Abstract
The cranial base is a component of the neurocranium and has a central role in the structural integration of the face, brain and vertebral column. Consequently, alteration in the shape of the human cranial base has been intimately linked with primate evolution and defective development is associated with numerous human facial abnormalities. Here we describe a novel recessive mutant mouse strain that presented with a domed head and fully penetrant cleft secondary palate coupled with defects in the formation of the underlying cranial base. Mapping and non-complementation studies revealed a specific mutation in Memo1 - a gene originally associated with cell migration. Expression analysis of Memo1 identified robust expression in the perichondrium and periosteum of the developing cranial base, but only modest expression in the palatal shelves. Fittingly, although the palatal shelves failed to elevate in Memo1 mutants, expression changes were modest within the shelves themselves. In contrast, the cranial base, which forms via endochondral ossification had major reductions in the expression of genes responsible for bone formation, notably matrix metalloproteinases and markers of the osteoblast lineage, mirrored by an increase in markers of cartilage and extracellular matrix development. Concomitant with these changes, mutant cranial bases showed an increased zone of hypertrophic chondrocytes accompanied by a reduction in both vascular invasion and mineralization. Finally, neural crest cell-specific deletion of Memo1 caused a failure of anterior cranial base ossification indicating a cell autonomous role for MEMO1 in the development of these neural crest cell derived structures. However, palate formation was largely normal in these conditional mutants, suggesting a non-autonomous role for MEMO1 in palatal closure. Overall, these findings assign a new function to MEMO1 in driving endochondral ossification in the cranium, and also link abnormal development of the cranial base with more widespread effects on craniofacial shape relevant to human craniofacial dysmorphology.
Keywords: Endochondral ossification, cranial base, cleft palate, palatogenesis, neural crest, MEMO1
Introduction
The human head skeleton is composed of two integrated components, the viscerocranium (also termed the splanchnocranium) and the neurocranium. The viscerocranium makes up the face and associated structures, while the neurocranium includes the cranial vault along with the underlying cranial base. The cranial base serves as a central integration point for development, growth, evolution and function of the craniofacial complex 1-3. Specifically, the cranial base provides a structural platform from which the cranial vault and viscerocranium grow. In addition, it houses the pituitary and sense organs of the head, links the cranium with the trunk via its connection with the vertebral column, and provides a structure through which the cranial vessels and nerves traverse 2. Thus, abnormalities in the size or flexion of the cranial base can impact overall facial shape and are often an underlying component of several human craniofacial defects 4-6. Moreover, changes in the morphology of the cranial base are a critical aspect of the alterations in primate facial shape and brain growth that occurred during human evolution 1,2. Despite the importance of the cranial base, there are only a limited number of animal model systems that provide information concerning its unique developmental program.
In mammals, the cranial base is derived from two major embryonic cell populations, the neural crest (anterior cranial base) and mesoderm of cranial paraxial and somitic origin (posterior cranial base) 7-9. This is in contrast to the cartilage and bone of the viscerocranium, which is largely neural crest cell derived 10,11. In addition, the cranial base develops through endochondral ossification, whereas the viscerocranium forms largely through intramembranous ossification 12. The cranial base initially develops from a series of cartilages that fuse to form the mature chondrocranium by about E16.5 of mouse development 7,13. Subsequently, a number of ossification centers arise in the midline resulting in a rostral-caudal progression of the presphenoid, basisphenoid and basioccipital bones of the cranial base by birth. Each pair of bones is separated by a cartilaginous synchondrosis that is important to allow continued growth of the cranial base until maturity 4,7. Therefore, the transition between cartilage and bone in the cranial base must be tightly regulated with both temporal and spatial resolution to allow normal growth and function. Generation of ossified bone during endochondral ossification is a well-documented process in which mesenchymal precursors first form a cartilage intermediate before being replaced by bone 14,15. As such, this process involves defined stages of cartilage maturation, vascular invasion, and subsequent mineralization by bone forming osteoblasts 14,15. In contrast, intramembranous ossification involves the conversion of mesenchymal precursors into bone forming osteoblasts directly, without a cartilage intermediate 14,15. Given these distinct developmental processes, unique gene-regulatory programs and biological mechanisms likely drive ossification of these two bone groupings in the head. Supporting this, differences in vascular invasion have been observed between endochondral and intramembranous bones of the human fetal head, and animal models exist with defects in a distinct subset of these craniofacial bones 16,17. However, given the final product of both mechanisms is ossified bone, there are also likely overlapping genetic programs involved, although the degree of dependency on these programs may be different between these two bone populations 12. Teasing out these dependencies may provide insight into the etiologies of various osteogenic disorders affecting subsets of bones in the craniofacial complex.
To identify genes and alleles affecting mouse craniofacial formation we have recently conducted a recessive N-ethyl-N-nitrosourea (ENU) screen 18. Here we report the phenotypic and molecular characterization of one mutant from this screen that displayed a combination of craniofacial defects including cleft secondary palate, a domed cranium, and cranial base defects. We identified the affected gene responsible for these defects as Memo1 (Mediator of Erbb2 Driven Cell Motility 1), which encodes a multi-functional protein originally linked with cell migration and metastasis in breast cancer 19-21. Mice homozygous for a Memo1 null mutation have previously been shown to die beginning at E13.5, accompanied by severe vasculature defects 22. However, no craniofacial defects were noted in these null embryos. Therefore, our analysis indicates a novel role for MEMO1 in craniofacial development, and further suggests that some instances of human cleft palate may arise secondary to defects in development of the cranial base.
Results
Identification and characterization of the F1-9-13FP mutant
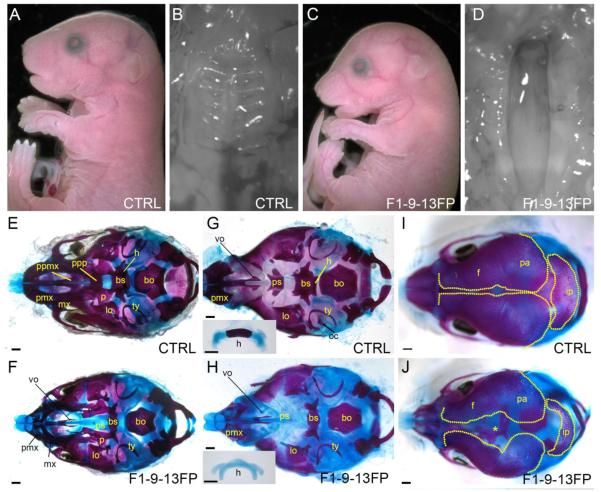
Following ENU treatment of C57BL/6J male mice, we generated founder males that were used in a three-generation cross to screen for recessive mutations that lead to defects in head morphology at embryonic day 18.5 (E18.5) 18. From this, we identified a mutant line that we originally termed F1-9-13FP in which ~25% of the offspring from heterozygous matings displayed a variety of craniofacial defects including a misshapen nasal bridge, a downward positioned cranium, a domed skull (Fig 1A, C), and a fully penetrant cleft palate (Fig 1B, D). When these mice were allowed to develop to term, they did not thrive, and displayed no milk in their stomachs, consistent with the cleft palate defect (Table 1).
Figure 1. The F1-9-13FP craniofacial phenotype.
(A-D) E18.5 control (A, B) or F1-9-13FP (C, D) mouse embryo, either shown in lateral view (A, C) or a ventral view of the secondary palate (B, D). (E-J) E18.5 craniofacial skeletal preparations of either a control (E, G, I) or F1-9-13FP mutant (F, H, J), shown in ventral views with the mandible removed (E, F), mandible plus palatal bones removed (G, H), or dorsal view of the developing calvaria (I, J). Inset in G and H is the hyoid skeletal element shown in isolation. Abbreviations: bs, basisphenoid; bo, basioccipital; f, frontal; h, hyoid; ip, interparietal; lo, lamina obturans; mx, maxillary; oc, otic capsule; p, palatine; pa, parietal; pmx, premaxillary; ppmx, palatal process of maxilla; ppp, palatal process of palatine; ps, presphenoid; ty, tympanic; vo,vomer. The "*" in panel J indicates the enlarged space between the frontal bones of the mutant. Scale bar: 500uM.
Table 1.
Summary of F1-9-13FP genotype frequencies at E18.5 and P21
| E18.5 | P21 | |||
|---|---|---|---|---|
| Genotype | Observed# | Expected | Observed& | Expected |
| wt/wt | 37 | 31.5 | 22 | 18 |
| wt/F1-9-13FP | 70 | 63 | 50 | 36 |
| F1-9-13FP/F1-9-13FP | 19* | 31.5 | 0 | 18 |
|
| ||||
| Total | 126 | χ2 = 0.035 | 72 | χ2 = 5.201E-06 |
Counts are based on > 95% C57BL/6J background
Counts are based on > 85% C57BL/6J background
gross external examination indicated that 19/19 displayed reported craniofacial phenotypes, while 7/19 had some evidence of vasculature defects including edema and purpura (data not shown)
χ2 analysis at E18.5 indicates that homozygous mutant numbers are lower than expected, suggesting some embryos are lost earlier in embryogenesis.
To probe the underlying skeletal organization in F1-9-13FP homozygous mutants, we processed embryos for bone and cartilage analysis. The bones of the head are mainly derived from either of two embryonic populations, the neural crest or mesoderm 7,11,13,23. In addition, the majority of the craniofacial bones are formed through intramembranous ossification, although a subset are formed through endochondral ossification 12,13. Initial examination of E18.5 craniofacial skeletons revealed that the most severely affected bones were those of neural crest cell origin, especially those that undergo endochondral ossification (Fig 1E-J). The endochondral bones of the cranial base - the presphenoid, basisphenoid, and the basioccipital - were of particular interest in this regard as fate mapping studies have shown that these bones and their cartilage templates are derived from neural crest cells, neural crest cells and cranial paraxial mesoderm, or somitic mesoderm, respectively 7-9. Notably, in F1-9-13FP homozygous mutants, the degree of hypoplasia of these bones correlated with the extent of their neural crest cell origin, with the presphenoid often absent (a small remnant remains in some instances), the basisphenoid very hypoplastic, and the basioccipital only slightly affected compared with controls (Fig 1G,H). In addition to the cranial base, other endochondral bones including the neural crest cell derived hyoid 11,13 and the mesoderm derived otic capsule 24, also failed to ossify in F1-9-13FP homozygous mutants (Fig 1G,H).
Although endochondral bones were the most severely affected, a subset of neural crest cell derived intramembranous elements of the viscerocranium, including the premaxillary, maxillary, and palatine bones were also hypoplastic in F1-9-13FP homozygous mutants as compared to controls (Fig 1E, F). Several additional craniofacial skeletal elements also showed defects, including the palatal processes of the palatine and maxillary bones, which were absent (Fig 1 E, F), and the bones of the calvaria, which were more widely separated and appeared less ossified overall in F1-9-13FP homozygous mutants than in controls (Fig 1I, J). In common with the cranial base, the majority of the bones of the trunk and appendicular skeleton are derived via endochondral ossification. The majority of these bones had a normal appearance in homozygous F1-9-13FP mutants, although we did note reduced ossification of several bones at the extremities, specifically the phalanges and the xiphoid process of the sternum (data not shown). Therefore, there does not appear to be a generalized defect in endochondral ossification in the mutants. Instead, our analysis indicates that the mutation present in the F1-9-13FP strain has the greatest impact on the neural crest cell derived endochondral bones of the head, and to a lesser extent, additional neural crest cell and mesoderm derived intramembranous craniofacial bones.
A point mutation in Memo1 (Mediator of Erbb2 driven cell motility 1) correlates with the F1-9-13FP phenotype
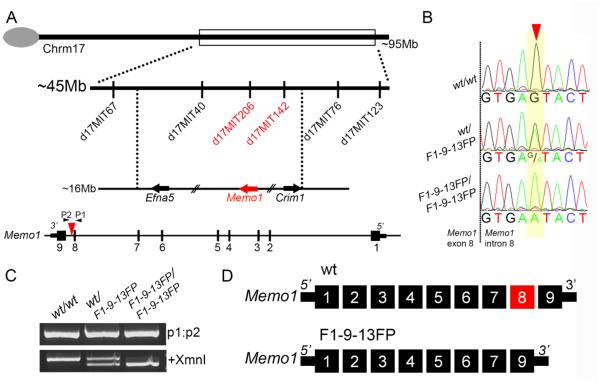
The ENU-mutagenized C57BL/6J founder male was outcrossed onto a wild type 129S1/SvImJ background for mapping and maintenance purposes. Meiotic recombination mapping enabled the approximate position of the mutation to be identified using polymorphic markers that distinguished between these two mouse strains. Initially, using markers spanning ~10 mega base pair (Mb) intervals throughout the mouse autosomes, the mutation was mapped to an ~45Mb region on chromosome 17 (between polymorphic markers D17Mit67 and D17Mit123) (Fig 2A). Subsequently, markers D17Mit40, D17Mit206, D17Mit142, and D17Mit76 were used to refine the position of the mutation to one of two possible 5-10Mb intervals. Next, genomic DNA from a mutant embryo was subjected to whole exome enrichment and sequencing. Inspection of the relevant intervals indicated that this mutant, as well as all subsequent mutants but not their littermate controls, were homozygous for a point mutation in an intron of Mediator of ErbB2 induced cell motility 1, or Memo1 (Fig 2B). This point mutation maps to the interval between the previously identified markers D17Mit206 and D17Mit142, consistent with Memo1 being the causative gene.
Figure 2. Mapping and exome sequencing identifies a point mutation in Mediator of ErbB2 driven cell motility 1 associated with the F1-9-13FP phenotype.
A) Top: Schematic of mouse chromosome 17 showing markers used to map the position of the ENU induced mutation in F1-9-13FP embryos. Bottom: Schematic of the Memo1 locus showing relative location of point mutation (red arrowhead in intron 8). Note direction of transcription is shown from right to left to match chromosomal arrangement. B) Representative chromatograms of sequencing results from genomic DNA of wild-type as well as F1-9-13FP heterozygous and homozygous embryos, with position of the single nucleotide transition in intron 8 of Memo1 shown by red arrowhead. C) Gel electrophoresis of PCR products generated using primers (P1 and P2 in Fig 2A) flanking the ENU induced mutation and genomic DNA from wild-type as well as F1-9-13FP heterozygous and homozygous embryos. Prior to size separation a portion of the PCR product was digested with XmnI restriction enzyme (lower gel). D) Schematic of the nine exon wild-type Memo1 mRNA transcript as well as the major mRNA splice product produced in F1-9-13FP mutants, namely an in-frame deletion of the exon 8 (for clarity, the orientation of the transcript has been reversed from that shown in A).
More specifically, the ENU-induced mutation was a single base-pair change in the splice donor of exon 8 of Memo1, converting the 5th nucleotide 3’ of the exon 8-intron 8 boundary from a guanine to an adenine (Fig 2B). Note that this mutation also creates a novel XmnI restriction site that can be used for genotyping confirmation (Fig 2B,C). Importantly, previous studies have identified this 5th position 3’ of an exon-intron boundary to be highly conserved and critical for proper mRNA splicing 25. To assess the potential consequence of this point mutation on mRNA splicing, RNA was harvested from F1-9-13FP mutants and control embryos, cDNA synthesized, and PCR carried out using primers flanking exon 8 of Memo1. Depending on the primer pair used for analysis, a variety of aberrant splice products were detected in the mutants and no wild-type product was produced (data not shown). Cloning and sequencing of these splice products indicated a variety of mis-splicing events, although the most prominent appeared to be an exclusion of exon 8, resulting in an in-frame fusion of exon 7 and 9 (Fig 2D and Supplemental Fig 1). These observations were consistent with Memo1 being the affected locus in F1-9-13FP mutants.
Genetic non-complementation and protein analysis indicates that F1-9-13FP mutants are MEMO1-null
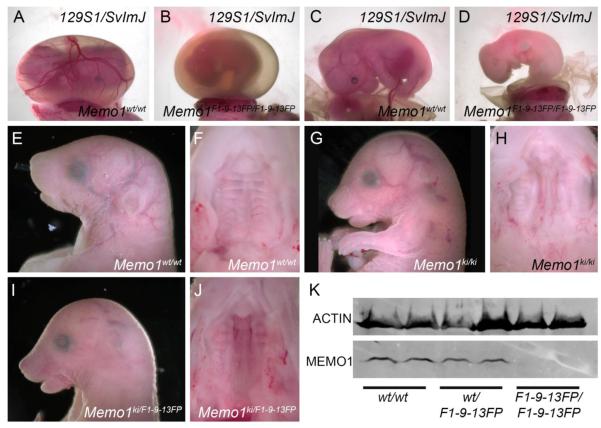
Previously, Memo1 null mutants were observed to die beginning at ~E13.5, accompanied by vascular abnormalities, but defects in craniofacial development were not reported 22. The differences in phenotype between Memo1 null and F1-9-13FP mutants suggest that if Memo1 is responsible for the F1-9-13FP phenotype either the phenotype is dependent on genetic background and/or the ENU-induced mutation generates a hypomorphic allele. To distinguish between these possibilities, we outcrossed the ENU allele, which was generated on a C57BL/6J background, onto the 129S1/SvImJ background (>95% 129S1/SvImJ), the strain used for the previous Memo1 mutant studies 22. During successive out-crossings, no F1-9-13FP(129S1/SvlmJ) homozygous mutants were identified at late embryonic stages (E18.5). However, Memo1 mutants were detected at earlier time-points (~E11.5-E14.5) and similar to earlier studies, these embryos displayed reduced vascular development within the yolk-sac (Fig 3A,B). In addition, in F1-9-13FP(129S1/SvlmJ) mutants, the embryo proper began to show defects in vasculogenesis, displaying a combination of unorganized blood vessel formation, general hemorrhaging and edema, associated with embryonic lethality evident around E12.5-E14.5 (Fig 3C,D and not shown).
Figure 3. F1-9-13FP generates a null mutation in Memo1 that is subject to genetic background effects.
(A-D) An E14.5 control (A, C) or Memo1m1Will mutant (B, D) on the 129S1/SvlmJ genetic background showing the intact yolk-sac (A, B) or a lateral view of the embryo proper (C, D). (E-J) E18.5 mouse embryos, of the genotypes indicated, from a largely C57BL/6J background showing lateral views of the head (E, G, I) or ventral views of the secondary palate after the mandible has been removed (F, H, J). (K) Western blot analysis of protein isolated from E13.5 heads of wild-type, heterozygous, or homozygous F1-9-13FP mutants, and probed with an anti-MEMO1 antibody (anti-ACTIN used as a loading control, one embryo per lane).
In a reciprocal experiment, we used available targeted ES-cells from the European Conditional Mouse Mutagenesis (EUCOMM) repository to generate mice with a Memo1ki allele (EUCOMM tm1b allele, referred to as Memo1ki from this point forward) lacking exon 4, but containing a LacZ expression cassette (Supplemental Fig 2C) 26. We subsequently generated Memo1ki/ki offspring on a largely C57BL/6J background (>80%) and then assessed their embryonic phenotype. Examination of Memo1ki/ki embryos at E18.5 identified a similar defect to that observed for F1-9-13FP mutants on the C57BL/6J background, namely a domed head, snout defects (Fig 3E, G) and clefting of the secondary palate (Fig 3F, H). Moreover, Memo1ki/ki mutants were unable to thrive after birth, as none were found at weanling stages (P21-P28). In addition, genetic non-complementation analysis was carried out between the Memo1ki allele and the F1-9-13FP allele on a largely C57BL/6J background and the resulting offspring assessed at E18.5. As anticipated, embryos harboring both the KI and ENU alleles displayed the aforementioned craniofacial defects, whereas heterozygous embryos were indistinguishable from control embryos (Fig 3I,J and not shown). Together, these findings support the hypothesis that the embryonic phenotypes associated with loss of MEMO1 are highly dependent on genetic background. The similarity between the Memo1ki/ki and F1-9-13FP mutant phenotypes further suggested that the ENU-induced mutation generated a null allele. We therefore assayed MEMO1 protein expression using a monoclonal antibody that recognizes the N-terminus of MEMO1 21. Western blot analysis of protein isolated from E13.5 heads of either the F1-9-13FP mutants (Fig 3K) or Memo1ki/ki mutants (Supplemental Fig 3) showed a complete absence of protein in both instances indicating that the ENU induced mutation likely generates a null allele. In sum, genetic complementation coupled with protein analysis confirms: 1. Memo1 is the affected locus in F1-9-13FP mutants (allele referred to as Memo1m1Will from this point forward); 2. genetic background alters the embryonic phenotype due to loss of MEMO1; and 3. the ENU-induced mutation generates a null allele.
Analysis of Memo1 expression during embryonic craniofacial development
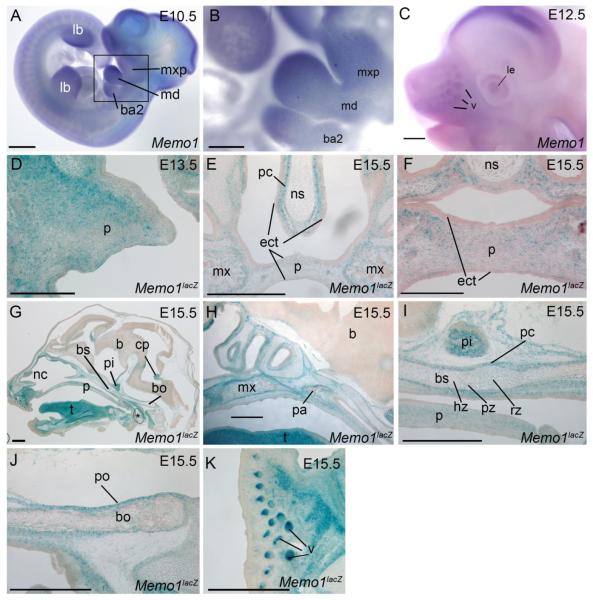
Expression analysis was next performed to gain insight into MEMO1’s role during normal craniofacial development. Previous reports have identified a generally ubiquitous expression pattern for Memo1 during embryonic development as well as expression within most adult organs 22,27. However, a more thorough analysis of Memo1 expression, specifically within the craniofacial region during key stages of development, has not been carried out. Therefore, using a combination of RNA in situ hybridization, the Memo1neo LacZ reporter line (EUCOMM tm1a allele, referred to as Memo1neo from this point forward, Supplemental Fig 2A), and global gene expression analysis, we more closely examined Memo1 expression specifically within the developing head, focusing most heavily on the secondary palate and cranial base, given the defects observed in these structures (Fig 1). RNA expression profiling of the facial prominences indicated that Memo1 was expressed in all three facial prominences, in both the ectoderm and mesenchyme, between E10.5-E12.5 (Supplemental Fig 4). Next, we employed whole mount analysis of Memo1 RNA expression to refine the spatial expression pattern (Fig 4 A-C). At E10.5 Memo1 RNA was detected throughout the facial prominences (frontonasal, maxillary, and mandibular) as well as in the 2nd branchial arch (Fig 4A, B). Interestingly, expression levels were not uniform, with highest signal seen at distal versus proximal locations in the facial prominences (Fig 4A, B). Elevated levels of Memo1 expression were also detected in the distal limb buds at E10.5, and to a lesser extent in a segmented pattern in the developing trunk (Fig 4A). At E12.5 Memo1 expression produced a more diffuse expression pattern in the developing facial prominences, with some enrichment in the developing lens and vibrissae (Fig 4C). For the analysis of later time-points, we utilized embryos heterozygous for the Memo1neo LacZ knock-in allele (Supplemental Fig 2A) to study specific regions of the developing head. With respect to the secondary palate, analysis of E13.5 Memo1neo/+ heads demonstrated β-galactosidase activity (i.e. LacZ expression) diffusely throughout the nasal side of the developing palatal shelf mesenchyme (Fig 4D). By E15.5, after the palatal shelves had fused, LacZ expression was reduced at the midline of the palate, but was enriched at the periphery near the developing maxillary bones, as well as locations internal to locations within these developing bones (Fig 4E, F). Expression was also detected in the presumptive perichondrium surrounding the nasal cartilage (Fig 4E). In contrast, expression in the ectoderm of the palatal shelves or nasal septum was far less prominent. Analysis of the cranial base at E15.5 revealed strong LacZ expression in the perichondrium and periosteum of the developing cartilage and bone, respectively (Fig 4G-J). Expression was also detected at lower levels in cells at various stages of chondrocyte differentiation (resting, proliferative, hypertrophic) (Fig 4I) as well as internal to locations within these developing cranial base bones (Fig 4I, J). Other notable sites of enriched LacZ expression in the head included the tongue (Fig 4G), pituitary gland (Fig 4G, I), vibrissae (Fig 4K), and the choroid plexus (Fig 4G). In summary, although the expression of Memo1 is extensive in the developing embryo, sites of enriched expression are evident within the facial prominences at E10.5. Furthermore, at later embryonic time-points Memo1 expression occurs in the mesenchyme of the palatal shelves and associated with bones and cartilage of the cranial base, maxilla, and nasal septum consistent with the occurrence of defects in many of these structures.
Figure 4. Embryonic expression of Memo1 during craniofacial development.
(A-C) Lateral views of embryos processed by whole-mount in situ hybridization using an anti-sense Memo1 riboprobe at E10.5 (A, B) and E12.5 (C). Note, (B) is boxed region in (A) shown at higher magnification. (D-K) b-galactosidase staining (blue) of frozen Memo1neo/+ sections at the time points indicated in either a frontal (D-F) or sagittal (G-K) plane, counterstained with nuclear fast red. A single palatal shelf is shown at E13.5, prior to fusion (D), or both shelves at E15.5 subsequent to secondary palatal fusion (E, F). Note, E is slightly more posterior than F. Additional images from E15.5 show a midsagittal plane of the entire embryonic head (G) or detailed images of the secondary palate and anterior cranial base (H), mid-cranial base (I), posterior cranial base (J), and snout (K). Orientation for (G-K) is rostral left, caudal right. Abbreviations: b, brain; ba2, branchial arch 2; bs, basisphenoid; bo, basioccipital; cp, choroid plexus; ect, ectoderm; hz, hypertrophic zone; lb, limb bud; le, lens; md, mandibular prominence; mx, maxilla; mxp, maxillary prominence; nc, nasal cavity; ns, nasal septum; p, palate; pa, palatine bone; pc, perichondrium; pi, pituitary gland; po, periosteum; pz, proliferative zone; rz, resting zone; t, tongue; v, vibrissae. Note, asterisk in G. highlights air bubble from processing. Scale bars: A, C, E, G-K: 500uM; B: 125uM; D, F: 200uM
Analysis of neural crest cell migration and palatogenesis in Memo1m1Will mutants
Gross phenotype assessment and skeletal analysis indicated major defects in the secondary palate and cranial base in Memo1m1Will mutants (Fig 1). To understand the mechanistic basis of these defects we conducted a variety of histological, molecular, and genetic studies on these two structures during development.
One common link between the secondary palate and the cranial base is that both have major contributions from neural crest cells. Moreover, MEMO1 has been previously linked with cell migration, a process that is also critical for neural crest cell contribution to the developing head 19,20,28. Therefore, we began by studying general neural crest cell development and migration in Memo1m1Will homozygous mutant mice. Tfap2a 29,30, Inka1 31, and Hoxa2 32 expression were used as markers of cranial neural crest cell migration and we determined that this process was equivalent between mutant and control embryos at E9.5 and E10.5 (Supplemental Fig 5A-D and not shown). Next, we employed anti-neurofilament immunostaining and Sox10 RNA expression to study the differentiation of neural crest cells into peripheral nervous system components, including the cranial ganglia. Again, we did not detect any differences between mutant and control embryos in this assay (Supplemental Fig 5F, G and not shown). To assess later stages of neural crest cell migration into the developing palatal shelves, we utilized the combination of a Wnt1-Cre transgene and a rosa-Tomato reporter line 33,34. Examination of labeled neural crest cells in the E13.5 palatal shelves of either control or Memo1-null embryos indicated successful population of the facial mesenchyme in both strains (Supplemental Fig 5H, I). Overall, these findings suggest that loss of MEMO1 does not cause a generalized defect in neural crest cell induction, migration or differentiation that would account for the observed craniofacial phenotypes.
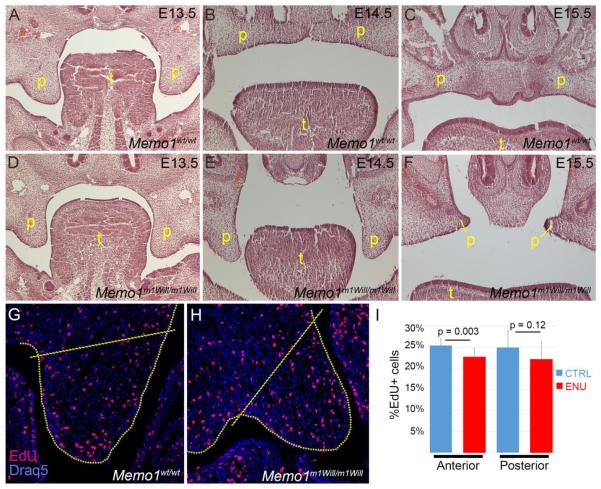
Next, we performed a histological analysis of palatogenesis in Memo1m1Will mutants and littermate controls to determine the step at which fusion failed. Serial frontal sections were H&E stained from E13.5 to E15.5, the time at which the palatal shelves normally elevate and fuse (Fig 5). At E13.5 in a wild-type embryo, the paired palatal shelves are growing parallel to the developing tongue (Fig 5A). Subsequently, by E14.5, the palatal shelves transition to a horizontal plane above the tongue and shortly thereafter the paired shelves meet at the midline and begin to fuse (Fig 5B). Finally, by E15.5 the epithelium at the midline of the fused palatal shelves (midline epithelial seam) is removed, resulting in a single contiguous layer of mesenchyme between the now fused shelves (Fig 5C). Memo1m1Will mutants were initially indistinguishable at E13.5 from controls, but by E14.5 the palatal shelves had failed to elevate into the horizontal plane in both anterior and posterior positions of the developing palate (Fig 5D,E and data not shown). By E15.5, the palatal shelves remained separate as unfused remnants resulting in the presence of a cleft palate (Fig 5F). Since neural crest cells were able to populate the shelves (Supplemental Fig 5H, I), we next considered that their failed elevation might result from altered neural crest cell differentiation or cell proliferation. However, changes in gene expression (measured using RNA-Seq analysis or the examination of specific marker expression in vivo) or cell proliferation (measured using EdU incorporation) were relatively modest (Fig 5G-I and Supplemental Fig 5E, J-Q). In addition, late stage analysis of EdU incorporation (E14.5) or cell death (TUNEL, E15.5) in the palate did not identify major changes between control and Memo1m1Will mutants (data not shown). Taken together, these results suggest that the failure of secondary palate closure in Memo1-null embryos either results from subtle defects within the shelves themselves or that it is secondary to developmental events occurring elsewhere in the Memo1-null mutant head.
Figure 5. Memo1m1Will mutant embryos undergo a failure of palatal shelf elevation and show a modest reduction in mesenchymal cell proliferation in the palatal shelves.
(A-F) H&E stained frontal sections midway through the developing palate of control (A-C) or Memo1m1Will mutant (D-F) embryos at the time point indicated. (G, H) Representative images of frontal sections through the anterior palatal shelf at E13.5 in a control (G) or Memo1m1Will mutant (H) embryo that have been processed for EdU immunoreactivity to mark proliferative cells. Area internal to dashed lines indicates region quantified in I. (I) Total number of proliferating cells relative to the total cell number in the palatal shelf mesenchyme was quantified between control and Memo1m1Will mutant embryos in both anterior and posterior positions of the developing palate. (N = 3 embryos per group, 10-12 individual palatal shelves [3-4 per embryo] per location). Abbreviations: p, palate; t, tongue.
Memo1m1Will mutants show normal chondrocranium development but reduction in regulators of endochondral ossification in the cranial base
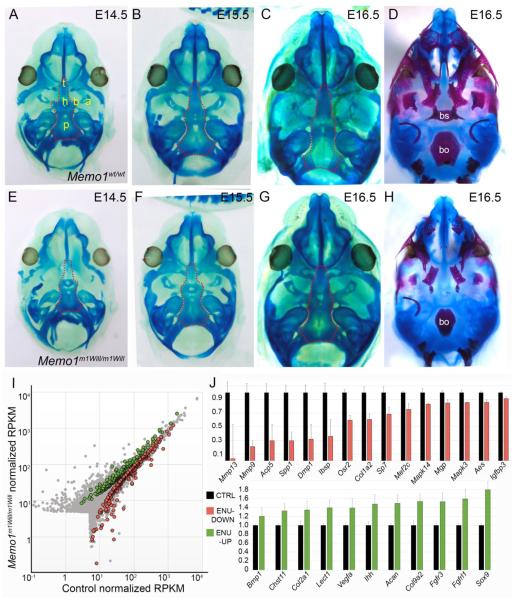
Next, to assess development of the cartilage anlage of the cranial base we stained control and Memo1m1Will mutant embryos with alcian blue (Fig 6A-H). At E14.5, cartilage deposition in the anterior cranial base is slightly delayed in Memo1m1Will mutants as compared to controls (Fig 6A,E). However, by E15.5, cartilage condensation and formation appears equivalent in the cranial base by gross examination in controls as compared to in Memo1m1Will mutants (Fig 6B,F). By E16.5, alcian blue negative regions are evident in the mid and posterior cranial base of a control embryo, corresponding to zones of ossification in the basisphenoid and basioccipital, respectively, as seen by alizarin red staining (bone stain) at a similar stage (Fig 6C,D). By contrast, in Memo1m1Will mutants at an identical developmental stage, the majority of the cranial base remained strongly stained with alcian blue (Fig 6G). This persistent staining corresponds with reduced ossification of the cranial base bones at a similar stage (Fig 6H). Overall, this analysis identified that development of the chondrocranium (cartilage condensation) proceeded largely unabated in Memo1m1Will mutants. However, following condensation, replacement of cartilage with ossified bone failed to occur at control levels.
Figure 6. Memo1m1Will mutants show normal chondrocranium development but reduction in regulators of endochondral ossification in the cranial base.
(A-H) Ventral views of the developing cranial base with mandible removed, in control (A-D) or Memo1m1Will mutants (E-H). Samples are stained with either alcian blue alone for cartilage (A-C, E-G), with the dotted outline highlighting the cranial base, or alcian blue plus alizarin red, marking cartilage and bone (D, H). (I) Scatter plot depicting average RPKM (reads per kilobase per million mapped reads) values of RNAseq conducted on RNA isolated from E15.5 cranial bases (similar to outlined area in B and F) of control (X-axis) or Memo1m1Will mutants (Y-axis) (3/group). Colored points represent transcripts that are statistically significant between groups (Green: up-regulated in Memo1m1Will, ~220 genes; and red: down-regulated ~300 genes). (J) Upper panel: histogram plotting normalized RPKM values of genes associated with endochondral ossification and significantly down-regulated in Memo1m1Will mutant cranial base samples as compared to controls. Lower panel: same as upper panel, but plotting significantly up-regulated genes from Memo1m1Will cranial base samples as compared to controls and associated with cartilage development. Error bars represent standard error calculated from 3 replicates. Abbreviations: a, ala temporalis cartilage; b, basitrabecular process; bo, basioccipital; bs, basisphenoid; h, hypophyseal cartilage; p, parachordal cartilage; t, trabecular cartilage (trabecular plate).
To assess the molecular changes associated with the difference in cranial base ossification we dissected cranial base tissue from both control and Memo1m1Will mutants at E15.5 (3/group, similar to outlined region in Fig 6B and F) and analyzed global gene expression differences by RNA-seq. In contrast to the small number of statistically significant differences observed for the palatal shelves, gene expression differences for the cranial base were far more pronounced. Thus, Memo1m1Will mutants showed ~300 genes significantly down-regulated and ~220 genes significantly upregulated (at p<0.05), as compared to controls (Fig 6I, Supplemental Table 1). Functional annotation of all genes showing significantly altered expression using DAVID 35 indicated that the most significantly affected categories encompassed the extracellular matrix, skeletal system development and mitochondrial function (Supplemental Table 1).
The degree of gene expression change was more pronounced in the down-regulated category, with ~20 genes showing a greater than 4-fold reduction in mapped transcripts in the mutant cranial base, while no genes were increased by more than 4-fold. Functional annotation clustering using genes most highly down-regulated (p < 0.05 and >1.35 fold-change, 139/520 significant genes) captured skeletal system and ossification categories (Supplemental Table 1). Prominent within the down-regulated category were genes involved in degradation of a collagen-based extracellular matrix (ECM) that are required for endochondral ossification, particular Mmp13 and Mmp9, which were decreased by >30 fold and ~5 fold respectively (Fig 6J, upper panel) 36-39. Several other genes involved with formation or modification of the extracellular matrix (Acp5/TRAP, Spp1, Dmp1, and Ibsp), as well as markers of the osteoblast lineage (Sp7/Osterix, Col1a2, and Osr2) were also expressed at significantly lower levels in the mutants, relative to controls (Fig 6J, upper panel) 40-46. Conversely, functional annotation clustering of the most significantly upregulated genes (p < 0.05 and >1.35 fold-change, 123/520 significant genes) captured categories associated with cartilage development, cartilage condensation, and ECM (Fig 6J, lower panel, Supplemental Table 1). These genes included those encoding proteins involved in cell signaling (Ihh, Fgfr3, Fgfrl1), transcription factors (Sox9), and cartilage ECM proteins (Acan/Aggrecan, Col2a1, Col9a1, Lect1) 47-54. In summary, global gene-expression profiling identified a significant reduction in expression of genes important for endochondral ossification, concomitant with an increase in expression of cartilage specific genes. These findings support those seen by the cartilage and bone staining (Fig 6), namely an inhibition of endochondral ossification at a stage subsequent to cartilage formation in Memo1m1Will mutants. In addition, the data indicate these changes are associated with alterations in the expression of genes involved in formation and degradation of the ECM, as well as respiration, suggesting a complex interplay of several physiological processes in driving MEMO1-dependent cranial base development.
Reduced endochondral ossification in Memo1m1Will mutants is associated with disrupted hypertrophic chondrocyte replacement, vascular invasion, and mineralization of the developing bone
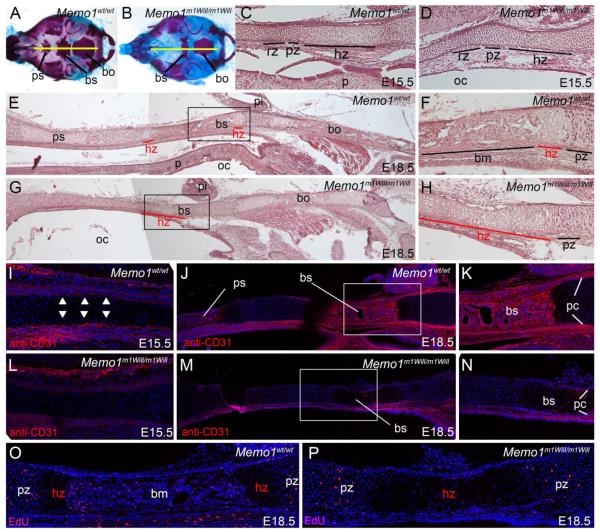
Chondrocranium analysis by histology and RNA expression profiling indicated a failure of Memo1m1Will mutants to transition from a cartilage template anlage to ossified bone (Fig 6). This transition has been well defined and includes precisely staged events marked by distinct cellular identities, including resting, proliferative, and hypertrophic chondrocytes, followed by vascular recruitment and subsequent invasion by proliferating osteoblasts, which promote bone mineralization 14,15. Therefore, we examined the differentiation status of the cartilage and its associated vasculature in greater detail in Memo1m1Will mutants versus controls using both histology and molecular marker analysis. To begin, we examined H&E midsagittal sections (Fig. 7A, B) through the cranial base at E15.5 and E18.5 in control and Memo1m1Will mutants (Fig 7C-H). In particular, we focused on the basisphenoid bone, as this element is clearly affected in Memo1m1Will mutants and its stages of ossification are well defined at these developmental stages. At E15.5, resting, proliferative, and hypertrophic cartilage zones were clearly demarcated in the cranial base of a control embryo (Fig 7C). Similar zones were evident at E18.5, although the hypertrophic chondrocyte zone was quickly replaced by mineralized bone matrix, forming the basisphenoid bone (Fig 7E, F). In contrast, at E15.5 in Memo1m1Will mutants, resting and proliferative zones were present, although the progression of hypertrophic chondrocytes was slightly perturbed, evident by these chondrocyte cells appearing less hypertrophic (enlarged) as compared to controls (Fig 7D). By E18.5, defects in cartilage to bone progression in the Memo1m1Will mutant cranial base became even more evident. Although Memo1m1Will mutants displayed grossly normal resting and proliferative zones, an abnormally elongated zone of hypertrophic chondrocytes was evident (Fig 7G, H). Concomitant with this exaggerated hypertrophic zone was a major reduction in matrix deposition within the region that would normally form the basisphenoid bone (Fig 7G, H). These findings suggest defects in cranial base ossification in Memo1m1Will mutants are associated with a general failure in conversion of hypertrophic chondrocytes to mineralized bone.
Figure 7. Reduced ossification in the cranial base of Memo1m1Will mutants is associated with an elongated hypertrophic chondrocyte zone and reduced vascular invasion.
Analysis of the cranial base skeleton at E15.5 (C, D, I, L) and E18.5 (A, B, E-H, J-K, M-P) for the genotypes indicated shown as either whole mount (A, B) or midsagittal sections (C-P). For all images, rostral is to the left and caudal to the right. (A, B) Craniofacial skeletons used to diagram approximate position of sections (yellow line) through the cranial base. Note, skeletons shown are E18.5, although similar sections were used at E15.5. (C, D) H&E stained sections, focusing on the developing basisphenoid bone. (E-H) H&E stained sections, focusing on the entire cranial base (E, G) or the basisphenoid bone alone (F, H). Note, F and H are higher magnification images of the boxed regions in E and G. (I-N) anti-CD31 stained frozen sections, focusing on the developing basisphenoid bone. Note, K and N are higher magnification images of the boxed regions in J and M. Arrowheads in (I) highlight perichondrial staining. (O, P) EdU staining, marking proliferating cells, in the basisphenoid bone and flanking cartilage. Note, for sections in I-P, nuclei were counterstained with DRAQ5. Abbreviations: bm, bone matrix; bo, basioccipital; bs, basisphenoid; hz, hypertrophic zone; oc, oral cavity; p, palate; pi, pituitary; pc, perichondrium; ps, presphenoid; pz, proliferative zone; rz, resting zone.
A key event in promoting the transition from hypertrophic chondrocytes to ossified bone is the ECM remodeling activity of MMP9 and MMP13, which have been shown to free ECM bound VEGFA within the hypertrophic chondrocyte zone, allowing diffusion of the ligand to the perichondrium/periosteum, thus promoting vascular invasion and bone ossification 55,56. Interestingly, expression profiling identified significant reductions in both Mmp9 and Mmp13 mRNA levels in the cranial base of Memo1m1Will mutants (Fig 6J).
Previous studies in long bones have shown that perturbation of MMP9/MMP13 activity can result in a similar elongated zone of hypertrophic chondrocytes associated with reduced vascular invasion of this zone 38. To assess the status of vascular recruitment into the hypertrophic zone of Memo1m1Will mutants versus controls, we examined mid-sagittal sections of E15.5 and E18.5 embryos via anti-CD31 immunoreactivity, which marks endothelial cells of the developing vasculature 57. In E15.5 control embryos, CD31+ cells surrounded the hypertrophic zone, in the perichondrium near the future basisphenoid bone (Fig 7I, arrowheads). By E18.5, this immunoreactivity was more prominent in the perichondrium and was seen throughout the mineralized portion of the basisphenoid bone (Fig 7J, K). By contrast, in Memo1m1Will mutants, reduced CD31+ cells were seen lining the hypertrophic zone at E15.5 (Fig 7L). This reduction in CD31+ cells in the perichondrium of the hypertrophic zone was still evident at E18.5 and coincided with a failure of vascular invasion of the elongated hypertrophic zone of cartilage that occurs in place of the normal basisphenoid bone matrix (Fig 7M, N). Coincident with reduced vascular invasion, there was also a drastic reduction in the amount of EdU+, proliferating cells (presumed osteoblasts) within the developing basisphenoid bone of Memo1m1Will mutants as compared to controls (Fig 7O, P). The observation that the vascular network does not surround the cartilage from E15.5 through E18.5 at levels similar to control embryos in the cranial base of the mutants, while general maturation of the embryo still occurs normally, also argues against the phenotype being caused by a generalized developmental delay. Instead, these results indicate that the loss of MEMO1 has a specific effect on the cranial base, with reduced mineralization of cranial base bones in Memo1m1Will mutants associated with a failure of hypertrophic chondrocyte removal and a loss of vascular invasion of these skeletal elements.
MEMO1 acts cell-autonomously within the cranial neural crest cells during cranial base ossification but not for closure of the secondary palate
The secondary palate is derived from both neural crest cells and ectoderm, while the cranial base is composed of both neural crest cells and mesoderm derivatives. Therefore, we next sought to determine whether MEMO1 functions autonomously in the shared cranial neural crest cell component to drive both cranial base and palate development. The Memo1neo allele was used as a starting point for generation of a conditional Memo1 allele (EUCOMM tm1c allele, termed Memo1flp from this point forward, Supplemental Fig 2B, see methods). Subsequently, mice homozygous for the Memo1flp allele (Memo1flp/flp) were bred with mice carrying the Memo1ki allele plus the Wnt1CRE transgene, allowing precise deletion of MEMO1 from the developing neural crest cells (Supplemental Fig 2D) 33.
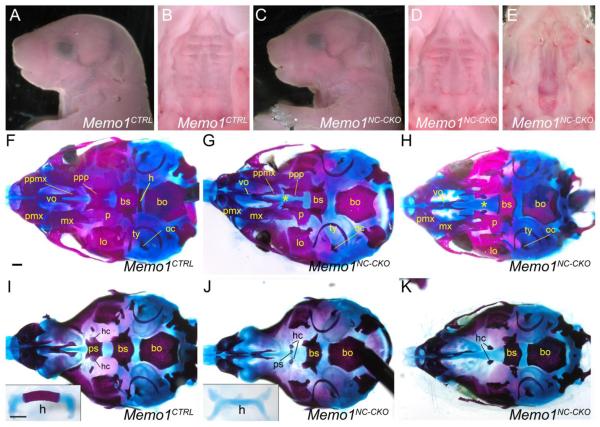
At E18.5, the gross external appearance of the Memo1 conditional knockout (Memo1NC-CKO) was similar to a milder version of the Memo1m1Will mutant phenotype, with a slightly dipped nasal bridge, and a domed skull (Fig 8A, C). However, with the exception of one mutant, the majority of these embryos did not have a cleft palate (9 of 10; Fig 8B, D, E). Similar to Memo1m1Will mutants, Memo1NC-CKO pups did not survive after birth, likely due to an inability to suckle as no milk was observed in the stomachs of mutants irrespective of the presence of an obvious cleft palate (data not shown). We next assessed the underlying skeleton of the mutants at E18.5 to determine the effect of the neural crest cell specific loss of MEMO1 on both the palate and the cranial base. For the majority of mice that did not have an overt cleft secondary palate we still noted hypoplastic development of both the palatal process of the palatine bone, and to a lesser extent the palatal process of the maxillary bone (Fig 8F,G). In contrast, the palatal bones in the single Memo1NC-CKO mutant that displayed a cleft palate more closely resembled those found in the Memo1m1Will mutant (Compare Fig 8H and 1F). With respect to the cranial base, the presphenoid is the only bone that is derived from the neural crest 7, and in common with the Memo1m1Will mutants this bone failed to ossify or was severely hypoplastic in the Memo1NC-CKO mutants (Fig 8I-K). In contrast, the basisphenoid was far less affected, displaying a mild rostral truncation, and the basioccipital had a normal appearance (Fig 8I-K). These latter two bones are derived from mesodermal cells (cranial paraxial and somitic, respectively) and their appearance was consistent with the lack of a direct neural crest cell influence on their phenotype. Examination of other bones in the cranial base and remaining craniofacial complex also largely supported a cell autonomous role for MEMO1 in the neural crest cells during skeletal development. Thus, the premaxilla was hypoplastic, and the hyoid failed to ossify in common with the Memo1m1Will mutants (Fig 8I,J). In contrast, mesodermally derived bones, including the two small bones formed from the hypochiasmatic cartilages that flank the presphenoid bone, and the otic capsules, were less affected. These bones were absent in the Memo1m1Will embryos but were visible in Memo1NC-CKO embryos (compare Fig 1H and Fig 8I-K). In addition, note that the single mutant that had a clear cleft secondary palate also had a more severely affected presphenoid (Fig 8K). Finally, regions corresponding to the neural crest cell derived metopic and coronal sutures were more widely spaced in Memo1NC-CKO as compared to controls (Supplemental Fig 6A, B). Taken together, these findings support a cell autonomous role for MEMO1 in the neural crest cells with respect to cranial base ossification, but also indicate that loss of MEMO1 in the mesenchyme of the palatal shelves may not be the underlying cause of overt cleft secondary palate in Memo1-null embryos.
Figure 8. MEMO1 has a cell-autonomous function within neural crest cells for cranial base ossification but not for palatal shelf fusion.
(A-E) E18.5 mouse embryo of the genotype indicated, either shown in lateral view (A and C) or a ventral view of the secondary palate (B, D, and E). (F-K) E18.5 craniofacial skeletal preparations of either control (F, I) or Memo1NC-CKO embryos (G, H, J, K), shown in ventral views with either the mandible (F-H) or mandible plus palatal bones (I-K) removed. Insets in I and J show the hyoid element in isolation. “*” denotes region of hypoplastic development of palatal processes (maxillary and palatine bones). Abbreviations: bs, basisphenoid; bo, basioccipital; h, hyoid; hc, hypochiasmatic cartilage; lo, lamina obturans; mx, maxilla; oc, otic capsule; p, palatine; pmx, premaxilla; ppmx, palatal process of maxilla; ppp, palatal process of palatine; ps, presphenoid; ty, tympanic ring; vo, vomer. Scale bars: 500uM.
Discussion
Here we identify Memo1, a gene that encodes a previously described receptor tyrosine kinase (RTK)-adapter molecule and reactive oxygen species (ROS) generating redox enzyme, as a novel effector in promoting bone ossification, most notably in the cranial base 21,22. The importance of MEMO1 in craniofacial development was initially identified through our recessive ENU-based forward genetic screen. Subsequently, non-complementation and protein analyses indicated that the ENU-induced mutation in Memo1 results in a null allele. Moreover, our studies show that the embryonic phenotypes are strongly dependent on mouse genetic background. In agreement with previous studies, on a 129 background, the loss of MEMO1 results in mid-gestation lethality 22. However, on a C57BL/6J background, Memo1-null mice can live to birth and display many craniofacial defects, particularly cleft secondary palate and alterations in the cranial base, a structure known to form through endochondral ossification. The cranial base has a major role in determining the morphology of the mammalian head, and the following features were consistently observed with respect to this central skeletal structure in Memo1 mutants. First, the cranial base anlage, the chondrocranium, formed appropriately in the absence of Memo1, although there was a slight initial delay in its development. Second, as chondrocranium development progressed, replacement by ossified bone was defective, resulting in a persistent cartilage template. This observation was supported molecularly by RNA expression profiling of the cranial base, which identified an increase in transcripts associated with cartilage biology and a concomitant decrease in ossification associated transcripts in Memo1 mutants. Third, reduced ossification coincided histologically with a loss of vascular invasion of the developing cranial base skeletal elements. Initially, this was seen as reduced vascular cells in the perichondrial lining of the cartilage hypertrophic zone and subsequently a loss of vascular invasion of the ossified bone itself. Fourth, the failure of bone mineralization was associated with an elongated zone of hypertrophic chondrocytes. Collectively, these observations suggest that MEMO1 facilitates the replacement of terminally differentiated chondrocytes by ossified bone in the cranial base, potentially through regulating vascular development.
In what germ layers, tissues, and cell types does MEMO1 function during cranial base ossification? Using the LacZ reporter allele to track Memo1 expression, we found most robust β-galactosidase activity in the perichondrium and periosteum of the chondrocranium. Less prominent β-galactosidase staining was observed in various zones of chondrocyte maturation, including resting, proliferative, and hypertrophic chondrocytes. In addition, punctate β-galactosidase activity was observed within developing bones of the cranial base (as well as intramembranous bones, such as the maxillary and palatine bone). These findings suggested that MEMO1 could function in multiple components of the developing cranial base. Therefore, we have employed Cre-recombinase mediated deletion of Memo1 to begin probing the tissue specific role of this gene in craniofacial development. Focusing on the neural crest, we have determined that MEMO1 has an autonomous role within this tissue during cranial base ossification, as the most anterior element, the presphenoid, failed to ossify. Given that neural crest cells supply the undifferentiated mesenchyme of the anterior cranial base 7, and thus all tissues of the cartilage anlage, the supporting perichondrium and periosteum, and bone forming cells (osteoblasts), MEMO1’s role is likely autonomous to one of these cell types. In this context, it is important to consider the complex interplay of mesoderm and neural crest cells in development of the cranial base. Thus, both the cartilage and bone of the presphenoid are neural crest cell-derived, whereas the basioccipital cartilage and bone is derived entirely from the somitic mesoderm. Between these elements, the basisphenoid has a more complex origin, with the cartilage anlage of the basisphenoid bone mainly neural crest cell-derived, whereas the bone originates from cranial paraxial mesoderm 7-9. Importantly, following neural crest cell-specific deletion of Memo1 the basisphenoid bone is relatively unperturbed, especially in comparison to the full null, where it is hypoplastic. This finding suggests that any defects in the neural crest cell-derived cartilage can be at least partially rescued if the bone (i.e. osteoblasts) itself is derived from the mesoderm and further supports a prominent role for MEMO1 at a “post-cartilage development” stage. Moreover, we note that additional bones of the cranial base can form normally in the neural crest cell-conditional mutant compared with the Memo1-null. Thus, the presence of bones derived from the hypochiasmatic cartilages in the conditional deletion, but not the null, highlights the mesodermal origin of these elements and further indicates that MEMO1 presumably functions in the mesoderm to regulate their ossification. In the future, tissue-specific deletion of Memo1 in additional domains should pinpoint the critical tissues/cell types in which this gene might function during cranial base ossification, including the mesoderm, cartilage, osteoblasts, and perichondrium. It is also possible that MEMO1 could function in the blood vessels associated with the developing cranial base, especially given the apparent vasculature defects observed when Memo1 is lost on the 129S1/SvImJ background 22. Indeed, the neural crest cells are known to provide pericytes to the craniofacial vasculature 58. However, we do not favor this possibility as previous studies targeting Memo1 with the endothelial-specific Tie2-Cre frequently resulted in mice that were viable in the post-natal period 22, implying that primary defects in the endothelial component of the vasculature is not a major source of the embryonic lethality associated with the craniofacial defects we observed in our neural crest-specific conditional mutants.
What is the function of MEMO1 in driving cranial base ossification? Based on previous roles identified for MEMO1 as well as the findings presented here, there are a variety of non-mutually exclusive models for how MEMO1 may drive ossification of the cranial base. First, MEMO1 may facilitate signaling downstream of an RTK during cranial base ossification. Specifically, analyses in other physiological and developmental processes have shown that MEMO1 mediates signaling downstream of a variety of RTKs, including ERBB2 19,59, EGFR 19, FGFR 19,27, IGF1R 59, and PDGFR 22. Interestingly, previous work has also identified a role for these signaling pathways, including their effectors such as ERK1/2, in driving endochondral ossification, either in the cranial base directly or other endochondral bones 60-65. VEGFR1 (Flt1) and/or VEGFR2 (Flk1) are also excellent RTK candidates that might function in concert with MEMO1 as disruption to VEGF signaling results in reduced ossification associated with decreased vascularity and an elongated hypertrophic zone, similar to Memo1 mutants 66-68. In addition, MEMO1 has been shown to interact with and influence downstream signaling from the estrogen receptor, another key pathway essential for bone development and homeostasis 59,69. Thus, one possibility is that the loss of MEMO1 would uncouple one or more RTKs or signaling pathways from its downstream function in driving ossification. In turn, lost or reduced signaling could ultimately lead to a reduction in expression of cartilage to bone promoting factors, such as MMP9 and MMP13, as has been described during ossification in EGFR mutants 70. A second plausible model for how MEMO1 may promote ossification is through its ability to generate ROS, a recently identified biochemical function for MEMO1 in other contexts 21, and a known mechanism driving endochondral ossification 71-73. ROS production during endochondral ossification has been shown to promote chondrocyte maturation and eventual apoptosis during bone replacement 71-73. In addition, ROS has been shown to drive mRNA expression of pro-osteogenic Mmp13 in chondrocytes 74, a gene which was also significantly down-regulated in the cranial base of Memo1 mutants. Physiological levels of ROS are also required for appropriate vasculogenesis, and alterations in ROS production could conceivably impact the distribution of blood vessels in the cranial base. Therefore, the loss of MEMO1 function in ROS production would affect many of the physiological pathways required for normal endochondral ossification. Future studies will be needed to address the biochemical function(s) of MEMO1 that is critical in regulating endochondral ossification.
Another major defect evident in Memo1 mutants is clefting of the secondary palate. MEMO1’s role during overt palatal closure largely appears to be extrinsic to this structure based on the following observations. First, despite a failure in palatal shelf elevation in Memo1 mutants, thorough analysis of the palate identified few molecular changes. Second, MEMO1 was only expressed at a moderate level within the palatal shelf mesenchyme directly, especially by E15.5. Third, conditional deletion of MEMO1 from neural crest cells, in the majority of cases, failed to recapitulate the cleft palate phenotype as seen in Memo1 null embryos. As such, palatal bones (maxillary and palatine) were less affected in conditional mutants, despite their neural crest cell origin 11,13,23. Preliminary data also show that loss of Memo1 in the ectoderm of the palatal shelves does not cause an overt clefting phenotype (EVO, unpublished observations). Taken together, these findings suggest that the overt cleft palate phenotype may be secondary to primary defects in other tissues, including the cranial base. However, additional testing will be required to test the validity of this model. It is important to note that conditional mutants still presented with pathological defects of the secondary palate, although these were milder than those seen in the null. Specifically, neural crest cell-specific deletion of Memo1 resulted in hypoplastic development of the palatal process of both the maxillary and palatine bones, consistent with a submucosal cleft palate. Importantly, there are currently few mouse models for this common type of human palatal defect but a submucosal cleft palate is present in Tbx22 mouse mutants, responsible for X-linked cleft palate and ankyloglossia in humans. Notably, these Tbx22 mutants die at birth due to an inability to suckle 75 similar to Memo1 neural crest conditional mutants. Future studies will need to address the full extent of MEMO1’s role during palatogenesis and suckling as well as how structures such as the cranial base and tongue – potentially via defects in hyoid ossification - may be influencing these events. Note that defects in the cranial base would provide an additional mechanism by which closure of the palate could be linked with abnormalities in an extrinsic developmental process, alongside a failure of descent of the tongue, as has previously been described 76. Teasing out these developmental relationships will be critical as cranial base defects have been associated with a variety of craniofacial anomalies, including isolated cleft palate. Indeed, the hypothesis that alterations in human cranial base morphology can affect development and clefting of the secondary palate has been the subject of much debate, and our studies provide strong support for such a mechanistic connection 4-6,77-80.
Finally, identification of MEMO1 as a novel effector of both ossification and palatal development implicates it as a potential component of human congenital disorders affecting palatogenesis and/or osteogenesis. To date, no mutations in MEMO1 have been identified in human orofacial clefting, although a case-parent trio study examining candidate genes for non-syndromic cleft palate, which focused on chromosome 2, identified evidence of linkage disequilibrium (LD) for markers located in SLC30A6 81. Interestingly, SLC30A6 is located 50-100Kb from MEMO1 (separated only by the genes SPAST and DPY30), raising the possibility that significant LD is a result of MEMO1, or MEMO1 cis-regulatory elements, in affected individuals. In the context of human osteogenic conditions, multiple studies on patients with low bone mineral density find significant linkage to chromosome 2p23-24, a locus that includes MEMO1 82-84. It is also interesting to note that cranial base anomalies are an important complication of osteogenesis imperfect (OI), associated with mutations in COL1A1, COL1A2, and SP7, and both Col1a2 and Sp7 are significantly down-regulated in the cranial base of Memo1 mutants 85-88. In conclusion, our findings suggest that MEMO1 is part of the regulatory hierarchy controlling the timing of cranial base ossification and palate formation. Thus, we predict that a reduction in human MEMO1 function would lead to the types of cranial base defects observed in OI, such as platybasia, and potentially cleft palate. Alternatively, an increase in MEMO1 activity would lead to premature fusion of the cranial base synchondroses and with associated defects in growth of the face and calvaria. Our studies, therefore suggest that MEMO1 be considered in the pathology related to disorders of ossification, in particular those affecting the cranial base, as well as in palatal clefting.
Materials and Methods
Mice
Male C57BL/6J mice treated with ENU were kindly provided by Monica Justice (Baylor College of Medicine, Houston, Texas, now at The Hospital for Sick Children [SickKids] in Toronto, Ontario). Additional C57BL/6J mice and 129S1/SvImJ mice were obtained from the Jackson Laboratory (Bar Harbor, ME). Wnt1-CRE mice were previously reported and obtained from Jackson Laboratory 33. The rosa-Tomato mouse strain was also previously reported and was a kind gift from the lab of Linda Barlow (University of Colorado - Denver, Aurora, CO) 34. This study was carried out in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. The protocol was approved by the Institutional Animal Care and Use Committee of the University of Colorado Denver. Noon on the day a copulatory plug was present was denoted as embryonic day 0.5. Yolk sacs or tail clips were used for genotyping. DNA for PCR was extracted using DirectPCR Lysis Reagent (Viagen Biotech. Inc, Los Angeles, CA) plus 10ug/ml Proteinase K (Roche, Basel, Switzerland) followed by heat inactivation at 85°C for 45 mins. Samples were then used directly for PCR-based genotyping using the primers listed in Supplemental Table 2 at a concentration of 200 nM using the Qiagen DNA polymerase kit, including the optional Q Buffer solution (Qiagen, Valencia, CA). XmnI used for further analysis of PCR products was obtained from New England Biolabs (Ipswich, MA).
Skeletal analysis
Bone and cartilage staining
Embryos were collected at appropriate time points and processed as previously described 89. Briefly, following euthanasia, skin and organs were removed and embryos were dehydrated in 95% ethanol until further processing. Subsequently, embryos were incubated in acetone (~2 days) followed by incubation in a solution of acetic acid (5%) and 70% ethanol containing alcian blue (0.3%), and alizarin red (0.1%), for 2-3 days with shaking at 37°C, for staining of bone and cartilage elements. Following staining, remaining soft tissue was dissolved in 2% KOH (~1-2 days) and then 1% KOH (~1-2 days) until properly cleared. Final skeletal preparations were stored at 4°C in 25% glycerol and 1% KOH.
Cartilage staining
Embryos were collected at appropriate time points and processed as previously described 90. Briefly, following euthanasia, embryos were fixed in Bouin’s solution at 4°C overnight. Following fixation, embryos were washed with repeated changes of a solution of 70% ethanol and 0.1% NH4OH, until all traces of Bouin’s were removed. Subsequently, tissue was permeabilized by two 1 hour washes in 5% acetic acid, followed by overnight incubation in a solution of alcian blue (0.05%) and acetic acid (5%). Following staining, embryos were washed twice with 5% acetic acid (~1hr each wash) and then twice with 100% MeOH (~1hr each wash). Finally, embryos were cleared with a solution consisting of one part benzyl alcohol and two parts benzyl benzoate (BABB).
Mapping and Exome sequencing
Meiotic mapping to localize the ENU mutation responsible for the F1-9-13FP phenotype was performed as previously described 91,92. Subsequently, mutations within this interval were identified by sequencing F1-9-13FP homozygous mutant genomic DNA using the NimbleGen Mouse Exome Capture Kit followed by HiSeq2000 NextGen sequencing with >30x coverage (Otogenetics Corporation, Norcross, GA). Following sequencing, reads were mapped to the reference mouse genome sequence (Mm9) with large-scale alignment software (gSNAP). Sequence calls for single-nucleotide polymorphisms were performed using the Broad’s Genome Analysis Toolkit (GATK). The resulting variants were filtered in a systematic way to further reduce false positive results. Subsequently, the program ANNOVAR was used to cross reference all the variants across several genetic variation databases (e.g., dbSNP, AVSIFT, etc.) to find those variants that were likely to be novel. Variants considered as putative mutations after data filtering and analysis were ‘reconfirmed’ with custom designed Sanger Sequencing methods.
Western blot analysis
For Western blot analysis, E13.5 embryonic heads were lysed in RIPA buffer containing protease inhibitors (Thermo Scientific™ Protease Inhibitor Cocktail, Thermo Scientific, Waltham, MA). Briefly, heads were first minced using a razor blade followed by further disruption using a tissue homogenizer (Pro200 homogenizer, PRO Scientific Inc, Oxford, CT). Following homogenization, samples were allowed to lyse on ice for ~30 minutes. After lysis, 6x Laemmli Buffer (plus β-mercaptoethanol) was added to 1x final concentration and samples stored at −80°C until use. On the day of use, samples were boiled at ~100°C for 10 minutes, spun at 13,000 rpm for 10 minutes, and then 30uL of lysate loaded per lane on a 12% stacking polyacrylamide gel and run at 100V. Once samples were resolved, they were transferred onto a nitrocellulose membrane overnight at 4°C at 40mA. Following transfer, membranes were blocked one hour with 3% powdered milk in TBST (TBSTM), and then incubated with the primary antibody, diluted in TBSTM, overnight at 4°C (anti-MEMO1, 1:1000) 21. Following primary antibody incubation, membranes were washed 4x20min with TBSTM and then incubated with an infrared (IR)-labeled secondary antibody for 1hr (1:20,000 in TBST, LI-COR Biosciences, Lincoln, NE). The membrane was then washed 4x20min with TBSTM, 5 min with PBS, and then imaged on a LI-COR Odyssey imager. Following imaging, as a loading control, an additional primary antibody was added to the membrane (anti-ACTIN, H-196, 1:1000, Santa Cruz Biotechnology, Inc., Dallas, TX) and a similar washing, secondary, and imaging procedure carried out.
EUCOMM alleles - ES cells, injection, chimera generation, breeding, genotyping
To generate the Memo1 null allele we obtained ES-cells (Memo1tm1a(EUCOMM)Wtsi) from the European Conditional Mouse Mutagenesis (EUCOMM) repository that harbored a “targeted trap” allele at the Memo1 locus, generated on the C57BL/6N background (this allele includes an inserted LacZ and neomycin selection cassette, downstream of a strong splice-acceptor, as well as loxP sites flanking exon 4 of Memo1, see Supplemental Fig 2A) 26. Using standard procedures, chimeric mice were generated using these ES-cells, and subsequently bred with outbred wild-type Black Swiss mice to obtain germ-line transmission, establishing a heterozygous line (EUCOMM tm1a allele, this allele referred to as Memo1neo, confirmed by PCR, see Supplemental Fig 2A). Subsequently, to generate a “null-allele” these mice were bred with mice harboring CRE-recombinase expressed ubiquitously under control of an Actin promoter 93, resulting in deletion of the neo-cassette as well as exon 4 of Memo1 in 25% of the generated offspring (although the LacZ-cassette was retained, EUCOMM tm1b allele, referred to as Memo1 knock-in, or Memo1ki, see Supplemental Fig 2C). The deletion of exon 4 in a subset of the offspring was confirmed by PCR from genomic DNA, followed by sequencing (Supplemental Fig 2C and not shown). The conditional allele was generated by breeding mice harboring the Memo1neo allele with mice ubiquitously expressing FLPe recombinase 94. Acquisition of both alleles in the generated offspring resulted in FLPe mediated excision of the LacZ-neomycin selection cassette, resulting in loxP sites flanking exon 4 of Memo1 (EUCOMM tm1c allele, referred to as Memo1flp, see Supplemental Fig 2B), confirmed by PCR. Once generated, alleles were maintained on the C57BL/6J background. All primers used for genotyping are listed in Supplemental Table 2.
In situ hybridization/Immunohistochemistry/Immunofluorescence/H&E staining
In situ hybridization was carried out essentially as previously described 95. For all probes listed, with the exception of Hoxa2 (a kind gift from Robb Krumlauf) 32, a unique fragment (primer sequences given upon request) was cloned into a TOPO vector (Life Technologies, Grand Island, NY), using cDNA synthesized from mouse embryonic mRNA, as a template. cDNA was generated using the SuperScript® III First-Strand Synthesis System (Life Technologies, Grand Island, NY), as per manufacturer's instructions. Sequence verified plasmids were linearized and antisense probes synthesized using an appropriate DNA-dependent RNA polymerase (T7/T3/SP6) and DIG RNA labeling mix (Roche, Basel, Switzerland).
Whole-mount immunohistochemistry was carried out essentially as described, replacing 10% horse serum with 5% milk as a blocking reagent 96. The anti-neurofilament antibody 97 was used at a 1:100 dilution (IgG clone 2H3, obtained from the Developmental Studies Hybridoma Bank, created by the NICHD of the NIH and maintained at The University of Iowa, Department of Biology, Iowa City, IA).
Section Immunofluorescence: Dissected samples were fixed overnight in 4% paraformaldehyde (PFA). Subsequent to genotyping, samples were taken through a series of 30% sucrose, 30% sucrose/50% OCT (Tissue-Tek® O.C.T. Compound, Electron Microscopy Sciences, Hatfield, PA), and 100% OCT, and frozen on dry ice. Sections were cut at 10µm on a Leica CM 1900 cryostat (Leica Biosystems Inc., Buffalo Grove, IL) and placed on charged slides. Post-sectioning, samples were processed using the M.O.M. kit (Vector Laboratories, Inc., Burlingame, CA), as per manufacturer’s instructions, with the following exceptions: H2O2 treatment and antigen retrieval were omitted and a fluorescent secondary antibody used in place of colorimetric detection. Following primary antibody incubation (CD31, IgG clone 2H8, 1:100) 57, samples were rinsed 2×10 min with PBS, and then incubated with a fluorescent secondary antibody (goat-anti-mouse 594) (Invitrogen, Carlsbad, CA) along with the nuclear counterstain, DRAQ5 (Abcam, Cambridge, UK), for ~30 min. Following this incubation, samples were rinsed 2×10 min in PBS and cover-slipped. For older staged embryos (E18.5), samples were first decalcified in 10% EDTA for ~5 days post-fixation, prior to embedding in sucrose and OCT.
Hematoxylin and Eosin (H&E) staining was carried out essentially as previously described 98. Briefly, samples were fixed overnight in 4% PFA. Following fixation, tissue was dehydrated in a graded series of ethanol and xylene and subsequently embedded in paraffin. After embedding, samples were sectioned with a Leica RM 2235 microtome at 10uM, onto charged glass slides. After drying, sections were processed for H&E staining using standard procedures.
β-galactosidase staining
β-galactosidase staining of frozen sections was carried out as previously described 99. Briefly, dissected samples were fixed for 30 minutes in 0.2% glutaraldehyde, soaked in 10% sucrose in PBS for 30 minutes, then PBS plus 2mM MgCl2, followed by 30% sucrose/50% OCT /2mM MgCl2 for at least two hours. Subsequently, tissue was embedded in OCT on dry-ice. Sections were cut at 10uM and mounted on charged glass slides. Following sectioning, samples were fixed in 0.2% glutaraldehyde for 10 minutes on ice, rinsed with 2mM MgCl2 in PBS, incubated in 2mM MgCl2 in PBS for 10 minutes on ice, incubated in detergent rinse solution (0.005% NP40, 0.01% sodium deoxycholate in PBS) for 10 minutes on ice, and then developed in LacZ staining solution (detergent rinse solution plus 1mg/ml X-Gal, Invitrogen/Life Technologies, Carlsbad, CA) at room temperature for two days in the dark. Following staining, sections were counterstained with Nuclear Fast Red (Vector Laboratories, Inc., Burlingame, CA).
Cell proliferation and cell death analysis
For EdU staining, staged pregnant mice were injected intraperitoneally with 150uL of 1mg/ml EdU in PBS, using the Click-iT® EdU Alexa Fluor® 594 Imaging Kit (Life Technologies/ThermoFisher Scientific). Approximately 1 hour later, embryos were removed and fixed in 4% PFA overnight at 4°C. Following fixation, tissue was dehydrated in a graded series of ethanol and xylene and subsequently embedded in paraffin. After embedding, samples were sectioned with a Leica RM 2235 microtome at 10uM, onto charged glass slides. After drying, sections were processed for EdU immunofluorescence as per manufacturer's instructions and subsequently counterstained with DRAQ5 in PBS (1:1000). Processed samples were imaged on a Leica TCS SP5 II confocal microscope and individual images taken for quantification. Images were scored blindly for both total cell number (DRAQ5+ cells) and total proliferative cell number (EdU+ cells) using the cell counting plugin for ImageJ 100, and percentage of proliferating cells calculated. Mean and standard deviation were calculated between groups and a standard Student’s t-test was used for calculating p-value. Analysis of cell death was carried out using the DeadEnd™ Fluorometric TUNEL System, as per manufacturer's instructions (Promega, Madison, WI).
RNAseq
For the palatal RNA-seq, E13.5 embryos, generated from a Memo1wt/m1Will in-cross (on a largely C57 background, >95%), were dissected in ice-cold PBS. Subsequently, palatal shelves were carefully dissected from each embryo and stored in RNAlater (Ambion/Life Technologies) until later use. Following genotyping, RNA was extracted from both palatal shelves (3 control and 3 mutant embryos = 6 palatal shelves/group) using the microRNA Purification Kit (Norgen Biotek Corp., Thorold, ON), following manufacturer's protocol.
Following elution, mRNA was further purified using the Qiagen RNAeasy Kit (Qiagen, Valencia, CA), according to manufacturer's protocol. Quality of extracted mRNA was assessed using DNA Analysis ScreenTape (Agilent Technologies, Santa Clara, CA) to ensure that it was of sufficient quality for library production. Following validation of extracted mRNA, cDNA libraries were generated using the Illumina TruSeq Stranded mRNA Sample Prep Kit (Illumina, San Diego, CA). Following library generation and subsequent quality control assessment, cDNA was sequenced using the Illumina HiSeq2500 platform and single-end reads (1x50). For RNAseq of cranial base samples, a similar approach was used, with the exception that E15.5 embryos were used and the cranial base processed rather than the palatal shelves. For functional annotation clustering of significantly upregulated and downregulated genes, DAVID 35, with default parameters, was used. Following functional annotation clustering, “annotation clusters” above a 2-fold enrichment threshold were considered.
Supplementary Material
Supplemental Figure 1. Diagram of predicted MEMO1 protein sequence in F1-9-13FP mutants. (A) Wild-type MEMO1 amino acid sequence. For clarity, residues encoded by exons 1-7 are colored red, exon 8 black (and underlined), and exon 9 blue. (B) Predicted major MEMO1 amino acid sequence in F1-9-13FP mutants, in which the ENU-induced point mutation results in an in-frame deletion of exon 8 (indicated by absence of black residues).
Supplemental Figure 2. Description of Memo1 EUCOMM alleles. (A-D) Schematics of the Memo1 alleles utilized in this study, as described by the EUCOMM repository 26. For each allele, the primers used for genotyping are depicted below the allele and a representative PCR genotyping result is shown following gel electrophoresis (with the exception of D). A) The Memo1neo allele (or “knock-out first” allele) includes a strong splice acceptor (SA) sequence 5’ of a LacZ reporter cassette, located in the third intron of the Memo1 locus. In addition, this allele retains the neomycin (neo) resistance cassette used to select for targeted ES-cells. B) The Memo1flp allele is generated after FLP-mediated recombination of the Memo1neo allele, resulting in deletion of sequence located between FRT sites, including both the LacZ and neo cassettes. The resulting Memo1flp allele includes loxP sites flanking exon 4 of the endogenous Memo1 locus. C) The Memo1ki allele is generated after CRE-mediated recombination of the Memo1neo allele, resulting in deletion of sequences located between loxP sites, including the neo cassette as well as exon 4 of endogenous Memo1, resulting in a disruption of the proper protein open reading frame, and loss of MEMO1 protein. Note, the Memo1ki allele retains the LacZ reporter cassette. D) The Memo1null allele is generated after CRE-mediated recombination of the Memo1flp allele. Following recombination, sequences located between loxP sites (flanking exon 4 of endogenous Memo1) are deleted, resulting in disruption to the proper MEMO1 open reading frame, and subsequent loss of MEMO1 protein.
Supplemental Figure 3. Loss of MEMO1 protein in Memo1ki/ki mutant embryos. Western blot analysis of total protein isolated from E13.5 mouse embryonic heads of a Memo1ki/wt in-cross. Each lane corresponds to a single embryonic head from the listed genotype (2 embryos per genotype). ACTIN was used as a loading control.
Supplemental Figure 4. mRNA expression levels of Memo1 during early stages of craniofacial development. Graph depicting Memo1 log2 expression levels in the developing facial prominences (nasal [left], maxillary [middle], and mandibular [right]) from E10.5 through E12.5. Relative Memo1 expression levels in the ectoderm (blue) and underlying mesenchyme (red) are shown based on microarray analysis, along with standard error bars (3 replicates). Note that for technical reasons, at E10.5, it was not possible to analyze the ectoderm of the frontonasal and maxillary prominences, nor the mesenchyme of the maxillary prominence (data are taken from Hooper et al, manuscript in preparation).
Supplemental Figure 5. Memo1m1Will mutants show unperturbed early neural crest cell development and palatal patterning. (A-D) Lateral view of an E9.5 wild-type (A, C) or Memo1m1Will mutant (B, D) embryo stained in whole mount for Tfap2a (A, B) or Inka1 (C, D) expression, labeling the neural crest cell streams migrating into the facial prominences. Dorsal at right, rostral at top. (E) Scatter plot of average RPKM values from RNAseq conducted on RNA isolated from E13.5 palatal shelves (3 pairs/group), comparing control (X-axis) versus Memo1m1Will mutants (Y-axis). Red and green shaded points are those that meet criteria for significance (see methods) and are either down-regulated (red, SIG-DOWN, 19/22) or up-regulated (green, SIG-UP, 3/22), respectively in the Memo1m1Will mutant. (F, G) Lateral view of E10.5 wild-type (F) or Memo1m1WIll mutant (G) embryo processed for anti-neurofilament immunoreactivity, labeling differentiated cranial ganglia (V, VII, and VIII). Dorsal at right, rostral at top. (H, I) Frontal sections of the developing palatal shelves at E13.5 of a control (H) or Memo1ki/ki mutant (I) harboring the Wnt1-CRE transgene and rosa-Tomato reporter, resulting in fluorescent labeling of neural crest cells (all pink/orange fluorescent cells are neural crest). Blue fluorescence is DRAQ5 counterstain of nuclei, most evident in the neural crest cell negative oral and tongue epithelium. (J-Q) Ventral view of the developing palate, of the genotype indicated, at E13.5 (J, K and N, O) or E14.5 (L, M and P, Q), and processed by in situ hybridization for either Shh (J-M) or Tbx22 expression (N-Q, arrowheads). Developing rugae are numbered in (J-M). Also note, in contrast to a control embryo, Tbx22 expression at E14.5 remains on in the Memo1m1Will posterior palate (arrowheads in Q), presumably as a result of the failure of palatal shelf fusion. Abbreviations: ba1, branchial arch 1; ba2, branchial arch 2; e, epithelium; fnp, frontonasal process; NF, neurofilament; ps, palatal shelf; t, tongue. Scale bars: 500uM.
Supplemental Figure 6. Cranial vault defects in Memo1NC-CKO embryos. (A, B) Dorsal view of both a control (A) and Memo1NC-CKO (B) E18.5 embryonic head, processed for skeletal stain, revealing the craniofacial calvaria. Asterisk denotes larger space in conditional mutants versus controls. Abbreviations: f, frontal bone; ip, interparietal bone; p, parietal bone.
Highlights.
Forward genetic screen isolates MEMO1 as a key factor in craniofacial development
MEMO1 drives cranial base and palatal development
High levels of Memo1 expression localized to bones in the developing head
MEMO1 shown to promote cartilage to bone turnover in the cranial base
MEMO1 functions autonomously during ossification of anterior cranial base
Acknowledgements
This work supported by Fellowship F32DE02370 from the National Institute of Dental and Craniofacial Research (EVO), in part by a Cleft Palate Foundation Research Support Grant (EVO) and an American Association of Anatomists Postdoctoral Fellowship (EVO); Grants R01 DE019843 and R01 HD081562 (TW); SNF 310030A-121574 and 310030B-138674 (NEH); and Department of Pediatrics, Children’s Hospital Colorado (LN, for ENU screen). LN was an investigator of the Howard Hughes Medical Institute. Bioinformatic analyses were supported in part by the Biostatistics/Bioinformatics Shared Resources of Colorado's NIH/NCI Cancer Center (support grant P30CA046934). We thank the Genomics and Microarray Core at UC-Denver for their assistance in RNA-seq experiments, Philip O’Neill (UC-Denver, Aurora, CO) for help in processing of RNA-seq data, Timothy P Nottoli (Yale University, New Haven, CT) for blastocyst injections, Monica Justice (originally Baylor College of Medicine, Houston, Texas, now at The Hospital for Sick Children [SickKids] in Toronto, Ontario) for initially supplying the ENU mutagenized sperm used in the recessive screen, and Irene Choi (UC-Denver, Aurora, CO) for mouse maintenance and DNA extraction from ENU mutants.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Lieberman DE, Pearson OM, Mowbray KM. Basicranial influence on overall cranial shape. Journal of human evolution. 2000;38:291–315. doi: 10.1006/jhev.1999.0335. doi:10.1006/jhev.1999.0335. [DOI] [PubMed] [Google Scholar]
- 2.Lieberman DE, Ross CF, Ravosa MJ. The primate cranial base: ontogeny, function, and integration. American journal of physical anthropology. 2000;Suppl 31:117–169. doi: 10.1002/1096-8644(2000)43:31+<117::aid-ajpa5>3.3.co;2-9. [DOI] [PubMed] [Google Scholar]
- 3.Martinez-Abadias N, et al. Pervasive genetic integration directs the evolution of human skull shape. Evolution; international journal of organic evolution. 2012;66:1010–1023. doi: 10.1111/j.1558-5646.2011.01496.x. doi:10.1111/j.1558-5646.2011.01496.x. [DOI] [PubMed] [Google Scholar]
- 4.Nie X. Cranial base in craniofacial development: developmental features, influence on facial growth, anomaly, and molecular basis. Acta odontologica Scandinavica. 2005;63:127–135. doi: 10.1080/00016350510019847. doi:10.1080/00016350510019847. [DOI] [PubMed] [Google Scholar]
- 5.Jahanbin A, Eslami N, Hoseini Zarch H, Kobravi S. Comparative evaluation of cranial base and facial morphology of cleft lip and palate patients with normal individuals in cone beam computed tomography. The Journal of craniofacial surgery. 2015;26:785–788. doi: 10.1097/SCS.0000000000001361. doi:10.1097/SCS.0000000000001361. [DOI] [PubMed] [Google Scholar]
- 6.Ye B, et al. A comparative cephalometric study for adult operated cleft palate and unoperated cleft palate patients. Journal of cranio-maxillo-facial surgery : official publication of the European Association for Cranio-Maxillo-Facial Surgery. 2015 doi: 10.1016/j.jcms.2015.04.015. doi:10.1016/j.jcms.2015.04.015. [DOI] [PubMed] [Google Scholar]
- 7.McBratney-Owen B, Iseki S, Bamforth SD, Olsen BR, Morriss-Kay GM. Development and tissue origins of the mammalian cranial base. Developmental biology. 2008;322:121–132. doi: 10.1016/j.ydbio.2008.07.016. doi:10.1016/j.ydbio.2008.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Couly GF, Coltey PM, Le Douarin NM. The triple origin of skull in higher vertebrates: a study in quail-chick chimeras. Development. 1993;117:409–429. doi: 10.1242/dev.117.2.409. [DOI] [PubMed] [Google Scholar]
- 9.Le Douarin N, Kalcheim C. The neural crest. 2nd Cambridge University Press; 1999. [Google Scholar]
- 10.Gross JB, Hanken J. Review of fate-mapping studies of osteogenic cranial neural crest in vertebrates. Developmental biology. 2008;317:389–400. doi: 10.1016/j.ydbio.2008.02.046. doi:10.1016/j.ydbio.2008.02.046. [DOI] [PubMed] [Google Scholar]
- 11.Jiang X, Iseki S, Maxson RE, Sucov HM, Morriss-Kay GM. Tissue origins and interactions in the mammalian skull vault. Developmental biology. 2002;241:106–116. doi: 10.1006/dbio.2001.0487. doi:10.1006/dbio.2001.0487. [DOI] [PubMed] [Google Scholar]
- 12.Percival CJ, Richtsmeier JT. Angiogenesis and intramembranous osteogenesis. Developmental dynamics : an official publication of the American Association of Anatomists. 2013;242:909–922. doi: 10.1002/dvdy.23992. doi:10.1002/dvdy.23992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rossant J, Tam PPL. Mouse development : patterning, morphogenesis, and organogenesis. 2002 Academic. [Google Scholar]
- 14.Yang Y. Skeletal morphogenesis during embryonic development. Critical reviews in eukaryotic gene expression. 2009;19:197–218. doi: 10.1615/critreveukargeneexpr.v19.i3.30. [DOI] [PubMed] [Google Scholar]
- 15.Franz-Odendaal TA. Induction and patterning of intramembranous bone. Frontiers in bioscience. 2011;16:2734–2746. doi: 10.2741/3882. [DOI] [PubMed] [Google Scholar]
- 16.Hayashi S, et al. Interface between intramembranous and endochondral ossification in human foetuses. Folia morphologica. 2014;73:199–205. doi: 10.5603/FM.2014.0029. doi:10.5603/FM.2014.0029. [DOI] [PubMed] [Google Scholar]
- 17.Chung UI, Kawaguchi H, Takato T, Nakamura K. Distinct osteogenic mechanisms of bones of distinct origins. Journal of orthopaedic science : official journal of the Japanese Orthopaedic Association. 2004;9:410–414. doi: 10.1007/s00776-004-0786-3. doi:10.1007/s00776-004-0786-3. [DOI] [PubMed] [Google Scholar]
- 18.Feng W, Choi I, Clouthier DE, Niswander L, Williams T. The Ptch1(DL) mouse: a new model to study lambdoid craniosynostosis and basal cell nevus syndrome-associated skeletal defects. Genesis. 2013;51:677–689. doi: 10.1002/dvg.22416. doi:10.1002/dvg.22416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marone R, et al. Memo mediates ErbB2-driven cell motility. Nature cell biology. 2004;6:515–522. doi: 10.1038/ncb1134. doi:10.1038/ncb1134. [DOI] [PubMed] [Google Scholar]
- 20.Meira M, et al. Memo is a cofilin-interacting protein that influences PLCgamma1 and cofilin activities, and is essential for maintaining directionality during ErbB2-induced tumor-cell migration. Journal of cell science. 2009;122:787–797. doi: 10.1242/jcs.032094. doi:10.1242/jcs.032094. [DOI] [PubMed] [Google Scholar]
- 21.MacDonald G, et al. Memo is a copper-dependent redox protein with an essential role in migration and metastasis. Science signaling. 2014;7:ra56. doi: 10.1126/scisignal.2004870. doi:10.1126/scisignal.2004870. [DOI] [PubMed] [Google Scholar]
- 22.Kondo S, et al. Memo has a novel role in S1P signaling and is [corrected] crucial for vascular development. PloS one. 2014;9:e94114. doi: 10.1371/journal.pone.0094114. doi:10.1371/journal.pone.0094114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Noden DM, Trainor PA. Relations and interactions between cranial mesoderm and neural crest populations. Journal of anatomy. 2005;207:575–601. doi: 10.1111/j.1469-7580.2005.00473.x. doi:10.1111/j.1469-7580.2005.00473.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thompson H, Ohazama A, Sharpe PT, Tucker AS. The origin of the stapes and relationship to the otic capsule and oval window. Developmental dynamics : an official publication of the American Association of Anatomists. 2012;241:1396–1404. doi: 10.1002/dvdy.23831. doi:10.1002/dvdy.23831. [DOI] [PubMed] [Google Scholar]
- 25.Weir M, Rice M. Ordered partitioning reveals extended splice-site consensus information. Genome Res. 2004;14:67–78. doi: 10.1101/gr.1715204. doi:10.1101/gr.1715204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Skarnes WC, et al. A conditional knockout resource for the genome-wide study of mouse gene function. Nature. 2011;474:337–342. doi: 10.1038/nature10163. doi:10.1038/nature10163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Haenzi B, et al. Loss of Memo, a novel FGFR regulator, results in reduced lifespan. FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 2014;28:327–336. doi: 10.1096/fj.13-228320. doi:10.1096/fj.13-228320. [DOI] [PubMed] [Google Scholar]
- 28.Zaoui K, Honore S, Isnardon D, Braguer D, Badache A. Memo-RhoA-mDia1 signaling controls microtubules, the actin network, and adhesion site formation in migrating cells. The Journal of cell biology. 2008;183:401–408. doi: 10.1083/jcb.200805107. doi:10.1083/jcb.200805107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang J, et al. Neural tube, skeletal and body wall defects in mice lacking transcription factor AP-2. Nature. 1996;381:238–241. doi: 10.1038/381238a0. doi:10.1038/381238a0. [DOI] [PubMed] [Google Scholar]
- 30.Mitchell PJ, Timmons PM, Hebert JM, Rigby PW, Tjian R. Transcription factor AP-2 is expressed in neural crest cell lineages during mouse embryogenesis. Genes & development. 1991;5:105–119. doi: 10.1101/gad.5.1.105. [DOI] [PubMed] [Google Scholar]
- 31.Reid BS, Sargent TD, Williams T. Generation and characterization of a novel neural crest marker allele, Inka1-LacZ, reveals a role for Inka1 in mouse neural tube closure. Developmental dynamics : an official publication of the American Association of Anatomists. 2010;239:1188–1196. doi: 10.1002/dvdy.22248. doi:10.1002/dvdy.22248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wilkinson DG, Bhatt S, Cook M, Boncinelli E, Krumlauf R. Segmental expression of Hox-2 homoeobox-containing genes in the developing mouse hindbrain. Nature. 1989;341:405–409. doi: 10.1038/341405a0. doi:10.1038/341405a0. [DOI] [PubMed] [Google Scholar]
- 33.Danielian PS, Muccino D, Rowitch DH, Michael SK, McMahon AP. Modification of gene activity in mouse embryos in utero by a tamoxifen-inducible form of Cre recombinase. Current biology : CB. 1998;8:1323–1326. doi: 10.1016/s0960-9822(07)00562-3. [DOI] [PubMed] [Google Scholar]
- 34.Madisen L, et al. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nature neuroscience. 2010;13:133–140. doi: 10.1038/nn.2467. doi:10.1038/nn.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huang da W, et al. Extracting biological meaning from large gene lists with DAVID. Current protocols in bioinformatics / editoral board, Andreas D. Baxevanis … [et al.] 2009 doi: 10.1002/0471250953.bi1311s27. Chapter 13, Unit 13 11. doi:10.1002/0471250953.bi1311s27. [DOI] [PubMed] [Google Scholar]
- 36.Vu TH, et al. MMP-9/gelatinase B is a key regulator of growth plate angiogenesis and apoptosis of hypertrophic chondrocytes. Cell. 1998;93:411–422. doi: 10.1016/s0092-8674(00)81169-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Inada M, et al. Critical roles for collagenase-3 (Mmp13) in development of growth plate cartilage and in endochondral ossification. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:17192–17197. doi: 10.1073/pnas.0407788101. doi:10.1073/pnas.0407788101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stickens D, et al. Altered endochondral bone development in matrix metalloproteinase 13-deficient mice. Development. 2004;131:5883–5895. doi: 10.1242/dev.01461. doi:10.1242/dev.01461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gustafsson E, et al. Role of collagen type II and perlecan in skeletal development. Annals of the New York Academy of Sciences. 2003;995:140–150. doi: 10.1111/j.1749-6632.2003.tb03217.x. [DOI] [PubMed] [Google Scholar]
- 40.Hayman AR, et al. Mice lacking tartrate-resistant acid phosphatase (Acp 5) have disrupted endochondral ossification and mild osteopetrosis. Development. 1996;122:3151–3162. doi: 10.1242/dev.122.10.3151. [DOI] [PubMed] [Google Scholar]
- 41.Holm E, Aubin JE, Hunter GK, Beier F, Goldberg HA. Loss of bone sialoprotein leads to impaired endochondral bone development and mineralization. Bone. 2015;71:145–154. doi: 10.1016/j.bone.2014.10.007. doi:10.1016/j.bone.2014.10.007. [DOI] [PubMed] [Google Scholar]
- 42.Padovano JD, Ramachandran A, Bahmanyar S, Ravindran S, George A. Bone-specific overexpression of DMP1 influences osteogenic gene expression during endochondral and intramembranous ossification. Connective tissue research. 2014;55 Suppl 1:121–124. doi: 10.3109/03008207.2014.923878. doi:10.3109/03008207.2014.923878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bouet G, et al. The impairment of osteogenesis in bone sialoprotein (BSP) knockout calvaria cell cultures is cell density dependent. PloS one. 2015;10:e0117402. doi: 10.1371/journal.pone.0117402. doi:10.1371/journal.pone.0117402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nakashima K, et al. The novel zinc finger-containing transcription factor osterix is required for osteoblast differentiation and bone formation. Cell. 2002;108:17–29. doi: 10.1016/s0092-8674(01)00622-5. [DOI] [PubMed] [Google Scholar]
- 45.Chipman SD, et al. Defective pro alpha 2(I) collagen synthesis in a recessive mutation in mice: a model of human osteogenesis imperfecta. Proceedings of the National Academy of Sciences of the United States of America. 1993;90:1701–1705. doi: 10.1073/pnas.90.5.1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kawai S, Yamauchi M, Wakisaka S, Ooshima T, Amano A. Zinc-finger transcription factor odd-skipped related 2 is one of the regulators in osteoblast proliferation and bone formation. Journal of bone and mineral research : the official journal of the American Society for Bone and Mineral Research. 2007;22:1362–1372. doi: 10.1359/jbmr.070602. doi:10.1359/jbmr.070602. [DOI] [PubMed] [Google Scholar]
- 47.St-Jacques B, Hammerschmidt M, McMahon AP. Indian hedgehog signaling regulates proliferation and differentiation of chondrocytes and is essential for bone formation. Genes & development. 1999;13:2072–2086. doi: 10.1101/gad.13.16.2072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Deng C, Wynshaw-Boris A, Zhou F, Kuo A, Leder P. Fibroblast growth factor receptor 3 is a negative regulator of bone growth. Cell. 1996;84:911–921. doi: 10.1016/s0092-8674(00)81069-7. [DOI] [PubMed] [Google Scholar]
- 49.Wiedemann M, Trueb B. Characterization of a novel protein (FGFRL1) from human cartilage related to FGF receptors. Genomics. 2000;69:275–279. doi: 10.1006/geno.2000.6332. doi:10.1006/geno.2000.6332. [DOI] [PubMed] [Google Scholar]
- 50.Akiyama H, Chaboissier MC, Martin JF, Schedl A, de Crombrugghe B. The transcription factor Sox9 has essential roles in successive steps of the chondrocyte differentiation pathway and is required for expression of Sox5 and Sox6. Genes & development. 2002;16:2813–2828. doi: 10.1101/gad.1017802. doi:10.1101/gad.1017802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Watanabe H, et al. Mouse cartilage matrix deficiency (cmd) caused by a 7 bp deletion in the aggrecan gene. Nature genetics. 1994;7:154–157. doi: 10.1038/ng0694-154. doi:10.1038/ng0694-154. [DOI] [PubMed] [Google Scholar]
- 52.Li SW, et al. Transgenic mice with targeted inactivation of the Col2 alpha 1 gene for collagen II develop a skeleton with membranous and periosteal bone but no endochondral bone. Genes & development. 1995;9:2821–2830. doi: 10.1101/gad.9.22.2821. [DOI] [PubMed] [Google Scholar]
- 53.Fassler R, et al. Mice lacking alpha 1 (IX) collagen develop noninflammatory degenerative joint disease. Proceedings of the National Academy of Sciences of the United States of America. 1994;91:5070–5074. doi: 10.1073/pnas.91.11.5070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nakamichi Y, et al. Chondromodulin I is a bone remodeling factor. Molecular and cellular biology. 2003;23:636–644. doi: 10.1128/MCB.23.2.636-644.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ortega N, Behonick D, Stickens D, Werb Z. How proteases regulate bone morphogenesis. Annals of the New York Academy of Sciences. 2003;995:109–116. doi: 10.1111/j.1749-6632.2003.tb03214.x. [DOI] [PubMed] [Google Scholar]
- 56.Ortega N, Behonick DJ, Werb Z. Matrix remodeling during endochondral ossification. Trends in cell biology. 2004;14:86–93. doi: 10.1016/j.tcb.2003.12.003. doi:10.1016/j.tcb.2003.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bogen SA, Baldwin HS, Watkins SC, Albelda SM, Abbas AK. Association of murine CD31 with transmigrating lymphocytes following antigenic stimulation. The American journal of pathology. 1992;141:843–854. [PMC free article] [PubMed] [Google Scholar]
- 58.Etchevers HC, Vincent C, Le Douarin NM, Couly GF. The cephalic neural crest provides pericytes and smooth muscle cells to all blood vessels of the face and forebrain. Development. 2001;128:1059–1068. doi: 10.1242/dev.128.7.1059. [DOI] [PubMed] [Google Scholar]
- 59.Jiang K, et al. Mediator of ERBB2-driven cell motility (MEMO) promotes extranuclear estrogen receptor signaling involving the growth factor receptors IGF1R and ERBB2. The Journal of biological chemistry. 2013;288:24590–24599. doi: 10.1074/jbc.M113.467837. doi:10.1074/jbc.M113.467837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fisher MC, Clinton GM, Maihle NJ, Dealy CN. Requirement for ErbB2/ErbB signaling in developing cartilage and bone. Development, growth & differentiation. 2007;49:503–513. doi: 10.1111/j.1440-169X.2007.00941.x. doi:10.1111/j.1440-169X.2007.00941.x. [DOI] [PubMed] [Google Scholar]
- 61.Britto JA, Evans RD, Hayward RD, Jones BM. From genotype to phenotype: the differential expression of FGF, FGFR, and TGFbeta genes characterizes human cranioskeletal development and reflects clinical presentation in FGFR syndromes. Plastic and reconstructive surgery. 2001;108:2026–2039. doi: 10.1097/00006534-200112000-00030. discussion 2040-2026. [DOI] [PubMed] [Google Scholar]
- 62.Rice DP, Rice R, Thesleff I. Fgfr mRNA isoforms in craniofacial bone development. Bone. 2003;33:14–27. doi: 10.1016/s8756-3282(03)00163-7. [DOI] [PubMed] [Google Scholar]
- 63.Matsushita T, et al. FGFR3 promotes synchondrosis closure and fusion of ossification centers through the MAPK pathway. Human molecular genetics. 2009;18:227–240. doi: 10.1093/hmg/ddn339. doi:10.1093/hmg/ddn339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Matsushita T, et al. Extracellular signal-regulated kinase 1 (ERK1) and ERK2 play essential roles in osteoblast differentiation and in supporting osteoclastogenesis. Molecular and cellular biology. 2009;29:5843–5857. doi: 10.1128/MCB.01549-08. doi:10.1128/MCB.01549-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Guntur AR, Rosen CJ. IGF-1 regulation of key signaling pathways in bone. Bonekey Rep. 2013;2:437. doi: 10.1038/bonekey.2013.171. doi:10.1038/bonekey.2013.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gerber HP, et al. VEGF couples hypertrophic cartilage remodeling, ossification and angiogenesis during endochondral bone formation. Nature medicine. 1999;5:623–628. doi: 10.1038/9467. doi:10.1038/9467. [DOI] [PubMed] [Google Scholar]
- 67.Zelzer E, et al. VEGFA is necessary for chondrocyte survival during bone development. Development. 2004;131:2161–2171. doi: 10.1242/dev.01053. doi:10.1242/dev.01053. [DOI] [PubMed] [Google Scholar]
- 68.Duan X, et al. Vegfa regulates perichondrial vascularity and osteoblast differentiation in bone development. Development. 2015;142:1984–1991. doi: 10.1242/dev.117952. doi:10.1242/dev.117952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Borjesson AE, Lagerquist MK, Windahl SH, Ohlsson C. The role of estrogen receptor alpha in the regulation of bone and growth plate cartilage. Cell Mol Life Sci. 2013;70:4023–4037. doi: 10.1007/s00018-013-1317-1. doi:10.1007/s00018-013-1317-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhang X, et al. Epidermal growth factor receptor (EGFR) signaling regulates epiphyseal cartilage development through beta-catenin-dependent and -independent pathways. The Journal of biological chemistry. 2013;288:32229–32240. doi: 10.1074/jbc.M113.463554. doi:10.1074/jbc.M113.463554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Morita K, et al. Reactive oxygen species induce chondrocyte hypertrophy in endochondral ossification. The Journal of experimental medicine. 2007;204:1613–1623. doi: 10.1084/jem.20062525. doi:10.1084/jem.20062525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Koretsi V, Kirschneck C, Proff P, Romer P. Expression of glutathione peroxidase 1 in the spheno-occipital synchondrosis and its role in ROS-induced apoptosis. European journal of orthodontics. 2015;37:308–313. doi: 10.1093/ejo/cju045. doi:10.1093/ejo/cju045. [DOI] [PubMed] [Google Scholar]
- 73.Kim KS, Choi HW, Yoon HE, Kim IY. Reactive oxygen species generated by NADPH oxidase 2 and 4 are required for chondrogenic differentiation. The Journal of biological chemistry. 2010;285:40294–40302. doi: 10.1074/jbc.M110.126821. doi:10.1074/jbc.M110.126821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ahmad R, Sylvester J, Ahmad M, Zafarullah M. Involvement of H-Ras and reactive oxygen species in proinflammatory cytokine-induced matrix metalloproteinase-13 expression in human articular chondrocytes. Archives of biochemistry and biophysics. 2011;507:350–355. doi: 10.1016/j.abb.2010.12.032. doi:10.1016/j.abb.2010.12.032. [DOI] [PubMed] [Google Scholar]
- 75.Pauws E, et al. Tbx22null mice have a submucous cleft palate due to reduced palatal bone formation and also display ankyloglossia and choanal atresia phenotypes. Human molecular genetics. 2009;18:4171–4179. doi: 10.1093/hmg/ddp368. doi:10.1093/hmg/ddp368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Parada C, et al. Disruption of the ERK/MAPK pathway in neural crest cells as a potential cause of Pierre Robin sequence. Development. 2015 doi: 10.1242/dev.125328. doi:10.1242/dev.125328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sandham A, Cheng L. Cranial base and cleft lip and palate. The Angle orthodontist. 1988;58:163–168. doi: 10.1043/0003-3219(1988)058<0163:CBACLA>2.0.CO;2. doi:10.1043/0003-3219(1988)058<0163:CBACLA>2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 78.Horswell BB, Gallup BV. Cranial base morphology in cleft lip and palate: a cephalometric study from 7 to 18 years of age. Journal of oral and maxillofacial surgery : official journal of the American Association of Oral and Maxillofacial Surgeons. 1992;50:681–685. doi: 10.1016/0278-2391(92)90095-h. discussion 686. [DOI] [PubMed] [Google Scholar]
- 79.Harris EF. Size and form of the cranial base in isolated cleft lip and palate. The Cleft palate-craniofacial journal : official publication of the American Cleft Palate-Craniofacial Association. 1993;30:170–174. doi: 10.1597/1545-1569_1993_030_0170_safotc_2.3.co_2. doi:10.1597/1545-1569(1993)030<0170:SAFOTC>2.3.CO;2. [DOI] [PubMed] [Google Scholar]
- 80.Ross RB. My friend the cranial base: why is it so normal? The Cleft palate-craniofacial journal : official publication of the American Cleft Palate-Craniofacial Association. 1993;30:511–512. doi: 10.1597/1545-1569_1993_030_0511_mftcbw_2.3.co_2. doi:10.1597/1545-1569(1993)030<0511:MFTCBW>2.3.CO;2. [DOI] [PubMed] [Google Scholar]
- 81.Beaty TH, et al. Analysis of candidate genes on chromosome 2 in oral cleft case-parent trios from three populations. Human genetics. 2006;120:501–518. doi: 10.1007/s00439-006-0235-9. doi:10.1007/s00439-006-0235-9. [DOI] [PubMed] [Google Scholar]
- 82.Devoto M, et al. First-stage autosomal genome screen in extended pedigrees suggests genes predisposing to low bone mineral density on chromosomes 1p, 2p and 4q. European journal of human genetics : EJHG. 1998;6:151–157. doi: 10.1038/sj.ejhg.5200169. doi:10.1038/sj.ejhg.5200169. [DOI] [PubMed] [Google Scholar]
- 83.Niu T, et al. A genome-wide scan for loci linked to forearm bone mineral density. Human genetics. 1999;104:226–233. doi: 10.1007/s004390050940. [DOI] [PubMed] [Google Scholar]
- 84.Wynne F, et al. Suggestive linkage of 2p22-25 and 11q12-13 with low bone mineral density at the lumbar spine in the Irish population. Calcified tissue international. 2003;72:651–658. doi: 10.1007/s00223-002-2086-2. doi:10.1007/s00223-002-2086-2. [DOI] [PubMed] [Google Scholar]
- 85.Pope FM, Nicholls AC, McPheat J, Talmud P, Owen R. Collagen genes and proteins in osteogenesis imperfecta. Journal of medical genetics. 1985;22:466–478. doi: 10.1136/jmg.22.6.466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Prockop DJ, et al. Mutations in osteogenesis imperfecta leading to the synthesis of abnormal type I procollagens. Annals of the New York Academy of Sciences. 1985;460:289–297. doi: 10.1111/j.1749-6632.1985.tb51176.x. [DOI] [PubMed] [Google Scholar]
- 87.Lapunzina P, et al. Identification of a frameshift mutation in Osterix in a patient with recessive osteogenesis imperfecta. American journal of human genetics. 2010;87:110–114. doi: 10.1016/j.ajhg.2010.05.016. doi:10.1016/j.ajhg.2010.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Cheung MS, et al. Cranial base abnormalities in osteogenesis imperfecta: phenotypic and genotypic determinants. Journal of bone and mineral research : the official journal of the American Society for Bone and Mineral Research. 2011;26:405–413. doi: 10.1002/jbmr.220. doi:10.1002/jbmr.220. [DOI] [PubMed] [Google Scholar]
- 89.McLeod MJ. Differential staining of cartilage and bone in whole mouse fetuses by alcian blue and alizarin red S. Teratology. 1980;22:299–301. doi: 10.1002/tera.1420220306. doi:10.1002/tera.1420220306. [DOI] [PubMed] [Google Scholar]
- 90.Jegalian BG, De Robertis EM. Homeotic transformations in the mouse induced by overexpression of a human Hox3.3 transgene. Cell. 1992;71:901–910. doi: 10.1016/0092-8674(92)90387-r. [DOI] [PubMed] [Google Scholar]
- 91.Garcia-Garcia MJ, et al. Analysis of mouse embryonic patterning and morphogenesis by forward genetics. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:5913–5919. doi: 10.1073/pnas.0501071102. doi:10.1073/pnas.0501071102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kasarskis A, Manova K, Anderson KV. A phenotype-based screen for embryonic lethal mutations in the mouse. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:7485–7490. doi: 10.1073/pnas.95.13.7485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lewandoski M, Meyers EN, Martin GR. Analysis of Fgf8 gene function in vertebrate development. Cold Spring Harbor symposia on quantitative biology. 1997;62:159–168. [PubMed] [Google Scholar]
- 94.Rodriguez CI, et al. High-efficiency deleter mice show that FLPe is an alternative to Cre-loxP. Nature genetics. 2000;25:139–140. doi: 10.1038/75973. doi:10.1038/75973. [DOI] [PubMed] [Google Scholar]
- 95.Simmons O, Bolanis EM, Wang J, Conway SJ. In situ hybridization (both radioactive and nonradioactive) and spatiotemporal gene expression analysis. Methods in molecular biology. 2014;1194:225–244. doi: 10.1007/978-1-4939-1215-5_12. doi:10.1007/978-1-4939-1215-5_12. [DOI] [PubMed] [Google Scholar]
- 96.Pabst O, Rummelies J, Winter B, Arnold HH. Targeted disruption of the homeobox gene Nkx2.9 reveals a role in development of the spinal accessory nerve. Development. 2003;130:1193–1202. doi: 10.1242/dev.00346. [DOI] [PubMed] [Google Scholar]
- 97.Dodd J, Morton SB, Karagogeos D, Yamamoto M, Jessell TM. Spatial regulation of axonal glycoprotein expression on subsets of embryonic spinal neurons. Neuron. 1988;1:105–116. doi: 10.1016/0896-6273(88)90194-8. [DOI] [PubMed] [Google Scholar]
- 98.Cardiff RD, Miller CH, Munn RJ. Manual hematoxylin and eosin staining of mouse tissue sections. Cold Spring Harbor protocols. 2014;2014:655–658. doi: 10.1101/pdb.prot073411. doi:10.1101/pdb.prot073411. [DOI] [PubMed] [Google Scholar]
- 99.Chai Y, et al. Fate of the mammalian cranial neural crest during tooth and mandibular morphogenesis. Development. 2000;127:1671–1679. doi: 10.1242/dev.127.8.1671. [DOI] [PubMed] [Google Scholar]
- 100.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nature methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1. Diagram of predicted MEMO1 protein sequence in F1-9-13FP mutants. (A) Wild-type MEMO1 amino acid sequence. For clarity, residues encoded by exons 1-7 are colored red, exon 8 black (and underlined), and exon 9 blue. (B) Predicted major MEMO1 amino acid sequence in F1-9-13FP mutants, in which the ENU-induced point mutation results in an in-frame deletion of exon 8 (indicated by absence of black residues).
Supplemental Figure 2. Description of Memo1 EUCOMM alleles. (A-D) Schematics of the Memo1 alleles utilized in this study, as described by the EUCOMM repository 26. For each allele, the primers used for genotyping are depicted below the allele and a representative PCR genotyping result is shown following gel electrophoresis (with the exception of D). A) The Memo1neo allele (or “knock-out first” allele) includes a strong splice acceptor (SA) sequence 5’ of a LacZ reporter cassette, located in the third intron of the Memo1 locus. In addition, this allele retains the neomycin (neo) resistance cassette used to select for targeted ES-cells. B) The Memo1flp allele is generated after FLP-mediated recombination of the Memo1neo allele, resulting in deletion of sequence located between FRT sites, including both the LacZ and neo cassettes. The resulting Memo1flp allele includes loxP sites flanking exon 4 of the endogenous Memo1 locus. C) The Memo1ki allele is generated after CRE-mediated recombination of the Memo1neo allele, resulting in deletion of sequences located between loxP sites, including the neo cassette as well as exon 4 of endogenous Memo1, resulting in a disruption of the proper protein open reading frame, and loss of MEMO1 protein. Note, the Memo1ki allele retains the LacZ reporter cassette. D) The Memo1null allele is generated after CRE-mediated recombination of the Memo1flp allele. Following recombination, sequences located between loxP sites (flanking exon 4 of endogenous Memo1) are deleted, resulting in disruption to the proper MEMO1 open reading frame, and subsequent loss of MEMO1 protein.
Supplemental Figure 3. Loss of MEMO1 protein in Memo1ki/ki mutant embryos. Western blot analysis of total protein isolated from E13.5 mouse embryonic heads of a Memo1ki/wt in-cross. Each lane corresponds to a single embryonic head from the listed genotype (2 embryos per genotype). ACTIN was used as a loading control.
Supplemental Figure 4. mRNA expression levels of Memo1 during early stages of craniofacial development. Graph depicting Memo1 log2 expression levels in the developing facial prominences (nasal [left], maxillary [middle], and mandibular [right]) from E10.5 through E12.5. Relative Memo1 expression levels in the ectoderm (blue) and underlying mesenchyme (red) are shown based on microarray analysis, along with standard error bars (3 replicates). Note that for technical reasons, at E10.5, it was not possible to analyze the ectoderm of the frontonasal and maxillary prominences, nor the mesenchyme of the maxillary prominence (data are taken from Hooper et al, manuscript in preparation).
Supplemental Figure 5. Memo1m1Will mutants show unperturbed early neural crest cell development and palatal patterning. (A-D) Lateral view of an E9.5 wild-type (A, C) or Memo1m1Will mutant (B, D) embryo stained in whole mount for Tfap2a (A, B) or Inka1 (C, D) expression, labeling the neural crest cell streams migrating into the facial prominences. Dorsal at right, rostral at top. (E) Scatter plot of average RPKM values from RNAseq conducted on RNA isolated from E13.5 palatal shelves (3 pairs/group), comparing control (X-axis) versus Memo1m1Will mutants (Y-axis). Red and green shaded points are those that meet criteria for significance (see methods) and are either down-regulated (red, SIG-DOWN, 19/22) or up-regulated (green, SIG-UP, 3/22), respectively in the Memo1m1Will mutant. (F, G) Lateral view of E10.5 wild-type (F) or Memo1m1WIll mutant (G) embryo processed for anti-neurofilament immunoreactivity, labeling differentiated cranial ganglia (V, VII, and VIII). Dorsal at right, rostral at top. (H, I) Frontal sections of the developing palatal shelves at E13.5 of a control (H) or Memo1ki/ki mutant (I) harboring the Wnt1-CRE transgene and rosa-Tomato reporter, resulting in fluorescent labeling of neural crest cells (all pink/orange fluorescent cells are neural crest). Blue fluorescence is DRAQ5 counterstain of nuclei, most evident in the neural crest cell negative oral and tongue epithelium. (J-Q) Ventral view of the developing palate, of the genotype indicated, at E13.5 (J, K and N, O) or E14.5 (L, M and P, Q), and processed by in situ hybridization for either Shh (J-M) or Tbx22 expression (N-Q, arrowheads). Developing rugae are numbered in (J-M). Also note, in contrast to a control embryo, Tbx22 expression at E14.5 remains on in the Memo1m1Will posterior palate (arrowheads in Q), presumably as a result of the failure of palatal shelf fusion. Abbreviations: ba1, branchial arch 1; ba2, branchial arch 2; e, epithelium; fnp, frontonasal process; NF, neurofilament; ps, palatal shelf; t, tongue. Scale bars: 500uM.
Supplemental Figure 6. Cranial vault defects in Memo1NC-CKO embryos. (A, B) Dorsal view of both a control (A) and Memo1NC-CKO (B) E18.5 embryonic head, processed for skeletal stain, revealing the craniofacial calvaria. Asterisk denotes larger space in conditional mutants versus controls. Abbreviations: f, frontal bone; ip, interparietal bone; p, parietal bone.