Abstract
Bacterial topoisomerase functions are required for regulation of DNA supercoiling and overcoming the DNA topological barriers that are encountered during many vital cellular processes. DNA gyrase and topoisomerase IV of the type IIA bacterial topoisomerase family are important clinical targets for antibacterial therapy. Topoisomerase I, belonging to the type IA topoisomerase family, has recently been validated as a potential antitubercular target. The topoisomerase I activity has been shown to be essential for bacterial viability and infection in a murine model of tuberculosis. Mixture-based combinatorial libraries were screened in this study to identify novel bacterial topoisomerase I inhibitors. Using positional-scanning deconvolution, selective small-molecule inhibitors of bacterial topoisomerase I were identified starting from a polyamine scaffold. Antibacterial assays demonstrated that four of these small-molecule inhibitors of bacterial topoisomerase I are bactericidal against Mycobacterium smegmatis and Mycobacterium tuberculosis. The MICs for growth inhibition of M. smegmatis increased with overexpression of recombinant M. tuberculosis topoisomerase I, consistent with inhibition of intracellular topoisomerase I activity being involved in the antimycobacterial mode of action.
INTRODUCTION
Antimicrobial resistance is becoming increasingly prevalent among microbial pathogens, limiting the availability of clinical treatment options for bacterial infections (1). It is imperative to develop novel classes of antibacterial compounds, preferably against a new target to avoid cross-resistance due to mutations in current targets. Tuberculosis (TB) affects 9.6 million people a year and causes 1.5 million deaths each year (2). The problem presented by multidrug resistance is illustrated by the 480,000 cases of multidrug-resistant TB (MDR-TB) that do not respond to first-line treatment drugs, with around 10% of these cases being extensively drug-resistant tuberculosis (XDR-TB) cases that are resistant to even some of the second-line drugs (2, 3). New combinations of anti-TB drugs are needed to treat the MDR-TB and XDR-TB cases.
Topoisomerases are essential in every organism for regulation of DNA topology so that vital cellular processes, including DNA replication, transcription, recombination, and repair, can proceed without hindrance (4, 5). Type IIA topoisomerases cut and rejoin a double strand of DNA during catalysis (6). DNA gyrase and topoisomerase IV are prokaryotic type IIA topoisomerases that have been extensively explored as validated targets for antibacterial therapy in the clinic (7, 8). At least one type IA topoisomerase is present in every bacterial pathogen to resolve topological barriers that require the cutting and rejoining of a single strand of DNA and passage of DNA through the transient break (9, 10). Topoisomerase I is the major type IA topoisomerase activity responsible for preventing excessive negative supercoiling in bacteria (11, 12). Recently, bacterial topoisomerase I has received much interest as a novel antibacterial drug target (9, 13). Escherichia coli topoisomerase I is the most extensively studied type IA topoisomerase, with crystal structures of the covalent cleavage complex (14) and full-length enzyme-DNA complex (15) available. Inhibition of E. coli topoisomerase I by endogenous polypeptide inhibitors (16, 17, 18, 19) can lead to cell killing, even though compensatory mutations could allow E. coli strains with topA deletions to be viable (20, 21). There is also evidence that topoisomerase I function is essential for a number of bacterial pathogens, including Streptococcus pneumoniae (22) and Helicobacter pylori (23). There is only one type IA topoisomerase encoded by the genomes of mycobacteria. Mycobacterium tuberculosis topoisomerase I (MtbTopI) has been demonstrated in genetic studies to be essential for viability in vitro (24, 25) and in vivo (25). Experimental data showed that the MIC of select small molecules against M. tuberculosis can be shifted by overexpression of topoisomerase I (25, 26), further validating topoisomerase I as a vulnerable target in M. tuberculosis for chemical inhibition.
Many of the small molecules identified previously as bacterial topoisomerase I inhibitors are DNA intercalators (22, 26–28) or minor groove binders (29, 30) that would not be attractive candidates for antibiotic development. Different molecular scaffolds need to be explored in the efforts to discover inhibitors of bacterial topoisomerase I. In this study, we explored the Torrey Pines Institute for Molecular Studies (TPIMS) scaffold-ranking libraries (31) that consist of more than 30 million small molecules for identification of novel bacterial topoisomerase I inhibitors. This approach was utilized successfully in the identification of antimicrobials that inhibit tyrosine recombinases and Holliday junction-resolving enzymes (32). Each scaffold library mixture contains only compounds with the same specific core scaffold, so only close structural analogues are contained in each scaffold library mixture. A single scaffold library mixture comprised of 345,600 compounds was selected based on inhibition of the relaxation activity of E. coli topoisomerase I at a 50-μg/ml scaffold library mixture concentration. The 345,600 compounds contained in the promising scaffold library mixture sample were then systematically formatted into 216 different mixture samples, a technique known as positional scanning (33). The results from screening these 216 samples identified R groups that correlated with potent activity against topoisomerase I and were used to design 80 individual compounds for synthesis. These individual compounds were assayed for inhibition of E. coli and M. tuberculosis topoisomerase I, as well as antibacterial activity. A subset of these compounds inhibited the growth of Mycobacterium smegmatis and M. tuberculosis. Overexpression of M. tuberculosis topoisomerase I in M. smegmatis increased the MIC of four of the topoisomerase I inhibitors, consistent with intracellular topoisomerase I activity being the target of their antimycobacterial mode of action.
MATERIALS AND METHODS
Topoisomerase enzymes.
Recombinant E. coli topoisomerase I (EcTopI) and M. tuberculosis topoisomerase I (MtbTopI), expressed in E. coli, were purified as described previously (34, 35). E. coli DNA gyrase was obtained from New England BioLabs. Human topoisomerase I and topoisomerase IIα were purchased from TopoGen.
Scaffold-ranking library.
The TPIMS scaffold-ranking library tested contains one sample for each of the 50 positional-scanning libraries. Each of these samples contains an approximately equal molar amount of each compound in that library. Sample 2229 in the scaffold-ranking library contains 345,600 polyamines in approximately equal molar amounts. These scaffold-ranking library samples can be prepared by mixing the cleaved products of the complete positional-scanning library, or they can be synthesized directly as a single mixture, as was the case with sample 2229 (31, 36–38).
Positional-scanning library 2229 and individual polyamines.
The polyamine positional-scanning library 2229 and all polyamine individual compounds were synthesized using the scheme described in Fig. S1 in the supplemental material (detailed chemical characterization for the individual compounds can be found in the supplemental material). The positional-scanning library contains 216 samples and incorporates both individual and mixtures of amino acids (R1 and R2) and carboxylic acid (R3). The synthetic technique facilitates the generation of information regarding the likely activity of individual compounds from the screening of the library (31, 33, 39). Equimolar isokinetic ratios have been previously determined and calculated for each of the amino and carboxylic acids utilized for the respective mixtures (40, 41). The polyamine library 2229 has a total diversity of 345,600 compounds (60 × 60 × 96 = 345,600). The R1 and R2 positions were each derived from 60 amino acids, and the R3 position was derived from 96 carboxylic acids.
Bacterial strains and plasmids.
Bacterial strains and plasmids used in this study are listed in Table 1. The M. tuberculosis topA gene encoding topoisomerase I (MtbTopI) was PCR amplified using primers containing BamHI and ScaI restriction sites and cloned into the TA cloning vector (pCR2.1-TOPO from Invitrogen), resulting in pTAmtop. Plasmid pKW08-Lx (gift of Brian Robertson, Addgene) is an E. coli-mycobacterium shuttle vector containing the luxAB genes under the control of a tetracycline-inducible promoter, TetRO (42). The BamHI-ScaI fragment of pKW08-Lx containing luxAB genes was replaced by the BamHI-ScaI fragment from pTAmtop containing the M. tuberculosis topA gene. The resulting plasmid, pTA-M+, expressed tetracycline-inducible MtbTopI. A control plasmid, pKW-nol, was also constructed when pKW08-Lx was religated following the removal of the luxAB genes. These plasmids were electroporated into M. smegmatis mc2155 separately, and the resulting strains were used in growth inhibition assays.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Genotype or description | Source (reference) |
|---|---|---|
| Strains | ||
| M. smegmatis mc2155 | ATCC | |
| Mtb:lux | M. tuberculosis CDC1551 containing the pMV306hsp+LuxG13 plasmid | This work |
| Plasmids | ||
| pKW08-Lx | Tetracycline-inducible expression plasmid | Gift from Brian Robertson (Addgene plasmid 25012) (42) |
| pTAmtop | PCR clone of topA gene coding for MtbTopI | This work |
| pTA-M+ | Luciferase gene in pKW08-Lx replaced by M. tuberculosis topA | This work |
| pTA-nol | pKW08-derived clone with no insert | This work |
| pMV306hsp+LuxG13 | Integrating E. coli-mycobacteria shuttle vector expressing an optimized luxCDABE operon | Gift from Brian Robertson and Siouxsie Wiles (Addgene plasmid 26161) (43) |
For analysis of the compounds' activity against M. tuberculosis, M. tuberculosis CDC1551 constitutively expressing the luxCDABE operon from an integrated pMV306hsp+LuxG13 plasmid carrying an aph kanamycin resistance cassette (the Mtb::lux strain) was cultured in Middlebrook 7H9 broth containing 0.5% glycerol, 0.05% Tween 80, and OADC supplement (oleic acid, albumin, dextrose, catalase) or 7H10 agar medium supplemented with OADC. Kanamycin (50 μg/ml) was added to broth cultures for selection where appropriate. pMV306hsp+LuxG13 was a gift from Brian Robertson and Siouxsie Wiles (Addgene plasmid 26161) (43).
Bacterial topoisomerase I relaxation activity inhibition assay.
The relaxation activity of bacterial topoisomerase I was assayed in a buffer containing 10 mM Tris-HCl, pH 8.0, 50 mM NaCl, 0.1 mg/ml gelatin, and 0.5 mM MgCl2. One microliter of the compounds dissolved in 5% dimethylformamide (DMF) or solvent alone was mixed with 10 μl of the reaction buffer containing 10 ng of enzyme before the addition of 9 μl of reaction buffer containing 160 ng of supercoiled pBAD/Thio plasmid DNA purified by cesium chloride gradient as the substrate. Following incubation of the mixtures at 37°C for 30 min, the reactions were terminated by the addition of 4 μl of 50% glycerol, 50 mM EDTA, and 0.5% (vol/vol) bromophenol blue and the mixtures were analyzed by agarose gel electrophoresis. The gels were stained in ethidium bromide and photographed under UV light.
DNA gyrase supercoiling inhibition assay.
DNA gyrase (obtained from New England BioLabs) supercoiling assays were carried out by mixing the compounds with 2 U of the enzyme in reaction buffer supplied by the manufacturer, followed by the addition of 300 ng of relaxed covalently closed circular DNA, for a final reaction mixture volume of 20 μl. The samples were incubated at 37°C for 30 min before the reactions were terminated by the addition of a buffer containing 5% SDS, 0.25% bromophenol blue, and 25% glycerol. The reaction mixtures were then analyzed by agarose gel electrophoresis.
Human topoisomerase I relaxation inhibition assay.
Human topoisomerase I (obtained from TopoGen) relaxation assays were carried out with 0.5 U of enzyme in reaction buffer supplied by the manufacturer. The enzyme was mixed with the indicated concentration of compound dissolved before 160 ng of supercoiled pBAD/Thio plasmid DNA was added in the same buffer, for a final volume of 20 μl. Following incubation of the mixtures at 37°C for 30 min, the reactions were terminated with a buffer containing 5% SDS, 0.25% bromophenol blue, and 25% glycerol, and the mixtures were analyzed by agarose gel electrophoresis.
Human topoisomerase IIα decatenation inhibition assay.
Human topoisomerase IIα (obtained from TopoGen) assays were carried out by adding the compounds to 180 ng of kinetoplast DNA (kDNA) in the buffer supplied by the manufacturer before the addition of 2 U of the enzyme. The samples were incubated for 15 min at 37°C before the addition of 4 μl of stop buffer containing 5% Sarkosyl, 0.25% bromophenol blue, and 25% glycerol. The reaction mixtures were then analyzed by electrophoresis in 1% agarose gels containing 0.5 μg/ml ethidium bromide before being photographed under UV light.
Assay of M. smegmatis growth inhibition and loss of viability.
Cells were cultured from individual colonies in Middlebrook 7H9 medium supplemented with 0.2% glycerol; 0.05% Tween 80; and 10% albumin, dextrose, sodium chloride (ADN) for 1 day. For the strains with plasmids, 50 μg/ml hygromycin B was included. The culture was then diluted 1:500 with the same medium without ADN and grown until the optical density at 600 nm (OD600) was between 0.6 and 0.7. After adjustment of OD600 to 0.5, the cultures were diluted 1:10, and 50 μl was added to clear round-bottom 96-well plates containing 50 μl of serially diluted compounds in the same medium. The plates were then incubated with shaking at 37°C for 48 h, with absorbance readings being taken approximately every 4 h. The MIC was determined to be the minimum compound concentration that prevented increase in absorbance over time.
To determine if the compounds found to inhibit growth of the mycobacteria are bactericidal, the cells from the MIC assays treated with compound concentrations above the MICs were plated on LB-hygromycin agar plates after treatment with the compounds and allowed to grow at 37°C. The colonies formed from 0.1 ml of cultures were counted after 3 days. The CFU values determined were divided by the CFU of cells before treatment to determine the ratio of loss of viability.
Assay of M. tuberculosis growth inhibition by bioluminescence and CFU assays.
Stock solutions of compounds 2471-12, 2471-24, and 2471-80 at 25 mM in 100% dimethyl sulfoxide (DMSO) were diluted in water to obtain a 5× working stock (500 μM in 2% DMSO). A stock solution of compound 2471-36 at 1.3 mM in 5% DMF was used directly for assays, diluted into medium to achieve the desired concentrations starting at 100 μM in 0.4% DMF. Ten-point 2-fold serial dilutions of compounds in sterile medium were done using a robotic liquid handler (BioTek Precision), with 1/5 volume (6 μl) of compound per well of solid white 384-well plates. Final concentrations of compounds ranged from 100 μM to 0.195 μM, with each one tested in triplicate. Cultures of the Mtb::lux strain at mid-log phase were diluted in 7H9 OADC to a final optical density (600 nm) of 0.01 before addition of 24 μl/well to plates containing compounds. After 5 days of incubation at 37°C, luminescence was measured using a Synergy H4 microplate reader (BioTek). Samples were taken from each well, serially diluted in phosphate-buffered saline (PBS) plus 0.05% Tween 80, and plated on 7H10 agar. Each 384-well plate contained 8 replicate wells of the following controls, which were also plated in triplicate for CFU counting: 0.4% DMSO and 10 μM, 1 μM, and 0.1 μM rifampin and isoniazid.
Resazurin-based cytotoxicity assay.
J774 macrophages were cultured in Dulbecco's minimal essential medium (DMEM) supplemented with 10% heat-inactivated fetal bovine serum. Cells (2.5 × 104/well) were seeded in black solid-bottom 384-well plates 6 h prior to addition of compounds and controls (0.4% DMSO or 0.4% DMF and 2% Triton X-100 [final concentrations]). Cell survival was determined after 24 h by resazurin reduction measurement. Addition of 0.02 mg/ml resazurin was followed by 4 h of incubation at 37°C and subsequent fluorescence reading (560 nm/590 nm) using a Synergy H4 (BioTek) plate reader. Data are presented as percent viability compared to the vehicle (0.4% DMSO or 0.4% DMF) control with 100% being noncytotoxic and 2% Triton X-100 control used to define 0% viable.
Anisotropy assay of DNA binding by MtbTopI.
Binding of MtbTopI to single-stranded DNA was assayed by change of anisotropy of a 59-base oligonucleotide modified with 6-carboxyfluorescein at the 3′ end (synthesized by Biosearch Technologies) as previously described (44). All measurements were performed at room temperature in 0.5 ml of binding buffer (50 mM Tris-HCl, pH 7.5, 100 mM NaCl, 0.1 mM EDTA). Excitation and emission wavelengths were 495 and 520 nm, respectively, with the excitation and emission slits set at 5 and 10 nm, respectively. Data were collected using the Advanced Reads program on a Varian Cary Eclipse fluorescence spectrophotometer.
RESULTS
Selection of a polyamine scaffold as a platform for inhibitors of bacterial topoisomerase I from screening of scaffold-ranking library mixtures.
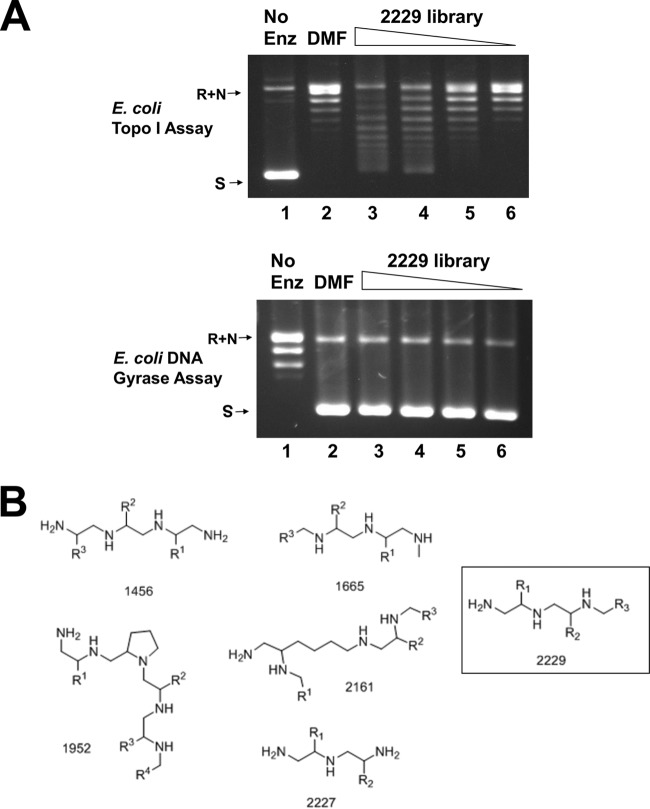
In order to identify novel inhibitors of bacterial topoisomerase I, 50 scaffold-ranking library mixtures were assayed at 100 μg/ml for inhibition of the relaxation activity of EcTopI. Scaffold-ranking library 2229 was the only library mixture that showed >50% inhibition of the EcTopI relaxation activity (Fig. 1A). This is not due to nonspecific inhibition of the topoisomerase activity by the positively charged polyamine structure, as the supercoiling activity of the type IIA topoisomerase DNA gyrase was not inhibited by library 2229 compounds at similar concentrations (Fig. 1A). There are five other polyamine scaffolds (Fig. 1B) among the 50 scaffold-ranking mixtures tested in this study, and only library 2229 was found to inhibit EcTopI relaxation activity at 100 μg/ml.
FIG 1.
Inhibition of E. coli topoisomerase I relaxation activity by scaffold-ranking library 2229. (A) Scaffold library mixture 2229 inhibits E. coli topoisomerase I but not DNA gyrase. Lanes 1, control reaction mixture with no enzyme added; lanes 2, DMF control; lanes 3 to 6, scaffold library mixture 2229 at concentrations of 100, 50, 25, and 12.5 μg/ml, respectively. (B) Structures of the six polyamine scaffolds.
Individual small-molecule inhibitors of bacterial topoisomerase I.
The 216 samples in the positional-scanning library 2229 were each assayed for inhibition of the relaxation activity of EcTopI. The effects of different R1, R2, and R3 substitutions in the positional-scanning library on the 50% inhibitory concentration (IC50) values for inhibition of EcTopI relaxation activity were analyzed. Several structure-activity relationship (SAR) trends are apparent within the positional-scanning data. At the R1 position, there is a preference for positively charged functional groups or large aromatics, which produce IC50s for inhibition of the relaxation activity of EcTopI in the low-micromolar range. Simple aliphatic substitutions at R1 produce compounds with IC50s greater than 100 μM. The positional-scanning data were used to direct the synthesis of individual compounds. In cases where SAR trends were observed (as for R1 described above), substitutions were selected to prepare those compounds most likely to be active. In the other instances, individual substitutions were selected to generate diversity around the scaffold. This information was utilized for the design of the 2471 series of 80 individual compounds. The IC50s for the relaxation activity of EcTopI for these 80 individual compounds were found to range from 1.25 μM to >160 μM (see Table S1 in the supplemental material). While the 18 individual 2471 series compounds with IC50s at 20 μM or less for inhibition of EcTopI relaxation activity all contain a naphthyl group at the R3 position (see Table S1), a number of additional compounds were made with alternative substitutions at R3 (i.e., dihydroxyphenyl, halogenated phenyls, aliphatic groups, etc.); however, all of these additional compounds exhibit IC50s greater than 20 μM for inhibition of EcTopI relaxation activity (see Table S1), indicating the importance of the naphthyl group in this position.
Inhibitors with antimycobacterial activity sensitive to overexpression of topoisomerase I.
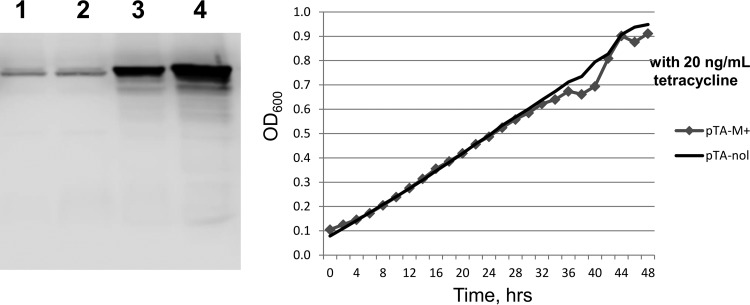
The 18 individual 2471 series compounds with IC50s at 20 μM or less did not show antibacterial activity when assayed against E. coli strains that include MG1655, AS17 (topAts) (45), and AS17 with a ΔtolC::Kan mutation (46) introduced by P1 phage transduction. Inhibition of MtbTopI relaxation activity was similar to inhibition of EcTopI (Table 2). A subset of these 18 compounds was able to inhibit the growth of M. smegmatis (Table 2). Transformants of M. smegmatis overexpressing recombinant MtbTopI or with the cloning vector only were used to investigate if topoisomerase I activity is involved in the antimycobacterial mode of action. Overexpression of recombinant MtbTopI was achieved by replacing the luciferase gene in pKW08-Lx plasmid with the MtbTopI gene. In the absence of tetracycline inducer, the level of topoisomerase I in M. smegmatis transformed by pTA-M+ was found by Western blot and densitometry analysis to be 6-fold higher than the topoisomerase I level in M. smegmatis transformed with control plasmid pTA-nol (Fig. 2). Addition of 20 ng/ml tetracycline can further increase the overexpression to ∼9-fold, but there was also an observable effect of growth inhibition from MtbTopI overexpression induced by tetracycline (Fig. 2). Therefore, the effect of MtbTopI overexpression on sensitivity to topoisomerase I inhibitors was tested in cultures grown in the absence of tetracycline.
TABLE 2.
Antimycobacterial activity and cytotoxicity of the most potent bacterial topoisomerase I inhibitors characterized in this study (IC50 of ≤20 μM)
| Compound | IC50 (μM) for topoisomerase I |
MIC (μM) for M. smegmatis mc2155 | |
|---|---|---|---|
| EcTopI | MtbTopI | ||
| 2471-18 | 1.25 | 1.25 | 50 |
| 2471-27 | 1.25 | 1.9 | 25 |
| 2471-15 | 1.9 | 0.625 | 50 |
| 2471-3 | 2.5 | 2.5 | 50 |
| 2471-6 | 2.5 | 1.25 | 25 |
| 2471-30 | 2.5 | 2.5 | 25 |
| 2471-80 | 2.5 | 5 | 6.25 |
| 2471-9 | 5 | 5 | 50 |
| 2471-33 | 5 | 5 | 50 |
| 2471-12 | 7.5 | 7.5 | 12.5 |
| 2471-21 | 7.5 | 10 | 50 |
| 2471-24 | 10 | 7.5 | 25 |
| 2471-36 | 20 | 15 | 25 |
| 2471-76 | 20 | 10 | 12.5 |
FIG 2.
Overexpression of recombinant MtbTopI in M. smegmatis. (Left) Western blot analysis of mycobacterial topoisomerase I levels. The whole-cell lysates of M. smegmatis transformed with pTA-nol (lanes 1 and 2) or pTA-M+ (lanes 3 and 4) cultured with no inducer added (lanes 1 and 3) or induced with 20 ng/ml tetracycline (lanes 2 and 4) were analyzed by Western blotting using rabbit polyclonal antibodies against MtbTopI. (Right) Growth of induced cultures monitored by absorbance at 600 nm.
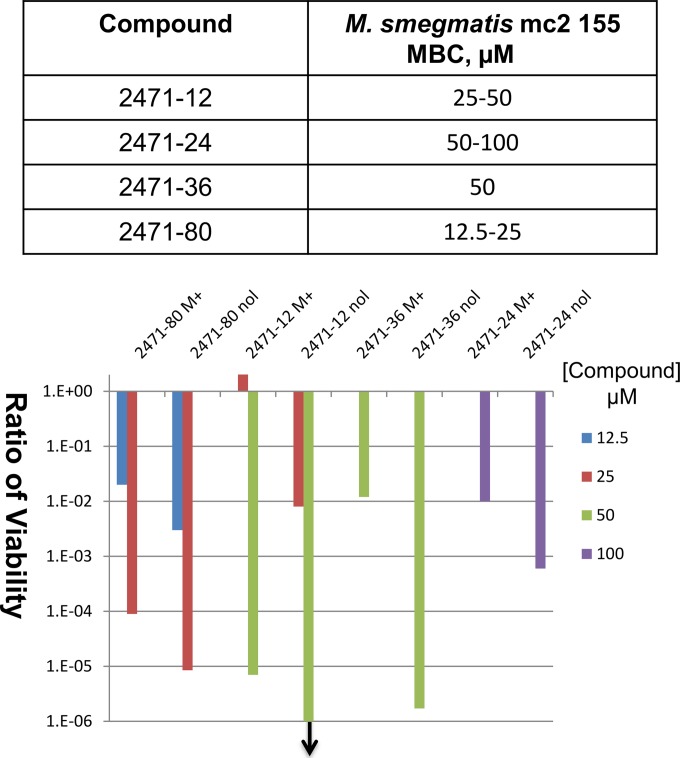
Growth inhibition assays showed that the MIC values against M. smegmatis for four of the compounds, 2471-12, 2471-24, 2471-36, and 2471-80, were increased by 2- to 4-fold with overexpression of recombinant MtbTopI (Table 3). The overexpression of MtbTopI has no effect on the MIC of ciprofloxacin, a fluoroquinolone drug that targets the related type II topoisomerase enzyme gyrase, thus showing that topoisomerase I overexpression did not have nonspecific effects on the growth inhibition even by a drug targeting a related enzyme. Determination of viable colony counts demonstrated that these compounds are bactericidal against M. smegmatis, and loss of viability after treatment with the compounds for 46 h was also less severe when recombinant MtbTopI was overexpressed (Fig. 3).
TABLE 3.
Effect of recombinant MtbTopI overexpression on MICs of select compounds against M. smegmatis mc2155

FIG 3.
Effect of MtbTopI overexpression on the bactericidal effect of selected topoisomerase I inhibitors. The minimal bactericidal concentration (MBC) values against M. smegmatis mc2155 are shown in the table. The loss of viability following treatment with compounds for 44 h was compared between transformants overexpressing MtbTopI (M+) from pTA-M+ and with control vector pTA-nol (nol). The downward arrow indicates that no viable colonies could be detected following 44 h of treatment with compound 2471-12 at a 50 μM concentration.
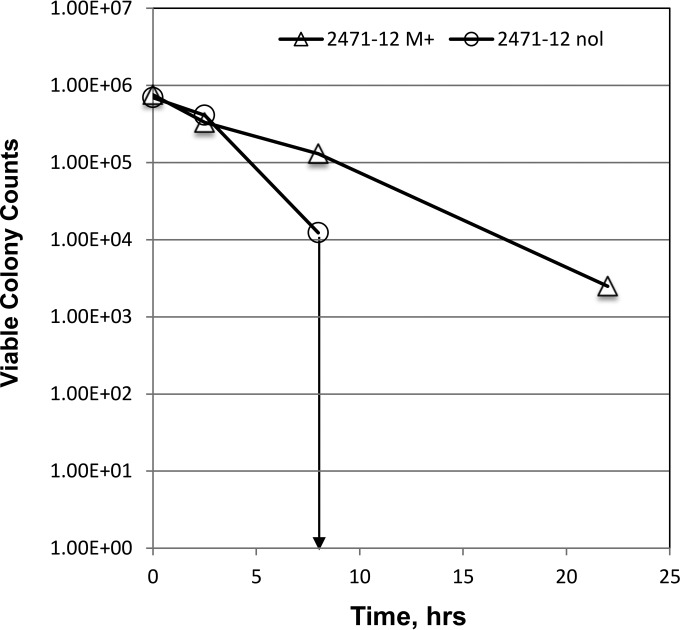
The time course of cell killing by 2471-12 at a 50 μM concentration (Fig. 4) showed rapid loss of viable colony counts following compound treatment. A rapid cell-killing mechanism is favorable for the quick reduction of bacterial burden and to limit the enrichment of new resistant mutants. At 22 h posttreatment, viable colonies were not observed for M. smegmatis transformed with control cloning vector (nol), while overexpression of MtbTopI in M. smegmatis (M+) provided significant protection against the rapid cell killing, supporting the involvement of topoisomerase I in a specific cell-killing mechanism.
FIG 4.
Effect of MtbTopI overexpression on rapid killing of M. smegmatis by inhibitor 2471-12. Viable colony counts from 0.1 ml of M. smegmatis transformed with either pTA-M+ (M+) overexpressing MtbTopI or pTA-nol (nol) control vector were determined prior to and following treatment with 50 μM compound 2471-12. The downward arrow indicates that no viable colonies could be detected at time points later than 8 h following addition of compound.
Bactericidal activity of TP-2471 polyamine analogues against M. tuberculosis.
Four TP-2471 analogues (2471-12, 2471-24, 2471-36, and 2471-80) with potent activity against purified MtbTopI and topoisomerase I-specific bactericidal activity against M. smegmatis were tested for antibacterial activity against M. tuberculosis using an autoluminescent strain of M. tuberculosis CDC1551 expressing an optimized bacterial luxCDABE operon (the Mtb::lux strain). Dose-response curves revealed that all four compounds had IC50s of ∼3 to 9 μM for 50% reduction in luminescence signal compared to controls, a relatively potent starting point for a hit series (Table 4). Validation of these results by CFU further revealed that 2471-80 exerted particularly potent dose-dependent bactericidal activity, resulting in ∼2- to 3-log-greater killing than 2471-12 or 2471-24 at 50 μM or higher concentrations (3- to 4-log reduction versus 1 log) (data not shown). Although additional medicinal-chemistry optimization is needed during hit-to-lead development, resazurin-based viability assays using J774 macrophages yielded 50% cytotoxicity concentrations (CC50s) of ∼40 μM for 2471-36 and 2471-80 and ∼100 μM for 2471-12 and 2471-24, corresponding to selectivity index values (SI = CC50/IC50) of >10 for three of the compounds (Table 4). Thus, TP-2471 compounds identified as specific inhibitors of the essential MtbTopI enzyme are able to penetrate the mycobacterial cell wall to access their intracellular target and kill M. tuberculosis.
TABLE 4.
Selective antimicrobial activity against M. tuberculosis
| Compound | IC50 (μM) for Mtb::lux straina | CC50 (μM) for J774b | SIc |
|---|---|---|---|
| 2471-12 | 6.8 | 104.3 | 15.4 |
| 2471-24 | 8.4 | 101.4 | 12 |
| 2471-36 | 3.3 | 36.49 | 11.2 |
| 2471-80 | 5.9 | 40.39 | 6.9 |
Drug concentration leading to 50% reduction in luminescence signal compared to controls.
Drug concentration leading to 50% loss of viability in J774 macrophage cell line after 24 h of exposure.
SI = CC50/IC50.
Specificity of selected bacterial topoisomerase I inhibitors.
Inhibition of MtbTopI relaxation activity by compounds 2471-12, 2471-24, 2471-36, and 2471-80 was comparable to inhibition of EcTopI. Inhibition of the supercoiling of DNA gyrase, relaxation activity of human topoisomerase I, and decatenation activity of human topoisomerase IIα required >10-fold-higher compound concentrations (Table 5). It is therefore unlikely that the inhibition observed for bacterial topoisomerase I is due to nonspecific interaction of the positively charged compounds with DNA.
TABLE 5.
IC50s for inhibition of topoisomerase activitiesa
| Compound | IC50 (μM) for enzyme: |
|||
|---|---|---|---|---|
| M. tuberculosis topoisomerase I | E. coli DNA gyrase | Human topoisomerase I | Human topoisomerase IIα | |
| 2471-12 | 7.5 | 160 | 80 | 80 |
| 2471-24 | 7.5 | >160 | >160 | 160 |
| 2471-36 | 15 | >160 | >160 | >160 |
| 2471-80 | 5 | 160 | 80 | 80 |
As an example, gel images of the topoisomerase inhibition assay for 2471-80 are shown in Fig. S2 in the supplemental material.
Inhibition of MtbTopI activity at a step following formation of enzyme-DNA complex.
Binding of MtbTopI to single-stranded DNA was monitored by the change in anisotropy of an oligonucleotide substrate modified with a fluorescence reporter at the 3′ end. Addition of 2471-12 and 2471-80 at their IC50s did not inhibit the binding of MtbTopI to this oligonucleotide substrate (see Fig. S3 in the supplemental material). The Kd (dissociation constant) derived from the anisotropy data was found to be decreased from 1.2 nM in the absence of inhibitor to 0.78 nM in the presence of 2471-80 at its IC50. These polyamine-based inhibitors likely interact with the enzyme-DNA complex to inhibit the overall relaxation activity at a step following the initial binding of enzyme to DNA.
DISCUSSION
Inhibitors against type IIA and type IB topoisomerases have been developed into important antibacterial and anticancer drugs. Bacterial type IA topoisomerases can potentially be high-value targets for general or pathogen-specific antibacterial therapy and merit attention because of the need for new antibiotics against a novel target. Different molecular scaffolds with structures not related to previously identified bacterial topoisomerase I inhibitors were explored in this study in order to discover novel chemical space for selective inhibition of type IA topoisomerase activity. Five polyamine scaffolds were represented in the 50 scaffold-ranking library mixtures screened initially for inhibition of EcTopI, and only scaffold 2229 was found to inhibit the relaxation activity of EcTopI. Using the positional-scanning approach, substitutions most favorable for inhibition of EcTopI relaxation activity were identified. Selectivity for type IA topoisomerase inhibition was observed both for the initial 2229 scaffold-ranking library mixture and subsequently for the individual compounds ranked highest for strong inhibition of EcTopI relaxation activity. These compounds showed comparable inhibition of MtbTopI activity. The results presented here demonstrate that combination of specific substitutions on this polyamine molecular scaffold can lead to compounds that represent a new class of bactericidal antimycobacterial agents with topoisomerase I as a target for their cellular mode of action.
The lack of whole-cell activity against E. coli observed for these inhibitors may be due to lack of compound uptake by E. coli. Alternatively, the presence of two type IA topoisomerases in that organism may be a factor. One of the goals of this study that has not been achieved was to identify compounds that can act as bacterial topoisomerase I poison inhibitors. The antibacterial activity of topoisomerase poison inhibitors results not from elimination of the topoisomerase activity but from corruption of the covalent topoisomerase reaction intermediate formed after DNA cleavage by topoisomerase. While the action of topoisomerase poison inhibitors would not require that topoisomerase function be essential, a catalytic inhibitor may be effective for growth inhibition only if the target topoisomerase is essential. Hence, E. coli may be less sensitive to reduction in topoisomerase I catalytic activity from a catalytic inhibitor because of the presence of topoisomerase III. Topoisomerase I is the only type IA topoisomerase in mycobacteria and has been validated as an antitubercular target. Our results showed that the TP-2471 inhibitors can trigger a rapid cell-killing mechanism involving topoisomerase I, even though these compounds are likely to be acting as catalytic inhibitors instead of topoisomerase poisons acting in a manner similar to that of quinolones against gyrase. These inhibitors provide useful starting points for further optimization of potency, selectivity, and other properties that are required for antitubercular drug leads. Our data so far on the TP-2471 class of inhibitors suggested that these compounds do not interact with the enzyme alone to prevent DNA binding. Studies are ongoing to determine the detailed mechanism of inhibition and cell killing.
Penetration and retention of individual compounds in bacterial cells would be important determinants of whether an enzyme inhibitor could possess whole-cell activity and may be factors that account for the lack of potent antimycobacterial activity observed for some of the compounds listed in Table 2 that have low IC50s for inhibition of MtbTopI. Other combinations in the substitutions on the polyamine scaffold could exert antimycobacterial activity via an unknown mechanism not related to topoisomerase I. The use of mycobacterial strains with different levels of topoisomerase I expression in cell-based assays would complement the enzyme-based assays in future optimization of other analogues that can target topoisomerase I for antimycobacterial activity. Future SAR studies taking into account MtbTopI target interactions and selective whole-cell bactericidal activity will allow us to identify compound features that optimally balance these two parameters critical for drug efficacy. Cocrystallization of compounds with topoisomerase I would elucidate the direct interactions between topoisomerase I and an individual compound. Selection of M. tuberculosis mutants resistant to these compounds and identification of causative mutations in topA would further validate MtbTopI as the target.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by grant R01AI069313 from the National Institutes of Health to Y.-C.T.-D. and funded in part through the Florida Drug Discovery Acceleration Program by the State of Florida, Department of Health.
None of the authors has any financial conflicts of interest to declare.
Funding Statement
REU participant Carlos Paz was supported by NSF-REU Site Grant CHE1156886 to FIU.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.00288-16.
REFERENCES
- 1.Friedman ND, Temkin E, Carmeli Y. 2015. The negative impact of antibiotic resistance. Clin Microbiol Infect doi: 10.1016/j.cmi.2015.12.002. [DOI] [PubMed] [Google Scholar]
- 2.Engstrom A. 2016. Fighting an old disease with modern tools: characteristics and molecular detection methods of drug-resistant Mycobacterium tuberculosis. Infect Dis (Lond) 48:1–17. doi: 10.3109/23744235.2015.1061205. [DOI] [PubMed] [Google Scholar]
- 3.Matteelli A, Roggi A, Carvalho AC. 2014. Extensively drug-resistant tuberculosis: epidemiology and management. Clin Epidemiol 6:111–118. doi: 10.2147/CLEP.S35839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen SH, Chan NL, Hsieh TS. 2013. New mechanistic and functional insights into DNA topoisomerases. Annu Rev Biochem 82:139–170. doi: 10.1146/annurev-biochem-061809-100002. [DOI] [PubMed] [Google Scholar]
- 5.Vos SM, Tretter EM, Schmidt BH, Berger JM. 2011. All tangled up: how cells direct, manage and exploit topoisomerase function. Nat Rev Mol Cell Biol 12:827–841. doi: 10.1038/nrm3228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schoeffler AJ, Berger JM. 2008. DNA topoisomerases: harnessing and constraining energy to govern chromosome topology. Q Rev Biophys 41:41–101. doi: 10.1017/S003358350800468X. [DOI] [PubMed] [Google Scholar]
- 7.Aldred KJ, Kerns RJ, Osheroff N. 2014. Mechanism of quinolone action and resistance. Biochemistry 53:1565–1574. doi: 10.1021/bi5000564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tomasic T, Masic LP. 2014. Prospects for developing new antibacterials targeting bacterial type IIA topoisomerases. Curr Top Med Chem 14:130–151. [DOI] [PubMed] [Google Scholar]
- 9.Tse-Dinh YC. 2009. Bacterial topoisomerase I as a target for discovery of antibacterial compounds. Nucleic Acids Res 37:731–737. doi: 10.1093/nar/gkn936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang JC. 2002. Cellular roles of DNA topoisomerases: a molecular perspective. Nat Rev Mol Cell Biol 3:430–440. doi: 10.1038/nrm831. [DOI] [PubMed] [Google Scholar]
- 11.Drlica K. 1992. Control of bacterial DNA supercoiling. Mol Microbiol 6:425–433. doi: 10.1111/j.1365-2958.1992.tb01486.x. [DOI] [PubMed] [Google Scholar]
- 12.Masse E, Drolet M. 1999. Relaxation of transcription-induced negative supercoiling is an essential function of Escherichia coli DNA topoisomerase I. J Biol Chem 274:16654–16658. doi: 10.1074/jbc.274.23.16654. [DOI] [PubMed] [Google Scholar]
- 13.Tse-Dinh YC. 2015. Targeting bacterial topoisomerase I to meet the challenge of finding new antibiotics. Future Med Chem 7:459–471. doi: 10.4155/fmc.14.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang Z, Cheng B, Tse-Dinh YC. 2011. Crystal structure of a covalent intermediate in DNA cleavage and rejoining by Escherichia coli DNA topoisomerase I. Proc Natl Acad Sci U S A 108:6939–6944. doi: 10.1073/pnas.1100300108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tan K, Zhou Q, Cheng B, Zhang Z, Joachimiak A, Tse-Dinh YC. 2015. Structural basis for suppression of hypernegative DNA supercoiling by E. coli topoisomerase I. Nucleic Acids Res 43:11031–11046. doi: 10.1093/nar/gkv1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yamaguchi Y, Inouye M. 2015. An endogenous protein inhibitor, YjhX (TopAI), for topoisomerase I from Escherichia coli. Nucleic Acids Res 43:10387–10396. doi: 10.1093/nar/gkv1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yigit H, Reznikoff WS. 1999. Escherichia coli DNA topoisomerase I copurifies with Tn5 transposase, and Tn5 transposase inhibits topoisomerase I. J Bacteriol 181:3185–3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yigit H, Reznikoff WS. 1998. Escherichia coli DNA topoisomerase I and suppression of killing by Tn5 transposase overproduction: topoisomerase I modulates Tn5 transposition. J Bacteriol 180:5866–5874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weinreich MD, Yigit H, Reznikoff WS. 1994. Overexpression of the Tn5 transposase in Escherichia coli results in filamentation, aberrant nucleoid segregation, and cell death: analysis of E. coli and transposase suppressor mutations. J Bacteriol 176:5494–5504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pruss GJ, Manes SH, Drlica K. 1982. Escherichia coli DNA topoisomerase I mutants: increased supercoiling is corrected by mutations near gyrase genes. Cell 31:35–42. doi: 10.1016/0092-8674(82)90402-0. [DOI] [PubMed] [Google Scholar]
- 21.DiNardo S, Voelkel KA, Sternglanz R, Reynolds AE, Wright A. 1982. Escherichia coli DNA topoisomerase I mutants have compensatory mutations in DNA gyrase genes. Cell 31:43–51. doi: 10.1016/0092-8674(82)90403-2. [DOI] [PubMed] [Google Scholar]
- 22.Garcia MT, Blazquez MA, Ferrandiz MJ, Sanz MJ, Silva-Martin N, Hermoso JA, de la Campa AG. 2011. New alkaloid antibiotics that target the DNA topoisomerase I of Streptococcus pneumoniae. J Biol Chem 286:6402–6413. doi: 10.1074/jbc.M110.148148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Suerbaum S, Brauer-Steppkes T, Labigne A, Cameron B, Drlica K. 1998. Topoisomerase I of Helicobacter pylori: juxtaposition with a flagellin gene (flaB) and functional requirement of a fourth zinc finger motif. Gene 210:151–161. doi: 10.1016/S0378-1119(98)00065-1. [DOI] [PubMed] [Google Scholar]
- 24.Ahmed W, Menon S, Godbole AA, Karthik PV, Nagaraja V. 2014. Conditional silencing of topoisomerase I gene of Mycobacterium tuberculosis validates its essentiality for cell survival. FEMS Microbiol Lett 353:116–123. doi: 10.1111/1574-6968.12412. [DOI] [PubMed] [Google Scholar]
- 25.Ravishankar S, Ambady A, Awasthy D, Mudugal NV, Menasinakai S, Jatheendranath S, Guptha S, Sharma S, Balakrishnan G, Nandishaiah R, Ramachandran V, Eyermann CJ, Reck F, Rudrapatna S, Sambandamurthy VK, Sharma UK. 2015. Genetic and chemical validation identifies Mycobacterium tuberculosis topoisomerase I as an attractive anti-tubercular target. Tuberculosis (Edinb) 95:589–598. doi: 10.1016/j.tube.2015.05.004. [DOI] [PubMed] [Google Scholar]
- 26.Godbole AA, Ahmed W, Bhat RS, Bradley EK, Ekins S, Nagaraja V. 2014. Inhibition of Mycobacterium tuberculosis topoisomerase I by m-AMSA, a eukaryotic type II topoisomerase poison. Biochem Biophys Res Commun 446:916–920. doi: 10.1016/j.bbrc.2014.03.029. [DOI] [PubMed] [Google Scholar]
- 27.Tang SC, Shapiro TA. 2010. Newly identified antibacterial compounds are topoisomerase poisons in African trypanosomes. Antimicrob Agents Chemother 54:620–626. doi: 10.1128/AAC.01025-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cheng B, Liu I, Tse-Dinh YC. 2007. Compounds with antibacterial activity that enhance DNA cleavage by bacterial DNA topoisomerase I. J Antimicrob Chemother 59:640–645. doi: 10.1093/jac/dkl556. [DOI] [PubMed] [Google Scholar]
- 29.Bansal S, Sinha D, Singh M, Cheng B, Tse-Dinh YC, Tandon V. 2012. 3,4-Dimethoxyphenyl bis-benzimidazole, a novel DNA topoisomerase inhibitor that preferentially targets Escherichia coli topoisomerase I. J Antimicrob Chemother 67:2882–2891. doi: 10.1093/jac/dks322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nimesh H, Sur S, Sinha D, Yadav P, Anand P, Bajaj P, Virdi JS, Tandon V. 2014. Synthesis and biological evaluation of novel bisbenzimidazoles as Escherichia coli topoisomerase IA inhibitors and potential antibacterial agents. J Med Chem 57:5238–5257. doi: 10.1021/jm5003028. [DOI] [PubMed] [Google Scholar]
- 31.Houghten RA, Pinilla C, Giulianotti MA, Appel JR, Dooley CT, Nefzi A, Ostresh JM, Yu Y, Maggiora GM, Medina-Franco JL, Brunner D, Schneider J. 2008. Strategies for the use of mixture-based synthetic combinatorial libraries: scaffold ranking, direct testing in vivo, and enhanced deconvolution by computational methods. J Comb Chem 10:3–19. doi: 10.1021/cc7001205. [DOI] [PubMed] [Google Scholar]
- 32.Rideout MC, Boldt JL, Vahi-Ferguson G, Salamon P, Nefzi A, Ostresh JM, Giulianotti M, Pinilla C, Segall AM. 2011. Potent antimicrobial small molecules screened as inhibitors of tyrosine recombinases and Holliday junction-resolving enzymes. Mol Divers 15:989–1005. doi: 10.1007/s11030-011-9333-2. [DOI] [PubMed] [Google Scholar]
- 33.Pinilla C, Appel JR, Blanc P, Houghten RA. 1992. Rapid identification of high affinity peptide ligands using positional scanning synthetic peptide combinatorial libraries. Biotechniques 13:901–905. [PubMed] [Google Scholar]
- 34.Narula G, Annamalai T, Aedo S, Cheng B, Sorokin E, Wong A, Tse-Dinh YC. 2011. The strictly conserved Arg-321 residue in the active site of Escherichia coli topoisomerase I plays a critical role in DNA rejoining. J Biol Chem 286:18673–18680. doi: 10.1074/jbc.M111.229450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Annamalai T, Dani N, Cheng B, Tse-Dinh YC. 2009. Analysis of DNA relaxation and cleavage activities of recombinant Mycobacterium tuberculosis DNA topoisomerase I from a new expression and purification protocol. BMC Biochem 10:18. doi: 10.1186/1471-2091-10-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Santos RG, Appel JR, Giulianotti MA, Edwards BS, Sklar LA, Houghten RA, Pinilla C. 2013. The mathematics of a successful deconvolution: a quantitative assessment of mixture-based combinatorial libraries screened against two formylpeptide receptors. Molecules 18:6408–6424. doi: 10.3390/molecules18066408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reilley KJ, Giulianotti M, Dooley CT, Nefzi A, McLaughlin JP, Houghten RA. 2010. Identification of two novel, potent, low-liability antinociceptive compounds from the direct in vivo screening of a large mixture-based combinatorial library. AAPS J 12:318–329. doi: 10.1208/s12248-010-9191-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu J, Zhang Y, Maida LE, Santos RG, Welmaker GS, LaVoi TM, Nefzi A, Yu Y, Houghten RA, Toll L, Giulianotti MA. 2013. Scaffold ranking and positional scanning utilized in the discovery of nAChR-selective compounds suitable for optimization studies. J Med Chem 56:10103–10117. doi: 10.1021/jm401543h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Houghten RA, Pinilla C, Appel JR, Blondelle SE, Dooley CT, Eichler J, Nefzi A, Ostresh JM. 1999. Mixture-based synthetic combinatorial libraries. J Med Chem 42:3743–3778. doi: 10.1021/jm990174v. [DOI] [PubMed] [Google Scholar]
- 40.Acharya AN, Ostresh JM, Houghten RA. 2002. Determination of isokinetic ratios necessary for equimolar incorporation of carboxylic acids in the solid-phase synthesis of mixture-based combinatorial libraries. Biopolymers 65:32–39. doi: 10.1002/bip.10206. [DOI] [PubMed] [Google Scholar]
- 41.Ostresh JM, Winkle JH, Hamashin VT, Houghten RA. 1994. Peptide libraries: determination of relative reaction rates of protected amino acids in competitive couplings. Biopolymers 34:1681–1689. doi: 10.1002/bip.360341212. [DOI] [PubMed] [Google Scholar]
- 42.Williams KJ, Joyce G, Robertson BD. 2010. Improved mycobacterial tetracycline inducible vectors. Plasmid 64:69–73. doi: 10.1016/j.plasmid.2010.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Andreu N, Zelmer A, Fletcher T, Elkington PT, Ward TH, Ripoll J, Parish T, Bancroft GJ, Schaible U, Robertson BD, Wiles S. 2010. Optimisation of bioluminescent reporters for use with mycobacteria. PLoS One 5:e10777. doi: 10.1371/journal.pone.0010777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Narula G, Tse-Dinh YC. 2012. Residues of E. coli topoisomerase I conserved for interaction with a specific cytosine base to facilitate DNA cleavage. Nucleic Acids Res 40:9233–9243. doi: 10.1093/nar/gks688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang Y, Lynch AS, Chen SJ, Wang JC. 2002. On the molecular basis of the thermal sensitivity of an Escherichia coli topA mutant. J Biol Chem 277:1203–1209. doi: 10.1074/jbc.M109436200. [DOI] [PubMed] [Google Scholar]
- 46.Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol 2:2006.0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.