Abstract
Carotenoids, which generate yellow, orange, and red colors, are crucial pigments in avian plumage. Investigations into genes associated with carotenoid-based coloration in avian species are important; however, such research is difficult because carotenoids cannot be synthetized in vertebrates as they are only derived from dietary sources. Here, the golden pheasant(Chrysolophus pictus)was used as a model in analysis of candidate gene expression profiles implicated in carotenoid binding and deposition. Using mass and Raman spectrometry to confirm the presence of carotenoids in golden pheasant feathers, we found C40H54O and C40H56O2 in feathers with yellow to red colors, and in the rachis of iridescent feathers. The global gene expression profiles in golden pheasant skins were analyzed by RNA-seq and all six carotenoid binding candidate genes sequenced were studied by real-time PCR. StAR4, GSTA2, Scarb1, and APOD in feather follicles showed different expressions in red breast and orange nape feathers compared with that of iridescent mantle feathers. Further comparison of golden pheasant yellow rump and Lady Amherst's pheasant(Chrysolophus amherstiae)white napefeathers suggested that GSTA2 and APOD played a potential role in carotenoid-based coloration in golden pheasant.
Keywords: Expression, Carotenoid coloration, Candidate genes, Golden pheasant, Feather
INTRODUCTION
Golden pheasant(Chrysolophus pictus)is a world-renowned avian species with particularly attractive and varied plumage. As one of the most prevalent integument pigments in birds, carotenoids are usually deposited in bare body parts and produce yellow, orange, and red coloration in feathers(usually in the distal parts). To date, around 30 different carotenoids have been identified in hundreds of avian species(Hill & McGraw, 2006; LaFountain et al., 2010). Furthermore, this class of pigment has been confirmed in 95 of 236 extant bird families, including three species of Galliformes, such as the golden pheasant(Thomas et al., 2014a). Unlike other pigments, such as melanin, carotenoids are difficult to study in regards to the genetics of bird coloration because knowledge of the molecular genetics of carotenoid-based coloration is incomplete(Roulin & Ducrest, 2013). Under this limitation, transportation, deposition, and metabolism-related proteins and their encoding genes have been studied in consideration of the dietary-derived characteristics of these pigments(Pointer et al., 2012; Walsh et al., 2012). In chicken skins, mutations of beta-carotene dioxygenase 2(BCDO2)inhibit gene expression and result in carotenoid accumulation(Eriksson et al., 2008). In bird feathers, carotenoids are bound directly to keratin or other proteins(Stradi et al., 1995; Mendes-Pinto et al., 2012), so it is generally accepted that carotenoid-binding protein(CBP)is the starting point to investigate carotenoid-based coloration of plumage at the molecular level.
A previous report reviewed 11 plausible candidate genes associated with carotenoid uptake, binding, and deposition, including steroidogenic acute regulatory protein 4(StAR4), glutathione S-transferase alpha 2(GSTA2), StAR-related lipid transfer domain containing 3(STARD3, also known as MLN64), scavenger receptor class B, member 1(Scarb1, also known as SR-BI), apolipoprotein D(APOD), and lipid storage droplet 2-like(PLIN)genes(Walsh et al., 2012). As a member of the StAR protein family, StAR4 specifically binds lutein with high affinity, and is a possible candidate for CBP in human retina(Bhosale et al., 2009). Scarb1 is involved in carotenoid transport into retinal pigment epithelium(RPE)cells, and gene silencing can result in a marked inhibition of carotenoid uptake(During et al., 2008). Although APOD is a carotenoid-binding gene in crustaceans and mammals, a paralogous gene has since been characterized in chickens, and is expressed in skins and developing feathers(Ganfornina et al., 2005). GSTA2 is the closest avian homologue of the carotenoid binding gene GSTP1 in mammalian retina and is involved in plumage dichromatism(Walsh et al., 2012; Zhang et al., 2014). It is predicted, therefore, that these genes might be highly expressed in carotenoid feathers.
Prior to pigment identification, we analyzed the expression levels of six carotenoid deposition genes in golden pheasant feather follicles, in which the corresponding feathers may or may not have contained carotenoids.
MATERIALS AND METHODS
Animals and samples
Three male artificially bred golden pheasants were used in this study. The birds were provided by the Yuanfeng Wildlife Farm(Jilin, China)and were housed in cages with free access to water and commercial food. This study was approved by the Institutional Animal Care and Use Committee of the Inner Mongolia University. For RNA extraction, skins and feather follicles were sampled. Briefly, feathers of various colors were plucked from the breast, nape, rump, and mantle of the body(Figure 1). Regrown feather shafts and skins were then collected and stored in liquid nitrogen(Poelstra et al., 2014). For pigment extraction, feathers were plucked from corresponding regions directly, then placed in envelopes and stored in the dark. Blood samples were drawn from the neck veins using vacuum blood collection tubes, with the samples then centrifuged in heparinized microcapillary tubes for 10 min at 1 500 g. The plasma was then removed and stored at -80 ℃ in 1.5 mL Eppendorf tubes until further analysis(McGraw et al., 2002a).
Figure 1.
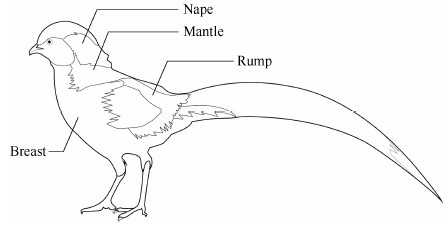
Feathers and feather follicles sampled from the breast, nape, mantle and rump of the golden pheasant
RNA-seq
The isolated mRNA from golden pheasant skins was fragmented and used for cDNA synthesis. The short cDNA fragments were treated following the manufacturer's instructions and sequenced using an Illumina Hiseq 2000(Illumina, San Diego, USA). The produced reads were assembled by mapping to the golden pheasant genome we sequenced and annotated based on homology prediction. The expression abundance of each assembled transcript was measured through reads per kilobase transcriptome per million mapped reads(RPKM)(Mortazavi et al., 2008). The expressions of each read between sample pairs were calculated according to RPKM values in different samples. A minimum two-fold difference was considered as an expression difference with a false discovery rate(FDR)≤0.05(Benjamini et al., 2001). GO enrichment analysis was performed using GO: TermFinder software(http://smd.stanford.edu/help/GO-TermFinder/GO_TermFinder_help.shtml/).
Carotenoids extraction
Feather and plasma carotenoid pigments were extracted, respectively, using previously published methods(McGraw et al., 2002b, 2005). In brief, feathers from various parts of the body were soaked in ethanol and hexane, in turn, in order to remove surface lipids. Feather vanes were trimmed into pieces and placed into 10 mL glass tubes. After that, 1 mL of acidified pyridine was infused to cover the pieces. The tubes were then filled with argon gas, capped tightly, and placed in a 95 ℃ water bath for 3 h until the solution became colorful. After the thermochemical procedures, the cooled tubes were added with 2 mL of pure water and 1 mL of hexane: tert-butyl methyl ether(1:1)to separate the carotenoid pigments from the solution. The mixtures were shaken strongly for 2 min and centrifuged at 3000 r/min for 5 min, with the supernatants then transferred to clean glass tubes, dried under nitrogen gas, and stored at -80 ℃ until use. For blood samples, the extraction procedures were the same as above(McGraw et al., 2002a). Ten microliters of plasma with 100 μL of ethanol were mixed in 1.5 mL Eppendorf tubes, followed by the addition of 100 μL of tert-butyl methyl ether. After being vortexed, the tubes were centrifuged at 3 000 r/min for 3 min, with the same treatment as that for plumage pigment extraction.
Mass spectrometry
The extracted pigments were used for molecular weight analysis using an HPLC-MS instrument composed of a LC-20AD system(Shimadzu, Japan)fitted with a WatersTM C18 column(5 μm, 150 mm×2.1 mm)and an LTQ Velos Orbitrap(Thermo, Germany). Samples were dissolved in methanol and injected into the instrument at 0.2 mL/min and analyzed in the positive mode with the capillary heated to 350 ℃ and at a cone voltage of 4.5 kV. A gradient solvent system was used for compound elution, with the percentage of acetonitrile varying from 5% to 95% corresponding to the methanoic acid solution(v=5%)in the first 11 min, before decreasing to 5% in the next 3 min, and finally being held isocratically for 1 min.
Raman spectroscopy
Feather samples were examined in a Labram HR1800 spectrometer(HORIBA Jobin Yvon, France)without pretreatment using an excitation laser source at 514 nm and 0.3 mW power. Single spectra were obtained using a 100× confocal objective and a grating of 600 lines/mm. Data were analyzed by LabSpec 5 software. Standards of lutein(Chromadex, USA)and zeaxanthin(Sigma, USA)were used in comparative analysis.
Real-time PCR
Sequences of candidate genes were obtained from the RNA-seq data. The predicted sequences were used to design primers on exon regions spanning introns using Primer Premier 5(Table 1). Housekeeping gene β-actin was used for normalization. Total RNA was extracted from the feather follicle samples taken from each of the three birds using RNAiso Plus reagent(Takara, Japan). cDNA preparation with quantitative real-time PCR for each candidate gene was performed using a PrimeScriptTM RT Reagent Kit with gDNA Eraser(Perfect Real Time)(Takara, Japan). Each sample was pooled with 5-10 feather follicles. Reactions were performed in an ABI PRISM 7500 Real-Time PCR System(Applied Biosystems, USA)with default parameters. Three replicates were used for each sample.
Table 1.
Primers used for real-time PCR
| Gene | Forward primer(5'-3') | Reverse primer(3'-5') | TM(℃) |
| StAR4 | CAAGCTATGAAGATGGGCTTC | ATCTGATTGCAAGGAATGCAC | 60 |
| GSTA2 | TCCCTTTTCAAGCAGCCGAT | GCTGCCAGGCTGCAAGAAT | 60 |
| STARD3 | GACCACGCACAGCCTGAA | AGGCTCTGGTGGATGAGGTA | 60 |
| Scarb1 | ACTTCTACAATGCTGACCCAA | AGCTTGATGGAGCAATTCAT | 60 |
| APOD | GCGTCCGCTTCAACTGGT | CGTGGGCATCATCTTGTCG | 60 |
| PLIN | CCATCCAAAGTGCCAAGAG | CAGGTCTGCTTGGGCTTC | 60 |
| β-actin | CTCCCTGATGGTCAAGTCAT | TGGATACCACAGGACTCCAT | 60 |
RESULTS
Carotenoid identification in golden pheasant plumage
Pigments were separated from golden pheasant plasma and feathers according to solubility. Both procedures result in carotenoid transfer into the upper organic phase and profile colors in it if this kind of pigment is present. We found yellow colors emerged in the mentioned phases after yellow rump, orange nape, and red breast feathers were treated. This indicated that yellow fat-soluble pigments were contained in these feathers, but not in the iridescent mantle feathers. The same phenomenon occurred in the plasma samples. To confirm the molecular weight of the compounds, the extracted pigments from plasma and plumage were analyzed by mass spectrometry. Search results in databases(METACYC, LIPID, KEGG)showed that C40H54O and C40H56O2 were present in all samples. According to previous publication, lutein and zeaxanthin are isomers with the formula C40H56O2, and echinenone, which is a common carotenoid found in plumage, is C40H54O(Hill & McGraw, 2006).
A single iridescent feather taken from the mantle region of the golden pheasant was examined by Raman spectroscopy for carotenoid identification, as described previously(Thomas et al., 2014a, b). We took two collection points at the rachis and barb, respectively, and found characteristic bands of carotenoids in the rachis at 1 500-1 535 /cm(identified as ν[C=C]), 1 145-1 165 /cm(identified as ν[C-C]), and 1 000-1 010 /cm(identified as δ[CH2])(Thomas et al., 2014b). For the barb, bands of eumelanin(Galván et al., 2013)were detected(Figure 2).
Figure 2.
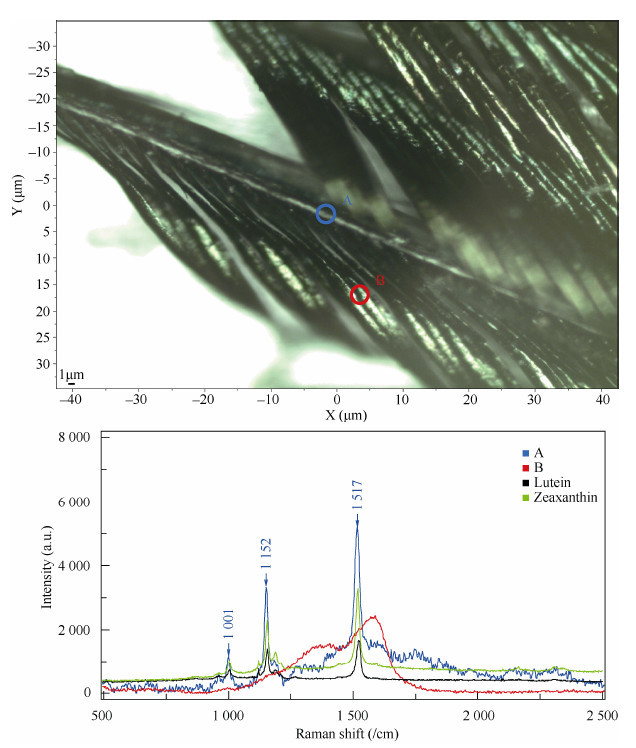
Detected points(above)and corresponding spectrograms(below)by Raman spectroscopy in an iridescent mantle feather of the golden pheasant
Transcriptome sequencing and differential gene expression profiling
Four libraries were constructed for golden pheasant skin transcriptome study, including the breast, nape, mantle, and rump. Raw data were sequenced by an Illumina Hiseq 2000 and filtered to produce a total of 23.83 Gb clean reads. We mapped the reads to the golden pheasant genome to assemble the transcripts for each library. A summary of the mapping results is shown in Table 2. Transcriptome data were deposited in the Sequence Read Archive(SRA)database under accession no. SRP075618.
Table 2.
Statistics of transcriptome sequencing data
| Breast | Nape | Mantle | Rump | |
| Total reads | 68 796 446 | 64 410 352 | 64 910 570 | 66 653 538 |
| Total mapped reads | 48 578 519 | 45 006 710 | 44 868 949 | 46 045 821 |
| Total mapped rate(%) | 70.61 | 69.87 | 69.12 | 69.08 |
| Unique mapped reads | 46 213 276 | 43 605 488 | 42 886 126 | 44 029 043 |
| Unique mapped rate(%) | 67.17 | 67.70 | 66.07 | 66.06 |
A total of 50 228 expressed genes were predicted. To find differences in gene expression between two kinds of color morphs, we compared the RPKM values of the transcripts in mantle skin to the other three parts. Results showed that 452, 748, and 532 genes were upregulated and 686, 1 185, and 710 genes were downregulated in the breast, nape and rump, respectively. The genes with significantly differential expression were enriched in 46 pathways, including the retinol metabolism pathway, which might be involved in carotenoid metabolism(Table S1). We enriched and overlapped the genes that showed preferential expression in carotenoid-containing tissues(Figure 3). Sixty-six genes were significantly expressed in all three carotenoid-containing tissues, of which 57 were functionally annotated(Table S2).
Table S1.
The KEGG enrichment pathways of differentially expressed genes
| Pathway names | P-value | Gene number |
| Breast vs Mantle | ||
| Asthma | 0.009529 | 7 |
| Rheumatoid arthritis | 0.010248 | 15 |
| Primary immunodeficiency | 0.010914 | 11 |
| Protein processing in endoplasmic reticulum | 0.015881 | 25 |
| Taste transduction | 0.017159 | 8 |
| Epstein-Barr virus infection | 0.026659 | 31 |
| Autoimmune thyroid disease | 0.029717 | 8 |
| Dorso-ventral axis formation | 0.035248 | 10 |
| Allograft rejection | 0.039137 | 7 |
| Phenylalanine metabolism | 0.042316 | 5 |
| Cell cycle | 0.049864 | 17 |
| Nape vs Mantle | ||
| Tuberculosis | 0.00248 | 50 |
| GABAergic synapse | 0.004119 | 28 |
| Drug metabolism - other enzymes | 0.006265 | 11 |
| NOD-like receptor signaling pathway | 0.006723 | 20 |
| Pentose and glucuronate interconversions | 0.009242 | 8 |
| Retrograde endocannabinoid signaling | 0.01119 | 29 |
| p53 signaling pathway | 0.011736 | 23 |
| Cell cycle | 0.013267 | 29 |
| Allograft rejection | 0.018443 | 11 |
| Morphine addiction | 0.02048 | 26 |
| Phagosome | 0.028967 | 41 |
| Dilated cardiomyopathy | 0.030255 | 50 |
| Retinol metabolism | 0.033083 | 11 |
| MAPK signaling pathway | 0.034771 | 56 |
| Ascorbate and aldarate metabolism | 0.035275 | 5 |
| African trypanosomiasis | 0.036597 | 12 |
| HTLV-I infection | 0.039812 | 51 |
| Vibrio cholerae infection | 0.043547 | 35 |
| Primary immunodeficiency | 0.046528 | 14 |
| GnRH signaling pathway | 0.046684 | 24 |
| Autoimmune thyroid disease | 0.048439 | 11 |
| Rump vs Mantle | ||
| Protein export | 0.002098 | 7 |
| Epstein-Barr virus infection | 0.002347 | 38 |
| MAPK signaling pathway | 0.003028 | 43 |
| Progesterone-mediated oocyte maturation | 0.007463 | 17 |
| NOD-like receptor signaling pathway | 0.010668 | 14 |
| HTLV-I infection | 0.012849 | 37 |
| Pyrimidine metabolism | 0.016391 | 19 |
| Bile secretion | 0.01703 | 15 |
| Endometrial cancer | 0.024112 | 11 |
| Morphine addiction | 0.024376 | 18 |
| Purine metabolism | 0.02743 | 28 |
| Drug metabolism - other enzymes | 0.030022 | 7 |
| GABAergic synapse | 0.035824 | 17 |
| Chemokine signaling pathway | 0.037094 | 30 |
Figure 3.
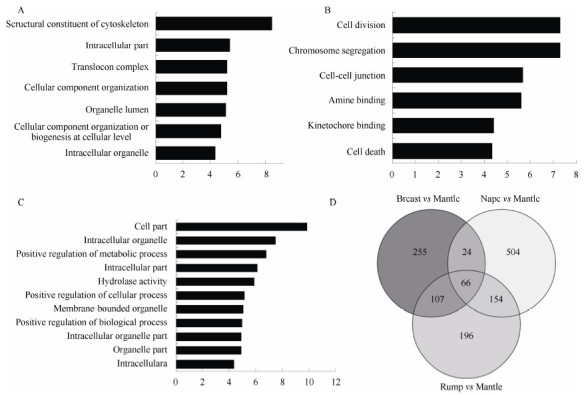
Analysis of differentially expressed genes(DEGs)upregulated in golden pheasant breast, nape, and rump compared with that in the mantle
Table S2.
The overlapped differentially expressed genes
| Gene ID | Gene name | Description in Nr/Nt |
| Pictus_GLEAN_10014658 | DISP1 | Protein dispatched homolog 1 |
| Pictus_GLEAN_10000108 | GART | Trifunctional purine biosynthetic protein adenosine-3 |
| Pictus_GLEAN_10010005 | Gdf15 | Growth/differentiation factor 15 |
| Pictus_GLEAN_10011222 | BFAR | Bifunctional apoptosis regulator |
| Pictus_GLEAN_10012474 | Slc5a7 | High affinity choline transporter 1 |
| Pictus_GLEAN_10003211 | Unknown | Transmembrane protein C9orf91 homolog |
| Pictus_GLEAN_10000348 | NFYA | Nuclear transcription factor Y subunit alpha |
| Pictus_GLEAN_10008738 | - | - |
| Pictus_GLEAN_10004368 | AIM1L | Absent in melanoma 1-like protein |
| Pictus_GLEAN_10005259 | Unknown | Antimicrobial peptide THP2 |
| Pictus_GLEAN_10012008 | ASIC2 | Acid-sensing ion channel 2 |
| Pictus_GLEAN_10012500 | STXBP5 | Syntaxin-binding protein 5 |
| Pictus_GLEAN_10014876 | COMMD8 | COMM domain-containing protein 8 |
| Pictus_GLEAN_10003225 | SH2D1A | SH2 domain-containing protein 1A |
| Pictus_GLEAN_10008440 | UVSSA | UV-stimulated scaffold protein A |
| Pictus_GLEAN_10001408 | Stk3 | Serine/threonine-protein kinase 3 |
| Pictus_GLEAN_10004559 | - | - |
| Pictus_GLEAN_10013092 | SLC29A1 | Equilibrative nucleoside transporter 1 |
| Pictus_GLEAN_10002330 | NFIX | Nuclear factor 1 X-type |
| Pictus_GLEAN_10015591 | ADAMTS6 | A disintegrin and metalloproteinase with thrombospondin motifs 6 |
| Pictus_GLEAN_10003672 | Pax5 | Paired box protein Pax-5 |
| Pictus_GLEAN_10016591 | - | - |
| Pictus_GLEAN_10003044 | STAG1 | Cohesin subunit SA-1 |
| Pictus_GLEAN_10014897 | - | - |
| Pictus_GLEAN_10004422 | MTERFD3 | mTERF domain-containing protein 3, mitochondrial |
| Pictus_GLEAN_10015534 | HEXB | Beta-hexosaminidase subunit beta |
| Pictus_GLEAN_10010980 | SMIM5 | Small integral membrane protein 5 |
| Pictus_GLEAN_10004217 | ALMS1 | Alstrom syndrome protein 1 |
| Pictus_GLEAN_10002454 | Rcan1 | Calcipressin-1 |
| Pictus_GLEAN_10001753 | PAF1 | RNA polymerase II-associated factor 1 homolog |
| Pictus_GLEAN_10009937 | RPE65 | Retinoid isomerohydrolase |
| Pictus_GLEAN_10016196 | mfhas1 | Malignant fibrous histiocytoma-amplified sequence 1 homolog |
| Pictus_GLEAN_10004574 | - | - |
| Pictus_GLEAN_10012457 | AK3 | GTP:AMP phosphotransferase, mitochondrial |
| Pictus_GLEAN_10012216 | PEMT | Phosphatidylethanolamine N-methyltransferase |
| Pictus_GLEAN_10014346 | - | - |
| Pictus_GLEAN_10011735 | EYA1 | Eyes absent homolog 1 |
| Pictus_GLEAN_10016118 | - | - |
| Pictus_GLEAN_10005108 | SCNN1A | Amiloride-sensitive sodium channel subunit alpha |
| Pictus_GLEAN_10013033 | Ssna1 | Sjoegren syndrome nuclear autoantigen 1 homolog |
| Pictus_GLEAN_10008204 | - | - |
| Pictus_GLEAN_10011762 | KLHL32 | Kelch-like protein 32 |
| Pictus_GLEAN_10013038 | Rab18 | Ras-related protein Rab-18 |
| Pictus_GLEAN_10013624 | IPO5 | Importin-5 |
| Pictus_GLEAN_10002914 | PCDH15 | Protocadherin-15 |
| Pictus_GLEAN_10010556 | Slc25a32 | Mitochondrial folate transporter/carrier |
| Pictus_GLEAN_10010087 | TENM3 | Teneurin-3 |
| Pictus_GLEAN_10009788 | EIF3J | Eukaryotic translation initiation factor 3 subunit J |
| Pictus_GLEAN_10003779 | C21orf58 | Uncharacterized protein C21orf58 |
| Pictus_GLEAN_10001877 | Cdkn2b | Cyclin-dependent kinase 4 inhibitor B |
| Pictus_GLEAN_10000247 | EFNA5 | Ephrin-A5 |
| Pictus_GLEAN_10010108 | - | - |
| Pictus_GLEAN_10013900 | COG5 | Conserved oligomeric Golgi complex subunit 5 |
| Pictus_GLEAN_10003784 | POFUT2 | GDP-fucose protein O-fucosyltransferase 2 |
| Pictus_GLEAN_10013330 | DUSP13 | Dual specificity protein phosphatase 13 |
| Pictus_GLEAN_10008736 | Dgkb | Diacylglycerol kinase beta |
| Pictus_GLEAN_10006805 | RAP1GAP | Rap1 GTPase-activating protein 1 |
| Pictus_GLEAN_10010383 | Pithd1 | PITH domain-containing protein 1 |
| Pictus_GLEAN_10007637 | isl2a | Insulin gene enhancer protein isl-2a |
| Pictus_GLEAN_10010311 | EIF2C4 | Protein argonaute-4 |
| Pictus_GLEAN_10012322 | LIF | Leukemia inhibitory factor |
| Pictus_GLEAN_10004513 | CCDC102A | Coiled-coil domain-containing protein 102A |
| Pictus_GLEAN_10015988 | DDB2 | DNA damage-binding protein 2 |
| Pictus_GLEAN_10011796 | NHLRC3 | NHL repeat-containing protein 3 |
| Pictus_GLEAN_10009917 | HTR1D | 5-hydroxytryptamine receptor 1D (Fragment) |
| Pictus_GLEAN_10006460 | Prss57 | Serine protease 57 |
Expression of candidate genes in golden pheasant feather follicles
To confirm the expressions of carotenoid binding related genes in feather follicles, six homologous candidate genes were determined using real-time PCR. Expression differences in developing feather follicles taken from the rump, breast, nape, and mantle regions of the male golden pheasant were determined and the expression patterns were compared with RNA-seq data(Figure 4). All candidate genes were expressed in all tissue samples. Comparison of the expression patterns of the six genes revealed no differences(P > 0.05)between yellow rump and iridescent mantle feather follicles. Compared with mantle tissue, there was no difference(P > 0.05)in the expression levels of STARD3 in the breast, nape and rump samples, and the expression of PLIN showed significant differences only in red breast feather follicles(P < 0.05). The remaining five genes were differentially expressed in red and orange plumage. For red breast plumage, expressions of StAR4 and PLIN showed significant differences(P < 0.05), while expressions of GSTA2, Scarb1, and APOD showed highly significant differences(P < 0.01). For orange nape plumage, expressions of StAR4 and APOD showed significant differences(P < 0.05), while expressions of GSTA2 and Scarb1 showed highly significant differences(P < 0.01). We also compared the expression levels of the candidate genes between golden pheasant yellow rump feathers and Lady Amherst's pheasantnape feathers, which are white with black stripes without carotenoid-based coloration. Data showed the expressions of GSTA2 and APOD were significantly different(Figure 5).
Figure 4.
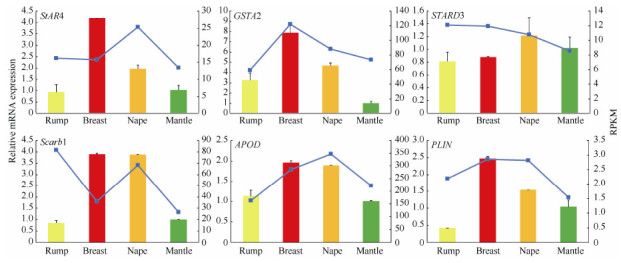
Expression levels of carotenoid candidate genes in different feather follicles of the golden pheasant(mean±SD)by real-time PCR
Figure 5.
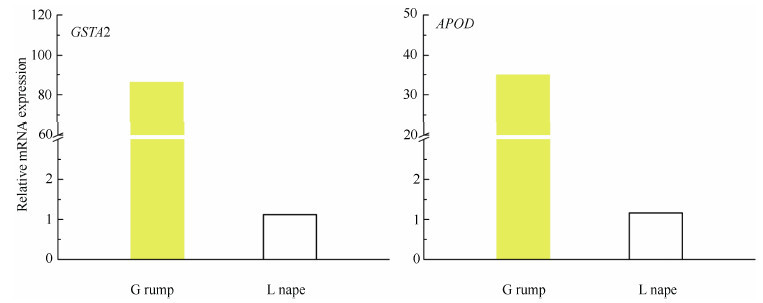
Expression levels using real-time PCR of carotenoid candidate genes in different feather follicles between golden pheasant(G)rump and Lady Amherst's pheasant(L)nape
DISCUSSION
Thermochemical techniques for carotenoid extraction are based on weakening the non-covalent hydrogen bonds that bind pigments to proteins, thereby subsequently releasing pigments into solution(McGraw et al., 2005). In our study on carotenoid separation from golden pheasant plumage, we found that the organic phases of red, orange, and yellow tested barbs were pigmented after treatment. This phenomenon suggested the presence of lipochrome in the feathers, with further mass spectrometry analysis confirming that these pigments were carotenoids. This conclusion was confirmed by searching molecular weights in databases(METACYC, LIPID, KEGG), with a focus on 40 specific carbon compounds. According to the common carotenoids documented in birds(Hill & McGraw, 2006), we suggest that the identified chemicals might be lutein, zeaxanthin, and echinenone. Therefore, it follows that at least two common dietary carotenoids(lutein and zeathanxin)exist in golden pheasant plumage.
As the plasma samples achieved the same results in carotenoid identification, we inferred that this kind of dietary-derived pigment was absorbed and transported into feather follicles. Consequently, we determined the expression of several carotenoid-binding genes in golden pheasant feather follicles and found some patterns were significantly different between carotenoid-containing samples and mantle samples, in which carotenoids were absent in barbs.
Genes related to carotenoid deposition did not show significant differential expression through RNA-seq analysis. Consequently, we performed real-time PCR and found that four genes(StAR4, GSTA2, Scarb1, and APOD)were highly expressed in red breast and orange nape feathers, suggesting their possible function in carotenoid deposition in these tissues of golden pheasant. The differences in expression indicate that these genes might play different roles in carotenoid coloration in different feathers. In addition, all candidate genes were expressed in all tissues, even iridescent mantle feather follicles, while, the expression levels in yellow rump feathers showed no differences compared with iridescent mantle feathers.To clarify this issue, we tested a mantle feather using Raman spectroscopy and found carotenoids in the rachis, but not in the barbs. This led to the hypothesis that the transported carotenoids might combine with some other factors in the mantle feather rachis, such as keratins(McGraw et al., 2003), and thus the expression of carotenoid-binding genes might not be the only reason for carotenoid deposition in these regions. Compared with the mantle feathers, GSTA2 and APOD were highly expressed in red breast and orange nape feathers, but not in yellow rump feathers in which carotenoids were detected. Therefore, a further comparison with white control feathers was carried out. Results indicated that the two genes might be important to carotenoid deposition in golden pheasant plumage.
The genes involved in carotenoid deposition in feathers are poorly known, yet are of substantial interest. The golden pheasant is an excellent species to address these questions, being one of the few Galliformes to express carotenoids in feather follicles. Six homologous candidate genes documented and investigated in Quelea quelea(Walsh et al., 2012)were studied in the golden pheasant in the present study. We found at least two candidate genes associated with carotenoid coloration, with differences in expressions raising the possibility that these genes might play different roles in golden pheasant plumage. Whether functional features of potential genes are species specific in golden pheasant or universal across avian taxa will spur the exploration of genetic and genomic investigations in future work.
ACKNOWLEDGMENTS
We owe many thanks to Gui-Cheng LI from the IMU for providing argon gas and other experimental material. We are also grateful to Tian-Yuan WANG from Yuanfeng Wildlife Farm for taking care of the experimental birds.
Funding Statement
This work was supported by the 2014 Fundamental Research Program from Science and Technology of the Inner Mongolia Autonomous Region of China
REFERENCES
- 1. Benjamini Y, Yekutieli D. 2001. The control of the false discovery rate in multiple testing under dependency. Annals of Statistics, 29 (4): 1165- 1188. [Google Scholar]
- 2. Bhosale P, Li BX, Sharifzadeh M, Gellermann W, Frederick JM, Tsuchida K, Bernstein PS. 2009. Purification and partial characterization of a lutein-binding protein from human retina. Biochemistry, 48 (22): 4798- 4807. [DOI] [PubMed] [Google Scholar]
- 3. During A, Doraiswamy S, Harrison EH. 2008. Xanthophylls are preferentially taken up compared with β-carotene by retinal cells via a SRBI-dependent mechanism. Journal of Lipid Research, 49 (8): 1715- 1724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Eriksson J, Larson G, Gunnarsson U, Bed'hom B, Tixier-Boichard M, StrÖmstedt L, Wright D, Jungerius A, Vereijken A, Randi E, Jensen P, Andersson L. 2008. Identification of the yellow skin gene reveals a hybrid origin of the domestic chicken. PLoS Genetics, 4 (2): e1000010- [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Galván I, Jorge A, Ito K, Tabuchi K, Solano F, Wakamatsu K. 2013. Raman spectroscopy as a non-invasive technique for the quantification of melanins in feathers and hairs. Pigment Cell & Melanoma Research, 26 (6): 917- 923. [DOI] [PubMed] [Google Scholar]
- 6. Ganfornina MD, Sánchez D, Pagano A, Tonachini L, Descalzi-Cancedda F, Martínez S. 2005. Molecular characterization and developmental expression pattern of the chicken apolipoprotein D gene: implications for the evolution of vertebrate lipocalins. Developmental Dynamics, 232 (1): 191- 199. [DOI] [PubMed] [Google Scholar]
- 7. Hill GE, McGraw KJ. 2006. Bird Coloration, vol. 1, Mechanisms and Measurements, Havard: Havard University Press; 177- 242. [Google Scholar]
- 8. LaFountain AM, Kaligotla S, Cawley S, Riedl KM, Schwartz SJ, Frank HA, Prum RO. 2010. Novel methoxy-carotenoids from the burgundy-colored plumage of the Pompadour Cotinga Xipholena punicea. Archives of Biochemistry and Biophysics, 504 (1): 142- 153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. McGraw KJ, Adkins-Regan E, Parker RS. 2002. a. Anhydrolutein in the zebra finch: a new, metabolically derived carotenoid in birds. Comparative Biochemistry and Physiology Part B-Biochemistry & Molecular Biology, 132 (4): 811- 818. [DOI] [PubMed] [Google Scholar]
- 10. McGraw KJ, Hill GE, Stradi R, Parker RS. 2002. b. The effect of dietary carotenoid access on sexual dichromatism and plumage pigment composition in the American goldfinch. Comparative Biochemistry and Physiology Part B-Biochemistry & Molecular Biology, 131 (2): 261- 269. [DOI] [PubMed] [Google Scholar]
- 11. McGraw KJ, Beebee MD, Hill GE, Parker RS. 2003. Lutein-based plumage coloration in songbirds is a consequence of selective pigment incorporation in to feathers. Comparative Biochemistry and Physiology Part B-Biochemistry & Molecular Biology, 135 (4): 689- 696. [DOI] [PubMed] [Google Scholar]
- 12. McGraw KJ, Hudon J, Hill GE, Parker RS. 2005. A simple and inexpensive chemical test for behavioral ecologists to determine the presence of carotenoid pigments in animal tissues. Behavioral Ecology & Sociobiology, 57 (4): 391- 397. [Google Scholar]
- 13. Mendes-Pinto MM, Lafountain AM, Stoddard MC, Prum RO, Frank HA, Robert B. 2012. Variation in carotenoid-protein interaction in bird feathers produces novel plumage coloration. Journal of the Royal Society Interface, 9 (77): 3338- 3350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. 2008. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nature Methods, 5 (7): 621- 628. [DOI] [PubMed] [Google Scholar]
- 15. Poelstra JW, Vijay N, Bossu CM, Lantz H, Ryll B, Müller I, Baglione V, Unneberg P, Wikelski M, Grabherr MG, Wolf JBW. 2014. The genomic landscape underlying phenotypic integrity in the face of gene flow in crows. Science, 344 (6190): 1410- 1414. [DOI] [PubMed] [Google Scholar]
- 16. Pointer MA, Prager M, Andersson S, Mundy NI. 2012. A novel method for screening a vertebrate transcriptome for genes involved in carotenoid binding and metabolism. Molecular Ecology Resources, 12 (1): 149- 159. [DOI] [PubMed] [Google Scholar]
- 17. Roulin A, Ducrest AL. 2013. Genetics of colouration in birds. Seminars in Cell & Developmental Biology, 24 (6-7): 594- 608. [DOI] [PubMed] [Google Scholar]
- 18. Stradi R, Celentano G, Rossi E, Rovati G, Pastore M. 1995. Carotenoids in bird plumage: Ⅰ.The carotenoid pattern in a series of palearctic carduelinae. Comparative Biochemistry and Physiology Part B-Biochemistry and Molecular Biology, 110 (1): 131- 143. [Google Scholar]
- 19. Thomas DB, McGraw KJ, Butler MW, Carrano MT, Madden O, James HF. 2014. a. Ancient origins and multiple appearances of carotenoid-pigmented feathers in birds. Proceedings of the Royal Society B-Biological Sciences, 281 (1788): 20140806- [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Thomas DB, McGraw KJ, James HF, Madden O. 2014. b. Non-destructive descriptions of carotenoids in feathers using Raman spectroscopy. Analytical Methods, 6 (5): 1301- 1308. [Google Scholar]
- 21. Walsh N, Dale J, McGraw KJ, Pointer MA Mundy NI. 2012. Candidate genes for carotenoid coloration in vertebrates and their expression profiles in the carotenoid-containing plumage and bill of a wild bird. Proceedings of the Royal Society B-Biological Sciences, 279 (1726): 58- 66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zhang GJ, Li C, Li QY, Li B, Larkin DM, Lee C, Storz JF, Antunes A, Greenwold MJ, Meredith RW, Ödeen A, Cui J, Zhou Q, Xu LH, Pan HL, Wang ZJ, Jin LJ, Zhang P, Hu H, Yang W, Hu J, Xiao J, Yang ZK, Liu Y, Xie QL, Yu H, Lian JM, Wen P, Zhang F, Li H, Zeng YL, Xiong ZJ, Liu SP, Zhou L, Huang ZY, An N, Wang J, Zheng QM, Xiong YQ, Wang GB, Wang B, Wang JJ, Fan Y, da Fonseca RR, Alfaro-Nññez A, Schubert M, Orlando L, Mourier T, Howard JT, Ganapathy G, Pfenning A, Whitney O, Rivas MV, Hara E, Smith J, Farré M, Narayan J, Slavov G, Romanov MN, Borges R, Machado JP, Khan I, Springer MS, Gatesy J, Hoffmann FG, Opazo JC, Håstad O, Sawyer RH, Kim H, Kim KW, Kim HJ, Cho S, Li N, Huang Y, Bruford MW, Zhan XJ, Dixon A, Bertelsen MF, Derryberry E, Warren W, Wilson RK, Li SB, Ray DA, Green RE, O'Brien SJ, Griffin D, Johnson WE, Haussler D, Ryder OA, Willerslev E, Graves GR, AlstrÖm P, Fjeldså J, Mindell DP, Edwards SV, Braun EL, Rahbek C, Burt DW, Houde P, Zhang Y, Yang HM, Wang J; Avian Genome Consortium, Jarvis ED, Gilbert MTP, Wang J. 2014. Comparative genomics reveals insights into avian genome evolution and adaptation. Science, 346 (6215): 1311- 1320. [DOI] [PMC free article] [PubMed] [Google Scholar]


