Abstract
Cyclopropavir (CPV) is a promising antiviral drug against human cytomegalovirus (HCMV). As with ganciclovir (GCV), the current standard for HCMV treatment, activation of CPV requires multiple steps of phosphorylation and is enantioselective. We hypothesized that the resulting CPV triphosphate (CPV-TP) would stereoselectively target HCMV DNA polymerase and terminate DNA synthesis. To test this hypothesis, we synthesized both enantiomers of CPV-TP [(+) and (−)] and investigated their action on HCMV polymerase. Both enantiomers inhibited HCMV polymerase competitively with dGTP, with (+)-CPV-TP exhibiting a more than 20-fold lower apparent Ki than (−)-CPV-TP. Moreover, (+)-CPV-TP was a more potent inhibitor than GCV-TP. (+)-CPV-TP also exhibited substantially lower apparent Km and somewhat higher apparent kcat values than (−)-CPV-TP and GCV-TP for incorporation into DNA by the viral polymerase. As is the case for GCV-TP, both CPV-TP enantiomers behaved as nonobligate chain terminators, with the polymerase terminating DNA synthesis after incorporation of one additional nucleotide. These results elucidate how CPV-TP acts on HCMV DNA polymerase and help explain why CPV is more potent against HCMV replication than GCV.
INTRODUCTION
Cyclopropavir (CPV), a methylenecyclopropane analogue (1) of 2′-deoxyguanosine, is currently undergoing phase I clinical safety studies as a potential drug against human cytomegalovirus (HCMV) (2–4), a common opportunistic pathogen responsible for diseases in multiple organ systems, particularly in immunocompromised patients and newborns (5). Previous mechanism of action studies have revealed that CPV, like ganciclovir (GCV), the first-line therapy for HCMV infection, is initially phosphorylated by the virus-encoded UL97 kinase (6), while conversion of CPV monophosphate (CPV-MP) to CPV triphosphate (CPV-TP) can be performed by cellular kinases (7, 8). Accordingly, certain HCMV UL97 mutations confer CPV resistance (9, 10). CPV inhibits viral DNA replication (3), and accumulation of CPV-TP has been detected during viral infection (11). As certain mutations affecting the catalytic subunit (UL54, Pol) of the HCMV DNA polymerase also confer CPV resistance, it has been hypothesized that CPV-TP targets HCMV Pol (12) and terminates viral DNA synthesis. However, whether and how CPV-TP acts to inhibit HCMV Pol have not been determined. In particular, whether CPV-TP inhibits competitively, whether it is a substrate for incorporation into DNA, and whether it causes chain termination are not known.
Interestingly, CPV is substantially more potent than GCV against HCMV replication in cell culture (2, 3). Some of this increased potency may be due to greater accumulation in infected cells of CPV-TP than GCV-TP at equivalently effective concentrations, which is consistent with more extensive phosphorylation of CPV than GCV by UL97 (6, 11). However, CPV is also more potent than GCV against HCMV lacking UL97 (3, 9, 13). We hypothesized that this could be due to more potent inhibition of HCMV Pol by CPV-TP than by GCV-TP.
The initial phosphorylation and the interaction of the triphosphate with viral polymerase are the most crucial steps in the determination of enantioselectivity of antiviral nucleoside analogues (14). Previous studies showed that the (+) enantiomers are the preferred substrates during enzymatic conversion of CPV to CPV-diphosphate by UL97 and GMP kinase (6, 7). Although there is precedent for stereoselective inhibition of a herpesvirus polymerase by GCV-TP (15), whether CPV-TP is also stereoselective in its activity against HCMV Pol has not been tested.
To investigate these questions, we synthesized CPV-TP in both enantiomeric forms and investigated their actions on HCMV Pol.
MATERIALS AND METHODS
Chemicals and reagents.
All solvents and reagents, unless otherwise indicated, were analysis-grade commercial products and were used as received. CPV-TP enantiomers were synthesized according to the routes presented in Fig. 1, as described below. CPV-TP enantiomers are soluble in water at a concentration of 5 mM, and their purity was confirmed using high-pressure liquid chromatography.
FIG 1.
Synthesis of CPV-TP enantiomers (+)-4 and (−)-4. The reaction conditions were as follows: (A) (a) (1) Me3SiCl, imidazole, pyridine; (2) I2; (b) (1) Bu3N, N,N′-carbonyldiimidazole; (2) MeOH. (3) (Bu3NH)4P2O7, MeOH; (4) NH4OH, 48 h, room temperature; (B) (a) Ac2O, AcONEt4, pyridine; (b) (1) Bu3N, N,N′-carbonyldiimidazole; (2) MeOH; (3) (Bu3NH)4P2O7, MeOH; (4) NH4OH, 3 h. According to the tentative configurational assignment of the corresponding CPV monophosphates (7), the CPV triphosphate (+)-4 can be assigned the S configuration and (−)-4 can be assigned the R configuration.
Chemical syntheses. (i) (Z)-9-{[2-(Acetoxymethyl)-2-(hydroxymethyl)cyclopropylidene]methyl}-N2-isobutyrylguanine phosphate (compound 3).
A mixture of phosphite 2 (88 mg, 0.2 mmol) and imidazole (67 mg, 8 mmol) in pyridine (5 ml) was sonicated at room temperature for 20 min. Trimethylsilyl chloride (0.5 ml, 4 mmol) was added dropwise with stirring. After 20 min, iodine (103 mg, 0.4 mmol) in pyridine (1 ml) was added, and the stirring was continued for 16 h. The solvent was evaporated in vacuo, and the residue was partitioned between water (30 ml) and dichloromethane (3 times, 30 ml each). The aqueous portion was lyophilized, and the residue was passed through Dowex-50 WX2 (H+) in water to give phosphate 3, 77 mg (84%). 31P nuclear magnetic resonance (NMR) indicated the presence of 17% deacetylated compound 3 (Ac replaced with H). 1H NMR (dimethyl sulfoxide [DMSO]-d6) δ 12.11, 11.72 (2s, 2H, NH), 8.21 (s, 1H, H8), 7.26 (s, 1H, H1′), 5.50 (bs, OH), 4.30, 4.02 (AB partially overlapped with s, 4H, J = 11.0 Hz, CH2OAc, CH2OP), 2.76 (m, 1H, CH of isobutyryl), 1.90 (s, 3H, CH3 of acetate), 1.65 (m, 2H, H3′), 1.10 (d, 6H, CH3 of isobutyryl, J = 6.7 Hz). 13C NMR 180.9, 170.6 (CO), 155.4, 149.1, 148.1, 137.0, 120.5 (purine), 118.1 (C2′), 112.6 (C1′), 66.9 (poorly resolved d, CH2OP), 65.4 (CH2OAc), 35.4 (CH of isobutyryl), 26.4 (d, J = 9.0 Hz, C4′), 21.1 (CH3 of acetate), 19.5 (CH3 of isobutyryl), 12.8 (C3′). 31P NMR −0.21 (7, 83%), 0.15 (phosphate 3, Ac=H, 17%). Negative electrospray ionization-mass spectroscopy (ESI-MS) 454 (100.0, M − H).
(ii) Cyclopropavir triphosphate (+)-4.
The mixture of phosphate 3 (40 mg, 0.09 mmol) and tributylamine (21 μl, 0.09 mmol) in dimethylformamide (DMF) (1 ml) was stirred at room temperature for 30 min. N,N′-Carbonyldiimidazole (71.6 mg, 0.44 mmol) was added, and the mixture was stirred for 3 h. The excess of N,N′-carbonyldiimidazole was removed by adding methanol (29 μl, 0.7 mmol) and stirring for 30 min. Tri-n-butylammonium pyrophosphate (620 mg, 1.67 mmol) was then added, and the mixture was stirred for 16 h at room temperature. The solvent was evaporated, the residue was dissolved in aqueous ammonia (30%, 50 ml), and the solution was stirred for 48 h at room temperature. The volatile components were evaporated, and the crude product was chromatographed on a DEAE Sephadex A-25/HCO3−, 20- by 1-cm column using a discontinuous gradient of NH4HCO3: 0.1 M, 0.15 M, 0.20 M, 0.25 M, and 0.3 M (50 ml of each). Appropriate fractions were combined and lyophilized to give triphosphate (+)-4 (12 mg, 24%) as a white solid. Molecular weight was 648.18 as determined spectrophotometrically for tetraammonium salt with 4.9 H2O using ε268 12,000 (pH 7.0) for cyclopropavir, [α]27D 17.2° (c 0.5, H2O). Storage of this material at −70°C or lower is recommended. 1H NMR (D2O) δ 8.30 (s, 1H, H8), 7.19 (s, 1H, H1′), 4.31, 3.98 (2dd, J = 10.5, 5.1 Hz, 2H, CH2OP), 3.84, 3.66 (AB, 2H, J = 11.8 Hz, CH2OH), 1.61, 1.55 (AB, 2H, J = 9.4 Hz, H3′). 31P NMR −4.83 (d, J = 19.8 Hz, Pγ), −10.18 (d, J = 18.7 Hz, Pα), −20.69 (t, J = 19.8 Hz, Pβ). Negative ESI-MS (methanol [MeOH]) 502 (M − H, 100.0), 422 (M − H2PO3, 49.1).
(iii) (Z)-9-{[2-(Acetoxymethyl)-2-(hydroxymethyl)cyclopropylidene]methyl}guanine phosphate (compound 6).
A mixture of phosphate12 (−)-5 (47 mg, 0.14 mmol) and tetraethylammonium acetate (352 mg, 1.37 mmol) in pyridine (0.5 ml) was stirred for 30 min at room temperature, whereupon the solvent was evaporated and the residue was dried in vacuum for 16 h. Pyridine (2 ml) and acetic anhydride (0.13 ml, 1.37 mmol) were added, and the mixture was stirred for 16 h at room temperature. The solvent was evaporated, and an aqueous solution of the residue was passed through a Dowex-50 WX2 200, H+ column to give product 6 (39 mg, 74%) as a white solid. 1H NMR (D2O) δ 8.79 (s, 1H, H8), 7.13 (s, 1H, H1′), 4.32, 3.95 (AB, 2H, J = 11.6 Hz, CH2OAc), 4.10, 3.73 (2dd, 2H, J = 4.9, 11.0 Hz, CH2OP), 1.82 (s, 3H, CH3), 1.62 (s, 2H, H3′). 13C NMR (D2O) 173.9 (CO), 155.9, 155.2, 149.3, 135.7, 124.1 (purine), 111.2, 109.5 (C1′, C2′), 67.4 (d, J = 4.8 Hz, CH2OP), 66.0 (CH2OAc), 26.0 (d, J = 10.0 Hz, C4′), 20.2 (CH3), 12.7 (C3′). 31P NMR 0.75. Negative ESI-MS 384 (100.0, M − H).
(iv) Cyclopropavir triphosphate (−)-4.
The procedure described for triphosphate (+)-4 was followed with phosphate 6 (18.65 mg, 0.05 mmol). The deacetylation was performed in aqueous ammonia (30%, 50 ml) for 3 h at room temperature. Further workup, including chromatography on a DEAE Sephadex A-25 column, followed the procedure described for triphosphate (+)-4 to give enantiomer (−)-4 (19.6 mg, 71%) as a white solid. A molecular weight of 596.69 was determined spectrophotometrically as described for triphosphate (+)-4 as a tetraammonium salt with 1.4 H2O. [α]27D −20.0° (c 0.5, H2O). For storage, see enantiomer (+)-4. 1H NMR, 31P NMR, and negative ESI-MS corresponded to those of (+)-4.
Purification of triphosphates (+)-4 and (−)-4.
Prolonged storage of triphosphates (+)-4 and (−)-4 leads to a partial decomposition to diphosphates. The crude triphosphate dissolved in a minimum amount of water was loaded onto a Polygram Cell 300 polyethyleneimine (PEI) cellulose thin-layer chromatography (TLC) plate 0.1 mm thick which was developed in a 1:1 mixture, LiCl (2 M)-HCO2H (2 M). The slower-moving triphosphate band was scraped off, and it was washed with water to remove LiCl. The triphosphate was then eluted with NH4HCO3 (0.6 M), and the eluate was lyophilized to give a white solid of (+)-4 or (−)-4.
Preparation and purification of HCMV Pol.
Wild-type (WT) HCMV Pol, expressed as a glutathione S-transferase (GST) fusion protein, was expressed, prepared, and purified as previously described (16).
Polymerase assays.
Four different analyses were performed: 50% inhibitory concentration (IC50) determinations, measurements of apparent Ki values, measurements of apparent Km and kcat values, and assays of chain termination.
IC50s for each enantiomer of CPV-TP were determined using a polymerase assay described previously (16), with some modifications. Briefly, all reactions were performed in 10-μl volumes and reaction mixtures contained 0.25 μM unlabeled hairpin primer template T1 (Integrated DNA Technologies), 6 nM HCMV Pol, 0.5 μM dGTP containing 0.1 μCi [α-32P]dGTP (PerkinElmer), (+)-CPV-TP with concentrations ranging from 0 to 200 μM or (−)-CPV-TP with concentrations ranging from 0 to 500 μM, 50 mM Tris (pH 8.0), 1 mM dithiothreitol (DTT), 100 mM KCl, and 40 μg/ml bovine serum albumin (BSA). Reactions were initiated by adding MgCl2 to 10 mM and quenched using 10 μl of stopping buffer (0.05% bromophenol blue, 0.05% xylene cyanol, and 10 mM EDTA in formamide) after incubation at 37°C for 10 min. The stopped reactions were analyzed on 20% denaturing polyacrylamide gels, and the gel images were quantified using a phosphorimager (Bio-Rad). IC50s were determined using GraphPad Prism version 6 software.
Apparent Ki values were determined with a similar assay, with some changes to meet the requirements for Michaelis-Menten kinetic analysis and to monitor dGTP incorporation using a filter-based method. Each 10-μl reaction mixture contained unlabeled 0.27 μM primer template T1; 1 nM HCMV Pol; various concentrations of dGTP containing 1% [α-32P]dGTP (specific activity, 2 Ci/mmol); and either 1, 4, or 8 μM (+)-CPV-TP, 30 μM (−)-CPV-TP, or 3 μM GCV-TP. The reaction without inhibitors was also tested to measure the apparent Km and kcat values for dGTP incorporation. All the reactions were linear for at least 8 min. Aliquots from the reaction mixtures were taken at 0, 1, 2, and 5 min; quenched with equal volumes of 0.2 M EDTA; and then loaded on DE81 ion exchange paper (Whatman). After two washes in 0.2 M Na2HPO4 and one wash in 95% ethanol, the amount of dGTP incorporated into the primer template was quantified using a scintillation counter (PerkinElmer), and the values were converted to rates. The data were fitted to a competitive inhibition model using GraphPad Prism (version 6) to generate apparent Ki values.
Apparent Km and kcat values for incorporation of CPV-TP enantiomers were measured using the same polymerase assay as that used for the IC50 analysis, but with some changes in conditions to meet the requirement for Michaelis-Menten kinetic analysis and the use of a radiolabeled primer template. Each 10-μl reaction mixture contained 0.27 μM primer template T1 radiolabeled at its 5′ end using [γ-32P]ATP and T4 polynucleotide kinase, 3 nM HCMV Pol for (+)-CPV-TP or 9 nM Pol for (−)-CPV-TP, various concentrations of (+)-CPV-TP ranging from 0 to 5 μM, or various concentrations of (−)-CPV-TP ranging from 0 to 50 μM. All the reactions were linear for 8 min and were stopped at 5 min. Kinetic parameters were measured using previously described methods (16, 17).
To assess chain termination, in each 10-μl polymerase reaction mixture, 0.25 μM radiolabeled primer template T1 was incubated with HCMV Pol at 7.2 nM, 36 nM, or 72 nM; a 2-fold molar excess of UL44ΔC290 (kindly provided by Gloria Komazin-Meredith, Harvard Medical School); 25 μM dGTP; 25 μM (+)-CPV-TP or 25 μM (−)-CPV-TP; and 25 μM dCTP/dATP/dTTP in the same buffer used for the IC50 analysis. The reactions were quenched after incubation at 37°C for 15 min and analyzed by electrophoresis on a 20% denaturing polyacrylamide gel, followed by quantification using a phosphorimager.
Measurement of intracellular concentrations of CPV-TP in HCMV-infected cells.
CPV-TP levels in human foreskin fibroblasts infected with HCMV strain Towne were measured at 120 h postinfection (hpi) using reverse-phase high-pressure liquid chromatography as described previously (11), except that the cells were treated with 0.5 μM radiolabeled CPV (the 50% effective concentration [EC50] rather than 2.5 μM [5× EC50]).
RESULTS
Synthesis of CPV-TP enantiomers.
Enantiomeric phosphates or intermediates thereof served as starting materials for synthesis of CPV-TP (+)-4 and (−)-4 (Fig. 1). Initial experiments with racemic CPV-MP (7, 18) using phosphoromorpholidate (19) or N,N′-carbonyldiimidazole (20) and inorganic pyrophosphate led to a complex mixture of products. This indicated the necessity of keeping one hydroxymethyl group protected during the synthesis. Thus, phosphite 2 available from enantiomeric (7) acetate (+)-1, an intermediate in the synthesis of (+)-CPV monophosphate, was oxidized to acetylated phosphate 3 in 84% yield (Fig. 1A). The reaction with N,N′-carbonyldiimidazole and inorganic pyrophosphate (20) afforded triphosphate (+)-4 in 24% yield after N,O-deacylation by NH4OH. A modified approach was employed for the synthesis of enantiomer (−)-4 (Fig. 1B). In order to improve the yield of the triphosphate by preventing the cyclization to cyclic phosphate and limiting the exposure to NH4OH in the final deacylation step, the unprotected CPV monophosphate (−)-5 (7) was acetylated using acetic anhydride and excess tetraethylammonium acetate in pyridine to give acetate 6 in 74% yield. Similar conditions that prevented cyclization to 2′,3′-cyclic phosphates were used for synthesis of 2′-O-acetylribonucleoside 3′-phosphates (21). Reaction of acetate 6 with N,N′-carbonyldiimidazole and inorganic pyrophosphate gave, after deacetylation by NH4OH, triphosphate (−)-4 in 71% yield. Although the yield of triphosphate is higher, it is necessary to prepare the deprotected starting phosphate first and protect the remaining hydroxymethyl group.
Inhibitory effect of CPV-TP on HCMV Pol.
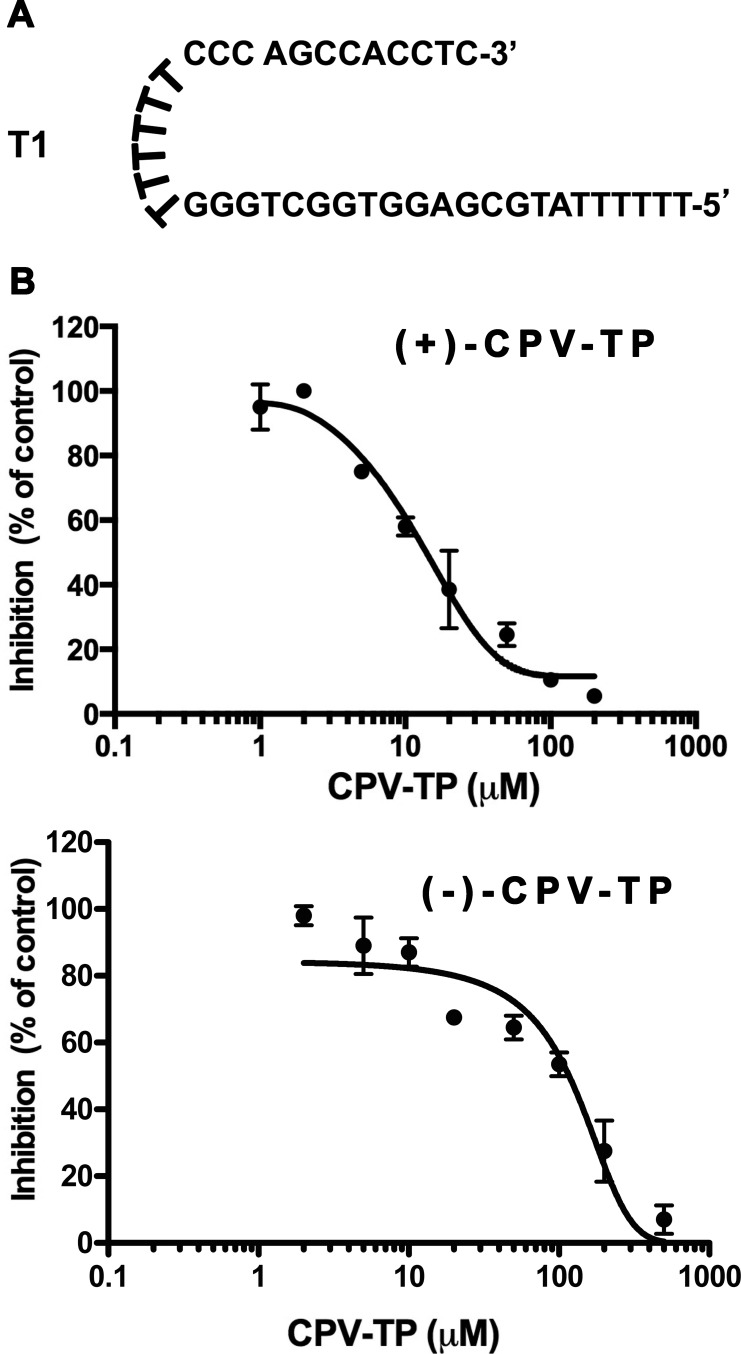
With two CPV-TP enantiomers in hand, we began to investigate their action on HCMV Pol. In an initial experiment, we incubated HCMV Pol and 32P-labeled dGTP with various concentrations of each of the CPV-TP enantiomers and tested dGTP incorporation by HCMV Pol using the primer template T1 (Fig. 2A) described previously (16) and then analyzed incorporation using polyacrylamide gels and autoradiography. This primer template accepts dGTP or a dGTP analogue as the first incorporated nucleotide opposite a C on the template strand. As shown in Fig. 2B, both of the enantiomers could inhibit HCMV Pol in a dose-dependent manner. The concentration that inhibited incorporation by 50% (IC50) obtained for (+)-CPV-TP was 12 μM, 10-fold lower than that of (−)-CPV-TP (120 μM), indicating that (+)-CPV-TP is more potent than (−)-CPV-TP in inhibiting HCMV Pol.
FIG 2.
Dose-dependent inhibition of HCMV Pol by CPV-TP enantiomers. (A) Primer template T1. (B) Inhibition of HCMV Pol incorporation of radiolabeled dGTP into T1 by various concentrations of CPV-TP enantiomers. Error bars show standard errors of the means.
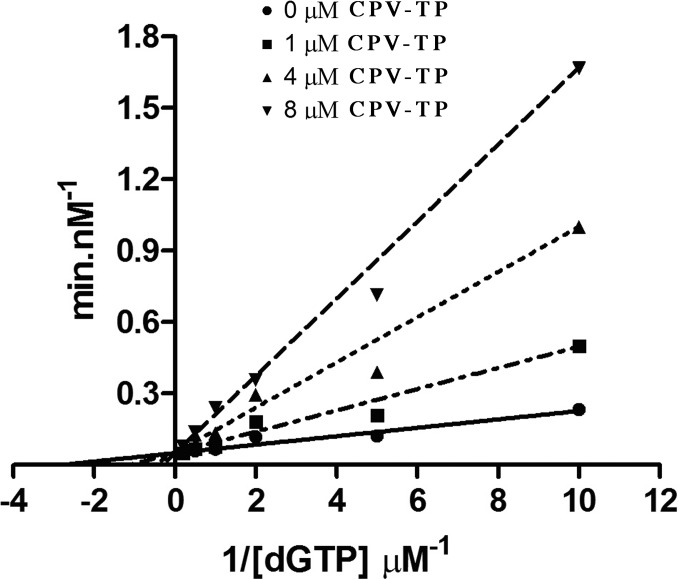
We next measured apparent Ki values for inhibition of dGTP incorporation by HCMV Pol, again using unlabeled primer template T1 and radiolabeled dGTP. As described previously (16), we took a steady-state approach, omitting the presumptive HCMV polymerase processivity subunit, UL44, to reduce any contribution of dissociation of polymerase from primer template to the rate of incorporation, and conducting the assays under Michaelis-Menten conditions. However, for these measurements, we used a filter-binding assay to measure incorporation, rather than the gel-based assay used previously. In this assay, HCMV Pol incorporated dGTP with an apparent Km of 1.1 μM and an apparent kcat of 50 min−1, both values roughly 2-fold higher than the values previously obtained using a labeled primer template, unlabeled dGTP, and polyacrylamide gel electrophoresis and autoradiography (16) (Table 1). We compared inhibition by GCV-TP, (+)-CPV-TP, and (−)-CPV-TP. Inhibition by all three drug triphosphates was, as expected, most consistent with a competitive model, based on fitting the data to that model versus other models and based on Lineweaver-Burk plots (example in Fig. 3). For (+)-CPV-TP, the probability of a competitive model was 99%, while the probabilities of noncompetitive and uncompetitive inhibition models were <1%. (+)-CPV-TP exhibited an apparent Ki value of 0.81 ± 0.2 μM. (This value is substantially lower than the IC50, most likely because the latter was determined using a higher concentration of Pol and a time point at which incorporation was no longer linear with time.) Consistent with its much higher IC50 than that of (+)-CPV-TP, (−)-CPV-TP exhibited a more than 20-fold higher apparent Ki value (18 ± 3.3 μM). Using this assay, we obtained an apparent Ki value for GCV-TP of 3.4 μM. This value is slightly lower than that obtained previously from an assay using radiolabeled primer template and unlabeled dGTP followed by polyacrylamide gel electrophoresis and autoradiography (16); this difference is likely explained by the previous assay overestimating the apparent Ki due to small amounts of incorporation of GCV-TP. Regardless, in the present assay, the apparent Ki for GCV-TP was about 4-fold higher than that of (+)-CPV-TP. Thus, (+)-CPV-TP was a more potent inhibitor of HCMV Pol than GCV-TP in this assay.
TABLE 1.
Apparent kinetic constants for substrates of HCMV Polc
| Substrate | Km, μM | kcat, min−1 |
|---|---|---|
| dGTPa | 0.44 ± 0.05 | 22 ± 0.70 |
| GCV-TPa | 5.5 ± 1.2 | 1.7 ± 0.17 |
| (+)-CPV-TPb | 0.51 ± 0.10 | 2.7 ± 0.2 |
| (−)-CPV-TPb | 15 ± 2.0 | 1.3 ± 0.1 |
Values for dGTP and GCV-TP are from reference 16.
Apparent Km values for CPV-TP enantiomers were determined by fitting data points to the Michaelis-Menten equation using GraphPad Prism (version 6). Apparent kcat values were determined by dividing apparent Vmax values by the enzyme concentrations.
Values are means ± standard errors on the basis of two independent replicates.
FIG 3.
Lineweaver-Burk plot for inhibition of dGTP incorporation by (+)-CPV-TP. Primer template T1 was incubated with HCMV Pol and various concentrations of dGTP containing [α-32P]dGTP in the presence of 0 μM (circles), 1 μM (squares), 4 μM (triangles), or 8 μM (inverted triangles) (+)-CPV-TP. The amount of dGTP incorporated into primer template in each reaction was quantified by scintillation counting, the values were converted to rates (nanomolar concentration per minute), and then the reciprocals of the concentrations and rates were plotted and the data were fitted (dashed lines) using GraphPad Prism (version 6).
Kinetic analysis of CPV-TP as a substrate for HCMV Pol.
Most antiviral nucleoside analogue triphosphates act as both inhibitor and substrate for viral polymerase. With the demonstration that CPV-TP is able to inhibit HCMV Pol, we further tested if CPV-TP is also a substrate of this enzyme. To investigate this, we measured incorporation of CPV-TP into DNA by HCMV Pol, using the same primer template T1, radiolabeled on its 5′ end, followed by polyacrylamide gel electrophoresis and autoradiography. We measured apparent Km and kcat values using the steady-state kinetic approach reported previously (16, 17) and used above, running all the experiments under the conditions required for Michaelis-Menten kinetic analysis. As shown in Table 1, the apparent Km value for (+)-CPV-TP was 0.51 ± 0.1 μM, very close to the apparent Ki value, and very close to the apparent Km obtained for dGTP using this assay, suggestive of high binding affinity between (+)-CPV-TP and HCMV Pol. The apparent kcat for (+)-CPV-TP was, however, roughly 10-fold lower than that of dGTP, suggesting that differences between the drug triphosphate and the natural nucleotide, such as a more rigid ring moiety and the lack of a 2′ position equivalent, mainly affect steps after binding. Consistent with its less potent inhibition of HCMV Pol, (−)-CPV-TP exhibited a ∼30-fold-higher apparent Km for HCMV Pol than did (+)-CPV-TP; however, there was only a 2-fold-lower apparent kcat. Notably, (+)-CPV-TP exhibited an apparent Km value almost 10-fold lower than that of GCV-TP and a slightly higher apparent kcat than that of GCV-TP, translating to an apparent 17-fold difference in kinetic efficiency. Thus, (+)-CPV-TP was a better substrate than GCV-TP for incorporation by HCMV Pol.
DNA termination induced by CPV-TP.
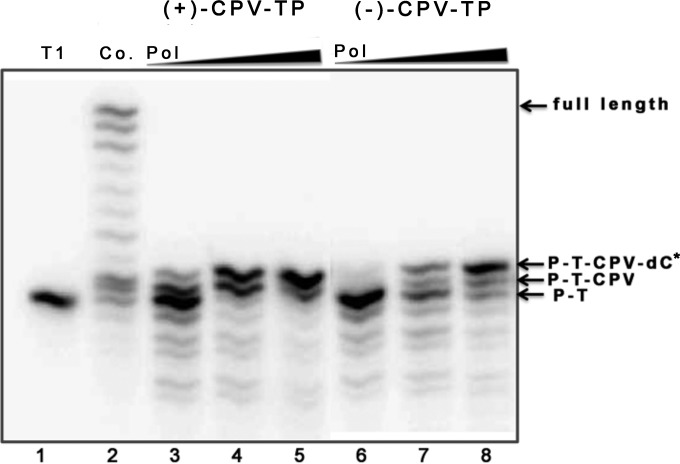
We wondered whether CPV-TP incorporation by HCMV Pol results in termination of DNA synthesis, as the drug triphosphate is not an obligate chain terminator; i.e., it has the equivalent of a 3′ hydroxyl moiety. To investigate this, we incubated radiolabeled primer template T1 with HCMV Pol in the presence of either (+)-CPV-TP; (−)-CPV-TP; or dGTP, dATP/dCTP/dTTP, and UL44, the presumed HCMV polymerase processivity subunit. In the presence of dGTP, as expected, the enzyme synthesized a variety of longer products, including ones corresponding to extension of the primer to the end of the template (full length) (Fig. 4, lane 2), and also produced some shorter products, as expected from its 3′-5′ exonuclease activity. For CPV-TP, as expected from the kinetic analysis, there was more efficient incorporation of the (+) enantiomer than the (−) enantiomer. Interestingly, in the presence of either enantiomer, HCMV Pol incorporated no more than one additional nucleotide after the drug triphosphate (n + 1 position) (Fig. 4), akin to the termination pattern induced by GCV-TP (16, 22–25), even when using higher concentrations of HCMV Pol (Fig. 4, lanes 3 to 8). There was also more accumulation of shorter 3′-5′ exonuclease products than when dGTP was present. In the absence of UL44, HCMV Pol alone also efficiently terminated DNA elongation after incorporating either of the CPV-TP enantiomers and an additional nucleotide (data not shown). Thus, like GCV-TP, CPV-TP is a nonobligate chain terminator of DNA synthesis by HCMV Pol; i.e., it causes chain termination despite having the equivalent of a 3′ hydroxyl group.
FIG 4.
DNA extension by HCMV DNA polymerase following CPV incorporation. Radiolabeled primer template T1 (Fig. 2A) was incubated with either dGTP, (+)-CPV-TP, or (−)-CPV-TP, dATP, dCTP, and dTTP, and various concentrations of HCMV Pol (7.2 nM, lanes 2, 3, and 6; 36 nM, lanes 4 and 7; 72 nM, lanes 5 and 8; increasing polymerase concentrations indicated as wedges above the panel) in the presence of UL44, and the products were analyzed by polyacrylamide gel electrophoresis and autoradiography. Lane 1, untreated radiolabeled T1; lane 2, DNA extension by HCMV Pol and UL44 in the presence of all four deoxynucleoside triphosphates without CPV-TP (Co.); lanes 3 to 5, DNA extension in the presence of (+)-CPV-TP; lanes 6 to 8, DNA extension in the presence of (−)-CPV-TP. The arrows at the right of the panel indicate the major species observed. P-T, unmodified primer template T1; P-T-CPV, T1 with either (+)-CPV-TP or (−)-CPV-TP added; P-T-CPV-dC*, T1 with either (+)-CPV-TP or (−)-CPV-TP and the next nucleotide added, the asterisk indicating that it has not been established that this residue is dC; full length, the largest product observed in lane 2.
Levels of CPV-TP in HCMV-infected cells treated with the 50% effective concentration of CPV.
It has previously been shown that following treatment of HCMV-infected cells with 2.5 μM CPV, which is 5 times the concentration that reduces viral replication by 50% (EC50), CPV-TP accumulates linearly with time, starting at 24 h postinfection (hpi) and reaching a level of 121 pmol/106 cells at 120 hpi (11). To assess the levels of CPV-TP at the EC50, we treated infected cells with 0.5 μM CPV (EC50). We found that CPV-TP levels at 120 hpi were 25.3 ± 6.7 pmol/106 cells. The relationship of these levels to the kinetic parameters determined above is discussed below.
DISCUSSION
CPV is a promising anti-HCMV drug. Previous work had established that CPV is phosphorylated to CPV-TP in HCMV-infected cells (11), that CPV inhibits HCMV DNA synthesis (3), and that mutations in the HCMV pol gene can confer CPV resistance (12). These results suggested that CPV-TP targets HCMV Pol to block viral DNA synthesis, but the action of CPV-TP on HCMV Pol had not been explored. In this study, we investigated the action of two enantiomeric forms of the triphosphate of CPV on HCMV Pol, comparing them with the triphosphate of the current standard treatment, GCV. We found that the (+) enantiomer was not only a more potent inhibitor of this enzyme than either the (−) enantiomer or GCV-triphosphate, it was also a better substrate for incorporation. As is true for GCV-triphosphate (16, 22–25), CPV-triphosphate proved to be a nonobligate chain terminator of DNA synthesis catalyzed by HCMV Pol with termination occurring after incorporation of the next nucleotide.
The identification of CPV resistance mutations in both the UL97 and pol genes (9, 10, 12) implies that both UL97 and Pol are important for the selectivity of CPV, much as they are for GCV (26). However, CPV exhibits more potent anti-HCMV activity than GCV (2, 3). Because higher levels of CPV-TP than GCV-TP were detected in HCMV-infected cells when CPV and GCV were used at equivalently effective concentrations, it was suggested that the higher potency of CPV may stem from more efficient phosphorylation of CPV by UL97 instead of more potent action of CPV-TP against HCMV Pol (11). In the present study, our results suggest that both UL97, which phosphorylates CPV more extensively than GCV, and Pol, which both is more potently inhibited by CPV-TP than GCV-TP and incorporates CPV-TP into DNA more efficiently than GCV-TP, are important for CPV's greater potency than GCV against wild-type (WT) HCMV replication (2). Our results further suggest that the greater potency of CPV than GCV against UL97-null HCMV (3) is due to more potent action of CPV-TP on HCMV Pol.
The HCMV UL97 kinase stereoselectively phosphorylates CPV to its (+)-monophosphate, and this enantiomer in turn is preferentially converted to its diphosphate by GMP kinase (6–8). Thus, it is likely that HCMV Pol is primarily exposed to the (+) enantiomer of CPV-TP. Here, successful synthesis of CPV-TP enantiomers allowed us to show that the (+) enantiomer is also a more potent inhibitor and better substrate of HCMV Pol. Thus, the (+) enantiomer is favored at multiple steps of antiviral action. This pattern of enantioselectivity is in line with the results of certain other methylenecyclopropane nucleoside analogues such as synguanol, whose (+) enantiomer is active against HCMV but whose (−) enantiomer is not (27–29).
It is interesting to compare the kinetic parameters for inhibition and incorporation of (+)-CPV-TP with the levels of CPV-TP found in infected cells following treatment with CPV at the EC50. Assuming an intracellular volume of 5 pl (30), this translates to a concentration of 5 μM at 120 hpi. Previous results (11) indicate that at the time when viral DNA replication starts (24 to 48 hpi) CPV-TP levels are 5- to 10-fold lower, which would correspond to a concentration of 0.5 to 1 μM. That range of concentrations is very similar to the apparent Ki and Km values that we determined for (+)-CPV-TP inhibition and incorporation, respectively.
Our current results show that CPV-TP targets HCMV Pol in a manner similar to that of GCV-TP: it competes with dGTP to inhibit HCMV Pol and also serves as a substrate of Pol whose incorporation results in chain termination after incorporation of one nucleotide (n + 1). GCV causes chain termination at the n + 1 position because the incorporated GCV prevents removal of the n + 1 nucleotide by the 3′-5′ exonuclease, while increasing the rate of removal of the n + 2 nucleotide by the exonuclease so that it at least matches the rate of extension of the n + 1-terminated primer (idling) (16). Because the methylenecyclopropane ring of CPV contains the equivalent of a 3′-hydroxyl group, it is not surprising that HCMV Pol is able to continue to synthesize DNA at least for one residue after CPV-TP incorporation. We have not identified the nucleotide incorporated after CPV-TP in our assays. The nucleotide that would Watson-Crick base-pair with the next position in the template is dCTP. However, given the rigid structure of CPV, we cannot exclude the possibility that this affects the structure of the primer so that other nucleotides can be incorporated at the n + 1 position.
Regardless, what happens next to prevent extension of the n + 1-terminated primer is not yet known. One possibility is that, as rapidly as the polymerase extends from the n + 1-terminated primer, the exonuclease excises the n + 2 nucleotide, similar to what is seen with GCV (16). We have also considered the possibility that incorporated CPV and/or the n + 1 nucleotide is degraded rapidly by the 3′-5′ exonuclease, which might explain the increased amounts of shorter exonuclease products in the presence of CPV-TP. However, we think that is unlikely, given that with increasing concentrations of DNA polymerase in the presence of CPV-TP, we observed both less accumulation of exonuclease products and greater accumulation of n + 1 chain-terminated products. That result is more consistent with the possibility that either the CPV-terminated primer template and/or the primer template terminated at the n + 1 position is a poor substrate for the 3′-5′ exonuclease, as observed for primer templates terminated at the n + 1 position following GCV-TP incorporation (16). Thus, we think it more likely that the increased amounts of exonuclease products in the presence of the CPV-TP enantiomers are due to their lower rate of incorporation relative to dGTP, thereby permitting more effective competition by the 3′-5′ exonuclease activity.
It is not clear which step of CPV-TP action is most important for CPV inhibition of viral replication. In the case of GCV, incorporation and chain termination are clearly important for drug action, as pol mutations that ablate exonuclease activity to permit full-length extension after incorporation of GCV confer resistance (16). Interestingly, such mutations do not confer CPV resistance (12). Why this occurs is unclear, but one speculation is that CPV incorporation into the primer might so drastically reduce the rate of extension of the primer by HCMV Pol that the absence of exonuclease activity has little impact. This and other possibilities are under investigation.
It is known that incorporation of nucleoside analogues into DNA is potentially mutagenic. Indeed, approved anti-HCMV nucleoside analogues exhibit significant genotoxicity (31). Although CPV can also be incorporated into DNA, so far, we have not detected any genotoxicity during preclinical toxicology studies (J. Brooks and T. L. Bowlin, unpublished data). A simple comparison of the structures of GCV, CPV, and dG reveals that GCV has 7 rotatable bonds, whereas CPV has only 5 and the natural nucleoside dG has 4. Thus, reduced entropy of CPV may help to explain its decreased genotoxicity relative to GCV.
The more potent antiviral activity, good bioavailability without prodrug modification (4), highly efficient phosphorylation by viral UL97 kinase, potent action on HCMV Pol, and lack of significant cross-resistance with GCV in clinical isolates endow CPV with very promising antiviral properties for the treatment or prophylaxis of HCMV diseases. Further examination of its mechanism of action and its continued development as an antiviral therapy appear warranted.
ACKNOWLEDGMENTS
H.C., T.L.B., J.Z., B.G.G., and D.M.C. conceived and designed the study; H.C., C.L., J.Z., and B.G.G. acquired data; H.C., C.L., J.Z., B.G.G., T.L.B., and D.M.C. analyzed and interpreted data; H.C., J.Z., and D.M.C. drafted the manuscript; and H.C., C.L., B.G.G., J.Z., T.L.B., and D.M.C. critically revised the draft and approved the final version.
We thank Gloria Komazin-Meredith for UL44 and for very helpful suggestions, L. M. Hrihorczuk (Wayne State University) for mass spectra, Ben Jagow (Drake University) for determining purity of the CPV-TP solutions, and John Drach for his helpful insights regarding the mechanism of action of CPV and enthusiastic support. We also thank Donald Moir (Microbiotix) for critical reading of the manuscript.
H.C. and T.L.B. work at Microbiotix, which is developing cyclopropavir. J.Z. is an inventor on patents regarding cyclopropavir and related compounds that have been licensed by Microbiotix.
Funding Statement
The content is solely the responsibility of the authors and does not necessarily represent the official views of the funding agencies.
REFERENCES
- 1.Zemlicka J. 2007. Methylenecyclopropane analogues of nucleosides as anti-herpes agents, p 113–165. In Clercq ED. (ed), Advances in antiviral drug design, 1st ed, vol 5 Elsevier Science, New York, NY. [Google Scholar]
- 2.Zhou S, Breitenbach JM, Borysko KZ, Drach JC, Kern ER, Gullen E, Cheng YC, Zemlicka J. 2004. Synthesis and antiviral activity of (Z)- and (E)-2,2-[bis(hydroxymethyl)cyclopropylidene]methylpurines and-pyrimidines: second-generation methylenecyclopropane analogues of nucleosides. J Med Chem 47:566–575. doi: 10.1021/jm030316s. [DOI] [PubMed] [Google Scholar]
- 3.Kern ER, Kushner NL, Hartline CB, Williams-Aziz SL, Harden EA, Zhou S, Zemlicka J, Prichard MN. 2005. In vitro activity and mechanism of action of methylenecyclopropane analogs of nucleosides against herpesvirus replication. Antimicrob Agents Chemother 49:1039–1045. doi: 10.1128/AAC.49.3.1039-1045.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bowlin TL, Brooks J, Zemlicka J. 2009. Preclinical pharmacokinetic, toxicokinetic and toxicology results for cyclopropavir, a promising new agent for the treatment of beta- and gamma-herpesviruses. Antiviral Res 82:A46–A47. doi: 10.1016/j.antiviral.2009.02.104. [DOI] [Google Scholar]
- 5.Mocarski ES, Shenk T, Griffiths PD, Pass RF. 2013. Cytomegaloviruses, p 1960–2014. In Knipe DM, Howley PM, Cohen JI, Griffin DE, Lamb RA, Martin MA, Racaniello VR, Roizman B (ed), Fields virology, 6th ed, vol 2 Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 6.Gentry BG, Kamil JP, Coen DM, Zemlicka J, Drach JC. 2010. Stereoselective phosphorylation of cyclopropavir by pUL97 and competitive inhibition by maribavir. Antimicrob Agents Chemother 54:3093–3098. doi: 10.1128/AAC.00468-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li C, Gentry BG, Drach JC, Zemlicka J. 2009. Synthesis and enantioselectivity of cyclopropavir phosphates for cellular GMP kinase. Nucleosides Nucleotides Nucleic Acids 28:795–808. doi: 10.1080/15257770903172720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gentry BG, Gentry SN, Jackson TL, Zemlicka J, Drach JC. 2011. Phosphorylation of antiviral and endogenous nucleotides to di- and triphosphates by guanosine monophosphate kinase. Biochem Pharmacol 81:43–49. doi: 10.1016/j.bcp.2010.09.005. [DOI] [PubMed] [Google Scholar]
- 9.Chou S, Bowlin TL. 2011. Cytomegalovirus UL97 mutations affecting cyclopropavir and ganciclovir susceptibility. Antimicrob Agents Chemother 55:382–384. doi: 10.1128/AAC.01259-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gentry BG, Vollmer LE, Hall ED, Borysko KZ, Zemlicka J, Kamil JP, Drach JC. 2013. Resistance of human cytomegalovirus to cyclopropavir maps to a base pair deletion in the open reading frame of UL97. Antimicrob Agents Chemother 57:4343–4348. doi: 10.1128/AAC.00214-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gentry BG, Drach JC. 2014. Metabolism of cyclopropavir and ganciclovir in human cytomegalovirus-infected cells. Antimicrob Agents Chemother 58:2329–2333. doi: 10.1128/AAC.02311-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chou S, Marousek G, Bowlin TL. 2012. Cyclopropavir susceptibility of cytomegalovirus DNA polymerase mutants selected after antiviral drug exposure. Antimicrob Agents Chemother 56:197–201. doi: 10.1128/AAC.05559-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.James SH, Hartline CB, Harden EA, Driebe EM, Schupp JM, Engelthaler DM, Keim PS, Bowlin TL, Kern ER, Prichard MN. 2011. Cyclopropavir inhibits the normal function of the human cytomegalovirus UL97 kinase. Antimicrob Agents Chemother 55:4682–4691. doi: 10.1128/AAC.00571-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zemlicka J. 2000. Enantioselectivity of the antiviral effects of nucleoside analogues. Pharmacol Ther 85:251–266. doi: 10.1016/S0163-7258(99)00062-5. [DOI] [PubMed] [Google Scholar]
- 15.Karkas JD, Germershausen J, Tolman RL, MacCoss M, Wagner AF, Liou R, Bostedor R. 1987. Stereochemical considerations in the enzymatic phosphorylation and antiviral activity of acyclonucleosides. I. Phosphorylation of 2′-nor-2′-deoxyguanosine. Biochim Biophys Acta 911:127–135. [DOI] [PubMed] [Google Scholar]
- 16.Chen H, Beardsley GP, Coen DM. 2014. Mechanism of ganciclovir-induced chain termination revealed by resistant viral polymerase mutants with reduced exonuclease activity. Proc Natl Acad Sci U S A 111:17462–17467. doi: 10.1073/pnas.1405981111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang L, Ishii KK, Zuccola H, Gehring AM, Hwang CBC, Hogle J, Coen DM. 1999. The enzymological basis for resistance of herpesvirus DNA polymerase mutants to acyclovir: relationship to the structure of α-like DNA polymerases. Proc Natl Acad Sci U S A 96:447–452. doi: 10.1073/pnas.96.2.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yan Z, Kern ER, Gullen E, Cheng YC, Drach JC, Zemlicka J. 2005. Nucleotides and pronucleotides of 2,2-bis(hydroxymethyl)methylenecyclopropane analogues of purine nucleosides: synthesis and antiviral activity. J Med Chem 48:91–99. doi: 10.1021/jm040149b. [DOI] [PubMed] [Google Scholar]
- 19.Moffatt JG. 1963. A general synthesis of nucleoside-5′ triphosphates. Can J Chem 42:599–604. [Google Scholar]
- 20.Hoard DE, Ott DG. 1965. Conversion of mono- and oligodeoxyribonucleotides to 5′-triphosphates. J Am Chem Soc 87:1785–1788. doi: 10.1021/ja01086a031. [DOI] [PubMed] [Google Scholar]
- 21.Rammler DH, Lapidot Y, Khorana HG. 1963. Studies on polynucleotides. XIX. The specific synthesis of C3″-C5″ inter-ribonucleotidic linkage. A new approach and its use in the synthesis of C3″-C5″-linked uridine oligonucleotides. J Am Chem Soc 85:1987–1997. [Google Scholar]
- 22.Reid R, Mar EC, Huang ES, Topal MD. 1988. Insertion and extension of acyclic, dideoxy, and ara nucleotides by herpesviridae, human α and human β polymerases. J Biol Chem 263:3898–3904. [PubMed] [Google Scholar]
- 23.Ilsley DD, Lee SH, Miller WH, Kuchta RD. 1995. Acyclic guanosine analogs inhibit DNA polymerases α, δ and ε with very different potencies and have unique mechanisms of action. Biochemistry 34:2504–2510. [DOI] [PubMed] [Google Scholar]
- 24.Frank KB, Chiou JF, Cheng YC. 1984. Interaction of herpes simplex virus-induced DNA polymerase with 9-(1,3-dihydroxy-2-propoxymethyl) guanine triphosphate. J Biol Chem 259:1566–1569. [PubMed] [Google Scholar]
- 25.Reardon JE. 1989. Herpes simplex virus type 1 and human DNA polymerase interactions with 2′-deoxyguanosine 5′-triphosphate analogues. J Biol Chem 264:19039–19044. [PubMed] [Google Scholar]
- 26.Coen DM, Richman DD. 2013. Antiviral agents, p 338–373. In Knipe DM, Howley PM, Cohen JI, Griffin DE, Lamb RA, Martin MA, Racaniello VR, Roizman B (ed), Fields virology, 6th ed, vol 1 Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 27.Chen X, Kern ER, Drach JC, Gullen E, Cheng YC, Zemlicka J. 2003. Structure-activity relationships of (S,Z)-2-aminopurine methylenecyclopropane analogues of nucleosides. variation of purine-6 substituents and activity against herpesviruses and hepatitis B virus. J Med Chem 46:1531–1537. doi: 10.1021/jm0205245. [DOI] [PubMed] [Google Scholar]
- 28.Qiu YL, Ksebati MB, Ptak RG, Fan BY, Breitenbach JM, Lin JS, Cheng YC, Kern ER, Drach JC, Zemlicka J. 1998. (Z)- and (E)-2-((Hydroxymethyl)cyclopropylidene)methyladenine and-guanine. New nucleoside analogues with a broad-spectrum antiviral activity. J Med Chem 41:10–23. [DOI] [PubMed] [Google Scholar]
- 29.Qiu YL, Geiser F, Kira T, Gullen E, Cheng YC, Ptak RG, Breitenbach JM, Drach JC, Hartline CB, Kern ER, Zemlicka J. 2000. Synthesis and enantioselectivity of the antiviral effects of (R,Z)-, (S,Z)-methylenecyclopropane analogues of purine nucleosides and phosphoralaninate prodrugs: influence of heterocyclic base, type of virus and host cells. Antiviral Chem Chemother 11:191–202. doi: 10.1177/095632020001100302. [DOI] [PubMed] [Google Scholar]
- 30.Mastrocola T, Lambert IH, Kramhøft B, Rugolo M, Hoffmann EK. 1993. Volume regulation in human fibroblasts: role of Ca2+ and 5-lipoxygenase products in the activation of the Cl- efflux. J Membr Biol 136:55–62. [DOI] [PubMed] [Google Scholar]
- 31.Coen DM, Schaffer PA. 2003. Antiherpesvirus drugs: a promising spectrum of new drugs and drug targets. Nat Rev Drug Discov 2:278–288. doi: 10.1038/nrd1065. [DOI] [PubMed] [Google Scholar]