Abstract
The human MET gene imparts a replicated risk for autism spectrum disorder (ASD), and is implicated in the structural and functional integrity of brain. MET encodes a receptor tyrosine kinase, MET, which plays a pleiotropic role in embryogenesis and modifies a large number of neurodevelopmental events. Very little is known, however, on how MET signaling engages distinct cellular events to collectively affect brain development in ASD-relevant disease domains. Here, we show that MET protein expression is dynamically regulated and compartmentalized in developing neurons. MET is heavily expressed in neuronal growth cones at early developmental stages and its activation engages small GTPase Cdc42 to promote neuronal growth, dendritic arborization, and spine formation. Genetic ablation of MET signaling in mouse dorsal pallium leads to altered neuronal morphology indicative of early functional maturation. In contrast, prolonged activation of MET represses the formation and functional maturation of glutamatergic synapses. Moreover, manipulating MET signaling levels in vivo in the developing prefrontal projection neurons disrupts the local circuit connectivity made onto these neurons. Therefore, normal time-delimited MET signaling is critical in regulating the timing of neuronal growth, glutamatergic synapse maturation and cortical circuit function. Dysregulated MET signaling may lead to pathological changes in forebrain maturation and connectivity, and thus contribute to the emergence of neurological symptoms associated with ASD.
INTRODUCTION
Normal brain development is dictated by a plethora of growth factors and their receptor protein tyrosine kinases 1. Molecular signaling via MET receptor tyrosine kinase (RTK), and its ligand hepatocyte growth factor (HGF), plays a pleiotropic role in the ontogenesis of multiple organs 2, 3. MET and HGF are also expressed in the developing nervous system of rodents, monkeys and humans 4–7, and are known to mediate a multitude of neurodevelopmental events including neural induction 8, neurotrophic and chemotrophic effects of developing axons 6, 9, and motogenic effects of specialized central neuron populations 10. Further corroborating its pleiotropic role in the nervous system, MET signaling also promotes neuronal survival 11, 12, and facilitates development of neuronal dendritic morphology under in vitro cultured conditions 13.
The human MET gene is a well-established risk factor for autism spectrum disorder 14–17, a highly heritable psychiatric condition defined by disrupted ontogeny of neural connectivity 18. Notably, a non-coding promoter variant of the human MET gene, the rs1858830 ‘C’ allele, which reduces MET transcription and protein translation, is associated with increased ASD risk 14, 15. The role of MET in autism etiology is further supported by its selective expression in brain structures involved in higher levels of cognition, social and language skills, and executive functions 4, 7. In addition, human MET gene transcription can be regulated by FOXP2 and MeCP2 7, 19, factors known to affect ASD-related circuits development in humans 20, 21. The rs1858830 ‘C’ risk allele also predicts atypical fMRI activation and deactivation patterns of human brain to social stimuli, and is correlated with reduced structural and functional connectivity in temporoparietal lobes 22, areas known to selectively express MET 4. Therefore, existing literature strongly suggests that MET signaling converges on cellular substrates relevant to ASD etiology.
Importantly, MET expression is tightly regulated at both spatial and temporal domains of the developing brain. Peak levels of MET expression in mice and monkeys coincide with a period of rapid postnatal neuronal growth and synaptogenesis 4, 5, but undergo precipitous down-regulation during synapse pruning and maturation stage. Currently, the functional significance of this time-delimited signaling is largely unknown. Whether down-regulation of MET expression is required for further development of glutamatergic synapse and refinement of circuit connections is an intriguing and open question. We and others have previously shown that in forebrain-specific Met conditional knockout mice, MET loss-of-function results in a region- and cell type- specific alterations in neuronal structure 5 and synaptic function 23, 24. In this study, we hypothesized that MET is capable of profoundly shaping neural morphology development and glutamatergic synapse formation, particularly in MET-expressing brain structures involved in higher cognitive function. We tested this hypothesis by investigating how altered MET signaling impacts the developmental trajectory of neuronal growth, maturation and synaptic connectivity in the mouse forebrain in search for a potential biological underpinning of the ASD risk conferred by MET.
MATERIALS AND METHODS
A detailed description can be found in Supplementary Information.
Mice
C57Bl6 mice were used for embryonic primary neuron cultures, prefrontal cortex (PFC) slice cultures and in utero electroporation studies. The day of vaginal plug detection was designated as E0.5 and the day of birth as P0. Dorsal pallial-specific conditional Met mutant mice (Metfx/fx; Emx1cre, ‘cKO’) were generated and genotyped using previously described protocols 5. In this model, cre-mediated excision of Met exon 16 leads to a loss-of-function ‘kinase-dead’ MET protein 25 (Figure 3a). For all studies, samples were coded so that the experimenter was blind to genotype. All procedures using mice were approved by the Institutional Animal Care and Use Committees of University of Southern California and the University of Arizona and conformed to NIH guidelines.
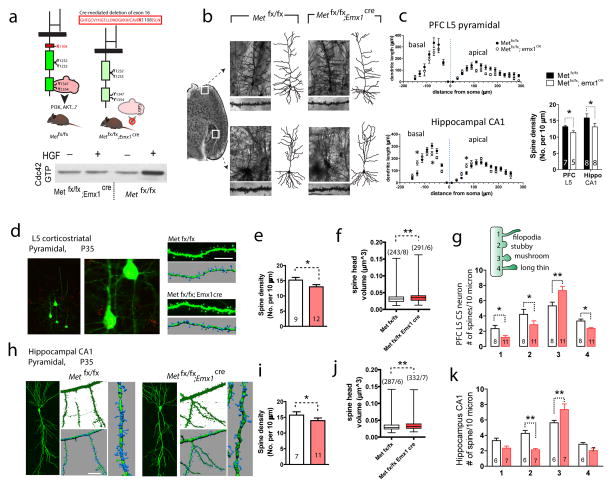
Figure 3. Conditional Met knockout leads to morphological alterations in dendritic and spine structures in forebrain neurons.
(a) Schematic representation of dorsal pallium specific Met cKO (Met fx/fx; emx1cre) mice. Cre-mediated excision of Exon 16, which encodes a critical ATP-binding site (Lys1108), inactivates the intracellular tyrosine kinase activity of MET and abrogates adaptor protein recruitment and downstream signaling. Consistently, HGF treatment in cKO PFC slices failed to induce an increase in pY1234/1235 (n = 2, quantification not shown). (b) Photomicrograph of Golgi-stained prefrontal (infra-limbic) L5 pyramidal neuron, and hippocampus CA1 neuron in Metfx/fx and cKO brains. A representative dendritic arbor reconstructed in 3D using Neurolucida for each neuron was shown on the right, and dendritic segments from which dendritic spine density was calculated were shown below the image. (c) Sholl analysis of PFC L5 and hippocampal CA1 neuron dendritic complexity. A significant reduction of dendritic length was found in PFC L5 (two-way ANOVA with Bonferroni post hoc test, F(1, 288) = 19.06. n = 7 neurons/3 mice for Metfx/fx and n = 8/4 for cKO. *p < 0.05). No significant overall difference was found for CA1 neurons (F(1, 315) = 0.003. n = 8 neurons/3 mice for Metfx/fx and n = 9/3 for cKO), while dendritic length was distributed differently (*p < 0.05). Dendritic spine density showed a significant reduction in both prefrontal L5 pyramidal neuron and CA1 pyramidal neuron in cKO brains (n = 10 neurons/3 mice, *p < 0.05, t test). (d) Dendritic spine morphology in P35 PFC L5 corticostriatal neurons, labeled by red retrograde ‘beads’, was reconstructed using Imaris and high-resolution confocal Z stack images. (e) Met cKO PFC L5 neurons showed a significant reduction in spine density (t19 = 3.67, * p < 0.05). (f) Dendritic spine head volume in cKO PFC L5 neurons is significantly larger (**p < 0.01). (g) Dendritic spines from cKO PFC L5 neurons show significantly increased number in ‘mushroom’ type (**p < 0.01), while significant decreased numbers in all other categories (* p < 0.05). Numbers labeled on bar denotes number of neurons, based on which > 500 spines were analyzed for each group. (h) Morphological reconstruction of CA1 neuron spines. (i–j) CA1 neurons from cKO mice show a significant reduction in spine density (i, t16 = 3.42, *p < 0.05), and a significant increase in dendritic spine head volume (j, **p < 0.01). (k) CA1 neuron dendritic spines from cKO mice show significant increased number in ‘mushroom’ type (**p < 0.01), and significant decreased numbers in the ‘stubby’ category (**p < 0.01).
DNA constructs, neuronal transfection and in utero electroporation (IUEP)
Met cDNA vector that expresses the full ORF of mice Met gene (NM_008591), the RNAi construct for Met, and their validity has been reported 24. Cdc42 RNAi sequence (GCAGTCACAGTTATGATTG) was cloned into pSuper and verified by co-transfection with Cdc42 cDNA in HEK293 cells (data not shown). Dominant negative Cdc42 (Cdc42-N17) was from D. Webb (Vanderbilt University). We co-transfected plasmids with pEGFP-C3 (‘GFP’) vector using calcium phosphate precipitation method 24. To target the developing layer 5 (L5) PFC neurons in vivo, IUEP experiments were carried out using time-pregnant mice at E13.5, similar to a previous report 26. The embryos’ right prefrontal region was targeted for electroporation. The offspring undergone IUEP were collected for electrophysiology recording and LSPS mapping studies at postnatal (P) 23–25.
Primary neuron culture and PFC slice cultures
Cortical or hippocampal neuron cultures were prepared from embryonic day E16.5 time pregnant C57BL/6 mice and grown on 12-mm glass coverslips at low densities (~10,000 cells/mm2), as described previously 24. Neurons were harvested at day in vitro (DIV) 5–28 for immunocytochemistry (ICC) labeling or electrophysiology recording. For organotypic cultures of cortical slices containing PFC, postnatal day 6–7 C57Bl6 mouse brains were sliced into 250 μm-thick sagittal sections using a vibratome (Leica V-1200S) under sterilized conditions. Slices were cultured on Millicell® cell culture inserts (Millipore, Billerica, MA) in DMEM/F12 medium supplemented with 10% FBS. HGF (50 ng/ml, Millipore) was added at 14 day in culture where necessary. Patch clamp recordings on L5 PFC neurons were conducted at day 22–25.
Growth cone fraction isolation, biochemistry, Western blot and immunocytochemistry
We isolated growth cone-enriched fraction of postnatal day 0 mouse cortices using the methods reported by Pfenninger et al. 27 (Figure 1F, Supplementary Information). Western blot analysis of tissue/cells and ICC staining were performed and quantified using standard methods 24. We micro-dissected PFC and hippocampus regions in brain slices and homogenized the tissues in cold NP40 cell lysis buffer (FNN0021, Life Technologies) supplemented 1:50 protease inhibitor cocktail (Sigma P8340) and 1 mM PMSF. Total proteins were quantified using a micro-BCA assay (Pierce). Sample proteins are separated on 9% SDS-polyacrylamide gels, transferred to PVDF membranes (Immobilon-P, Millipore), incubated with primary antibodies overnight at 4 °C, and followed by incubation with HRP-conjugated secondary antibodies (Promega, Madison, WI) for 2 h at RT. Enhanced chemiluminescence method (ECL Plus, GE Healthcare) was used to quantify the signal. Optical density of immunoreactive bands was quantified by densitometry using Image J. All antibodies and their sources are listed on Supplementary Information. To probe activated (GTP-bound) Cdc42, we used a Cdc42 activation assay kit (Millipore, 17-441) according to manufacturer’s directions.
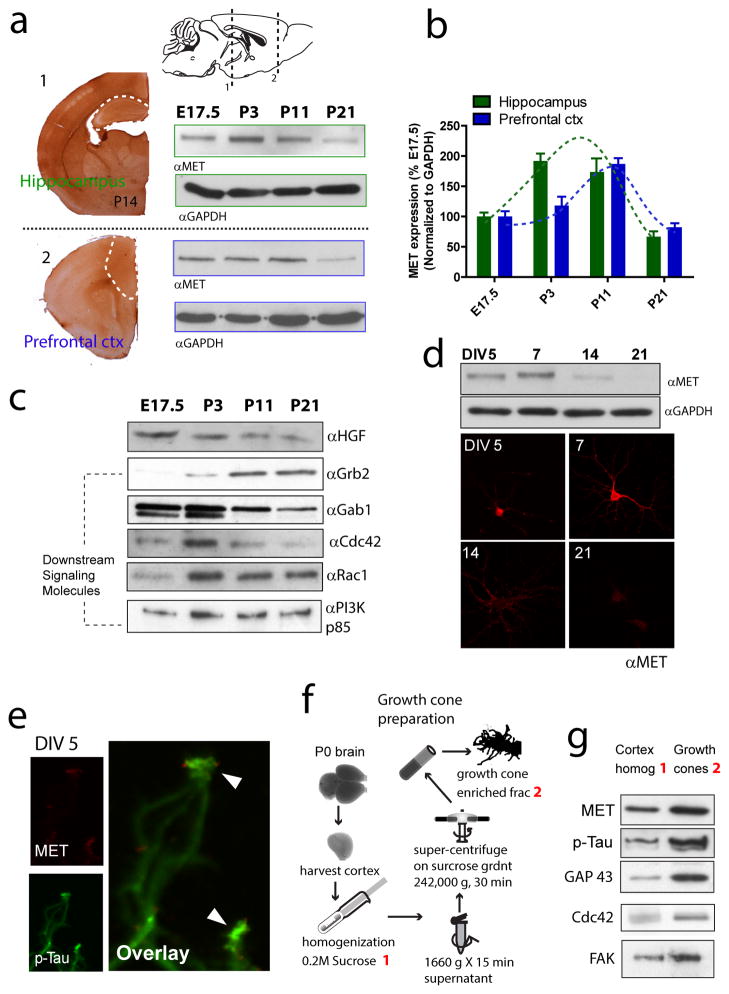
Figure 1. Temporal profile of MET expression and growth cone enrichment in the developing forebrain.
(a) IHC staining in P14 coronal slices reveals strong immunoreactivity in the hippocampus and PFC (delineated by white dotted lines). MET protein expression in the hippocampus and PFC was probed by Western blot analysis at E17.5, P3, P11 and P21. (b) Quantification of a. Peak expression of MET protein (normalized to GAPDH levels) occurs around P3-11, and declines precipitously thereafter. (c) Expression levels of the MET receptor ligand, HGF, and molecules potentially mediating downstream signaling events (Gab1, Grb2, Cdc42, Rac1 and PI3K/P85) (n = 2). (d) Western blot analysis (n = 3) and immunocytochemistry staining of MET protein in cultured low-density hippocampal neurons during in vitro development. (e) High resolution confocal imaging reveals strong MET immunoreactivity localized to axon growth cone tip (n = 4 experiments). (f) Diagram depicting isolation of growth cone fractions from P0 mouse brain. (g) Western blot analysis of MET reveals enriched protein levels at the growth cone, similar to other proteins known to be concentrated in growth cones (p-Tau, GAP43, Cdc42 and FAK) during early neuronal development (n = 3, quantification not shown).
Acute PFC slice preparation, drug treatment, patch clamp whole cell recording
To test the signaling competency of MET protein in the developing mouse brain, we adopted an ex vivo live brain slice preparation. Acute sagittal brain slices at postnatal day 9–10 containing PFC were sliced and kept alive in artificial cerebrospinal fluid (ACSF). Slices were treated with HGF (50 ng/ml, 30min) at 35°C with or without MET kinase inhibitors PHA665752 or SU11274, or PI3K inhibitors Wortmanin or LY294002. PFC tissues from these slices were then dissected, and subjected to Western blot analysis or small GTPase activation assay.
Miniature excitatory postsynaptic currents (mEPSCs) were recorded in cultured cortical neurons and L5 PFC neurons in cultured slices using whole cell patch clamp technique. Neurons or slices were perfused with ACSF containing (in mM), 126 NaCl, 2.5 KCl, 26 NaHCO3, 2 CaCl2, 1 MgCl2, 1.25 NaH2PO4, and 10 glucose, saturated with 95% O2 and 5% CO2. Neurons were visualized under an Olympus BX51WI microscope equipped with infrared DIC optics and epifluorescence. The patch electrode (4–6 MΩ) contains (in mM): 130 K-gluconate, 4 KCl, 2 NaCl, 10 HEPES, 4 ATP-Mg, 0.3 GTP-Na, 1 EGTA and 14 phosphocreatine (pH 7.2, 295 mOsm). In experiments where AMPA/NMDAR current ratio and ifenprodil sensitivity were measured, the electrode contained (in mM): 125 Cs gluconate, 5 tetraethylammonium–Cl, 10 Hepes, 8 NaCl, 5 QX314-Cl, 4 Mg2+-ATP, 2.5 CsCl, 0.3 Na3GTP, 0.2 EGTA, 10 phosphocreatine, and adjusted to pH 7.2, 280–290 mOsm. For all whole cell experiments, series resistance (Rs) was monitored, only stable (<15% change) cells with Rs < 25 MΩ throughout the recordings were included. All chemical ligands used in this study, including Wortmanin, LY294002, PHA665732, SU11274, ifenprodil, R-CPP and MNI-caged glutamate were purchased from Tocris/R&D Systems.
Laser scanning photostimulation (LSPS) for cortical circuit mapping
For LSPS mapping of prefrontal circuits, essentially the same protocol was reported previously 23,28 (Supplementary Information). Pups born to IUEP dams were sacrificed at P22-25. Sagittal slices containing the right PFC were made. Individual GFP-expressing PFC L5 neurons were selected for patch clamp recording and LSPS mapping based on anatomical landmarks delineated by the online Allen Brain Atlas (prelimbic area, layer 5). Slices were perfused in modified ACSF (4mM Ca2+, 4mM Mg2+), with 0.2mM MNI-caged glutamate and 5μM R-CPP. Electrophysiological signals were amplified with a Multiclamp 700B amplifier (Molecular Devices) and acquired with BNC 6259 data acquisition boards (National Instruments) under control of Ephus software (http://www.ephus.org). Signals were digitized and acquired at 10 kHz. LSPS mapping on slices was performed at RT in a customized recording chamber mounted on a motorized stage (Sutter MPC-78). Neuronal soma selected for recording was at least 50 μm below the slice surface to minimize truncation of dendritic structures. LSPS mapping combined with glutamate uncaging were performed with a 4× objective lens (NA 0.8; Olympus) and 20 mW, 1-ms UV laser (355 nm; DPSS Lasers) pulses. Digital images and stimulation locations were registered using a CCD camera (Retiga 2000DC, Qimaging). Neuronal responses to laser uncaging/mapping were analyzed offline with custom Matlab routines.
Neuronal morphological analysis
In some experiments, a rapid Golgi staining kit (FD Neurotechnologies) was used to reveal neuronal dendritic morphology. P35-38 brains were fixed for 10–12 days, sectioned into 250 μm slices, and further processed according to the manufacturer’s protocol. Dendritic arbors were traced using Neurolucida (MicroBrightField). Morphometric parameters were extracted using Neurolucida explorer, similar to that described previously 29. To reconstruct spines in PFC L5 corticostriatal (CS) or CA1 neurons from Met cKO mice and their littermate controls, we used biocytin injection to reveal morphology. For CS neurons, we first injected 50 nanoliter retrograde red fluorescent latex microspheres (‘beads’, Lumafluor Inc.) into left dorsal striatum, then prepared right PFC slice at 24 h or later. L5 beads-positive CS neurons were dialyzed (500 pA holding current for 10 min) with 1% biocytin dissolved in the potassium-based internal electrode solution. Slices were then fixed by 4% PFA overnight followed by avidin-Alexa488 (Life technologies) conjugation for 24 h. Confocal Z stacks were acquired from secondary apical branches 150–200 μm away from soma using a Zeiss 710 microscope (63x oil objective, NA 1.4, with 2x digital zoom). We used Imaris software (Bitplane, V7.01) to measure spine parameters and classify them into ‘filopodia’, ‘stubby’, ‘mushroom’ and ‘long thin’ categories using Imaris default spine classification module 30.
Statistical Analysis
Quantitative results were expressed as mean ± SEM. Data were processed using Microsoft Excel, Matlab and GraphPad Prism 5.0. Sample size was estimated by power analysis using an R script that takes pre-specified effect size, type I and II errors as input arguments. The same algorithm was used to estimate sample size of mice. Student t test or one-way/two-way analysis of variations (ANOVA) was used when data passed normality test. Potential data outliers were identified as those differ at least three standard deviations from the mean and excluded from analysis where applicable. For IUEP experiments, mice were grouped randomly. Sample size n represents number of independent treatments obtained from at least three mice. Kolmogorov-Smirnov (K-S) test was used to compare cumulative distribution of mEPSC amplitude and inter-event intervals. Spine head size (non-normal distribution) comparison was made using non-parametric Mann-Whitney test. Dunnett’s multiple comparisons or Bonferroni post hoc test was used for between-group comparisons after ANOVA reveals overall significance. p < 0.05 was considered statistically significant.
RESULTS
Developmental MET expression in mouse forebrain and sub-cellular localization in growth cones
The developing mouse forebrain is known to transiently express MET 5. We started by probing the levels of the MET protein and its putative molecular signaling constituents across multiple prenatal and postnatal developmental stages. We focused on two forebrain regions, the PFC and the hippocampus, areas implicated in psychiatric conditions of developmental origin 31. Immunohistochemistry (IHC) labeling using specific MET antibody (sc-8057) revealed strong MET immunoreactivity at P14 in both PFC and hippocampus (Figure 1a). Next, protein samples were collected from PFC and hippocampus in live brain slices at embryonic day (E) 17.5 and postnatal days (P) 3, 11 and 21 to probe MET levels using Western blot. MET protein expression in both PFC and hippocampus peaks during P3 to P11, and significantly declines by P21 (Figure 1a and b, n = 3). MET ligand, HGF, and other proteins implicated in MET signaling pathway, including adaptor proteins Gab1 and Grb2, potential downstream signaling molecules PI3K, small GTPases Cdc42 and Rac1 are all abundantly expressed in PFC tissue across E17.5-P21 (Figure 1c, n = 2, quantification not shown). The temporal regulation of MET expression seems to be cell autonomous, as Western blot analysis and immunocytochemistry staining of primary cultured hippocampal neurons also displayed a similar temporally-regulated expression pattern (Figure 1d).
Importantly, confocal imaging revealed strong MET immunoreactivity at the tip of growth cones (Figure 1e), indicating MET-mediated signaling may be functionally coupled with molecular machinery operating at the growth cone, and may account for HGF-stimulated neuronal growth 13. To further confirm this sub-cellular distribution of MET during early neuronal growth stage, we prepared growth cone fractions from P0 mouse cortex (Figure 1f) 27, and found MET protein is indeed enriched in the growth cone fraction, along with other known growth cone-enriched proteins (p-Tau, GAP43, Cdc42 and FAK) (Figure 1g). We therefore hypothesize that the growth cone localization of MET is indicative for a functional role in mediating early neuronal growth.
MET-induced neuronal growth effects depend on its kinase activity and downstream activation of small GTPase Cdc42
In epithelial cells, MET is known to signal through adaptor proteins leading to an activation of small GTPases, including Rap1, and Rho family small GTPases RhoA, Cdc42 and Rac1, which are responsible for the formation of filopodia and lamellipodia 32–34. In neurons, activation of Rho family small GTPases, such as Cdc42, is critical for dendritic and spine morphogenesis, axon growth, and motility 35, 36. We hypothesized that Rho family small GTPases may be activated and mediate MET effects on early neuronal growth. We first tested the competency of MET signaling in an ex vivo brain slice preparation, utilizing the endogenous MET and its native signaling components. Acute live PFC slices were prepared from P9-10 mice and treated with HGF (50 ng/ml, 30min) or saline (Figure 2a). We found HGF induces tyrosine phosphorylation of MET at Y1234/1235, an event that is critical in mediating MET-induced growth in peripheral cell types 2. HGF induced pY1234/1235 can be partially antagonized by 200nM PHA665752, an MET kinase activity inhibitor (Figure 2b), but cannot be blocked by 100nM Wortmanin, a PI3K inhibitor (Figure 2c). We next tested whether the Rho family small GTPases are activated in neurons. Both ex vivo P9-10 PFC slices (n = 4) and in vitro cultured cortical neurons (DIV11, n = 3) were treated with HGF (50ng/ml, 30min), and probed for activated, GTP-bound form of Cdc42 (see Methods). We consistently observed a dramatic increase in Cdc42-GTP (Figure 2d). These results for the first time revealed the involvement of small GTPases following MET activation in neurons. Furthermore, Cdc42 activation depends on both MET kinase activity and PI3K function, as both PHA665752 (200nM) and Wortmanin (100nM) blocks the increase of Cdc42-GTP following HGF treatment (Figure 2e, n = 4). To overcome the potential issue of specificity of kinase inhibitors, we also tested another MET inhibitor SU11274 (10, 50 nM) and PI3K inhibitor LY294002 (5, 20 μM) on HGF-induced Cdc42 activation. Both of these inhibitors reduced Cdc42-GTP in a dose-dependent manner (Figure S1).
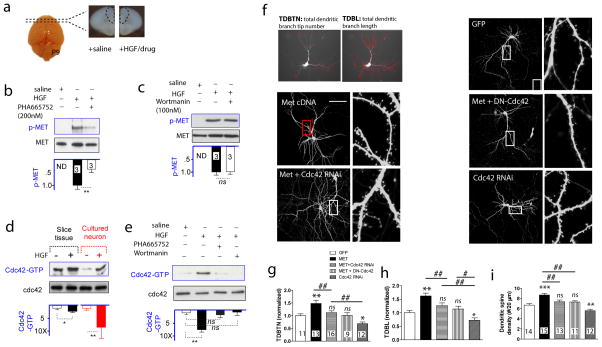
Figure 2. MET signaling activates the small GTPase Cdc42, which mediates its effects on dendritic and spine morphogenesis.
(a) Schematic illustration of an ex vivo prefrontal brain slice preparation (coronal sections from P9 mice) used for HGF treatment to initiate MET signaling. (b) MET activation, using p-MET (Y1234/1235) level as a surrogate, was induced by HGF, and blocked by MET kinase activity inhibitor PHA665752 (200 nM) (**p < 0.01. ND, not detected). (c) The PI3K inhibitor, Wortmanin (100 nM), did not affect p-MET levels, indicating PI3K activity was not required MET activation. (d) In both P9-10 ex vivo slices and DIV11 cortical neuron culture preparations, HGF (50 ng/ml) induces Cdc42 activation, measured by PAK P21 domain pull-down of the GTP-bound Cdc42. In contrast, no change of overall Cdc42 level was observed (*p < 0.05, **p < 0.01). (e) Cdc42 activation was blocked by either PHA665752 or Wortmanin, indicating it is dependent on MET kinase activity and downstream to PI3K activation (**p < 0.01). (f) Neuronal morphometric measurement using TDBTN and TDBL, and representative images of dendritic and spine morphology from cultured neurons transfected with Met cDNA, or in combination with Cdc42 loss-of -function plasmids (DN-Cdc42 or Cdc42 RNAi). (g) TDBTN quantification (normalized to GFP control neurons) reveals MET OE significantly increased TDBTN, which is significantly blocked by either DN-Cdc42 or Cdc42 RNAi co-transfection. Note Cdc42 RNAi alone significantly reduced TDBTN (*p < 0.05, **p < 0.01 compared with GFP, ns, not significant. ##p < 0.01. Number of neurons quantified listed on bar). (h) MET OE increases TDBL, an effect that is also blocked by co-transfection with DN-Cdc42 or Cdc42 RNAi (*p < 0.05, **p < 0.01, #p < 0.05, ##p < 0.01). (i) Enhanced MET signaling increases dendritic spine density, which is blocked by Cdc42 antagonism (**p < 0.01, ##p < 0.01, ns, not significant).
Previous studies indicate that MET affects neuronal growth, dendritic and spine morphology development 13, 24. We next tested whether Cdc42 mediate some of these effects. Cultured cortical neurons at DIV11 were transfected with Met cDNA or in combination with a dominant negative form of Cdc42 (T17N, DN-Cdc42), or Cdc42 interference RNA (Cdc42 RNAi) to knockdown endogenous Cdc42. GFP vector was co-transfected to reveal morphology. Quantification of neuronal morphology was done at DIV25 (Figure 2f). We quantified total dendritic branch tip number (TDBTN), total dendritic branch length (TDBL) and spine density for each group 37. We found that Met cDNA over-expression (MET OE) significantly increased TDBTN (F(4, 56) = 10.73, p < 0.001, one-way ANOVA with Dunnett’s post hoc test), an effect that was significantly blocked by co-transfection with DN-Cdc42 or Cdc42 RNAi (Figure 2g). Notably, Cdc42 RNAi alone significantly reduced TDBTN (p < 0.05), underscoring the importance of Cdc42 in dendritic morphology development 38. Similar conclusion was obtained when neurons were quantified using TDBL (F(4, 56) = 12.01, p < 0.001) (Figure 2h). In addition, we found MET OE significantly increased dendritic spine density (F(4, 60) = 15.4, p < 0.001), an effect that was also significantly blocked by DN-Cdc42 or Cdc42 RNAi (Figure 2i). Further corroborating these findings, MET RNAi-induced reduction of dendritic length and spine density was reversed by elevated Cdc42 activity, as a constitutive active form of Cdc42(V12) 38 completely antagonized MET effect (Figure S2). Taken together, these results reveal a novel molecular mechanism involving Cdc42 downstream to MET signaling that mediates MET effect on dendritic and spine development.
Dorsal pallial-specific conditional knockout of MET impacts neuronal morphology development
Previous studies show that MET is expressed by discrete subtypes of long-projecting neurons of the forebrain, including prefrontal cortex and the hippocampus CA1 4, 5. We next investigated the effect of MET loss-of-function on neuronal morphology during in vivo development using a dorsal pallial-specific Met conditional knockout (cKO) mice model (Metfx/fx; Emx1cre; see Methods) 5. Consistent with MET loss-of-function in cKO and the role of Cdc42 in mediating MET effects, HGF activation in P10 cKO slices did not lead to Cdc42 activation compared with its effects in Metfx/fx control slices (Figure 3a, n = 2). We next conducted a detailed morphological investigation on the projection neurons in Golgi-stained brain sections from juvenile (P35-38) cKO mice and their Metfx/fx littermate controls. We selected PFC L5 neurons and CA1 neurons for morphological reconstruction (Figure 3b). Sholl analysis 39 revealed an overall significant reduction of dendritic length at both apical and basal dendritic structure in cKO PFC L5 neurons (Figure 3c, n = 7 neurons for each genotype. F(1,288) = 19.1 for the genotype effect, p < 0.001). For CA1 neurons, although no significant genotype effects was observed (F(1,315) = 0.003) on the overall dendritic length, dendritic branching pattern was significantly altered in cKO. We also observed a significant reduction in the dendritic spine density in cKO neurons, quantified in secondary branches located 100–200 μm from soma (p < 0.05, for both PFC L5 and CA1 neurons, Student t test).
Because Golgi staining underestimates spine density, and is incapable of measuring spine size quantitatively 40, we used biocytin to fill PFC L5 and CA1 neurons, followed by avidin-Alexa488 to reveal fine spine morphology. To ensure the same neuronal type is selected for comparison, we injected red fluorescent ‘beads’ 23 in contralateral dorsal striatum to label the crossing corticostriatal (CS) neurons, and then selected beads-positive L5 CS neurons for spine morphology studies using Imaris reconstruction from confocal Z stack images (Figure 3d). Quantitative analysis revealed that cKO neurons exhibited significantly reduced spine density (Figure 3e. 13.4 ± 0.5 spines per 10 μm dendrite, compared with 15.3 ± 0.7 in Met fx/fx control. t20 = 4.23, p < 0.05) and enlarged spine head volume (Figure 3f, p < 0.01). In cKO, significantly fewer spines on L5 CS neurons falls into classification of ‘filopodia’ (1.21 ± 0.21 vs 2.35 ± 0.40 spines per 10 μm dendrite, p < 0.05), ‘stubby’ (2.86 ± 0.41 vs 4.21 ± 0.56, p < 0.05) and ‘long thin’ (2.38 ± 0.13 vs 3.35 ± 0.21, p < 0.05); while more spines are classified into ‘mushroom’ type (7.32 ± 0.44 vs 5.21 ± 0.48, p < 0.01) (Figure 3g), which are putative mature type of spines 41.
Next, we analyzed CA1 pyramidal neurons (Figure 3h), and found that spine density in cKO neurons is lower overall (Figure 3i. 13.8 ± 0.73 spines per 10 μm dendrite, compared with 15.7 ± 0.9 in Metfx/fx controls. p < 0.05). Similar to L5 CS neurons, we found that spine head volume in CA1 neurons is significantly enlarged (Figure 3j, p< 0.01). A significant increase in mature ‘mushroom’ type spine density (7.35 ± 0.62 vs 5.63 ± 0.32. p < 0.01) and a decrease in immature ‘stubby’ type spines (2.13 ± 0.16 vs 4.26 ± 0.38, p < 0.01) were observed in cKO neurons (Figure 3k). These morphology results indicate enlarged spine size at two major forebrain structures following MET loss-of-function. Considering that spine size and geometry are highly correlated with glutamatergic synapse function 42, these results strongly implicate altered synaptic function in these two forebrain structures at the development stage examined.
Prolonged MET signaling represses glutamatergic synapse formation and functional maturation
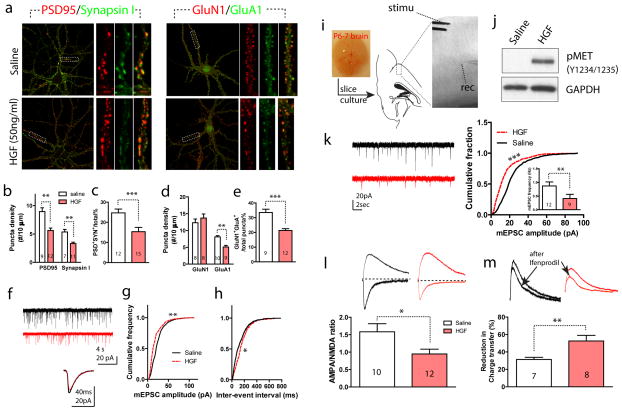
MET protein expression levels are tightly regulated across the temporal domain during forebrain development, with peak expression coinciding the principal periods of neurite outgrowth and synaptogenesis, followed rapid down-regulation thereafter 5 (Figure 1). The functional significance of down-regulation is not clear. We prolonged MET signaling by adding HGF during a period when MET protein is undergoing down-regulation. We tested this prolonged HGF effect on synapse formation in cultured cortical neurons and cortical slices containing the PFC region. HGF (50 ng/ml) was added to cultured cortical neuron at DIV14. We then used ICC to label two pairs of excitatory synapse markers (synapsin I + PSD95; GluN1 + GluA1) at DIV25. Each protein marker produced discrete punctate labeling, potentially reflecting a synaptic site. Putative functional synapses are represented by co-labeling of puncta with either marker pair (Figure 4a). Quantitative analysis (Figure 4b) reveals that HGF treatment significantly reduces the puncta density of both synapsin I and PSD95 (number of Synapin I+ puncta per 10 μm, saline, 5.35 ± 0.44; HGF, 3.32 ± 0.24. p < 0.01. PSD+ puncta, saline, 8.95 ± 0.67; HGF, 5.62 ± 0.44. p < 0.01). In addition, the fraction of putative functional synapse in HGF-treated neurons (15.4 ± 2.1%) was significantly lower than that from saline-treated control neurons (24.7 ± 1.9%) (Figure 4c, p < 0.001, t test). Similar conclusion was made when synapse density was quantified using GluN1/GluA1 co-labeling. HGF treatment significantly reduces the puncta density of GluA1 (Figure 4d. saline, 8.11 ± 0.35; HGF, 5.12 ± 0.44. p < 0.01), without altering the density of GluN1 labeling. Functional synapse defined by co-labeling of GluN1/GluA1 was also significantly lower in HGF treated neurons (21.3 ± 1.2%) compared with saline treated control neurons (33.5 ± 2.2%) (Figure 4e. p < 0.001, t-test). This reduced functional synapse was consistent with electrophysiological findings from these neurons (Figure 4f); compared with saline-treated neurons, mEPSCs from neurons with prolonged HGF treatment were generally of smaller amplitude (Figure 4g, p < 0.01) and longer inter-event intervals (Figure 4h, p < 0.05).
Figure 4. Prolonged HGF/MET signaling reduces functional glutamatergic synapse formation and represses its maturation.
(a) Representative images of immunocytochemistry staining of DIV25 cultured cortical neurons with or without HGF treatment. Two pairs of synapse markers, the post-synaptic protein PSD95 / presynaptic protein synapsin I; and the NMDA receptor subunit GluN1 / AMPA receptor subunit GluA1 were used to estimate functional synapse (colocalized puncta in yellow). (b) Quantification of staining in a. Neurons from HGF treated groups show significantly reduced puncta density in both PSD95 and synapsin I staining (**p < 0.01). HGF also significantly reduced the proportion of putative functional synapse number, defined by PSD95+/Synapsin I+ puncta (c. t25 = 7.73, ***p < 0.001). (d) HGF-treated neurons display reduced GluA1+ puncta density (t17 = 4.33, **p < 0.01). (e) HGF also significantly reduced the proportion of putative functional synapse number, defined by GluN1+/GluA1+ puncta (t19 = 7.84, ***p < 0.001). (f) Representative mEPSC recording from a control- (saline) and HGF-treated neuron. Below, averaged mEPSC from a 1-min epoch was superimposed for comparison. (g) Compared with saline-treated neurons, HGF-treated neurons displayed significantly reduced mEPSC amplitude distribution (n > 2500 events/6 neurons for both groups, **p < 0.01, K-S test) and increased inter-event intervals (h, n > 2000 events/8 neurons, *p < 0.05, K-S test). (i) Illustration of cultured brain slices containing the PFC region, with a L5 neuron targeted for whole cell patch clamp recording, and L2/3 was electrically stimulated to obtain monosynaptic responses. (j) HGF treatment for in cultured slices leads to prolonged MET activation, measured by increased pY1234/1235 (n = 2, quantification not shown). (k) Representative mEPSC recording from a L5 PFC neuron in control (saline) and HGF-treated cultured slices. Quantification reveals that HGF-treatment significantly reduced mEPSC amplitude (n > 3000 events/9 neurons for each group, ***p < 0.001, K-S test) and frequency (t19 = 5.21, **p < 0.01). (l) HGF-treated L5 neurons showed significantly reduced A:N current ratio (t20 = 3.89, *p < 0.05) compared with saline-treated control neurons. (m) HGF-treated L5 neurons showed significantly increased blockade of NMDAR current by ifenprodil (t13 = 4.21, **p < 0.01) compared with saline treated control neurons.
We next prepared cultures of organotypic brain slices containing the PFC region from P6-7 mice. This preparation has the advantage of preserving the local three-dimensional structure of native neuron types and the laminar connections within and between cortical layers 43, which allows us to test the development of synaptic properties in intact cortical circuits by stimulating L2/3 and recording in the synaptically connected L5 neurons (Figure 4i). HGF (50ng/ml) was added in the treatment group at DIV14, and experiments were carried out at DIV22-25. This treatment increases MET pY1234/1235 signal when measured at DIV22 (Figure 4j), suggesting elevated MET signaling persists the duration of HGF application. Analysis of the mEPSC recorded from L5 PFC neurons (Figure 4k) revealed that HGF significantly reduced the mEPSC amplitude (p < 0.001) and frequency (p < 0.01) compared with saline treated neurons. We next used whole cell patch clamp to record AMPA/NMDA receptor current ratio (A:N) by stimulating L2/3 and obtaining a monosynaptic response in L5 neurons. We found that A:N ratio is significantly reduced in HGF treated cells (HGF, 1.05 ± 0.15; saline, 1.58 ±.23. t21 = 6.4, p < 0.01). The functional maturation of cortical circuits also entails NMDA subunit switching, with GluN2A gradually replacing GluN2B 44. We quantified synaptic NMDAR currents contributed by GluN2B by probing sensitivity of NMDAR-EPSC to ifenprodil (3μM), a GluN2B-selective antagonist 45. We found that ifenprodil caused a 52.6 ± 6.4% reduction in NMDAR-mediated charge transfer in HGF-treated L5 PFC neurons, which is significantly higher than that from saline treated L5 cells (31.4 ± 2.4%, t13= 8.3, p < 0.01. Figure 4m). These results, together with decreased A:N ratio and mEPSC amplitude and frequency, and reduced functional synapse proportions following HGF treatment, strongly argue that prolonged MET signaling represses glutamatergic synapse maturation.
Single neuron gain- or loss-of-function of MET alters intra-cortical circuit connectivity
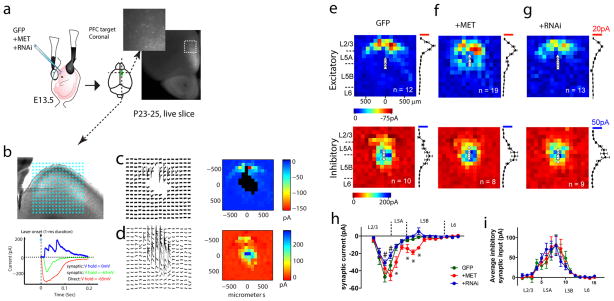
Based on the significant effects observed on neuron morphology, we next asked whether altering MET signaling in single neurons during normal in vivo development leads to cell-autonomous changes in local cortical network connectivity. IUEP was used to deliver MET OE or RNAi manipulations with GFP to the developing L5 neurons in the right frontal lobe of E13.5 mouse embryos (see Supplementary Information). P23-25 mice offspring were selected for electrophysiology and LSPS mapping studies in L5 projection neurons (Figure 5a). We found that neither MET OE nor RNAi significantly altered L5 neuron membrane properties (Supplementary Figure S3). We next used LSPS mapping 23, 28 to quantify both excitatory and inhibitory synaptic inputs made onto these L5 neurons. As expected, L5 PFC neurons receive primary excitatory synaptic inputs from L2/3, and inhibitory synaptic inputs from local sources adjacent to the recorded neuron soma (Figure 5b–5d, recorded from a L5 PFC neuron IUEP with GFP). Multiple excitatory and inhibitory LSPS maps from at least 5 mice were collected from MET OE, RNAi or GFP control neurons. Analysis of pooled maps (Figure 5e–i) revealed that compared with GFP control neurons, MET OE neurons received significantly increased L2/3 excitatory synaptic inputs (Figure 5f, 5h. p < 0.05 for the MET OE effect); while RNAi neurons displayed a pronounced reduction in L2/3 synaptic drive (Figure 5g, 5h. p < 0.05 for MET knockdown effects). Strikingly, the increased synaptic connectivity onto MET OE neurons primarily came from L5 (Figure 5f, 5h. p < 0.01. Bonferroni post hoc test), which is not a significant synaptic input source to L5 neurons in the anterior frontal cortex under normal development conditions 23 or in control neurons IUEP with GFP (Figure 5e). We next analyzed local inhibitory synaptic inputs onto these L5 MET OE, RNAi and GFP neurons. We did not detect significant treatment effects on the overall inhibitory inputs onto these neurons (Figure 5i. p > 0.05). Therefore, single neuron manipulations in MET signaling during in vivo development are capable of disrupting local excitatory synaptic circuits without altering inhibitory connectivity.
Figure 5. Single neuron alternation in MET signaling during in vivo development disrupts local cortical circuit connectivity.
(a) Experimental paradigm. C57Bl6 embryos were electroporated with MET OE, RNAi plasmids in combination with GFP, or control GFP alone at E13.5. A DIC image of live coronal slice superimposed with the GFP channel was shown to denote PFC L5 neuron targeting by IUEP. (b) Illustration of a sagittal PFC slice prepared from IUEP mice, from which L5 PFC neurons were selected for LSPS mapping. A stimulation grid of 16*16 was overlaid to denote stimulation locations. Glutamate uncaging using UV laser pulses produces three types of responses, a direct response (red trace) when glutamate was uncaged at a location close to the soma or dendrites; a synaptic circuit response (green) when glutamate was uncaged and activated a synaptically connected presynaptic neuronal population; or an inhibitory response (voltage clamped at 0 mV) in the form of outward current from synaptically connected interneurons. (c) An example of a matrix of excitatory synaptic input responses to a L5 PFC neuron electroporated with GFP, in response to glutamate uncaging at locations indicated in b. Note direct responses were not plotted. The pseudo-color representation (i.e. ‘map’) of the location and strength of these responses was shown to the right. (d) An inhibitory response map was collected at 0 mV holding potential following the excitatory map. (e) Averaged excitatory (n = 12 neurons/4 mice) maps from L5 neurons with GFP electroporation. For each individual map, the 16 columns of responses were binned into L2/3, L5A, L5B and L6, and the averaged EPSC and IPSC was calculated for each column. Mean and standard errors of the pooled maps were plotted to the right of the average map. Averaged inhibitory inputs (n = 10/4) and their laminar distribution were shown in the panel below. (f) Averaged excitatory (top panel, n = 19/3) and inhibitory (n = 8/3) maps for L5 PFC neurons electroporated with Met cDNA. (g) Averaged excitatory (top panel, n = 13/4) and inhibitory (n = 9/4) maps for L5 PFC neurons electroporated with MET RNAi. (h) Pooled excitatory synaptic inputs to all groups of L5 neurons as a function of their laminar location. Two-way ANOVA revealed a significant treatment effect (F(2, 656) = 15.8), with significantly increased input strength in MET OE neurons at L5A and L5B (*p < 0.05, Bonferroni post hoc test). In contrast, RNAi neurons showed significantly reduced synaptic inputs from L2/3 (#p < 0.05, Bonferroni post hoc test). (i) Pooled inhibitory synaptic inputs to all groups of L5 neurons as a function of their laminar location. No significant difference was detected for the treatment effects on laminar distribution of inhibitory inputs (F(2, 384) = 0.09. p > 0.05).
DISCUSSION
Functional variants of human MET gene have been linked to differential ASD risks, but the underlying mechanism connecting genetic susceptibility and disease phenotype is not established. In this study, we investigated the role of MET RTK in neural development and circuit function. The major findings of our study are as follows. First, we ascertain the temporal expression patterns of MET protein in two mice forebrain regions relevant to autism pathogenesis, and uncover that MET is concentrated in a sub-cellular domain (growth cone) conducive to early neuronal growth. Second, we show that MET signaling leads to activation of small GTPase Cdc42, which accounts for aspects of MET-induced neuronal growth and dendritic morphology development. Third, we find that prolonged MET signaling in developing cortical neurons and slice cultures represses functional glutamatergic synapse formation and maturation. Last, mosaic genetic manipulation of MET expression in single neuron during in vivo prefrontal cortex development leads to changes of intra-cortical connectivity, indicative of altered cortical information processing in a circuit highly relevant to ASD pathogenesis.
Disruptions to the molecular mechanisms controlling excitatory synapse structure and function, often seen with deleterious mutations in synaptic proteins 46–49, have been suggested to underlie neurodevelopmental disorders affecting cognition, including intellectual disability and ASD 50, 51. Unlike many other synaptic proteins implicated in ASD, MET protein expression spans a long developmental time line and is functionally pleiotropic. Both MET mRNA and protein become abundant at late embryonic stage, and persist through early post-weaning time in mice, with precipitous down-regulation thereafter 4, 5. Therefore, peak expression of MET coincides the prime period of neurite growth, synaptogenesis and circuit formation 52. In adult mouse brain, however, a lower levels of MET is located at both presynaptic and postsynaptic compartments 53, 54, and is capable of modulating mature synapse function and plasticity 55. Therefore, the impact of MET on the developing brain can be multifaceted, prolonged, and profound. This is in agreement with accumulating evidence that time delimited disruption of molecular signaling events during development can have long-term effects on brain function 56, 57.
Our study for the first time established that MET engages small GTPase Cdc42 as growth-associated mechanisms in early neuronal development. This early dendritic morphology development may be a prerequisite for later emergence of dendritic spines and synaptic signaling macromolecules, including glutamate receptors and their downstream targets. We show that MET OE is sufficient to increase dendritic complexity and spine density, a process that is blocked by Cdc42 antagonism. The fact that Cdc42 is activated downstream of MET in a PI3K- and MET kinase activity-dependent manner delineates a previously unrecognized pathway operating in developing neurons and circuits. Cdc42 activation has been shown to promote dendritic protrusion and spine morphogenesis 38, 58, and insufficient signaling through this pathway may well account for reduced spine density in CA1 neurons following MET RNAi 24. The fact that MET is concentrated at the growth cone, and responds to HGF by activating small GTPase Cdc42, is also consistent with a recent ASD genetic study reporting an enrichment of genetic variations that disrupts “functional gene sets involved in cellular proliferation, projection and motility, and GTPase/Ras signaling” 59.
It is unclear from the current study what downstream mechanism following MET activation may operate to suppress glutamatergic synapse maturation. These potential mechanisms may be negatively coupled with pre- and postsynaptic assembly, inhibition of AMPAR recruitment to the synapse to maintain the proportion of ‘silent synapses’ (Liao 1995), and suppression of GluN2B>GluN2A subunit switching with the implication of altered cortical plasticity. Therefore, normal developmental down-regulation of MET signaling beyond early postnatal stage may release a developmental brake that inhibits synapse maturation. LSPS functional mapping revealed that with reduced developmental MET expression, L5 PFC neurons receive less synaptic inputs. In comparison, enhanced MET expression leads to more synaptic connections overall, specifically from ‘ectopic’ laminar locations. It is unclear how MET gain- and loss-of-function in single cortical neurons may cell-autonomously affect local cortical laminar connectivity. This altered connectivity could be the combinatory effects of altered dendritic spine density, synapse maturation, or activity-dependent synaptic pruning processes, as manipulating MET in CA1 neurons also alters dendritic morphology, spine size and density, and maturation timing 24. Nonetheless, because the topography of synaptic drive onto these cortical projection neurons is significantly changed, one can expect altered precision and specificity of cortical information processing and altered cortical output resulting from disrupted MET signaling levels.
How does alteration of a single protein cause such dramatic morphological and functional changes in neurons? The current study and previous works 13, 24, 29, 60 clearly indicate MET as a key factor for dendritic and spine development. Dendritic and spine morphology are known to be regulated by multiple growth factors and are key determinants of neurotransmitter signaling efficiency and critical constituents of circuit function 1, 61. For instance, spine size and geometry is well-correlated with its glutamate receptor content and functional maturity 42. A large spine head size is more mature, yet lacks the plasticity and motility of an immature spine, which more likely serves to promote initial circuit wiring and experience-shaped rewiring of neuronal circuits during development 62. Thus, a faulty developmental program that causes earlier spine maturation may hardwire the circuits earlier and render them resistant to later phases of experience-dependent refinement. Interestingly, this “accelerated maturation” hypothesis has been suggested in mice with haploinsufficiency of SynGAP1 57 or over-expression of another autism risk gene NLGN1 in mice 63.
In summary, our study provides important new insight into the role MET receptor tyrosine kinase in mammalian CNS development, and how dysregulation of MET signaling alters neural development and circuit function. Our study also supports the findings of a recent study that implicates an enrichment of ASD risk genes related to growth factor signaling 64, the hypothesized aberrant growth trajectories in the ASD brain 65, and the coalescence of functional ASD gene networks in cortical projection neurons 66. Although future work will be required to fully understand how loss- and gain- of MET signaling engages downstream signaling cascades impinging on the developing brain to alter the risk for ASD, we show that changes in MET expression profoundly affect neuronal structure and function, formation and maturation of excitatory synapses, and circuit connectivity in the disease-related brain regions and functional domains. The roles of MET described here may help inform how altered developmental signaling pathway give rise to cortical circuit dysfunction that potentially contribute to the development of ASD-related pathophysiology.
Supplementary Material
Acknowledgments
This work was supported by a National Institute of Mental Health (NIMH) grants K99MH087628, R00MH087628, and an Institute for Mental Health Research Grant and to S.Q. We thank Dr. Daping Fan (University of South Carolina) for his expert guidance in RNAi constructs.
Footnotes
CONFLICT OF INTEREST
The authors declare no conflict of interest.
References
- 1.Park H, Poo MM. Neurotrophin regulation of neural circuit development and function. Nat Rev Neurosci. 2013;14(1):7–23. doi: 10.1038/nrn3379. [DOI] [PubMed] [Google Scholar]
- 2.Naldini L, Vigna E, Ferracini R, Longati P, Gandino L, Prat M, et al. The tyrosine kinase encoded by the MET proto-oncogene is activated by autophosphorylation. Mol Cell Biol. 1991;11(4):1793–1803. doi: 10.1128/mcb.11.4.1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Uehara Y, Minowa O, Mori C, Shiota K, Kuno J, Noda T, et al. Placental defect and embryonic lethality in mice lacking hepatocyte growth factor/scatter factor. Nature. 1995;373(6516):702–705. doi: 10.1038/373702a0. [DOI] [PubMed] [Google Scholar]
- 4.Judson MC, Amaral DG, Levitt P. Conserved subcortical and divergent cortical expression of proteins encoded by orthologs of the autism risk gene MET. Cereb Cortex. 2011;21(7):1613–1626. doi: 10.1093/cercor/bhq223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Judson MC, Bergman MY, Campbell DB, Eagleson KL, Levitt P. Dynamic gene and protein expression patterns of the autism-associated met receptor tyrosine kinase in the developing mouse forebrain. J Comp Neurol. 2009;513(5):511–531. doi: 10.1002/cne.21969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Maina F, Hilton MC, Ponzetto C, Davies AM, Klein R. Met receptor signaling is required for sensory nerve development and HGF promotes axonal growth and survival of sensory neurons. Genes Dev. 1997;11(24):3341–3350. doi: 10.1101/gad.11.24.3341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mukamel Z, Konopka G, Wexler E, Osborn GE, Dong H, Bergman MY, et al. Regulation of MET by FOXP2, genes implicated in higher cognitive dysfunction and autism risk. J Neurosci. 2011;31(32):11437–11442. doi: 10.1523/JNEUROSCI.0181-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Streit A, Stern CD, Thery C, Ireland GW, Aparicio S, Sharpe MJ, et al. A role for HGF/SF in neural induction and its expression in Hensen’s node during gastrulation. Development. 1995;121(3):813–824. doi: 10.1242/dev.121.3.813. [DOI] [PubMed] [Google Scholar]
- 9.Ebens A, Brose K, Leonardo ED, Hanson MG, Jr, Bladt F, Birchmeier C, et al. Hepatocyte growth factor/scatter factor is an axonal chemoattractant and a neurotrophic factor for spinal motor neurons. Neuron. 1996;17(6):1157–1172. doi: 10.1016/s0896-6273(00)80247-0. [DOI] [PubMed] [Google Scholar]
- 10.Giacobini P, Messina A, Wray S, Giampietro C, Crepaldi T, Carmeliet P, et al. Hepatocyte growth factor acts as a motogen and guidance signal for gonadotropin hormone-releasing hormone-1 neuronal migration. J Neurosci. 2007;27(2):431–445. doi: 10.1523/JNEUROSCI.4979-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bai L, Lennon DP, Caplan AI, DeChant A, Hecker J, Kranso J, et al. Hepatocyte growth factor mediates mesenchymal stem cell-induced recovery in multiple sclerosis models. Nat Neurosci. 2012;15(6):862–870. doi: 10.1038/nn.3109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Davey F, Hilton M, Davies AM. Cooperation between HGF and CNTF in promoting the survival and growth of sensory and parasympathetic neurons. Mol Cell Neurosci. 2000;15(1):79–87. doi: 10.1006/mcne.1999.0803. [DOI] [PubMed] [Google Scholar]
- 13.Gutierrez H, Dolcet X, Tolcos M, Davies A. HGF regulates the development of cortical pyramidal dendrites. Development. 2004;131(15):3717–3726. doi: 10.1242/dev.01209. [DOI] [PubMed] [Google Scholar]
- 14.Campbell DB, D’Oronzio R, Garbett K, Ebert PJ, Mirnics K, Levitt P, et al. Disruption of cerebral cortex MET signaling in autism spectrum disorder. Ann Neurol. 2007;62(3):243–250. doi: 10.1002/ana.21180. [DOI] [PubMed] [Google Scholar]
- 15.Campbell DB, Sutcliffe JS, Ebert PJ, Militerni R, Bravaccio C, Trillo S, et al. A genetic variant that disrupts MET transcription is associated with autism. Proc Natl Acad Sci U S A. 2006;103(45):16834–16839. doi: 10.1073/pnas.0605296103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jackson PB, Boccuto L, Skinner C, Collins JS, Neri G, Gurrieri F, et al. Further evidence that the rs1858830 C variant in the promoter region of the MET gene is associated with autistic disorder. Autism Res. 2009;2(4):232–236. doi: 10.1002/aur.87. [DOI] [PubMed] [Google Scholar]
- 17.Thanseem I, Nakamura K, Miyachi T, Toyota T, Yamada S, Tsujii M, et al. Further evidence for the role of MET in autism susceptibility. Neurosci Res. 2010;68(2):137–141. doi: 10.1016/j.neures.2010.06.014. [DOI] [PubMed] [Google Scholar]
- 18.Geschwind DH, Levitt P. Autism spectrum disorders: developmental disconnection syndromes. Curr Opin Neurobiol. 2007;17(1):103–111. doi: 10.1016/j.conb.2007.01.009. [DOI] [PubMed] [Google Scholar]
- 19.Plummer JT, Evgrafov OV, Bergman MY, Friez M, Haiman CA, Levitt P, et al. Transcriptional regulation of the MET receptor tyrosine kinase gene by MeCP2 and sex-specific expression in autism and Rett syndrome. Transl Psychiatry. 2013;3:e316. doi: 10.1038/tp.2013.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Konopka G, Bomar JM, Winden K, Coppola G, Jonsson ZO, Gao F, et al. Human-specific transcriptional regulation of CNS development genes by FOXP2. Nature. 2009;462(7270):213–217. doi: 10.1038/nature08549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wood L, Gray NW, Zhou Z, Greenberg ME, Shepherd GM. Synaptic circuit abnormalities of motor-frontal layer 2/3 pyramidal neurons in an RNA interference model of methyl-CpG-binding protein 2 deficiency. J Neurosci. 2009;29(40):12440–12448. doi: 10.1523/JNEUROSCI.3321-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rudie JD, Hernandez LM, Brown JA, Beck-Pancer D, Colich NL, Gorrindo P, et al. Autism-associated promoter variant in MET impacts functional and structural brain networks. Neuron. 2012;75(5):904–915. doi: 10.1016/j.neuron.2012.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Qiu S, Anderson CT, Levitt P, Shepherd GM. Circuit-specific intracortical hyperconnectivity in mice with deletion of the autism-associated Met receptor tyrosine kinase. J Neurosci. 2011;31 (15):5855–5864. doi: 10.1523/JNEUROSCI.6569-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Qiu S, Lu Z, Levitt P. MET receptor tyrosine kinase controls dendritic complexity, spine morphogenesis, and glutamatergic synapse maturation in the hippocampus. J Neurosci. 2014;34 (49):16166–16179. doi: 10.1523/JNEUROSCI.2580-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bladt F, Riethmacher D, Isenmann S, Aguzzi A, Birchmeier C. Essential role for the c-met receptor in the migration of myogenic precursor cells into the limb bud. Nature. 1995;376(6543):768–771. doi: 10.1038/376768a0. [DOI] [PubMed] [Google Scholar]
- 26.Niwa M, Kamiya A, Murai R, Kubo K, Gruber AJ, Tomita K, et al. Knockdown of DISC1 by in utero gene transfer disturbs postnatal dopaminergic maturation in the frontal cortex and leads to adult behavioral deficits. Neuron. 2010;65(4):480–489. doi: 10.1016/j.neuron.2010.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pfenninger KH, Ellis L, Johnson MP, Friedman LB, Somlo S. Nerve growth cones isolated from fetal rat brain: subcellular fractionation and characterization. Cell. 1983;35(2 Pt 1):573–584. doi: 10.1016/0092-8674(83)90191-5. [DOI] [PubMed] [Google Scholar]
- 28.Xu X, Callaway EM. Laminar specificity of functional input to distinct types of inhibitory cortical neurons. J Neurosci. 2009;29(1):70–85. doi: 10.1523/JNEUROSCI.4104-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Judson MC, Eagleson KL, Wang L, Levitt P. Evidence of cell-nonautonomous changes in dendrite and dendritic spine morphology in the met-signaling-deficient mouse forebrain. J Comp Neurol. 2010;518(21):4463–4478. doi: 10.1002/cne.22467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pathania M, Davenport EC, Muir J, Sheehan DF, Lopez-Domenech G, Kittler JT. The autism and schizophrenia associated gene CYFIP1 is critical for the maintenance of dendritic complexity and the stabilization of mature spines. Transl Psychiatry. 2014;4:e374. doi: 10.1038/tp.2014.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gordon JA. Oscillations and hippocampal-prefrontal synchrony. Curr Opin Neurobiol. 2011;21(3):486–491. doi: 10.1016/j.conb.2011.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Royal I, Lamarche-Vane N, Lamorte L, Kaibuchi K, Park M. Activation of cdc42, rac, PAK, and rho-kinase in response to hepatocyte growth factor differentially regulates epithelial cell colony spreading and dissociation. Mol Biol Cell. 2000;11(5):1709–1725. doi: 10.1091/mbc.11.5.1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sakkab D, Lewitzky M, Posern G, Schaeper U, Sachs M, Birchmeier W, et al. Signaling of hepatocyte growth factor/scatter factor (HGF) to the small GTPase Rap1 via the large docking protein Gab1 and the adapter protein CRKL. J Biol Chem. 2000;275(15):10772–10778. doi: 10.1074/jbc.275.15.10772. [DOI] [PubMed] [Google Scholar]
- 34.Takaishi K, Sasaki T, Kato M, Yamochi W, Kuroda S, Nakamura T, et al. Involvement of Rho p21 small GTP-binding protein and its regulator in the HGF-induced cell motility. Oncogene. 1994;9 (1):273–279. [PubMed] [Google Scholar]
- 35.Hall A, Lalli G. Rho and Ras GTPases in axon growth, guidance, and branching. Cold Spring Harb Perspect Biol. 2010;2(2):a001818. doi: 10.1101/cshperspect.a001818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Newey SE, Velamoor V, Govek EE, Van Aelst L. Rho GTPases, dendritic structure, and mental retardation. J Neurobiol. 2005;64(1):58–74. doi: 10.1002/neu.20153. [DOI] [PubMed] [Google Scholar]
- 37.Peng YR, He S, Marie H, Zeng SY, Ma J, Tan ZJ, et al. Coordinated changes in dendritic arborization and synaptic strength during neural circuit development. Neuron. 2009;61(1):71–84. doi: 10.1016/j.neuron.2008.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tashiro A, Minden A, Yuste R. Regulation of dendritic spine morphology by the rho family of small GTPases: antagonistic roles of Rac and Rho. Cereb Cortex. 2000;10(10):927–938. doi: 10.1093/cercor/10.10.927. [DOI] [PubMed] [Google Scholar]
- 39.Sholl DA. Dendritic organization in the neurons of the visual and motor cortices of the cat. J Anat. 1953;87(4):387–406. [PMC free article] [PubMed] [Google Scholar]
- 40.Shen H, Sesack SR, Toda S, Kalivas PW. Automated quantification of dendritic spine density and spine head diameter in medium spiny neurons of the nucleus accumbens. Brain Struct Funct. 2008;213(1–2):149–157. doi: 10.1007/s00429-008-0184-2. [DOI] [PubMed] [Google Scholar]
- 41.Ethell IM, Pasquale EB. Molecular mechanisms of dendritic spine development and remodeling. Prog Neurobiol. 2005;75(3):161–205. doi: 10.1016/j.pneurobio.2005.02.003. [DOI] [PubMed] [Google Scholar]
- 42.Matsuzaki M, Ellis-Davies GC, Nemoto T, Miyashita Y, Iino M, Kasai H. Dendritic spine geometry is critical for AMPA receptor expression in hippocampal CA1 pyramidal neurons. Nat Neurosci. 2001;4(11):1086–1092. doi: 10.1038/nn736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McAllister AK, Lo DC, Katz LC. Neurotrophins regulate dendritic growth in developing visual cortex. Neuron. 1995;15(4):791–803. doi: 10.1016/0896-6273(95)90171-x. [DOI] [PubMed] [Google Scholar]
- 44.Monyer H, Burnashev N, Laurie DJ, Sakmann B, Seeburg PH. Developmental and regional expression in the rat brain and functional properties of four NMDA receptors. Neuron. 1994;12 (3):529–540. doi: 10.1016/0896-6273(94)90210-0. [DOI] [PubMed] [Google Scholar]
- 45.Harlow EG, Till SM, Russell TA, Wijetunge LS, Kind P, Contractor A. Critical period plasticity is disrupted in the barrel cortex of FMR1 knockout mice. Neuron. 2010;65(3):385–398. doi: 10.1016/j.neuron.2010.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Durand CM, Betancur C, Boeckers TM, Bockmann J, Chaste P, Fauchereau F, et al. Mutations in the gene encoding the synaptic scaffolding protein SHANK3 are associated with autism spectrum disorders. Nat Genet. 2007;39(1):25–27. doi: 10.1038/ng1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gilman SR, Iossifov I, Levy D, Ronemus M, Wigler M, Vitkup D. Rare de novo variants associated with autism implicate a large functional network of genes involved in formation and function of synapses. Neuron. 2011;70(5):898–907. doi: 10.1016/j.neuron.2011.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hamdan FF, Daoud H, Piton A, Gauthier J, Dobrzeniecka S, Krebs MO, et al. De novo SYNGAP1 mutations in nonsyndromic intellectual disability and autism. Biol Psychiatry. 2011;69(9):898–901. doi: 10.1016/j.biopsych.2010.11.015. [DOI] [PubMed] [Google Scholar]
- 49.Tabuchi K, Blundell J, Etherton MR, Hammer RE, Liu X, Powell CM, et al. A neuroligin-3 mutation implicated in autism increases inhibitory synaptic transmission in mice. Science. 2007;318(5847):71–76. doi: 10.1126/science.1146221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Penzes P, Cahill ME, Jones KA, VanLeeuwen JE, Woolfrey KM. Dendritic spine pathology in neuropsychiatric disorders. Nat Neurosci. 2011;14(3):285–293. doi: 10.1038/nn.2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zoghbi HY, Bear MF. Synaptic dysfunction in neurodevelopmental disorders associated with autism and intellectual disabilities. Cold Spring Harb Perspect Biol. 2012;4(3) doi: 10.1101/cshperspect.a009886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fiala JC, Feinberg M, Popov V, Harris KM. Synaptogenesis via dendritic filopodia in developing hippocampal area CA1. J Neurosci. 1998;18(21):8900–8911. doi: 10.1523/JNEUROSCI.18-21-08900.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Eagleson KL, Milner TA, Xie Z, Levitt P. Synaptic and extrasynaptic location of the receptor tyrosine kinase met during postnatal development in the mouse neocortex and hippocampus. J Comp Neurol. 2013;521(14):3241–3259. doi: 10.1002/cne.23343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tyndall SJ, Walikonis RS. The receptor tyrosine kinase Met and its ligand hepatocyte growth factor are clustered at excitatory synapses and can enhance clustering of synaptic proteins. Cell Cycle. 2006;5(14):1560–1568. doi: 10.4161/cc.5.14.2918. [DOI] [PubMed] [Google Scholar]
- 55.Akimoto M, Baba A, Ikeda-Matsuo Y, Yamada MK, Itamura R, Nishiyama N, et al. Hepatocyte growth factor as an enhancer of nmda currents and synaptic plasticity in the hippocampus. Neuroscience. 2004;128(1):155–162. doi: 10.1016/j.neuroscience.2004.06.031. [DOI] [PubMed] [Google Scholar]
- 56.Ansorge MS, Zhou M, Lira A, Hen R, Gingrich JA. Early-life blockade of the 5-HT transporter alters emotional behavior in adult mice. Science. 2004;306(5697):879–881. doi: 10.1126/science.1101678. [DOI] [PubMed] [Google Scholar]
- 57.Clement JP, Aceti M, Creson TK, Ozkan ED, Shi Y, Reish NJ, et al. Pathogenic SYNGAP1 mutations impair cognitive development by disrupting maturation of dendritic spine synapses. Cell. 2012;151(4):709–723. doi: 10.1016/j.cell.2012.08.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Luo L. Actin cytoskeleton regulation in neuronal morphogenesis and structural plasticity. Annu Rev Cell Dev Biol. 2002;18:601–635. doi: 10.1146/annurev.cellbio.18.031802.150501. [DOI] [PubMed] [Google Scholar]
- 59.Pinto D, Pagnamenta AT, Klei L, Anney R, Merico D, Regan R, et al. Functional impact of global rare copy number variation in autism spectrum disorders. Nature. 2010;466(7304):368–372. doi: 10.1038/nature09146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tyndall SJ, Patel SJ, Walikonis RS. Hepatocyte growth factor-induced enhancement of dendritic branching is blocked by inhibitors of N-methyl-D-aspartate receptors and calcium/calmodulin-dependent kinases. J Neurosci Res. 2007;85(11):2343–2351. doi: 10.1002/jnr.21390. [DOI] [PubMed] [Google Scholar]
- 61.Xu B, Zang K, Ruff NL, Zhang YA, McConnell SK, Stryker MP, et al. Cortical degeneration in the absence of neurotrophin signaling: dendritic retraction and neuronal loss after removal of the receptor TrkB. Neuron. 2000;26(1):233–245. doi: 10.1016/s0896-6273(00)81153-8. [DOI] [PubMed] [Google Scholar]
- 62.Konur S, Yuste R. Imaging the motility of dendritic protrusions and axon terminals: roles in axon sampling and synaptic competition. Mol Cell Neurosci. 2004;27(4):427–440. doi: 10.1016/j.mcn.2004.07.005. [DOI] [PubMed] [Google Scholar]
- 63.Dahlhaus R, Hines RM, Eadie BD, Kannangara TS, Hines DJ, Brown CE, et al. Overexpression of the cell adhesion protein neuroligin-1 induces learning deficits and impairs synaptic plasticity by altering the ratio of excitation to inhibition in the hippocampus. Hippocampus. 2010;20(2):305–322. doi: 10.1002/hipo.20630. [DOI] [PubMed] [Google Scholar]
- 64.Wittkowski KM, Sonakya V, Bigio B, Tonn MK, Shic F, Ascano M, et al. A novel computational biostatistics approach implies impaired dephosphorylation of growth factor receptors as associated with severity of autism. Transl Psychiatry. 2014;4:e354. doi: 10.1038/tp.2013.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Berg JM, Geschwind DH. Autism genetics: searching for specificity and convergence. Genome Biol. 2012;13(7):247. doi: 10.1186/gb-2012-13-7-247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Parikshak NN, Luo R, Zhang A, Won H, Lowe JK, Chandran V, et al. Integrative functional genomic analyses implicate specific molecular pathways and circuits in autism. Cell. 2013;155(5):1008–1021. doi: 10.1016/j.cell.2013.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.