Abstract
Retrotransposons are mutagenic units able to move within the genome. Despite many defenses deployed by the host to suppress potentially harmful activities of retrotransposons, these genetic units have found ways to meld with normal cellular functions through processes of exaptation and domestication. The same host mechanisms targeting transposon mobility allow for expansion and rewiring of gene regulatory networks on an evolutionary time scale. Recent works demonstrating retrotransposon activity during development, cell differentiation and neurogenesis shed new light on unexpected activities of transposable elements. Moreover, new technological advances illuminated subtler nuances of the complex relationship between retrotransposons and the host genome, clarifying the role of retroelements in evolution, development and impact on human disease.
INTRODUCTION
Transposable elements (TE) are genomic units able to move within the genome of virtually all organisms [1]. More than half of our genome and likely over two-thirds of it [2] consists of TEs or their ancient relatives. Notably, in some plants such as maize, gene coding regions are just small islands “floating in a sea of retrotransposons” [3]. Transposons were discovered in maize and described as “controlling elements” by Barbara McClintock in the late 1940s [4]. TEs were considered “genomic junk” [5] until more recent works highlighted the substantial impact of mobile elements on shaping the genome and potentially rewiring its control [6-8]. Previous reviews give comprehensive historical analysis of the different perspectives, considering transposable elements either “controlling elements” with major functions in genome regulation, “selfish DNA” owing only to their selfish purpose of expansion [9] or, more recently “both mutualistic and extreme parasites” [6].
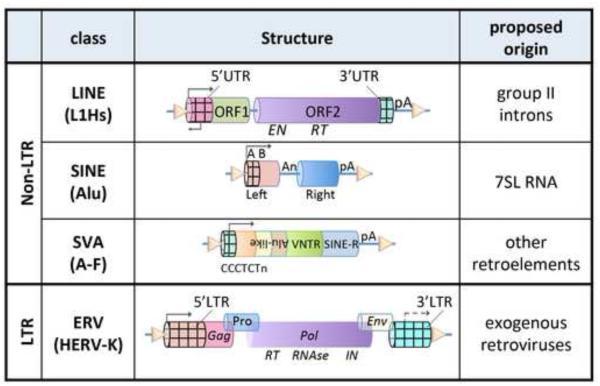
TEs are usually subdivided into two major classes: retrotransposons (class I) that use a “copy and paste” process for their replication and expansion and DNA transposons (class II) that use a “cut and paste” mechanism. Of these, only retrotransposons are active in the modern human genome and represent a prominent force of genomic evolution [6,10], although other mammals, notably certain bat taxa, have much more diverse TE populations, including active DNA transposons [11,12]. Retrotransposons’ classification and molecular features are summarized in Figure 1; see previous reviews [6,13-15]. Retrotransposons are classified into two categories: LTR (Long Terminal Repeat)-retrotransposons [16], and non-LTR retrotransposons including LINEs (Long Interspersed Nucleotide Elements), SINEs (Short Interspersed Nucleotide Elements) and in humans, SVAs (SINE-VNTR-Alu elements). LINEs, SVAs and LTR-elements are transcribed by RNA polymerase II while SINEs are transcribed by RNA polymerase III. Retroelement RNA is post-transcriptionally retrotranscribed and the cDNA is integrated into a new genomic location, a process called retrotransposition. The LINE elements are the only autonomous retrotransposons; SINEs and SVA elements depend on LINE-1 machinery for retrotransposition. Retrotransposons have distinct evolutionary histories. LTR endogenous retroviruses are clearly evolved from ancient viral infections of the germ-line and are maintained vertically in the germ line. Endogenous retroviruses encode Pro, Gag, Pol and sometime Env-like proteins like their exogenous cousins. Non-LTR retrotransposons like L1 are thought to have a common ancestor with group II introns, which often encode a reverse-transcriptase and can self-splice; they are likely ancestral to the modern spliceosome [17]. The Alu SINEs derive from cellular 7SL RNA, the RNA subunit of the signal recognition particle [18]. SVA elements are composite “patchworks” originating from distinct retroelements. Retroelements’ distinct origins underlie substantial differences in life-cycles, functional behavior and host-interactions; these differences have to be taken into account when retrotransposons are considered collectively.
Figure 1. Schematic of human retrotransposons.
Retrotransposons (class I transposons) are subclassified into two categories: LTR (Long Terminal Repeats)-retrotransposons, similar to exogenous retroviruses and further divided into multiple sub-families [16], and non-LTR retrotransposons which include LINEs (i.e. L1Hs), SINEs (i.e. Alus) and in humans, SVAs (SINE-VNTR-Alu elements, themselves subdivided into classes A-F). Retroelements are thought to have evolved differently and their proposed origin is reported. The transcriptionally active domains of the different retroelements are also indicated with checkered cylinders (see text). The triangles indicate target site duplications (TSD). The inverted “Alu-like” tag in the figure indicates the inverted orientation of these domains in SVA elements. Abbreviations: UTR= untranslated regions; ORF=open reading frame; EN=endonuclease domain; RT=reverse transcriptase domain; An= A-rich domain; pA=poly A; A B= domains essential for SINE transcription; VNTR= variable number target repeats; Pro= protease; Gag= group-specific antigen (coat protein) gene; Pol=polymerase (reverse transcriptase); Env=envelope gene; IN=integrase.
The repetitive nature of TEs makes them challenging to map onto a reference genome especially in the age of short read DNA sequencing. Therefore, despite their abundance in animals and plant genomes, the study of TEs, their evolution and behaviour and ultimately their impact on the host has lagged behind. Recently, technological advances in bioinformatics [19] including creation of comprehensive databases of annotated repetitive elements such as RepBase [20] and Dfam [21], incredible advances in DNA sequencing including long-read methods [22,23] and clearer knowledge of the genome from an ever-expanding variety of organisms (i.e. [24]), have injected new technological power into “transposonology”.
In recent years, the discovery of retrotransposon activity in somatic cells of the brain or their expression in specific stages of development and cell differentiation [23,25-28] raised the possibility of an actual beneficial role conferred by retrotransposon activity on the host, for example in neuronal plasticity [24,27,29]. Additionally, the study of somatic insertions in cancer [30-32] and the strive to elucidate the role of active retrotransposons in human pathologies [33,34], underscore the importance of retrotransposons not only on an evolutionary time scale but also in more dynamic and sometimes deleterious processes. These include epigenetic control and transcriptional regulation, cell differentiation and reprogramming [28], cancer initiation and progression [14], as well as processes like normal aging [35-37].
Here we briefly cover the consolidated impact of retrotransposons on genome architecture and genome evolution with particular focus on human retrotransposons and new findings validating a more dynamic impact on retrotransposon-induced regulation such as epigenetic and gene transcriptional regulation. These more recently identified effects of transposon mobilization may be less “disruptive” and imply a more subtle reshaping of genome control as opposed to gross effects on its structural organization. The more recent data supporting “positive” effects of retrotransposon activation will be discussed in light of the rediscovered view of retrotransposons as major drivers of genome evolution, a concept postulated by McClintock and by Britten and Davidson [38,39] more than a half-century ago.
RETROTRANSPOSON-INDUCED STRUCTURAL GENOMIC REORGANIZATION AND GENETIC INSTABILITY
Retrotransposon-induced genetic rearrangements
Because of their repetitive nature, retrotransposons are a source of chromatin instability and genomic rearrangements with deleterious consequences [15,40]. In the human genome, insertional inactivation and other genome rearrangements lead to a wide spectrum of genetic diseases including hemophilia, thalassemia and muscular dystrophy [41]. Retroelement-induced genetic rearrangements can be passive (due to the repetitive nature of TE) or active (directly caused by retrotransposition events) and of several types (reviewed in [13,15,40,42]): (I) non-allelic homologous recombination [41] mainly driven by Alu elements in humans, (II) insertional mutagenesis due to the “hopping” of retrotransposons within gene coding sequences; it causes diverse effects on target gene expression depending on intragenic location, orientation, length of the inserted sequence and other factors, (III) 3’ and 5’ transduction during which flanking genomic regions can be co-retrotransposed with the retroelements [43], (IV) trans-mediated mobilization of RNAs by “template switch” as is common with U6 RNA or by “template choice” as for the creation of processed pseudogenes (for more details see [44]).
Retrotransposon-induced changes in genome topology
Numerous lines of evidence demonstrated the organization of chromatin into nuclear domains [45] able to affect genome regulation and gene expression [46]. Heterochromatization of repeats through the processes described below have an effect on the topological distribution of genomic regions [46-49] and on the 3D organization of chromatin, likely through CTCF/cohesin binding to TEs [50,51]. It has been shown that at least 40% of the CTCF binding sites in the mouse genome (22.8% in human) are derived from SINEs elements [51]. The actual percentages are likely to be substantially higher thanks to ancient transposition events that can no longer be recognized due to mutational erosion. However, direct evidence for such retroelement-dependent reorganization is still lacking. Chromatin conformational studies using e.g. Hi-C focused on retrotransposons and their relevance in the evolution of genomic looping and long-range interactions could add a new dimension to the established relevance of TEs to the diaspora of TSS and TF binding sites discussed below [8,51]. It would be interesting to compare the topological distribution of common and species-specific retrotransposons in nuclei of cells from closely and distantly related organisms to evaluate the relevance of retroelements to extant genome architecture.
RETROTRANSPOSON-INDUCED CHANGES IN GENOME REGULATION
Transposon-induced changes in gene expression
Most genome scale work on retrotransposons examines TEs and flanking sequences in genomes of model organisms. This approach overlooks those insertions selected against during evolution that likely had the strongest effect on neighboring sequences. The majority of retrotransposon insertions are unsurprisingly found in non-coding or intronic regions. The effect of these insertions is usually thought to be neutral or affecting processes like alternative splicing, premature termination, long-range interactions or the creation of new regulatory regions. Han et al. [42,52] proposed a model according to which antisense LINE-1 insertion in an intron decreases RNA polymerase II processivity, reducing transcription rate of the genes in which L1 is inserted. This model called the “rheostat hypothesis” was demonstrated in vitro but direct evidence for it is limited. More recently, methylation status of intronic TEs in Arabidopsis thaliana was correlated with lower transcription of genes with TE insertions [53].
A classical example of retrotransposon dependent gene regulation in mice is the agouti gene (A). The efficiency in silencing an IAP (Intracisternal A-type Particle) element upstream of this gene correlates with a range of coat colors from yellow when the IAP is completely silenced to dark brown when the IAP is active [54].
A rigorous comparison of whole genome RNA expression with DNA sequencing identifying novel sites of insertion of in vitro expressed and “trackable” retroelements (i.e. recoded retroelements easily distinguishable from endogenous sequences [55]) will help answer these questions. Also, more systematic knowledge about the influence of stress or environmental cues on epigenetic control of retrotransposons as well as impact of transposons on phenotypic plasticity is still lacking. The stochastic and sometime incomplete nature of epigenetic silencing of retrotransposons may help explain and model complex systems such as cancer progression, lineage differentiation and brain complexity.
Epigenetic control and retrotransposon repression
Repetitive element mobilization represents a “dangerous” process for the host cell/organism when viewed from an individual perspective. Indeed, a clear “arms race” exists between retrotransposons and host defense mechanisms [56,57]. Conversely, it has been suggested that epigenetic control of the genome (a process likely rooted in transposon control, see below) paradoxically favored retroelement expansion by inhibiting excessive homologous recombination [58]. However, several mechanisms such as DNA and histone methylation and RNAi, actively suppress retrotransposon expression. The epigenetic mechanisms controlling retroelements may well follow retrotransposons during their movement “around” the genome and thereby modify the epigenetic control of retrotransposition targeted loci [59,60]. Below we describe ways of retrotransposon repression that contributed to sculpt the modern genome and its regulatory mechanisms.
Repression by cellular environment
An important factor that played an essential role in promoting retrotransposon expansion was probably the more permissive transcript survival environment of the eukaryotic cytoplasm, promoted by the 5’ cap/3’ polyA structure. On the other hand, cytoplasmic retrotransposons with longer mRNA half-life had to deal with the inhibitory effect of the nuclear membrane, which may represent a primitive defense against retrotransposition [61]. It has been proposed that disruption of the nuclear membrane during mitosis may be necessary for the entrance of the retrotransposon RNP particles into the nucleus [62]. The mechanisms that mediate nuclear translocation of retrotransposons are still unknown despite the obvious relevance to retrotransposon life-cycle/activity.
Repression by DNA methylation
DNA methylation is essential to control transposon repression in the germline and undifferentiated cells [63,64]. Recent studies suggest that LTR hypomethylation and activation of HERV-K and HERV-H endogenous retroviruses during early developmental stages directly contributes to pluripotency maintenance [25,26]. In the case of HERV-H, it can provide binding sites for TFs that mediate expression of pluripotency transcripts. Tellingly, HERV-H transcripts were also shown to function as lncRNAs important to maintain pluripotency [65] and HERV-K was shown to protect potentially vulnerable early embryonic cells from exogenous virus infection, suggesting exaptation.
Interestingly, CpG islands created by de novo somatic retrotransposition were shown to be hypomethylated, implying an inability of differentiated cells to silence newly mobilized elements [59]. Moreover, hypomethylated CpG islands create graded influence of hypomethylation on nearby CpGs, a phenomenon termed “sloping shores”. Because newly inserted retrotransposons created sloping shores, previously shown to influence neighbouring gene expression, it is likely that retrotransposition events in somatic cells influence gene expression of flanking regions by modifying their methylation status.
Repression by histone modifications
Histone modifications are also essential for retroelement repression particularly in undifferentiated cells [66,67]. G9a [68], Eset/Setdb1 [69,70], KAP1/ZNF proteins [37,71] and Lsd1/ KDM1A [72] repress retrotransposons in embryonic stem (ES) cells. Jenuwein and colleagues [73] showed that in mouse ES cells, Suv39h histone methyltransferase is recruited specifically to intact, full length LINE-1. Recent studies support the idea that retrotransposon and heterochromatin repression is initiated by random recruitment of TFs such as Pax3/9, ZNF proteins and homeodomain TFs [74]. Low-level mRNA transcribed upon random recruitment of these TFs may mediate silencing of repetitive element regions in undifferentiated cells [73,75]. Moreover, long non-coding RNAs (lncRNAs) can mediate HP1 and H3K9me3 independent recruitment of the H4K20me3 methyl transferase enzyme Suv4-20h2 onto non-pericentric or telomeric IAP retroelements in quiescent and terminally differentiated cells [76].
Histone deacetylation is also important for LINE-1 retrotransposition suppression in human embryonic carcinoma cells [77].
These studies collectively demonstrate that retrotransposons are targeted by several epigenetic modifications fundamental for establishment and maintenance of heterochromatin and that could have enabled rewiring of transcriptome regulation through retroelement mobility [54]. A better understanding of the key players in retrotransposon repression will certainly shed light on basic unanswered questions about the molecular mechanisms necessary for the establishment and maintenance of heterochromatin repression.
Repression by RNA interference (RNAi) and piRNAs
RNA interference is another layer of control that host organisms use to down-regulate retrotransposons [78-80]. Of the known RNA interference pathways (siRNA, miRNA, piRNA, rasiRNA, endo-siRNA) retrotransposons seem to involve a complex combination of DICER-dependent and -independent RNAi responses [80,81]. It has also been proposed that the miRNAs evolved from TEs [82]. Interestingly, piRNA-mediated silencing of TEs can spread to adjacent genes, affecting their expression in D. melanogaster [60]. Intriguingly, in Drosophila germline stem cells (GSC) establishment of heterochromatin by SETDB1 was shown to be essential for expression of piRNA targeting transposable elements [70] supporting a intertwining of transposon expression and host cell chromatin regulation.
Overall, the existence of diverse RNAi mechanisms targeting retrotransposons implies that RNAi control is another genomic process “expanded” way beyond retrotransposon control and that has been exapted and rewired by the host cells in response to TE activity.
RETROTRANSPOSON-INDUCED GENETIC INNOVATION
Retrotransposons can also impact gene regulation simply by inserting their own intrinsic regulatory sequences (promoters, cryptic splice sites, terminators, enhancers and insulators) in new genomic loci upon retrotransposition (Fig. 1)[6]. These regulatory elements can disrupt expression and structure of genes located near or within retrotransposition sites.
Alternative splicing broadens the diversity of protein repertoire produced from a “fixed” genome. Retrotransposition into an intron can alter its splicing through exon skipping, alternative donor or acceptor splice sites, intron retention [10,83] and exonization [84]. The LINE1 retrotransposon (L1) was shown to contain numerous functional splice acceptor and donor sites. L1 mRNA processing through splicing that renders the spliced retrotransposon inactive was proposed to serve as a host defense mechanism against excessively burdensome L1 transcription [85]. Unpublished data from our laboratory also support this hypothesis.
Retrotransposon promoter/enhancer sequences have donated regulatory elements pervasively to many genes and many if not all such sequences are targeted by several host signals and proteins. Despite many predicted transcription factor (TF) binding sites can be mapped on the L1 5’UTR, and on endogenous retroviral LTRs [86], few proteins have been directly shown to regulate retroelement transcription. RUNX3 [87], MeCP2 [88], p53 [89], SRY [90], Sp1 and Sp3 [91], YY1 [92,93] and more recently Oct4, Sox2, Nanog and KLF4 [25,26,94] are proteins demonstrated to mediate retrotransposon transcription. Recent work demonstrated recruitment of SIRT6 protein to the 5’UTR promoter of L1 and its repression through ribosylation of KAP1. Interestingly, SIRT6 recruitment and repression function decreases with aging, perhaps by redistribution of SIRT6 proteins on DNA damage sites in aged animals or senescent cells [37]. Future efforts should focus on elucidating TFs responsible for retrotransposon transcription in a more comprehensive manner.
Several studies show that retrotransposon regulatory units were expanded by being scattered genomewide through retrotransposition events and were subsequently “rewired” evolutionarily to provide many tissue specific gene regulatory elements (promoters/enhancers) [95]. Regulatory features (i.e. promoter or enhancer regions) of many retroelements have been shown to be co-opted by the host cells (exaptation)[96]. C-GATE is a publicly available catalogue of known putative and directly characterized transposons exapted by their host organisms [97]. These observations led to the hypothesis of relevant evolutionary importance for retroelement activity, e.g. in evolution of humans from the least common ancestor with other great apes [98].
Also, some LTR-retrotransposon derived proteins have been directly incorporated into host cellular processes in a phenomenon defined as “transposon domestication” [99,100]. The phenomenon of domestication/exaptation provides a framework for understanding the fundamental roles played by TEs in shaping genomic evolution in several organisms. These phenomena support the idea of a strong evolutionary benefit in retrotransposon mobilization although this must always be balanced with the clear negative effect at the level of the individual [7-9,51]. According to this viewpoint transposons are “dormant genetic units” with mutagenic and regulatory potential ready to be set into action and mobilized for adaptation to environmental stresses [101] (Figure 2). The concept of “genomic shock” initially hypothesized by Barbara McClintock finally found substantial supporting evidence in more recent studies showing that perhaps the majority of DNA regulatory regions (promoters, enhancers, TF binding sites) evolved from mobilization of TEs. Through various approaches, it has been shown that at least 20% of evolutionary conserved regulatory regions (TSS, enhancers or some TF binding sites) are derived from TEs [8,51,102]. These very comprehensive studies clearly demonstrate evolutionary relevance of retrotransposon mobility to the rewiring and selection of the most “fit” gene networks.
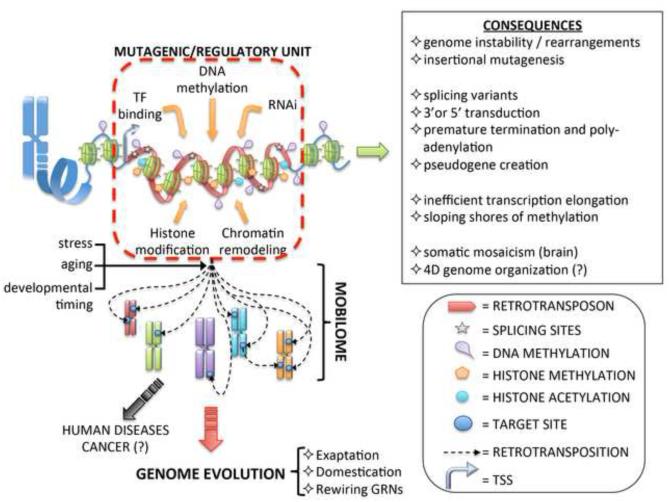
Figure 2. Retrotransposons shape genome regulation.
Retrotransposons contain several DNA controlling elements (transcription/enhancer domain, splicing signals, transcription factor (TF) binding sites, repression signals etc.) mobilized as part of retrotransposon activity in “jumping around” the genome. The immediate effect of retrotransposon activity is usually deleterious for the host cells (see top right insert and [10,13]) and in humans may lead to diseases such as cancer (black arrow). From an evolutionary standpoint retrotransposons can be defined as mutagenic units able to rewire and expand gene regulatory networks (GRNs) (red arrow). Stimuli such as stress, aging and specific developmental cues induce retrotransposon mobilization. Upon jumping, retrotransposon functional units can be exapted by the cell and in some cases retrotransposon features can be “domesticated” and incorporated into host cell functions, such as stem and germ cell regulation.
Despite the well-substantiated nature of transposons as “controlling elements”, the stress-induced activation of TEs is still not mechanistically well-characterized [103,104]. Stress activation (i.e. ionizing radiation, DNA damage, nitrogen starvation, severe adenine starvation or heat shock, adenovirus infection and cycloheximide treatment) has been shown for Ty1 in yeast and/or for SINEs and LINE-1 in human cells, but the TFs regulating such activation are mostly unknown. Recent studies demonstrated activation of retroelement activity upon circadian and aging stress [35,105,106]. A genome-wide catalog of factors and signals affecting transcription of L1 and other retrotransposons would be very valuable.
Moreover, domestication and exaptation can also help understanding the more recently described “advantageous” cellular effects of retroelements mobilization. For example, active retrotransposition upon environmental cues such as exercise has been demonstrated in hippocampus [27], an area with high adult neurogenic potential. This observation suggests a potential role of retrotransposition in the expansion of neuronal diversity in response to external stimuli. Controversy over the extent of retrotransposition activity in brain challenges such a mechanistic role [29,107-110]. In line with such ideas, recent work also demonstrated a fundamental role of L1 expression in fetal oocyte attrition, the process of prenatal elimination of most oocytes [111]. As mention above, HERV-K and HERV-H reactivation have been shown to play a role in maintaining pluripotency in ES cells [25,26,65]. Interestingly, certain retroelements are also reactivated in iPS cells demonstrating that the process of reprogramming and resetting of pluripotency induces and perhaps requires TE expression [112,113].
These studies suggest that exaptation of retrotransposons regulatory elements during cell development and differentiation induces inevitable reactivation of the corresponding retroelements still active in the genome during those same developmental states (as for HERV-H exaptation [26]). Domestication of retroelements proteins (as for HERV-K Env protein [25]), on the other hand, created a more direct need for expression of retroelements during specific cell stages. Finally, somatic reactivation of retroelements in tissues like the brain or oocyte during attrition may represent a type of domestication in the broader sense of the term, as reactivation and mobilization of specific retroelements may facilitate general processes like programmed cell death and neuronal plasticity.
IMPLICATIONS AND PERSPECTIVES
The newly gained information about retroelements made possible by great technological advances in bioinformatics and deep sequencing leaves us with many new questions. How does genome plasticity conferred by retrotransposons respond to different type of environmental stresses and what are the molecular mechanisms driving this stress-induced response? What is the impact of retroelement mobility in processes like cancer, cellular reprogramming and aging? What is the molecular relevance of retrotransposon activity in tissues like the brain or developing germ cells in which retrotransposons are not completely repressed? The more recent perspectives on the subject seem to suggest that in these contexts, TE activity can no longer be considered simply due to spurious and uncontrolled loss of regulation because of the newly identified “beneficial” roles conferred by retrotransposons that suggest the existence of retroelement functions co-opted and “safely” modulated by the host cell. Arguably, these views leave open the idea of “symbiotic retrotransposons” however antithetical this may seem to a dyed in the wool “selfish gene” devotee [9]. Thus, what may seem like TE activation during vulnerable windows in the organism development may in fact have provided an opportunity for exaptation of TEs for very specific cellular functions. It is even possible that controlled retrotransposition might provide a selective advantage in a very defined context (i.e. neuronal plasticity or pluripotency maintenance). Targeted genome engineering methods based on the CRISPR/Cas9 nuclease, may help answer these questions, providing clean in-vivo systems for studies of retrotransposon impact. It is now plausible to imagine construction of cells or organisms completely lacking active retrotransposons [114] and therefore determine their role in processes like cell differentiation, neurogenesis, aging, tumorigenic proliferation or genome stability.
The relevance of retrotransposons and TEs to nuclear architecture and 3D genome structure is still underdeveloped. Heterochromatin compartmentalization in distinct nuclear territories and the increasingly recognized importance of nuclear chromatin topology in processes like gene repression and activation hint at a potentially important role in genome architecture for retrotransposons, one of the major components of heterochromatin. Is retrotransposon mobility able to induce topological restructuring of the genome? Could alteration of retrotransposon repression do so? Are there phenotypic/functional consequences of retrotransposon activity that can be explained by an alteration of nuclear architecture? These questions are still open and surely poised to be answered soon.
Overall, the recent and more “dynamic” and nuanced view of transposons, demonstrates the enormous relevance of repetitive elements to genome control. From an evolutionary standpoint it is fair to consider the modern genome of several, if not all organisms, as a simple “snapshot” of their complex and ever-changing mobilome. The newly proposed “positive” cellular effects of retrotransposons can be explained considering that these effects evolved randomly from the activity of retroelements and have been fixed genomically because of the positive consequences they fortuitously offered to the host organism. The emergence of these apparent retrotransposon-dependent evolutionary “advantages” may help explain “windows” of reactivation that are not only tolerated by the host but actually create opportunities for evolution and adaptation of new functions. In this view the role of retrotransposon activity in human diseases can be considered a failed attempt towards evolutionary advance/adaptation (in the case of genetic disorders) or a “misuse” of the evolutionarily powerful but dangerous weapon represented by TEs (in the case of reactivation of retroelements in cancer).
Acknowledgements
We thank Neta Agmon, Leslie Mitchell and Emily Adney for the stimulating discussion and constructive criticism of the manuscript. Research on retrotransposons in our laboratory is supported by NIH grants P50GM107632 and 1R01CA161210.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Bibliography
- 1.Munoz-Lopez M, Garcia-Perez JL. DNA transposons: nature and applications in genomics. Curr Genomics. 2010;11:115–128. doi: 10.2174/138920210790886871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.de Koning AP, Gu W, Castoe TA, Batzer MA, Pollock DD. Repetitive elements may comprise over two-thirds of the human genome. PLoS Genet. 2011;7:e1002384. doi: 10.1371/journal.pgen.1002384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bushman F. Lateral DNA transfer : mechanisms and consequences. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2002. [Google Scholar]
- 4.Ravindran S. Barbara McClintock and the discovery of jumping genes. Proc Natl Acad Sci U S A. 2012;109:20198–20199. doi: 10.1073/pnas.1219372109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ohno S. So much "junk" DNA in our genome. Brookhaven Symp Biol. 1972;23:366–370. [PubMed] [Google Scholar]
- 6.Rebollo R, Romanish MT, Mager DL. Transposable elements: an abundant and natural source of regulatory sequences for host genes. Annu Rev Genet. 2012;46:21–42. doi: 10.1146/annurev-genet-110711-155621. (**) Outstanding review covering the history and development of the different interpretations and views of TEs as sources of genome plasticity in the human population. [DOI] [PubMed] [Google Scholar]
- 7.Lynch VJ, Leclerc RD, May G, Wagner GP. Transposon-mediated rewiring of gene regulatory networks contributed to the evolution of pregnancy in mammals. Nat Genet. 2011;43:1154–1159. doi: 10.1038/ng.917. (*) The transcriptomes of human, armadillo and opossum endometrial cells were compared to identify genes that had a key role in placenta evolution. This analysis identified the insulator function of the DNA transposon MER20 as an important contributor in the development of pregnancy in placental mammals. [DOI] [PubMed] [Google Scholar]
- 8.Faulkner GJ, Kimura Y, Daub CO, Wani S, Plessy C, Irvine KM, Schroder K, Cloonan N, Steptoe AL, Lassmann T, et al. The regulated retrotransposon transcriptome of mammalian cells. Nat Genet. 2009;41:563–571. doi: 10.1038/ng.368. (**) Human and mouse Cap Analysis Gene Expression (CAGE) libraries were used to show that many transcription start sites (TSS) occurred within repetitive elements (mainly retroelements). Analysis of the TE derived TSS revealed a tissue specific and "sharp" transcription initiation from these elements, which were also shown to be more active in developmental and cancerous tissues. [DOI] [PubMed] [Google Scholar]
- 9.Dawkins R. The selfish gene. 30th anniversary. Oxford University Press; Oxford ; New York: 2006. pp. xxiii–360. p. (**) This classic book that inspired a generation of TE biologists explores the idea of "selfish" genetic units that, during natural selection, can paradoxically lead to "cooperation of self-interested" clusters of genes that are mutually compatible. [Google Scholar]
- 10.Cordaux R, Batzer MA. The impact of retrotransposons on human genome evolution. Nat Rev Genet. 2009;10:691–703. doi: 10.1038/nrg2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tang Z, Zhang HH, Huang K, Zhang XG, Han MJ, Zhang Z. Repeated horizontal transfers of four DNA transposons in invertebrates and bats. Mob DNA. 2015;6:3. doi: 10.1186/s13100-014-0033-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thomas J, Phillips CD, Baker RJ, Pritham EJ. Rolling-circle transposons catalyze genomic innovation in a mammalian lineage. Genome Biol Evol. 2014;6:2595–2610. doi: 10.1093/gbe/evu204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goodier JL, Kazazian HH., Jr. Retrotransposons revisited: the restraint and rehabilitation of parasites. Cell. 2008;135:23–35. doi: 10.1016/j.cell.2008.09.022. (*) Comprehensive review of retroelements, their features and biology mainly focused on non-LTR retrotransposons LINEs and SINEs. [DOI] [PubMed] [Google Scholar]
- 14.Burns KH, Boeke JD. Human transposon tectonics. Cell. 2012;149:740–752. doi: 10.1016/j.cell.2012.04.019. (*) Comprehensive review of human active retroelements, their regulation, and new advances in the identification of retroelements and somatic insertions in the genome. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Prak ET, Kazazian HH., Jr. Mobile elements and the human genome. Nat Rev Genet. 2000;1:134–144. doi: 10.1038/35038572. [DOI] [PubMed] [Google Scholar]
- 16.Blomberg J, Benachenhou F, Blikstad V, Sperber G, Mayer J. Classification and nomenclature of endogenous retroviral sequences (ERVs): problems and recommendations. Gene. 2009;448:115–123. doi: 10.1016/j.gene.2009.06.007. [DOI] [PubMed] [Google Scholar]
- 17.Zimmerly S, Semper C. Evolution of group II introns. Mob DNA. 2015;6:7. doi: 10.1186/s13100-015-0037-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ullu E, Weiner AM. Human genes and pseudogenes for the 7SL RNA component of signal recognition particle. EMBO J. 1984;3:3303–3310. doi: 10.1002/j.1460-2075.1984.tb02294.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Makalowski W, Pande A, Gotea V, Makalowska I. Transposable elements and their identification. Methods Mol Biol. 2012;855:337–359. doi: 10.1007/978-1-61779-582-4_12. [DOI] [PubMed] [Google Scholar]
- 20.Bao W, Kojima KK, Kohany O. Repbase Update, a database of repetitive elements in eukaryotic genomes. Mob DNA. 2015;6:11. doi: 10.1186/s13100-015-0041-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wheeler TJ, Clements J, Eddy SR, Hubley R, Jones TA, Jurka J, Smit AF, Finn RD. Dfam: a database of repetitive DNA based on profile hidden Markov models. Nucleic Acids Res. 2013;41:D70–82. doi: 10.1093/nar/gks1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chaisson MJ, Huddleston J, Dennis MY, Sudmant PH, Malig M, Hormozdiari F, Antonacci F, Surti U, Sandstrom R, Boitano M, et al. Resolving the complexity of the human genome using single-molecule sequencing. Nature. 2015;517:608–611. doi: 10.1038/nature13907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Richardson SR, Morell S, Faulkner GJ. L1 retrotransposons and somatic mosaicism in the brain. Annu Rev Genet. 2014;48:1–27. doi: 10.1146/annurev-genet-120213-092412. [DOI] [PubMed] [Google Scholar]
- 24.Albertin CB, Simakov O, Mitros T, Wang ZY, Pungor JR, Edsinger-Gonzales E, Brenner S, Ragsdale CW, Rokhsar DS. The octopus genome and the evolution of cephalopod neural and morphological novelties. Nature. 2015;524:220–224. doi: 10.1038/nature14668. (*) Analysis of the octopus genome, which shows a clear arms-race between host defense (ZNFs protein expansion) and retroelement activity, consistent with previous analysis of the human genome. In octopus, as in human, retroelements (SINEs) are highly expressed in neural tissues. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grow EJ, Flynn RA, Chavez SL, Bayless NL, Wossidlo M, Wesche DJ, Martin L, Ware CB, Blish CA, Chang HY, et al. Intrinsic retroviral reactivation in human preimplantation embryos and pluripotent cells. Nature. 2015;522:221–225. doi: 10.1038/nature14308. (**) HERVK is shown to be actively expressed during normal human embryogenesis. The HERVK protein Rec is sufficient to inhibit exogenous retroviruses infection and modulate the ribosome occupancy of several cellular RNAs. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang J, Xie G, Singh M, Ghanbarian AT, Rasko T, Szvetnik A, Cai H, Besser D, Prigione A, Fuchs NV, et al. Primate-specific endogenous retrovirus-driven transcription defines naive-like stem cells. Nature. 2014;516:405–409. doi: 10.1038/nature13804. (**) In human embryonic stem cells or induced pluripotent stem cells the HERVH retrotransposon is highly expressed and plays a key role in maintenance of pluripotency, likely through recruitment of LBP9 and other pluripotency transcription factors. [DOI] [PubMed] [Google Scholar]
- 27.Muotri AR, Zhao C, Marchetto MC, Gage FH. Environmental influence on L1 retrotransposons in the adult hippocampus. Hippocampus. 2009;19:1002–1007. doi: 10.1002/hipo.20564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tohonen V, Katayama S, Vesterlund L, Jouhilahti EM, Sheikhi M, Madissoon E, Filippini-Cattaneo G, Jaconi M, Johnsson A, Burglin TR, et al. Novel PRD-like homeodomain transcription factors and retrotransposon elements in early human development. Nat Commun. 2015;6:8207. doi: 10.1038/ncomms9207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Upton KR, Gerhardt DJ, Jesuadian JS, Richardson SR, Sanchez-Luque FJ, Bodea GO, Ewing AD, Salvador-Palomeque C, van der Knaap MS, Brennan PM, et al. Ubiquitous L1 mosaicism in hippocampal neurons. Cell. 2015;161:228–239. doi: 10.1016/j.cell.2015.03.026. (**) Single cell retrotransposon capture sequencing (RC-seq) was applied to human glia, hippocampal and cortical neurons to explore the features and characteristics of somatic LINE-1 insertions in neuronal cells. At least one retrotransposition event for every hippocampal neuron was detected as compared to a much lower frequency in glia. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hancks DC, Kazazian HH., Jr. Active human retrotransposons: variation and disease. Curr Opin Genet Dev. 2012;22:191–203. doi: 10.1016/j.gde.2012.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Solyom S, Kazazian HH., Jr. Mobile elements in the human genome: implications for disease. Genome Med. 2012;4:12. doi: 10.1186/gm311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rodic N, Steranka JP, Makohon-Moore A, Moyer A, Shen P, Sharma R, Kohutek ZA, Huang CR, Ahn D, Mita P, et al. Retrotransposon insertions in the clonal evolution of pancreatic ductal adenocarcinoma. Nat Med. 2015 doi: 10.1038/nm.3919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhao K, Du J, Han X, Goodier JL, Li P, Zhou X, Wei W, Evans SL, Li L, Zhang W, et al. Modulation of LINE-1 and Alu/SVA retrotransposition by Aicardi-Goutieres syndrome-related SAMHD1. Cell Rep. 2013;4:1108–1115. doi: 10.1016/j.celrep.2013.08.019. (*) In this work the authors identify the Deoxynucleoside Triphosphate Triphosphohydrolase SAMHD1 protein as potent cellular factor that suppresses LINE-1 activity in dividing cells. SAMDH1 dNTPase activity was shown to be dispensible for LINE-1 suppression but necessary for HIV-1 and HBV replication. Interestingly, SAMHD1 point mutantions from Aicardi Goutieres patients display compromised ability to suppress LINE-1 activity. This observations are in line with other published works that suggest the possible involvement of LINE-1 expression in autoimmune desorders as Aicardi-Goutieres syndrome. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim Y, Tarallo V, Kerur N, Yasuma T, Gelfand BD, Bastos-Carvalho A, Hirano Y, Yasuma R, Mizutani T, Fowler BJ, et al. DICER1/Alu RNA dysmetabolism induces Caspase-8-mediated cell death in age-related macular degeneration. Proc Natl Acad Sci U S A. 2014;111:16082–16087. doi: 10.1073/pnas.1403814111. (*) This study further elucidates the molecular mechanism that lead to age-related macular degeneration. This disease was shown to be caused by deficiency in DICER1 enzyme and consequent accumulation of Alu RNA. This accumulation induces caspase 8 mediated apoptosis through activation of NLRP3 inflasmmosome and IL18 signaling. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.De Cecco M, Criscione SW, Peterson AL, Neretti N, Sedivy JM, Kreiling JA. Transposable elements become active and mobile in the genomes of aging mammalian somatic tissues. Aging (Albany NY) 2013;5:867–883. doi: 10.18632/aging.100621. (*) This work shows an increased reactivation of mouse retroelements (LINE-1, B1 , B2 and MuD) in aged mice, an effect that can be mitigated through calorie restriction. Higher L1 and MusD expression was also detected in age-related cancers (naturally occurring lymphoma and hepatocellular carcinoma) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gorbunova V, Boeke JD, Helfand SL, Sedivy JM. Human Genomics. Sleeping dogs of the genome. Science. 2014;346:1187–1188. doi: 10.1126/science.aaa3177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Van Meter M, Kashyap M, Rezazadeh S, Geneva AJ, Morello TD, Seluanov A, Gorbunova V. SIRT6 represses LINE1 retrotransposons by ribosylating KAP1 but this repression fails with stress and age. Nat Commun. 2014;5:5011. doi: 10.1038/ncomms6011. (*) This work shows a novel regulation of LINE-1 retrotransposon. SIRT6 protein binds the LINE-1 promoter and mono-ADP ribosylates the KAP1 protein facilitating its repressive activity through HP1 recruitment. Upon aging and DNA damage induction SIRT6 is depleted from LINE-1 loci leading to derepression of these retroelements. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McClintock B. Chromosome organization and genic expression. Cold Spring Harb Symp Quant Biol. 1951;16:13–47. doi: 10.1101/sqb.1951.016.01.004. [DOI] [PubMed] [Google Scholar]
- 39.Britten RJ, Davidson EH. Gene regulation for higher cells: a theory. Science. 1969;165:349–357. doi: 10.1126/science.165.3891.349. [DOI] [PubMed] [Google Scholar]
- 40.Beck CR, Garcia-Perez JL, Badge RM, Moran JV. LINE-1 elements in structural variation and disease. Annu Rev Genomics Hum Genet. 2011;12:187–215. doi: 10.1146/annurev-genom-082509-141802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Deininger PL, Batzer MA. Alu repeats and human disease. Mol Genet Metab. 1999;67:183–193. doi: 10.1006/mgme.1999.2864. [DOI] [PubMed] [Google Scholar]
- 42.Han JS, Boeke JD. LINE-1 retrotransposons: modulators of quantity and quality of mammalian gene expression? Bioessays. 2005;27:775–784. doi: 10.1002/bies.20257. (**) Review formalizing the "reostat hypothesis", which states that retroelements can affect gene transcription through modulation of RNA polymerase processivity. [DOI] [PubMed] [Google Scholar]
- 43.Pickeral OK, Makalowski W, Boguski MS, Boeke JD. Frequent human genomic DNA transduction driven by LINE-1 retrotransposition. Genome Res. 2000;10:411–415. doi: 10.1101/gr.10.4.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Garcia-Perez JL, Doucet AJ, Bucheton A, Moran JV, Gilbert N. Distinct mechanisms for trans-mediated mobilization of cellular RNAs by the LINE-1 reverse transcriptase. Genome Res. 2007;17:602–611. doi: 10.1101/gr.5870107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y, Hu M, Liu JS, Ren B. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485:376–380. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cavalli G, Misteli T. Functional implications of genome topology. Nat Struct Mol Biol. 2013;20:290–299. doi: 10.1038/nsmb.2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Meuleman W, Peric-Hupkes D, Kind J, Beaudry JB, Pagie L, Kellis M, Reinders M, Wessels L, van Steensel B. Constitutive nuclear lamina-genome interactions are highly conserved and associated with A/T-rich sequence. Genome Res. 2013;23:270–280. doi: 10.1101/gr.141028.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Olins AL, Ishaque N, Chotewutmontri S, Langowski J, Olins DE. Retrotransposon Alu is enriched in the epichromatin of HL-60 cells. Nucleus. 2014;5:237–246. doi: 10.4161/nucl.29141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mamillapalli A, Pathak RU, Garapati HS, Mishra RK. Transposable element 'roo' attaches to nuclear matrix of the Drosophila melanogaster. J Insect Sci. 2013;13:111. doi: 10.1673/031.013.11101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Merkenschlager M, Odom DT. CTCF and cohesin: linking gene regulatory elements with their targets. Cell. 2013;152:1285–1297. doi: 10.1016/j.cell.2013.02.029. [DOI] [PubMed] [Google Scholar]
- 51.Sundaram V, Cheng Y, Ma Z, Li D, Xing X, Edge P, Snyder MP, Wang T. Widespread contribution of transposable elements to the innovation of gene regulatory networks. Genome Res. 2014;24:1963–1976. doi: 10.1101/gr.168872.113. (**) The analysis of ChIP-seq peaks derived from 26 pairs of orthologous human/mouse TFs and several epigenetic marks showed that about 20% of transcription factor binding sites derived from transposable element expansion. A particular interest is given to CTCF binding sites, which are highly enriched in retroelements and provides a possible link to the evolution of long-range genetic interactions of transposable elements. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Han JS, Szak ST, Boeke JD. Transcriptional disruption by the L1 retrotransposon and implications for mammalian transcriptomes. Nature. 2004;429:268–274. doi: 10.1038/nature02536. [DOI] [PubMed] [Google Scholar]
- 53.Le TN, Miyazaki Y, Takuno S, Saze H. Epigenetic regulation of intragenic transposable elements impacts gene transcription in Arabidopsis thaliana. Nucleic Acids Res. 2015;43:3911–3921. doi: 10.1093/nar/gkv258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Whitelaw E, Martin DI. Retrotransposons as epigenetic mediators of phenotypic variation in mammals. Nat Genet. 2001;27:361–365. doi: 10.1038/86850. (**) In this commentary article the authors hypothesize an important role of retrotransposons and retrotransposon silencing in genomic plasticity and in the establishment of distinct epigenotypes in otherwise genetically identical organisms. The importance of RNA mediated suppression and the idea possible relevance of retrotransposons in disease susceptibility is also discussed. [DOI] [PubMed] [Google Scholar]
- 55.An W, Dai L, Niewiadomska AM, Yetil A, O'Donnell KA, Han JS, Boeke JD. Characterization of a synthetic human LINE-1 retrotransposon ORFeus-Hs. Mob DNA. 2011;2:2. doi: 10.1186/1759-8753-2-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Friedli M, Trono D. The Developmental Control of Transposable Elements and the Evolution of Higher Species. Annu Rev Cell Dev Biol. 2015;31:429–451. doi: 10.1146/annurev-cellbio-100814-125514. [DOI] [PubMed] [Google Scholar]
- 57.Jacobs FM, Greenberg D, Nguyen N, Haeussler M, Ewing AD, Katzman S, Paten B, Salama SR, Haussler D. An evolutionary arms race between KRAB zinc-finger genes ZNF91/93 and SVA/L1 retrotransposons. Nature. 2014;516:242–245. doi: 10.1038/nature13760. (**) A classical example of the host/retrotransposon arms race whereby the authors show that ZNF91 and ZNF93 have evolved to repress specific retroelements and show counter-evolution of these retrotransposons to evade ZNF mediated repressions. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fedoroff NV. Presidential address. Transposable elements, epigenetics, and genome evolution. Science. 2012;338:758–767. doi: 10.1126/science.338.6108.758. [DOI] [PubMed] [Google Scholar]
- 59.Grandi FC, Rosser JM, Newkirk SJ, Yin J, Jiang X, Xing Z, Whitmore L, Bashir S, Ivics Z, Izsvak Z, et al. Retrotransposition creates sloping shores: a graded influence of hypomethylated CpG islands on flanking CpG sites. Genome Res. 2015;25:1135–1146. doi: 10.1101/gr.185132.114. (**) This work shows that in somatic cells newly retrotranspososed LINE1 elements are hypomethylated, creating "sloping shores" of hypomethylation in the flanking regions and possibly affecting gene expression. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lee YC. The Role of piRNA-Mediated Epigenetic Silencing in the Population Dynamics of Transposable Elements in Drosophila melanogaster. PLoS Genet. 2015;11:e1005269. doi: 10.1371/journal.pgen.1005269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Martin W, Koonin EV. Introns and the origin of nucleus-cytosol compartmentalization. Nature. 2006;440:41–45. doi: 10.1038/nature04531. [DOI] [PubMed] [Google Scholar]
- 62.Xie Y, Mates L, Ivics Z, Izsvak Z, Martin SL, An W. Cell division promotes efficient retrotransposition in a stable L1 reporter cell line. Mob DNA. 2013;4:10. doi: 10.1186/1759-8753-4-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Crichton JH, Dunican DS, Maclennan M, Meehan RR, Adams IR. Defending the genome from the enemy within: mechanisms of retrotransposon suppression in the mouse germline. Cell Mol Life Sci. 2014;71:1581–1605. doi: 10.1007/s00018-013-1468-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Li Z, Dai H, Martos SN, Xu B, Gao Y, Li T, Zhu G, Schones DE, Wang Z. Distinct roles of DNMT1-dependent and DNMT1-independent methylation patterns in the genome of mouse embryonic stem cells. Genome Biol. 2015;16:115. doi: 10.1186/s13059-015-0685-2. (*) This study shows a more complex mechanism of retroelement methylation than previously thought. DNMT3a/b and DNMT1 co-operate for methylation maintenance of retrotransposon elements. Further, in some instances gene expression is intriguingly activated by demethylation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lu X, Sachs F, Ramsay L, Jacques PE, Goke J, Bourque G, Ng HH. The retrovirus HERVH is a long noncoding RNA required for human embryonic stem cell identity. Nat Struct Mol Biol. 2014;21:423–425. doi: 10.1038/nsmb.2799. [DOI] [PubMed] [Google Scholar]
- 66.Hutnick LK, Huang X, Loo TC, Ma Z, Fan G. Repression of retrotransposal elements in mouse embryonic stem cells is primarily mediated by a DNA methylation-independent mechanism. J Biol Chem. 2010;285:21082–21091. doi: 10.1074/jbc.M110.125674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Leung DC, Lorincz MC. Silencing of endogenous retroviruses: when and why do histone marks predominate? Trends Biochem Sci. 2012;37:127–133. doi: 10.1016/j.tibs.2011.11.006. [DOI] [PubMed] [Google Scholar]
- 68.Leung DC, Dong KB, Maksakova IA, Goyal P, Appanah R, Lee S, Tachibana M, Shinkai Y, Lehnertz B, Mager DL, et al. Lysine methyltransferase G9a is required for de novo DNA methylation and the establishment, but not the maintenance, of proviral silencing. Proc Natl Acad Sci U S A. 2011;108:5718–5723. doi: 10.1073/pnas.1014660108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Matsui T, Leung D, Miyashita H, Maksakova IA, Miyachi H, Kimura H, Tachibana M, Lorincz MC, Shinkai Y. Proviral silencing in embryonic stem cells requires the histone methyltransferase ESET. Nature. 2010;464:927–931. doi: 10.1038/nature08858. [DOI] [PubMed] [Google Scholar]
- 70.Rangan P, Malone CD, Navarro C, Newbold SP, Hayes PS, Sachidanandam R, Hannon GJ, Lehmann R. piRNA production requires heterochromatin formation in Drosophila. Curr Biol. 2011;21:1373–1379. doi: 10.1016/j.cub.2011.06.057. (*) This study performed in Drosophila using germline stem cells, clearly demonstrates an intimate correlation between transposon repression/activation, host genome regulation and stem cell fate. The authors elegantly show that transcription of piRNA requires the activity of the SETDB1 H3K9 methyl transferase. Moreover, the authors show that transposon activation triggers differentiation of germline stem cells maybe through activation of a DNA damage response pathaway due to TE transposition. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rowe HM, Jakobsson J, Mesnard D, Rougemont J, Reynard S, Aktas T, Maillard PV, Layard-Liesching H, Verp S, Marquis J, et al. KAP1 controls endogenous retroviruses in embryonic stem cells. Nature. 2010;463:237–240. doi: 10.1038/nature08674. [DOI] [PubMed] [Google Scholar]
- 72.Macfarlan TS, Gifford WD, Agarwal S, Driscoll S, Lettieri K, Wang J, Andrews SE, Franco L, Rosenfeld MG, Ren B, et al. Endogenous retroviruses and neighboring genes are coordinately repressed by LSD1/KDM1A. Genes Dev. 2011;25:594–607. doi: 10.1101/gad.2008511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bulut-Karslioglu A, De La Rosa-Velazquez IA, Ramirez F, Barenboim M, Onishi-Seebacher M, Arand J, Galan C, Winter GE, Engist B, Gerle B, et al. Suv39h-dependent H3K9me3 marks intact retrotransposons and silences LINE elements in mouse embryonic stem cells. Mol Cell. 2014;55:277–290. doi: 10.1016/j.molcel.2014.05.029. (**) The SUv39h histone methyltransferase is shown to mainly target ERVs and LINEs. Specifically, Suv39h is recruited to the 5'UTR of full-length and active LINE1 elements in embryonic stem cells. In committed cells the Suv39h recruited elements are repressed by DNA methylation, which compensates for the decrease in H3K9me3. [DOI] [PubMed] [Google Scholar]
- 74.Bulut-Karslioglu A, Perrera V, Scaranaro M, de la Rosa-Velazquez IA, van de Nobelen S, Shukeir N, Popow J, Gerle B, Opravil S, Pagani M, et al. A transcription factor-based mechanism for mouse heterochromatin formation. Nat Struct Mol Biol. 2012;19:1023–1030. doi: 10.1038/nsmb.2382. [DOI] [PubMed] [Google Scholar]
- 75.Luteijn MJ, Ketting RF. PIWI-interacting RNAs: from generation to transgenerational epigenetics. Nat Rev Genet. 2013;14:523–534. doi: 10.1038/nrg3495. [DOI] [PubMed] [Google Scholar]
- 76.Bierhoff H, Dammert MA, Brocks D, Dambacher S, Schotta G, Grummt I. Quiescence-induced LncRNAs trigger H4K20 trimethylation and transcriptional silencing. Mol Cell. 2014;54:675–682. doi: 10.1016/j.molcel.2014.03.032. [DOI] [PubMed] [Google Scholar]
- 77.Garcia-Perez JL, Morell M, Scheys JO, Kulpa DA, Morell S, Carter CC, Hammer GD, Collins KL, O'Shea KS, Menendez P, et al. Epigenetic silencing of engineered L1 retrotransposition events in human embryonic carcinoma cells. Nature. 2010;466:769–773. doi: 10.1038/nature09209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ha H, Song J, Wang S, Kapusta A, Feschotte C, Chen KC, Xing J. A comprehensive analysis of piRNAs from adult human testis and their relationship with genes and mobile elements. BMC Genomics. 2014;15:545. doi: 10.1186/1471-2164-15-545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Buchon N, Vaury C. RNAi: a defensive RNA-silencing against viruses and transposable elements. Heredity (Edinb) 2006;96:195–202. doi: 10.1038/sj.hdy.6800789. [DOI] [PubMed] [Google Scholar]
- 80.Svoboda P. Renaissance of mammalian endogenous RNAi. FEBS Lett. 2014;588:2550–2556. doi: 10.1016/j.febslet.2014.05.030. [DOI] [PubMed] [Google Scholar]
- 81.Faulkner GJ. Retrotransposon silencing during embryogenesis: dicer cuts in LINE. PLoS Genet. 2013;9:e1003944. doi: 10.1371/journal.pgen.1003944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Roberts JT, Cardin SE, Borchert GM. Burgeoning evidence indicates that microRNAs were initially formed from transposable element sequences. Mob Genet Elements. 2014;4:e29255. doi: 10.4161/mge.29255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ayarpadikannan S, Lee HE, Han K, Kim HS. Transposable element-driven transcript diversification and its relevance to genetic disorders. Gene. 2015;558:187–194. doi: 10.1016/j.gene.2015.01.039. [DOI] [PubMed] [Google Scholar]
- 84.Huda A, Bushel PR. Widespread Exonization of Transposable Elements in Human Coding Sequences is Associated with Epigenetic Regulation of Transcription. Transcr Open Access. 2013;1 doi: 10.4172/2329-8936.1000101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Belancio VP, Roy-Engel AM, Deininger P. The impact of multiple splice sites in human L1 elements. Gene. 2008;411:38–45. doi: 10.1016/j.gene.2007.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Thornburg BG, Gotea V, Makalowski W. Transposable elements as a significant source of transcription regulating signals. Gene. 2006;365:104–110. doi: 10.1016/j.gene.2005.09.036. [DOI] [PubMed] [Google Scholar]
- 87.Yang N, Zhang L, Zhang Y, Kazazian HH., Jr. An important role for RUNX3 in human L1 transcription and retrotransposition. Nucleic Acids Res. 2003;31:4929–4940. doi: 10.1093/nar/gkg663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Muotri AR, Marchetto MC, Coufal NG, Oefner R, Yeo G, Nakashima K, Gage FH. L1 retrotransposition in neurons is modulated by MeCP2. Nature. 2010;468:443–446. doi: 10.1038/nature09544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Harris CR, Dewan A, Zupnick A, Normart R, Gabriel A, Prives C, Levine AJ, Hoh J. p53 responsive elements in human retrotransposons. Oncogene. 2009;28:3857–3865. doi: 10.1038/onc.2009.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Tchenio T, Casella JF, Heidmann T. Members of the SRY family regulate the human LINE retrotransposons. Nucleic Acids Res. 2000;28:411–415. doi: 10.1093/nar/28.2.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Fedorov AV, Lukyanov DV, Podgornaya OI. Identification of the proteins specifically binding to the rat LINE1 promoter. Biochem Biophys Res Commun. 2006;340:553–559. doi: 10.1016/j.bbrc.2005.12.040. [DOI] [PubMed] [Google Scholar]
- 92.Becker KG, Swergold GD, Ozato K, Thayer RE. Binding of the ubiquitous nuclear transcription factor YY1 to a cis regulatory sequence in the human LINE-1 transposable element. Hum Mol Genet. 1993;2:1697–1702. doi: 10.1093/hmg/2.10.1697. [DOI] [PubMed] [Google Scholar]
- 93.Athanikar JN, Badge RM, Moran JV. A YY1-binding site is required for accurate human LINE-1 transcription initiation. Nucleic Acids Res. 2004;32:3846–3855. doi: 10.1093/nar/gkh698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kunarso G, Chia NY, Jeyakani J, Hwang C, Lu X, Chan YS, Ng HH, Bourque G. Transposable elements have rewired the core regulatory network of human embryonic stem cells. Nat Genet. 2010;42:631–634. doi: 10.1038/ng.600. [DOI] [PubMed] [Google Scholar]
- 95.Cohen CJ, Lock WM, Mager DL. Endogenous retroviral LTRs as promoters for human genes: a critical assessment. Gene. 2009;448:105–114. doi: 10.1016/j.gene.2009.06.020. [DOI] [PubMed] [Google Scholar]
- 96.Brosius J, Gould SJ. On "genomenclature": a comprehensive (and respectful) taxonomy for pseudogenes and other "junk DNA". Proc Natl Acad Sci U S A. 1992;89:10706–10710. doi: 10.1073/pnas.89.22.10706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Rebollo R, Farivar S, Mager DL. C-GATE - catalogue of genes affected by transposable elements. Mob DNA. 2012;3:9. doi: 10.1186/1759-8753-3-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Marchetto MC, Narvaiza I, Denli AM, Benner C, Lazzarini TA, Nathanson JL, Paquola AC, Desai KN, Herai RH, Weitzman MD, et al. Differential L1 regulation in pluripotent stem cells of humans and apes. Nature. 2013;503:525–529. doi: 10.1038/nature12686. (**) In this work the authors compare human to non human primates (Chimpanzee and Bonobo) iPS cells finding a higher restriction of retrotransposition activity in human cells. This lower activity was shown to be dependent, at least in part, by the upregulation of PIWIL2 and APOBEC3B protein expression in human iPS cells compared to NHP iPS cells. Based on these comparison the authors suggested that L1 mobility have a strong adaptative significance and it could have been involved in differentially shaping the genomes of humans and NHPs. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Kaneko-Ishino T, Ishino F. The role of genes domesticated from LTR retrotransposons and retroviruses in mammals. Front Microbiol. 2012;3:262. doi: 10.3389/fmicb.2012.00262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Henke C, Strissel PL, Schubert MT, Mitchell M, Stolt CC, Faschingbauer F, Beckmann MW, Strick R. Selective expression of sense and antisense transcripts of the sushi-ichi-related retrotransposon--derived family during mouse placentogenesis. Retrovirology. 2015;12:9. doi: 10.1186/s12977-015-0138-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Boeke JD. Retrotransposons. RNA Genetics. 1987:59–103. II: Retroviruses, Viroids, and RNA Recombination. [Google Scholar]
- 102.Lindblad-Toh K, Garber M, Zuk O, Lin MF, Parker BJ, Washietl S, Kheradpour P, Ernst J, Jordan G, Mauceli E, et al. A high-resolution map of human evolutionary constraint using 29 mammals. Nature. 2011;478:476–482. doi: 10.1038/nature10530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kim C, Rubin CM, Schmid CW. Genome-wide chromatin remodeling modulates the Alu heat shock response. Gene. 2001;276:127–133. doi: 10.1016/s0378-1119(01)00639-4. [DOI] [PubMed] [Google Scholar]
- 104.Li TH, Schmid CW. Differential stress induction of individual Alu loci: implications for transcription and retrotransposition. Gene. 2001;276:135–141. doi: 10.1016/s0378-1119(01)00637-0. [DOI] [PubMed] [Google Scholar]
- 105.Belancio VP, Blask DE, Deininger P, Hill SM, Jazwinski SM. The aging clock and circadian control of metabolism and genome stability. Front Genet. 2014;5:455. doi: 10.3389/fgene.2014.00455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.deHaro D, Kines KJ, Sokolowski M, Dauchy RT, Streva VA, Hill SM, Hanifin JP, Brainard GC, Blask DE, Belancio VP. Regulation of L1 expression and retrotransposition by melatonin and its receptor: implications for cancer risk associated with light exposure at night. Nucleic Acids Res. 2014;42:7694–7707. doi: 10.1093/nar/gku503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Evrony GD, Cai X, Lee E, Hills LB, Elhosary PC, Lehmann HS, Parker JJ, Atabay KD, Gilmore EC, Poduri A, et al. Single-neuron sequencing analysis of L1 retrotransposition and somatic mutation in the human brain. Cell. 2012;151:483–496. doi: 10.1016/j.cell.2012.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Evrony GD, Lee E, Mehta BK, Benjamini Y, Johnson RM, Cai X, Yang L, Haseley P, Lehmann HS, Park PJ, et al. Cell lineage analysis in human brain using endogenous retroelements. Neuron. 2015;85:49–59. doi: 10.1016/j.neuron.2014.12.028. (**) This report takes advantage of high-coverage whole-genome sequencing of single neurons coupled with single cell transposable element analyzer (scTea) for lineage tracing of neurons in human brain. This approach confirmed somatic retrotransposition at different stages of neuronal development as also reported by the same group in 2012 [107]. The rate of retrotransposition extrapolated from this analysis is quite low (less than 0.6 unique somatic insertions per cells) and in contrast with the one reported by Upton et. al [29] (13.7 somatic L1 insertions per hippocampal neuron) and Coufal et al. [110] (80 somatic L1 insertions per brain cell) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Reilly MT, Faulkner GJ, Dubnau J, Ponomarev I, Gage FH. The role of transposable elements in health and diseases of the central nervous system. J Neurosci. 2013;33:17577–17586. doi: 10.1523/JNEUROSCI.3369-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Coufal NG, Garcia-Perez JL, Peng GE, Yeo GW, Mu Y, Lovci MT, Morell M, O'Shea KS, Moran JV, Gage FH. L1 retrotransposition in human neural progenitor cells. Nature. 2009;460:1127–1131. doi: 10.1038/nature08248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Malki S, van der Heijden GW, O'Donnell KA, Martin SL, Bortvin A. A role for retrotransposon LINE-1 in fetal oocyte attrition in mice. Dev Cell. 2014;29:521–533. doi: 10.1016/j.devcel.2014.04.027. (*) LINE-1 is shown to play a key role in the activation of fetal oocyte attrition (FOA), highlighting a "positive" effect of retrotranspon activation. Elevated expression of LINE-1 induces oocyte elimination. FOA is suggested to serve to eliminate oocytes with excessive L1 expression. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Wissing S, Munoz-Lopez M, Macia A, Yang Z, Montano M, Collins W, Garcia-Perez JL, Moran JV, Greene WC. Reprogramming somatic cells into iPS cells activates LINE-1 retroelement mobility. Hum Mol Genet. 2012;21:208–218. doi: 10.1093/hmg/ddr455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Friedli M, Turelli P, Kapopoulou A, Rauwel B, Castro-Diaz N, Rowe HM, Ecco G, Unzu C, Planet E, Lombardo A, et al. Loss of transcriptional control over endogenous retroelements during reprogramming to pluripotency. Genome Res. 2014;24:1251–1259. doi: 10.1101/gr.172809.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Yang L, Guell M, Niu D, George H, Lesha E, Grishin D, Aach J, Shrock E, Xu W, Poci J, et al. Genome-wide inactivation of porcine endogenous retroviruses (PERVs) Science. 2015 doi: 10.1126/science.aad1191. [DOI] [PubMed] [Google Scholar]