Abstract
Mobile elements comprise a major fraction of most mammalian genomes. To protect their fitness and stability, hosts must keep mobile elements in check in their germline. In most tissues mobile element insertions are decorated with chromatin modifications suggestive of transcriptional silencing. However, germline cells undergo massive chromatin reprogramming events, which erase repressive chromatin marks and necessitate de novo re-establishment of silencing. How do host genomes achieve the discrimination necessary for this de novo silencing? A series of recent studies have revealed aspects of the multi-pronged strategy that mammalian genomes use to identify and silence mobile elements. These strategies include the use of small RNA-guides, of specialized DNA-binding protein adaptors and of proteins that repair chromatin discontinuities caused by retroelement insertions. Genetic analyses reveal the importance of these mechanisms of protection, each of which specializes in silencing mobile elements of different evolutionary ages. Together, these strategies allow mammalian genomes to withstand the high burden of their parasites.
Retroelement expression and silencing in the germline
Retroelements, selfish genetic elements that mobilize via an RNA intermediate, comprise a large fraction of mammalian genomes. The most successful mammalian retroelements are non-LTR retrotransposons of the LINE-1 family (Long Interspersed Nuclear Element), which occupy ~20% and ~17% of the mouse and human genome respectively. LINE-1 also help propagate non-autonomous SINE (Short Interspersed Nuclear Element) retroposons, which comprise 8% and ~11% of the mouse and human genome respectively. The final types of retroelements commonly found in mammalian genomes are LTR-retrotransposons, or endogenous retroviruses (ERVs), which are largely derived from infectious retroviruses. ERVs constitute ~10% of both mouse and human genomes. Together, these three elements dwarfs the total protein-coding gene compendium (roughly 1.2%). By over-replicating themselves via “copy-paste” retrotransposition cycle, retroelements selfishly increase their copy number within the host genomes. This is essential for them to stave off mutational extinction, but can be deleterious to the host.
The mobilization of active retroelements is inherently mutagenic to host genomes, which use a variety of strategies to prevent mobilization. In this review, we focus on repression of retroelement transcription, the earliest step in their mobilization. In differentiated cells, most retroelement insertions are decorated by chromatin modifications incompatible with transcription initiation, or heterochromatin, such as the methylation of lysines 9 and 27 of histone H3 (H3K9me and H3K27me respectively) and DNA methylation, whereas they are depleted for chromatin marks associated with transcriptional activation such as histone acetylation [1]. Thus proteins that mediate these DNA or histone modifications i.e., DNA methyl-transferases (DNMTs), histone methyl-transferases (HMTs) or histone deacetylases (HDACs), are crucial for the maintenance of retroelement repression.
From an evolutionary perspective, retrotransposition in differentiated somatic cells is not an effective strategy for the retroelement since it might deleteriously affect host fitness without increasing retroelement copy numbers in genomes of the host’s descendants. Instead, evolutionarily successful retroelements ensure their retrotransposition in germline cells, which transmit their genomes to subsequent generations. Intriguingly, mammalian germline cells undergo unique chromatin transitions that could make them vulnerable to retroelement mobilization.
The mammalian germline includes not only germ cells (or reproductive cells) but also cells of the early zygote and the inner cell mass (from which embryonic stem (ES) cells are derived). Together these cells transmit to their descendants all the information necessary to complete embryogenesis. This unique developmental potency is reset at each generation in a process that is referred to as ‘epigenetic reprogramming’. Once during pre-implantation development, and again following the induction of primordial germ cell (PGCs), repressive chromatin marks that typically repress retroelements are erased genome-wide and have to be de novo reestablished (reviewed in [2,3]). This lowering of the guard could transiently allow transcription and thereby proliferation of retroelements.
Indeed, there is abundant evidence for increased retroelement transcription in germline cells. For instance, adult oocytes and early mouse embryos have a high degree of ERV transcription [4–6]. Similarly, retroelement transcript and proteins have been observed in developing mouse germ cells [7,8], a finding nicely corroborated by recent transcriptome analysis of sorted mouse and human primordial germ cells [9,10]. Furthermore, transcriptome analyses revealed that discrete sets of LTR retrotransposon families were sequentially activated during human pre-implantation development indicative of a high degree of adaptation to this environment [11]. This activation is so stereotypical that ERV transcripts expressed during human and mouse embryogenesis can be used as precise markers of toti- and pluri-potent cells both in vitro as well as in vivo [12–15].
Despite these observations, there remains limited evidence about whether epigenetic reprogramming directly causes retroelements activation due to erasure of repressive chromatin marks. With some exceptions [16,17], global retrotransposon reactivation displayed when disrupting HMTs or DNMTs could be the result of broad pleiotropic effects on chromatin patterns. However, recent studies in induced pluripotent stem cells (iPSCs), or embryonic stem cells (ESCs) do suggest that the germline chromatin state is more permissive to retroelements activity than in somatic cells. For instance, upon reprogramming to a more germline-like state, iPSCs dramatically upregulate LINE-1 expression and retrotransposition relative to the parental cell lines they were derived from [18].
Retroelement control: recognizing the enemy
In somatic cells, the propagation and replenishment of repressive chromatin likely maintains retroelement silencing. The findings that retroelements are activated in germline cells might suggest that the germline is a fairly promiscuous environment for retroelement transcription. However, that would be an over-simplification. Closer analyses of published transcriptomes suggest that only a selected number of retroelement subfamilies are actually activated, whereas the vast majority is silent. How is retroelement silencing achieved in germline cells in spite of the wholesale erasure of chromatin marks? How are the repressive chromatin marks established following this erasure?
Ultimately, the challenge in maintaining retroelement control is about discriminating potentially harmful retroelements from host sequences. Any silencing mechanism has also to take into account that retroelements can evolve rapidly to adapt to host transcription factor repertoires and become integral parts of cell-type specific regulatory networks. Indeed, retroelements account for a large portion of bound sites for some transcription factors [19–22]. For example, it was recently shown that the primate specific LTR7 element (belonging to HERVHs ERVs) provide multiple OCT4, NANOG and LBP9 binding-sites that are essential for the maintenance of human pluripotent stem cells in culture [15]. Mammalian genomes therefore have the challenging task of distinguishing and silencing threatening retroelement but not those that have been coopted as host sequences.
Retroelement families, which flourished in our distant ancestors, have been under the control of silencing pathways for more generations and acquired mutations leading them to pose less of an imminent threat to the germline than more recently active families. Pathways silencing older elements will typically involve passive spreading or maintenance of chromatin modifications; they are part of the canonical regulatory toolkit of our cells. In contrast, pathways monitoring recently integrated copies of actively replicating retrotransposon families need to use targeting mechanisms to deposit silencing modifications to the correct locations de novo. In response, retroelements are under severe evolutionary pressure to devise ‘escape strategies’. Thus, not only do host strategies need to be efficient and discriminative, they also need to be evolutionarily flexible to adapt to new threats to the germline. Here, we highlight three different strategies that mammalian germline cells use to achieve this control (Figure 1).
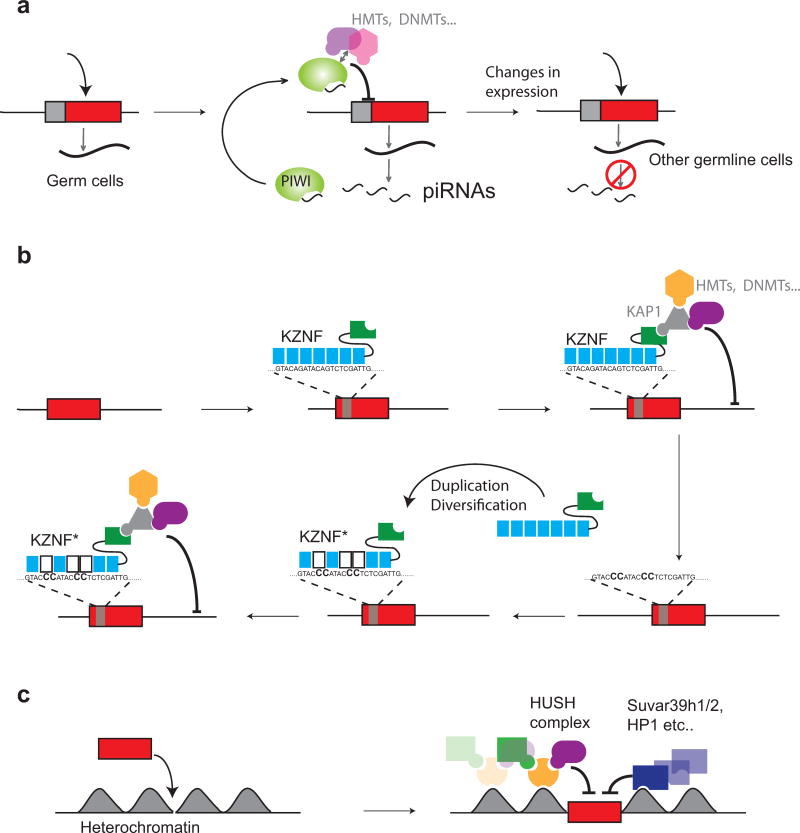
Figure 1. Three layers of retrotransposon control in the mammalian germline.
(a) piRNA-mediated silencing in male germ cells. Retroelement insertions expressed in germ cells (left panel) are processed into piRNAs, which bind to PIWI proteins (middle panel). In turn, these piRNA-PIWI complexes guide the deposition of repressive chromatin marks on individual retroelement insertions. New families of retroelements can escape the piRNA pathway by changing their promoter sequences (white bars) and avoid expression in germ cells (right panel). (b) An evolutionary arms race between host KZNF proteins and retroelements. KZNF proteins bind to individual insertions via their ZNF domain (blue boxes) and trigger silencing by recruiting KAP1, HMTs and DNMTs via their KRAB domain (green box). New retroelements evade KZNFs by changing (or deleting) this binding site. Upon KZNF gene duplication and ZNF diversification a new KZNF (KZNF*) can regain control over the new element. (c) A positioneffect-variegation (PEV) like mechanism for silencing retroelements. Upon the insertion of a retroelement into a heterochromatin domain (left panel), chromatin-binding complexes (e.g., HUSH complex proteins) can read and repair these scars by ‘spreading’ heterochromatin and establish effective silencing over the element (right panel).
Finger on the pulse: the piRNA pathway mediates silencing of the youngest retroelements in germ cells
The discovery of the role of piwi proteins and their associated piRNAs (PIWI interacting RNAs) in Drosophila and in mice revealed a widespread mechanism of germline defense conserved in most animal species. Studies in these two model organisms uncovered the key steps that allow this RNA-based silencing mechanism to detect retroelements, but also some important differences. For instance, whereas flies rely on transposon graveyards (piRNA clusters) to combat young and old transposon invasions in the ovary, mammalian piRNAs adapt to new transposons independently of piRNA cluster formation. In developing mammalian male germ cells, abundantly expressed retroelements are processed into a pool of small RNAs that, once bound to PIWI proteins, guide these complexes to complementary target sequences [23,24]. Mouse PIWI proteins MILI (PIWIL2) and MIWI2 (PIWIL4) begin their expression soon after epigenetic reprogramming; this expression pattern is conserved in humans [10]. Mice knocked out for either MILI or MIWI2 are fully sterile and display retrotransposon derepression [25–27].
The earliest indication that the piRNA pathway contributes to transcriptional silencing came from studies showing that piRNAs, in complex with MIWI2, translocated to the nucleus of PGCs [23]. Furthermore, DNA methylation patterns were compromised in germ cells of mutant animals [23–25,28]. High-throughput sequencing of MILI mutant germ cells revealed that DNA methylation and H3K9 methylation are unaltered over the bulk of retroelement insertions but specifically lost over the promoters of a small number of retroelements, which were members of the youngest and most active subfamilies [29,30]. These data indicate that piRNAs are absolutely required to detect threatening retroelement insertions and target them for de novo heterochromatin formation in PGCs, although there are still unknowns about how information from piRNAs is relayed back to chromatin modification. Aside from its role in male germ cells, PIWI proteins have been reported to affect human L1 regulation in induced pluripotent stem cells [31]. However, its role during the embryonic stages of the germline cycle or in female germ cells remains mysterious [32]. It is likely that other pathways regulate retroelement silencing in these tissues.
By using an RNA-based detection and targeting mechanism, the piRNA pathway is the best strategy to respond to new retroelement insertions and new subfamily invasions with little-to-no a priori ‘education’. The sole expression of the retroelement could suffice to trigger the response. To escape this silencing, retroelements might be under selection to rapidly alter their cis-regulatory elements to ensure they are not expressed in germ cells where they would be under surveillance, but instead expressed in other cells of the germline (Figure 1a). It is conceivable that such ‘escape strategies’ could explain the rapid diversification seen in regulatory regions of L1s (5′UTRs) and ERVs (regulatory LTR regions) during retroelement evolution [33]; such diversification could also occur during ERV evolution although this remains unexplored [34].
Stalking their prey: DNA binding proteins shadow and silence active retroelement lineages
Parallel to the RNA-based silencing of the piRNA pathway is the DNA sequence-based recognition and silencing of retroelements by host KZNF DNA-binding proteins (for Krüppel associated box (KRAB) containing C2H2-zinc-finger (ZNF)). These modular proteins recruit the universal co-repressor KAP1 (KRAB associated protein 1, also called TRIM28 or TIF1β) through their KRAB domain and bind DNA via an array of ZNFs. By virtue of sampling new DNA-binding sequences via ZNF diversification, KZNF target various subfamilies of retroelements. In turn, KAP1 recruits HMTs and DNMTs to retroelement insertions to promote the deposition of repressive marks [35–37].
This paradigm of KZNF-mediated retroelement silencing was elucidated by pioneering work showing how the KZNF protein ZFP809 silenced genomic insertions of the Murine Leukemia Virus (MLV) in mouse ES cells via the recognition of its primer-binding site (PBS) [38]. Recent genome-wide analyses of ZFP809-deficient mouse demonstrated how this silencing extended to other retroelements with a similar PBS [39]. Similar to ZFP809, many KZNFs are expressed during the germline cycle and have the potential to silence many retroelement families ([10,40]) although there are only a handful of examples where the association between a retrotransposon sequence and a unique KZNF has been demonstrated [41–44].
Unlike the more immediate piRNA response, KZNF-based targeting is evolutionarily slower to respond to new retroelement families, since it requires gene duplication and adaptive evolution of DNA-binding preferences. However, once the appropriate specificity is achieved, KZNF proteins can globally repress copies of a given subfamily. If the targeted sequence is essential for retrotransposition, as it is in the ZFP809/MLV interaction, it is more difficult for the retroelement to ‘escape’ detection [38]. KZNF genes can also effectively target intermediate aged retroelements, which are less of a threat to the host. This sets up an evolutionary arms race, in which DNA binding proteins recurrently duplicate and adapt to new retroelement features without losing control over old ones or causing deleterious ‘self’-recognition, whereas retroelements evolve to evade this recognition (Figure 1b). Indeed, the tempo of KZNF gene duplication nicely mirrors the tempo retroelement family diversification [45]. A recent illustration of this arms-race emerged from the study of specific KZNFs from ES cells that target LINE-1s and SVA (SINE-VNTR-Alu) retroelements. In this study, both gain of specific KZNF DNA-binding affinity as well as retroelement escape via target sequence deletion was shown to have occurred in recent primate history [42].
Although KAP1 coordinates the deposition of repressive chromatin marks over many retroelement families in mouse and human ES cells [46–50], it may recruit distinct effectors to different retroelement classes. For instance, silencing of Class I and II ERVs is strictly dependent on the HMT SetDB1 (or ESET) whereas ERVL and LINE-1 retroelements appear to be specifically regulated by the G9a and Suv39h1/2 HMTs respectively [51–54]. Although SETDB1 silencing has been proposed to be dependent on deposition of the histone variant H3.3 [55], the role of this variant histone in ERV silencing still remains to be fully clarified [56]. These studies further suggest that there may be additional regulators beyond KZNF proteins that specify retroelement targeting by KAP1, for example, the transcription factor YY1 required for LTR retroelement silencing in ES cells [57].
Neighborhood watch: looking for signs of recent vandalism
Both the piRNA and KZNF-based defense rely on recognizing retroelement sequence features. However, retroelement insertions also have the effect of disrupting the contiguous chromatin context into which they insert. A recent study demonstrated that this discontinuity can be recognized as a chromatin ‘scar’ and recruits a novel chromatin-associated HUSH (human silencing hub) complex that spreads repressive marks onto the newly inserted retroelement (Figure 1c) [58]. In this study, a genetic screen was carried out to find genes required for the silencing of an LTR-based reporter vector introduced in human haploid cell lines. This approach identified a complex composed of the chromodomain protein MPP8 (M-Phase Phosphoprotein 8), which interacts with G9a and DNMT3a [59], the epigenetic modifier Fam208a/TASOR (Transgene Activation SuppressOR) [60] and Periphilin 1 (PPHLN1) [61]. Through its interaction with heterochromatin, HUSH directly recruits Setdb1 to mediate the deposition of H3K9me over both new insertions of the retroelement reporter in heterochromatin, as well as over hundreds of additional genomic regions [58]. Thus, HUSH simultaneously appears to perform the dual role of repairing heterochromatin scars and spreading heterochromatin over a subset of retroelements. Interestingly, Fam208a/TASOR knockout mice are embryonic lethal and MPP8 is essential for spermatogenesis suggesting that the HUSH complex could perform its function over the entire germline cycle [60,62].
The principle of ‘chromatin scars’ as a means to identify new genomic insertions is not exclusive to the HUSH complex and might be shared among other proteins identified in recent genetic screens for establishment or maintenance of retroelement silencing [63–65]. Among these, the DAXX protein, which is a histone H3.3 chaperone, is particularly intriguing because it appears to directly engage with the integrase protein of the incoming retrovirus to mediate epigenetic silencing of the resulting provirus [66]. Additionally, the HMTs Suv39h1/2 and the heterochromatin-associated proteins of the HP1 family can interact with pre-existing heterochromatin domains and consolidate retrotransposon silencing in ES cells and germ cells [51,54,67,68]. Going even beyond heterochromatin, there might be other diagnostic ‘scars’ or chromatin discontinuities introduced by retroelement insertions, such as DNA repair-associated signatures [69] which may be utilized by host genomes to deposit specific histone or DNA modifications over subsets of retroelement insertions. For example, retroelement-directed arginine methylation of histone tails by PRMT5 [70] or retroelements DNA methylation regulated by the helicase LSH ([71]) still lack clear described targeting mechanisms in the germline.
In terms of the host-retroelement conflict, repairing chromatin ‘scars’ provides only an insertion-specific defense, but this defense is not dependent on target specificity like piRNAs or KZNFs. It is therefore not clear how retroelements could evade such silencing except to avoid inserting in certain chromatin neighborhoods.
Summary
We have discussed a few of the molecular pathways used to recognize and silence retroelement insertions in the mammalian germline. On the surface, they appear as redundant pathways, often mediating the deposition of similar chromatin modifications or targeting similar retrotransposon families. However, if one looks deeper, they each act on different evolutionary strata of retrotransposon insertions. RNA-guided pathways are tuned to the youngest evolutionary stratum of retroelements; the expression of these young, active retroelements in germ cells is also their vulnerability. In contrast, DNA-binding proteins evolve rapidly via both gene duplication and adaptive evolution to act as adaptors to target the silencing of the intermediate stratum. Finally, complexes repairing ‘chromatin scars’ also help silence new retroelement insertions independent of sequence determinants. A sobering thought is that although each of these molecular pathways has operated for hundreds of millions of years, our genomes are still vulnerable to retroelement activity, as demonstrated by the high genomic burden of retroelement insertions into mammalian genomes.
Acknowledgments
We thank Ines Anna Drinnenberg, Janet Young and Sarah Zanders for helpful comments. We are supported by a postdoctoral fellowship from the Damon Runyon Cancer Research Foundation (DRG 2192-14 to AM) and by a grant from the Mathers Foundation and NIH grant R01-GM074108 (to HSM). HSM is an Investigator of the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Bibliography
- 1.Consortium EP. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Reik W, Dean W, Walter J. Epigenetic reprogramming in mammalian development. Science. 2001;293:1089–1093. doi: 10.1126/science.1063443. [DOI] [PubMed] [Google Scholar]
- 3.Surani MA, Hayashi K, Hajkova P. Genetic and epigenetic regulators of pluripotency. Cell. 2007;128:747–762. doi: 10.1016/j.cell.2007.02.010. [DOI] [PubMed] [Google Scholar]
- 4.Brulet P, Kaghad M, Xu YS, Croissant O, Jacob F. Early differential tissue expression of transposon-like repetitive DNA sequences of the mouse. Proc Natl Acad Sci U S A. 1983;80:5641–5645. doi: 10.1073/pnas.80.18.5641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kigami D, Minami N, Takayama H, Imai H. MuERV-L is one of the earliest transcribed genes in mouse one-cell embryos. Biol Reprod. 2003;68:651–654. doi: 10.1095/biolreprod.102.007906. [DOI] [PubMed] [Google Scholar]
- 6.Peaston AE, Evsikov AV, Graber JH, de Vries WN, Holbrook AE, Solter D, Knowles BB. Retrotransposons regulate host genes in mouse oocytes and preimplantation embryos. Dev Cell. 2004;7:597–606. doi: 10.1016/j.devcel.2004.09.004. [DOI] [PubMed] [Google Scholar]
- 7.Branciforte D, Martin SL. Developmental and cell type specificity of LINE-1 expression in mouse testis: implications for transposition. Mol Cell Biol. 1994;14:2584–2592. doi: 10.1128/mcb.14.4.2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dupressoir A, Heidmann T. Germ line-specific expression of intracisternal A-particle retrotransposons in transgenic mice. Mol Cell Biol. 1996;16:4495–4503. doi: 10.1128/mcb.16.8.4495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Seisenberger S, Andrews S, Krueger F, Arand J, Walter J, Santos F, Popp C, Thienpont B, Dean W, Reik W. The dynamics of genome-wide DNA methylation reprogramming in mouse primordial germ cells. Mol Cell. 2012;48:849–862. doi: 10.1016/j.molcel.2012.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10*.Tang WW, Dietmann S, Irie N, Leitch HG, Floros VI, Bradshaw CR, Hackett JA, Chinnery PF, Surani MA. A Unique Gene Regulatory Network Resets the Human Germline Epigenome for Development. Cell. 2015;161:1453–1467. doi: 10.1016/j.cell.2015.04.053. In-depth methylome and transcriptome analysis during epigenetic reprogramming of human primordial germ cells (PGCs) as well as extensive comparison of KZNF expression between human PGCs and embryonic stem cells leads to detailed analysis of retrotransposon methylation and expression in human PGCs. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goke J, Lu X, Chan YS, Ng HH, Ly LH, Sachs F, Szczerbinska I. Dynamic transcription of distinct classes of endogenous retroviral elements marks specific populations of early human embryonic cells. Cell Stem Cell. 2015;16:135–141. doi: 10.1016/j.stem.2015.01.005. [DOI] [PubMed] [Google Scholar]
- 12.Kelley D, Rinn J. Transposable elements reveal a stem cell-specific class of long noncoding RNAs. Genome Biol. 2012;13:R107. doi: 10.1186/gb-2012-13-11-r107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lu X, Sachs F, Ramsay L, Jacques PE, Goke J, Bourque G, Ng HH. The retrovirus HERVH is a long noncoding RNA required for human embryonic stem cell identity. Nat Struct Mol Biol. 2014;21:423–425. doi: 10.1038/nsmb.2799. [DOI] [PubMed] [Google Scholar]
- 14.Macfarlan TS, Gifford WD, Driscoll S, Lettieri K, Rowe HM, Bonanomi D, Firth A, Singer O, Trono D, Pfaff SL. Embryonic stem cell potency fluctuates with endogenous retrovirus activity. Nature. 2012;487:57–63. doi: 10.1038/nature11244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang J, Xie G, Singh M, Ghanbarian AT, Rasko T, Szvetnik A, Cai H, Besser D, Prigione A, Fuchs NV, et al. Primate-specific endogenous retrovirus-driven transcription defines naive-like stem cells. Nature. 2014;516:405–409. doi: 10.1038/nature13804. [DOI] [PubMed] [Google Scholar]
- 16.Malki S, van der Heijden GW, O’Donnell KA, Martin SL, Bortvin A. A role for retrotransposon LINE-1 in fetal oocyte attrition in mice. Dev Cell. 2014;29:521–533. doi: 10.1016/j.devcel.2014.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17*.Zamudio N, Barau J, Teissandier A, Walter M, Borsos M, Servant N, Bourc’his D. DNA methylation restrains transposons from adopting a chromatin signature permissive for meiotic recombination. Genes Dev. 2015;29:1256–1270. doi: 10.1101/gad.257840.114. Demonstration that DNA methylation over retrotransposons prevents the formation of SPO11-dependent crossovers during male meiosis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Friedli M, Turelli P, Kapopoulou A, Rauwel B, Castro-Diaz N, Rowe HM, Ecco G, Unzu C, Planet E, Lombardo A, et al. Loss of transcriptional control over endogenous retroelements during reprogramming to pluripotency. Genome Res. 2014;24:1251–1259. doi: 10.1101/gr.172809.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bourque G, Leong B, Vega VB, Chen X, Lee YL, Srinivasan KG, Chew JL, Ruan Y, Wei CL, Ng HH, et al. Evolution of the mammalian transcription factor binding repertoire via transposable elements. Genome Res. 2008;18:1752–1762. doi: 10.1101/gr.080663.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kunarso G, Chia NY, Jeyakani J, Hwang C, Lu X, Chan YS, Ng HH, Bourque G. Transposable elements have rewired the core regulatory network of human embryonic stem cells. Nat Genet. 2010;42:631–634. doi: 10.1038/ng.600. [DOI] [PubMed] [Google Scholar]
- 21.Schmidt D, Schwalie PC, Wilson MD, Ballester B, Goncalves A, Kutter C, Brown GD, Marshall A, Flicek P, Odom DT. Waves of retrotransposon expansion remodel genome organization and CTCF binding in multiple mammalian lineages. Cell. 2012;148:335–348. doi: 10.1016/j.cell.2011.11.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sundaram V, Cheng Y, Ma Z, Li D, Xing X, Edge P, Snyder MP, Wang T. Widespread contribution of transposable elements to the innovation of gene regulatory networks. Genome Res. 2014;24:1963–1976. doi: 10.1101/gr.168872.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Aravin AA, Sachidanandam R, Bourc’his D, Schaefer C, Pezic D, Toth KF, Bestor T, Hannon GJ. A piRNA pathway primed by individual transposons is linked to de novo DNA methylation in mice. Mol Cell. 2008;31:785–799. doi: 10.1016/j.molcel.2008.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kuramochi-Miyagawa S, Watanabe T, Gotoh K, Totoki Y, Toyoda A, Ikawa M, Asada N, Kojima K, Yamaguchi Y, Ijiri TW, et al. DNA methylation of retrotransposon genes is regulated by Piwi family members MILI and MIWI2 in murine fetal testes. Genes Dev. 2008;22:908–917. doi: 10.1101/gad.1640708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Carmell MA, Girard A, van de Kant HJ, Bourc’his D, Bestor TH, de Rooij DG, Hannon GJ. MIWI2 is essential for spermatogenesis and repression of transposons in the mouse male germline. Dev Cell. 2007;12:503–514. doi: 10.1016/j.devcel.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 26.Kuramochi-Miyagawa S, Kimura T, Ijiri TW, Isobe T, Asada N, Fujita Y, Ikawa M, Iwai N, Okabe M, Deng W, et al. Mili, a mammalian member of piwi family gene, is essential for spermatogenesis. Development. 2004;131:839–849. doi: 10.1242/dev.00973. [DOI] [PubMed] [Google Scholar]
- 27.Kuramochi-Miyagawa S, Kimura T, Yomogida K, Kuroiwa A, Tadokoro Y, Fujita Y, Sato M, Matsuda Y, Nakano T. Two mouse piwi-related genes: miwi and mili. Mech Dev. 2001;108:121–133. doi: 10.1016/s0925-4773(01)00499-3. [DOI] [PubMed] [Google Scholar]
- 28.Aravin AA, Sachidanandam R, Girard A, Fejes-Toth K, Hannon GJ. Developmentally regulated piRNA clusters implicate MILI in transposon control. Science. 2007;316:744–747. doi: 10.1126/science.1142612. [DOI] [PubMed] [Google Scholar]
- 29*.Molaro A, Falciatori I, Hodges E, Aravin AA, Marran K, Rafii S, McCombie WR, Smith AD, Hannon GJ. Two waves of de novo methylation during mouse germ cell development. Genes Dev. 2014;28:1544–1549. doi: 10.1101/gad.244350.114. Whole genome bisulfite sequencing comparison of MILI KO vs WT germ cells demonstrates that de novo DNA methylation of retroelements is mostly piRNA-independent, except for the most recent insertions of the youngest retroelement subfamilies. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30*.Pezic D, Manakov SA, Sachidanandam R, Aravin AA. piRNA pathway targets active LINE1 elements to establish the repressive H3K9me3 mark in germ cells. Genes Dev. 2014;28:1410–1428. doi: 10.1101/gad.240895.114. Chromatin-immunoprecipitation (ChIP)-seq analyses of H3K9me3 in MIWI2 mutant germ cells find that piRNAs direct the deposition of this mark over a small set of potentially active LINE-1 elements. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31**.Marchetto MC, Narvaiza I, Denli AM, Benner C, Lazzarini TA, Nathanson JL, Paquola AC, Desai KN, Herai RH, Weitzman MD, et al. Differential L1 regulation in pluripotent stem cells of humans and apes. Nature. 2013;503:525–529. doi: 10.1038/nature12686. Gene expression and retrotransposon activity analysis of iPS cells derived from human, chimpanzee and bonobo suggest that PIWI proteins play a role in L1 silencing in human cells. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Roovers EF, Rosenkranz D, Mahdipour M, Han CT, He N, Chuva de Sousa Lopes SM, van der Westerlaken LA, Zischler H, Butter F, Roelen BA, et al. Piwi proteins and piRNAs in mammalian oocytes and early embryos. Cell Rep. 2015;10:2069–2082. doi: 10.1016/j.celrep.2015.02.062. [DOI] [PubMed] [Google Scholar]
- 33.Khan H, Smit A, Boissinot S. Molecular evolution and tempo of amplification of human LINE-1 retrotransposons since the origin of primates. Genome Res. 2006;16:78–87. doi: 10.1101/gr.4001406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mager DL, Stoye JP. Mammalian Endogenous Retroviruses. Microbiol Spectr. 2015;3 doi: 10.1128/microbiolspec.MDNA3-0009-2014. MDNA3-0009-2014. [DOI] [PubMed] [Google Scholar]
- 35.Ivanov AV, Peng H, Yurchenko V, Yap KL, Negorev DG, Schultz DC, Psulkowski E, Fredericks WJ, White DE, Maul GG, et al. PHD domain-mediated E3 ligase activity directs intramolecular sumoylation of an adjacent bromodomain required for gene silencing. Mol Cell. 2007;28:823–837. doi: 10.1016/j.molcel.2007.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Quenneville S, Verde G, Corsinotti A, Kapopoulou A, Jakobsson J, Offner S, Baglivo I, Pedone PV, Grimaldi G, Riccio A, et al. In embryonic stem cells, ZFP57/KAP1 recognize a methylated hexanucleotide to affect chromatin and DNA methylation of imprinting control regions. Mol Cell. 2011;44:361–372. doi: 10.1016/j.molcel.2011.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schultz DC, Ayyanathan K, Negorev D, Maul GG, Rauscher FJ., 3rd SETDB1: a novel KAP-1-associated histone H3, lysine 9-specific methyltransferase that contributes to HP1-mediated silencing of euchromatic genes by KRAB zinc-finger proteins. Genes Dev. 2002;16:919–932. doi: 10.1101/gad.973302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wolf D, Goff SP. Embryonic stem cells use ZFP809 to silence retroviral DNAs. Nature. 2009;458:1201–1204. doi: 10.1038/nature07844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wolf G, Yang P, Fuchtbauer AC, Fuchtbauer EM, Silva AM, Park C, Wu W, Nielsen AL, Pedersen FS, Macfarlan TS. The KRAB zinc finger protein ZFP809 is required to initiate epigenetic silencing of endogenous retroviruses. Genes Dev. 2015;29:538–554. doi: 10.1101/gad.252767.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Corsinotti A, Kapopoulou A, Gubelmann C, Imbeault M, Santoni de Sio FR, Rowe HM, Mouscaz Y, Deplancke B, Trono D. Global and stage specific patterns of Kruppel-associated-box zinc finger protein gene expression in murine early embryonic cells. PLoS One. 2013;8:e56721. doi: 10.1371/journal.pone.0056721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Guallar D, Perez-Palacios R, Climent M, Martinez-Abadia I, Larraga A, Fernandez-Juan M, Vallejo C, Muniesa P, Schoorlemmer J. Expression of endogenous retroviruses is negatively regulated by the pluripotency marker Rex1/Zfp42. Nucleic Acids Res. 2012;40:8993–9007. doi: 10.1093/nar/gks686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42**.Jacobs FM, Greenberg D, Nguyen N, Haeussler M, Ewing AD, Katzman S, Paten B, Salama SR, Haussler D. An evolutionary arms race between KRAB zinc-finger genes ZNF91/93 and SVA/L1 retrotransposons. Nature. 2014;516:242–245. doi: 10.1038/nature13760. Characterization of human KZNFs regulating L1 and SVA SINE elements. Elegant demonstration of the ‘evolutionary arms-race’ hypothesis that DNA binding specificity of these KZNFs was shaped by retrotransposon diversification in primates. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Najafabadi HS, Mnaimneh S, Schmitges FW, Garton M, Lam KN, Yang A, Albu M, Weirauch MT, Radovani E, Kim PM, et al. C2H2 zinc finger proteins greatly expand the human regulatory lexicon. Nat Biotechnol. 2015;33:555–562. doi: 10.1038/nbt.3128. [DOI] [PubMed] [Google Scholar]
- 44.Tan X, Xu X, Elkenani M, Smorag L, Zechner U, Nolte J, Engel W, Pantakani DV. Zfp819, a novel KRAB-zinc finger protein, interacts with KAP1 and functions in genomic integrity maintenance of mouse embryonic stem cells. Stem Cell Res. 2013;11:1045–1059. doi: 10.1016/j.scr.2013.07.006. [DOI] [PubMed] [Google Scholar]
- 45.Thomas JH, Schneider S. Coevolution of retroelements and tandem zinc finger genes. Genome Res. 2011;21:1800–1812. doi: 10.1101/gr.121749.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Castro-Diaz N, Ecco G, Coluccio A, Kapopoulou A, Yazdanpanah B, Friedli M, Duc J, Jang SM, Turelli P, Trono D. Evolutionally dynamic L1 regulation in embryonic stem cells. Genes Dev. 2014;28:1397–1409. doi: 10.1101/gad.241661.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Matsui T, Leung D, Miyashita H, Maksakova IA, Miyachi H, Kimura H, Tachibana M, Lorincz MC, Shinkai Y. Proviral silencing in embryonic stem cells requires the histone methyltransferase ESET. Nature. 2010;464:927–931. doi: 10.1038/nature08858. [DOI] [PubMed] [Google Scholar]
- 48.Rowe HM, Friedli M, Offner S, Verp S, Mesnard D, Marquis J, Aktas T, Trono D. De novo DNA methylation of endogenous retroviruses is shaped by KRAB-ZFPs/KAP1 and ESET. Development. 2013;140:519–529. doi: 10.1242/dev.087585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rowe HM, Jakobsson J, Mesnard D, Rougemont J, Reynard S, Aktas T, Maillard PV, Layard-Liesching H, Verp S, Marquis J, et al. KAP1 controls endogenous retroviruses in embryonic stem cells. Nature. 2010;463:237–240. doi: 10.1038/nature08674. [DOI] [PubMed] [Google Scholar]
- 50.Turelli P, Castro-Diaz N, Marzetta F, Kapopoulou A, Raclot C, Duc J, Tieng V, Quenneville S, Trono D. Interplay of TRIM28 and DNA methylation in controlling human endogenous retroelements. Genome Res. 2014;24:1260–1270. doi: 10.1101/gr.172833.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bulut-Karslioglu A, De La Rosa-Velazquez IA, Ramirez F, Barenboim M, Onishi-Seebacher M, Arand J, Galan C, Winter GE, Engist B, Gerle B, et al. Suv39h-dependent H3K9me3 marks intact retrotransposons and silences LINE elements in mouse embryonic stem cells. Mol Cell. 2014;55:277–290. doi: 10.1016/j.molcel.2014.05.029. [DOI] [PubMed] [Google Scholar]
- 52.Di Giacomo M, Comazzetto S, Sampath SC, Sampath SC, O’Carroll D. G9a co-suppresses LINE1 elements in spermatogonia. Epigenetics Chromatin. 2014;7:24. doi: 10.1186/1756-8935-7-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu S, Brind’Amour J, Karimi MM, Shirane K, Bogutz A, Lefebvre L, Sasaki H, Shinkai Y, Lorincz MC. Setdb1 is required for germline development and silencing of H3K9me3-marked endogenous retroviruses in primordial germ cells. Genes Dev. 2014;28:2041–2055. doi: 10.1101/gad.244848.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54*.Maksakova IA, Thompson PJ, Goyal P, Jones SJ, Singh PB, Karimi MM, Lorincz MC. Distinct roles of KAP1, HP1 and G9a/GLP in silencing of the two-cell-specific retrotransposon MERVL in mouse ES cells. Epigenetics Chromatin. 2013;6:15. doi: 10.1186/1756-8935-6-15. RNA-seq and H3K9me3 ChIP analyses of various chromatin regulators in mouse ES cells show non-overlapping roles in ERV silencing. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Elsasser SJ, Noh KM, Diaz N, Allis CD, Banaszynski LA. Histone H3.3 is required for endogenous retroviral element silencing in embryonic stem cells. Nature. 2015;522:240–244. doi: 10.1038/nature14345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jang CW, Shibata Y, Starmer J, Yee D, Magnuson T. Histone H3.3 maintains genome integrity during mammalian development. Genes Dev. 2015;29:1377–1392. doi: 10.1101/gad.264150.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schlesinger S, Lee AH, Wang GZ, Green L, Goff SP. Proviral silencing in embryonic cells is regulated by Yin Yang 1. Cell Rep. 2013;4:50–58. doi: 10.1016/j.celrep.2013.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58**.Tchasovnikarova IA, Timms RT, Matheson NJ, Wals K, Antrobus R, Gottgens B, Dougan G, Dawson MA, Lehner PJ. GENE SILENCING. Epigenetic silencing by the HUSH complex mediates position-effect variegation in human cells. Science. 2015;348:1481–1485. doi: 10.1126/science.aaa7227. Forward genetic screen in haploid cell lines characterizing genes involved in maintaining silencing of retroelement insertions in heterochromatin reveals a novel role for the HUSH complex in silencing retroelements via SetDB1 recruitment. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chang Y, Sun L, Kokura K, Horton JR, Fukuda M, Espejo A, Izumi V, Koomen JM, Bedford MT, Zhang X, et al. MPP8 mediates the interactions between DNA methyltransferase Dnmt3a and H3K9 methyltransferase GLP/G9a. Nat Commun. 2011;2:533. doi: 10.1038/ncomms1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Harten SK, Bruxner TJ, Bharti V, Blewitt M, Nguyen TM, Whitelaw E, Epp T. The first mouse mutants of D14Abb1e (Fam208a) show that it is critical for early development. Mamm Genome. 2014;25:293–303. doi: 10.1007/s00335-014-9516-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Huh JW, Kim TH, Yi JM, Park ES, Kim WY, Sin HS, Kim DS, Min DS, Kim SS, Kim CB, et al. Molecular evolution of the periphilin gene in relation to human endogenous retrovirus m element. J Mol Evol. 2006;62:730–737. doi: 10.1007/s00239-005-0109-0. [DOI] [PubMed] [Google Scholar]
- 62.Murata K, Sato S, Haruta M, Goshima T, Chiba Y, Takahashi S, Sharif J, Koseki H, Nakanishi M, Shimada M. Physical interaction between MPP8 and PRC1 complex and its implication for regulation of spermatogenesis. Biochem Biophys Res Commun. 2015;458:470–475. doi: 10.1016/j.bbrc.2015.01.122. [DOI] [PubMed] [Google Scholar]
- 63.Poleshko A, Einarson MB, Shalginskikh N, Zhang R, Adams PD, Skalka AM, Katz RA. Identification of a functional network of human epigenetic silencing factors. J Biol Chem. 2010;285:422–433. doi: 10.1074/jbc.M109.064667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Poleshko A, Palagin I, Zhang R, Boimel P, Castagna C, Adams PD, Skalka AM, Katz RA. Identification of cellular proteins that maintain retroviral epigenetic silencing: evidence for an antiviral response. J Virol. 2008;82:2313–2323. doi: 10.1128/JVI.01882-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yang BX, El Farran CA, Guo HC, Yu T, Fang HT, Wang HF, Schlesinger S, Seah YF, Goh GY, Neo SP, et al. Systematic Identification of Factors for Provirus Silencing in Embryonic Stem Cells. Cell. 2015;163:230–245. doi: 10.1016/j.cell.2015.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Greger JG, Katz RA, Ishov AM, Maul GG, Skalka AM. The cellular protein daxx interacts with avian sarcoma virus integrase and viral DNA to repress viral transcription. J Virol. 2005;79:4610–4618. doi: 10.1128/JVI.79.8.4610-4618.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Brown JP, Bullwinkel J, Baron-Luhr B, Billur M, Schneider P, Winking H, Singh PB. HP1gamma function is required for male germ cell survival and spermatogenesis. Epigenetics Chromatin. 2010;3:9. doi: 10.1186/1756-8935-3-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sripathy SP, Stevens J, Schultz DC. The KAP1 corepressor functions to coordinate the assembly of de novo HP1-demarcated microenvironments of heterochromatin required for KRAB zinc finger protein-mediated transcriptional repression. Mol Cell Biol. 2006;26:8623–8638. doi: 10.1128/MCB.00487-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Shalginskikh N, Poleshko A, Skalka AM, Katz RA. Retroviral DNA methylation and epigenetic repression are mediated by the antiviral host protein Daxx. J Virol. 2013;87:2137–2150. doi: 10.1128/JVI.02026-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70*.Kim S, Gunesdogan U, Zylicz JJ, Hackett JA, Cougot D, Bao S, Lee C, Dietmann S, Allen GE, Sengupta R, et al. PRMT5 protects genomic integrity during global DNA demethylation in primordial germ cells and preimplantation embryos. Mol Cell. 2014;56:564–579. doi: 10.1016/j.molcel.2014.10.003. Analysis of PRMT5 mutant mice indicates that H2A/H4R3me2 is essential for LINE-1 silencing during epigenetic reprogramming in the germline. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yu W, McIntosh C, Lister R, Zhu I, Han Y, Ren J, Landsman D, Lee E, Briones V, Terashima M, et al. Genome-wide DNA methylation patterns in LSH mutant reveals derepression of repeat elements and redundant epigenetic silencing pathways. Genome Res. 2014;24:1613–1623. doi: 10.1101/gr.172015.114. [DOI] [PMC free article] [PubMed] [Google Scholar]