Abstract
With growing environmental pressures placed on our marine habitats there is concern that the prevalence and severity of diseases affecting marine organisms will increase. Yet relative to terrestrial systems, we know little about the underlying causes of many of these diseases. Moreover, factors such as saprophytic colonizers and a lack of baseline data on healthy individuals make it difficult to accurately assess the role of specific microbial pathogens in disease states. Emerging evidence in the field of medicine suggests that a growing number of human diseases result from a microbiome imbalance (or dysbiosis), questioning the traditional view of a singular pathogenic agent. Here we discuss the possibility that many diseases seen in marine systems are, similarly, the result of microbial dysbiosis and the rise of opportunistic or polymicrobial infections. Thus, understanding and managing disease in the future will require us to also rethink definitions of disease and pathogenesis for marine systems. We suggest that a targeted, multidisciplinary approach that addresses the questions of microbial symbiosis in both healthy and diseased states, and at that the level of the holobiont, will be key to progress in this area.
Keywords: dysbiosis, marine diseases, opportunistic pathogens, microbial interactions, microbiome, holobiont
Introduction
Almost two decades on, the wealth of microbial biodiversity uncovered by the molecular ecology revolution continues to amaze. Microbiome studies are being reported at a rapid rate, and include ecosystems ranging from the deep oceans to the upper troposphere (DeLeon-Rodriguez et al., 2013; Inagaki et al., 2015). It is now widely accepted that microorganisms have important functions (albeit often not well understood), not only for our personal health, but also for the survival of our planet (Alivisatos et al., 2015).
In the marine environment, microorganisms constitute over 90% of the living biomass where they are essential for nutrient cycling and form intimate associations (symbiosis) with all other marine organisms. Microbial symbioses in marine environments can take many forms, and today one might argue that the excellent textbook cases of a single-symbiont to a single-host organism [e.g., Euprymna scolopes and Vibrio fischeri (McFall-Ngai, 2014)] are rare. Rather, most interactions between microorganisms and their host/s involve multiple partners that work in multifaceted ways to impact host development and physiology (Wahl et al., 2012), such as in the case for sponges (Webster and Thomas, 2016), corals (Rädecker et al., 2015), and algae (Egan et al., 2013; Abby et al., 2014; Amin et al., 2015).
However microbial–host interactions are not always beneficial and marine organisms are reported to suffer from a variety of disease symptoms, often as a result of a yet unknown etiology (Munn, 2006; Bourne et al., 2009; Gachon et al., 2010; Morado, 2011; Egan et al., 2014). Disease events in the marine environment not only impact directly on the host population, but can also result in ecosystem-wide impacts due to, for example, the mass mortality of keystone species (Burge et al., 2013). These events are predicted to increase with global climate change and elevating anthropogenic pressures (Gattuso et al., 2015). Hence, there is an urgent need to generate data that speak to both the causes and the environmental factors mitigating disease in marine systems. As a guide to understand the complexity of marine diseases, marine scientists may need to look to advances in the human microbiome field. In the past decade, research into human disease has suggested that many chronic diseases (including skin, bowel, and lung disorders) are driven by a disturbance (or shift) in the natural microbiome [i.e., dysbiosis (see Table 1 for definition)] rather than a singular etiological agent (Althani et al., 2015). Here, we propose that many diseases in marine systems can also be viewed from the point of microbial dysbiosis, analogous to some chronic diseases in humans. However, while a conceptual framework can be gained from studies of dysbiosis in the medical field, the nature of marine systems and the inherent differences between humans and marine organisms may necessitate a specific set of approaches and working models.
Table 1.
Definition of terminology commonly used to describe microbial–host interactions in the context of host health and disease.
| Term | Definition | Further reading |
|---|---|---|
| Commensal | A host-associated organism that does not induce host damage upon colonization | Casadevall and Pirofski, 2000 |
| Disease | The health outcome of an organism where normal function is impaired after damage (often induced by a microbe(s)) has occurred. | Casadevall and Pirofski, 2003 |
| Dysbiosis | A microbial community shift that has a negative impact on the host. | Petersen and Round, 2014 |
| Holobiont | A host organism and the entirety of its microbial community, under normal conditions, and in the absence of disease. | Rosenberg et al., 2007 |
| Koch’s postulates | A microorganism-centric methodology used to demonstrate a causal relationship between a pathogen and a disease. The postulates commonly cited are: the pathogen must be present in each case of the disease and absent from healthy individuals; and when isolated in pure culture and used to experimentally infect an individual, the pathogen must induce the disease. | Kaufmann and Schaible, 2005 |
| Opportunistic pathogen | An organism that is capable of causing damage to a host under specific conditions, but may also exist as a commensal on the same host. | Casadevall and Pirofski, 2000 |
| Polymicrobial infection | Disease as a result of the co-infection by multiple microorganisms | Hajishengallis and Lamont, 2016 |
| Saprophyte | An organism that lives and proliferates on dead or already diseased hosts. Often a secondary invader or opportunist. | Willey et al., 2008 |
| Virulence | A relative measure of a microorganism’s ability to induce disease on a host. | Méthot and Alizon, 2014 |
| ‘Dual role’ virulence factor | A microbial trait (molecule, protein, etc.) that has direct/indirect roles in both environmental survival/persistence and host disease progression. | Vezzulli et al., 2008 |
Are Emerging Diseases in Marine Systems Related to Dysbiosis and Opportunistic Pathogens?
The last decade has seen an increase in the reporting of disease syndromes of cultured and natural populations of marine organisms including fish, seagrass, seaweeds, corals, and other invertebrates (Harvell et al., 2002, 2004; Lafferty et al., 2004; Ward and Lafferty, 2004; Bourne et al., 2009; Gachon et al., 2010). However, in many cases the causative agent/s of the disease are unknown; and indeed debate over the importance of microbial pathogens in relatively well-studied marine diseases continues (Hoegh-Guldberg et al., 2007; Rosenberg et al., 2009; Gachon et al., 2010; Burge et al., 2013).
The first likely explanation for the uncertainty over the microbial involvement in particular marine diseases is due to the difficulty in distinguishing pathogens from other saprophytic bacteria that proliferate on a diseased or decaying host (Burge et al., 2013; Egan et al., 2014). Adding to this uncertainty is the fact that in marine systems, without continued monitoring, it is often difficult to determine the early (and likely asymptomatic) stages of infection. Thus, it is possible that the commonly reported disease symptoms (e.g., bleaching, spotting, or rotting) for marine organisms are representative of late stages of disease and the proliferation of secondary invaders. Indeed, many features attributed to potential pathogen/s, such as the ability to colonize and degrade host tissue, could also be utilized by saprophytic organism/s, further obscuring the identity of the true primary pathogen/s.
A second explanation is that disease is not the result of a single microbial agent, but rather a consortium of microbial species (i.e., polymicrobial) with each member playing a distinct role in the pathogenic process. The black band disease (BBD) of corals is arguably, to date, the best model for polymicrobial disease in marine systems, with a complex microbial community consisting of cyanobacteria, heterotrophic and sulfur-cycling bacteria, as well as archaeal members implicated in disease progression (Sato et al., 2016).
A third possibility is that some microbial pathogens exist as part of a host’s native microbiome, and under certain conditions, i.e., host senesce, can switch from a commensal to a pathogenic lifestyle. For example, the marine bacterium Phaeobacter gallaeciensis BS107 produces antibacterial and growth promoting compounds that are beneficial to its host microalga, Emiliania huxleyi; however, upon detection of algal breakdown products, the beneficial bacterium is converted into an opportunistic pathogen through the production of algaecide compounds (Seyedsayamdost et al., 2011). In such cases it can be difficult to apply classical measurements of disease causality, such as Koch’s postulates (see Table 1 for definition), which assume that the causative agent is always associated with disease while being completely absent in healthy individuals (Koch, 1882; Kaufmann and Schaible, 2005).
Finally, a forth and often overlooked explanation is that disease results from the proliferation of any number of opportunistic pathogens when there is dysbiosis in the host microbiome as a result of environmental pressures and/or host stress (Lesser et al., 2007; Burge et al., 2013; Egan et al., 2014) (Figure 1). Microbial dysbiosis has emerged as an explanation for several acute and chronic human diseases, including atopic dermatitis of the skin (Brüssow, 2015), gastrointestinal disorders (Clemente et al., 2012; Parekh et al., 2015), periodontal disease (Meyle and Chapple, 2015), and inflammatory respiratory diseases (Mahdavinia et al., 2016). In these examples, scientists and medical professionals suggest that a combination of environmental, immune, and host genetic immune factors, together with microbiome disturbance, play a role in the establishment and severity of these diseases (Legatzki et al., 2014; Butto et al., 2015; Donaldson et al., 2016; Hajishengallis and Lamont, 2016). For example, lower microbial diversity and depletion of commensal bacteria in the gut is associated with the development of asthma and Crohn’s Disease, implicating a role for microbial dysbiosis in these now common diseases (Frank et al., 2007; Candela et al., 2012; Abrahamsson et al., 2014). Further, there is a growing appreciation for the complex interplay between beneficial species, opportunistic pathogens, environmental factors and the immune system, and the influence that these interactions have on human health outcomes, including the onset of chronic disease (Mazmanian et al., 2008; Chow and Mazmanian, 2010; Butto et al., 2015).
FIGURE 1.
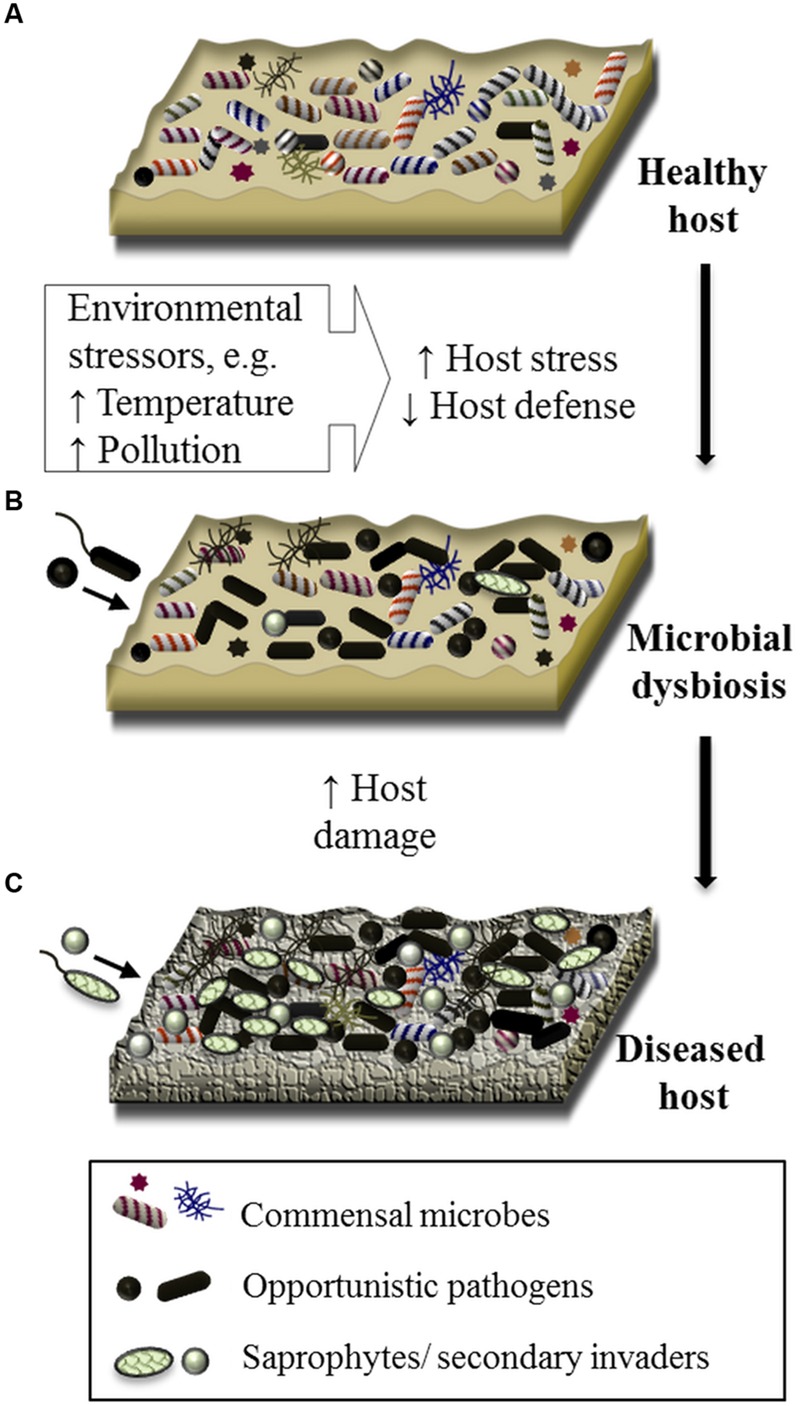
Schematic of the proposed involvement of microbial dysbiosis, opportunistic pathogens and polymicrobial interactions in the disease of marine organisms. The surface of a healthy marine living organism (healthy host) is colonized by a consortium of commensal microorganisms, which may include bacteria, archaea, microbial eukaryotes, and viruses (A). Under conditions of elevated environmental pressure, the host can become stressed negatively impacting the host defence mechanisms. On this susceptible host, opportunistic pathogens and/or pathogenic consortia (i.e., polymicrobial infection), originating from the healthy host or from the surrounding environment, proliferate, resulting in a state of microbial dysbiosis (B). Microbial pathogens along with saprophytes or secondary colonizers further cause damage to the host resulting in visible disease (C).
While less understood, there is evidence for dysbiosis syndromes in marine systems. Meres et al. (2012) proposed microbial dysbiosis and the emergence of opportunistic pathogens (including the previously reported Aquimarina spp. pathogens) as a mitigating factor in the epizootic shell disease (ESD) of lobsters. Likewise, analysis of microbial communities associated with healthy and diseased Montastraea corals indicated multiple etiological factors, including polymicrobial infections, are likely to be responsible for BBD (Miller and Richardson, 2015) and white-plague disease (Cook et al., 2013). Studies investigating the impact of environmental change on marine hosts have given further insight to the role of microbial dysbiosis and disease. Exposure to stressful environmental conditions results in distinct shifts in microbial community assemblages that can be detected prior to any physical sign of disease in corals (Webster et al., 2013; Closek et al., 2014), seaweeds (Fernandes et al., 2012; Stratil et al., 2013) and sponges (Webster et al., 2008; Fan et al., 2013) (Figure 1). In some cases, these shifts have been characterized by an increase in groups of pathogenic bacteria (e.g., Vibrio spp. (Tout et al., 2015), Roseobacter species (Zozaya-Valdes et al., 2015)) and/or an increase in the abundance of virulence-related functional genes (Vega Thurber et al., 2009; Littman et al., 2011; Fernandes et al., 2012). This may be considered early evidence for dysbiosis and the enrichment of bacteria with pathogenic potential. Thus, a disease framework centered around stress-induced dysbiosis and the emergence of opportunistic pathogens would not only explain inconsistencies in laboratory infection assays and pathogen detection in characterized diseases (e.g., Rosenberg et al., 2009; Meres et al., 2012; Cook et al., 2013; Joyner et al., 2015; Zozaya-Valdes et al., 2015), but would also help to understand the etiology of yet undefined syndromes.
A Call for a Fresh Approach to Characterize and Manage Marine Dysbiosis and Disease
Historically, studies of host–pathogen interactions have centered on the detection and control of the infectious agent/s. For decades researchers have studied the detailed molecular mechanisms that enable pathogenic microorganisms to infect their host and cause disease. These studies have resulted in a wealth of knowledge, including the identification of growth and virulence factors [e.g., toxins, enzymes, and host colonization factors (Finlay and Falkow, 1997)]. There is no doubt about the value of such an approach to disease control, especially where an understanding of pathogenesis has facilitated the development of vaccines and antibiotics to treat highly infectious pathogens such as those responsible for measles, diphtheria, and typhoid. However, if dysbiosis, opportunistic pathogens and/ or polymicrobial infections are responsible for disease, a paradigm shift in the way we think about host health and microbial symbiosis is required.
The importance of this paradigm shift has recently been recognized by medical microbiologists with calls to “ditch the term pathogen” (Casadevall and Pirofski, 2014; Altmann and Boyton, 2015) and to place greater emphasis on the complex interplay between the host and its microbiome (Méthot and Alizon, 2014). The importance of a less ‘pathogen-centric’ view of disease is now being recognized (Casadevall and Pirofski, 2015) and recent studies have also questioned the value of using common virulence genes alone as predictors of a bacterium’s ability to cause disease (de Lorenzo, 2015; Hilker et al., 2015; Wassenaar and Gunzer, 2015). Indeed, many virulence traits in well-studied pathogens have secondary roles [or ‘dual function roles’ (see Table 1 for definition)] in persistence and stress adaptation outside the host environment (Casadevall et al., 2003; Vezzulli et al., 2008). Therefore, the discovery of a high proportion of virulence genes in the genomes of ‘non-pathogenic’ marine bacteria (Thomas et al., 2008; Persson et al., 2009; Gennari et al., 2012) and the dual function of known virulence factors (Erken et al., 2013; Gardiner et al., 2014) further speaks to the need for a broader contextual view of disease in the marine environment.
One of the first challenges in understanding the role of dysbiosis in marine diseases is obtaining suitable baseline data on what is considered a “healthy” microbiome. While the past decade has seen an increase in marine microbiome studies, inherent natural variation, inconsistency in sample collection and different approaches to data analysis means it is difficult to compare data across individual studies. Thus, there is an urgent need for larger spatial and temporal scale microbiome studies, particularly of species that are habitat-forming (e.g., macroalgae, corals), important to food security (e.g., farmed finfish and invertebrate species), or at high risk of disease (e.g., threatened or endangered species). For example, by analyzing the microbial diversity associated with over 250 samples covering a large spatial distribution of the reef forming kelp Ecklonia radiata, Marzinelli et al. (2015) identified a consistent microbiome for healthy individuals. However, when significant deviations in the kelp microbiome occurred, they could be correlated with visible signs of disease. Thus, with finer temporal sampling, microbial community data may be an effective environmental monitoring tool to detect microbiome changes as predictors for disease outcomes.
Despite the decreasing cost of characterizing microbial communities via sequencing, the scale required to adequately obtain the baseline data remains prohibitive to most individual research laboratories. Therefore, continued engagement of community and philanthropic supported initiatives will be essential. Such large scale microbiome initiatives have proven to be successful strategies for the analysis of oceanic microorganisms [e.g., GOS (Yooseph et al., 2007) and TARA (Pesant et al., 2015)], the human microbiome1 and other environmental systems [(e.g., Earth Microbiome Project2 (EMP)], with recent calls to move to an international microbiome initiative (IMI) that encompasses all habitats and is not limited by national boarders (Dubilier et al., 2015). We suggest that such global pattern analysis should be extended to include temporal studies of the typical microbiome of host species to gain insight into the emergence of dysbiosis and disease.
To accurately discover and manage the factors that contribute to dysbiosis and disease in marine systems requires a true holobiont approach (i.e., host and associated microbiome as one entity see Table 1). As stated above, such an approach needs to go beyond correlating health with the presence of specific groups of bacteria. Indeed, studies of vibriosis in oysters are finding that diseased animals are often colonized by diverse populations of Vibrio strains, rather than single virulent isolates (Lemire et al., 2015). Such findings further highlight the need to move beyond simple characterization of single pathogens and towards an understanding of how the host and its microbiome respond to, and interact with, each other, and how these interactions are influenced by environmental factors. Microbiologists, ecologists, environmental scientists, system biologists, and host physiologists will need to work closely together and occasionally step out of their comfort zone. For example, molecular microbiologists will need to become less reductionist and accept that the answers maybe more complex and variable than they feel confident with. Likewise (macro-) ecologists will need to embrace the concept that the organisms they study are not functioning on their own, but are influenced by millions of microorganisms that are not always simple to measure or control.
Compared to the medical field the understanding of dysbiosis in marine systems is in its infancy. In the last 10 years, over 1020 articles have been published on microbial dysbiosis in humans alone (using keywords “dysbiosis human”, PubMed.gov search 10.02.16). However a similar search on marine organisms (using keywords “marine dysbiosis”) retrieved only three publications. Whilst one can argue that the scientific breakthroughs in the medical field are due to the significantly larger research support (in terms of numbers of scientists and research funding), an important consideration is that this research effort is going towards one model organism, i.e., humans. This is in contrast to work on marine symbiosis, where relatively smaller amounts of money are spread across a large range of host species. Thus, in order for marine symbiosis research to come close to the equivalent depth of understanding of dysbiosis and disease as in the medical field, basic principles may need to first be studied collectively on fewer model species. The choice of model host/s should be made by the science community taking into consideration accessibility, ecological context, available metadata, as well as their ability to grow in aquaria and be experimentally manipulated. Indeed, some excellent marine model organisms exists to study microbial interactions and/or disease in macroalgae [e.g., Ectocarpus siliculosus (Zambounis et al., 2013) and Delisea pulchra (Harder et al., 2012)], invertebrates [e.g., oysters Crassostrea gigas (Le Roux et al., 2016); and cnidarians such as species of Aiptasia (Weis et al., 2008) and Hydra (Bosch, 2014)], vertebrates (e.g., zebrafish Danio rerio (Rawls et al., 2006), and medaka Oryzias latipes (Schartl, 2014)), and these systems should be further explored.
In addition it is important to consider how far the current theoretical framework of disease (predominately established for terrestrial systems) can be applied to marine systems. Indeed, McCallum et al. (2004) raised this question over 10 years ago and argued that, while the same theory may be applicable to marine systems, fundamental differences between marine and terrestrial systems may limit its usefulness. For example, marine systems will require their own set of disease models and management strategies that take into account the different life histories of marine and terrestrial organisms, the different modes of dispersal and recruitment of both microorganisms and their hosts (i.e., the “open” nature of marine systems), and differences in the impact of environmental change on marine and terrestrial systems (McCallum et al., 2004; Pollock et al., 2011; Arnold et al., 2012; Garren and Azam, 2012; Wahl et al., 2012; Burge et al., 2014).
Conclusions and Perspectives
Ever since the early descriptions of the infectious agents by Robert Koch in the 1800s, the study of microbial diseases has been overshadowed by the desire to discover and eliminate the causative pathogen. However, as our understanding of microbial ecology and host–microbial interactions improve it is becoming increasingly clear that the relationship between a host and its microbiome is complex, and the lines between commensal microorganisms and pathogens are often blurred.
Next generation ‘–omic’ technologies are living up to the promise of providing the data resolution and scale required to understand many of these complex interactions. However, major questions remain unanswered, including: How are microbiomes established and maintained? What specific functions do microbiomes play in host resilience, and how do changes in microbial community structure impact host health and disease? Answers to these questions and the need to rethink the definitions of disease are particularly pertinent to marine scientists today in the face of environmental change and increasing observations of disease inflicting marine organisms.
Author Contributions
SE and MG both made a direct contribution to planning and writing of the manuscript and both have approved it for publication.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors would like to thank the Australian Research Council and the Centre for Marine Bio-Innovation for research support. We also thank A/Prof. Torsten Thomas and Adam Bournazos for helpful discussion and editing of the final manuscript.
Footnotes
References
- Abby S. S., Touchon M., De Jode A., Grimsley N., Piganeau G. (2014). Bacteria in Ostreococcus tauri cultures – friends, foes or hitchhikers? Front. Microbiol. 5:505 10.3389/fmicb.2014.00505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abrahamsson T. R., Jakobsson H. E., Andersson A. F., Bjorksten B., Engstrand L., Jenmalm M. C. (2014). Low gut microbiota diversity in early infancy precedes asthma at school age. Clin. Exp. Aller. 44 842–850. 10.1111/cea.12253 [DOI] [PubMed] [Google Scholar]
- Alivisatos A. P., Blaser M. J., Brodie E. L., Chun M., Dangl J. L., Donohue T. J., et al. (2015). Microbiome. A unified initiative to harness Earth’s microbiomes. Science 350 507–508. 10.1126/science.aac8480 [DOI] [PubMed] [Google Scholar]
- Althani A., Marei H., Hamdi W. S., Nasrallah G. K., El Zowalaty M. E., Al Khdor S., et al. (2015). Human microbiome and its association with health and diseases. J. Cell. Physiol. 9999 1–7. 10.1002/jcp.25284 [DOI] [PubMed] [Google Scholar]
- Altmann D., Boyton R. (2015). Replace ‘pathogens’ with ‘perceptogens.’ Nature 518:35 10.1038/518035c [DOI] [PubMed] [Google Scholar]
- Amin S. A., Hmelo L. R., Van Tol H. M., Durham B. P., Carlson L. T., Heal K. R., et al. (2015). Interaction and signalling between a cosmopolitan phytoplankton and associated bacteria. Nature 522 98–101. 10.1038/nature14488 [DOI] [PubMed] [Google Scholar]
- Arnold T., Mealey C., Leahey H., Miller A. W., Hall-Spencer J. M., Milazzo M., et al. (2012). Ocean acidification and the loss of phenolic substances in marine plants. PLoS ONE 7:e35107 10.1371/journal.pone.0035107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch T. C. G. (2014). Rethinking the role of immunity: lessons from Hydra. Trends Immunol. 35 495–502. 10.1016/j.it.2014.07.008 [DOI] [PubMed] [Google Scholar]
- Bourne D. G., Garren M., Work T. M., Rosenberg E., Smith G. W., Harvell C. D. (2009). Microbial disease and the coral holobiont. Trends Microbiol. 17 554–562. 10.1016/j.tim.2009.09.004 [DOI] [PubMed] [Google Scholar]
- Brüssow H. (2015). Turning the inside out: the microbiology of atopic dermatitis. Environ. Microbiol. Advan. 10.1111/1462-2920.13050 [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- Burge C. A., Eakin C. M., Friedman C. S., Froelich B., Hershberger P. K., Hofmann E. E., et al. (2014). “Climate change influences on marine infectious diseases: Implications for management and society,” in Annual Review of Marine Science Vol. 6 eds Carlson C. A., Giovannoni S. J. (Palo Alto, CA: Annual Reviews; ) 249–277. [DOI] [PubMed] [Google Scholar]
- Burge C. A., Kim C. J. S., Lyles J. M., Harvell C. D. (2013). Special issue oceans and humans health: the ecology of marine opportunists. Microb. Ecol. 65 869–879. 10.1007/s00248-013-0190-7 [DOI] [PubMed] [Google Scholar]
- Butto L. F., Schaubeck M., Haller D. (2015). Mechanisms of microbe-host interaction in Crohn’s disease: dysbiosis vs. Pathobiont selection. Front. Immunol. 6:555 10.3389/fimmu.2015.00555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Candela M., Rampelli S., Turroni S., Severgnini M., Consolandi C., De Bellis G., et al. (2012). Unbalance of intestinal microbiota in atopic children. BMC Microbiol. 12:95 10.1186/1471-2180-12-95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casadevall A., Pirofski L. A. (2000). Host-pathogen interactions: basic concepts of microbial commensalism, colonization, infection, and disease. Infect. Immun. 68 6511–6518. 10.1128/iai.68.12.6511-6518.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casadevall A., Pirofski L. A. (2003). The damage-response framework of microbial pathogenesis. Nat. Rev. Microbiol. 1 17–24. 10.1038/nrmicro732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casadevall A., Pirofski L. A. (2014). Ditch the term pathogen. Nature 516 165–166. 10.1038/516165a [DOI] [PubMed] [Google Scholar]
- Casadevall A., Pirofski L. A. (2015). What is a host? Incorporating the microbiota into the damage-response framework. Infect. Immun. 83 2–7. 10.1128/iai.02627-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casadevall A., Steenbergen J. N., Nosanchuk J. D. (2003). ‘Ready made’ virulence and ‘dual use’ virulence factors in pathogenic environmental fungi - the Cryptococcus neoformans paradigm. Curr. Opin. Microbiol. 6 332–337. 10.1016/s1369-5274(03)00082-1 [DOI] [PubMed] [Google Scholar]
- Chow J., Mazmanian S. K. (2010). A pathobiont of the microbiota balances host colonization and intestinal inflammation. Cell Host Microbe 7 265–276. 10.1016/j.chom.2010.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemente J. C., Ursell L. K., Parfrey L. W., Knight R. (2012). The impact of the gut microbiota on human health: an integrative view. Cell 148 1258–1270. 10.1016/j.cell.2012.01.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Closek C. J., Sunagawa S., Desalvo M. K., Piceno Y. M., Desantis T. Z., Brodie E. L., et al. (2014). Coral transcriptome and bacterial community profiles reveal distinct yellow band disease states in Orbicella faveolata. ISME J. 8 2411–2422. 10.1038/ismej.2014.85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook G. M., Rothenberger J. P., Sikaroodi M., Gillevet P. M., Peters E. C., Jonas R. B. (2013). A comparison of culture-dependent and culture-independent techniques used to characterize bacterial communities on healthy and white plague-diseased corals of the Montastraea annularis species complex. Coral Reefs 32 375–388. 10.1007/s00338-012-0989-6 [DOI] [Google Scholar]
- de Lorenzo V. (2015). Pseudomonas aeruginosa: the making of a pathogen. Environ. Microbiol. 17 1–3. 10.1111/1462-2920.12620 [DOI] [PubMed] [Google Scholar]
- DeLeon-Rodriguez N., Lathem T. L., Rodriguez R. L., Barazesh J. M., Anderson B. E., Beyersdorf A. J., et al. (2013). Microbiome of the upper troposphere: species composition and prevalence, effects of tropical storms, and atmospheric implications. Proc. Natl. Acad. Sci. U.S.A. 110 2575–2580. 10.1073/pnas.1212089110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson G. P., Lee S. M., Mazmanian S. K. (2016). Gut biogeography of the bacterial microbiota. Nat. Rev. Microbiol. 14 20–32. 10.1038/nrmicro3552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubilier N., Mcfall-Ngai M., Zhao L. (2015). Create a global microbiome effort. Nature 526 631–634. 10.1038/526631a [DOI] [PubMed] [Google Scholar]
- Egan S., Fernandes N. D., Kumar V., Gardiner M., Thomas T. (2014). Bacterial pathogens, virulence mechanism and host defence in marine macroalgae. Environ. Microbiol. 16 925–938. 10.1111/1462-2920.12288 [DOI] [PubMed] [Google Scholar]
- Egan S., Harder T., Burke C., Steinberg P., Kjelleberg S., Thomas T. (2013). The seaweed holobiont: understanding seaweed-bacteria interactions. FEMS Microbiol. Rev. 37 462–476. 10.1111/1574-6976.12011 [DOI] [PubMed] [Google Scholar]
- Erken M., Lutz C., Mcdougald D. (2013). The rise of pathogens: predation as a factor driving the evolution of human pathogens in the environment. Microb. Ecol. 65 860–868. 10.1007/s00248-013-0189-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan L., Liu M., Simister R., Webster N. S., Thomas T. (2013). Marine microbial symbiosis heats up: the phylogenetic and functional response of a sponge holobiont to thermal stress. ISME J. 7 991–1002. 10.1038/ismej.2012.165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes N., Steinberg P., Rusch D., Kjelleberg S., Thomas T. (2012). Community structure and functional gene profile of bacteria on healthy and diseased thalli of the red seaweed Delisea pulchra. PLoS ONE 7:e50854 10.1371/journal.pone.0050854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finlay B. B., Falkow S. (1997). Common themes in microbial pathogenicity revisited. Microbiol. Mol. Biol. Rev. 61 136–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank D. N., Amand A. L. S., Feldman R. A., Boedeker E. C., Harpaz N., Pace N. R. (2007). Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc. Natl. Acad. Sci. U.S.A. 104 13780–13785. 10.1073/pnas.0706625104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gachon C. M. M., Sime-Ngando T., Strittmatter M., Chambouvet A., Kim G. H. (2010). Algal diseases: spotlight on a black box. Trends Plant Sci. 15 633–640. 10.1016/j.tplants.2010.08.005 [DOI] [PubMed] [Google Scholar]
- Gardiner M., Hoke D. E., Egan S. (2014). An ortholog of the Leptospira interrogans lipoprotein LipL32 aids in the colonization of Pseudoalteromonas tunicata to host surfaces. Front. Microbiol. 5:323 10.3389/fmicb.2014.00323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garren M., Azam F. (2012). New directions in coral reef microbial ecology. Environ. Microbiol. 14 833–844. 10.1111/j.1462-2920.2011.02597.x [DOI] [PubMed] [Google Scholar]
- Gattuso J. P., Magnan A., Bille R., Cheung W. W. L., Howes E. L., Joos F., et al. (2015). Contrasting futures for ocean and society from different anthropogenic CO2 emissions scenarios. Science 349 45–55. 10.1126/science.aac4722 [DOI] [PubMed] [Google Scholar]
- Gennari M., Ghidini V., Caburlotto G., Lleo M. M. (2012). Virulence genes and pathogenicity islands in environmental Vibrio strains nonpathogenic to humans. FEMS Microbiol. Ecol. 82 563–573. 10.1111/j.1574-6941.2012.01427.x [DOI] [PubMed] [Google Scholar]
- Hajishengallis G., Lamont R. J. (2016). Dancing with the stars: how choreographed bacterial interactions dictate nososymbiocity and give rise to keystone pathogens, accessory pathogens, and pathobionts. Trends Microbiol. Adv. 24 477–489. 10.1016/j.tim.2016.02.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harder T., Campbell A. H., Egan S., Steinberg P. D. (2012). Chemical mediation of ternary interactions between marine holobionts and their environment as exemplified by the red alga Delisea pulchra. J. Chem. Ecol. 38 442–450. 10.1007/s10886-012-0119-5 [DOI] [PubMed] [Google Scholar]
- Harvell C. D., Mitchell C. E., Ward J. R., Altizer S., Dobson A. P., Ostfeld R. S., et al. (2002). Ecology - Climate warming and disease risks for terrestrial and marine biota. Science 296 2158–2162. 10.1126/science.1063699 [DOI] [PubMed] [Google Scholar]
- Harvell D., Aronson R., Baron N., Connell J., Dobson A., Ellner S., et al. (2004). The rising tide of ocean diseases: unsolved problems and research priorities. Front. Ecol. Environ. 2:375–382. 10.2307/3868363 [DOI] [Google Scholar]
- Hilker R., Munder A., Klockgether J., Losada P. M., Chouvarine P., Cramer N., et al. (2015). Interclonal gradient of virulence in the Pseudomonas aeruginosa pangenome from disease and environment. Environ. Microbiol. 17 29–46. 10.1111/1462-2920.12606 [DOI] [PubMed] [Google Scholar]
- Hoegh-Guldberg O., Mumby P. J., Hooten A. J., Steneck R. S., Greenfield P., Gomez E., et al. (2007). Coral reefs under rapid climate change and ocean acidification. Science 318 1737–1742. 10.1126/science.1152509 [DOI] [PubMed] [Google Scholar]
- Inagaki F., Hinrichs K. U., Kubo Y., Bowles M. W., Heuer V. B., Hong W. L., et al. (2015). Deep biosphere. Exploring deep microbial life in coal-bearing sediment down to ∼2.5 km below the ocean floor. Science 349 420–424. 10.1126/science.aaa6882 [DOI] [PubMed] [Google Scholar]
- Joyner J. L., Sutherland K. P., Kemp D. W., Berry B., Griffin A., Porter J. W., et al. (2015). Systematic analysis of white pox disease in acropora palmata of the florida keys and role of Serratia marcescens. Appl. Environ. Microbiol. 81 4451–4457. 10.1128/aem.00116-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann S. H., Schaible U. E. (2005). 100th anniversary of robert koch’s nobel prize for the discovery of the tubercle bacillus. Trends Microbiol. 13 469–475. 10.1016/j.tim.2005.08.003 [DOI] [PubMed] [Google Scholar]
- Koch R. (1882). Die aetiologie der tuberculose (Nach einem in der physiologischen Gesellschaft zu Berlin am 24.März cr. gehaltenem Vortrage). Berl. Klin. Wochenschr. 19:221. [PubMed] [Google Scholar]
- Lafferty K. D., Porter J. W., Ford S. E. (2004). Are diseases increasing in the ocean? Annu. Rev. Ecol. Evol. Syst. 35 31–54. 10.1146/annurev.ecolsys.35.021103.105704 [DOI] [Google Scholar]
- Le Roux F., Wegner K. M., Polz M. F. (2016). Oysters and Vibrios as a model for disease dynamics in wild animals. Trends Microbiol. Adv. 10.1016/j.tim.2016.03.006 [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- Legatzki A., Rosler B., Von Mutius E. (2014). Microbiome diversity and asthma and allergy risk. Curr. Allergy Asthma Rep. 14:466 10.1007/s11882-014-0466-0 [DOI] [PubMed] [Google Scholar]
- Lemire A., Goudenège D., Versigny T., Petton B., Calteau A., Labreuche Y., et al. (2015). Populations, not clones, are the unit of Vibrio pathogenesis in naturally infected oysters. ISME J. 9 1523–1531. 10.1038/ismej.2014.233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesser M. P., Bythell J. C., Gates R. D., Johnstone R. W., Hoegh-Guldberg O. (2007). Are infectious diseases really killing corals? Alternative interpretations of the experimental and ecological data. J. Exp. Mar. Biol. Ecol. 346 36–44. 10.1016/j.jembe.2007.02.015 [DOI] [Google Scholar]
- Littman R., Willis B. L., Bourne D. G. (2011). Metagenomic analysis of the coral holobiont during a natural bleaching event on the Great Barrier Reef. Environ. Microbiol. Rep. 3 651–660. 10.1111/j.1758-2229.2010.00234.x [DOI] [PubMed] [Google Scholar]
- Mahdavinia M., Keshavarzian A., Tobin M. C., Landay A. L., Schleimer R. P. (2016). A comprehensive review of the nasal microbiome in chronic rhinosinusitis (CRS). Clin. Exp. Aller. 46 21–41. 10.1111/cea.12666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzinelli E. M., Campbell A. H., Valdes E. Z., Verges A., Nielsen S., Wernberg T., et al. (2015). Continental-scale variation in seaweed host-associated bacterial communities is a function of host condition, not geography. Environ. Microbiol. 17 4078–4088. 10.1111/1462-2920.12972 [DOI] [PubMed] [Google Scholar]
- Mazmanian S. K., Round J. L., Kasper D. L. (2008). A microbial symbiosis factor prevents intestinal inflammatory disease. Nature 453 620–625. 10.1038/nature07008 [DOI] [PubMed] [Google Scholar]
- McCallum H. I., Kuris A., Harvell C. D., Lafferty K. D., Smith G. W., Porter J. (2004). Does terrestrial epidemiology apply to marine systems? Trends Ecol. Evol. 19 585–591. 10.1016/j.tree.2004.08.009 [DOI] [Google Scholar]
- McFall-Ngai M. J. (2014). “The importance of microbes in animal development: lessons from the squid-Vibrio symbiosis,” in Annual Review of Microbiology ed. Gottesman S. (Palo Alto, CA: Annual Reviews; ) 177–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meres N. J., Ajuzie C. C., Sikaroodi M., Vemulapalli M., Shields J. D., Gillevet P. M. (2012). Dysbiosis in epizootic shell disease of the American lobster (Homarus americanus). J. Shellfish Res. 31 463–472. 10.2983/035.031.0206 [DOI] [Google Scholar]
- Méthot P. O., Alizon S. (2014). What is a pathogen? Toward a process view of host-parasite interactions. Virulence 5 775–785. 10.4161/21505594.2014.960726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyle J., Chapple I. (2015). Molecular aspects of the pathogenesis of periodontitis. Periodontol 2000 7–17. 10.1111/prd.12104 [DOI] [PubMed] [Google Scholar]
- Miller A. W., Richardson L. L. (2015). Emerging coral diseases: a temperature-driven process? Mar. Ecol. 36 278–291. 10.1111/maec.12142 [DOI] [Google Scholar]
- Morado J. F. (2011). Protistan diseases of commercially important crabs: a review. J. Invertebr. Pathol. 106 27–53. 10.1016/j.jip.2010.09.014 [DOI] [PubMed] [Google Scholar]
- Munn C. B. (2006). Viruses as pathogens of marine organisms - from bacteria to whales. J. Mar. Biol. Assoc. 86 453–467. 10.1017/s002531540601335x [DOI] [Google Scholar]
- Parekh P. J., Balart L. A., Johnson D. A. (2015). The influence of the gut microbiome on obesity, metabolic syndrome and gastrointestinal disease. Clin. Transl. Gastroenterol. 6:e91 10.1038/ctg.2015.16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persson O. P., Pinhassi J., Riemann L., Marklund B. I., Rhen M., Normark S., et al. (2009). High abundance of virulence gene homologues in marine bacteria. Environ. Microbiol. 11 1348–1357. 10.1111/j.1462-2920.2008.01861.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pesant S., Not F., Picheral M., Kandels-Lewis S., Le Bescot N., Gorsky G., et al. (2015). Open science resources for the discovery and analysis of Tara Oceans data. Sci. Data 2 150023–150023. 10.1038/sdata.2015.23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen C., Round J. L. (2014). Defining dysbiosis and its influence on host immunity and disease. Cell. Microbiol. 16 1024–1033. 10.1111/cmi.12308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollock F. J., Morris P. J., Willis B. L., Bourne D. G. (2011). The urgent need for robust coral disease diagnostics. PLoS Pathog. 7:e1002183 10.1371/journal.ppat.1002183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rädecker N., Pogoreutz C., Voolstra C. R., Wiedenmann J., Wild C. (2015). Nitrogen cycling in corals: the key to understanding holobiont functioning? Trends Microbiol. 23 490–497. 10.1016/j.tim.2015.03.008 [DOI] [PubMed] [Google Scholar]
- Rawls J. F., Mahowald M. A., Ley R. E., Gordon J. I. (2006). Reciprocal gut microbiota transplants from zebrafish and mice to germ-free recipients reveal host habitat selection. Cell 127 423–433. 10.1016/j.cell.2006.08.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg E., Koren O., Reshef L., Efrony R., Zilber-Rosenberg I. (2007). The role of microorganisms in coral health, disease and evolution. Nat. Rev. Microbiol. 5 355–362. 10.1038/nrmicro1635 [DOI] [PubMed] [Google Scholar]
- Rosenberg E., Kushmaro A., Kramarsky-Winter E., Banin E., Yossi L. (2009). The role of microorganisms in coral bleaching. ISME J. 3 139–146. 10.1038/ismej.2008.104 [DOI] [PubMed] [Google Scholar]
- Sato Y., Civiello M., Bell S. C., Willis B. L., Bourne D. G. (2016). Integrated approach to understanding the onset and pathogenesis of black band disease in corals. Environ. Microbiol. 18 752–765. 10.1111/1462-2920.13122 [DOI] [PubMed] [Google Scholar]
- Schartl M. (2014). Beyond the zebrafish: diverse fish species for modeling human disease. Dis. Model. Mech. 7 181–192. 10.1242/dmm.012245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seyedsayamdost M. R., Case R. J., Kolter R., Clardy J. (2011). The Jekyll-and-hyde chemistry of Phaeobacter gallaeciensis. Nat. Chem. 3 331–335. 10.1038/nchem.1002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stratil S. B., Neulinger S. C., Knecht H., Friedrichs A. K., Wahl M. (2013). Temperature-driven shifts in the epibiotic bacterial community composition of the brown macroalga Fucus vesiculosus. Microbiologyopen 2 338–349. 10.1002/mbo3.79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas T., Evans F. F., Schleheck D., Mai-Prochnow A., Burke C., Penesyan A., et al. (2008). Analysis of the Pseudoalteromonas tunicata genome reveals properties of a surface-associated life style in the marine environment. PLoS ONE 3:e3252 10.1371/journal.pone.0003252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tout J., Siboni N., Messer L. F., Garren M., Stocker R., Webster N. S., et al. (2015). Increased seawater temperature increases the abundance and alters the structure of natural Vibrio populations associated with the coral Pocillopora damicornis. Front. Microbiol. 6:432 10.3389/fmicb.2015.00432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vega Thurber R., Willner-Hall D., Rodriguez-Mueller B., Desnues C., Edwards R. A., Angly F., et al. (2009). Metagenomic analysis of stressed coral holobionts. Environ. Microbiol. 11 2148–2163. 10.1111/j.1462-2920.2009.01935.x [DOI] [PubMed] [Google Scholar]
- Vezzulli L., Guzman C. A., Colwell R. R., Pruzzo C. (2008). Dual role colonization factors connecting Vibrio cholerae’s lifestyles in human and aquatic environments open new perspectives for combating infectious diseases. Curr. Opin. Biotechnol. 19 254–259. 10.1016/j.copbio.2008.04.002 [DOI] [PubMed] [Google Scholar]
- Wahl M., Goecke F., Labes A., Dobretsov S., Weinberger F. (2012). The second skin: ecological role of epibiotic biofilms on marine organisms. Front. Microbiol. 3:292 10.3389/fmicb.2012.00292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward J. R., Lafferty K. D. (2004). The elusive baseline of marine disease: are diseases in ocean ecosystems increasing? PLoS Biol. 2:E120 10.1371/journal.pbio.0020120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wassenaar T. M., Gunzer F. (2015). The prediction of virulence based on presence of virulence genes in E. coli may not always be accurate. Gut Pathog. 7:15 10.1186/s13099-015-0062-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster N. S., Cobb R. E., Negri A. P. (2008). Temperature thresholds for bacterial symbiosis with a sponge. ISME J. 2 830–842. 10.1038/ismej.2008.42 [DOI] [PubMed] [Google Scholar]
- Webster N. S., Negri A. P., Flores F., Humphrey C., Soo R., Botte E. S., et al. (2013). Near-future ocean acidification causes differences in microbial associations within diverse coral reef taxa. Environ. Microbiol. Rep. 5 243–251. 10.1111/1758-2229.12006 [DOI] [PubMed] [Google Scholar]
- Webster N. S., Thomas T. (2016). The sponge hologenome. mBio 7:e00135 10.1128/mBio.00135-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weis V. M., Davy S. K., Hoegh-Guldberg O., Rodriguez-Lanetty M., Pringe J. R. (2008). Cell biology in model systems as the key to understanding corals. Trends Ecol. Evol. 23 369–376. 10.1016/j.tree.2008.03.004 [DOI] [PubMed] [Google Scholar]
- Willey J. M., Sherwood L., Woolverton C. J., Prescott L. M. (2008). Prescott, Harley, and Klein’s Microbiology. New York, NY: McGraw-Hill Higher Education. [Google Scholar]
- Yooseph S., Sutton G., Rusch D. B., Halpern A. L., Williamson S. J., Remington K., et al. (2007). The sorcerer II global ocean sampling expedition: expanding the universe of protein families. PLoS Biol. 5:e16 10.1371/journal.pbio.0050016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zambounis A., Strittmatter M., Gachon C. M. M. (2013). Chronic stress and disease resistance in the genome model marine seaweed Ectocarpus siliculosus. Aquat. Bot. 104 147–152. 10.1016/j.aquabot.2012.07.008 [DOI] [Google Scholar]
- Zozaya-Valdes E., Egan S., Thomas T. (2015). A comprehensive analysis of the microbial communities of healthy and diseased marine macroalgae and the detection of known and potential bacterial pathogens. Front. Microbiol. 6:146 10.3389/fmicb.2015.00146 [DOI] [PMC free article] [PubMed] [Google Scholar]


