Abstract
Metallo-β-lactamases (MBLs) confer resistance to carbapenems, and their increasing global prevalence is a growing clinical concern. To elucidate the mechanisms by which these enzymes recognize and hydrolyze carbapenems, we solved 1.4 to 1.6 Å crystal structures of SMB-1 (Serratia metallo-β-lactamase 1), a subclass B3 MBL, bound to hydrolyzed carbapenems (doripenem, meropenem, and imipenem). In these structures, SMB-1 interacts mainly with the carbapenem core structure via elements in the active site, including a zinc ion (Zn-2), Q157[113] (where the position in the SMB-1 sequence is in brackets after the BBL number), S221[175], and T223[177]. There is less contact with the carbapenem R2 side chains, strongly indicating that SMB-1 primarily recognizes the carbapenem core structure. This is the first report describing how a subclass B3 MBL recognizes carbapenems. We also solved the crystal structure of SMB-1 in complex with the approved drugs captopril, an inhibitor of the angiotensin-converting enzyme, and 2-mercaptoethanesulfonate, a chemoprotectant. These drugs are inhibitors of SMB-1 with Ki values of 8.9 and 184 μM, respectively. Like carbapenems, these inhibitors interact with Q157[113] and T223[177] and their thiol groups coordinate the zinc ions in the active site. Taken together, the data indicate that Q157[113], S221[175], T223[177], and the two zinc ions in the active site are key targets in the design of SMB-1 inhibitors with enhanced affinity. The structural data provide a solid foundation for the development of effective inhibitors that would overcome the carbapenem resistance of MBL-producing multidrug-resistant microbes.
INTRODUCTION
The increasing prevalence of metallo-β-lactamases (MBLs) among pathogenic Gram-negative bacteria is becoming a major clinical concern because these enzymes hydrolyze most of the β-lactams used daily in clinical settings, including carbapenems (1). MBLs are classified into three subclasses, B1, B2, and B3, based on the primary zinc-binding motifs, which are essential for enzymatic activity (2). The genes for subclass B1 MBLs such as IMP-, VIM-, and NDM-type enzymes are often located on transferable plasmids and are widely distributed among clinical isolates of Enterobacteriaceae, Pseudomonas spp., and Acinetobacter spp. (3). On the other hand, subclass B3 MBL-encoding genes are generally found in the chromosomes of bacteria such as Stenotrophomonas maltophilia and Chryseobacterium spp. However, B3 MBLs, including AIM-1 and SMB-1 (Serratia metallo-β-lactamase 1) enzymes, were recently reported to have been horizontally acquired by Pseudomonas aeruginosa and Serratia marcescens, respectively (4, 5). Thus, it is very likely that B3 MBLs will soon spread among pathogenic bacteria.
Notably, B3 MBLs in environmental bacteria have catalytic properties different from those found in pathogenic bacterial species. For example, BJP-1 from Bradyrhizobium japonicum (6), FEZ-1 from Fluoribacter gormanii (7), and THIN-B from Janthinobacterium lividum (8) hydrolyze carbapenems with kcat/Km ratios of <106 s−1/M, while SMB-1 (4), AIM-1 (5), and L1 from S. maltophilia (9) degrade carbapenems more efficiently, with kcat/Km ratios of >106 s−1/M. One way to explain the differences between these enzymatic activities is to investigate their substrate preferences through X-ray crystallography. For B3 MBLs, only the structure of L1 MBL in complex with hydrolyzed moxalactam has been solved (10), although the native structures of SMB-1, AIM-1, L1, BJP-1, and FEZ-1 are available (11–15). Therefore, to gain further insight into substrate binding and hydrolysis by B3 MBLs, we determined the precise structure of SMB-1 in complex with hydrolyzed carbapenems.
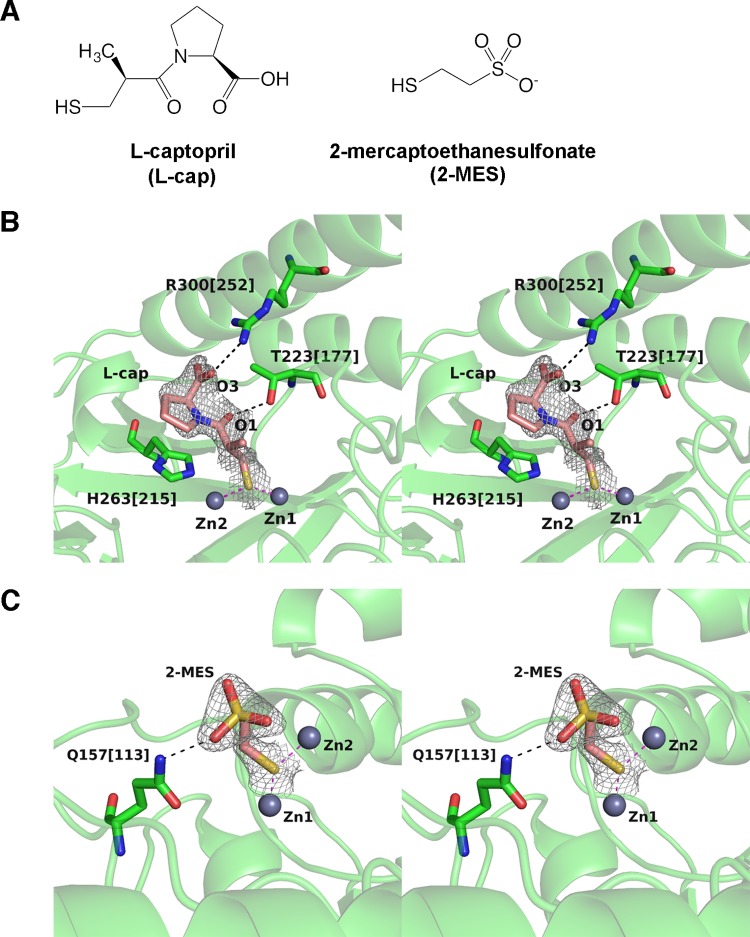
In addition, clinically useful MBL inhibitors are urgently needed to restore the efficacy of β-lactams. Notably, it has been demonstrated that some approved drugs possessing thiol groups inhibit MBLs (16–18). These drugs include l-captopril, an inhibitor of the angiotensin-converting enzyme (19). In a preliminarily study, we found that 2-mercaptoethanesulfonate (2-MES), which also contains a thiol group, may inhibit SMB-1. 2-MES is used as a chemoprotectant to minimize the undesirable side effects of ifosfamide or cyclophosphamide. To investigate the mode of inhibition, we also determined the structure of SMB-1 in complex with l-captopril and 2-MES.
MATERIALS AND METHODS
Protein expression and purification.
SMB-1 was expressed in and purified from Escherichia coli BL21(DE3)pLysS as described previously (11, 20), with slight modifications. Briefly, SMB-1 was purified by cation-exchange chromatography, dialyzed for 16 h at 4°C against 50 mM Tris-HCl (pH 7.5) containing 1.8 M ammonium sulfate, and loaded onto a HiTrap Phenyl HP column (GE Healthcare). After elution with 50 mM Tris-HCl (pH 7.5), the protein was buffer exchanged into 20 mM HEPES-NaOH (pH 7.5) and concentrated to 60 mg/ml with an Amicon Ultra Centrifugal Filter (Millipore).
Crystallization.
To obtain enzyme-substrate complexes, 60 mg/ml SMB-1 was mixed 1:1 with 20 mg/ml carbapenem, incubated at 20°C for 16 h, and crystallized, respectively, in 0.2 M ammonium sulfate, 0.15 M Tris-HCl (pH 8.5), and 30% (wt/vol) polyethylene glycol (PEG) 4000 for doripenem; in 0.2 M lithium sulfate, 0.1 M Tris-HCl (pH 8.5), and 20% (wt/vol) PEG 4000 for meropenem; and in 0.2 M lithium sulfate, 0.1 M Tris-HCl (pH 8.5), and 20% (wt/vol) PEG 5000 monomethyl ether for imipenem.
To obtain enzyme-inhibitor complexes, native crystals were soaked for 1 or 3 days in reservoir solutions supplemented with 10 mM 2-MES and 10 mM l-captopril, respectively. For soaking with 2-MES, native crystals were first obtained in 1 M lithium chloride, 0.1 M morpholineethanesulfonic acid (pH 6.0), and 26% (wt/vol) PEG 6000. For soaking with l-captopril, native crystals were initially grown under the same conditions as crystals of SMB-1–doripenem.
Data collection and refinement.
X-ray diffraction data were collected at the Photon Factory (Tsukuba, Japan) and the Aichi Synchrotron Radiation Center (Aichi, Japan), processed, and scaled with HKL-2000 (21) or iMosfilm/SCALA (22, 23). Structures were solved by molecular replacement in MOLREP (24) as implemented in CCP4 (25) by using the structure of native SMB-1 (Protein Data Bank [PDB] accession no. 3VPE), which we previously determined. Coot (26) and REFMAC5 (27) were used to build and refine models. The quality of final models was assessed with RAMPAGE (28). Coordinates and molecular library files for hydrolyzed substrates were constructed with Sketcher in CCP4 (25). Data collection and refinement statistics are listed in Table 1.
TABLE 1.
Data collection and refinement statistics
| Parameter | hDPM | hMPM | hIPM | l-Captopril | 2-MES |
|---|---|---|---|---|---|
| Data collection statistics | |||||
| Beamline | BL2S1 (AichiSR) | BL2S1 (AichiSR) | NE3A (PF-AR) | NW12A (PF-AR) | NW12A (PF-AR) |
| Wavelength (Å) | 1.12 | 1.12 | 1.00 | 1.00 | 1.00 |
| Resolution range (Å) | 40.70–1.39 (1.47–1.39)b | 40.62–1.39 (1.47–1.39) | 40.58–1.57 (1.65–1.57) | 50.00–2.02 (2.05–2.02) | 50.00–2.10 (2.14–2.10) |
| Space group | P1 | P1 | P1 | P31 | P31 |
| Unit cell dimensions | |||||
| a (Å) | 37.01 | 36.90 | 37.03 | 68.10 | 67.11 |
| b (Å) | 40.92 | 41.17 | 41.46 | 68.10 | 67.11 |
| c (Å) | 45.32 | 45.32 | 45.44 | 49.11 | 46.72 |
| α (°) | 107.57 | 107.95 | 108.32 | 90.00 | 90.00 |
| β (°) | 102.56 | 102.46 | 102.78 | 90.00 | 90.00 |
| γ (°) | 107.01 | 106.85 | 106.28 | 120.00 | 120.00 |
| No. of unique reflections | 39,572 (5,662) | 39,458 (5,793) | 30,412 (4,382) | 16,635 (844) | 13,471 (677) |
| Redundancy | 1.9 (1.9) | 1.9 (1.9) | 3.3 (3.4) | 11.0 (10.7) | 11.6 (11.5) |
| Completeness (%) | 86.6 (84.7) | 86.2 (86.0) | 93.6 (92.7) | 99.6 (99.1) | 100.0 (100.0) |
| Rmergea (%) | 4.0 (7.2) | 4.6 (6.3) | 4.2 (7.9) | 7.1 (30.6) | 8.1 (29.8) |
| Mean I/σ〈I〉 | 12.8 (6.7) | 11.2 (7.1) | 19.0 (11.9) | 64.2 (17.1) | 42.5 (10.7) |
| Refinement statistics | |||||
| Rworking (%) | 13.6 | 13.7 | 12.2 | 21.2 | 15.0 |
| Rfree (%) | 15.8 | 16.3 | 15.5 | 28.2 | 21.4 |
| Ramachandran statistics (%) | |||||
| Favored | 97.3 | 97.3 | 97.3 | 92.6 | 96.9 |
| Allowed | 2.7 | 2.7 | 2.7 | 7.0 | 3.1 |
| Disallowed | 0 | 0 | 0 | 0.4 | 0 |
| Avg B factors (Å2) | |||||
| Protein | 9.3 | 8.5 | 6.5 | 40.3 | 22.7 |
| Ligand/ion | 18.1 | 19.5 | 15.7 | 60.1 | 29.4 |
| Water | 20.5 | 19.3 | 19.0 | 37.2 | 26.4 |
| RMSD | |||||
| Bond length (Å) | 0.012 | 0.012 | 0.011 | 0.008 | 0.009 |
| Bond angle (°) | 1.589 | 1.562 | 1.433 | 1.120 | 1.163 |
Rmerge = ∑hkl∑i|Ii(hkl) − <I(hkl)>|/∑hkl∑iIi(hkl), where Ii(hkl) is the observed intensity for reflection for hkl and <I(hkl)> is the average intensity calculated for reflection hkl from replicate data.
Values in parentheses are for the highest-resolution shell.
Molecular modeling.
Doripenem was docked against rigid side chains in the SMB-1 active site with Autodock version 4.2 by using default parameters (29). To generate the base SMB-1 model for this simulation, water and ligand molecules were removed from the structure of SMB-1 in complex with hydrolyzed doripenem [hDPM; PDB accession no. 5B15]. A 60- by 60- by 60-point grid box was set with Autogrid, centered between the two zinc ions in the active site. A total of 100 docked poses were generated with a population size of 150, a maximum number of energy evaluations of 25 million, a maximum number of generations of 27,000, a gene mutation rate of 0.02, and a crossover rate of 0.8.
Kinetic parameters.
Steady-state constants Km and kcat were determined according to previously published methods (4). Ki was determined with 100 μM nitrocefin (Oxoid) as the reporter substrate. Ki values were calculated according to [E]/[E]0 = Ki/(Ki + [I]), where [E], [E]0, and [I] are the active enzyme, total enzyme, and inhibitor concentrations, respectively.
Susceptibility testing.
MICs were determined by the broth dilution method according to the guidelines of the Clinical and Laboratory Standards Institute (30).
Amino acid numbering.
Amino acid residues are numbered according to our previous report (11), by using class B β-lactamase (BBL) numbering as a reference (2). For amino acids in enzymes other than SMB-1, only the BBL number is indicated, e.g., R37. For amino acids in SMB-1, the position in the SMB-1 sequence is indicated in brackets after the BBL number, e.g., R37[3].
Protein structure accession numbers.
Atomic coordinates and structure factors of SMB-1 in complex with hDPM (accession no. 5B15), hydrolyzed meropenem (hMPM; accession no. 5AXO), hydrolyzed imipenem (hIPM; accession no. 5B1U), l-captopril (accession no. 5AYA), and 2-MES (accession no. 5AXR) have been deposited in the PDB.
RESULTS AND DISCUSSION
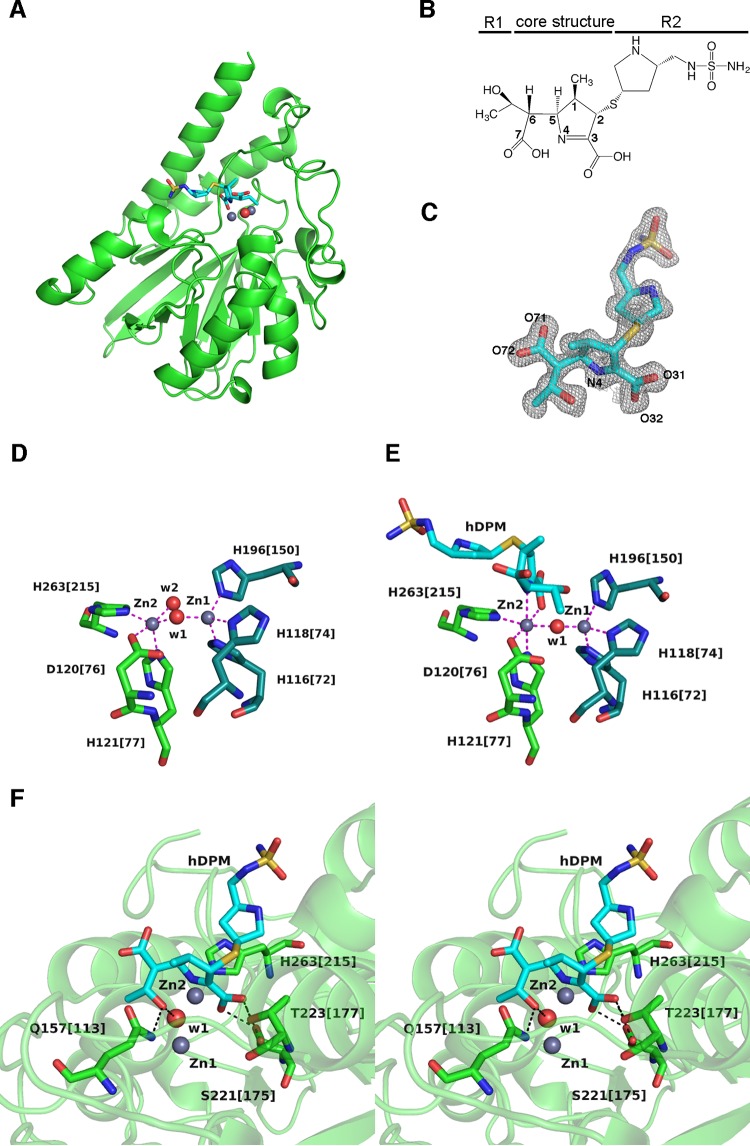
To obtain structures of enzyme-substrate complexes, we crystallized mixtures of SMB-1 and carbapenems. In this manner, we determined the structure of SMB-1 in complex with hDPM at 1.4 Å (Fig. 1A). This crystal is in space group P1, with one molecule per asymmetric unit (Table 1). The overall structure is very similar to that of native SMB-1 (PDB accession no. 3VPE) with a root mean square deviation (RMSD) of 0.6 Å in Cα atoms. Like other carbapenems, hDPM contains a small 1-hydroxyethyl group at the R1 side chain and a relatively bulky side chain at R2 (Fig. 1B). The hDPM molecule is bound to the active site and is observed in a 2|Fo|-|Fc| electron density map contoured at 1 σ (Fig. 1C). A |Fo|-|Fc| electron density map contoured at 2 σ is shown in Fig. S1 in the supplemental material. The geometry around the C-2 atom of the bound hDPM molecule is tetrahedral rather than planar, indicating that hDPM is bound as a Δ1-pyrroline tautomer (Fig. 1B). Two zinc ions (Zn-1 and Zn-2), bridged by a hydroxide oxygen atom (w1), were observed in the active site, just as in the native structure (Fig. 1D and E) (11). In both the hDPM-bound (Fig. 1E) and native (Fig. 1D) structures, Zn-1 is coordinated via a distorted tetrahedron formed by H116[72], H118[74], H196[150], and w1. On the other hand, Zn-2 is coordinated via a distorted square pyramid in the native structure and via a distorted octahedron formed by H263[215], D120[76], w1, the C-3 carboxylate oxygen O-32 (Fig. 1C), H121[77], and N-4 in the hDPM-bound structure. The distances between Zn-2 and N-4 and between Zn-2 and C-3 carboxylate oxygen O-32 are 2.3 and 2.2 Å, respectively. Similarly strong bonds among Zn-2, N-4, and C-3 carboxylate oxygen were observed in structures of NDM-1 complexed with hMPM (see Fig. S2 in the supplemental material) (19). Thus, these bonds appear to universally stabilize hydrolyzed carbapenems bound to dizinc MBLs. These bonds may also stabilize the anion intermediate, which contains the deprotonated nitrogen (N-4) atom of the pyrroline ring as the leaving group, suggesting that cleavage of the β-lactam ring occurs prior to protonation of the anionic nitrogen atom (31).
FIG 1.
(A) Ribbon representation of SMB-1 (green) bound to hDPM (cyan sticks). Zinc ions and the water molecule are depicted as gray and red spheres, respectively. (B) Chemical structure of hDPM. (C) 2|Fo|-|Fc| electron density map contoured at 1.0 σ (gray mesh). hDPM is shown as cyan (carbon), ocher (sulfur), red (oxygen), and blue (nitrogen) sticks. (D) Coordination of zinc ions in native SMB-1. Zinc ions and water molecules are depicted as gray and red spheres, respectively. Amino acids coordinating Zn-1 are Zn-2 are illustrated as dark green and green sticks, and coordination bonds are shown as magenta dashed lines. The image was adapted from the structure with PDB accession no. 3VPE. (E) Coordination of zinc ions in hDPM-bound SMB-1. hDPM is illustrated as in panel C, while other elements are rendered as in panel D. (F) Interactions other than coordination bonds between SMB-1 and hDPM. Amino acids are shown as green (carbon), red (oxygen), and blue (nitrogen) sticks, while hDPM is illustrated as in panel C. Water molecule w1 and zinc ions Zn-1 and Zn-2 are shown as in panel A. Hydrogen bonds are shown as black dashed lines.
Additional interactions between hDPM and SMB-1 occur mainly within 6 Å of the central zinc ions in the active site (Fig. 1F). The pyrrolidine ring of the R2 side chain rests approximately parallel against the imidazole side chain of H263[215], resulting in CH-π interaction with distances between atoms of less than 4.0 Å. An oxygen atom from the 1-hydroxyethyl group of R1 is hydrogen bonded at 2.6 Å to the hydroxide oxygen in w1. The C-3 carboxylate oxygens O-31 and O-32 are bound by the OG hydroxyl group of S221[175] at 2.8 Å, although an alternate conformation with 0.5 occupancy was observed for this hydroxyl group. C-3 carboxylate oxygen O-31 additionally interacts with the OG1 hydroxyl group of T223[177] at 2.7 Å. The NE2 side chain nitrogen of Q157[113] is hydrogen bonded at 3.0 Å to the oxygen atom of the 1-hydroxyethyl group of R1. The C-6 carboxylate group, which is generated after a water molecule attacks the carbonyl carbon of the β-lactam ring, was oriented toward to the solvent side.
Notably, the orientations of the 1-hydroxyethyl and C-6 carboxylate groups after hydrolysis are different in our model from those in NDM-1 complexed with hMPM, in which the C-6 carboxylate group is oriented toward the central zinc ions (see Fig. S2 in the supplemental material) (19, 32). The latter orientation is considered to reflect one intermediate state before product release, in light of the accepted mechanism of β-lactam hydrolysis by MBLs (10, 12, 32). On the other hand, we appear to have captured a predicted subsequent state, in which the C-5–C-6 bond has been rotated, as noted for B2 MBLs (33). This rotation may be due to the energetically more favorable reorientation of the carboxylate group toward the solvent and away from Zn-1. In addition, we suppose that the positioning of the 1-hydroxyethyl group toward Zn-1 might also be energetically and/or conformationally more favorable after the substrate has been depleted, at which point released products might rebind to the active site. In any case, our structure well reflects the catalytic state before the hydrolyzed carbapenem is released.
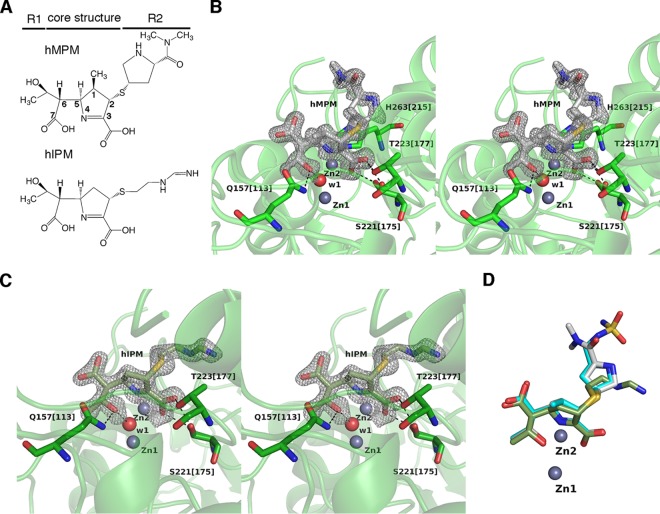
Crystals of SMB-1 in complex with hMPM and hIPM (Fig. 2A) diffracted to 1.4 and 1.6 Å, respectively. Both hMPM and hIPM molecules are observable in a 2|Fo|-|Fc| electron density map contoured at 1 σ (Fig. 2B and C) and are bound to the active site in a fashion very similar to that of hDPM (Fig. 2D). |Fo|-|Fc| electron density maps contoured at 2 σ are shown in Fig. S1 in the supplemental material. For instance, the positions of the 1-hydroxyethyl and carboxylate groups are spatially the same and coordinative bonds among Zn-2, N-4, and C-3 carboxylate oxygen are present in all complexes. The pyrroline ring takes the Δ1 form. In addition, there are no hydrogen bonds toward the R2 side chain. The similarity in the binding mode was not entirely unexpected, since the structural differences among hDPM, hMPM, and hIPM are small and typically consist of the functional group at R2 (Fig. 1B and 2A) and the presence (hDPM and hMPM) or absence (hIPM) of a C-1 methyl group in the five-membered ring.
FIG 2.
(A) Chemical structures of hMPM and hIPM. (B) Interactions between SMB-1 and hMPM. Amino acids are shown as green (carbon), red (oxygen), and blue (nitrogen) sticks, while hMPM is shown as silver (carbon), ocher (sulfur), red (oxygen), and blue (nitrogen) sticks. 2|Fo|-|Fc| electron density map contoured at 1.0 σ (gray mesh). Zinc ions and the water molecule are depicted as gray and red spheres, respectively. Black and magenta dashed lines indicate hydrogen and coordination bonds, respectively. (C) Interactions between SMB-1 and hIPM. hIPM is illustrated as light green (carbon), ocher (sulfur), red (oxygen), and blue (nitrogen) sticks. Other elements are illustrated as in panel B. (D) Superposition of hDPM (cyan), hMPM (silver), and hIPM (light green) bound to SMB-1. Zinc ions are depicted as spheres.
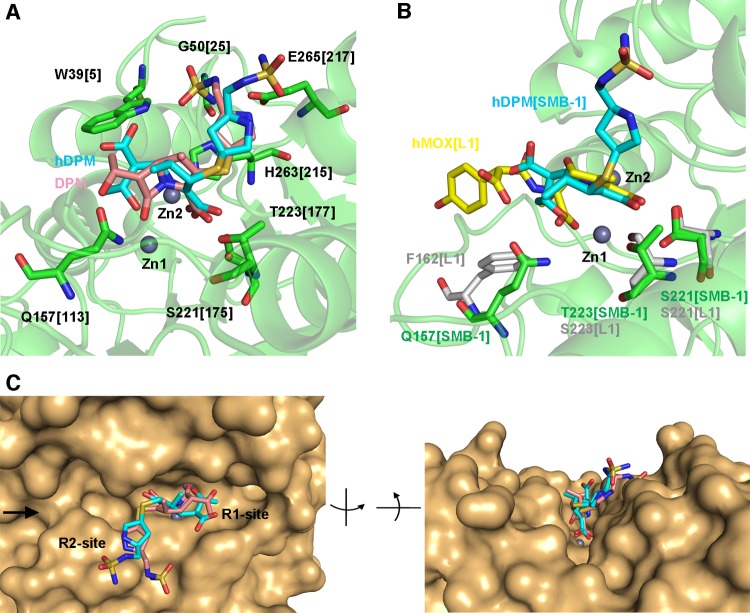
The structures of enzymes bound to hydrolyzed carbapenems indicate that the amino acids Q157[113], S221[175], and T223[177], as well as the two zinc ions, are involved in recognizing hydrolyzed carbapenems (Fig. 1F and 2B and C). This implies that these elements may contribute to the recognition of substrate carbapenems as well. To test this hypothesis, we docked doripenem into SMB-1 (Fig. 3A) and observed interactions between Zn-1 and the carbonyl oxygen of the β-lactam ring, as well as between Zn-2 and N-4/C-3 carboxylate oxygen. C-3 carboxylate oxygens are bound by the hydroxyl oxygens of S221[175] and T223[177]. Further, NE2 and OE1 of Q157[113] bind, respectively, the carbonyl oxygen and the oxygen atom in the 1-hydroxyethyl group at R1. Thus, amino acids Q157[113], S221[175], and T223[177] appear to recognize carbapenems.
FIG 3.
(A) Interaction between doripenem (DPM) and SMB-1, as simulated in Autodock. Amino acid residues that bind hDPM and DPM are illustrated as green sticks, while DPM and hDPM are shown as pink and cyan sticks, respectively. Zinc ions are shown as gray spheres. (B) Superposition of SMB-1 (green) and L1 (silver) (PDB accession no. 2AIO) bound to hDPM (cyan) and hydrolyzed moxalactam (hMOX; yellow), respectively. Zinc ions are illustrated as gray spheres. (C) Closeup view of the molecular surface of the SMB-1 active site with hDPM (cyan) and DPM (pink) bound. The image on the right is the same field as that on the left but viewed from the side marked with a black arrow.
We have previously demonstrated that a Q157A[113] substitution increases the Km values for ceftazidime (4.5-fold) and cefotaxime (7-fold) (11). In addition, an in silico model of AIM-1 bound to hydrolyzed cefoxitin suggests that NE2 of this residue may possibly bind the carbonyl oxygen of the β-lactam ring (12). While we did not obtain structures of SMB-1 in complex with cephalosporins, it is possible that Q157[113] in SMB-1 also recognizes the carbonyl oxygen in cephalosporins (11).
Another possible role for Q157[113] may be to stabilize hydrolysis products such as the anion intermediate via interactions with the C-6 carboxylate oxygens that form after hydrolysis of the β-lactam ring. Although the C-6 carboxylate group of hydrolyzed carbapenems is too far away from Q157[113] in our structure and the 1-hydroxyethyl group is near Q157[113] instead (Fig. 1F and 2B and C), the arrangement of these groups would be exchanged during β-lactam hydrolysis according to the generally accepted mechanism (10, 12, 32). Thus, C-6 carboxylate oxygens would interact with Q157[113] during hydrolysis but would subsequently be occluded by the 1-hydroxyethyl group. Indeed, Q157 in AIM-1 is predicted to interact with the carboxylate oxygens of hydrolyzed cefoxitin (12). Therefore, we conclude that Q157[113], without being essential, enhances the enzymatic activity of SMB-1 by recognizing β-lactams and/or stabilizing the hydrolyzed products. The role of this residue appears to be similar to that of N220 in NDM-1, the nitrogen atoms of which stabilize the substrate and the hydrolyzed products (19).
Binding of S221[175] and T223[177] to C-3 carboxylate oxygens O-31 and O-32 of the five-membered ring was observed in the structure of SMB-1 in complex with hydrolyzed carbapenems, as well as in simulated models of SMB-1 bound to doripenem (Fig. 1F, 2B and C, and 3A). Similarly, corresponding amino acids S221 and S223 in the enzyme L1 also contact O-31 and O-32 (Fig. 3B) (10). Considering that the B3 MBLs AIM-1, BJP-1, and FEZ-1 also contain amino acids that correspond to positions 221 and 223, these residues appear to universally bind carboxylate oxygens in the five- or six-membered rings of β-lactams in the same manner as residue K224 in B1 MBLs (34). Thus, these amino acids would be suitable targets for developing effective inhibitors of the hydrolytic activity of B3 MBLs, as will be discussed.
In the SMB-1 structure complexed with hydrolyzed carbapenems, the core structure has lower temperature factors (13.0 Å in hDPM, 16.4 Å in hMPM, and 11.4 Å in hIPM) than the R2 side chain (30.5 Å in hDPM, 29.9 Å in hMPM, and 21.8 Å in hIPM). Furthermore, SMB-1 has comparable Km and kcat values for doripenem (80 μM and 217 s−1), meropenem (144 μM and 604 s−1), and imipenem (133 μM and 518 s−1). These results indicate that recognition and hydrolysis depend largely on the carbapenem core structure (11). The tip of the R2 side chain in doripenem was shown by simulation to interact with W39[5], G50[25], and E265[217] (Fig. 3A). However, the mode by which SMB-1 recognizes the R2 side chain is unclear. In any case, recognition of the R2 side chain does not seem to be as critical as the interactions formed by the core structure.
A closeup view of the molecular surface around the substrate-binding cleft is depicted in Fig. 3C. The active site in SMB-1 is shallow and unshielded and thus will even accommodate β-lactams with bulky R1 and R2 side chains. NDM-1, a subclass B1 MBL, has a similarly shallow and unshielded cleft and thus interacts with a variety of β-lactams, including penicillins, cephalosporins, and carbapenems (19, 32). Therefore, structural features of the cleft around the active site may explain the ability of MBLs to hydrolyze a wide range of substrates with bulky side chains.
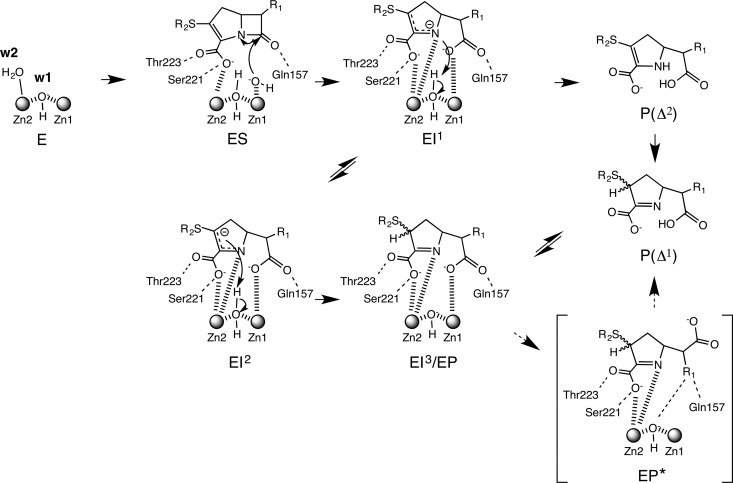
The proposed mechanism of carbapenem hydrolysis by SMB-1, which comprises a branched pathway to account for carbapenem tautomerization, is illustrated in Fig. 4, on the basis of the catalytic reaction advocated by Meini et al. (34, 35), which shows that subclass B1 and B3 MBLs hydrolyze carbapenems via quite similar catalytic mechanisms. In native SMB-1, the hydroxyl anion w1 bridges two zinc ions and the apical water molecule w2, which is stabilized next to Zn-2. In the product-bound state, the C-3 carboxylate oxygen assumes the place of w2 (Fig. 1D and E) (11). However, w2 may migrate to the void left by the nucleophilic w1 molecule as it attacks the carbonyl carbon of the β-lactam ring. Two intermediates, EI1 and EI2, are thus generated. EI1 contains an anionic N-4 atom tightly stabilized by Zn-2 and subsequently protonated by w2, to generate tautomer Δ2. In EI2, the negative charge on C-3 is localized and protonated, yielding the enzyme-product complex (EI3/EP). After protonation, w2 may be regenerated as the hydroxide required for nucleophilic attack. In our structures with hydrolyzed carbapenem, the S atom was not situated against the plane of the pyrroline ring, indicating that the double bond would be presented between N-4 and C-3, as shown in EP (tautomer Δ1). These results indicate that the hydrolyzed carbapenem in our structures is in the tautomer Δ1 form and is derived from only one branch of the proposed catalytic mechanism for SMB-1. The spatial position of C-6 carboxylate and the R1 side chain resembles that in EP*, a state in which the C-5–C-6 bond has been rotated before the product is released.
FIG 4.
Proposed mechanism of carbapenem hydrolysis by SMB-1. E, SMB-1 enzyme; S, substrate carbapenem; I1, I2, and I3, carbapenem intermediates formed during hydrolysis; P, hydrolyzed product; P*, hydrolyzed product observed in our structures. The EP* form observed in our structures is bracketed.
In addition, we have confirmed by crystallization that l-captopril binds SMB-1 (Fig. 5A and B; see Fig. S1 in the supplemental material). This crystal diffracted to 2.0 Å and is in space group P31 with one molecule in the asymmetric unit (Table 1). The structure is not significantly different from that of the native enzyme, with an RMSD of 0.3 Å in Cα atoms. The thiolate group S in l-captopril is bridged to Zn-1 and Zn-2 at 2.3 and 2.4 Å, respectively (Fig. 5B), and occupies the space typically occupied by w1 (Fig. 1D). Further interactions include hydrogen bonds between negatively charged carboxylate O-3 and positively charged NH-1 in R300[252] (3.0 Å) and between carbonyl O-1 and hydroxyl OG-1 in T223[117] (2.8 Å). The l-proline ring binds in a manner similar to that of the pyrrolidine ring of the R2 side chain in carbapenems and is approximately parallel to the imidazole side chain of H263[215].
FIG 5.
(A) Chemical structures of l-captopril and 2-MES. (B) Mode of binding between l-captopril and SMB-1. The gray mesh is a 2|Fo|-|Fc| electron density map contoured at 1.0 σ. l-Captopril is illustrated as pink (carbon), ocher (sulfur), red (oxygen), and blue (nitrogen) sticks, while SMB-1 amino acids are shown as green sticks. Zinc ions are shown as gray spheres. Coordination and hydrogen bonds are shown in magenta and black dashed lines, respectively. (C) Mode of binding between 2-MES and SMB-1. The image is drawn in the same scheme as that in panel B.
Superposition of SMB-1 and L1 bound to l- and d-captopril, respectively, demonstrates that the interaction between zinc ions and the thiol group is conserved, while other interactions are not (see Fig. S3 in the supplemental material). l-Captopril binds SMB-1 with a Ki of 8.9 ± 0.5 μM, while d-captopril binds L1 with a Ki of 20 μM at neutral pH (36). This difference may be partially due to the number of predicted hydrogen bonds. Indeed, SMB-1 has two potential hydrogen bond interactions (O-3–R300[252] and O-1–T223[177]) in addition to the coordination of thiolate in l-captopril to two zinc ions, while L1 has one such additional bond, O-3–S223. T223[177] in SMB-1 is in the same position as S223 in L1, and both interact with captopril, as well as other β-lactam substrates. Because other B3 MBLs, such as AIM-1 and BJP-1, have corresponding amino acids, this position should be a key target in any attempt to develop universal inhibitors against subclass B3 MBLs.
Similarly, a crystal that diffracted to 2.1 Å directly demonstrated that 2-MES binds to the SMB-1 active site (Fig. 5A and C; see Fig. S1 in the supplemental material). The Ki value was determined to be 184 ± 8 μM. The crystal belonged to space group P31 with one molecule in the asymmetric unit (Table 1). As shown in SMB-1 bound to l-captopril, the thiol group in 2-MES coordinates Zn-1 and Zn-2 over 2.0 and 2.3 Å, respectively, and replaces the catalytic water (Fig. 5C). Additionally, the sulfate oxygen interacts with NE2 of Q157[113]. Both l-captopril and 2-MES reduced the MIC of meropenem for recombinant E. coli expressing SMB-1 (see Table S1 in the supplemental material). Therefore, structures of the enzyme-inhibitor complexes are potential scaffolds for the design of SMB-1 inhibitors.
Conclusion.
In this paper, we report the crystal structure of SMB-1, a B3 MBL, in complex with three different hydrolyzed carbapenems. The amino acid residues Q157[113], S221[175], and T223[177], which are within 6 Å of the central zinc ions in the active site, recognize hydrolyzed carbapenems and, potentially, substrate carbapenems. Although Q157[113] is not strictly conserved, it enhances SMB-1 catalysis through interaction with carbonyl and carboxylate oxygens in substrates and hydrolyzed products, respectively.
In addition, we demonstrate that the approved drugs l-captopril, an inhibitor of angiotensin-converting enzyme that is used to treat hypertension, and 2-MES, a chemoprotectant, inhibit SMB-1 through interaction with the same residues and zinc atoms that contact hydrolyzed carbapenems. We have also previously demonstrated a similar inhibitory mechanism for mercaptoacetate (11). Taken together, Q157[113], S221[175], and T223[177], as well as the two zinc ions in the active site, are promising targets for inhibitors of SMB-1. Furthermore, docking studies by Liu et al. indicated that inhibitors derived from amino acid thioesters contact amino acid in L1 at position 221 (37). This amino acid is strictly conserved in B3 MBLs and thus is a strong potential drug target.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by grants from the Japanese Ministry of Education, Culture, Sports, Science and Technology (Wakate A and Wakate B), the Japanese Ministry of Health, Labor, and Welfare (H24-Shinkou-Ippan-010), the Japan Initiative for Global Research Network on Infectious Diseases from the Japan Agency for Medical Research and Development (AMED), and the Ohyama Health Foundation.
We thank staff at the Photon Factory (Tsukuba, Japan), at the Aichi Synchrotron Radiation Center (Aichi, Japan), and at the Division for Medical Research Engineering, Nagoya University Graduate School of Medicine, for their technical support.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AAC.03108-15.
REFERENCES
- 1.Cornaglia G, Giamarellou H, Rossolini GM. 2011. Metallo-β-lactamases: a last frontier for β-lactams? Lancet Infect Dis 11:381–393. doi: 10.1016/S1473-3099(11)70056-1. [DOI] [PubMed] [Google Scholar]
- 2.Garau G, García-Sáez I, Bebrone C, Anne C, Mercuri P, Galleni M, Frere JM, Dideberg O. 2004. Update of the standard numbering scheme for class B β-lactamases. Antimicrob Agents Chemother 48:2347–2349. doi: 10.1128/AAC.48.7.2347-2349.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nordmann P, Naas T, Poirel L. 2011. Global spread of carbapenemase-producing Enterobacteriaceae. Emerg Infect Dis 17:1791–1798. doi: 10.3201/eid1710.110655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wachino J, Yoshida H, Yamane K, Suzuki S, Matsui M, Yamagishi T, Tsutsui A, Konda T, Shibayama K, Arakawa Y. 2011. SMB-1, a novel subclass B3 metallo-β-lactamase, associated with ISCR1 and a class 1 integron, from a carbapenem-resistant Serratia marcescens clinical isolate. Antimicrob Agents Chemother 55:5143–5149. doi: 10.1128/AAC.05045-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yong D, Toleman MA, Bell J, Ritchie B, Pratt R, Ryley H, Walsh TR. 2012. Genetic and biochemical characterization of an acquired subgroup B3 metallo-β-lactamase gene, blaAIM-1, and its unique genetic context in Pseudomonas aeruginosa from Australia. Antimicrob Agents Chemother 56:6154–6159. doi: 10.1128/AAC.05654-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stoczko M, Frere JM, Rossolini GM, Docquier JD. 2006. Postgenomic scan of metallo-β-lactamase homologues in rhizobacteria: identification and characterization of BJP-1, a subclass B3 ortholog from Bradyrhizobium japonicum. Antimicrob Agents Chemother 50:1973–1981. doi: 10.1128/AAC.01551-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mercuri PS, Bouillenne F, Boschi L, Lamotte-Brasseur J, Amicosante G, Devreese B, van Beeumen J, Frere JM, Rossolini GM, Galleni M. 2001. Biochemical characterization of the FEZ-1 metallo-β-lactamase of Legionella gormanii ATCC 33297T produced in Escherichia coli. Antimicrob Agents Chemother 45:1254–1262. doi: 10.1128/AAC.45.4.1254-1262.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Docquier JD, Lopizzo T, Liberatori S, Prenna M, Thaller MC, Frere JM, Rossolini GM. 2004. Biochemical characterization of the THIN-B metallo-β-lactamase of Janthinobacterium lividum. Antimicrob Agents Chemother 48:4778–4783. doi: 10.1128/AAC.48.12.4778-4783.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Paton R, Miles RS, Amyes SG. 1994. Biochemical properties of inducible β-lactamases produced from Xanthomonas maltophilia. Antimicrob Agents Chemother 38:2143–2149. doi: 10.1128/AAC.38.9.2143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Spencer J, Read J, Sessions RB, Howell S, Blackburn GM, and Gamblin SJ. 2005. Antibiotic recognition by binuclear metallo-β-lactamases revealed by X-ray crystallography. J Am Chem Soc 127: 14439–14444. doi: 10.1021/ja0536062. [DOI] [PubMed] [Google Scholar]
- 11.Wachino J, Yamaguchi Y, Mori S, Kurosaki H, Arakawa Y, Shibayama K. 2013. Structural insights into the subclass B3 metallo-β-lactamase SMB-1 and the mode of inhibition by the common metallo-β-lactamase inhibitor mercaptoacetate. Antimicrob Agents Chemother 57:101–109. doi: 10.1128/AAC.01264-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Leiros HK, Borra PS, Brandsdal BO, Edvardsen KS, Spencer J, Walsh TR, Samuelsen O. 2012. Crystal structure of the mobile metallo-β-lactamase AIM-1 from Pseudomonas aeruginosa: insights into antibiotic binding and the role of Gln157. Antimicrob Agents Chemother 56:4341–4353. doi: 10.1128/AAC.00448-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Simm AM, Higgins CS, Carenbauer AL, Crowder MW, Bateson JH, Bennett PM, Clarke AR, Halford SE, Walsh TR. 2002. Characterization of monomeric L1 metallo-β-lactamase and the role of the N-terminal extension in negative cooperativity and antibiotic hydrolysis. J Biol Chem 277:24744–24752. doi: 10.1074/jbc.M201524200. [DOI] [PubMed] [Google Scholar]
- 14.García-Sáez I, Mercuri PS, Papamicael C, Kahn R, Frere JM, Galleni M, Rossolini GM, Dideberg O. 2003. Three-dimensional structure of FEZ-1, a monomeric subclass B3 metallo-β-lactamase from Fluoribacter gormanii, in native form and in complex with d-captopril. J Mol Biol 325:651–660. doi: 10.1016/S0022-2836(02)01271-8. [DOI] [PubMed] [Google Scholar]
- 15.Docquier JD, Benvenuti M, Calderone V, Stoczko M, Menciassi N, Rossolini GM, Mangani S. 2010. High-resolution crystal structure of the subclass B3 metallo-β-lactamase BJP-1: rational basis for substrate specificity and interaction with sulfonamides. Antimicrob Agents Chemother 54:4343–4351. doi: 10.1128/AAC.00409-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mojica MF, Mahler SG, Bethel CR, Taracila MA, Kosmopoulou M, Papp-Wallace KM, Llarrull LI, Wilson BM, Marshall SH, Wallace CJ, Villegas MV, Harris ME, Vila AJ, Spencer J, Bonomo RA. 2015. Exploring the role of residue 228 in substrate and inhibitor recognition by VIM metallo-β-lactamases. Biochemistry 54:3183–3196. doi: 10.1021/acs.biochem.5b00106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yamaguchi Y, Jin W, Matsunaga K, Ikemizu S, Yamagata Y, Wachino J, Shibata N, Arakawa Y, Kurosaki H. 2007. Crystallographic investigation of the inhibition mode of a VIM-2 metallo-β-lactamase from Pseudomonas aeruginosa by a mercaptocarboxylate inhibitor. J Med Chem 50:6647–6653. doi: 10.1021/jm701031n. [DOI] [PubMed] [Google Scholar]
- 18.Klingler FM, Wichelhaus TA, Frank D, Cuesta-Bernal J, El-Delik J, Muller HF, Sjuts H, Gottig S, Koenigs A, Pos KM, Pogoryelov D, Proschak E. 2015. Approved drugs containing thiols as inhibitors of metallo-β-lactamases: strategy to combat multidrug-resistant bacteria. J Med Chem 58:3626–3630. doi: 10.1021/jm501844d. [DOI] [PubMed] [Google Scholar]
- 19.King DT, Worrall LJ, Gruninger R, Strynadka NC. 2012. New Delhi metallo-β-lactamase: structural insights into β-lactam recognition and inhibition. J Am Chem Soc 134:11362–11365. doi: 10.1021/ja303579d. [DOI] [PubMed] [Google Scholar]
- 20.Wachino J, Yamaguchi Y, Mori S, Yamagata Y, Arakawa Y, Shibayama K. 2012. Crystallization and preliminary X-ray analysis of the subclass B3 metallo-β-lactamase SMB-1 that confers carbapenem resistance. Acta Crystallogr Sect F Struct Biol Cryst Commun 68:343–346. doi: 10.1107/S1744309112004691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Otwinowski Z, Minor W. 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol 276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 22.Battye TG, Kontogiannis L, Johnson O, Powell HR, Leslie AG. 2011. iMOSFLM: a new graphical interface for diffraction-image processing with MOSFLM. Acta Crystallogr D Biol Crystallogr 67:271–281. doi: 10.1107/S0907444910048675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Evans PR. 1997. SCALA, continuous scaling program. Joint CCP4 ESF-EACBM Newsl 33:22–24. [Google Scholar]
- 24.Vagin A, Teplyakov A. 2010. Molecular replacement with MOLREP. Acta Crystallogr. D Biol Crystallogr 66:22–25. doi: 10.1107/S0907444909042589. [DOI] [PubMed] [Google Scholar]
- 25.Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AG, McCoy A, McNicholas SJ, Murshudov GN, Pannu NS, Potterton EA, Powell HR, Read RJ, Vagin A, Wilson KS. 2011. Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr 67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Emsley P, Cowtan K. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol Crystallogr 60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 27.Murshudov GN, Vagin AA, and Dodson EJ. 1997. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr 53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 28.Lovell SC, Davis IW, Arendall WB III, de Bakker PI, Word JM, Prisant MG, Richardson JS, Richardson DC. 2003. Structure validation by Calpha geometry: phi, psi and Cbeta deviation. Proteins 50:437–450. doi: 10.1002/prot.10286. [DOI] [PubMed] [Google Scholar]
- 29.Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS, and Olson AJ. 2009. AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J Comput Chem 30:2785–2791. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Clinical and Laboratory Standards Institute. 2012. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically; approved standard—ninth edition. Document M07-A9. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 31.Page MI, Badarau A. 2008. The mechanisms of catalysis by metallo beta-lactamases. Bioinorg Chem Appl 2008:576297. doi: 10.1155/2008/576297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Feng H, Ding J, Zhu D, Liu X, Xu X, Zhang Y, Zang S, Wang DC, Liu W. 2014. Structural and mechanistic insights into NDM-1 catalyzed hydrolysis of cephalosporins. J Am Chem Soc 136:14694–14697. doi: 10.1021/ja508388e. [DOI] [PubMed] [Google Scholar]
- 33.Karsisiotis AI, Damblon CF, Roberts GC. 2014. A variety of roles for versatile zinc in metallo-β-lactamases. Metallomics 6:1181–1197. doi: 10.1039/c4mt00066h. [DOI] [PubMed] [Google Scholar]
- 34.Meini MR, Llarrull LI, and Vila AJ. 2015. Overcoming differences: the catalytic mechanism of metallo-β-lactamases. FEBS Lett 589:3419–3432. doi: 10.1016/j.febslet.2015.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gatti DL. 2012. Biapenem inactivation by B2 metallo β-lactamases: energy landscape of the post-hydrolysis reactions. PLoS One 7:e30079. doi: 10.1371/journal.pone.0030079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nauton L, Kahn R, Garau G, Hernandez JF, Dideberg O. 2008. Structural insights into the design of inhibitors for the L1 metallo-β-lactamase from Stenotrophomonas maltophilia. J Mol Biol 375:257–269. doi: 10.1016/j.jmb.2007.10.036. [DOI] [PubMed] [Google Scholar]
- 37.Liu XL, Shi Y, Kang JS, Oelschlaeger P, Yang KW. 2015. Amino acid thioester derivatives: a highly promising scaffold for the development of metallo-β-lactamase L1 inhibitors. ACS Med Chem Lett 6:660–664. doi: 10.1021/acsmedchemlett.5b00098. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.