOI] [PubMed] [Google Scholar]
- 8.Przewloka MR, et al. Molecular analysis of core kinetochore composition and assembly in Drosophila melanogaster. PloS one. 2007;2:e478. doi: 10.1371/journal.pone.0000478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Przewloka MR, et al. Searching for Drosophila Dsn1 kinetochore protein. Cell cycle. 2009;8:1292–1293. doi: 10.4161/cc.8.8.8159. [DOI] [PubMed] [Google Scholar]
- 10.Schleiffer A, et al. CENP-T proteins are conserved centromere receptors of the Ndc80 complex. Nature cell biology. 2012;14:604–613. doi: 10.1038/ncb2493. [DOI] [PubMed] [Google Scholar]
- 11.Drinnenberg IA, et al. Recurrent loss of CenH3 is associated with independent transitions to holocentricity in insects. eLife. 2014:3. doi: 10.7554/eLife.03676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Akiyoshi B, Gull K. Discovery of unconventional kinetochores in kinetoplastids. Cell. 2014;156:1247–1258. doi: 10.1016/j.cell.2014.01.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Akiyoshi B, Gull K. Evolutionary cell biology of chromosome segregation: insights from trypanosomes. Open biology. 2013;3:130023. doi: 10.1098/rsob.130023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Malik HS, Henikoff S. Major evolutionary transitions in centromere complexity. Cell. 2009;138:1067–1082. doi: 10.1016/j.cell.2009.08.036. [DOI] [PubMed] [Google Scholar]
- 15.Roach KC, et al. Rapid evolution of centromeres and centromeric/kinetochore proteins. In: Rama Singh JXRK, editor. Evolution in the Fast Lane: Rapidly Evolving Genes & Genetic Systems. Oxford University Press; 2012. pp. 83–93. [Google Scholar]
- 16.Palmer DK, et al. Purification of the centromere-specific protein CENP-A and demonstration that it is a distinctive histone. Proceedings of the National Academy of Sciences of the United States of America. 1991;88:3734–3738. doi: 10.1073/pnas.88.9.3734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Earnshaw WC, Rothfield N. Identification of a family of human centromere proteins using autoimmune sera from patients with scleroderma. Chromosoma. 1985;91:313–321. doi: 10.1007/BF00328227. [DOI] [PubMed] [Google Scholar]
- 18.Buchwitz BJ, et al. A histone-H3-like protein in C. elegans. Nature. 1999;401:547–548. doi: 10.1038/44062. [DOI] [PubMed] [Google Scholar]
- 19.Dawson SC, et al. The CenH3 histone variant defines centromeres in Giardia intestinalis. Chromosoma. 2007;116:175–184. doi: 10.1007/s00412-006-0091-3. [DOI] [PubMed] [Google Scholar]
- 20.Stoler S, et al. A mutation in CSE4, an essential gene encoding a novel chromatin-associated protein in yeast, causes chromosome nondisjunction and cell cycle arrest at mitosis. Genes & development. 1995;9:573–586. doi: 10.1101/gad.9.5.573. [DOI] [PubMed] [Google Scholar]
- 21.Henikoff S, et al. Heterochromatic deposition of centromeric histone H3-like proteins. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:716–721. doi: 10.1073/pnas.97.2.716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Talbert PB, Henikoff S. Histone variants--ancient wrap artists of the epigenome. Nature reviews Molecular cell biology. 2010;11:264–275. doi: 10.1038/nrm2861. [DOI] [PubMed] [Google Scholar]
- 23.Malik HS, Henikoff S. Phylogenomics of the nucleosome. Nat Struct Biol. 2003;10:882–891. doi: 10.1038/nsb996. [DOI] [PubMed] [Google Scholar]
- 24.Postberg J, et al. The evolutionary history of histone H3 suggests a deep eukaryotic root of chromatin modifying mechanisms. BMC evolutionary biology. 2010;10:259. doi: 10.1186/1471-2148-10-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Baldauf SL. The deep roots of eukaryotes. Science. 2003;300:1703–1706. doi: 10.1126/science.1085544. [DOI] [PubMed] [Google Scholar]
- 26.Zubacova Z, et al. Histone H3 Variants in Trichomonas vaginalis. Eukaryot Cell. 2012;11:654–661. doi: 10.1128/EC.00006-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Derelle R, et al. Bacterial proteins pinpoint a single eukaryotic root. Proceedings of the National Academy of Sciences of the United States of America. 2015;112:E693–699. doi: 10.1073/pnas.1420657112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Heckmann S, et al. Holocentric chromosomes of Luzula elegans are characterized by a longitudinal centromere groove, chromosome bending, and a terminal nucleolus organizer region. Cytogenetic and genome research. 2011;134:220–228. doi: 10.1159/000327713. [DOI] [PubMed] [Google Scholar]
- 29.Monen J, et al. Differential role of CENP-A in the segregation of holocentric C. elegans chromosomes during meiosis and mitosis. Nature cell biology. 2005;7:1248–1255. doi: 10.1038/ncb1331. [DOI] [PubMed] [Google Scholar]
- 30.Dumont J, et al. A kinetochore-independent mechanism drives anaphase chromosome separation during acentrosomal meiosis. Nature cell biology. 2010;12:894–901. doi: 10.1038/ncb2093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Muscat CC, et al. Kinetochore-independent chromosome segregation driven by lateral microtubule bundles. eLife. 2015;4:e06462. doi: 10.7554/eLife.06462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhong CX, et al. Centromeric retroelements and satellites interact with maize kinetochore protein CENH3. The Plant cell. 2002;14:2825–2836. doi: 10.1105/tpc.006106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gascoigne KE, et al. Induced ectopic kinetochore assembly bypasses the requirement for CENP-A nucleosomes. Cell. 2011;145:410–422. doi: 10.1016/j.cell.2011.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rago F, et al. Distinct organization and regulation of the outer kinetochore KMN network downstream of CENP-C and CENP-T. Current biology : CB. 2015;25:671–677. doi: 10.1016/j.cub.2015.01.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Talbert PB, et al. Adaptive evolution of centromere proteins in plants and animals. Journal of biology. 2004;3:18. doi: 10.1186/jbiol11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Milks KJ, et al. Dissection of CENP-C-directed centromere and kinetochore assembly. Molecular biology of the cell. 2009;20:4246–4255. doi: 10.1091/mbc.E09-05-0378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Screpanti E, et al. Direct binding of Cenp-C to the Mis12 complex joins the inner and outer kinetochore. Current biology : CB. 2011;21:391–398. doi: 10.1016/j.cub.2010.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Przewloka MR, et al. CENP-C is a structural platform for kinetochore assembly. Current biology : CB. 2011;21:399–405. doi: 10.1016/j.cub.2011.02.005. [DOI] [PubMed] [Google Scholar]
- 39.Carroll CW, et al. Dual recognition of CENP-A nucleosomes is required for centromere assembly. The Journal of cell biology. 2010;189:1143–1155. doi: 10.1083/jcb.201001013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kato H, et al. A conserved mechanism for centromeric nucleosome recognition by centromere protein CENP-C. Science. 2013;340:1110–1113. doi: 10.1126/science.1235532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Falk SJ, et al. Chromosomes. CENP-C reshapes and stabilizes CENP-A nucleosomes at the centromere. Science. 2015;348:699–703. doi: 10.1126/science.1259308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nishino T, et al. CENP-T provides a structural platform for outer kinetochore assembly. The EMBO journal. 2013;32:424–436. doi: 10.1038/emboj.2012.348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hori T, et al. CCAN makes multiple contacts with centromeric DNA to provide distinct pathways to the outer kinetochore. Cell. 2008;135:1039–1052. doi: 10.1016/j.cell.2008.10.019. [DOI] [PubMed] [Google Scholar]
- 44.Nishino T, et al. CENP-T-W-S-X forms a unique centromeric chromatin structure with a histone-like fold. Cell. 2012;148:487–501. doi: 10.1016/j.cell.2011.11.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Takeuchi K, et al. The centromeric nucleosome-like CENP-T-W-S-X complex induces positive supercoils into DNA. Nucleic acids research. 2014;42:1644–1655. doi: 10.1093/nar/gkt1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Amano M, et al. The CENP-S complex is essential for the stable assembly of outer kinetochore structure. The Journal of cell biology. 2009;186:173–182. doi: 10.1083/jcb.200903100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Singh TR, et al. MHF1-MHF2, a histone-fold-containing protein complex, participates in the Fanconi anemia pathway via FANCM. Molecular cell. 2010;37:879–886. doi: 10.1016/j.molcel.2010.01.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tanaka K, et al. CENP-C functions as a scaffold for effectors with essential kinetochore functions in mitosis and meiosis. Developmental cell. 2009;17:334–343. doi: 10.1016/j.devcel.2009.08.004. [DOI] [PubMed] [Google Scholar]
- 49.Foltz DR, et al. The human CENP-A centromeric nucleosome-associated complex. Nature cell biology. 2006;8:458–469. doi: 10.1038/ncb1397. [DOI] [PubMed] [Google Scholar]
- 50.Casola C, et al. Convergent domestication of pogo-like transposases into centromere-binding proteins in fission yeast and mammals. Molecular biology and evolution. 2008;25:29–41. doi: 10.1093/molbev/msm221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.d’Alencon E, et al. Characterization of a CENP-B homolog in the holocentric Lepidoptera Spodoptera frugiperda. Gene. 2011;485:91–101. doi: 10.1016/j.gene.2011.06.007. [DOI] [PubMed] [Google Scholar]
- 52.Mateo L, Gonzalez J. Pogo-like transposases have been repeatedly domesticated into CENP-B-related proteins. Genome biology and evolution. 2014;6:2008–2016. doi: 10.1093/gbe/evu153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nakagawa H, et al. Fission yeast CENP-B homologs nucleate centromeric heterochromatin by promoting heterochromatin-specific histone tail modifications. Genes & development. 2002;16:1766–1778. doi: 10.1101/gad.997702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Clarke L. Centromeres: proteins, protein complexes, and repeated domains at centromeres of simple eukaryotes. Current opinion in genetics & development. 1998;8:212–218. doi: 10.1016/s0959-437x(98)80143-3. [DOI] [PubMed] [Google Scholar]
- 55.Zaratiegui M, et al. CENP-B preserves genome integrity at replication forks paused by retrotransposon LTR. Nature. 2011;469:112–115. doi: 10.1038/nature09608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cam HP, et al. Host genome surveillance for retrotransposons by transposon-derived proteins. Nature. 2008;451:431–436. doi: 10.1038/nature06499. [DOI] [PubMed] [Google Scholar]
- 57.Masumoto H, et al. A human centromere antigen (CENP-B) interacts with a short specific sequence in alphoid DNA, a human centromeric satellite. The Journal of cell biology. 1989;109:1963–1973. doi: 10.1083/jcb.109.5.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Miga KH, et al. Centromere reference models for human chromosomes X and Y satellite arrays. Genome research. 2014;24:697–707. doi: 10.1101/gr.159624.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Goldberg IG, et al. Surprising deficiency of CENP-B binding sites in African green monkey alpha-satellite DNA: implications for CENP-B function at centromeres. Molecular and cellular biology. 1996;16:5156–5168. doi: 10.1128/mcb.16.9.5156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fachinetti D, et al. DNA Sequence-Specific Binding of CENP-B Enhances the Fidelity of Human Centromere Function. Developmental cell. 2015;33:314–327. doi: 10.1016/j.devcel.2015.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Henikoff S, et al. The centromere paradox: stable inheritance with rapidly evolving DNA. Science. 2001;293:1098–1102. doi: 10.1126/science.1062939. [DOI] [PubMed] [Google Scholar]
- 62.Chmatal L, et al. Centromere strength provides the cell biological basis for meiotic drive and karyotype evolution in mice. Current biology : CB. 2014;24:2295–2300. doi: 10.1016/j.cub.2014.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Marshall OJ, Choo KH. Putative CENP-B paralogues are not present at mammalian centromeres. Chromosoma. 2012;121:169–179. doi: 10.1007/s00412-011-0348-3. [DOI] [PubMed] [Google Scholar]
- 64.Fishman L, Saunders A. Centromere-associated female meiotic drive entails male fitness costs in monkeyflowers. Science. 2008;322:1559–1562. doi: 10.1126/science.1161406. [DOI] [PubMed] [Google Scholar]
- 65.Daniel A. Distortion of female meiotic segregation and reduced male fertility in human Robertsonian translocations: consistent with the centromere model of co-evolving centromere DNA/centromeric histone (CENP-A) Am J Med Genet. 2002;111:450–452. doi: 10.1002/ajmg.10618. [DOI] [PubMed] [Google Scholar]
- 66.Fishman L, Kelly JK. Centromere-associated meiotic drive and female fitness variation in Mimulus. Evolution. 2015;69:1208–1218. doi: 10.1111/evo.12661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Dawe RK, Henikoff S. Centromeres put epigenetics in the driver’s seat. Trends in biochemical sciences. 2006;31:662–669. doi: 10.1016/j.tibs.2006.10.004. [DOI] [PubMed] [Google Scholar]
- 68.Ross BD, et al. Stepwise evolution of essential centromere function in a Drosophila neogene. Science. 2013;340:1211–1214. doi: 10.1126/science.1234393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Basilico F, et al. The pseudo GTPase CENP-M drives human kinetochore assembly. eLife. 2014;3:e02978. doi: 10.7554/eLife.02978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Okada M, et al. The CENP-H-I complex is required for the efficient incorporation of newly synthesized CENP-A into centromeres. Nature cell biology. 2006;8:446–457. doi: 10.1038/ncb1396. [DOI] [PubMed] [Google Scholar]
- 71.Izuta H, et al. Comprehensive analysis of the ICEN (Interphase Centromere Complex) components enriched in the CENP-A chromatin of human cells. Genes to cells : devoted to molecular & cellular mechanisms. 2006;11:673–684. doi: 10.1111/j.1365-2443.2006.00969.x. [DOI] [PubMed] [Google Scholar]
- 72.Hornung P, et al. A cooperative mechanism drives budding yeast kinetochore assembly downstream of CENP-A. The Journal of cell biology. 2014;206:509–524. doi: 10.1083/jcb.201403081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hori T, et al. CENP-O class proteins form a stable complex and are required for proper kinetochore function. Molecular biology of the cell. 2008;19:843–854. doi: 10.1091/mbc.E07-06-0556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kagawa N, et al. The CENP-O complex requirement varies among different cell types. Chromosome Res. 2014 doi: 10.1007/s10577-014-9404-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Perpelescu M, Fukagawa T. The ABCs of CENPs. Chromosoma. 2011;120:425–446. doi: 10.1007/s00412-011-0330-0. [DOI] [PubMed] [Google Scholar]
- 76.Tromer E, et al. Widespread Recurrent Patterns of Rapid Repeat Evolution in the Kinetochore Scaffold KNL1. Genome biology and evolution. 2015;7:2383–2393. doi: 10.1093/gbe/evv140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Umbreit NT, et al. Kinetochores require oligomerization of Dam1 complex to maintain microtubule attachments against tension and promote biorientation. Nature communications. 2014;5:4951. doi: 10.1038/ncomms5951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Welburn JP, et al. The human kinetochore Ska1 complex facilitates microtubule depolymerization-coupled motility. Developmental cell. 2009;16:374–385. doi: 10.1016/j.devcel.2009.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Buttrick GJ, Millar JB. Ringing the changes: emerging roles for DASH at the kinetochore-microtubule Interface. Chromosome Res. 2011;19:393–407. doi: 10.1007/s10577-011-9185-8. [DOI] [PubMed] [Google Scholar]
- 80.Schmidt JC, et al. The kinetochore-bound Ska1 complex tracks depolymerizing microtubules and binds to curved protofilaments. Developmental cell. 2012;23:968–980. doi: 10.1016/j.devcel.2012.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Vleugel M, et al. Evolution and function of the mitotic checkpoint. Developmental cell. 2012;23:239–250. doi: 10.1016/j.devcel.2012.06.013. [DOI] [PubMed] [Google Scholar]
- 82.Abrieu A, et al. Mps1 is a kinetochore-associated kinase essential for the vertebrate mitotic checkpoint. Cell. 2001;106:83–93. doi: 10.1016/s0092-8674(01)00410-x. [DOI] [PubMed] [Google Scholar]
- 83.Lauze E, et al. Yeast spindle pole body duplication gene MPS1 encodes an essential dual specificity protein kinase. The EMBO journal. 1995;14:1655–1663. doi: 10.1002/j.1460-2075.1995.tb07154.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Espeut J, et al. Natural Loss of Mps1 Kinase in Nematodes Uncovers a Role for Polo-like Kinase 1 in Spindle Checkpoint Initiation. Cell reports. 2015;12:58–65. doi: 10.1016/j.celrep.2015.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Civril F, et al. Structural analysis of the RZZ complex reveals common ancestry with multisubunit vesicle tethering machinery. Structure. 2010;18:616–626. doi: 10.1016/j.str.2010.02.014. [DOI] [PubMed] [Google Scholar]
- 86.Griffis ER, et al. Spindly, a novel protein essential for silencing the spindle assembly checkpoint, recruits dynein to the kinetochore. The Journal of cell biology. 2007;177:1005–1015. doi: 10.1083/jcb.200702062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Williams BC, et al. Zwilch, a new component of the ZW10/ROD complex required for kinetochore functions. Molecular biology of the cell. 2003;14:1379–1391. doi: 10.1091/mbc.E02-09-0624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Furuyama S, Biggins S. Centromere identity is specified by a single centromeric nucleosome in budding yeast. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:14706–14711. doi: 10.1073/pnas.0706985104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Sullivan LL, et al. Genomic size of CENP-A domain is proportional to total alpha satellite array size at human centromeres and expands in cancer cells. Chromosome Res. 2011;19:457–470. doi: 10.1007/s10577-011-9208-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.McEwen BF, et al. Kinetochore fiber maturation in PtK1 cells and its implications for the mechanisms of chromosome congression and anaphase onset. The Journal of cell biology. 1997;137:1567–1580. doi: 10.1083/jcb.137.7.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Melters DP, et al. Holocentric chromosomes: convergent evolution, meiotic adaptations, and genomic analysis. Chromosome Res. 2012;20:579–593. doi: 10.1007/s10577-012-9292-1. [DOI] [PubMed] [Google Scholar]
- 92.Gassmann R, et al. An inverse relationship to germline transcription defines centromeric chromatin in C. elegans. Nature. 2012;484:534–537. doi: 10.1038/nature10973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Steiner FA, Henikoff S. Holocentromeres are dispersed point centromeres localized at transcription factor hotspots. eLife. 2014;3:e02025. doi: 10.7554/eLife.02025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Marques A, et al. Holocentromeres in Rhynchospora are associated with genome-wide centromere-specific repeat arrays interspersed among euchromatin. Proceedings of the National Academy of Sciences of the United States of America; 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Deans C, Maggert KA. What do you mean, “epigenetic”? Genetics. 2015;199:887–896. doi: 10.1534/genetics.114.173492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Voullaire LE, et al. A functional marker centromere with no detectable alpha-satellite, satellite III, or CENP-B protein: activation of a latent centromere? American journal of human genetics. 1993;52:1153–1163. [PMC free article] [PubMed] [Google Scholar]
- 97.Masumoto H, et al. The role of CENP-B and alpha-satellite DNA: de novo assembly and epigenetic maintenance of human centromeres. Chromosome Res. 2004;12:543–556. doi: 10.1023/B:CHRO.0000036593.72788.99. [DOI] [PubMed] [Google Scholar]
- 98.Henikoff JG, et al. A unique chromatin complex occupies young alpha-satellite arrays of human centromeres. Science advances. 2015:1. doi: 10.1126/sciadv.1400234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Heun P, et al. Mislocalization of the Drosophila centromere-specific histone CID promotes formation of functional ectopic kinetochores. Developmental cell. 2006;10:303–315. doi: 10.1016/j.devcel.2006.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Muller S, Almouzni G. A network of players in H3 histone variant deposition and maintenance at centromeres. Biochimica et biophysica acta. 2014;1839:241–250. doi: 10.1016/j.bbagrm.2013.11.008. [DOI] [PubMed] [Google Scholar]
- 101.McKinley KL, Cheeseman IM. Polo-like kinase 1 licenses CENP-A deposition at centromeres. Cell. 2014;158:397–411. doi: 10.1016/j.cell.2014.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Foltz DR, et al. Centromere-specific assembly of CENP-a nucleosomes is mediated by HJURP. Cell. 2009;137:472–484. doi: 10.1016/j.cell.2009.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Stoler S, et al. Scm3, an essential Saccharomyces cerevisiae centromere protein required for G2/M progression and Cse4 localization. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:10571–10576. doi: 10.1073/pnas.0703178104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Camahort R, et al. Scm3 is essential to recruit the histone h3 variant cse4 to centromeres and to maintain a functional kinetochore. Molecular cell. 2007;26:853–865. doi: 10.1016/j.molcel.2007.05.013. [DOI] [PubMed] [Google Scholar]
- 105.Dunleavy EM, et al. HJURP is a cell-cycle-dependent maintenance and deposition factor of CENP-A at centromeres. Cell. 2009;137:485–497. doi: 10.1016/j.cell.2009.02.040. [DOI] [PubMed] [Google Scholar]
- 106.Sanchez-Pulido L, et al. Common ancestry of the CENP-A chaperones Scm3 and HJURP. Cell. 2009;137:1173–1174. doi: 10.1016/j.cell.2009.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Mellone BG, et al. Assembly of Drosophila centromeric chromatin proteins during mitosis. PLoS Genet. 2011;7:e1002068. doi: 10.1371/journal.pgen.1002068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Schittenhelm RB, et al. Detrimental incorporation of excess Cenp-A/Cid and Cenp-C into Drosophila centromeres is prevented by limiting amounts of the bridging factor Cal1. Journal of cell science. 2010;123:3768–3779. doi: 10.1242/jcs.067934. [DOI] [PubMed] [Google Scholar]
- 109.Chen CC, et al. CAL1 is the Drosophila CENP-A assembly factor. The Journal of cell biology. 2014;204:313–329. doi: 10.1083/jcb.201305036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Phansalkar R, et al. Evolutionary insights into the role of the essential centromere protein CAL1 in Drosophila. Chromosome Res. 2012;20:493–504. doi: 10.1007/s10577-012-9299-7. [DOI] [PubMed] [Google Scholar]
- 111.Lacoste N, et al. Mislocalization of the centromeric histone variant CenH3/CENP-A in human cells depends on the chaperone DAXX. Molecular cell. 2014;53:631–644. doi: 10.1016/j.molcel.2014.01.018. [DOI] [PubMed] [Google Scholar]
- 112.Maddox PS, et al. Functional genomics identifies a Myb domain-containing protein family required for assembly of CENP-A chromatin. The Journal of cell biology. 2007;176:757–763. doi: 10.1083/jcb.200701065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Hayashi T, et al. Mis16 and Mis18 are required for CENP-A loading and histone deacetylation at centromeres. Cell. 2004;118:715–729. doi: 10.1016/j.cell.2004.09.002. [DOI] [PubMed] [Google Scholar]
- 114.Subramanian L, et al. Eic1 links Mis18 with the CCAN/Mis6/Ctf19 complex to promote CENP-A assembly. Open biology. 2014;4:140043. doi: 10.1098/rsob.140043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Fujita Y, et al. Priming of centromere for CENP-A recruitment by human hMis18alpha, hMis18beta, and M18BP1. Developmental cell. 2007;12:17–30. doi: 10.1016/j.devcel.2006.11.002. [DOI] [PubMed] [Google Scholar]
- 116.An S, et al. Mis16 Independently Recognizes Histone H4 and the CENP-A(Cnp1)-Specific Chaperone Scm3sp. Journal of molecular biology. 2015;427:3230–3240. doi: 10.1016/j.jmb.2015.08.022. [DOI] [PubMed] [Google Scholar]
- 117.Moree B, et al. CENP-C recruits M18BP1 to centromeres to promote CENP-A chromatin assembly. The Journal of cell biology. 2011;194:855–871. doi: 10.1083/jcb.201106079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Lermontova I, et al. Arabidopsis kinetochore null2 is an upstream component for centromeric histone H3 variant CenH3 deposition at centromeres. The Plant cell. 2013;25:3389–3404. doi: 10.1105/tpc.113.114736. [DOI] [PMC free article] [PubMed] [Google Scholar]