Summary
Cells in our body can induce hundreds of antiviral genes following virus sensing, many of which remain largely uncharacterized. CEACAM1 has been previously shown to be induced by various innate systems; however, the reason for such tight integration to innate sensing systems was not apparent. Here, we show that CEACAM1 is induced following detection of HCMV and influenza viruses by their respective DNA and RNA innate sensors, IFI16 and RIG-I. This induction is mediated by IRF3, which bound to an ISRE element present in the human, but not mouse, CEACAM1 promoter. Furthermore, we demonstrate that, upon induction, CEACAM1 suppresses both HCMV and influenza viruses in an SHP2-dependent process and achieves this broad antiviral efficacy by suppressing mTOR-mediated protein biosynthesis. Finally, we show that CEACAM1 also inhibits viral spread in ex vivo human decidua organ culture.
Graphical Abstract

Highlights
-
•
IFI16 and RIG-I sensing of HCMV and influenza viruses induces expression of CEACAM1
-
•
IRF3 directly binds the CEACAM1 promoter and mediates the induction of CEACAM1
-
•
CEACAM1 suppresses mTOR activation and virus production in an SHP2-dependent manner
-
•
CEACAM1 suppresses HCMV spread in human ex vivo organ culture model of infection
Cells are equipped with intracellular sensing apparatuses, such as IFI16 and RIG-I. Vitenshtein et al. show that in response to innate cellular sensing of HCMV and influenza viruses by these sensors, CEACAM1 expression is induced by the infected cells to suppress virus production through autoregulation of protein biosynthesis in an SHP2-dependent manner.
Introduction
Non-immune cells play a critical role in the host response to viral infection and are equipped with intrinsic sensing and antiviral mechanisms that confer a broad resistance to a variety of viruses (Desmet and Ishii, 2012, Honda et al., 2006, Stetson and Medzhitov, 2006). A prominent sensor that detects viral DNA is IFI16, which has been shown to detect infection by human cytomegalovirus (HCMV) (Li et al., 2013). Cells are also equipped with RNA sensing systems, such as RIG-I, that detect a host of viruses, among them the influenza virus (Kato et al., 2006). Many of these systems converge on IRF3 (Desmet and Ishii, 2012), which is recognized as the master regulator of the first line of antiviral defense (Hiscott, 2007). IRF3 is constitutively expressed in almost all cells and is poised to rapidly undergo phosphorylation, dimerization, and translocation to the nucleus. Once in the nucleus, IRF3 initiates a limited set of primary antiviral genes such as IFN-β, which upon secretion orchestrates the larger secondary wave of cellular antiviral genes (Honda et al., 2006). Surprisingly, while the ability of cells to activate effective antiviral mechanisms has been known for over half a century (Isaacs and Lindenmann, 1957), most of the antiviral genes remain largely uncharacterized.
Carcinoembryonic antigen-related cell adhesion molecule 1 (CEACAM1) forms homophilic interactions which deliver inhibitory signals through the SHP1 (mainly in hematopoietic cells) and SHP2 phosphatase through CEACAM1s’ immunoreceptor tyrosine based inhibitory motifs (ITIMs) (Gray-Owen and Blumberg, 2006, Huber et al., 1999, Müller et al., 2009, Nouvion et al., 2010). Studies have shown that CEACAM1 expression is induced very rapidly on endothelial and epithelial cells by the NF-κB pathway following TLR4-dependent sensing of N. gonorrhoeae infection (Muenzner et al., 2001). CEACAM1 has also been shown to be induced by IFN-γ, which is selectively secreted by T and NK cells, resulting in direct binding of IRF1 to an interferon-stimulated response element (ISRE) in the CEACAM1 promoter (Chen et al., 1996).
Whereas the N. gonorrhoeae-mediated induction of CEACAM1 has been shown to benefit the pathogen by facilitating its binding and infection, it remains unclear why CEACAM1 has been wired directly to pathogen sensing systems in non-immune cells. In the current study, we demonstrate that CEACAM1 is rapidly induced by innate sensing of extremely divergent viral pathogens, the HCMV and influenza viruses, and functions as a broad suppressor of viral infection.
Results
IFI16-Mediated Sensing of HCMV Infection Induces Expression of CEACAM1
We observed that CEACAM1 expression is induced on the surface of HFF cells 24 hr post infection (HPI) with HCMV laboratory strains AD169 and TB40/E (Figure 1A) or with a clinical isolate (Figure S1A). Analysis of transcription kinetics of newly synthesized CEACAM1 mRNA showed an extremely rapid response that was visible at 1–2 hr following infection (Figure 1B). Since a replication-defective UV-inactivated virus, which can infect cells, but not induce de novo viral gene transcripts, induced CEACAM1 (Figure S1B, left histogram), we concluded that a component in the virion was involved in CEACAM1 upregulation, but ruled out the involvement of secreted factors since supernatant transfer did not induce CEACAM1 (Figure S1B, right histograms). Additionally, CEACAM1 mRNA was induced even upon protein translation inhibition (Figure S1C), demonstrating that cellular machinery mediating the induction was pre-formed. Importantly, transfecting viral DNA, UV-treated or not, robustly induced the expression of CEACAM1 (Figure 1C). This prompted us to check whether IFI16, the innate sensor of HCMV DNA, was involved in the induction of CEACAM1 by knocking this sensor down. Infecting HFFs expressing IFI16 specific small hairpin (sh)RNAs, with HCMV, resulted in decreased CEACAM1 induction (Figure 1E). This indicated that IFI16 played a role in the induction of CEACAM1. The decreased induction was specific to lack of IFI16, as the cells were still fully responsive to other stimuli such as polyI:C (an RNA analog that triggers the RNA sensors; Desmet and Ishii, 2012), which induced CEACAM1 in these cells (Figure S1D). The involvement of the RNA sensing RIG-I pathway is studied below. The ARPE-19 cell line, which is also permissive to HCMV infection, has low levels of IFI16, and indeed, CEACAM1 was not induced upon infection (Figure 1F). Thus, we overexpressed in these cells the two dominant isoforms of IFI16 (that differ in the length of the hinge region between the DNA sensing domains; Unterholzner et al., 2010; and may affect viral sensing efficiency). Importantly, overexpression of the IFI16 isoforms (TB40-IFI16IsoB/IsoC) was sufficient to induce CEACAM1 (Figure 1F) during HCMV infection, directly indicating that sensing of HCMV DNA by IFI16 mediates this process.
Figure 1.
HCMV DNA Induces IFI16-Mediated CEACAM1 Expression during HCMV Infection
(A) FACS staining for CEACAM1 expression at 8 and 24 HPI, on mock (Mock) or HFF cells infected with HCMV (HCMV) strains TB40/E (left) and AD169 (right) at moi 3.
(B) Real-time PCR quantification of newly synthesized CEACAM1 transcripts in infected and 4-Thiouridine pulsed HFF cells at the designated HPI.
(C) Analysis of CEACAM1 expression on HFF cells transfected with 1 μg/ml of purified TB40/E DNA (DNA) or on HFF cells transfected with 1 μg/ml purified UV-inactivated TB40/E DNA (UV-DNA). The Mock is HFF cells treated with transfection reagent only.
(D) Intracellular FACS analysis of IFI16 expression in TB40/E infected HFF cells (moi 3) stably transduced with two shRNAs targeting IFI16 (αIFI16.1/2, empty black and red histograms) or control (Control, empty gray histogram) sequence.
(E) Quantification by FACS of the fold induction of CEACAM1 expression following infection with the TB40/E virus on cells from (D) at 3 DPI. The dotted line represents basal mean fluorescence intensity (MFI) level on mock treated control cells that was set as 1 and to which the fold increase in expression was compared to.
(F) Analysis of CEACAM1 expression in mock (Mock) or TB40/E (TB40, moi 3) infected ARPE-19 cells that were stably transduced with control empty vector (empty gray histogram) and two IFI16 isoforms B (IFI16IsoB, empty black histogram) and C (IFI16IsoC, empty red histogram). The background staining was conducted with isotype matched control antibody (background, filled gray histogram).
(A and C) The empty black and red histograms depict staining for CEACAM1 following indicated treatments (HCMV, A, DNA, or UV-DNA, C) and empty gray histogram depicts CEACAM1 staining on mock treated cells (Mock).
(A, C, D, and F) The filled gray histogram indicates background staining with control isotype matched antibody (Bckgnd). The figures show one representative experiment out of seven (A) or three (C, D, and F) performed.
(B and E) An average ±SD of triplicates are presented (∗p < 0.05 and ∗∗p < 0.01).
IRF3 Mediates CEACAM1 Expression
Using the IFI16-transfectant ARPE-19 cells, we next saw that knock down of IRF3, downstream mediator of IFI16, (Figure 2A) disrupted the upregulation of CEACAM1 during infection (Figure 2B). This effect was specific to IRF3, and other pathways such as IFN-γ, which induces expression of class I MHC in an IRF3-independent manner, remained fully operational (Figure S2A). Previous studies have characterized an interferon stimulated response element (ISRE) in the CEACAM1 promoter that binds IRF1 in response to stimulation with IFN-γ (Chen et al., 1996). Using the MatInspector algorithm (Cartharius et al., 2005), we identified this ISRE to potentially be compatible with the IRF3 binding sequence (Figure S2B). To investigate this ISRE, we conducted reporter assays in which 600 bp of the wild-type CEACAM1 promoter sequence and a promoter sequence mutated in the ISRE site predicted to be bound by IRF3 (IRF3Mut; Figure S2B) were fused to luciferase. While the wild-type promoter sequence mediated a strong induction of luciferase activity following HCMV infection (Figure 2C), the mutation entirely abrogated all promoter activity (Figure 2C). To check whether IRF3 bound the CEACAM1 promoter directly, we performed chromatin immunoprecipitation (ChIP) of IRF3. We observed that pull down of IRF3 co-precipitated enriched CEACAM1 promoter sequences in HCMV infected compared to mock cells, as quantified by real-time PCR (Figure 2D), demonstrating that IRF3 bound the promoter directly.
Figure 2.
CEACAM1 Induction Is Directly Mediated by IRF3 and Occurs Also by RIG-I Sensing of Influenza Virus Infection
(A) Assessment by western blot of IRF3 levels in TB40/E infected ARPE-19IFI16IsoB cells that underwent IRF3 targeted shRNA knockdown by two clones (αIRF3.1/2) or were transduced with control shRNA (Control, set as 100%). The upper image depicts the quantification of IRF3 levels in the lower image, normalized to GAPDH.
(B) The TB40/E infected (3 DPI) ARPE-19IFI16IsoB cells from (A) were analyzed by FACS for induction of CEACAM1.
(C) Relative luciferase activity in ARPE-19IFI16IsoB cells, following infection with TB40/E (moi 3). The cells were transfected with either naive firefly luciferase encoding vector (No Promoter), luciferase fused downstream to a 600 bp genomic sequence encoding the WT CEACAM1 promoter (WT pCEACAM1), or luciferase fused to the CEACAM1 promoter mutated in the predicted IRF3 binding site (IRF3Mut pCEACAM1). The results were normalized to co-transfected renilla luciferase and were compared to “No promoter” which was set as 1.
(D) Real-time PCR quantification of CEACAM1 promoter DNA sequences in anti-IRF3 ChIP of mock (Mock) and TB40/E (TB40) infected HFF cells at 4 HPI.
(E) FACS staining of A549 cells for CEACAM1 expression following infection with PR8 virus strain at moi 3 (empty black histogram) compared to mock treated cells (empty gray histogram) or isotype matched control antibody (Bckgnd, filled gray histogram).
(F) Western blot analysis for RIG-I (αRIG-I) and IRF3 (αIRF3) expression in PR8-infected A549 cells expressing two shRNAs targeting RIG-I (αRIG-I.1/2), IRF3 (αIRF3.1/2), or control sequence (Control). The upper image is expression quantification relative to GAPDH levels.
(G and H) A549 cells infected by PR8 (3 DPI) expressing two clones of shRNAs anti-RIG-I (αRIG-I.1/2) (G) and anti-IRF3 (αIRF3.1/2) (same cells from F) (H) were examined by FACS for the induction of CEACAM1expression.
(I) Abundance of CEACAM1 promoter sequences in anti-IRF3 ChIP of A549 cells following infection with PR8 (PR8) or mock treatment (Mock) at 3 HPI and as quantified by real-time PCR.
(B, G, and H) The induction of CEACAM1 was calculated as the fold increase in MFI of CEACAM1 staining in shRNA-expressing cells compared to the mock treated control shRNA transduced cells, which was set as 1 (dotted line).
(D and I) Presented values are the sequence abundance relative to the abundance in the sample prior to the ChIP (percent of input) and normalized to a quantified control region upstream to the CEACAM1 promoter. The data presented are a representative of three (A–D, F, and I), six (E), an average ±SD of three (G and H) independent experiments performed, and an average ±SD of three triplicates (B–D, F, and I) (∗∗p < 0.01 and ∗∗∗p < 0.001).
Notably, the sequence and position of the ISRE in the CEACAM1 promoter was conserved in higher primate species, although no such site was identified in the murine promoter (Figure S2C). In line with this, no significant mCEACAM1 induction was observed 48 HPI in various MCMV-permissive cell lines infected with murine CMV (strain C3X) (Figure S2D). The antibody used was fully functional and readily detected expression of mouse CEACAM1 in the PD1.6 cell line (Figure S2E).
CEACAM1 Is Induced during Influenza Virus Infection
IRF3 is a component in numerous innate sensing cascades (Figure S3A). Additionally, CEACAM1 expression was seen to be upregulated following sensing of polyI:C (Figure S1D). Therefore, we next checked whether CEACAM1 is induced in response to RNA viruses such as influenza. The influenza virus can efficiently infect only a limited number of cell lines in vitro, one of which is lung epithelial A549 (but not HFF or ARPE-19 cells; Li et al., 2009; that we used to study HCMV). Notably, infecting A549 cells with the influenza A PR8 virus strain led to a robust CEACAM1 cell surface expression (Figure 2E). Knock down of the RIG-I sensor, the RNA sensor that was shown to detect the influenza A virus (Kato et al., 2006) (Figure 2F), showed that the induction was dependent on this sensor (Figure 2G). This decreased activity was specific to knock down of RIG-I since transfecting UV-inactivated TB40/E DNA to these cells still showed a fully operational DNA sensing response that led to a robust induction of CEACAM1 (Figure S3B). Similar to IFI16, RIG-I also operates through IRF3 (Figure S3A; see Desmet and Ishii, 2012) and during CEACAM1 induction, since knock down of IRF3 (Figure 2F) significantly inhibited its expression during the influenza virus infection (Figure 2H). Once more, the decreased response was specific to knock down of IRF3, as all cells were fully responsive to IFN-γ, which induces an alternative IRF3-independent pathway (Figure S3C). Furthermore, IRF3 ChIP analysis demonstrated direct binding of IRF3 to the CEACAM1 promoter in PR8 infected A549 cells that had enriched promoter sequences (Figure 2I). Thus, because CEACAM1 expression is paired to IRF3, it is robustly induced by diverse innate sensing systems in response to different viruses.
CEACAM1 Suppresses HCMV and Influenza Virus Production
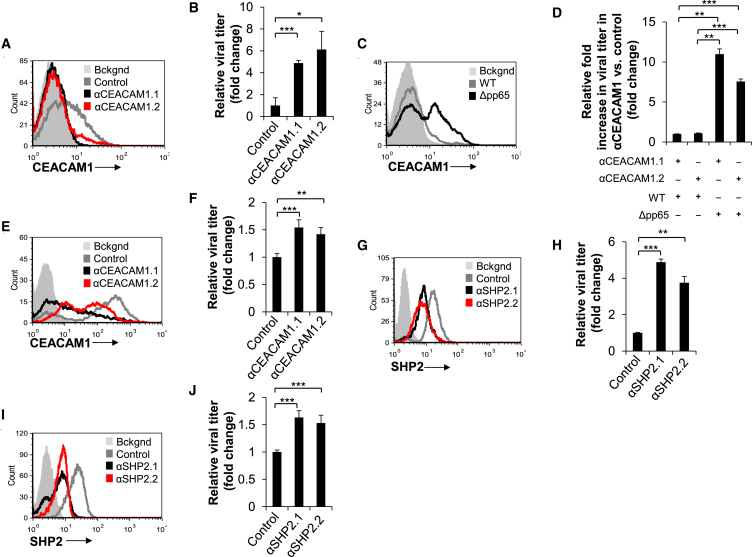
Next, we pursued to explore why CEACAM1 is upregulated by innate immune systems. To examine whether CEACAM1 affects viral infection, we silenced CEACAM1 expression in HFF cells (Figure 3A) and then infected these cells with HCMV. Importantly, we observed that CEACAM1 suppresses HCMV, as silencing its knockdown led to a significant elevation in virus production (Figure 3B).
Figure 3.
CEACAM1 Expression Suppresses Viral Replication through SHP2
(A and G) HFF cells were stably transduced with two shRNAs against CEACAM1 (A, αCEACAM1.1/2), SHP2 (G, αSHP2.1 and αSHP2.1/2), or with a control (Control) shRNA. Knockdown was confirmed by extra- and intracellular FACS staining for CEACAM1 and SHP2, respectively, following infection with TB40/E at moi 3.
(B and H) Plaque assay-based quantification of viral load in the supernatants of infected cells expressing CEACAM1 (B) or SHP2 (H) specific shRNAs (same cells from A and G, respectively).
(C) Cell surface expression of CEACAM1 on HFF cells following infection (moi 0.5) with either a WT (AD169, empty gray histogram) or pp65-deleted virus (Δpp65, empty black histogram). No staining was observed staining AD169 infected cells with isotype matched IgG (Bckgnd, filled gray histogram).
(D) Fold increase in viral titer in HFF cells (same cells from A) expressing two shRNAs against CEACAM1 (αCEACAM1.1/2) compared to viral titer in supernatant of cells expressing control shRNA. The cells were infected with either a WT (WT) or pp65-deleted (Δpp65) AD169 virus. The fold increase in cells expressing the αCEACAM1.1 shRNA clone infected with a WT virus was set as 1.
(E and I) A549 cells were subjected to control (Control), CEACAM1 (E, αCEACAM1.1/2), or SHP2 (I, αSHP2.1/2) specific shRNA-mediated knock down of two clones as confirmed by FACS staining, following PR8 infection at moi 3.
(F and J) Following infection with the PR8 virus (moi 3) of cells from (E) and (I) expressing CEACAM1 or SHP2 specific shRNAs, respectively, the influenza viral titer in supernatant was determined by an ELISA based assay. The amount of the virus present in the control transduced cells (B, F, H, and J) was set as 1. The experiments shown are a representative of three (A–D, G, and H) and five (E, F, I, and J) independent experiments with similar results and an average ±SD of four (B, D, and H) or eight (F and J) replicates (∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001).
HCMV is a master of immune evasion and has recently been shown to use its pp65 protein to evade the IFI16 innate cellular antiviral responses (Li et al., 2013). We speculated that HCMV might also be using the pp65 protein to evade the full antiviral capacity of CEACAM1. In line with this, while infecting HFF cells at a low moi with wild-type (WT) HCMV virus that did not induce expression of CEACAM1 (compared to higher moi in previous experiments), infection with HCMV virus deleted in the pp65 protein (Δpp65) led to a robust induction (Figure 3C). Given that CEACAM1 is induced more efficiently on cells infected by the Δpp65 as compared to WT HCMV, infecting CEACAM1 knockdown cells with the Δpp65 virus could demonstrate the full antiviral capacity of CEACAM1. To test this, we compared the fold change increase in viral titer in HFF cells that underwent CEACAM1 knockdown following infection with either a WT or Δpp65 virus. In the absence of pp65, the Δpp65 virus had a 10-fold higher fold increase in viral titer following CEACAM1 knockdown than that of the WT virus (Figure 3D). Strikingly, CEACAM1 also suppressed the influenza virus, as knock down of its expression in A549 cells (Figure 3E) led to a consistent increase in influenza viral titer when infecting these cells (Figure 3F).
Next, we investigated the mechanism of CEACAM1-mediated inhibition of virus production. Previous studies have established that the SHP2 phosphatase delivers the CEACAM1-inhibitory signals in non-immune cells (Huber et al., 1999, Müller et al., 2009, Nouvion et al., 2010). We observed CEACAM1 to operate through SHP2 during infection with HCMV and influenza, as it co-immunoprecipitated with CEACAM1 in HFF and A549 cells infected by these viruses (Figures S3D and S3E). Indeed, CEACAM1 suppression of viral titer was dependent on SHP2, as knocking down of SHP2 (Figures 3G and 3I) recapitulated the knock down of CEACAM1 and resulted in increased viral production during HCMV infection (Figure 3H) and influenza virus infection (Figure 3J). Notably, knocking down SHP2 in ARPE-19 cells, which do not induce expression of CEACAM1 upon infection (Figure 1F), did not have any affect on HCMV viral titer (Figures S3F and S3G). This indicated that SHP2 antiviral function was CEACAM1 dependent.
CEACAM1 Suppresses Viral Production by Regulating Mammalian Target of Rapamycin-Mediated Cellular Protein Translation
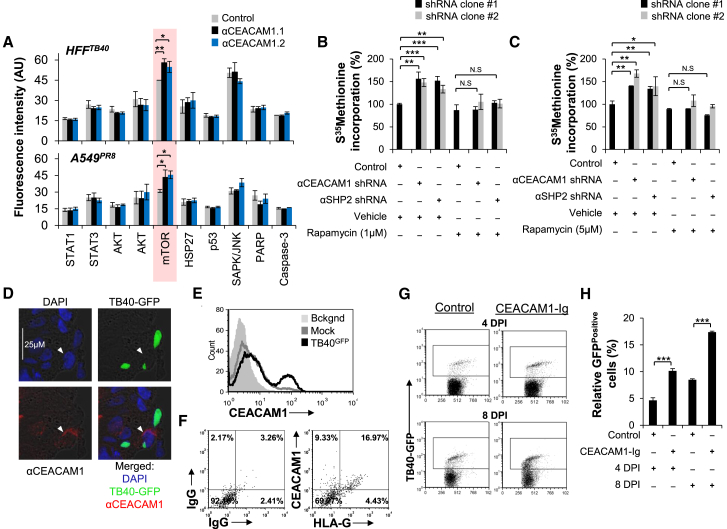
To understand how CEACAM1 inhibits virus production, we used a fluorescent phospho-specific antibody array to quantify the phosphorylation status of key components in signaling pathways in HFF and A549 cells, in which CEACAM1 expression was knocked down using two shRNAs and then infected with HCMV and influenza, respectively (Figure 4A). We observed the upregulation of an activating Serine2448 phosphorylation of mammalian target of rapamycin (mTOR) that was common during infections with both viruses (Figure 4A). mTOR is a key regulator of global cellular protein translation levels, and increased activity of this modulator could lead to higher rates of protein biosynthesis, which may facilitate the observed increase in viral production (Figure 3). Investigating this option, we observed that HFF and A549 cells stably expressing CEACAM1 and SHP2-targeting shRNAs, and infected with HCMV (Figure 4B) or influenza virus (Figure 4C), exhibited a higher protein production capacity. The observed increase in protein production was dependent on mTOR activity, as blocking mTOR with its specific inhibitor, rapamycin, prevented this increase in cells that underwent either CEACAM1 or SHP2-knockdown during both HCMV (Figure 4B) and influenza virus (Figure 4C) infections. We also observed that in reciprocal experiments, overexpression of CEACAM1 (Figure S4A) and SHP2 (Figure S4C) in HFF cells led to a complementary effect of suppressed global protein production during HCMV infection (Figures S4B and S4D). Similarly, overexpression of CEACAM1 (Figure S4E) and SHP2 (Figure S4G) in influenza virus-infected A549 cells was also seen to significantly suppress cellular protein production capacity (Figures S4F and S4H). As viruses critically depend on the cellular protein production machinery for their replication, we thus concluded that CEACAM1 suppresses HCMV and influenza virus infections by regulating mTOR activation and subsequent levels of cellular protein translation.
Figure 4.
CEACAM1 Suppresses Cellular Protein Translation through Downregulation of mTOR-Activating Phospho-Serine2448 and Suppresses HCMV Dissemination in Human Ex Vivo Organ Culture
(A) Stably knocked down CEACAM1 (black and blue columns, αCEACAM1.1/2) or stably expressed control shRNA (gray columns, Control shRNA) in HFF (HFFTB40) and A549 cells (A549PR8) was studied for its effects on the phosphorylation of key cellular kinases following 3 days of infection with TB40/E or PR8, respectively, at moi 3. The cell lysates were prepared and then probed on an antibody array with specific antibodies for the phosphorylated isoforms of the kinases listed. Using a far-red array scanner, levels of phospho-proteins were then quantified based on measurement of fluorescence emission.
(B and C) HFF cells infected with TB40/E (moi 3, B) and A549 cells infected with PR8 (moi 3, C) stably expressing control (Control), CEACAM1 (αCEACAM1.1/2), or SHP2 (αSHP2.1/2) specific shRNAs were analyzed for total cell protein production capacity by assessing the rate of [35S]Methionine incorporation. The cells were also administered with rapamycin at the indicated doses (Rapamycin) or with DMSO only as control treatment (Vehicle). The CPM values of cells expressing the control shRNA were set as 100%.
(D) Confocal microscopic analysis of ex vivo organ cultured decidua at 2 DPI, which underwent infection with TB40/EGFP. The tissue sections were stained for cell nucleus with DAPI, CEACAM1 (αCEACAM1), and cells infected with a virus were visualized by the presence of GFP (TB40-GFP). The arrowhead depicts the HCMV infected GFPPositive cell co-expressing CEACAM1.
(E) Isolated cells from TB40/EGFP infected (TB40/EGFP, black empty histogram) and mock treated (Mock, gray empty histogram) decidua organ culture were stained for CEACAM1 expression. The TB40/E infected cells stained with an isotype-matched control antibody served as background (Bckgnd, gray filled histogram) staining.
(F) Cells isolated from TB40/E-infected decidua were stained with either isotype-matched control IgGs (left) or dually stained for CEACAM1 and HLA-G expression (right).
(G and H) FACS dotplot of isolated cells from TB40/EGFP (TB40-GFP)-infected decidua organ cultures treated either with CEACAM1-Ig (CEACAM1-Ig) or control Ig fusion protein (Control) as determined by FACS at 4 DPI and 8 DPI by (G). The number of GFPPositive cells in the gated area in the histogram was quantified in (H). The data are an average of three (A and C) or representative of five (B, D, and E) or three (F–H) independent experiments, and an average ±SD of three (A and H) or six (B and C) replicates (not significant: N.S., ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001).
CEACAM1 Suppresses HCMV Infection in Human Ex Vivo Organ Culture
Our final goal was to assess the physiological role of CEACAM1 in vivo. However, since mouse CEACAM1 is not induced following infection (as it does not contain the IRF3 binding site; Figures S2C and S2D), we proceeded with a human ex vivo decidua (a tissue that is naturally targeted by HCMV infections in its human host) organ culture model for HCMV infection (Figure S4I). We observed that following infection with a GFP encoding TB40/E virus (TB40/EGFP), some GFPPositive infected cells also co-expressed CEACAM1, while others did not (Figure 4D). Staining mock and HCMV infected single cell homogenized decidual tissues showed that following infection, CEACAM1 was also being induced in the ex vivo organ culture (Figure 4E). To analyze which cells in the decidua upregulate CEACAM1 following infection, we stained the decidua organ cultures for HLA-G, a marker for trophoblasts (Kovats et al., 1990). We observed that 26.3% of HCMV-infected decidual cells were CEACAM1Positive (9.33% CEACAM1PositiveHLA-GNegative added to 16.97% CEACAM1PositiveHLA-GPositive cells), of which the majority (64.5%) were HLA-GPositive 64.5% (16.97% of HLA-GPositiveCEACAM1Positive out of 26.3% CEACAM1Positive cells) (Figure 4F).
Finally, to test what degree of involvement CEACAM1 may have in control of viral spread in the decidua culture, we used a CEACAM1-Ig fusion protein, composed of the extracellular domain of CEACAM1 fused to human IgG1 and can block CEACAM1 function (Markel et al., 2004). To confirm the blocking function of CEACAM1-Ig, we used the BWCEACAM1 mouse thymoma reporter cell line, in which a construct of an extracellular human CEACAM1 domain fused to an intracellular mouse zeta chain was stably expressed (Figure S4J). These cells secrete IL-2 upon crosslinking of cell surface CEACAM1, as occurred when they were co-cultured with a CEACAM1 expressing transfectant (721.221CEACAM1), but not parental (721.221Parental) cell line (Figure S4K), given that CEACAM1 binds homotypically. However, applying CEACAM1-Ig, and not a control fusion protein, blocked the binding of CEACAM1 and led to a marked decrease in the activation and IL-2 secretion by these cells (Figure S4K).
Next, we used the CEACAM1-Ig in the HCMV-infected decidua organ cultures and determined the degree of HCMV infection by the number of GFPPositive cells. This treatment did not cause any observable changes in tissue structure or distribution of CEACAM1 expression (data not shown). Importantly, however, we observed that blocking CEACAM1 interactions by using CEACAM1-Ig led to an increased viral dissemination as seen by an increase in the number of GFP expressing cells at 4 and 8 days (maximal days to sustain viable culture) post infection (Figure 4G; summarized in Figure 4H). These results show ex vivo evidence that CEACAM1 plays an important systemic role in suppression of HCMV dissemination in infected human tissues.
Discussion
In the current study, we demonstrate that CEACAM1 is strongly integrated to innate cellular pathogen sensing systems by direct binding of IRF3 to the CEACAM1 promoter following sensing of HCMV by IFI16 and influenza virus by RIG-I. Given that IRF3 is a master regulator that is activated by a wide range of innate systems, we propose that CEACAM1 is induced in response to a very diverse spectrum of viruses. Following its expression, CEACAM1 functions as an antiviral suppressor of both HCMV and the influenza virus. Due to the lack of an in vivo model, we also demonstrate that CEACAM1 is induced and plays a significant role in controlling viral spread using a human ex vivo decidual organ culture model for HCMV infection that we have developed (Weisblum et al., 2011). Viruses, on the other hand, have highly honed mechanisms to evade antiviral responses, and we show that HCMV employs the recently demonstrated pp65 immunoevasin (Li et al., 2013) to evade the full capacity of CEACAM1 expression and antiviral suppression. Similarly, the influenza virus subverts cellular antiviral systems with a key viral protein, NS1 (Mibayashi et al., 2007), that we suspect is used to subvert full CEACAM1 functionality.
We further demonstrate that CEACAM1 and SHP2 suppressed mTOR activity, a central rheostat that dictates global levels of cellular translation (Buchkovich et al., 2008). Consequently, this led to suppression of protein production in HFF and A549 cells that were infected with HCMV and the influenza virus, respectively. Since all viruses are fundamentally dependent on cellular protein biosynthesis machinery for replication, blocking such cellular machinery by CEACAM1 is a strategy that could potentially be effective against a very broad range of viruses. Although viruses strongly manipulate the translation machinery to maintain a functionality (Clippinger et al., 2011, Moorman et al., 2008, Walsh and Mohr, 2011), both HCMV and influenza viruses are known to depend on mTOR activity during specific phases of their life cycle (Burgui et al., 2007, Clippinger et al., 2011). This renders them susceptible to translation regulating mechanisms such as mediated by CEACAM1/SHP2. Although relieving the CEACAM1/SHP2 autoregulation mechanisms led to an increase in translation and viral production, it is hard to say whether this can be attributed to the bolstering of the mTOR-dependent stages in their lifecycle or additive mTOR activity to pre-existing translation capacity maintained by the viruses. It is also noteworthy that previous studies have found, in complementation to the current study, that SHP2 functions as a regulator of mTOR (Marin et al., 2008, Schramm et al., 2012, Zito et al., 2007). However, given that SHP2 is a tyrosine phosphatase and mTOR is activated via serine residue, this indicates that SHP2 is an upstream indirect pathway activator.
The co-evolution of viruses and their hosts has led to the development of highly diverse and sophisticated cellular defensive mechanisms and viral counter-measures, examples of both are presented here. Due to homophilic binding and self-activation, we describe here the development of a cellular mechanism that enables an immediate and broadly effective antiviral response by upregulation of a single protein, CEACAM1.
Experimental Procedures
Viruses, Infections, Titrations, Fluorescence-Activated Cell Sorting, Antibody Array, Transfections, and BW Assay
HCMV, MCMV, and influenza viruses were grown, titrated, and used to infect cells by standard procedures. Titrations were based on plaque assays for TB40/E or ELISA based method for PR8. Stable transduction of CEACAM1, IFI16, SHP2 transfectants, and shRNA clones (Sigma-Aldrich) was based on lentiviral and retroviral expression systems. Fluorescence-activated cell sorting (FACS) staining was standard protocols. For intracellular staining, a methanol fixation based method was used. PathScan antibody array was performed according to manufacturer’s instructions (Cell Signaling). Viral DNA was transfected at 1 μg/ml. For BW assay, a 3 day 1:3 E:T ration was used with Ig-fusion concentration of 5 μg/ml followed by IL-2 measurement (BioLegend). See also the Supplemental Information.
Real-Time PCR, shRNAs, and Luciferase Assay
Newly synthesized transcript analysis was previously described (Halenius et al., 2011). All shRNAs and control scrambled sequence transduction was performed according to manufacturer’s instructions (Sigma). Luciferase reporter assay was based on the pGL4.14 firefly reporter and pRL-CMV renilla loading control luciferase vectors See also the Supplemental Information.
ChIP, Coimmunoprecipitation, and Protein Translation Assay
ChIP was performed on the basis of Nelson et al. (2006) using an anti-IRF3 antibody (Santa Cruz). Coimmunoprecipitation (coIP) was performed by pull-down with anti-CEACAM1 5F4 antibody (provided by R.S.B.) conjugated to protein G sepharose (Santa Cruz) and probing with anti-SHP2 (Santa Cruz) according to manufacturer’s instructions. For translation analysis, starved cells were then administered with [35S]Methionine. See also the Supplemental Information.
Statistical Analysis
Statistical significance was determined by Student’s t test. p value of less than 0.05 was considered significant and indicated in figure and figure legends as Not significant: N.S., ∗p < 0.05, ∗∗p < 0.01, or ∗∗∗p < 0.001.
Author Contributions
A.V. performed most of the experiments and headed the project. S.H. and A.H. performed analysis of newly synthesized CEACAM1 mRNA kinetics. Y.W., E.O.-D., P.T., Y.B., Y.B.-O., N.S.-G., J.E., R.O., J.T., G.M., R.S.B., S.J., H.A., and R.K. provided critical reagents. H.H. and D.G.W. provided experimental advice and critical reagents. O.M. supervised the project.
Acknowledgments
This study was supported by the European Research Council under the European Union’s Seventh Framework Programme (FP/2007-2013)/ERC Grant Agreement no. 320473-BacNK. Additional support came from the Israel Science Foundation, the GIF foundation, the ICRF professorship grant, and the Helmholtz Israel grant (all to O.M.). This work was also supported by grants from the Israel Science Foundation (grant no. 275/13); the Israeli Ministry of Health (grant no. 10998); the European Union Seventh Framework Programme 562 FP7/2012-2016 (grant agreement no. 316655); and the Samueli Institute Brain Mind & Healing Center, under a contract from the US Army Medical Research and Materiel Command (USAMRAA award no. W81XWH-10-1-0938), within the TATRC portfolio of Integrative Medicine Research. We would like to kindly thank Beatrix Steer and Franziska Thomas for their assistance in studying the dynamics of MCMV and mCEACAM1 and Ihab Ansari for assistance with the ChIP experiments. O.M. is a Crown Professor of Molecular Immunology.
Published: June 2, 2016
Footnotes
Supplemental Information includes Supplemental Experimental Procedures and four figures and can be found with this article online at http://dx.doi.org/10.1016/j.celrep.2016.05.036.
Supplemental Information
References
- Buchkovich N.J., Yu Y., Zampieri C.A., Alwine J.C. The TORrid affairs of viruses: effects of mammalian DNA viruses on the PI3K-Akt-mTOR signalling pathway. Nat. Rev. Microbiol. 2008;6:266–275. doi: 10.1038/nrmicro1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgui I., Yángüez E., Sonenberg N., Nieto A. Influenza virus mRNA translation revisited: is the eIF4E cap-binding factor required for viral mRNA translation? J. Virol. 2007;81:12427–12438. doi: 10.1128/JVI.01105-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartharius K., Frech K., Grote K., Klocke B., Haltmeier M., Klingenhoff A., Frisch M., Bayerlein M., Werner T. MatInspector and beyond: promoter analysis based on transcription factor binding sites. Bioinformatics. 2005;21:2933–2942. doi: 10.1093/bioinformatics/bti473. [DOI] [PubMed] [Google Scholar]
- Chen C.J., Lin T.T., Shively J.E. Role of interferon regulatory factor-1 in the induction of biliary glycoprotein (cell CAM-1) by interferon-gamma. J. Biol. Chem. 1996;271:28181–28188. doi: 10.1074/jbc.271.45.28181. [DOI] [PubMed] [Google Scholar]
- Clippinger A.J., Maguire T.G., Alwine J.C. The changing role of mTOR kinase in the maintenance of protein synthesis during human cytomegalovirus infection. J. Virol. 2011;85:3930–3939. doi: 10.1128/JVI.01913-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desmet C.J., Ishii K.J. Nucleic acid sensing at the interface between innate and adaptive immunity in vaccination. Nat. Rev. Immunol. 2012;12:479–491. doi: 10.1038/nri3247. [DOI] [PubMed] [Google Scholar]
- Gray-Owen S.D., Blumberg R.S. CEACAM1: contact-dependent control of immunity. Nat. Rev. Immunol. 2006;6:433–446. doi: 10.1038/nri1864. [DOI] [PubMed] [Google Scholar]
- Halenius A., Hauka S., Dölken L., Stindt J., Reinhard H., Wiek C., Hanenberg H., Koszinowski U.H., Momburg F., Hengel H. Human cytomegalovirus disrupts the major histocompatibility complex class I peptide-loading complex and inhibits tapasin gene transcription. J. Virol. 2011;85:3473–3485. doi: 10.1128/JVI.01923-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiscott J. Triggering the innate antiviral response through IRF-3 activation. J. Biol. Chem. 2007;282:15325–15329. doi: 10.1074/jbc.R700002200. [DOI] [PubMed] [Google Scholar]
- Honda K., Takaoka A., Taniguchi T. Type I interferon [corrected] gene induction by the interferon regulatory factor family of transcription factors. Immunity. 2006;25:349–360. doi: 10.1016/j.immuni.2006.08.009. [DOI] [PubMed] [Google Scholar]
- Huber M., Izzi L., Grondin P., Houde C., Kunath T., Veillette A., Beauchemin N. The carboxyl-terminal region of biliary glycoprotein controls its tyrosine phosphorylation and association with protein-tyrosine phosphatases SHP-1 and SHP-2 in epithelial cells. J. Biol. Chem. 1999;274:335–344. doi: 10.1074/jbc.274.1.335. [DOI] [PubMed] [Google Scholar]
- Isaacs, A., and Lindenmann, J. (1957). Virus interference. I. The interferon. Proceedings of the Royal Society of London Series B, Containing papers of a Biological character Royal Society 147, 258–267. [DOI] [PubMed]
- Kato H., Takeuchi O., Sato S., Yoneyama M., Yamamoto M., Matsui K., Uematsu S., Jung A., Kawai T., Ishii K.J. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature. 2006;441:101–105. doi: 10.1038/nature04734. [DOI] [PubMed] [Google Scholar]
- Kovats S., Main E.K., Librach C., Stubblebine M., Fisher S.J., DeMars R. A class I antigen, HLA-G, expressed in human trophoblasts. Science. 1990;248:220–223. doi: 10.1126/science.2326636. [DOI] [PubMed] [Google Scholar]
- Li I.W., Chan K.H., To K.W., Wong S.S., Ho P.L., Lau S.K., Woo P.C., Tsoi H.W., Chan J.F., Cheng V.C. Differential susceptibility of different cell lines to swine-origin influenza A H1N1, seasonal human influenza A H1N1, and avian influenza A H5N1 viruses. J. Clin. Virol. 2009;46:325–330. doi: 10.1016/j.jcv.2009.09.013. [DOI] [PubMed] [Google Scholar]
- Li T., Chen J., Cristea I.M. Human cytomegalovirus tegument protein pUL83 inhibits IFI16-mediated DNA sensing for immune evasion. Cell Host Microbe. 2013;14:591–599. doi: 10.1016/j.chom.2013.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin T.M., Clemente C.F., Santos A.M., Picardi P.K., Pascoal V.D., Lopes-Cendes I., Saad M.J., Franchini K.G. Shp2 negatively regulates growth in cardiomyocytes by controlling focal adhesion kinase/Src and mTOR pathways. Circ. Res. 2008;103:813–824. doi: 10.1161/CIRCRESAHA.108.179754. [DOI] [PubMed] [Google Scholar]
- Markel G., Achdout H., Katz G., Ling K.L., Salio M., Gruda R., Gazit R., Mizrahi S., Hanna J., Gonen-Gross T. Biological function of the soluble CEACAM1 protein and implications in TAP2-deficient patients. Eur. J. Immunol. 2004;34:2138–2148. doi: 10.1002/eji.200425021. [DOI] [PubMed] [Google Scholar]
- Mibayashi M., Martínez-Sobrido L., Loo Y.M., Cárdenas W.B., Gale M., Jr., García-Sastre A. Inhibition of retinoic acid-inducible gene I-mediated induction of beta interferon by the NS1 protein of influenza A virus. J. Virol. 2007;81:514–524. doi: 10.1128/JVI.01265-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moorman N.J., Cristea I.M., Terhune S.S., Rout M.P., Chait B.T., Shenk T. Human cytomegalovirus protein UL38 inhibits host cell stress responses by antagonizing the tuberous sclerosis protein complex. Cell Host Microbe. 2008;3:253–262. doi: 10.1016/j.chom.2008.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muenzner P., Naumann M., Meyer T.F., Gray-Owen S.D. Pathogenic Neisseria trigger expression of their carcinoembryonic antigen-related cellular adhesion molecule 1 (CEACAM1; previously CD66a) receptor on primary endothelial cells by activating the immediate early response transcription factor, nuclear factor-kappaB. J. Biol. Chem. 2001;276:24331–24340. doi: 10.1074/jbc.M006883200. [DOI] [PubMed] [Google Scholar]
- Müller M.M., Klaile E., Vorontsova O., Singer B.B., Obrink B. Homophilic adhesion and CEACAM1-S regulate dimerization of CEACAM1-L and recruitment of SHP-2 and c-Src. J. Cell Biol. 2009;187:569–581. doi: 10.1083/jcb.200904150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson J.D., Denisenko O., Bomsztyk K. Protocol for the fast chromatin immunoprecipitation (ChIP) method. Nat. Protoc. 2006;1:179–185. doi: 10.1038/nprot.2006.27. [DOI] [PubMed] [Google Scholar]
- Nouvion A.L., Oubaha M., Leblanc S., Davis E.C., Jastrow H., Kammerer R., Breton V., Turbide C., Ergun S., Gratton J.P., Beauchemin N. CEACAM1: a key regulator of vascular permeability. J. Cell Sci. 2010;123:4221–4230. doi: 10.1242/jcs.073635. [DOI] [PubMed] [Google Scholar]
- Schramm C., Fine D.M., Edwards M.A., Reeb A.N., Krenz M. The PTPN11 loss-of-function mutation Q510E-Shp2 causes hypertrophic cardiomyopathy by dysregulating mTOR signaling. Am. J. Physiol. Heart Circ. Physiol. 2012;302:H231–H243. doi: 10.1152/ajpheart.00665.2011. [DOI] [PubMed] [Google Scholar]
- Stetson D.B., Medzhitov R. Type I interferons in host defense. Immunity. 2006;25:373–381. doi: 10.1016/j.immuni.2006.08.007. [DOI] [PubMed] [Google Scholar]
- Unterholzner L., Keating S.E., Baran M., Horan K.A., Jensen S.B., Sharma S., Sirois C.M., Jin T., Latz E., Xiao T.S. IFI16 is an innate immune sensor for intracellular DNA. Nat. Immunol. 2010;11:997–1004. doi: 10.1038/ni.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh D., Mohr I. Viral subversion of the host protein synthesis machinery. Nat. Rev. Microbiol. 2011;9:860–875. doi: 10.1038/nrmicro2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisblum Y., Panet A., Zakay-Rones Z., Haimov-Kochman R., Goldman-Wohl D., Ariel I., Falk H., Natanson-Yaron S., Goldberg M.D., Gilad R. Modeling of human cytomegalovirus maternal-fetal transmission in a novel decidual organ culture. J. Virol. 2011;85:13204–13213. doi: 10.1128/JVI.05749-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zito C.I., Qin H., Blenis J., Bennett A.M. SHP-2 regulates cell growth by controlling the mTOR/S6 kinase 1 pathway. J. Biol. Chem. 2007;282:6946–6953. doi: 10.1074/jbc.M608338200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.