Abstract
Infections caused by multidrug-resistant Acinetobacter baumannii are a major public health problem, and polymyxins are often the last line of therapy for recalcitrant infections by such isolates. The pharmacokinetics of the two clinically used polymyxins, polymyxin B and colistin, differ considerably, since colistin is administered as an inactive prodrug that undergoes slow conversion to colistin. However, the impact of these substantial pharmacokinetic differences on bacterial killing and resistance emergence is poorly understood. We assessed clinically relevant polymyxin B and colistin dosage regimens against one reference and three clinical A. baumannii strains in a dynamic one-compartment in vitro model. A new mechanism-based pharmacodynamic model was developed to describe and predict the drug concentrations and viable counts of the total and resistant populations. Rapid attainment of target concentrations was shown to be critical for polymyxin-induced bacterial killing. All polymyxin B regimens achieved peak concentrations of at least 1 mg/liter within 1 h and caused ≥4 log10 killing at 1 h. In contrast, the slow rise of colistin concentrations to 3 mg/liter over 48 h resulted in markedly reduced bacterial killing. A significant (4 to 6 log10 CFU/ml) amplification of resistant bacterial populations was common to all dosage regimens. The developed mechanism-based model explained the observed bacterial killing, regrowth, and resistance. The model also implicated adaptive polymyxin resistance as a key driver of bacterial regrowth and predicted the amplification of preexisting, highly polymyxin-resistant bacterial populations following polymyxin treatment. Antibiotic combination therapies seem the most promising option for minimizing the emergence of polymyxin resistance.
INTRODUCTION
The rise of multidrug-resistant (MDR) Gram-negative pathogens, such as Acinetobacter baumannii, presents a global threat to public health (1, 2). The number of antibiotics that remain effective for the treatment of infections caused by these pathogens continues to dwindle at an alarming rate, with few novel alternatives in late-phase clinical development (3). This has driven a resurgence in the use of polymyxins to treat recalcitrant infections that are resistant to most or all other currently available antibiotics (4). The two clinically used polymyxins, colistin and polymyxin B, differ substantially in clinical pharmacokinetics (PK) due to differences in administered form. Although colistin and polymyxin B have long been regarded as equivalent, clinical studies in critically ill patients have revealed considerable differences in their PK (5–12). Given the importance of polymyxins as last-line therapy, developing innovative dosage regimens for polymyxins will be critical to maintaining their efficacy against MDR pathogens.
Clinically, polymyxin B is administered in its active form whereas colistin (polymyxin E) is administered as its inactive prodrug colistin methanesulfonate (CMS), which subsequently undergoes slow conversion to colistin in patients (13). CMS is eliminated rapidly via the renal route in parallel with its gradual conversion to colistin, and consequently a large proportion of CMS is eliminated as CMS prior to conversion (14). Even when CMS loading doses are used, the low rate of conversion from CMS to colistin leads to delayed attainment of effective colistin concentrations (8). Conversely, administration of polymyxin B in its active form allows target concentrations to be rapidly attained.
A clear association between the timely initiation of appropriate antimicrobial therapy and patient survival has been established for conditions such as ventilator-associated pneumonia and septic shock (15–17). In light of this relationship, the differences in the concentration-versus-time profiles of polymyxin B and colistin may have a substantial impact on bacterial killing and clinical outcomes. The differences in bacterial killing and the emergence of resistance for polymyxin B and colistin during the initial stages of polymyxin therapy with clinically relevant dosage regimens have not been investigated.
The first aim of this study was to investigate, in A. baumannii, the effect of polymyxin concentration-versus-time profiles from standard and innovative (e.g., front-loading or augmented loading dose) dosage regimens on bacterial killing and emergence of resistance during the initial stages of polymyxin therapy. The second aim was to quantitatively characterize and predict the impact of polymyxin dosage regimens on antimicrobial activity and the emergence of resistance by developing a novel, mechanism-based mathematical model. To address these aims, experimental data from a dynamic one-compartment in vitro infection model (IVM) were integrated with pharmacokinetic/pharmacodynamic (PK/PD) modeling, enabling the interrogation and prediction of bacterial killing and regrowth for clinically relevant polymyxin dosage regimens.
MATERIALS AND METHODS
Bacterial strains and isolates.
One reference strain and three previously reported (18–20) clinical isolates were examined in this study. These were A. baumannii ATCC 19606 (MIC, 0.5 mg/liter), FADDI-AB008 (MIC, 0.5 mg/liter; described in reference 19 as isolate 8), FADDI-AB030 (MIC, 0.5 mg/liter; described in reference 20 as strain 248-01-C.248), and AB307-0294 (MIC, 1 mg/liter). The polymyxin B and colistin MIC values were identical for each of the strains. A. baumannii ATCC 19606 and FADDI-AB008 have previously been identified as being heteroresistant (19), consisting of predominantly polymyxin-susceptible bacteria but with a small subpopulation of polymyxin-resistant bacteria (21). Population analysis profiles (PAPs) for A. baumannii strain AB307-0294 showed evidence of heteroresistance, while there was no evidence for heteroresistance in susceptible strain FADDI-AB030. In A. baumannii ATCC 19606, polymyxin resistance can occur via the modification of lipid A with phosphoethanolamine (22) or the loss of lipopolysaccharide (LPS) from the outer membrane (23). In the case of FADDI-AB008, only one mechanism of polymyxin resistance has been reported, namely, the loss of LPS from the outer membrane (23).
Comparison of clinically relevant colistin and polymyxin B dosage regimens.
An IVM was used to simulate clinically relevant pharmacokinetic profiles for polymyxin B and colistin. The experimental setup for the IVM did not employ a filter and was identical to that used in previously published studies (24), except where described below. For each dosage regimen and bacterial strain, an elimination half-life (t1/2) of 11.6 h and an average steady-state concentration (Css,avg) of 3 mg/liter were simulated in the IVM, guided by the results of previously published population PK studies in critically ill patients (5, 8, 9, 25). Specifically, the elimination half-life was chosen to be within the range of values reported (9 to 13 h) for colistin formed from CMS and polymyxin B. Colistin and polymyxin B are approximately 50% bound in plasma of critically ill patients (9, 26, 27). The Css,avg of 3 mg/liter chosen for the studies in the IVM was selected to represent the upper region of clinically achievable unbound concentrations of each of the polymyxins. For colistin, an unbound Css,avg of 3 mg/liter represents the accumulation of colistin observed by Garonzik et al. (5) for a patient with moderate renal impairment (creatinine clearance, ∼50 ml/min/1.73 m2) receiving a daily maintenance CMS dose of ∼300 to 360 mg colistin base activity (CBA). A loading dose to more rapidly attain a concentration of 3 mg/liter was not included because 300 to 360 mg CBA is in the vicinity of the upper limit of the recommended loading dose (28). For polymyxin B, an unbound Css,avg of 3 mg/liter falls within the 90th percentile for a patient receiving a maintenance dose of 3 mg/kg per day reported by Sandri et al. (9).
The dosage regimens examined in the IVM (Fig. 1) were as follows: the gradual accumulation of colistin with a half-life of 11.6 h, adapted from previously reported (8) concentration-time profiles predicted for the initiation of therapy with CMS with no loading doses (regimen R1); polymyxin B, 1-h infusion every 12 h with no loading dose (R2); regimen R2 with a loading dose to rapidly achieve the steady-state concentration (R3); and regimen R2 initiated with an augmented loading dose to achieve an initial peak concentration of 6 mg/liter polymyxin B base with a subsequent decline to a Css,avg of 3 mg/liter (R4).
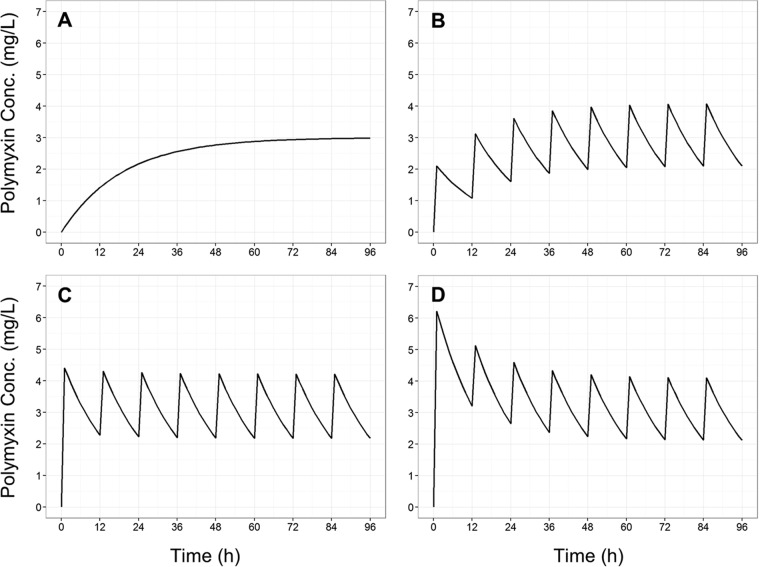
FIG 1.
Concentration-time profiles for colistin base (A) and polymyxin B base (B to D) of simulated (t1/2 = 11.6 h, Css,avg = 3 mg/liter) clinically relevant polymyxin dosage regimens. (A) Gradual rise of colistin (as seen in patients following CMS administration); (B) polymyxin B, 1-h infusion every 12 hours with no loading dose; (C) polymyxin B, 1-h infusion every 12 hours with a conventional loading dose; (D) polymyxin B, 1-h infusion every 12 hours with an augmented loading dose.
The volume and temperature of the central reservoir were 80 ml and 37°C, respectively, and cation-adjusted Mueller-Hinton broth was circulated at a rate of 4.8 ml/h to achieve an elimination half-life of 11.6 h. Viable counting was performed on drug-free and drug-containing (6.6 mg/liter polymyxin B base) plates at 0, 0.5, 1, 2, 4, 8, 11, 13, 23, 25, 26, 28, 47, 49, 50, 52, 71, 73, 74, 76, and 96 h. The PAPs were performed at 0, 23, 47, 71, and 96 h on polymyxin B-containing plates (1.7, 3.3, and 6.6 mg/liter polymyxin B base). Polymyxin B and colistin concentrations were measured using a previously described and validated liquid chromatography-tandem mass spectrometry assay (29).
Mechanism-based modeling.
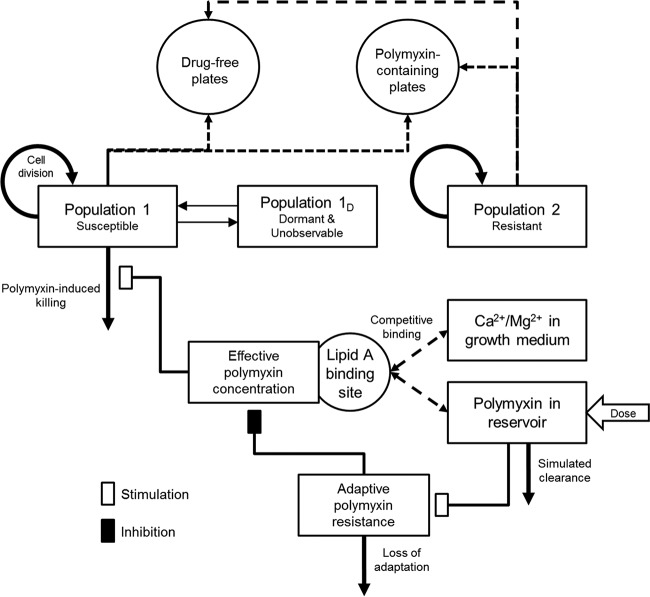
A mathematical model was developed and fitted to the experimental data for each strain to correlate the time course of polymyxin concentrations with bacterial killing and the emergence of resistance (Fig. 2). The PK of polymyxin B or colistin in the IVM was described by a one-compartment model with first-order elimination. The mechanism-based PD model contained growth of multiple bacterial populations, polymyxin target site binding, bacterial killing, and adaptive polymyxin resistance (30, 31).
FIG 2.
Model structure of the developed mechanism-based model for polymyxin-mediated bacterial killing and the dynamics of constitutive and adaptive polymyxin resistance. The washout of bacteria from the IVM has not been included in this diagram for clarity.
Bacterial growth model.
For each A. baumannii strain, bacterial cells were partitioned into 3 populations: polymyxin-susceptible (CFUS, equation 1), polymyxin-resistant (CFUR, equation 2), and dormant (i.e., nonreplicating or extremely slowly replicating; PopD, equation 3) bacteria. Replication of polymyxin-susceptible and -resistant bacteria was modeled as a first-order process, with separate growth rate constants for each population. The total bacterial population for each strain was constrained by a logistic growth term that defined the maximum population size (32).
| (1) |
| (2) |
| (3) |
The parameters in these equations are explained in detail in Table 1 and Fig. 2. The resistant population was assumed to be totally resistant, and therefore, equation 2 lacked a term for killing. Polymyxin-susceptible bacteria were allowed to transition, in both directions, into and out of a dormant state via a first-order process that rendered these cells both tolerant to polymyxin-induced killing and also nonobservable by viable counting. The presence of viable but nonculturable bacterial cells has been documented previously (33, 34); however, the impact of this phenomenon on polymyxin activity and the emergence of resistance has not been elucidated.
TABLE 1.
Population parameter estimates of the fitted models for each examined A. baumannii strain
| Parameter (symbol and/or unit) | Mean (SE) for Acinetobacter baumannii strain: |
|||
|---|---|---|---|---|
| ATCC 19606 | AB307-0294 | FADDI-AB008 | FADDI-AB030 | |
| Initial inoculum for CFUtotal (log10 CFU/ml) | 6.34 (3.14%) | 6.14 (1.87%) | 5.93 (4.14%) | 6.17 (3.02%) |
| Initial inoculum for CFUR (log10 CFU/ml) | −0.249 (95.2%) | −0.503 (34.6%) | −0.44 (73%) | −0.0466 (6.13%) |
| Maximal bacterial population (CFUmax, log10 CFU/ml) | 7.35 (1.04%) | 8.05 (1.12%) | 7.66 (1.07%) | 7.82 (1.56%) |
| Mean turnover time CFUS (min) | 75.2 (21.1%) | 32 (10.7%) | 35.2 (11.5%) | 27.7 (10.2%) |
| Mean turnover time CFUR (min) | 71.4 (12.3%) | 81.0 (6.79%) | 48.1 (9.58%) | 313 (9.85%) |
| Transition rate constant to dormancy (kSD, h−1) | 0.00635 (70%) | 0.0367 (42.9%) | 0.00246 (44.5%) | 0.000916 (102%) |
| Transition rate constant out of dormancy (kDS, h−1) | 3.19 (24.2%) | 0.0332 (47.1%) | 5.45 (38.9%) | 7.05 (15.7%) |
| Hill coefficient for polymyxin binding to LPS (Hillbinding) | 3.06 (16.2%) | 4.13 (30.3%) | 3.25 (68.2%) | 4.83 (18.3%) |
| EC50b for polymyxin binding to LPS (EC50) | 0.0483 (8.25%) | 0.0989 (9.76%) | 0.0237 (19.7%) | 0.0173 (7.17%) |
| Maximal polymyxin-induced killing rate constant (Killmax, h−1) | 100 (fixed) | 100 (fixed) | 100 (fixed) | 100 (fixed) |
| Hill coefficient for polymyxin-induced killing (Hillkilling) | 3.85 (20%) | 2.79 (9.64%) | 1.75 (13%) | 19.5 (14.9%) |
| EC50 for polymyxin-induced killing (KillC50, mg/liter) | 0.521 (22.5%) | 0.561 (22.3%) | 0.693 (23.9%) | 0.516 (13.1%) |
| Maximal fold reduction in the effective polymyxin concn due to adaptive resistance (Smax) | 300 (fixed) | 300 (fixed) | 300 (fixed) | 300 (fixed) |
| Adaptation rate constant (kadapt, h−1) | 7.20 (16.5%) | 14.2 (12.9%) | 8.09 (16.3%) | 15.2 (10.5%) |
| Maximal fitness cost associated with resistance (Ginhib,max) | NEa | NE | 0.994 (9.43%) | 0.991 (5.07%) |
| In vitro model flow rate (CL, liters/h) | 0.00434 (10.4%) | 0.00506 (9.14%) | 0.00585 (9.97%) | 0.00476 (8.98%) |
| In vitro model reservoir vol (V, liters) | 0.111 (13.5%) | 0.101 (13.6%) | 0.119 (9.87%) | 0.133 (17.6%) |
NE, not estimated: parameters were not included for estimation in the model, as the experimental data did not support the inclusion of this feature for these strains.
EC50, 50% effective concentration.
Polymyxin activity.
A previously reported model was used to describe the binding of polymyxin to its initial bactericidal target, the lipid A moiety of LPS on the bacterial outer membrane (30). This model accounts for the competitive displacement of bound divalent cations (Fbound,cations; Ca2+ and Mg2+) from lipid A by polymyxins (equations 4 and 5). The effective polymyxin concentration (Cpolymyxin,eff) was then calculated, accounting for the occupancy of the lipid A binding site (Fpolymyxin,eff) and the concentration of divalent cations in cation-adjusted Mueller-Hinton broth (equation 6). Bacterial killing by polymyxin (Killpolymyxin,eff) was described by a Hill equation (equation 7) that is commonly used to describe antimicrobial activity (35, 36).
| (4) |
| (5) |
| (6) |
| (7) |
Polymyxin resistance.
Resistance to polymyxins was characterized as either constitutive or adaptive. Constitutive polymyxin resistance was assumed to be complete (i.e., no bacterial killing). Therefore, the killing rate constant of the resistant population was zero. Adaptive resistance (also called tolerance) affected the polymyxin-susceptible bacterial population and reduced the effective polymyxin concentration (equation 6). In the model, adaptive resistance (Radaptive) was described in a previously reported (31) turnover model for adaptation (equations 8 and 9), accounting for effective polymyxin concentration (Stim). The term Smax represents the maximal fold reduction in the effective polymyxin concentration due to adaptive resistance. This approach is closely aligned with previous reports on the mechanism of adaptive polymyxin resistance, where lipid A modifications (with nonanionic moieties such as arabinose/phosphoethanolamine) lead to an attenuation of its electrostatic interaction with polymyxins (37, 38). In two of the four A. baumannii strains (FADDI-AB008 and FADDI-AB030), the experimental data supported the inclusion of a fitness cost (Fcost) associated with adaptive polymyxin resistance that slowed the replication rate of the susceptible population, depending on the rate of adaptation (equation 10).
| (8) |
| (9) |
| (10) |
Observation model.
The mathematical model outlined is premised on the number of colonies observed in standard viable counting on drug-free (CFUagar,drug-free) and drug-containing (CFUagar,drug-containing) plates being representative of bacterial population dynamics. We had to develop a new observation model, since current PK/PD models could not explain the gradual emergence of antibiotic-resistant bacteria in the absence of killing. This novel model was implemented to capture the contribution of both constitutive and adaptive resistance to the number of colonies formed on polymyxin-containing (6.6 mg/liter) agar plates. In this model, the viable count on polymyxin-containing agar plates was equal to the sum of the polymyxin-resistant population and the modeled proportion of polymyxin-susceptible bacteria that remain viable following a 24-h exposure to polymyxin at 6.6 mg/liter in agar (equation 12). The rate of bacterial killing (equation 7) used to calculate the proportion of bacteria that remained viable during incubation was assumed to be the killing rate at the time of sampling, remaining unchanged during incubation.
| (11) |
| (12) |
Parameter estimation, model qualification, and simulations.
For each A. baumannii strain, PK/PD model parameters were simultaneously estimated in S-ADAPT (version 1.57) (39) using a Monte Carlo parametric expectation maximization algorithm (i.e., importance sampling pmethod = 4 in S-ADAPT). SADAPT-TRAN was used to facilitate modeling analysis (40, 41). To assess the significance and overall contribution of each model component, alternate simplified models that lacked the respective model feature or parameter were fit to the experimental data. The change in the objective function (−2 log-likelihood) value was then used to determine the importance of the model component that was simplified.
The final model was then used to compare bacterial killing, bacterial regrowth, and the emergence of resistance associated with previously reported concentration-time profiles for colistin (8) and polymyxin B (9). For colistin, the unbound concentration-time profile used in the simulation mimicked that predicted by Plachouras et al. (8) following the administration of a 12-MU CMS loading dose (∼360 mg CBA) followed by 4.5 MU (∼140 mg CBA) every 12 hours in a patient with normal creatinine clearance. In the case of polymyxin B, the unbound concentration-time profile representative of the 50th percentile of Monte Carlo simulations reported by Sandri et al. (9) following a loading dose of 2 mg/kg of body weight (2-h infusion) and a daily maintenance dose of 1.25 mg/kg (1-h infusion) was used. The unbound concentrations for both polymyxins were calculated based upon 50% plasma protein binding (9, 26, 27). Model simulations were performed in Berkeley Madonna using the population mean parameter estimates based on the experimental data for polymyxin-heteroresistant A. baumannii strain AB307-0294.
RESULTS
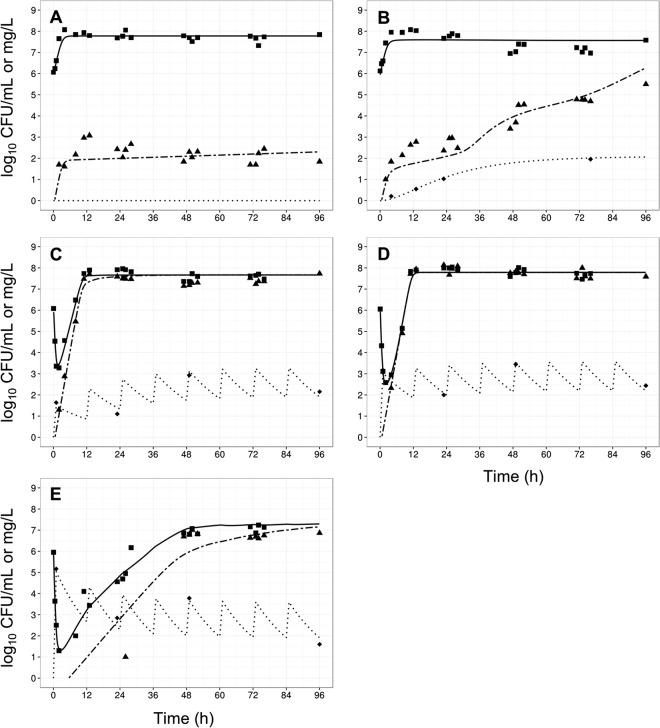
The gradual accumulation of colistin arising from regimen R1 (Fig. 1A) resulted in little antibacterial activity against all the examined A. baumannii strains (Fig. 3B, 4B, 5B, and 6B). Indeed, overall the time course of CFU per milliliter for each strain was similar to that for the corresponding growth control (panels A in each of Fig. 3 to 6). The maximum observed bacterial killing of ∼1 log10 CFU/ml was achieved between 47 and 49 h after commencing treatment (Fig. 6), despite achieving colistin base concentrations greater than 1 mg/liter (equivalent to 1.26 mg/liter colistin sulfate) within 24 h. Further, amplification of polymyxin-resistant bacterial populations was observed in the PAPS after approximately 48 h of colistin exposure in all examined strains. At the conclusion of treatment, a significant proportion of the bacterial population was polymyxin resistant. Analysis of the PAPs revealed a modest difference (<3 log10 CFU/ml) between viable counts on drug-free and 6.6-mg/liter polymyxin B plates for all A. baumannii strains except ATCC 19606, where the difference was ∼4 log10 CFU/ml.
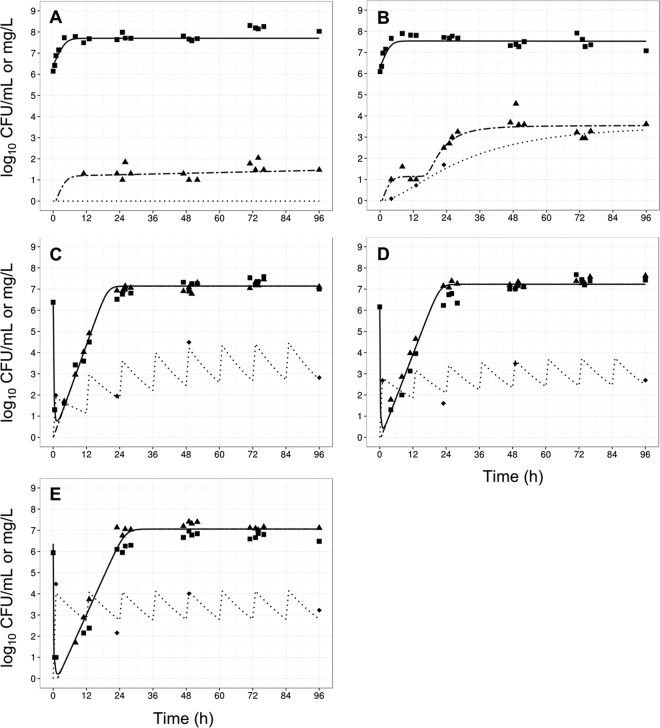
FIG 3.
Observed data (points) and individual model fits (lines) for total viable bacteria (■ and thick solid line), resistant bacteria (▲ and thick dashed line), and polymyxin concentrations (◆, dotted line) for control (A), gradual rise of colistin (R1) (B), polymyxin B with no loading dose (R2) (C), a conventional loading dose (R3) (D), and augmented loading dose (R4) (E) against A. baumannii strain ATCC 19606.
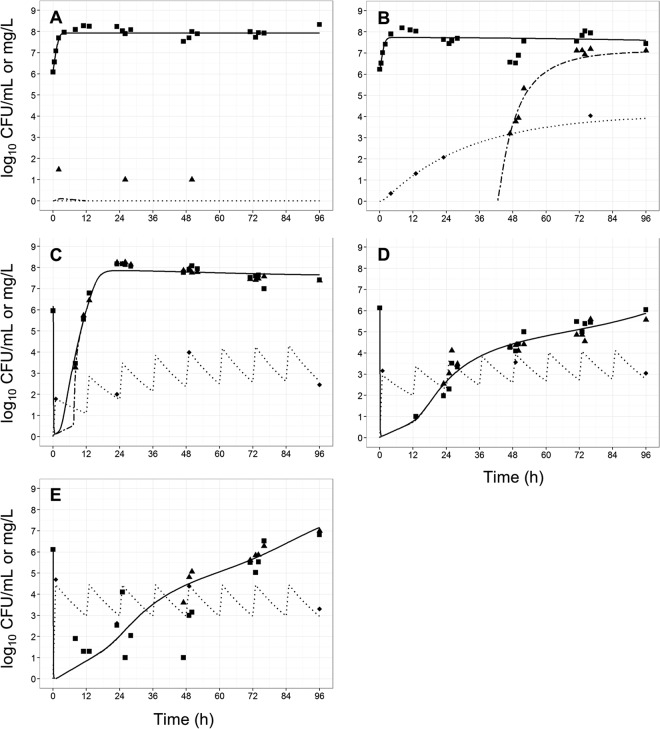
FIG 4.
Observed data (points) and individual model fits (lines) for total viable bacteria (■ and thick solid line), resistant bacteria (▲ and thick dashed line), and polymyxin concentrations (◆, dotted line) for control (A), gradual rise of colistin (R1) (B), polymyxin B with no loading dose (R2) (C), a conventional loading dose (R3) (D), and augmented loading dose (R4) (E) against A. baumannii strain AB307-0294.
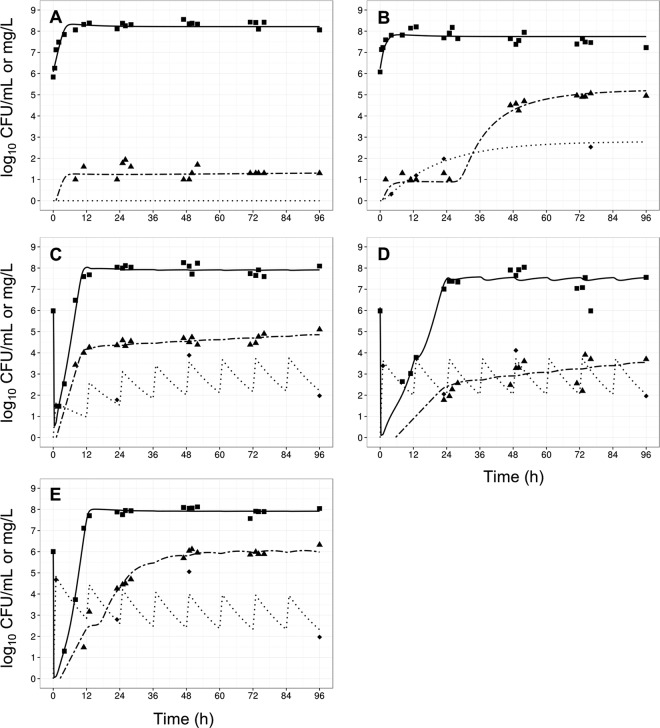
FIG 5.
Observed data (points) and individual model fits (lines) for total viable bacteria (■ and thick solid line), resistant bacteria (▲ and thick dashed line), and polymyxin concentrations (◆, dotted line) for control (A), gradual rise of colistin (R1) (B), polymyxin B with no loading dose (R2) (C), a conventional loading dose (R3) (D), and augmented loading dose (R4) (E) against A. baumannii strain FADDI-AB008.
FIG 6.
Observed data (points) and individual model fits (lines) for total viable bacteria (■ and thick solid line), resistant bacteria (▲ and thick dashed line), and polymyxin concentrations (◆, thick dotted line) for control (A), gradual rise of colistin (R1) (B), polymyxin B with no loading dose (R2) (C), a conventional loading dose (R3) (D), and augmented loading dose (R4) (E) against A. baumannii strain FADDI-AB030.
In contrast, all three polymyxin B dosage regimens (Fig. 1) displayed good initial antibacterial activity against the examined A. baumannii strains, achieving >4 log10 CFU/ml killing within 1 h of treatment initiation (panels C to E in each of Fig. 3 to 6). However, bacterial regrowth was observed at 11 to 13 h after the initiation of therapy. By 96 h, bacterial density was >7 log10 CFU/ml for all dosage regimens and strains examined. The bacterial population following regrowth was polymyxin resistant, with a <1-log10 CFU/ml difference in viable counts at 96 h between drug-free and polymyxin B-containing (6.6 mg/liter) plates for all strains except AB307-0294 (Fig. 4). Against A. baumannii strains FADDI-AB008 and FADDI-AB030, administration of the regimen with an augmented loading dose of polymyxin B (R4, Fig. 5E and 6E) resulted in delayed and slow bacterial regrowth compared to that observed with R2 (Fig. 5C and 6C). The administration of loading doses did not materially affect the regrowth of A. baumannii strains ATCC 19606 and AB307-0294 (panels E versus C in Fig. 3 and 4).
The developed mechanism-based PK/PD model (Fig. 2) described well the kinetics of bacterial killing and regrowth of A. baumannii on antibiotic-free and antibiotic-containing agar plates (Table 1; Fig. 3 to 6). To improve numerical stability during the estimation, the maximal bacterial killing rate (Killmax) was fixed to 100 h−1 (equivalent to 11 log10 killing in 15 min) and the maximal attenuation in killing (i.e., extent of adaptive resistance; Smax) to 300. Model qualification demonstrated the significant contribution (P < 0.05) of each component to model fits, with the objective function (−2 log likelihood) worsening with the omission of any individual component for at least 2 of 4 strains (Table 2).
TABLE 2.
Model validation results, showing the change in −2 log likelihood associated with modifying and deleting model features
| Model features | Resulta for Acinetobacter baumannii strain: |
|||
|---|---|---|---|---|
| ATCC 19606 | AB307-0294 | FADDI-AB008 | FADDI-AB030 | |
| Simplified model | ||||
| Contribution of susceptible bacteria to viable counts on drug-containing plates, equation 12 | NSa | 77.6 | NS | 96.5 |
| Transition of susceptible bacteria into a dormant, nonculturable, polymyxin-resistant state, equation 3 | 30.6 | 65.9 | 11.6 | 109.8 |
| Constitutively highly polymyxin-resistant bacteria, equation 2 | 488.5 | 391.7 | 338.0 | 281.3 |
| Adaptive resistance, equations 8 and 9 | 82.3 | —b | — | — |
| Alternative model specification | ||||
| Linear relationship between polymyxin concn and bacterial killing | 73.9 | 136.6 | — | 128.9 |
| Three transit compartments for adaptive resistance (instead of one; equation 9) | NS | 38.1 | NS | 51.3 |
| Five transitory compartments for adaptive resistance (instead of one; equation 9) | NS | 74.6 | 24.4 | 50.8 |
NS, not significant: the data did not support inclusion of this feature in the model for the respective strain.
—, feature lacking: the models that lacked this feature could not be robustly estimated for the respective strain(s), most likely since the simplified model provided very poor curve fits. This strongly suggested that the respective model feature was beneficial and should be included.
Examination of the modeled bacterial population dynamics revealed that adaptive resistance was a primary mode of polymyxin resistance enabling bacterial regrowth during polymyxin treatment. The mechanism-based model also strongly implicated the involvement of viable but nonculturable bacterial cells in bacterial regrowth during polymyxin treatment; deletion of this model feature resulted in a substantial worsening in the objective function (Table 2).
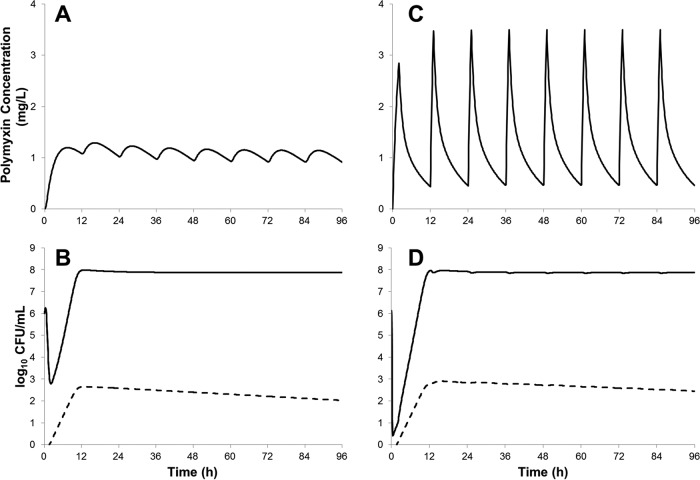
Simulations conducted using the model fitted for A. baumannii AB307-0294 suggested that unbound colistin concentration-time profiles achieved with the use of aggressive CMS loading doses (Fig. 7A) would result in ∼3 log10 CFU/ml killing (Fig. 7B). In contrast, the simulated polymyxin B dosage regimen (Fig. 7C) resulted in more-rapid and ∼2-log10 CFU/ml-greater bacterial killing than that obtained with the CMS dosage regimen (Fig. 7D). Regrowth of polymyxin-resistant bacteria due to adaptive resistance, as well as the amplification of constitutively polymyxin-resistant subpopulations (∼0.001% of total bacteria; ∼1,000-fold amplification), was predicted within less than 24 h with both dosage regimens.
FIG 7.
Model simulation results for A. baumannii strain AB307-0294 (initial inoculum, 106 CFU/ml) treated with unbound concentration-time profiles adapted from population PK studies (8, 9) of colistin following CMS administration in a patient with normal renal function (360 mg colistin base activity loading dose, 140 mg every 12 h maintenance dose) (A) and a polymyxin B dosage regimen representing the 50th percentile of critically ill patients receiving a 2-mg/kg polymyxin B loading dose, followed by 1.25 mg/kg every 12 h (C). The dynamics of initially susceptible (adaptive resistance) (solid line) and constitutively resistant (dashed line) bacterial populations are shown separately (B and D).
DISCUSSION
The resurgence in clinical usage of polymyxins and their importance as “last-line” therapeutic agents for the treatment of infections caused by MDR Gram-negative organisms necessitates investigations into the factors that influence polymyxin activity and the development of resistance. Colistin and polymyxin B, the two clinically used polymyxins, share similar in vitro activity but differ substantially in the time course of concentrations achieved in the hours, and potentially days, following initiation of therapy due to their administered forms (7). In this report, we have employed a dynamic in vitro PK/PD model and mathematical modeling to explore the effects of differences in concentration-time profiles on the activity of polymyxins and the development of polymyxin resistance due to amplification of preexisting resistant mutants and adaptive resistance (42, 43). The developed mathematical model implemented a new observation model that linked adaptive resistance to colony formation on drug-containing plates, extending the utility of previous models of adaptive resistance (31, 44).
The one-compartment IVM was able to simulate clinically relevant polymyxin B and colistin concentration-time profiles, as seen in previously published studies (5, 8, 9). It should be noted that the polymyxin concentrations investigated in the IVM were in the region of the upper limits of unbound polymyxin concentrations seen in critically ill patients, to account for the previously observed resilience of A. baumannii to treatment with colistin (45). For colistin, an unbound Css,avg of 3 mg/liter would be possible only in a patient with moderate renal function receiving ∼300 to 360 mg CBA each day; patients with higher renal function achieve substantially lower concentrations (5). Of great concern was our finding that the gradual rise in colistin concentrations to a Css,avg of 3 mg/liter enabled the acquisition of polymyxin resistance to outpace the accumulation of colistin, leading to little bacterial killing over the treatment period. In contrast, all the polymyxin B dosage regimens showed good bacterial killing during the initial stages of treatment, likely as a result of achieving bactericidal concentrations prior to the onset of adaptive polymyxin resistance. Notably, the emergence of polymyxin-resistant bacterial populations was substantial following treatment with either colistin or polymyxin B dosage regimens, despite significant differences in the magnitude of bacterial killing. Taken together, these findings show the clear link between the rapid attainment of therapeutic polymyxin concentrations and bacterial killing and the alarming rate at which resistance to polymyxin develops.
In this study, polymyxin B loading doses (conventional and augmented) were found to slow the rate of bacterial regrowth for FADDI-AB030 (conventional and augmented loading doses) and FADDI-AB008 (augmented loading dose only). A similarly clear relationship between the use of loading doses and improved suppression of bacterial regrowth was not seen in the remaining A. baumannii strains, ATCC 19606 and AB307-0294. The mechanistic basis of this observed slowing in the bacterial regrowth rate in A. baumannii when treated with polymyxin B loading doses remains to be elucidated; however, it has been suggested that heteroresistance plays a significant role in determining the rate at which antimicrobial resistance emerges (21).
Despite the growing body of clinical evidence surrounding the beneficial effect of rapid attainment of therapeutic antibiotic concentrations (15–17) and the demonstration of superior activity of polymyxin B loading doses in vitro against some strains in the current study, the benefits of polymyxin B loading doses have not been thoroughly investigated clinically. Clearly, the implications of such dosage regimens on nephrotoxicity should be carefully considered. The key drivers of polymyxin-induced nephrotoxicity are the subject of continuing research (46–49), and findings from those studies will be integral to the development of clinical guidelines for safe and effective polymyxin B dosing.
Current literature suggests that polymyxin resistance in A. baumannii is driven by either the modification of the lipid A moiety of LPS or the loss of LPS from the outer membrane (22, 23, 37, 50). In other bacterial species, these changes to the bacterial outer membrane arise due to preexisting mutations or adaptive responses to polymyxins (38). The dynamics between constitutively polymyxin-resistant (mutants) and adaptively polymyxin-resistant bacterial populations during polymyxin therapy have not been previously investigated. In line with recently published recommendations (51), the new mechanism-based model described in this report built upon prior work (35, 36, 52) on modeling antimicrobial activity and incorporated both of these resistance mechanisms, in addition to reversible dormancy. The model was able to describe well the observed viable counts on both antibiotic-free and antibiotic-containing agar plates (Fig. 3 to 6).
Analysis of the bacterial population dynamics as described by the mechanism-based model pointed to adaptive resistance as a key factor driving bacterial regrowth following treatment with clinically relevant dosage regimens in the IVM. Further, dormant, nonculturable bacterial cells were identified as a likely contributor to bacterial regrowth following polymyxin treatment. Exclusion of dormant cells from the model resulted in implausible parameter estimates for bacterial replication times (<15 min) to describe the bacterial regrowth following polymyxin-induced bacterial killing. Dormant and slow-growing bacterial cells have been reported to be involved in penicillin resistance in Escherichia coli (53). Further investigations that enable direct observation of bacterial cells (e.g., single-cell microfluidic microscopy) will be required to examine this phenomenon in A. baumannii during polymyxin treatment.
The use of a CMS loading dose to hasten the attainment of therapeutic colistin concentrations has been previously proposed (8, 11, 27, 54, 55). It should be noted that the reported disposition of formed colistin following CMS administration shows substantial variance; a recent report on the disposition of CMS/colistin in critically ill patients has described the rapid formation of colistin following CMS administration (55). However, these findings are contrary to previous reports from multiple investigators across several continents (5, 8, 27, 56). The reasons for the divergence in results remain to be elucidated, and interbrand variability has been hypothesized to be a key contributor (55). A concentration-time profile representative of the more commonly reported pharmacokinetics of CMS/colistin in patients with normal renal function was examined using the mechanism-based model fitted on data from A. baumannii AB307-0294 (Fig. 7). Compared to a polymyxin B dosage regimen with a loading dose as proposed by Sandri et al. (9), the CMS regimen was slow to achieve polymyxin concentrations of >1 mg/liter (cf. ∼4 h for CMS and 1 h for polymyxin B).
Mathematical model simulations indicated that the CMS regimen was likely to result in 2 log10 CFU/ml less bacterial killing (i.e., ∼3 log10 versus ∼5 log10) compared to the polymyxin B dosage regimen (Fig. 7). Model simulations for colistin and polymyxin B also predicted the amplification of constitutively polymyxin-resistant bacteria, in line with the previously identified link between the isolation of heteroresistant A. baumannii and prior treatment with colistin in critically ill patients (57). Heteroresistance in A. baumannii has been documented previously (19, 20, 45), and the clinical impact of these highly polymyxin-resistant bacteria remains to be fully established. However, it seems highly possible that these resistant mutants contribute to clinical failure (21). Conclusive evidence of these phenomena will require molecular confirmation of the modalities of polymyxin resistance mechanisms. Although optimization of polymyxin monotherapy remains as an important goal, a worrying trend in the experimental and simulation data was the regrowth of polymyxin-resistant bacteria across all strains and dosage regimens, a finding common to numerous previously published investigations into polymyxin activity against Gram-negative pathogens in vitro (45, 58, 59). These results suggest that simply optimizing polymyxin monotherapy is unlikely to be adequate to prevent the emergence of resistance and that polymyxin combination therapies must be explored as a strategy to combat multidrug resistance in A. baumannii.
A potential limitation of this study, and a key consideration, is the lack of a functioning immune system in the applied dynamic in vitro experimental model. Studies into granulocyte-mediated bacterial killing of Pseudomonas aeruginosa suggest that a reduction in bacterial burden to ≤6 log10 CFU/ml is likely to prevent saturation of bacterial killing by granulocytes and improve clinical outcomes (60, 61). The in vitro and in silico data from the present study point to bacterial killing of >3 log10 CFU/ml for polymyxin B dosage regimens with loading doses, suggesting that adequate polymyxin dosage regimens will reduce the bacterial burden sufficiently to prevent saturation of granulocyte-mediated killing. Translation of these findings into treatment options for critically ill patients with partially suppressed immune systems will require further investigation and consideration of other clinical factors.
In conclusion, this study demonstrated that clinically relevant concentration-versus-time profiles of colistin resulted in slower and less-extensive bacterial killing than that seen with polymyxin B dosage regimens that rapidly attained target concentrations. These findings demonstrate the advantages of polymyxin B over colistin (administered as CMS) with regard to rapid target concentration attainment and antibacterial activity. Mechanism-based modeling of bacterial populations also showed the important influence of the time course of polymyxin concentration over the first several hours of therapy on the population dynamics between adaptive and constitutively resistant bacterial cells. This information will be critical in the optimization of polymyxin therapy against A. baumannii as well as the rational design of synergistic combination therapies containing polymyxins.
ACKNOWLEDGMENTS
We declare that we have no conflict of interest.
Funding Statement
This work was supported by the National Institute of Allergy and Infectious Diseases at the National Institutes of Health (award numbers R01AI079330 and R01AI111990 to R.L.N., J.L., B.T.T., A.F., and J.B.B. and R01AI098771 to J.L. and R.L.N.). J.L. is an Australian National Health and Medical Research Council (NHMRC) Senior Research Fellow. J.B.B. is the recipient of an NHMRC Career Development Fellowship (APP1084163). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Allergy and Infectious Diseases or the National Institutes of Health.
REFERENCES
- 1.Peleg AY, Seifert H, Paterson DL. 2008. Acinetobacter baumannii: emergence of a successful pathogen. Clin Microbiol Rev 21:538–582. doi: 10.1128/CMR.00058-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Walker B, Barrett S, Polasky S, Galaz V, Folke C, Engstrom G, Ackerman F, Arrow K, Carpenter S, Chopra K, Daily G, Ehrlich P, Hughes T, Kautsky N, Levin S, Maler KG, Shogren J, Vincent J, Xepapadeas T, de Zeeuw A. 2009. Environment. Looming global-scale failures and missing institutions. Science 325:1345–1346. doi: 10.1126/science.1175325. [DOI] [PubMed] [Google Scholar]
- 3.Infectious Diseases Society of America (IDSA), Spellberg B, Blaser M, Guidos RJ, Boucher HW, Bradley JS, Eisenstein BI, Gerding D, Lynfield R, Reller LB, Rex J, Schwartz D, Septimus E, Tenover FC, Gilbert DN. 2011. Combating antimicrobial resistance: policy recommendations to save lives. Clin Infect Dis 52(Suppl 5):S397–S428. doi: 10.1093/cid/cir153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bergen PJ, Landersdorfer CB, Lee HJ, Li J, Nation RL. 2012. ‘Old’ antibiotics for emerging multidrug-resistant bacteria. Curr Opin Infect Dis 25:626–633. doi: 10.1097/QCO.0b013e328358afe5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Garonzik SM, Li J, Thamlikitkul V, Paterson DL, Shoham S, Jacob J, Silveira FP, Forrest A, Nation RL. 2011. Population pharmacokinetics of colistin methanesulfonate and formed colistin in critically ill patients from a multicenter study provide dosing suggestions for various categories of patients. Antimicrob Agents Chemother 55:3284–3294. doi: 10.1128/AAC.01733-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zavascki AP, Goldani LZ, Li J, Nation RL. 2007. Polymyxin B for the treatment of multidrug-resistant pathogens: a critical review. J Antimicrob Chemother 60:1206–1215. doi: 10.1093/jac/dkm357. [DOI] [PubMed] [Google Scholar]
- 7.Nation RL, Velkov T, Li J. 2014. Colistin and polymyxin B: peas in a pod, or chalk and cheese? Clin Infect Dis 59:88–94. doi: 10.1093/cid/ciu213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Plachouras D, Karvanen M, Friberg LE, Papadomichelakis E, Antoniadou A, Tsangaris I, Karaiskos I, Poulakou G, Kontopidou F, Armaganidis A, Cars O, Giamarellou H. 2009. Population pharmacokinetic analysis of colistin methanesulfonate and colistin after intravenous administration in critically ill patients with infections caused by gram-negative bacteria. Antimicrob Agents Chemother 53:3430–3436. doi: 10.1128/AAC.01361-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sandri AM, Landersdorfer CB, Jacob J, Boniatti MM, Dalarosa MG, Falci DR, Behle TF, Bordinhao RC, Wang J, Forrest A, Nation RL, Li J, Zavascki AP. 2013. Population pharmacokinetics of intravenous polymyxin B in critically ill patients: implications for selection of dosage regimens. Clin Infect Dis 57:524–531. doi: 10.1093/cid/cit334. [DOI] [PubMed] [Google Scholar]
- 10.Sandri AM, Landersdorfer CB, Jacob J, Boniatti MM, Dalarosa MG, Falci DR, Behle TF, Saitovitch D, Wang J, Forrest A, Nation RL, Zavascki AP, Li J. 2013. Pharmacokinetics of polymyxin B in patients on continuous venovenous haemodialysis. J Antimicrob Chemother 68:674–677. doi: 10.1093/jac/dks437. [DOI] [PubMed] [Google Scholar]
- 11.Karaiskos I, Friberg LE, Pontikis K, Ioannidis K, Tsagkari V, Galani L, Kostakou E, Baziaka F, Paskalis C, Koutsoukou A, Giamarellou H. 2015. Colistin population pharmacokinetics after application of a loading dose of 9 MU colistin methanesulfonate (CMS) in critically ill patients. Antimicrob Agents Chemother 59:7240–7248. doi: 10.1128/AAC.00554-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Karvanen M, Plachouras D, Friberg LE, Paramythiotou E, Papadomichelakis E, Karaiskos I, Tsangaris I, Armaganidis A, Cars O, Giamarellou H. 2013. Colistin methanesulfonate and colistin pharmacokinetics in critically ill patients receiving continuous venovenous hemodiafiltration. Antimicrob Agents Chemother 57:668–671. doi: 10.1128/AAC.00985-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bergen PJ, Li J, Rayner CR, Nation RL. 2006. Colistin methanesulfonate is an inactive prodrug of colistin against Pseudomonas aeruginosa. Antimicrob Agents Chemother 50:1953–1958. doi: 10.1128/AAC.00035-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li J, Milne RW, Nation RL, Turnidge JD, Smeaton TC, Coulthard K. 2004. Pharmacokinetics of colistin methanesulphonate and colistin in rats following an intravenous dose of colistin methanesulphonate. J Antimicrob Chemother 53:837–840. doi: 10.1093/jac/dkh167. [DOI] [PubMed] [Google Scholar]
- 15.Kumar A, Roberts D, Wood KE, Light B, Parrillo JE, Sharma S, Suppes R, Feinstein D, Zanotti S, Taiberg L, Gurka D, Kumar A, Cheang M. 2006. Duration of hypotension before initiation of effective antimicrobial therapy is the critical determinant of survival in human septic shock. Crit Care Med 34:1589–1596. doi: 10.1097/01.CCM.0000217961.75225.E9. [DOI] [PubMed] [Google Scholar]
- 16.Kumar A, Ellis P, Arabi Y, Roberts D, Light B, Parrillo JE, Dodek P, Wood G, Kumar A, Simon D, Peters C, Ahsan M, Chateau D, Cooperative Antimicrobial Therapy of Septic Shock Database Research Group. 2009. Initiation of inappropriate antimicrobial therapy results in a fivefold reduction of survival in human septic shock. Chest 136:1237–1248. doi: 10.1378/chest.09-0087. [DOI] [PubMed] [Google Scholar]
- 17.Luna CM, Aruj P, Niederman MS, Garzon J, Violi D, Prignoni A, Rios F, Baquero S, Gando S, Grupo Argentino de Estudio de la Neumonia Asociada al Respirador. 2006. Appropriateness and delay to initiate therapy in ventilator-associated pneumonia. Eur Respir J 27:158–164. doi: 10.1183/09031936.06.00049105. [DOI] [PubMed] [Google Scholar]
- 18.Adams MD, Goglin K, Molyneaux N, Hujer KM, Lavender H, Jamison JJ, MacDonald IJ, Martin KM, Russo T, Campagnari AA, Hujer AM, Bonomo RA, Gill SR. 2008. Comparative genome sequence analysis of multidrug-resistant Acinetobacter baumannii. J Bacteriol 190:8053–8064. doi: 10.1128/JB.00834-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li J, Rayner CR, Nation RL, Owen RJ, Spelman D, Tan KE, Liolios L. 2006. Heteroresistance to colistin in multidrug-resistant Acinetobacter baumannii. Antimicrob Agents Chemother 50:2946–2950. doi: 10.1128/AAC.00103-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yau W, Owen RJ, Poudyal A, Bell JM, Turnidge JD, Yu HH, Nation RL, Li J. 2009. Colistin hetero-resistance in multidrug-resistant Acinetobacter baumannii clinical isolates from the Western Pacific region in the SENTRY antimicrobial surveillance programme. J Infect 58:138–144. doi: 10.1016/j.jinf.2008.11.002. [DOI] [PubMed] [Google Scholar]
- 21.El-Halfawy OM, Valvano MA. 2015. Antimicrobial heteroresistance: an emerging field in need of clarity. Clin Microbiol Rev 28:191–207. doi: 10.1128/CMR.00058-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Beceiro A, Llobet E, Aranda J, Bengoechea JA, Doumith M, Hornsey M, Dhanji H, Chart H, Bou G, Livermore DM, Woodford N. 2011. Phosphoethanolamine modification of lipid A in colistin-resistant variants of Acinetobacter baumannii mediated by the pmrAB two-component regulatory system. Antimicrob Agents Chemother 55:3370–3379. doi: 10.1128/AAC.00079-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moffatt JH, Harper M, Harrison P, Hale JD, Vinogradov E, Seemann T, Henry R, Crane B, Michael F, Cox AD, Adler B, Nation RL, Li J, Boyce JD. 2010. Colistin resistance in Acinetobacter baumannii is mediated by complete loss of lipopolysaccharide production. Antimicrob Agents Chemother 54:4971–4977. doi: 10.1128/AAC.00834-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Deris ZZ, Yu HH, Davis K, Soon RL, Jacob J, Ku CK, Poudyal A, Bergen PJ, Tsuji BT, Bulitta JB, Forrest A, Paterson DL, Velkov T, Li J, Nation RL. 2012. The combination of colistin and doripenem is synergistic against Klebsiella pneumoniae at multiple inocula and suppresses colistin resistance in an in vitro pharmacokinetic/pharmacodynamic model. Antimicrob Agents Chemother 56:5103–5112. doi: 10.1128/AAC.01064-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zavascki AP, Goldani LZ, Cao G, Superti SV, Lutz L, Barth AL, Ramos F, Boniatti MM, Nation RL, Li J. 2008. Pharmacokinetics of intravenous polymyxin B in critically ill patients. Clin Infect Dis 47:1298–1304. doi: 10.1086/592577. [DOI] [PubMed] [Google Scholar]
- 26.Cheah SE, Wang J, Nguyen VT, Turnidge JD, Li J, Nation RL. 2015. New pharmacokinetic/pharmacodynamic studies of systemically administered colistin against Pseudomonas aeruginosa and Acinetobacter baumannii in mouse thigh and lung infection models: smaller response in lung infection. J Antimicrob Chemother doi: 10.1093/jac/dkv267. [DOI] [PubMed] [Google Scholar]
- 27.Mohamed AF, Karaiskos I, Plachouras D, Karvanen M, Pontikis K, Jansson B, Papadomichelakis E, Antoniadou A, Giamarellou H, Armaganidis A, Cars O, Friberg LE. 2012. Application of a loading dose of colistin methanesulfonate in critically ill patients: population pharmacokinetics, protein binding, and prediction of bacterial kill. Antimicrob Agents Chemother 56:4241–4249. doi: 10.1128/AAC.06426-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Directorate for the Quality of Medicines and HealthCare of the Council of Europe. 2014. Monographs for colistimethate sodium and colistin sulfate, p 1960–1962. European Pharmacopoeia 8.0. Directorate for the Quality of Medicines and HealthCare of the Council of Europe, Strasbourg, France. [Google Scholar]
- 29.Cheah SE, Bulitta JB, Li J, Nation RL. 2014. Development and validation of a liquid chromatography-mass spectrometry assay for polymyxin B in bacterial growth media. J Pharm Biomed Anal 92:177–182. doi: 10.1016/j.jpba.2014.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bulitta JB, Yang JC, Yohonn L, Ly NS, Brown SV, D'Hondt RE, Jusko WJ, Forrest A, Tsuji BT. 2010. Attenuation of colistin bactericidal activity by high inoculum of Pseudomonas aeruginosa characterized by a new mechanism-based population pharmacodynamic model. Antimicrob Agents Chemother 54:2051–2062. doi: 10.1128/AAC.00881-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bulitta JB, Ly NS, Landersdorfer CB, Wanigaratne NA, Velkov T, Yadav R, Oliver A, Martin L, Shin BS, Forrest A, Tsuji BT. 2015. Two mechanisms of killing of Pseudomonas aeruginosa by tobramycin assessed at multiple inocula via mechanism-based modeling. Antimicrob Agents Chemother 59:2315–2327. doi: 10.1128/AAC.04099-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yano Y, Oguma T, Nagata H, Sasaki S. 1998. Application of logistic growth model to pharmacodynamic analysis of in vitro bactericidal kinetics. J Pharm Sci 87:1177–1183. doi: 10.1021/js9801337. [DOI] [PubMed] [Google Scholar]
- 33.Oliver JD. 2005. The viable but nonculturable state in bacteria. J Microbiol 43:93–100. [PubMed] [Google Scholar]
- 34.Oliver JD. 2010. Recent findings on the viable but nonculturable state in pathogenic bacteria. FEMS Microbiol Rev 34:415–425. doi: 10.1111/j.1574-6976.2009.00200.x. [DOI] [PubMed] [Google Scholar]
- 35.Schuck EL, Dalhoff A, Stass H, Derendorf H. 2005. Pharmacokinetic/pharmacodynamic (PK/PD) evaluation of a once-daily treatment using ciprofloxacin in an extended-release dosage form. Infection 33(Suppl 2):S22–S28. [DOI] [PubMed] [Google Scholar]
- 36.Tam VH, Kabbara S, Vo G, Schilling AN, Coyle EA. 2006. Comparative pharmacodynamics of gentamicin against Staphylococcus aureus and Pseudomonas aeruginosa. Antimicrob Agents Chemother 50:2626–2631. doi: 10.1128/AAC.01165-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Arroyo LA, Herrera CM, Fernandez L, Hankins JV, Trent MS, Hancock RE. 2011. The pmrCAB operon mediates polymyxin resistance in Acinetobacter baumannii ATCC 17978 and clinical isolates through phosphoethanolamine modification of lipid A. Antimicrob Agents Chemother 55:3743–3751. doi: 10.1128/AAC.00256-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Olaitan AO, Morand S, Rolain JM. 2014. Mechanisms of polymyxin resistance: acquired and intrinsic resistance in bacteria. Front Microbiol 5:643. doi: 10.3389/fmicb.2014.00643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bauer RJ, Guzy S, Ng C. 2007. A survey of population analysis methods and software for complex pharmacokinetic and pharmacodynamic models with examples. AAPS J 9:E60–E83. doi: 10.1208/aapsj0901007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bulitta JB, Bingolbali A, Shin BS, Landersdorfer CB. 2011. Development of a new pre- and post-processing tool (SADAPT-TRAN) for nonlinear mixed-effects modeling in S-ADAPT. AAPS J 13:201–211. doi: 10.1208/s12248-011-9257-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bulitta JB, Landersdorfer CB. 2011. Performance and robustness of the Monte Carlo importance sampling algorithm using parallelized S-ADAPT for basic and complex mechanistic models. AAPS J 13:212–226. doi: 10.1208/s12248-011-9258-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bulitta JB, Landersdorfer CB, Forrest A, Brown SV, Neely MN, Tsuji BT, Louie A. 2011. Relevance of pharmacokinetic and pharmacodynamic modeling to clinical care of critically ill patients. Curr Pharm Biotechnol 12:2044–2061. doi: 10.2174/138920111798808428. [DOI] [PubMed] [Google Scholar]
- 43.Nielsen EI, Friberg LE. 2013. Pharmacokinetic-pharmacodynamic modeling of antibacterial drugs. Pharmacol Rev 65:1053–1090. doi: 10.1124/pr.111.005769. [DOI] [PubMed] [Google Scholar]
- 44.Mohamed AF, Nielsen EI, Cars O, Friberg LE. 2012. Pharmacokinetic-pharmacodynamic model for gentamicin and its adaptive resistance with predictions of dosing schedules in newborn infants. Antimicrob Agents Chemother 56:179–188. doi: 10.1128/AAC.00694-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tan CH, Li J, Nation RL. 2007. Activity of colistin against heteroresistant Acinetobacter baumannii and emergence of resistance in an in vitro pharmacokinetic/pharmacodynamic model. Antimicrob Agents Chemother 51:3413–3415. doi: 10.1128/AAC.01571-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Phe K, Lee Y, McDaneld PM, Prasad N, Yin T, Figueroa DA, Musick WL, Cottreau JM, Hu M, Tam VH. 2014. In vitro assessment and multicenter cohort study of comparative nephrotoxicity rates associated with colistimethate versus polymyxin B therapy. Antimicrob Agents Chemother 58:2740–2746. doi: 10.1128/AAC.02476-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Azad MA, Finnin BA, Poudyal A, Davis K, Li J, Hill PA, Nation RL, Velkov T, Li J. 2013. Polymyxin B induces apoptosis in kidney proximal tubular cells. Antimicrob Agents Chemother 57:4329–4335. doi: 10.1128/AAC.02587-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Akajagbor DS, Wilson SL, Shere-Wolfe KD, Dakum P, Charurat ME, Gilliam BL. 2013. Higher incidence of acute kidney injury with intravenous colistimethate sodium compared with polymyxin B in critically ill patients at a tertiary care medical center. Clin Infect Dis 57:1300–1303. doi: 10.1093/cid/cit453. [DOI] [PubMed] [Google Scholar]
- 49.Abdelraouf K, Braggs KH, Yin T, Truong LD, Hu M, Tam VH. 2012. Characterization of polymyxin B-induced nephrotoxicity: implications for dosing regimen design. Antimicrob Agents Chemother 56:4625–4629. doi: 10.1128/AAC.00280-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chin CY, Gregg KA, Napier BA, Ernst RK, Weiss DS. 2015. A PmrB-regulated deacetylase required for lipid A modification and polymyxin resistance in Acinetobacter baumannii. Antimicrob Agents Chemother 59:7911–7914. doi: 10.1128/AAC.00515-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jacobs M, Gregoire N, Couet W, Bulitta JB. 2016. Distinguishing antimicrobial models with different resistance mechanisms via population pharmacodynamic modeling. PLoS Comput Biol 12:e1004782. doi: 10.1371/journal.pcbi.1004782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gregoire N, Raherison S, Grignon C, Comets E, Marliat M, Ploy MC, Couet W. 2010. Semimechanistic pharmacokinetic-pharmacodynamic model with adaptation development for time-kill experiments of ciprofloxacin against Pseudomonas aeruginosa. Antimicrob Agents Chemother 54:2379–2384. doi: 10.1128/AAC.01478-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fridman O, Goldberg A, Ronin I, Shoresh N, Balaban NQ. 2014. Optimization of lag time underlies antibiotic tolerance in evolved bacterial populations. Nature 513:418–421. doi: 10.1038/nature13469. [DOI] [PubMed] [Google Scholar]
- 54.Dalfino L, Puntillo F, Mosca A, Monno R, Spada ML, Coppolecchia S, Miragliotta G, Bruno F, Brienza N. 2012. High-dose, extended-interval colistin administration in critically ill patients: is this the right dosing strategy? A preliminary study. Clin Infect Dis 54:1720–1726. doi: 10.1093/cid/cis286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gregoire N, Mimoz O, Megarbane B, Comets E, Chatelier D, Lasocki S, Gauzit R, Balayn D, Gobin P, Marchand S, Couet W. 2014. New colistin population pharmacokinetic data in critically ill patients suggesting an alternative loading dose rationale. Antimicrob Agents Chemother 58:7324–7330. doi: 10.1128/AAC.03508-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Koomanachai P, Landersdorfer CB, Chen G, Lee HJ, Jitmuang A, Wasuwattakul S, Sritippayawan S, Li J, Nation RL, Thamlikitkul V. 2014. Pharmacokinetics of colistin methanesulfonate and formed colistin in end-stage renal disease patients receiving continuous ambulatory peritoneal dialysis. Antimicrob Agents Chemother 58:440–446. doi: 10.1128/AAC.01741-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hawley JS, Murray CK, Jorgensen JH. 2008. Colistin heteroresistance in acinetobacter and its association with previous colistin therapy. Antimicrob Agents Chemother 52:351–352. doi: 10.1128/AAC.00766-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bergen PJ, Bulitta JB, Forrest A, Tsuji BT, Li J, Nation RL. 2010. Pharmacokinetic/pharmacodynamic investigation of colistin against Pseudomonas aeruginosa using an in vitro model. Antimicrob Agents Chemother 54:3783–3789. doi: 10.1128/AAC.00903-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bergen PJ, Li J, Nation RL, Turnidge JD, Coulthard K, Milne RW. 2008. Comparison of once-, twice- and thrice-daily dosing of colistin on antibacterial effect and emergence of resistance: studies with Pseudomonas aeruginosa in an in vitro pharmacodynamic model. J Antimicrob Chemother 61:636–642. doi: 10.1093/jac/dkm511. [DOI] [PubMed] [Google Scholar]
- 60.Drusano GL, Liu W, Fikes S, Cirz R, Robbins N, Kurhanewicz S, Rodriquez J, Brown D, Baluya D, Louie A. 2014. Interaction of drug- and granulocyte-mediated killing of Pseudomonas aeruginosa in a murine pneumonia model. J Infect Dis 210:1319–1324. doi: 10.1093/infdis/jiu237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Drusano GL, Vanscoy B, Liu W, Fikes S, Brown D, Louie A. 2011. Saturability of granulocyte kill of Pseudomonas aeruginosa in a murine model of pneumonia. Antimicrob Agents Chemother 55:2693–2695. doi: 10.1128/AAC.01687-10. [DOI] [PMC free article] [PubMed] [Google Scholar]