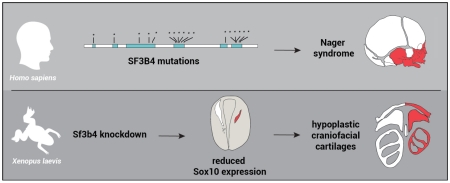
Abstract
Mandibulofacial dysostosis (MFD) is a human developmental disorder characterized by defects of the facial bones. It is the second most frequent craniofacial malformation after cleft lip and palate. Nager syndrome combines many features of MFD with a variety of limb defects. Mutations in SF3B4 (splicing factor 3b, subunit 4) gene, which encodes a component of the pre-mRNA spliceosomal complex, were recently identified as a cause for Nager syndrome, accounting for 60% of affected individuals. Nothing is known about the cellular pathogenesis underlying Nager type MFD. Here we describe the first animal model for Nager syndrome, generated by knocking down Sf3b4 function in Xenopus laevis embryos, using morpholino antisense oligonucleotides. Our results indicate that Sf3b4-depleted embryos show reduced expression of the neural crest genes sox10, snail2 and twist at the neural plate border, associated with a broadening of the neural plate. This phenotype can be rescued by injection of wild-type human SF3B4 mRNA but not by mRNAs carrying mutations that cause Nager syndrome. At the tailbud stage, morphant embryos had decreased sox10 and tfap2a expression in the pharyngeal arches, indicative of a reduced number of neural crest cells. Later in development, Sf3b4-depleted tadpoles exhibited hypoplasia of neural crest-derived craniofacial cartilages, phenocopying aspects of the craniofacial skeletal defects seen in Nager syndrome patients. With this animal model we are now poised to gain important insights into the etiology and pathogenesis of Nager type MFD, and to identify the molecular targets of Sf3b4.
Keywords: Neural crest, Sf3b4, Splicing factor, BMP, Nager syndrome, Xenopus, Craniofacial
Graphical Abstract
Introduction
The neural crest is a multipotent population of migratory cells unique to the vertebrate embryo. Neural crest cells form at the neural plate border during gastrulation, and around the time of neural tube closure, delaminate from the neuroepithelium to produce a diverse array of cell types, contributing to multiple organ systems. In the head, cranial neural crest cells originate from the mesencephalon and the rhombencephalon. They migrate in the periphery to populate the first, second and third pharyngeal/branchial arches, where they develop into craniofacial skeletal elements specific for each pharyngeal arch (Trainor and Krumlauf, 2000; Santagati and Rijli, 2003; Minoux and Rijli, 2010). The first pharyngeal arch has two components, the maxillary prominence forming the maxillary, zygomatic and temporal bones; and the mandibular prominence forming the middle ear bones, malleus and incus, and the mandible. The second pharyngeal arch gives rise to the third middle ear ossicle (stapes), styloid process and lesser horn of the hyoid, while the greater horn of the hyoid is derived from the third pharyngeal arch.
Mandibulofacial dysostosis (MFD) is a human condition involving defects in structures derived from the first and second pharyngeal arches (Passos-Bueno et al., 2009). It is characterized by hypoplasia of the facial bones, particularly the maxilla, mandible, and zygomatic complex, associated with malformation of the outer ears and middle ear ossicles (Wieczorek, 2013; Trainor and Andrews, 2013). It is the second most frequent craniofacial malformation after cleft lip and palate. One of the best-understood forms of MFD is Treacher Collins syndrome (OMIM #154500; Dixon, 1996). It is most commonly caused by mutations in one of three genes encoding factors implicated in ribosomal gene transcription TCOF1, POLR1C and POLR1D (Treacher Collins Syndrome Collaborative Group, 1996; Dauwerse et al., 2011). Mouse models of Treacher Collins syndrome indicate that this condition is caused by an early depletion of neural crest progenitors, leading to a reduced number of neural crest cells migrating to the first and second pharyngeal arches, resulting in malformation of the skeletal elements derived from these arches (Dixon et al., 2006; Jones et al., 2008; Trainor, 2010).
Acrofacial dysostosis combines many features of MFD with a variety of limb anomalies. The most common form of acrofacial dysostosis is Nager syndrome (OMIM #154400). In addition to craniofacial malformations, these patients present preaxial limb defects, typically hypoplasia or absence of the thumbs. With less than 100 cases described in the literature the exact prevalence of Nager syndrome is not known (Schlieve et al., 2012). Most cases are sporadic, however autosomal-dominant (Hall, 1989; Alysworth et al., 1991; McDonald and Gorski., 1993) and autosomal-recessive (Chemke et al. 1988; Kennedy and Teebi, 2004; Nur et al., 2013) forms of the disease have been reported. Mutations in SF3B4 (Splicing factor 3b, subunit 4) gene were recently identified as a cause of Nager syndrome, accounting for approximately 60% of the cases (Bernier et al., 2012; Czeschik et al., 2013; Petit et al., 2014). This finding is consistent with an older report describing a child diagnosed with Nager syndrome with a deletion of chromosomal region 1q12-q21, a region that comprises the SF3B4 gene (Waggoner et al., 1999). SF3B4 encodes spliceosome-associated protein 49 (SAP49), a component of the pre-mRNA spliceosomal complex, the machinery that accurately recognizes and removes the introns from the pre-mRNAs to produce mature mRNAs. The spliceosome consists of five U1, U2, U4/U6 and U5 small nuclear ribonucleoproteins (snRNPs), and a large number of associated proteins (Will and Luhrmann, 2011). During assembly of the U2 pre-spliceosomal complex, SAP49 binds to the pre-mRNA just upstream of the branch point sequence and plays a crucial role in tethering the U2 snRNP to the branch site during the splicing process (Champion-Arnaud and Reed, 1994). Mutations in genes encoding other components of the spliceosome, such as EFTUD2 (Lines et al., 2012), SNRPB (Lynch et al., 2014) and TXNL4A (Wieczorek et al, 2014) also cause craniofacial disorders, suggesting that defects in mRNA processing may underlie the etiology of some forms of MFD (Lehalle et al., 2015).
Nothing is known about the cellular and molecular mechanisms underlying Nager type MFD, therefore the generation of animal models is absolutely critical to investigate the pathogenesis of this condition and the role of SF3B4 in the presentation of the disease. Here we report the characterization of the first animal model for Nager type MFD obtained by specifically knocking down Sf3b4 function in Xenopus laevis embryos.
Materials and Methods
Plasmid constructs
The clone for Xenopus laevis Sf3b4 (accession # BC045264) was purchased from ThermoFisher Scientific (Waltham, MA). The open reading frame (ORF) of Xenopus laevis sf3b4 (Xsf3b4) was amplified by PCR using the following primers: forward, 5’-ATCGATGCCACCATGGCGGCTGGGCCGATA-3’ and reverse, 5’-CTCGAGTCACTGTGTCATGGGGAGCCT-3’. The 1134 bp PCR product was sequenced and inserted into the ClaI and XhoI sites of pCS2+ expression vector. The clone for Human SF3B4 (accession # BC004273) was purchased from GE Dharmacon (Lafayette, CO). Human SF3B4 (HSF3B4) ORF was amplified by PCR using the following primers: forward, 5’-ATCGATGCCACCATGGCTGCCGGGCCGATCTCC-3’ and reverse, 5’-TCTAGATTACTGAGGGAGAGGGCCTCG-3’. The 1274 bp PCR product was sequenced and inserted into the ClaI and XbaI sites of pCS2+, it is referred as pCS2+HSF3B4. Site-directed mutagenesis was used to generate constructs with single mutations in HSF3B4 coding region based on the 18 unique mutations identified in Nager syndrome patients (Bernier et al., 2012). We selected a frameshift mutation (c.88delT), predicted to affect the RNA recognition motifs (RRMs), and a nonsense mutation (c.625C>T). The QuickChange II site-directed mutagenesis kit (Agilent Technologies; Wilmington, DE) was used according to the manufacturer instructions using pCS2+HSF3B4 as template and the following primer sets: 5’-CATCTGCAAACAGCTAATGAGGGCGATCAGCCTG-3’ and 5’-CAGGCTGATCGCCCTCATTAGCTGTTTGCAGATG-3’ for the c.625C>T mutation, 5’-GGAGAAACAGTTCCCCAGCAGCGGTTCACT-3’ and 5’-AGTGAACCGCTGCTGGGGAACTGTTTCTCC-3’ for the c.88delT mutation. Mutations were confirmed by sequencing and the resulting proteins analyzed by Western blot after expression in Xenopus embryos.
Embryos, injections and explants culture
Xenopus laevis embryos were staged according to Nieuwkoop and Faber (1967) and raised in 0.1× NAM (Normal Amphibian Medium; Slack and Forman, 1980). This study was performed in accordance with the recommendations of the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. The procedures were approved by New York University Institutional Animal Care and Use Committee, under animal protocol # 150201. mRNAs were synthesized in vitro using the Ambion Message Machine kit (Austin, TX). Control (CoMO), Sf3b4 translation (Sf3b4MO; ATATCGGCCCAGCCGCCATTTTCAC), and splice (Sf3b4MO2; GCCAATAACCTGTGAGGAAAAAGAGC) blocking morpholino antisense oligonucleotides (MOs) were purchased from GeneTools (Philomath, OR). MOs and mRNAs were injected in one blastomere at the 2-cell stage and embryos were analyzed by in situ hybridization. To identify the injected side, 500 pg of β-galactosidase mRNA was coinjected as a lineage tracer. For animal explant experiments, both blastomere at the 2-cell stage were injected in the animal pole region with noggin mRNA (100 pg) alone or in combination with Xsf3b4 or HSF3B4 mRNA (1 ng), explants were dissected at the late blastula stage, cultured for 8 hours in NAM 0.5× and processed for in situ hybridization.
Lineage tracing and whole-mount in situ hybridization
Embryos at the appropriate stage were fixed in MEMFA stained for Red-Gal (Research Organics; Cleveland, OH) to visualize the lineage tracer (β-gal mRNA) and processed for in situ hybridization. Antisense digoxygenin-labeled probes (Genius kit; Roche, Indianapolis IN) were synthesized using template cDNA encoding sf3b4, snail2 (Mayor et al. 1995), sox10 (Aoki et al., 2003), tfap2a (Luo et al., 2003), sox2 (Mizuseki et al., 1998) and msx1 (Suzuki et al., 1997). Whole-mount in situ hybridization was performed as previously described (Harland, 1991).
Cartilage staining
Alcian blue staining of stage 45 tadpoles was performed as described in Berry et al., (1998). Briefly, tadpoles were fixed overnight in 10% formaldehyde, rinsed in tap water, skinned and eviscerated. Tadpoles were then dehydrated and stained in Alcian blue (Sigma-Aldrich; St. Louis, MO) for 12 hours. After several rinses in 95% ethanol, tadpoles were rehydrated and macerated in 2% potassium hydroxide. Specimens were then transferred successively in 20%, 40%, 60% and 80% glycerol in 2% potassium hydroxide. The ethmoid plate was dissected out and specimens mounted flat under a coverslip in 80% glycerol, and individually photographed on a Nikon SZX9 stereomicroscope. For each affected tadpole, the surface areas of the Meckel’s, ceratohyal and branchial cartilages on the injected side were measured using ImageJ and compared to the same structures on the contralateral side to define a percentage reduction.
TUNEL and proliferation assays
TUNEL staining was carried out as described (Hensey and Gautier, 1998). Sf3b4MO2-injected albinos embryos fixed in MEMFA were rehydrated in PBT and washed in TdT buffer (Roche, Indianapolis IN) for 30 min. End labeling was carried out overnight at room temperature in TdT buffer containing 0.5 μM DIG-dUTP and 150 U/ml TdT (Roche, Indianapolis IN). Embryos were then washed for 2 hours at 65°C in PBS/1 mM EDTA. DIG was detected with anti-DIG Fab fragments conjugated to alkaline phosphatase (Roche, Indianapolis IN; 1:2000), and the chromogenic reaction performed using NBT/BCIP (Roche, Indianapolis IN). For phosphohistone H3 detection (Saka and Smith, 2001), fixed albinos embryos were incubated successively in α-phosphohistone H3 antibody (Upstate Biotechnology, 06-570; 1 μg/ml) and an anti-rabbit IgG secondary antibody conjugated to alkaline phosphatase (ThermoFisher Scientific, G-21079; 1:1000). Alkaline phosphatase activity was revealed using NBT/BCIP (Roche, Indianapolis IN). To quantify changes in cell death and proliferation, embryos were individually photographed and the number of TUNEL- and pHH3-positive cells in the dorsal ectoderm was counted manually on the control and injected sides of each embryo. Student’s t-test was used to define statistically significant differences.
Western blot analysis
For western blots, embryos were injected at the 2-cell stage and cultured to stage 15. Pools of 10 embryos were homogenized in lysis buffer (0.5% Triton X-100, 10mM Tris–HCI at pH 7.5, 50mM NaCl, 1mM EDTA), containing Halt™ Protease Inhibitor Cocktail (ThermoFisher Scientific; Waltham, MA). After two consecutive centrifugations to eliminate lipids, the lysate was concentrated on an Amicon Ultra Centrifugal Filter (Merck Millipore; Billerica, MA), 10 μl of the concentrated lysate was resolved on a 10% NuPAGE Bis-Tris gel and transferred onto a PVDF membrane using the iBlot system (Invitrogen). Blots were subsequently incubated overnight with one of the following primary antibodies: anti Sf3b4 polyclonal antibody (ThermoFisher Scientific, PA5-28441; 1:500 dilution), anti α-tubulin antibody (Sigma Aldrich, T9026; 1:500 dilution), anti phospho-Smad1/5/8 antibody (Cell Signaling Technology, 13820; 1:500 dilution) or anti R-Smad1/5/8 antibody (Santa Cruz Technology, sc-6031-R; 1:250 dilution). The blots were then washed and incubated with anti-rabbit or anti-mouse IgG coupled to horseradish peroxidase (Santa Cruz Biotechnology; 1:5000 dilution). Peroxidase activity was detected with the Western Blotting Luminol Reagent (Santa Cruz Biotechnology) and imaged on a ChemiDoc MP Biorad gel documentation system (Hercules, CA). Membranes were stripped using Restore Western Blot Stripping Buffer (ThermoFisher Scientific) according to the manufacturer recommendations.
RT-PCR analysis
Total RNAs were extracted from 3-5 embryos using the RNeasy micro RNA isolation kit (Qiagen; Valencia, CA). The RNA samples were digested with RNase-free DNase I before RT-PCR. The amount of RNA isolated was quantified by measuring the optical density using a Nanodrop spectrophotometer (Nanodrop Technologies; Wilmington, DE). The reactions were performed using the One Step RT-PCR kit (Qiagen; Valencia CA) according to the manufacturer’s instructions using primers for Sox9, So×10, Twist, Pa×3, Keratin and EF1α (Hong and Saint-Jeannet, 2007) and the QuantiTect SYBR green RT-PCR kit (Qiagen; Valencia, CA) on an Eco Real-Time PCR System (Illumina; San Diego, CA). The reaction mixture consisted of 10 μl of QuantiTect SYBR Green RT-PCR Master Mix, 500 nM of forward and reverse primers and 60 ng of template RNA in a total volume of 20 μl. Cycling conditions were as follows: denaturation at 95°C (3 sec), annealing at 55°C (4 sec) and extension at 72°C (12 sec). By optimizing primers and reaction conditions a single specific product was amplified as confirmed by melting curve analysis. Each reaction included a control without template and a standard curve of serial dilution (in 10-fold increments) of test RNAs. For conventional RT-PCR, 1 μg of total RNAs was reverse transcribed using the Superscript VILO cDNA Synthesis Kit (Invitrogen; Grand Island, NY) and 1 μl of cDNA was amplified using PuReTaq™ ready-to-go PCR Beads (GE Healthcare Bio-Sciences Pittsburgh, PA) using primers for sf3b4 (F: GTGGAGTTTCTGAGTGAAGAGG; R: ACAGGAGGAAATGTGGGAATAC), Snail2 (F: CTGGTCAAGAAACACTTCAACAC; R: ATCTGTCTGCGAATGCTCTG), so×10 (F: GAAGTAGAGATGAGCCCTGTTG; R: GGCTGGTACTTGAGCTTTCT), twist (F: AGTCTGAGTAACAGCGAGGA; R: CTGTAAGACCTGGCAGAGAAAG), pax3 (F: CAGCTGGGTGGAGTGTTTAT; R: CTGTGGGTATTGGTATCCTGTG), sox9 (F: AAGCTGGCAGACCAATATCC; R: AAGGTTTCGATGGTGGAGATG) and odc (F: ACATGGCATTCTCCCTGAAG; R: TGGTCCCAAGGCTAAAGTTG). The PCR conditions were as follows: denaturation 94 °C (30 s), annealing at 55 °C (30 s), and extension at 72 °C (30 s), 30 cycles for sf3b4, snail2, so×10, twist, sox9 or pa×3, and 25 cycles for odc.
Results
Xenopus laevis Sf3b4
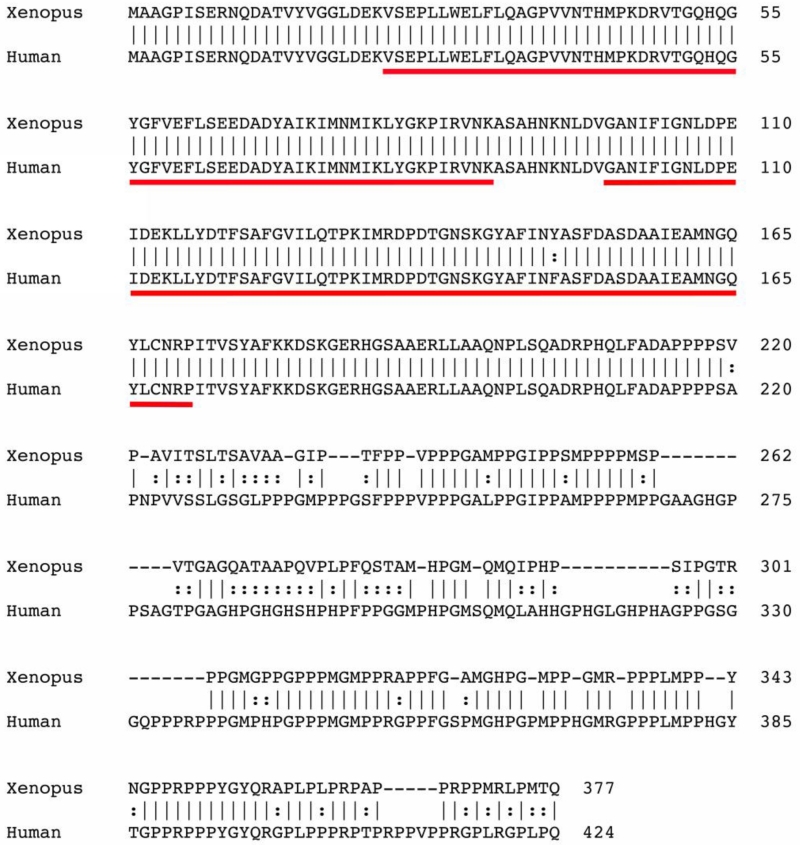
SF3B4 encodes spliceosome-associated protein 49 (SAP49), one of seven core proteins of the spliceosomal complex implicated in the removal of introns from the pre-mRNAs (Will and Lurhmann, 2011). Structurally, SAP49 contains two adjacent N-terminal RNA recognition motifs (RRMs) that bind directly to the pre-mRNA. Xenopus laevis Sf3b4 possesses an open reading frame encoding 377 amino acids (Fig 1). At the amino acid level, Xenopus laevis Sf3b4 shares over 80% identity with human SF3B4, with close to 100% identity at the levels of the RRMs (Fig 1).
Figure 1.
Sequence and structure comparison of Xenopus and Human SF3B4. The predicted amino acid sequences from Xenopus laevis and human, SF3B4 were aligned using ClustalW. The two RNA recognition motifs (RRM) are underlined.
Developmental expression of sf3b4
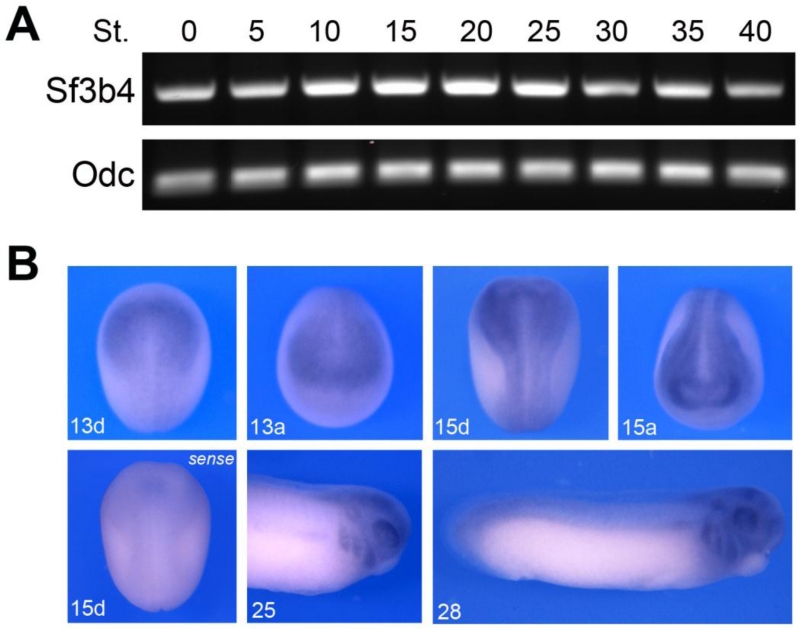
To analyze the temporal expression of Xenopus sf3b4 we performed RT-PCR on RNA isolated at different embryonic stages (Fig 2A). sf3b4 is maternally expressed (stages 0-5) and detected at all stages examined, up to the tadpole stage (stage 40). To determine the spatial expression pattern of sf3b4, we performed whole-mount in situ hybridization on embryos at different stages of development. Antisense and control sense probes were used to establish specificity of the signal (Fig 2B). sf3b4 transcripts are detected at the neurula stage (stages 13-15) and appear to be enriched dorsally and anteriorly (Fig 2B). At the tailbud stage (stage 25 and 28) Sf3b4 is detected in the branchial arches and in the developing eyes (Fig 2B). Upon longer chromogenic reaction, more intense staining was detected throughout the embryos (not shown), suggesting that sf3b4 is broadly expressed at early embryonic stages, which is not surprising given its role in a global process like pre-mRNA splicing.
Figure 2.
Developmental expression of sf3b4. (A) RT-PCR analysis of sf3b4 developmental expression. The embryonic stages (St.) are according to Nieuwkoop and Faber (1967). Ornithine decarboxylase (odc) is shown as a loading control. (B) By whole-mount in situ hybridization sf3b4 is enriched dorsally at neurula stage (stage 13 and 15). At tailbud stage (stage 25 and 28) sf3b4 is detected in the branchial arches and the developing eyes. Dorsal views (d), anterior to top. Anterior views (a), dorsal to top. Lateral view, anterior to right, dorsal to top (stage 25 and 28).
Sf3b4 is required for neural crest formation
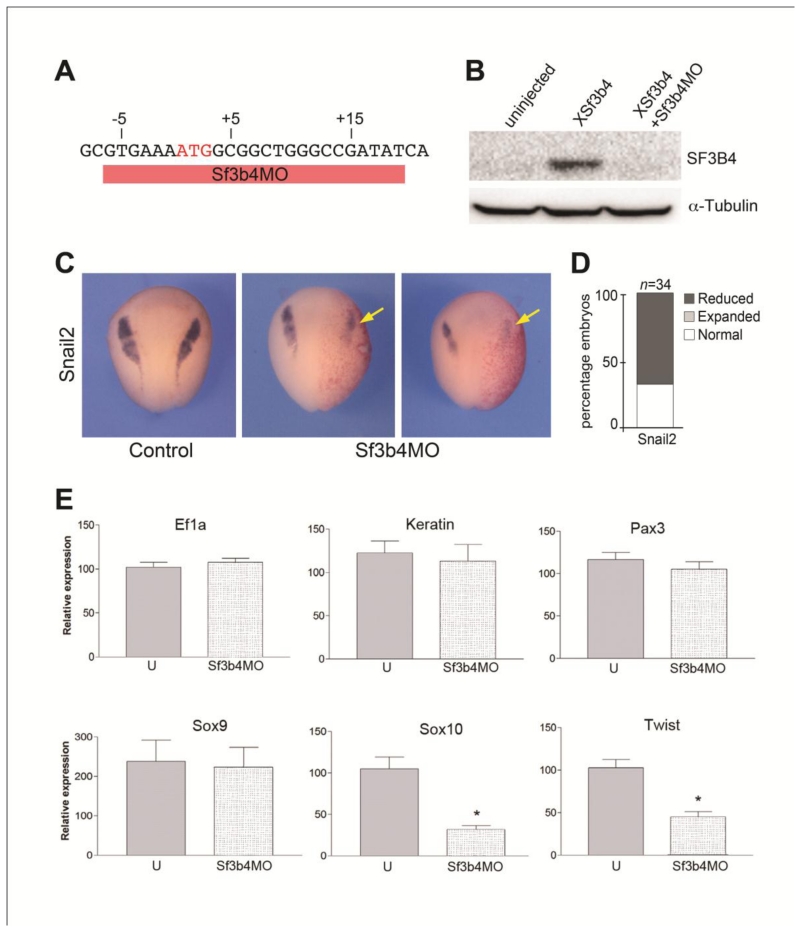
Mouse models of Treacher Collins syndrome suggest that this condition is caused by an early depletion of cranial neural crest progenitors (Dixon et al., 2006; Jones and et al., 2008). To determine whether MFD in Nager syndrome is also due to abnormal neural crest development in the head we used a translation blocking morpholino antisense oligonucleotide (Sf3b4MO; Fig 3A) to interfere with Sf3b4 function in Xenopus embryos. Western blot analysis of embryos coinjected with Xenopus sf3b4 mRNA and Sf3b4MO showed that Sf3b4MO blocks Sf3b4 protein accumulation in vivo (Fig 3B). Unilateral injection of Sf3b4MO (40 ng) in one cell at the 2-cell stage resulted in a marked decrease in Snail2 expression, a gene restricted to early neural crest progenitors (Fig 3C,D), suggesting that the production of neural crest progenitors is affected in the absence of Sf3b4 function. We expanded this analysis by looking at a broader range of genes by real-time RT-PCR (qPCR). These genes include: ef1a, a housekeeping gene, keratin expressed in the epidermis, and four transcription factors expressed in neural crest progenitors pax3, sox9, sox10 and twist. Embryos were injected with Sf3b4MO in both cells at the 2-cell stage, collected at the early neurula stage and analyzed by qPCR. We found that the housekeeping gene (ef1a) and epidermal keratin were unaffected in morphant embryos (Fig 3E). Interestingly, among the neural crest genes we observed differential response upon Sf3b4 knockdown. While pax3 and sox9 expression levels were largely unchanged, the expression levels of sox10 and twist were significantly reduced as compared to uninjected control (Fig 3E). In the gene regulatory network controlling neural crest formation pax3 and sox9 are acting up-stream of sox10 and twist (Betancur et al., 2010) raising the possibility that either Sf3b4 regulates sox10/twist pre-mRNA splicing, or Sf3b4 regulates the pre-mRNA splicing of factor(s) acting up-stream of sox10/twist and down-stream of pax3/sox9.
Figure 3.
Sf3b4MO, a translation blocking morpholino prevents snail2, sox10 and twist expression. (A) Design of the translation blocking morpholino antisense oligonucleotide. Sf3b4MO (red bar) targets the ATG start site of Xenopus Sf3b4 mRNA. (B) Western blot using lysates from embryos injected with XSf3b4 mRNA alone or in combination with Sf3b4MO, shows that Sf3b4MO blocks SF3B4 protein accumulation in vivo. α-tubulin is shown as a loading control. (C) Snail2 expression is reduced in Sf3b4MO-injected (40 ng) embryos (arrows). Dorsal views, anterior to top. The injected side (arrows) is indicated by the lineage tracer (Red-Gal). (D) The graph is a quantification of the results from three independent experiments. The number of embryos analyzed (n) is indicated on the top of the bar. (E) Gene expression analysis (qPCR) of control (U) and Sf3b4MO-injected embryos. Values are normalized to ef1α and presented as mean ± s.e.m.; (*) p < 0.002 (Student’s t-test), from five independent samples.
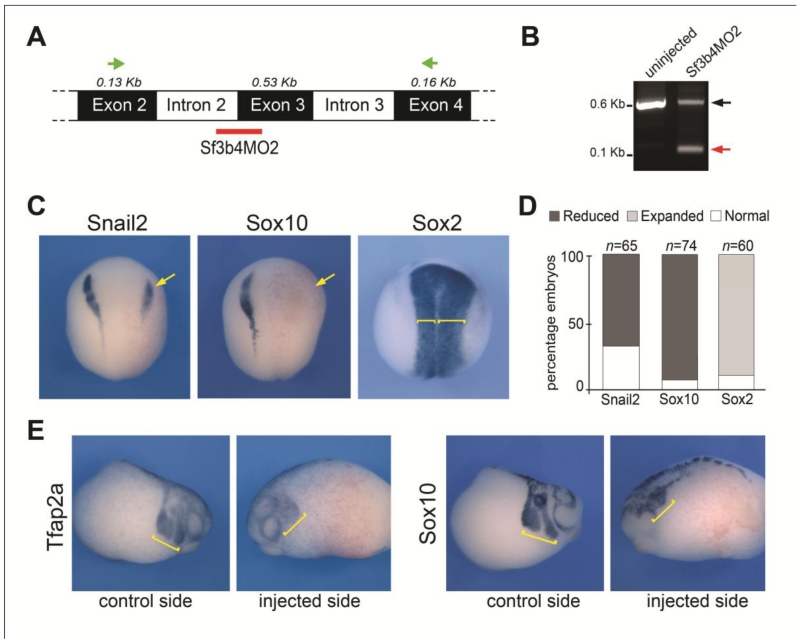
To confirm the specificity of the Sf3b4 knockdown phenotype, we used a second MO (Sf3b4MO2) that specifically interferes with sf3b4 pre-mRNA splicing by targeting the intron 2-exon 3 junction (Fig 4A), resulting in the production of a shorter transcript, due to the loss of exon 3 (Fig 4B). The phenotype of Sf3b4MO2-injected embryos was identical to the phenotype generated by injection of the translation blocking MO, with a strong decrease of snail2 and sox10 expression at early neurula stage (Fig 4C,D). In addition we found that the expression domain of the neural plate gene, sox2, appeared broader on the injected side, consistent with the loss of the neural plate border (Fig 4C,D). At the tailbud stage, as neural crest cells migrate to the pharyngeal arches, we analyzed by in situ hybridization two genes expressed in migrating neural crest cells, tfap2a and sox10. We found that the vast majority of the embryos had reduced tfap2a and sox10 expression indicative of a decreased number of neural crest cells migrating to the pharyngeal arches (Fig 4E). The similarity of the two knockdown phenotypes, which are interfering with sf3b4 translation and splicing, respectively, provides strong evidence for a specific requirement of Sf3b4 during neural crest progenitors formation.
Figure 4.
Sf3b4MO2, a splice blocking morpholino interferes with neural crest formation. (A) The PCR primers to analyze sf3b4 pre-mRNA splicing are indicated (green arrows). The splice blocking morpholino (Sf3b4MO2) targets intron2/exon3 junction (red bar). (B) In Sf3b4MO2-injected embryos a shorter Sf3b4 transcript is detected (red arrow) due to loss of exon 3. (C) At neurula stage the expression of snail2 and sox10 is reduced (arrows) in Sf3b4MO2-injected embryos (30 ng), while sox2 expression is broadened (brackets). Dorsal views, anterior to top. The injected side (right) is indicated by the lineage tracer (Red-Gal). (D) The graph is a quantification of the results from four independent experiments. The number of embryos analyzed (n) is indicated on the top of each bar. (E) At tailbud stage tfap2a and sox10 expression is reduced in the pharyngeal arches (brackets). Lateral views.
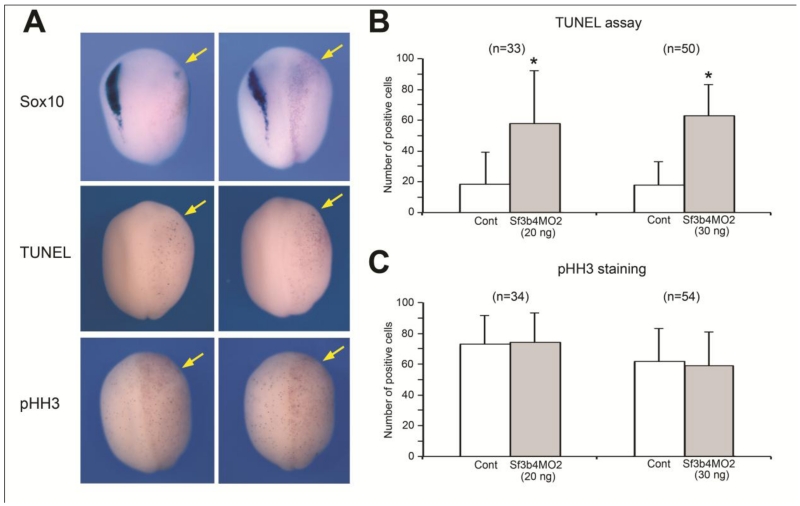
As the expression of a number of neural crest transcriptional regulators was reduced following Sf3b4 depletion, we examined whether their reduction could be due to an increase in cell death in this cell population. Embryos injected with Sf3b4MO2 (20 ng or 30 ng) were allowed to develop until stage 15 when apoptosis was assessed by TUNEL staining. We observed a highly significant increase in the number of TUNEL-positive nuclei on the Sf3b4-depleted versus control sides of these embryos (Fig 5A-B). Similarly, we examined whether the changes in gene expression could be a consequence of altered cell proliferation/cell cycle progression by analyzing the number of cells immunopositive for phosphor-histone H3 (pHH3). We found no difference in number of mitotic cells on the Sf3b4-depleted versus control sides of these embryos (Fig 5A-C). Altogether these results suggest that the reduction in neural crest genes expression in Sf3b4-depleted embryos reflects a loss of neural crest progenitors through a mechanism that involves primarily cell death.
Figure 5.
Sf3b4 depletion results in increased cell-death in the ectoderm. (A) In situ hybridization of Sf3b4MO2-injected embryos (30 ng) at stage 15 showing loss of sox10 expression on the injected side (upper panels). Sf3b4MO2 injection significantly increases the number of TUNEL-positive cells (middle panels). Sf3b4MO2 injection does not affect the levels of cell proliferation as visualized by phospho-histone H3 (pHH3) immunostaining (lower panels). Dorsal views, anterior to top. The injected side (right) is indicated by the lineage tracer (Red-Gal; arrows). (B-C) Graph illustrating the quantification of the number of TUNEL-positive (B) and pHH3-positive (C) cells on the control (Cont) and injected sides in embryos that received unilateral injection of Sf3b4MO2 (20 ng or 30 ng as indicated). The number of embryos analyzed (n=) is indicated on the top of each graph. Values are presented as mean ± s.e.m.; (*) p-value <0.0001 (Student’s t-test).
Human SF3B4 can rescue the phenotype of Sf3b4-depleted embryos
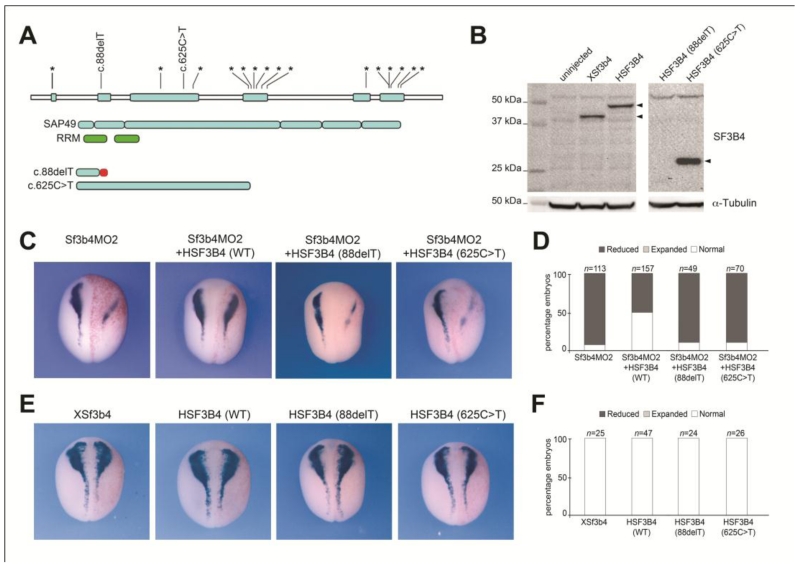
An important validation of Xenopus Sf3b4-depleted embryos as a model for Nager syndrome rests on the ability to rescue the morphant phenotype by expression of human SF3B4 (HSF3B4). Here we compared the ability of wild type (WT) HSF3B4 mRNA, and mRNAs carrying mutations that cause Nager syndrome (Bernier et al., 2012), at restoring sox10 expression in Sf3b4-depleted embryos. Using site-directed mutagenesis we generated two mutant mRNAs, c.88delT and c.625C>T (Fig 6A,B). Injection of 10 pg of WT HSF3B4 mRNA was sufficient to rescue sox10 expression close to normal levels in approximately 50% of morphant embryos (Fig 6C,D), while the same dose of mutant mRNAs were unable to restore Sox10 expression in Sf3b4-depleted embryos (Fig 6C,D). Interestingly, single injection of WT or mutant HSF3B4 mRNAs had no effect on sox10 expression, even for high doses (2-4 ng) of injected mRNAs (Fig 6E,F; not shown).
Figure 6.
Loss of Sox10 expression in Sf3b4-depleted embryos is rescued by human SF3B4. (A) Distribution of the 18 unique mutations in SF3B4 gene (*) that cause Nager syndrome (Bernier et al., 2012). Using site directed mutagenesis we generated two constructs that duplicate the human frameshift mutation in exon 2 (c.88delT) and a nonsense mutation in exon3 (c.625C>T). Domain structure of SF3B4/SAP49 (blue boxes) is indicated with the position of the RRMs (green boxes). The predicted translational effect of the two SF3B4 mutations used in this study is shown. The red bar indicates altered amino acid sequence. Modified from Bernier et al., (2012). (B) Western blot using lysate from embryos injected with 2 ng of Xsf3b4, HSF3B4, 88delT or 625C>T mRNAs. α-tubulin is shown as a loading control. 88delT mRNA is expected to generate a short product (~4.0 kDa) not recognized by Sf3b4 antibody. (C) Wild-type (WT) HSF3B4 restore sox10 expression in Sf3b4-depleted embryos, while the mutated mRNAs are unable to do so. Dorsal views, anterior to top. The injected side (right) is indicated by the lineage tracer (Red-Gal). (D) The graph is a quantification of the results from four independent experiments. The number of embryos analyzed (n) is indicated on the top of each bar. (E) Injections of Xsf3b4, HSF3B4 mRNAs and its mutated versions had no effect on sox10 expression. Dorsal views, anterior to top. The injected side (right) is indicated by the lineage tracer (Red-Gal). (F) The graph is a quantification of the results. The number of embryos analyzed (n) is indicated on the top of each bar.
Sf3b4-depleted embryos develop an abnormal craniofacial skeleton
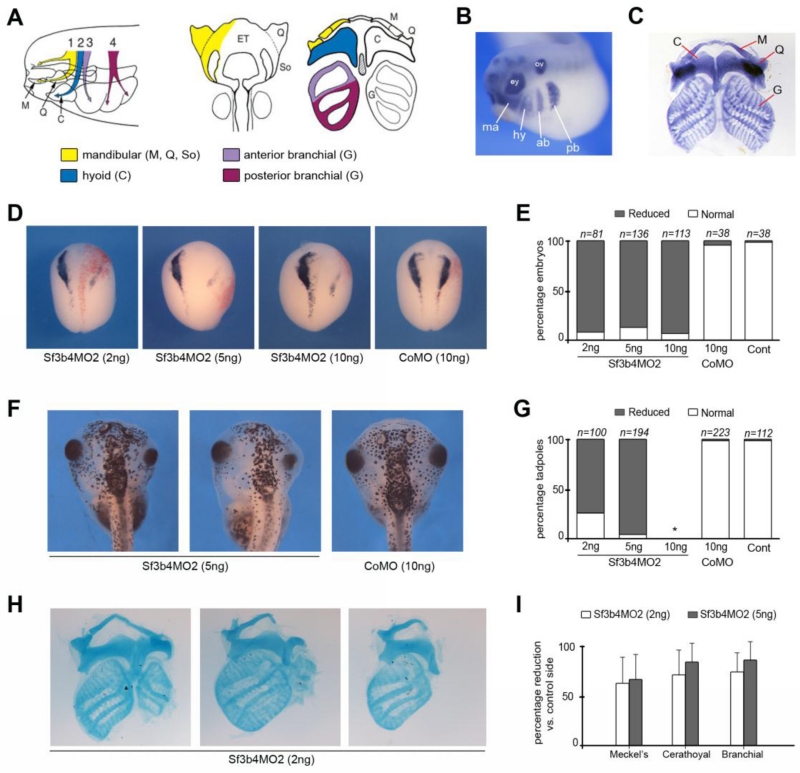
We next analyzed the long-term consequences of Sf3b4 depletion by documenting the development of neural crest-derived skeletal elements in morphant and control embryos. In Xenopus, at the tailbud stage cranial neural crest cells migrate as four individual streams towards the pharyngeal arches, the mandibular, hyoid, anterior and posterior branchial neural crest (Fig 7A-B). In the first pharyngeal arch, the mandibular neural crest cells contribute to the palatoquadrate (upper jaw) and the Meckel’s (lower jaw) cartilages; in the second arch the hyoid neural crest cells contribute to the ceratohyal cartilage; while in the third and fourth arches, the branchial neural crest cells contribute to the anterior and posterior regions of the gills cartilages, respectively (Fig 7A,C; Sadaghiani and Thiebaud, 1987).
Figure 7.
Long-term consequences of Sf3b4 depletion on craniofacial structures formation. (A) Migration and contribution of the neural crest to cranial skeletal elements. The mandibular neural crest segment (1; yellow) contributes to Meckel’s (M), palatoquadrate (Q), and subocular (So) cartilages; the hyoid neural crest segment (2; blue) contributes to the ceratohyal (C) cartilages; the anterior (3; purple) and posterior (4; red) branchial neural crest segments contribute to the cartilages of the gills (G). Modified from Sadaghiani and Thiebaud (1987) and Balzinger et al., (2005). (B) By in situ hybridization Sox9 is detected in the four streams of neural crest cells migrating towards the pharyngeal arches, the mandibular (ma), hyoid (hy), anterior branchial (ab) and posterior branchial (pb) segments. Lateral view, anterior to left. ey: eye; ov: otic vesicle. Modified from Lee and Saint-Jeannet, (2011). (C) Alcian blue-stained craniofacial skeletal elements from a stage 45 tadpole. Modified from O’Donnell et al., (2006). (D) At neurula stage the expression of sox10 is reduced in Sf3b4MO2-injected embryos. Dorsal views, anterior to top. The injected side (right) is indicated by the lineage tracer (Red-Gal). (E) The graph is a quantification of the results from seven independent experiments. The number of embryos analyzed (n) is indicated on the top of each bar. (F) Gross morphology of Sf3b4MO2 (5 ng) and CoMO (10 ng)-injected tadpoles. Dorsal views, anterior to top. (G) The graph is a quantification of the results from four independent experiments. The number of tadpoles analyzed (n) is indicated on the top of each bar. (*) Embryos injected with 10 ng Sf3b4MO2 did not survive to the tadpole stage. (H) Examples of flat-mounts of Alcian blue-stained skeletal elements from stage 45 tadpoles injected with Sf3b4MO2 (2 ng). Similar phenotypes were observed with 5 ng Sf3b4MO2. Injected side is on the right. (I) The graph is a quantification of the results from three independent experiments. The size of Meckel’s, ceratohyal and branchial cartilages of affected tadpoles was measured and compared to the same cartilage on the contralateral side to define the percentage reduction. Error bars indicate mean ± s.e.m. Sf3b4MO2 (2 ng), n=69 and Sf3b4MO2 (5 ng), n=38.
In this set of experiments, Sf3b4-depleted embryos were cultured for several days and analyzed at stage 45 for craniofacial defects. Since embryos injected with 40 ng of MO failed to survive beyond tailbud stage (not shown), embryos were injected with lower doses of MOs: 2ng, 5ng and 10ng for Sf3b4MO2, and 10ng for a control MO (CoMO). At stage 15, a subset of these embryos were fixed, and analyzed by in situ hybridization to ascertain that they had reduced sox10 expression. Sibling embryos were allowed to develop up to stage 45 and analyzed for gross craniofacial defects. For all doses of Sf3b4MO2, the vast majority of these embryos had reduced sox10 expression at stage 15 (Fig 7D,E), while sox10 expression was unaffected in CoMO-injected embryos (Fig 7D,E). Embryos injected with 10 ng Sf3b4MO2 did not survive to stage 45 and became necrotic at tailbud stage, however a vast majority of tadpoles that received lower doses of Sf3b4MO2 (2 ng and 5 ng) exhibited a reduction of the craniofacial structures on the injected side, while CoMO-injected tadpoles were unaffected (Fig 7F,G). To characterize and quantify the nature of these defects, the head of affected tadpoles were dissected and processed for Alcian blue staining to visualize the craniofacial skeletal elements (Spokony et al., 2002; O’Donnell et al., 2006). For each affected tadpole, the size of the affected Meckel’s, ceratohyal and branchial cartilages was measured and compared to the cartilage on the contralateral side (Fig. 7H,I). All three cartilages were equally affected, ranging from relatively mild reductions in size to complete loss of the corresponding cartilage as pictured in Fig 6H. It is important to point out that the general pattern of pigment cells (another neural crest derivative) was largely unperturbed in these tadpoles (Fig 7F), indicating that Sf3b4 primarily regulates the formation of cranial neural crest derivatives.
Pre-mRNA splicing versus non-canonical activity of Sf3b4
Because Sf3b4 is an essential component of the spliceosome and Sf3b4-depleted tadpoles show hypoplastic cranial neural crest derivatives, we examined the pre-mRNA processing of a subset of neural crest transcriptional regulators critical for neural crest formation namely, sox10, snail2, twist, sox9 and pax3. These genes represent potential molecular targets of Sf3b4. We performed RT-PCR on Sf3b4MO2 and CoMO injected embryos using primers designed to amplify the coding region of these genes, since defective pre-mRNA splicing is expected to result in intron retention. For all 5 genes examined we did not observe any increase in transcripts size (Supplementary Fig S1), suggesting that the selected genes are not targets of Sf3b4 splicing activity. However, we cannot exclude that the pre-mRNA processing of other genes more upstream in the neural crest gene regulatory network are affected. It is also possible that some of the aberrant transcripts may have been targeted for nonsense-mediated mRNA decay (Chang et al., 2007), this is especially true for Sox10, for which the normal transcripts are largely absent. Only a more global analysis of Sf3b4 knockdown on pre-mRNA processing will permit the identification of the true targets of Sf3b4 splicing activity.
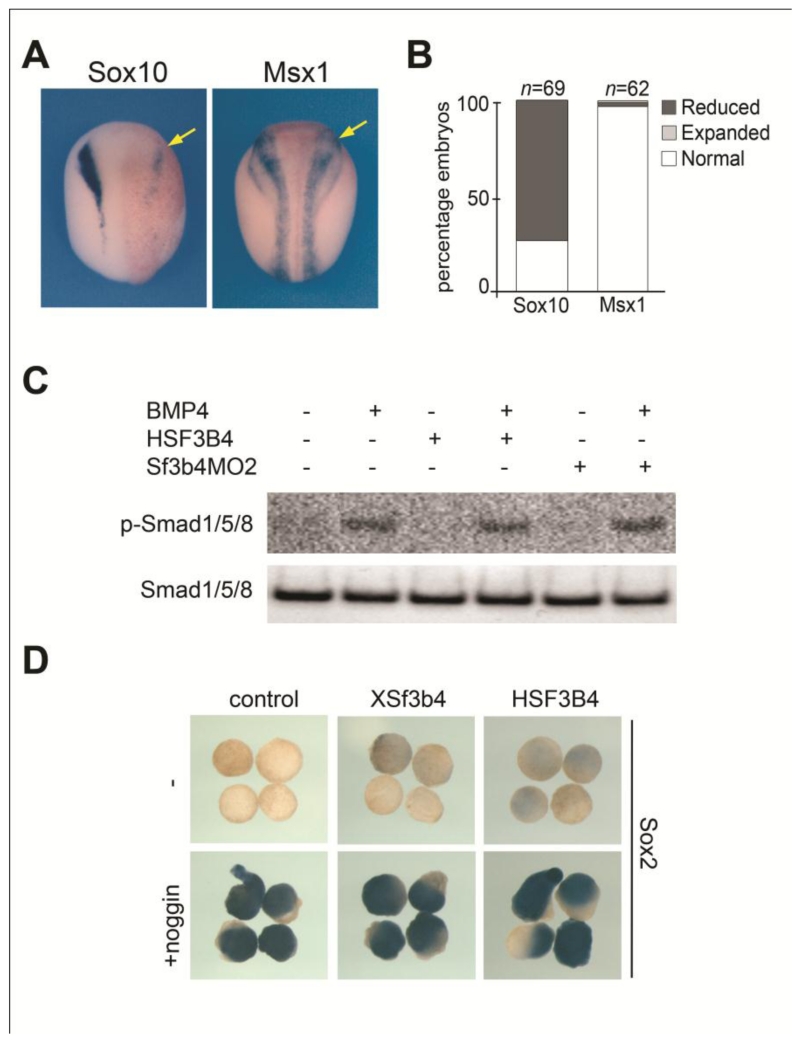
A couple of studies have linked SF3B4/SAP49 to bone morphogenetic protein (BMP) signaling. Sf3b4 was isolated in a yeast two-hybrid screen as a factor interacting with the intracellular domain of BMP receptor IA (BMPR-IA; Nishanian and Waldman, 2004) and was shown to interfere with BMP activity in tissue culture (Watanabe et al., 2007). To address a potential function of Sf3b4 in BMP signaling we first analyzed by in situ hybridization the consequences of Sf3b4 depletion on msx1 expression, a direct BMP target (Suzuki et al., 1997). We found no changes in msx1 expression at the neural plate border of morphant embryos, while sibling embryos had marked reduced sox10 expression (Fig 8A-B). Next, we evaluated whether Sf3b4 overexpression (HSF3B4 mRNA injection) or depletion (Sf3b4MO2 injection) could alter BMP4-mediated phosphorylation of Smad1/5/8 in the embryo. Western blots analysis indicates that Sf3b4 gain-of-function and loss-of-function had no impact on the levels of phosphorylated or unphosphorylated Smad1/5/8 in embryos expressing BMP4 (Fig 8C). Inhibition of BMP signaling by the BMP antagonist Noggin is implicated in neuralization of the ectoderm. We next analyzed the impact of Sf3b4 overexpression (XSf3b4 or HSF3B4 mRNA injection) on Noggin-mediated neural induction in dissected ectoderm explants. We found that the induction of sox2 by Noggin was unperturbed by XSf3b4 or HSF3B4 expression (Fig 8D). Here, using three different assays we were unable to establish a link between Sf3b4 and BMP signaling, suggesting that the cause for Sf3b4-mediated MFD may not depend on this non-canonical activity of Sf3b4.
Figure 8.
Non-canonical function of Sf3b4 in BMP signaling. (A) At neurula stage the expression of msx1 is unaffected in Sf3b4MO2-injected embryos (arrows), while sox10 expression is reduced in sibling embryos. Dorsal views, anterior to top. The injected side (right) is indicated by the lineage tracer (Red-Gal). (B) The graph is a quantification of the results from three independent experiments. The number of embryos analyzed (n) is indicated on the top of each bar. (C) Western blot using lysate from embryos injected bilaterally with HSF3B4 mRNA (1 ng) or Sf3B4MO2 (10 ng) alone or in combination with BMP4 mRNA (1 ng) to analyze the levels of phosphorylated and unphosphorylated Smad1/5/8 at stage 10. HSF3B4 and Sf3B4MO2 have no impact on BMP4-mediated Smad1/5/8 phosphorylation (p-Smad1/5/8) and do not affect the levels of unphosphorylated Smad1/5/8. (D) By in situ hybridization, sox2 expression in animal explants neuralized by the BMP antagonist Noggin is unaffected by injection of Xsf3b4 or HSF3B4 mRNA (1 ng each).
Discussion
Here we report the characterization of the first animal model for Nager type MFD obtained by specifically knocking down Sf3b4 function in Xenopus laevis embryos. We show that in the absence of Sf3b4 function neural crest progenitors formation is affected, and Sf3b4-depleted tadpoles phenocopy aspects of the craniofacial skeletal defects seen in Nager syndrome patients. This animal model has the strong potential to provide important novel insights into the etiology and pathogenesis of Nager type MFD.
SF3B4 encodes spliceosome-associated protein 49 (SAP49), a component of the spliceosomal complex essential for the accurate excision of introns from pre-mRNAs (Will and Luhrmann, 2011). Mutations in SF3B4 have been positively identified as a cause of Nager syndrome in three independent cohorts (Bernier et al., 2012; Czeschik et al., 2013; Petit et al., 2014), accounting for approximately 60% of the cases. It is very intriguing that disruption of a global process, like pre-mRNA splicing, may result in such a restricted and specific phenotype, affecting primarily craniofacial and limbs development. This could be explained by the fact that some embryonic tissues or developmental stage may have a higher demand for spliceosomal activity. This is for example the case for a spliceosome-associated disorder such as retinitis pigmentosa in which the retina is the tissue extremely sensitive to changes in spliceosomal activity (Tanakovic et al., 2011; Linder et al., 2011); mutations in PRPF4 (pre-mRNA processing factor 4) have been recently identified as a major cause for autosomal dominant retinitis pigmentosa (Chen et al., 2014). Therefore, in the context of Nager type MFD, we can speculate that the neural plate border may have a similar high demand in spliceosomal proteins around the time of neural crest progenitors specification. Interestingly, mutations in genes encoding other components of the spliceosome have been linked to craniofacial disorders. They include mutations in EFTUD2 (Lines et al., 2012), SNRPB (Lynch et al., 2014) and TXNL4A (Wieczorek et al, 2014), which cause related but distinct syndromes known as MFD with microcephaly (OMIM #610536), cerebro-costo-mandibular syndrome (OMIM #117650), and Burn-McKeown syndrome (OMIM #608572) respectively, suggesting that spliceosomopathies may underlie the etiology of some forms of MFD (Lehalle et al., 2015).
Zebrafish mutants carrying a mutation in another component of the SF3B complex, Sf3b1, have defects in several neural crest lineages, including cranial neural crest derivatives (An and Henion, 2012). Moreover, this study reported that the pre-mRNA processing of two neural crest transcriptional regulators, snai1b and sox9b, was disrupted in these mutant embryos (An and Henion, 2012), indicating that interference with the function of the SF3B complex can affect the pre-mRNA splicing of neural crest-specific transcripts. Interestingly, snail2 and sox9 pre-mRNA processing was not significantly affected in Sf3b4-depleted Xenopus embryos, suggesting that loss of Sf3b4 may affect a different subset of targets, however the basis for this difference remains unclear. sox10 transcripts were largely undetectable in morphant embryos, raising the possibility that aberrant transcripts may have been targeted for nonsense-mediated mRNA decay (Chang et al., 2007). Alternatively, factors acting upstream of Sox10 may represent the true targets of Sf3b4 splicing activity. Evaluating the global impact of Sf3b4 knockdown on pre-mRNA processing by comparing transcripts from wild type and Sf3b4-depleted embryos using RNA-Seq will provide important information regarding the potential targets of Sf3b4 splicing activity. The mRNA processing of factors involved in neural crest induction, pluripotency, proliferation, and/or survival are potential candidate target genes.
Several years ago, Sf3b4 was isolated in a yeast two-hybrid screen as a factor that interacts with the intracellular domain of BMPR-IA (Nishanian and Waldman, 2004; Watanabe et al., 2007). In tissue culture, overexpression of Sf3b4 was shown to inhibit BMP2-mediated osteogenic and chondrocytic differentiation (Watanabe et al., 2007). While no follow up studies have confirmed or expanded these observations, the possibility remains that SF3B4 may cause Nager syndrome through a mechanism that involves regulation of BMP-mediated osteochondral differentiation, in addition to its role in pre-mRNA processing. In the mouse, BMP pathway loss- or gain-of-function in the neural crest lineage result in an undersized first pharyngeal arch and truncated skeletal elements, indicating that BMP signaling levels are essential in the control of craniofacial development (Bonilla-Claudio et al., 2012). Similarly, BMP signaling is essential for limb development and chondrogenesis (Pignatti et al., 2014). BMP signaling is also an essential regulator of cell fate decisions in the embryonic ectoderm, where high and low levels of BMP signaling promote epidermis and neural plate fate, respectively, while intermediate levels of BMP signaling are required for neural crest formation (Stuhlmiller and Garcia-Castro, 2012; Bae and Saint-Jeannet, 2014). While this non-canonical activity of Sf3b4 in the regulation of BMP signaling has been proposed as a potential disease mechanism in Nager syndrome patients, we were unable to identify a link between Sf3b4 and BMP signaling in our system.
Mouse models of Treacher Collins syndrome indicates that this condition is caused by an early depletion of neural crest progenitors through a mechanism that involves increased apoptosis and reduced proliferation (Dixon et al., 2006; Jones and et al., 2008). In our model of Nager syndrome, the reduction in neural crest genes expression in Sf3b4-depleted embryos is associated with an increase in the rate of apoptosis in the ectoderm, without any reduction in the proliferative capacity of these cells, suggesting that MFD in both conditions share a common mechanism.
MFD is a condition involving defects in structures derived from the first and second pharyngeal arches. The craniofacial defects observed in Sf3b4-depleted embryos were not exclusively restricted to derivatives of the first and second pharyngeal arches, since the branchial cartilages which arise from the third and fourth arches were also affected at a similar frequency, suggesting that in Xenopus interference with Sf3b4 function may affect a broader range of cranial neural crest derivatives. However, a number of Nager syndrome patients also exhibit ventricular septal defects or tetralogy of Fallot (Bernier et al., 2012; Czeschik et al., 2013; Petit et al., 2014), two conditions often associated with abnormal cardiac neural crest development (Hutson and Kirby, 2007; Chin et al., 2012), a segment of neural crest cells that migrate through the third and fourth pharyngeal arches in mammals.
In conclusion, our observations provide an important link between the craniofacial defects seen in Nager syndrome patients and neural crest progenitors formation, and establish Sf3b4-depleted Xenopus embryos as a powerful model to analyze the pathogenesis of Nager syndrome. Future studies will determine the precise mechanisms underlying the root cause of Nager type MFD, and identify the targets of SF3B4.
Supplementary Material
Highlights.
-
-
Xenopus is a powerful model for studying craniofacial development and disease.
-
-
Nager syndrome is a human craniofacial disorder due to mutations in SF3B4.
-
-
Sf3b4-depleted Xenopus embryos have reduced neural crest genes expression.
-
-
Sf3b4-depleted tadpoles phenocopy aspects of the human craniofacial defects.
Acknowledgements
We are grateful to Dr. Thomas Sargent for reagents and to Mona Herold for helping with some injections. This work was supported by a grant from the National Institutes of Health to J-P S-J (R01-DE014212).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
A. D., H. J-G and J-P. S-J. designed the experiments, prepared the figures and wrote the manuscript. A. D., H. J-G., J. A. G., C-S. H. and J-P. S-J. performed the experiments and analyzed the data. All authors have read and approved the final manuscript.
References
- Aylsworth AS, Lin AE, Friedman PA. Nager acrofacial dysostosis: male-to-male transmission in 2 families. Am. J. Med. Genet. 1991;41:83–88. doi: 10.1002/ajmg.1320410121. [DOI] [PubMed] [Google Scholar]
- An M, Henion PD. The zebrafish sf3b1b460 mutant reveals differential requirements for sf3b1 pre-mRNA processing gene during neural crest development. Int. J. Dev. Biol. 2012;56:223–237. doi: 10.1387/ijdb.113383ma. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki Y, Saint-Germain N, Gyda M, Magner-Fink E, Lee YH, Credidio C, Saint-Jeannet J-P. Sox10 regulates the development of neural crest-derived melanocytes in Xenopus. Dev. Biol. 2003;259:19–33. doi: 10.1016/s0012-1606(03)00161-1. [DOI] [PubMed] [Google Scholar]
- Bae C-J, Saint-Jeannet J-P. Induction and Specification of Neural Crest Cells: Extracellular Signals and Transcriptional Switches. In: Trainor P, editor. Neural Crest Cells: Evolution, Development and Disease. Academic Press; 2014. pp. 27–49. [Google Scholar]
- Baltzinger M, Ori M, Pasqualetti M, Nardi I, Rijli FM. Hoxa2 knockdown in Xenopus results in hyoid to mandibular homeosis. Dev. Dyn. 2005;234:858–867. doi: 10.1002/dvdy.20567. [DOI] [PubMed] [Google Scholar]
- Bernier FP, Caluseriu O, Ng S, et al. Haploinsufficiency of SF3B4, a component of the pre-mRNA spliceosomal complex, causes Nager syndrome. Am. J. Hum. Genet. 2012;90:925–933. doi: 10.1016/j.ajhg.2012.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry DL, Rose CS, Remo BF, Brown DD. The expression pattern of thyroid hormone response genes in remodeling tadpole tissues defines distinct growth and resorbtion gene expression programs. Dev. Biol. 1998;203:24–35. doi: 10.1006/dbio.1998.8975. [DOI] [PubMed] [Google Scholar]
- Betancur P, Bronner-Fraser M, Sauka-Spengler T. Assembling neural crest regulatory circuits into a gene regulatory network. Annu. Rev. Cell Dev. Biol. 2010;26:581–603. doi: 10.1146/annurev.cellbio.042308.113245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonilla-Claudio M, Wang J, Bai Y, Klysik E, Selever J, Martin JF. Bmp signaling regulates a dose-dependent transcriptional program to control facial skeletal development. Development. 2012;140:709–719. doi: 10.1242/dev.073197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Champion-Arnaud P, Reed R. The prespliceosome components SAP 49 and SAP 145 interact in a complex implicated in tethering U2 snRNP to the branch site. Genes Dev. 1994;8:1974–1983. doi: 10.1101/gad.8.16.1974. [DOI] [PubMed] [Google Scholar]
- Chang Y-F, Saadi Imam J, Wilkinson MF. The nonsense-mediated decay RNA surveillance Pathway. Ann. Rev. Biochem. 2007;76:51–74. doi: 10.1146/annurev.biochem.76.050106.093909. [DOI] [PubMed] [Google Scholar]
- Chen X, Liu Y, Sheng X, Tam POS, Zhao K, Chen X, Rong W, Liu Y, Liu X, Pan X, Chen LJ, Zhao Q, Vollrath D, Pang CP, Zhao C. PRPF4 mutations cause autosomal dominant retinitis pigmentosa. Hum. Mol. Genet. 2014;23:2926–2939. doi: 10.1093/hmg/ddu005. [DOI] [PubMed] [Google Scholar]
- Chemke J, Mogilner BM, Ben-Itzhak I, et al. Autosomal recessive inheritance of Nager acrofacial dysostosis. J. Med. Genet. 1988;25:230–232. doi: 10.1136/jmg.25.4.230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin AJ, Saint-Jeannet J-P, Lo CW. How insights from cardiovascular developmental biology have impacted the care of infants and children with congenital heart disease. Mech. Dev. 2012;129:75–97. doi: 10.1016/j.mod.2012.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czeschik JC, Voigt C, Alanay Y, Albrecht B, et al. Clinical and mutation data in 12 patients with the clinical diagnosis of Nager syndrome. Hum. Genet. 2013;132:885–898. doi: 10.1007/s00439-013-1295-2. [DOI] [PubMed] [Google Scholar]
- Dauwerse JG, Dixon J, Seland S, et al. Mutations in genes encoding subunits of RNA polymerases I and III cause Treacher Collins syndrome. Nat Genet. 2011;43:20–22. doi: 10.1038/ng.724. [DOI] [PubMed] [Google Scholar]
- Dixon MJ. Treacher Collins syndrome. Hum. Molec. Genet. 1996;5:1391–1396. doi: 10.1093/hmg/5.supplement_1.1391. [DOI] [PubMed] [Google Scholar]
- Dixon J, Jones NC, Sandell LL, Jayasinghe SM, Crane J, Rey J-P, Dixon M, Trainor PA. Tcof1/Treacle is required for neural crest cell formation and proliferation deficiencies that cause craniofacial abnormalities. Proc. Natl. Acad. Sci. USA. 2006;103:13403–13408. doi: 10.1073/pnas.0603730103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall BD. Nager acrofacial dysostosis: Autosomal dominant inheritance in mild to moderately affected mother and lethally affected phocomelic son. Am. J. Med. Genet. 1989;33:94–397. doi: 10.1002/ajmg.1320330321. [DOI] [PubMed] [Google Scholar]
- Harland RM. In situ hybridization: an improved whole-mount method for Xenopus embryos. Meth. Cell Biol. 1991;36:685–695. doi: 10.1016/s0091-679x(08)60307-6. [DOI] [PubMed] [Google Scholar]
- Hensey C, Gautier J. Programmed cell death during Xenopus development: a spatio-temporal analysis. Dev. Biol. 1998;203:36–48. doi: 10.1006/dbio.1998.9028. [DOI] [PubMed] [Google Scholar]
- Hong C-S, Saint-Jeannet J-P. The activity of Pax3 and Zic1 regulates three distinct cell fates at the neural plate border. Mol. Biol. Cell. 2007;18:2192–2202. doi: 10.1091/mbc.E06-11-1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutson MR, Kirby ML. Model systems for the study of heart development and disease. Cardiac neural crest and conotruncal malformations. Semin. Cell Dev. Biol. 2007;18:101–110. doi: 10.1016/j.semcdb.2006.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones NC, Lynn ML, Gaudenz K, Sakai D, Aoto K, Rey JP, Glynn EF, Ellington L, Du C, Dixon J, Dixon MJ, Trainor PA. Prevention of the neurocristopathy Treacher Collins syndrome through inhibition of p53 function. Nat. Med. 2008;14:125–133. doi: 10.1038/nm1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy SJ, Teebi AS. Newly recognized autosomal recessive acrofacial dysostosis syndrome resembling Nager syndrome. Am. J. Med. Genet. A. 2004;129:73–76. doi: 10.1002/ajmg.a.30113. [DOI] [PubMed] [Google Scholar]
- Lee Y-H, Saint-Jeannet J-P. Sox9 function in craniofacial development and disease. Genesis. 2011;49:200–208. doi: 10.1002/dvg.20717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehalle D, Wieczorek D, Zechi-Ceide RM, Passos-Bueno MR, Lyonnet S, Amiel J, Gordon CT. A review of craniofacial disorders caused by spliceosomal defects. Clin. Genet. 2015 doi: 10.1111/cge.12596. DOI:10.1111/cge.12596. [DOI] [PubMed] [Google Scholar]
- Linder B, Dill H, Hirmer A, Brocher J, Lee GP, Mathavan S, Bolz HJ, Winkler C, Laggerbauer B, Fischer U. Systemic splicing factor deficiency causes tissue-specific defects: a zebrafish model for retinitis pigmentosa. Hum. Mol. Genet. 2011;20:368–377. doi: 10.1093/hmg/ddq473. [DOI] [PubMed] [Google Scholar]
- Lines M, Huang L, Schwartzentruber J, et al. Haploinsufficiency of a spliceosomal GTPase encoded by EFTUD2 causes mandibulofacial dysostosis with microcephaly. Am. J. Hum. Genet. 2012;90:369–377. doi: 10.1016/j.ajhg.2011.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo T, Lee Y-H, Saint-Jeannet J-P, Sargent TD. Induction of neural crest in Xenopus by transcription factor AP2alpha. Proc. Natl. Acad. Sci. (USA) 2003;100:532–537. doi: 10.1073/pnas.0237226100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch DC, Revil T, Schwartzentruber J, et al. Disrupted auto-regulation of the spliceosomal complex gene SNRPB causes cerebro-costo-mandibular syndrome. Nat. Commun. 2014;5:4483. doi: 10.1038/ncomms5483. doi: 10.1038/ncomms5483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayor R, Morgan R, Sargent M. Induction of the prospective neural crest of Xenopus. Development. 1995;121:767–777. doi: 10.1242/dev.121.3.767. [DOI] [PubMed] [Google Scholar]
- McDonald MT, Gorski JL. Nager acrofacial dysostosis. J. Med. Genet. 1993;30:779–782. doi: 10.1136/jmg.30.9.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minoux M, Rijli FM. Molecular mechanisms of cranial neural crest cell migration and patterning in craniofacial development. Development. 2010;137:2605–2621. doi: 10.1242/dev.040048. [DOI] [PubMed] [Google Scholar]
- Mizuseki K, Kishi M, Matsui M, Nakanishi S, Sasai Y. Xenopus Zic-related-1 and Sox-2, two factors induced by chordin, have distinct activities in the initiation of neural induction. Development. 1998;125:579–587. doi: 10.1242/dev.125.4.579. [DOI] [PubMed] [Google Scholar]
- Nieuwkoop PD,, Faber J. Normal table of Xenopus laevis (Daudin) North Holland Publishing Company; Amsterdam: 1967. [Google Scholar]
- Nishanian TG, Waldman T. Interaction of the BMPR-IA tumor suppressor with developmentally relevant splicing factor. Bioch. Bioph. Res. Comm. 2004;323:91–97. doi: 10.1016/j.bbrc.2004.08.060. [DOI] [PubMed] [Google Scholar]
- Nur BG, Bernier FP, Oztekin O, Kardele F, Kalay S, Parboosingh JS, Mihci E. Possible autosomal recessive inheritance in an infant with acrofacial dysostosis similar to Nager syndrome. Am. J. Med. Genet. Part A. 2013;161:2311–2315. doi: 10.1002/ajmg.a.36051. [DOI] [PubMed] [Google Scholar]
- O’Donnell M, Hong CS, Huang X, Delnicki RJ, Saint-Jeannet JP. Functional analysis of Sox8 during neural crest development in Xenopus. Development. 2006;133:3817–3826. doi: 10.1242/dev.02558. [DOI] [PubMed] [Google Scholar]
- Passos-Bueno MR, Ornelas CC, Fanganiello RD. Syndromes of the first and second pharyngeal arches: A Review. Am. J. Med. Genet. Part A. 2009;149:1853–1859. doi: 10.1002/ajmg.a.32950. [DOI] [PubMed] [Google Scholar]
- Petit F, Escande F, Jourdain AS, Porchet N, et al. Nager syndrome: confirmation of SF3B4 haploinsufficiency as the major cause. Clin. Genet. 2014;86:246–251. doi: 10.1111/cge.12259. [DOI] [PubMed] [Google Scholar]
- Pignatti E, Zeller R, Zuniga A. To BMP or not to BMP during vertebrate limb bud development. Sem. Cell Dev. 2014;32:119–127. doi: 10.1016/j.semcdb.2014.04.004. [DOI] [PubMed] [Google Scholar]
- Sadaghiani B, Thiebaud CH. Neural crest development in the Xenopus laevis embryo, studied by interspecific transplantation and scanning electron microscopy. Dev. Biol. 1987;124:91–110. doi: 10.1016/0012-1606(87)90463-5. [DOI] [PubMed] [Google Scholar]
- Saka Y, Smith JC. Spatial and temporal patterns of cell division during early Xenopus embryogenesis. Dev Biol. 2001;229:307–318. doi: 10.1006/dbio.2000.0101. [DOI] [PubMed] [Google Scholar]
- Santagati F, Rijli F. Cranial neural crest and the building of the vertebrate head. Nature Reviews Neuroscience. 2003;4:806–818. doi: 10.1038/nrn1221. [DOI] [PubMed] [Google Scholar]
- Schlieve T, Almusa M, Miloro M, Kolokythas A. Temporomandibular joint replacement for ankylosis correction in Nager syndrome: Case report and review of the literature. J. Oral Maxillofac. Surg. 2012;70:616–625. doi: 10.1016/j.joms.2011.02.053. [DOI] [PubMed] [Google Scholar]
- Slack JM, Forman D. An interaction between dorsal and ventral regions of the marginal zone in early amphibian embryos. J. Embryol. Exp. Morphol. 1980;56:283–299. [PubMed] [Google Scholar]
- Spokony RF, Aoki Y, Saint-Germain N, Magner-Fink E, Saint-Jeannet J-P. The transcription factor Sox9 is required for cranial neural crest development in Xenopus. Development. 2002;129:421–432. doi: 10.1242/dev.129.2.421. [DOI] [PubMed] [Google Scholar]
- Stuhlmiller TJ, Garcia-Castro MI. Current perspectives of the signaling pathways directing neural crest induction. Cell Mol. Life Sci. 2012;69:3715–i3737. doi: 10.1007/s00018-012-0991-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki A, Ueno N, Hemmati-Brivanlou A. Xenopus msx1 mediates epidermal induction and neural inhibition by BMP4. Development. 1997;124:3037–3044. doi: 10.1242/dev.124.16.3037. [DOI] [PubMed] [Google Scholar]
- Tanackovic G, Ransijn A, Thibault P, Abou Elela S, Klinck R, Berson EL, Chabot B, Rivolta C. PRPF mutations are associated with generalized defects in spliceosome formation and pre-mRNA splicing in patients with retinitis pigmentosa. Hum. Mol. Genet. 2011;20:2116–2130. doi: 10.1093/hmg/ddr094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trainor PA. Craniofacial birth defects: the role of neural crest cells in the etiology and pathogenesis of Treacher Collins syndrome and the potential for prevention. Am. J. Med. Genet. Part A. 2010;152:2984–2994. doi: 10.1002/ajmg.a.33454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trainor PA, Krumlauf R. Patterning the cranial neural crest: hindbrain segmentation and Hox gene plasticity. Nature Rev. Neurosc. 2000;1:116–124. doi: 10.1038/35039056. [DOI] [PubMed] [Google Scholar]
- Trainor PA, Andrews BT. Facial Dysostoses: etiology, pathogenesis and Management. Am. J. Med. Genet. Part C. 2013;163:283–294. doi: 10.1002/ajmg.c.31375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treacher Collins Syndrome Collaborative Group Positional cloning of a gene involved in the pathogenesis of Treacher Collins syndrome. Nat Genet. 1996;12:130–136. doi: 10.1038/ng0296-130. [DOI] [PubMed] [Google Scholar]
- Waggoner DJ, Ciske DJ, Dowton SB, Watson MS. Deletion of 1q in a patient with acrofacial dysostosis. Am. J. Med. Genet. 1999;82:301–304. doi: 10.1002/(sici)1096-8628(19990212)82:4<301::aid-ajmg5>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- Watanabe H, Shionyu M, Kimura T, Kimata K, Watanabe H. Splicing factor 3b subunit 4 binds BMPR-IA and inhibits osteochondral cell differentiation. J. Biol. Chem. 2007;282:20728–20738. doi: 10.1074/jbc.M703292200. [DOI] [PubMed] [Google Scholar]
- Wieczorek D. Human facial dysostoses. Clin. Genet. 2013;83:499–510. doi: 10.1111/cge.12123. [DOI] [PubMed] [Google Scholar]
- Wieczorek D, Newman WG, Wieland T, et al. Compound heterozygosity of low-frequency promoter deletions and rare loss-of-function mutations in TXNL4A causes Burn-McKeown syndrome. Am. J. Hum. Genet. 2014;95:698–707. doi: 10.1016/j.ajhg.2014.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Will CL, Luhrmann R. Spliceosome structure and function. Cold Spring Harb. Perspect. Biol. 2011;3:a003707. doi: 10.1101/cshperspect.a003707. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.