Abstract
Though current functional genomic screening systems are useful for investigating human susceptibility to chemical toxicity, they have limitations. Well-established, high-throughput yeast mutant screens identify only evolutionarily conserved processes. RNA interference can be applied in human cells but is limited by incomplete gene knockout and off-target effects. Human haploid cell screening is advantageous as it requires knockdown of only a single copy of each gene. A human haploid cell mutant library (KBM7-Mu), derived from a chronic myeloid leukemia (CML) patient, was recently developed and has been used to identify genes that modulate sensitivity to infectious agents and pharmaceutical drugs. Here, we sought to improve the KBM7-Mu screening process to enable efficient screening of environmental chemicals. We developed a semi-solid medium based screening approach that cultures individual mutant colonies from chemically resistant cells, faster (by 2–3 weeks) and with less labor than the original liquid medium-based approach. As proof of principle, we identified genetic mutants that confer resistance to the carcinogen formaldehyde (FA, 12 genes, 18 hits) and the CML chemotherapeutic agent imatinib (6 genes, 13 hits). Validation experiments conducted on KBM7 mutants lacking each of the 18 genes confirmed resistance of 6 FA mutants (CTC1, FCRLA, GOT1, LPR5, M1AP, and MAP2K5) and 1 imatinib-resistant mutant (LYRM9). Despite the improvements to the method, it remains technically challenging to limit false positive findings. Nonetheless, our findings demonstrate the broad applicability of this optimized haploid approach to screen toxic chemicals to identify novel susceptibility genes and gain insight into potential mechanisms of toxicity.
Keywords: functional genomics, KBM7, haploid, formaldehyde, imatinib.
Functional genomic screening is a useful approach to identify genes and pathways involved in response to toxicants and to discover candidate human susceptibility genes (Gaytan and Vulpe, 2014). This is achieved through knockdown or knockout of all nonessential genes in a cell of interest, and identification of genes and pathways associated with survival following toxicant exposure. For many years, such screens were conducted in yeast, due to the availability of haploid strains and mutant clones for all nonessential yeast genes, and the development of high-throughput methodologies and bioinformatics analyses (North and Vulpe, 2010; Winzeler et al., 1999). Through yeast screens, we and others identified genes and toxicity mechanisms involved in resistance to toxic metabolites of arsenic (Jo et al., 2009a), benzene (North et al., 2011b), benzo[a]pyrene (O’Connor et al., 2012), and formaldehyde (FA) (de Graaf et al., 2009; North et al., 2011a), and we validated the roles of human orthologs of yeast genes associated with benzene (Galvan et al., 2008; Ren et al., 2009, 2011b) and arsenic (Jo et al., 2009b; Ren et al., 2011a) toxicity in functional studies in mammalian cell lines using RNA interference (RNAi)-based knockdown. However, the ability to decipher human biological responses through yeast functional genomic screening is limited to evolutionarily conserved genes and processes. The establishment of the near-haploid KBM7 cell line (Carette et al., 2009) and development of a library of mutants by retrovirally mediated insertional mutagenesis has enabled the screening of haploid human cells directly (Burckstummer et al., 2013; Carette et al., 2009). Currently, over 3396 human haploid mutant clones are available, covering almost one-third of the expressed genome (Burckstummer et al., 2013).
Multiple KBM7 screening studies have identified genes and mechanisms that modulate host responses to viral and bacterial toxins (Carette et al., 2009, 2011a; Reiling et al., 2011; Timms et al., 2013; van den Boomen et al., 2014) and therapeutic agents (Birsoy et al., 2013; Carette et al., 2009; Chen et al., 2014; Reiling et al., 2013; Winter et al., 2014). It was recently used to identify novel susceptibility genes involved in the response to the pesticide and environmental contaminant chlorpyrifos (Zhu et al., 2015). We believe that the approach could be used for more widespread screening of chemicals but currently it is limited by experimental inefficiencies. The original screening approach utilized liquid culture medium and required limited dilution and subcloning to select individual mutant clones, which made the screening protocol lengthy and inefficient (Carette et al., 2009). Further, the KBM7 cells can revert to diploidy over time and mutants can arise spontaneously. Here, we sought to reduce the screening time and improve the efficiency by using semi-solid medium and enriching for haploid, mutant cells.
As proof of principle of our improved approach, we screened the KBM7 mutant library for genes whose absence confers resistance to FA, a widespread environmental toxin (Tang et al., 2009; Zhang et al., 2009) and Group 1 human carcinogen (IARC, 2006), and imatinib, a chemotherapeutic agent. There is a limited understanding of the mechanisms of FA toxicity and determinants of susceptibility. Previously, through yeast screening, we identified several yeast genes required for FA tolerance, whose human orthologs are involved in FA metabolism and multiple types of DNA repair (North et al., 2011a). Others have identified genes involved in DNA damage response using a chicken DT40 B lymphoblast functional genomic screening system (Ridpath et al., 2007; Yamazoe et al., 2004), and we validated the role of 1 such identified gene, FANCD2, in resistance to FA toxicity in human lymphoblast cells (Ren et al., 2013). Here, we sought to identify additional human genes involved in the response to FA using a complementary KBM7 screening approach. Further, KBM7 is of particular interest as it is a hematopoietic cell line, and we have found hematotoxicity associated with FA exposure in mouse (Ye et al., 2013; Zhang et al., 2013b) and human studies (Lan et al., 2015; Seow et al., 2015; Zhang et al., 2010).
Imatinib (marketed as Gleevec or Glivec) is a tyrosine-kinase inhibitor used in the treatment of multiple cancers, most notably Philadelphia chromosome-positive (Ph+) chronic myelogenous leukemia (CML). Imatinib acts by blocking constitutively activated breakpoint cluster region-Abelson murine leukemia viral oncogene homolog 1 (BCR-ABL) tyrosine kinase activity, leading to suppressed cell proliferation and eventual death (Goldman and Melo, 2003). We chose imatinib as a positive control as KBM7 cells were derived from blast-crisis CML (Kotecki et al., 1999) and a few factors governing resistance to imatinib previously have been identified in KBM7 mutant library screening (Carette et al., 2009).
Here, we sought to optimize functional genomic screening in KBM7 cells and, as proof of principle of the approach, to screen the mutant library for gene mutants that modulate resistance to FA and imatinib. Resistance was measured as survival and growth (colony-formation) following exposure.
MATERIALS AND METHODS
Cell Culture and Enrichment of Haploid KBM7 Cells
Wild-type KBM7 cells and a library of mutant cells (KBM7-Mu) were obtained from Dr Jan Carette at the Whitehead Institute of Biomedical Research, Cambridge, Massachusetts (Burckstummer et al., 2013; Carette et al., 2009). The library comprises clones covering 3396 genes, almost one-third of the expressed genome (Burckstummer et al., 2013). The cells were grown in Iscove’s modified Dulbecco’s medium (IMDM) with 10% heat-inactivated fetal bovine serum and 1% Penicillin-Streptomycin and 5% CO2 (Carette et al., 2009).
Chemical Treatment and Selection of Optimal Concentrations
Use of appropriate treatment concentrations is critical in lethality-based screens to identify the most important genes responsible for survival. Too high a concentration can kill both wild-type and mutant cells, while too low a concentration can generate too many hits and make it difficult to select those truly important for survival. Therefore, we conducted 4-day acute toxicity studies in KBM7 cells with a wide range of concentrations of FA (20–120 μM) and imatinib (0.01–10 μM) in liquid culture medium. Methanol-free FA ampules (16% wt/vol) and imatinib mesylate were purchased from Fisher Scientific. Stock solutions were prepared in PBS such that 1/100 volume was added to the media to achieve the desired concentration. PBS was added to the vehicle controls. The cells were plated at 20 000 per well in a 96-well culture plate, incubated at 37°C for 24 h and then treated with FA or imatinib. Cell viability was determined by trypan blue after 24 and 72 h. The range-finding experiments were repeated twice for FA and 4 times for imatinib, with 2 replicates per chemical concentration.
Optimization of Haploid Screening
We followed the haploid screening protocol described by Carette et al. (2009) to identify mutants resistant to FA and Imatinib, with the following modifications as summarized in Supplementary Figure 1.
Selection of haploid cells and gene-trap mutants by fluorescence-activated cell sorting
In long-term culture (over 12 weeks), the near haploid KBM7 cell line can become diploid (Kotecki et al., 1999). We used serial fluorescence-activated cell sorting (FACS) to conservatively select haploid cells according to DNA content by 4′,6-diamidino-2-phenylindole (DAPI) staining (Supplementary Figure 1A). Aliquots of sorted cells (90%–99% haploid) were cryopreserved and thawed cells were used in the screening experiments.
The library of mutants was generated by Carette et al. (2009) using gene-trap retroviruses that contain green fluorescent protein (GFP) as a marker gene. However, cells could also become resistant to chemical exposures through spontaneous genetic or epigenetic gene inactivation. To decrease the potential for false positive gene hits in the screening process, haploid mutant cells were further purified by GFP expression using FACS (Supplementary Figure 1B).
Screening in semi-solid medium
KBM7-Mu library cells (20 000) were seeded in 0.5 ml viscous ClonaCell-TCS semi-solid medium containing 9% methylcellulose, IMDM, serum, and bovine serum albumin (Stem Cell Technologies, Vancouver, British Columbia, Canada) in 12-well plates. Cells were incubated at 37°C for 24 h and then treated with FA or imatinib (Supplementary Figure 1C). After 2–3 weeks, individual colonies were picked out and expanded in liquid culture medium (2 × 12 ml in separate flasks) for 2–3 days for downstream processing (Supplementary Figure 1D).
Identification of target genes and retroviral insertional sites
Genomic DNA was isolated from the cells in each expanded colony (Supplementary Figure 1E) using QIAamp DNA Mini Kits (Qiagen Inc, Valencia, California). The presence of mutant cells was further confirmed by amplification of the gene trap vector by PCR. DNA from colonies with insertions were digested with MseI (New England Biolabs, Ipswich, Massachusetts) and linear DNA fragments containing both vector and target gene fragments were self-ligated using T4 DNA ligase (New England Biolabs) to form circular products. Inverse PCR was performed to amplify the DNA product containing the target gene fragment for 1 or 2 rounds of PCR until a single band of around 650–800 bp was visible on a 1% agarose gel. The protocol was essentially the same as that of Carette et al., 2009, with the exception of PCR primers which were redesigned to eliminate predicted primer-dimer formation: forward: 5′-CGA CCC CGT CAG GAT ATG TG-3′ and reverse: 5′-CGG GTG TTC AGA ACT CGT CA-3′. PCR products were purified using Qiaquick Gel Extraction kits (Qiagen). If more than 1 band was present, eg, due to the presence of more than 1 gene-trap vector per mutant cell, each band was purified separately. Purified PCR products were analyzed by Sanger sequencing (Supplementary Figure 1F). Briefly, 100 ng DNA/1000 bp PCR product and 0.8 pmol sequencing primer (5′-CGC AGG CGC AAA CAT TAG AT-3′) were mixed in a total volume of 13 µl ddH2O and submitted to University of California Berkeley’s DNA Sequencing Facility. The host resistance genes conferring survival in each mutant colony were identified using Basic Local Alignment Search Tool (BLAST) from National Center for Biotechnology Information (NCBI) and BLAST-like alignment tool (BLAT) from the University of California Santa Cruz (UCSC) Genome Browser (Supplementary Figure 1F).
Validation of Resistance in Mutant Clones Compared With Wild-Type Cells
We confirmed the findings by comparing cell proliferation in mutant clones with that in wild-type KBM7 cells in 2 kinds of validation experiment. We prioritized genes with multiple hits (different mutant clones or screens) and directly performed a full validation, with treatment at 8 doses and 4 time points (over 4 days). For genes with only 1 hit, we first conducted a preliminary validation at 2 doses and a single time point (3 days), followed by a full validation only if the preliminary validation findings were statistically significant. In each case, 2 to 3 independent experiments, with 2 replicates per dose were conducted. We did not perform full validation for all mutant clones, particularly the single-hit mutants, as it is labor intensive, generating 64 datasets for each mutant, and requires a large number of cells.
Cell Proliferation Inhibition Assay
Expanded mutant colony cells were treated with FA (0, 20, 40, 60, 80, 90, 100, or 120 μM) and imatinib (0, 0.1, 0.2, or 0.5 μM) for up to 96 h and cell proliferation data were collected 72 h after treatment. Briefly, dead cells were stained with trypan blue Vi-CELL XR reagent pack (Beckman Coulter, Inc, Fullerton, California) and the cell viability data were analyzed by a Vi-CELL XR cell viability analyzer (Beckman). The final cell proliferation data were calculated as a percentage (%) of vehicle (PBS) control treatments.
Flow Cytometry-Based Cell Death Assay
In validation assays of some FA mutants, cell death was evaluated by a flow cytometry-based method as well as by trypan blue. Briefly, after treatment with 0 or 90 μM of FA for 48 h, cells were washed and stained with using a LIVE/DEAD Fixable Violet Dead Cell Stain Kit (Life Technologies, Eugene, Oregon) according to the manufacturer’s protocol. The BD LSR Fortessa flow cytometer (BD Biosciences, San Jose, California) was used for cellular acquisition of up to 10 000 total singlet events per sample, and results were analyzed using FACSDiva Version 6.2 Flow Cytometry Analysis software (BD Biosciences).
mRNA Expression by qPCR
Total RNA was isolated from cells using the RNeasy Mini Kit (Qiagen) according to the manufacturer’s protocol. RNA concentration was determined by absorbance at A260 and RNA purity was determined by A260/A280. cDNA templates were generated using 1 μg of total RNA in 20 μl reactions using High-Capacity cDNA Reverse Transcription Kits from Applied Biosystems, Inc (Foster City, California) according to the manufacturer’s protocol. qPCR was performed in a 20 μl reactions with 4 ng of cDNA template and 900 nM primer using a SsoFast EvaGreen supermix (Bio-Rad Laboratories Inc, Hercules, California) according to the manufacturer’s protocol. Primers for candidate genes of interest were designed and verified by the NCBI online tool Primer-BLAST and synthesized by Integrated DNA Technologies, Inc (Coralville, Iowa): Low-density lipoprotein receptor-related protein 5 (LRP5): Forward- 5′-CCA AGC GAG CCT TTC TAC AC -3′, Reverse- 5′-TCT AGC GGG TCG TAG TCG AT-3′, meiosis 1 associated protein (M1AP): Forward- 5′-AAC CCC TTG CAT GTT CAA AGC-3′, Reverse- 5′-AGT CTT GCA GGG ATG CTT TCT C-3′, and ribosomal protein L13a (RPL13a, a housekeeping gene): forward- 5′-CCT GGA GGA GAA GAG GAA AGA GA-3′, reverse-5′ TTG AGG ACC TCT GTG TAT TTG TCA A-3′.
The PCR program included incubation at 95° for 10 min followed by 40 cycles of 95° for 30 s, 55° for 30 s, and 72° for 1 min and was performed on a CFX96 real-time PCR detection system (Bio-Rad Laboratories Inc). A melting curve analysis was carried out after the reaction to check for primer dimers, nonspecific binding, or other contamination. The relative gene expression levels were calculated using the relative standard curve method. The target gene expression levels were adjusted to RPL13a. Final results were presented as fold changes whereby the adjusted expression level of each target gene in the KBM7 treatment group was divided by that in the wild-type group.
Protein Level Measured by Western Blot
LRP5 protein level was qualitatively measured by Western blotting. Briefly, cell pellets were washed with ice-cold PBS and lysed directly in 4X Laemmli sample buffer (277.8 mM Tris-HCl, pH 6.8, 4.4% LDS, 44.4% (wt/vol) glycerol, 0.02% bromophenol blue) at approximately 2 × 106–1 × 107 cells per ml. The samples were heated in a boiling water bath for 5 min and stored at −80°C. Samples of 100 000 or 150 000 cells per lane were electrophoretically resolved in 4%–15% denaturing polyacrylamide gel and then transferred to the polyvinylidene fluoride membrane using Bio-Rad Mini-PROTEAN and Mini-Trans Blot systems (Bio-Rad Laboratories, Inc). The membranes were immunoblotted using primary antibodies against LRP5 (Cell Signaling Technology, Inc, Danvers, Massachusetts) and Laminin as an internal reference protein (Santa Cruz Biotechnology, Inc Santa Cruz, California). The protein signal was determined using enhanced chemiluminescence on the ChemiDoc XRS+ system (Bio-Rad Laboratories, Inc). Data were analyzed by IMAGE J software (V1.43, National Institute of Health, United States).
Transient Gene Knockdown by RNAi
The use of RNAi to block LRP5 expression in transient transfections was facilitated by the cloning of short hairpin RNA (shRNA) sequences using the BLOCK-iT U6 RNAi Entry Vector kit (Life Technologies, Grand Island, New York). Briefly, oligonucleotides (Bjorklund et al., 2007) targeting the human LRP5 gene, and the control-scrambled DNA, which has similar DNA structure but no homology to any known human sequence, were synthesized by DNA Technologies, Inc and annealed together to generate double-stranded oligonucleotides (ds oligos). The sequences for shRNAs and control-scrambled DNA are as follows:
RNAi LRP5 sense 5′-CAC CGC CTG CAT GGA CTG AGG AAC GTC AAA CGA ATT TGA CGT TCC TCA GTC CAT GCA GG-3′; RNAi LRP5 antisense 5′-AAA ACC TGC ATG GAC TGA GGA ACG TCA AAT TCG TTT GAC GTT CCT CAG TCC ATG CAG GC-3′; Control-scrambled sense 5′-CAC CGG CAC CTG AGA GAA CGG CTA TCA TAG CGA ACT ATG ATA GCC GTT CTC TCA GGT GC-3′; and Control-scrambled antisense 5′-AAA AGC ACC TGA GAG AAC GGC TAT CAT AGT TCG CTA TGA TAG CCG TTC TCT CAG GTG CC-3′.
The ds oligos were cloned into the pENTR/U6 vector provided in the kit according to the manufacturer’s protocol. The vector constructs that express shRNAs were transiently transfected into KBM7 Wild-type cells using Lipofectamine 2000 (Life Technologies) according the manufacturer’s protocol. After 4 days of transfection with either LRP5 shRNA or scrambled control vectors, cells were treated with FA for 4 days. In a recovery study, the cells were transfected as above for 4 days, washed and grown for an additional week in normal media (without transfection reagents), and then treated with FA. Cell proliferation was compared between the LRP5 RNAi group and the scrambled control group.
Statistical Analysis
Data were analyzed by Student’s t test and P values <0.05 were considered statistically significant. Data are expressed as mean ± SD.
RESULTS
Through multiple method improvements described in detail in Materials and Methods, we increased the efficiency of the screening protocol.
Enrichment of Haploid Cells and Gene-Trap Mutants
We performed preliminary enrichment steps on the KBM7 wild-type and mutant cells (Burckstummer et al., 2013; Carette et al., 2009). First, using FACS to select haploid cells according to DNA content by DAPI staining (Supplementary Figure 1A), we maintained haploidy at over 90% in the cultures for at least 18 passages (Supplementary Figure 2) compared with approximately 84% haploidy in the original protocol (Carette et al., 2009). Second, true haploid mutant cells were enriched by GFP expression using FACS (Supplementary Figure 1B). Thus, cells populations highly enriched for both haploid status and GFP expression were used in chemical screening experiments.
Use of Semi-Solid Medium
In the original screening protocol (Carette et al., 2009), mutant colony generation and isolation are challenging because the cells are cultured in liquid medium. If more than 1 colony survives per well after treatment, limited dilution and subcloning are necessary and take 2–3 weeks. This is technically challenging and limits the efficiency of the screening process. We optimized the screening process to utilize semi-solid medium (containing 9% methylcellulose), in which the cells and resulting colonies are immobilized during screening, enabling easier manipulation. Both protocols require 2–3 weeks for screening but use of semi-solid media removes the need for subcloning to generate single colonies and therefore reduces the overall protocol by 2–3 weeks.
We confirmed that the presence of methylcellulose during treatment did not attenuate FA toxicity, by seeding cells in semi-solid medium immediately after the addition of FA or 48 h after the addition of FA.
Selection of Optimal Chemical Treatment Concentrations
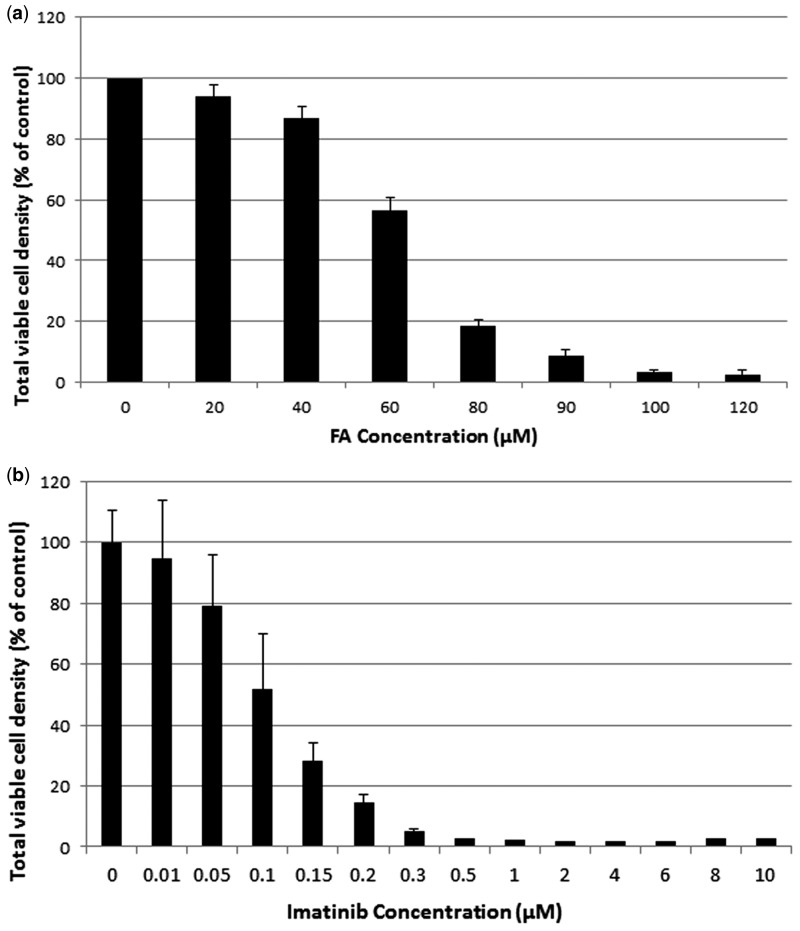
Based on the cytotoxicity data from the 4-day acute toxicity studies performed with the KBM7-Mu cells in liquid media, we selected 80, 85, and 90 μM for FA (Figure 1A) and 0.8, 1, 1.5, and 2 μM for imatinib (Figure 1B) for the screening experiments because these concentrations inhibited cell proliferation. For example, 72 h after treatment, 80 and 90 μM FA suppressed proliferation by about 80% and 90%, respectively, and imatinib at 1 and 2 μM each suppressed proliferation by close to 100%. We used more than 1 critical concentration of each chemical to minimize false negative results and increase the number of surviving mutants.
FIG. 1.
Selection of critical screening concentrations of formaldehyde (FA) and imatinib. Viable cell density as a percentage of untreated controls 72 h after treatment in liquid culture medium are shown for (A) 20–120 μM FA and (B) 0.01–10 μM imanitib. Data and SD are shown from 2 FA experiments and 4 imatinib experiments, each with 2 replicates per chemical concentration.
Identification of Candidate Gene Mutants Conferring Resistance to FA and Imatinib
Enriched haploid KBM7-Mu cells were exposed to FA and imatinib in 12-well plates in 3 separate screens and a total of 99 and 129 surviving colonies were identified, respectively, after 2–3 weeks. Colonies were then expanded in liquid culture for 2–3 days, genomic DNA was extracted, and DNA from colonies positive for gene trap insertion were subjected to inverse PCR and Sanger sequencing of individual PCR products to identify the insertion sites (Supplementary Figure 1E). The original PCR primers (Carette et al., 2009) were redesigned to eliminate predicted primer-dimer formation.
A total of 12 candidate gene mutants (18 colonies) from FA and 6 gene mutants (13 colonies) from imatinib were identified based on the growth of 1 or as many as 6 independent resistant clonal colonies in 1–3 independent experiments (Table 1). The remaining colonies did not lead to the identification of candidate genes for various reasons, including lack of mutant plasmid or insertion; failure to generate a PCR product or generation of multiple bands; or uninformative sequence product.
TABLE 1.
Gene Mutants That Confer Resistance to FA and Imatinib Identified by Haploid Screening
| Chemical | No. Resistant Colonies |
Total Hits | Validation Confirmed |
|||
|---|---|---|---|---|---|---|
| Screen 1 | Screen 2 | Screen 3 | Preliminary | Full | ||
| FA | ||||||
| CTC1 (NM_025099.5) | 1 | 1 | YES | YES | ||
| FCRLA(NM_032738.3) | 1 | 1 | YES | YES | ||
| GOT1 (NM_002079.2) | 1 | 2 | 3 | nd | YES | |
| LRP5 (NM_002335.3) | 1 | 1 | 2 | nd | YES | |
| M1AP (NM_001281296.1 | 1 | 2 | 3 | nd | YES | |
| MAP2K5(NM_145160.2) | 1 | 1 | YES | YES | ||
| IGF2BP3(NM_006547.2) | 1 | 1 | 2 | nd | NO | |
| NOD1(NM_006092.2) | 1 | 1 | NO | nd | ||
| ITGB1(NM_002211.3) | 1 | 1 | NO | nd | ||
| RYBP(NM_012234.6) | 1 | 1 | NO | nd | ||
| NOTCH1(NM_017617.3) | 1 | 1 | NO | nd | ||
| NKIRAS2(NC_000003.12) | 1 | 1 | NO | nd | ||
| Sub total colony | 4 | 8 | 6 | 18 | ||
| Imatinib | ||||||
| CASP10(NM_001206524.1) | 1 | 1 | YES | NO | ||
| CUX1(NM_001202545.2) | 2 | 2 | nd | NO | ||
| NF1(NM_001042492.2) | 2 | 2 | nd | NO | ||
| LYRM9(NC_000017.11) | 3 | 2 | 1 | 6 | nd | YES |
| ZPBP(NM_001159878.1) | 1 | 1 | NO | nd | ||
| CEBPG(NM_001252296.1) | 1 | 1 | NO | nd | ||
| Sub total colony | 10 | 2 | 1 | 13 | ||
nd, not done; not necessary because there are ≥ 2 hits so a full validation was directly conducted; NO, the validation was conducted but findings were not statistically significant; and YES, the validation was conducted and findings were not statistically significant.
Validation of FA-Resistant Gene Mutants
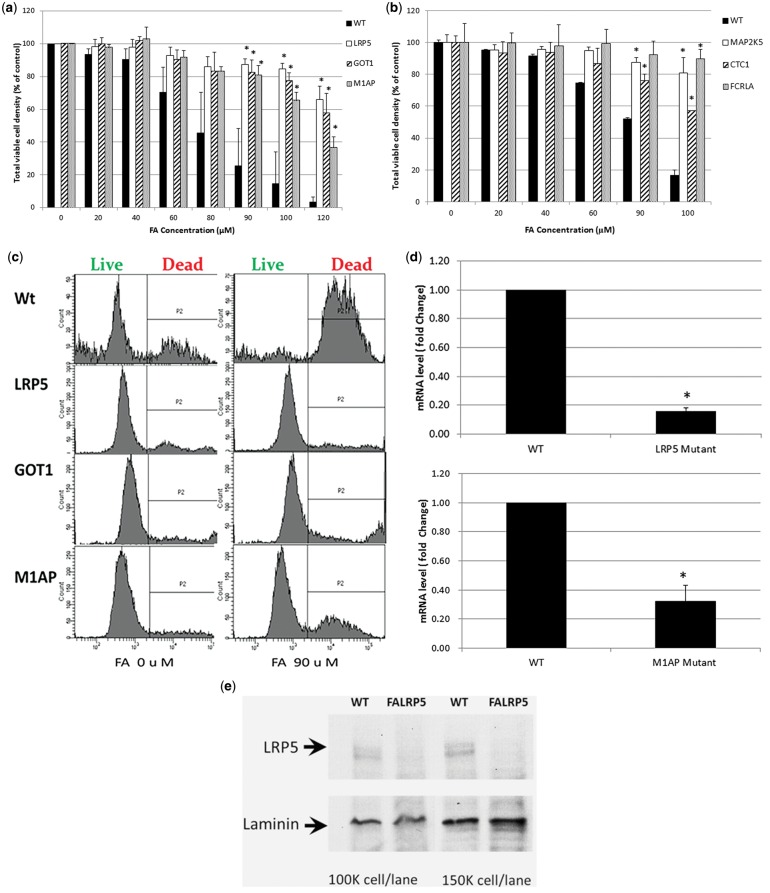
Validation experiments were conducted to confirm the resistance of the mutant clones to FA. Expanded colony cells were cultured and treated with 0–120 μM FA for 96 h. As listed in Table 1 and shown in Figures 2A and 2B, 6 mutants, CST telomerase maintenance complex component 1 (CTC1), CTC1, Fc Receptor-Like A (FCRLA), glutamic oxoloacetic transaminase 1 (GOT1), LRP5, M1AP, and mitogen-activated protein kinase kinase 5 [MAP2K5]) were confirmed as resistant to FA by full validation, as measured by cell proliferation in mutant cells compared with wild-type KBM7 cells. LRP5, GOT1, and M1AP mutants, identified through multiple hits, also exhibited decreased cell death compared with wild-type cells after treatment with FA for 72 h (Figure 2C). However, the IGF2BBP3 mutant (identified in 2 independent screens) was not resistant by full validation. Prior to the full validation, 3 mutants with single hits, CTC1, FCRLA, and MAP2K5 were also confirmed with preliminary validation. We did not test the remainder of single-hit mutant clones further since they did not show their in preliminary validations (Table 1).
FIG. 2.
Validation of resistance to FA of select mutant clones. A, Viable cell density of lipoprotein receptor-related protein 5 (LRP5), glutamic-oxaloacetic transaminase 1 (GOT1), and meiosis 1 associated protein (M1AP) mutants and wild-type KBM7 cells, 72 h after treatment with FA. Average (and SD) of 3 replicate experiments shown as a percentage of untreated controls cells; B, Viable cell density of mitogen-activated protein kinase kinase 5 (MAP2K5), CTC1, and FCRLA mutants and wild-type KBM7 cells 72 h after treatment with FA. Average and SD of 2 replicate experiments shown as a percentage of untreated controls cells; C, Representative data (from 1 of 2 experiments, 2 replicates per experiment) showing live/dead LRP5, GOT1, and M1AP mutants and wild-type KBM7 cells, after treatment with FA (90 µM) for 72 h, compared with untreated cells; D, Validation of knockdown of LRP5 and M1AP mRNA in the mutant cells by RT-PCR normalized to the housekeeping gene ribosomal protein L13a. Data (fold change and SD) are averaged from 4 experiments and 3 experiments, respectively, 2 replicates per experiment; E, Validation of LRP5 protein knockdown in the mutant cells by Western blot. Representative data from 1 of 2 experiments shown. *P < .05 by Student’s t test.
We confirmed knockdown of LRP5 and M1AP by up to 80% in their respective mutant cell cultures by reverse-transcription (RT)-PCR (Figure 2D). Availability of an LRP5 antibody allowed us to measure the LRP5 protein level by Western blot (Figure 2E), results of which showed a clear inhibition of LRP5 protein. We attempted to validate the findings by RNAi in wild-type KBM7 cells (Supplementary Figure 3). Compared with a scrambled siRNA control, cells with transient loss of LRP5 exhibited enhanced survival (viability) 48 and 72 h after treatment with 100 μM FA, that reverted to similar levels following recovery from the transient knockdown. However, the enhanced survival (10%–15% viability) was much less pronounced than that of the expanded gene-trap KBM7 clone (80% viability relative to wild-type KBM7 cells, Figure 2A) treated with the same concentration of FA. This may be attributable to the low siRNA transfection efficiencies achieved with KBM7 cells (at most 50%, data not shown) that are typical of blood cells.
Validation of Imatinib-Resistant Gene Mutants
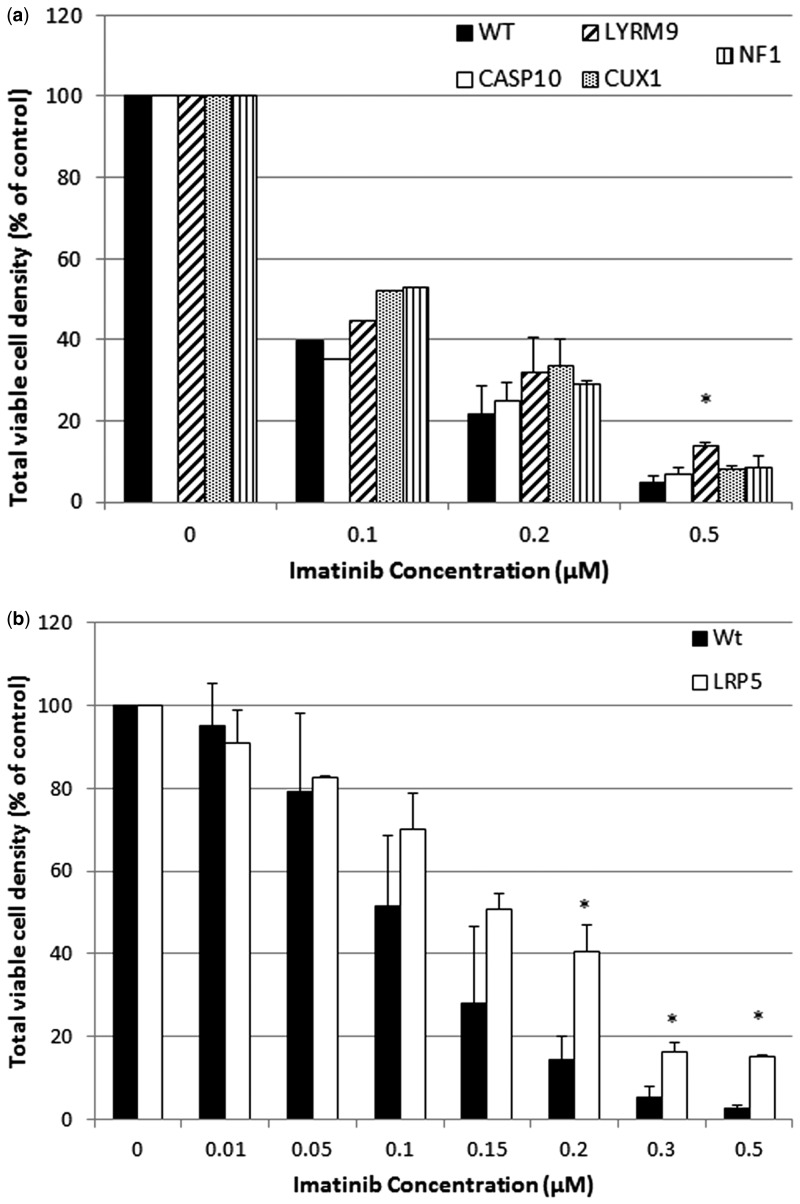
As listed in Table 1, resistance to imatinib of mutant clones representing 2 genes was confirmed either by preliminary (Caspase 10, CASP10) or full validation (LYR motif containing 9, LYRM9). LYRM9 showed the strongest response as seen in the data from the full validation experiments shown in Figure 3A. The remaining 2 mutant clones that survived screening , zona pellucida binding protein (ZPBP) and CCAAT/enhancer binding protein, gamma (CEBPG), were not resistant in preliminary validation and were not tested further. Although LRP5 was not identified as an imatinib resistance gene by screening, LRP5-mutant cells also showed significant resistance to imatinib treatment compared with the KBM7 wild-type cells (Figure 3B).
FIG. 3.
Validation of resistance to imatinib of select mutant clones. A, Viable cell density of Caspase 10 (CASP10), LYR motif containing 9 (LYRM9), CUX1, and neurofibromin 1 mutants and wild-type KBM7 cells 72 h after treatment with imatinib. Average (and SD) of 2 replicate experiments, 2 replicates per experiment, shown as a percentage of untreated controls cells. B, Viable cell density of LRP5 mutant cells and wild-type KBM7 cells 72 h after treatment with imatinib. Average (and SD) of 2 replicate experiments, 2 replicates per experiment, shown as a percentage of untreated controls cells. *P < .05 by Student’s t test.
DISCUSSION
To improve the efficiency of human haploid KBM7 screening, we optimized the original protocol (Carette et al., 2009) through several modifications. As proof of principle, we sought to identify gene mutants important for the response to FA and imatinib across a narrow range of concentrations selected to minimize false positives and maximize the number of resistant mutant colonies. Despite the optimization and streamlining of the screening protocol, it is still challenging to limit the false positive rate. Many resistant mutant colonies did not have identifiable genes and some mutant clones with identifiable genes could not be confirmed as resistant based on their proliferation relative to wild-type cells. Nonetheless, we identified candidate genes that modulate response to FA and imatinib.
Identification of Genes Involved in FA Response
Validation of FA resistance was achieved for 6 of 12 candidate mutants identified (Table 1) across a range of concentrations in in vitro treatment experiments in expanded colony cells. LRP5 was identified in 2 clones in 2 separate screening experiments. LRP5 is a coreceptor in the Wnt/β-catenin signaling pathway and plays important roles in embryonic and hematopoietic development. Wnt signaling is activated in acute myeloid leukemia (AML), leukemic stem cells (LSC), and myeloid blast crisis of CML and is thought to support the development and maintenance of myeloid LSC (Heidel et al., 2015). Translocations involving LRP5 were reported in 2 AML patients (Sarova et al., 2011). A mixture of volatile organic compound including FA was reported to alter Wnt signaling in mouse lung in vivo (Wang et al., 2014a) and high levels of FA altered expression of genes involved in Wnt signaling in the nasal epithelium of exposed rats (Andersen et al., 2010). To the best of our knowledge, Wnt signaling in hematopoietic cells has not been examined in association with FA exposure.
GOT1 was identified as a candidate resistance gene in 3 mutant clones in 2 separate screening experiments. GOT1 is a cytoplasmic pyridoxal phosphate-dependent enzyme that plays roles in amino acid metabolism and the urea and tricarboxylic acid cycles. It is an important regulator of levels of glutamate, the major excitatory neurotransmitter of the vertebrate central nervous system, thus influencing brain neuroprotection. FA is a well-established neurotoxin and has been proposed to be a contributor to brain neurodegeneration (Tulpule and Dringen, 2013). In primary rat astrocytes, FA has been shown to decrease glutamate transporter expression, inhibiting glutamate uptake (Song et al., 2010). In the context of leukemia, ETV6/GOT1 fusions, resulting from t(10;12) (q24;p13) translocation, have been described in myelodysplastic syndrome (Janssen et al., 2006; Struski et al., 2008).
M1AP was identified as a candidate resistance gene in 3 independent mutant clones in 2 separate screening experiments. It encodes a protein that is likely to function in progression of meiosis and its relevance to leukemia or FA toxicity is unknown.
CTC1, FCRLA, and MAP2K5 were each identified in a single clone. CTC1 is a component of the CST (CTC1-STN1-TEN1) complex, which plays an essential role in several aspects of telomere replication (Gu et al., 2012; Huang et al., 2012; Kasbek et al., 2013; Miyake et al., 2009; Stewart et al., 2012; Surovtseva et al., 2009; Wang et al., 2012) and facilitates recovery from many forms of exogenous DNA damage (Wang et al., 2014b). Defects in telomere biology can lead to genomic instability and are implicated in aging as well as in tumorigenesis and sporadic aplastic anemia (Gramatges and Bertuch, 2013). In hematopoietic cancer cells, telomere length has been associated with disease progression, prognosis, and outcomes (Jones et al., 2012). Telomere shortening has also been observed in Fanconi anemia, an inherited bone marrow failure syndrome associated with defects in DNA repair mechanisms (Gramatges and Bertuch, 2013) and susceptibility to certain cancers and sensitivity to DNA-DNA crosslinking agents including FA (Kennedy and D’Andrea, 2005). We and others have identified and validated some of the factors that counteract DNA damage induced by FA using functional genomic screening in chicken cells (Ren et al., 2013; Ridpath et al., 2007; Yamazoe et al., 2004) and yeast (de Graaf et al., 2009; North et al., 2011a). As FA has not been reported to alter telomere length or biology, further studies are needed to investigate the role of FA in telomere biology, genomic instability, and leukemia.
FCRLA is a B cell-specific protein in the FcR family (Davis et al., 2002; Facchetti et al., 2002; Maltais et al., 2006; Mechetina et al., 2002) that may bind to Ig and regulate Ig maturation and/or secretion (Reshetnikova et al., 2012; Santiago et al., 2011; Wilson et al., 2010). Though we found that occupational exposure to FA did not alter B cell counts (Hosgood et al., 2013), others have reported increased (Jia et al., 2014) or decreased (Costa et al., 2013) B cell numbers. To our knowledge, no human studies have examined B cell differentiation or function in response to FA exposure. Increased levels of IgM and IgA, and decreased levels of IgG, were induced in rats exposed to FA (Sapmaz et al., 2015). Additional functional studies are needed to establish the relationship between FA exposure, immunoglobulin production, and disease.
MAP2K5 is a signaling module in the MAPK pathway that is activated by stress stimuli and has been proposed to play a role in the pathology of cancer. MAP2K5 interacts with and activates the MAPK7/ERK5 pathway that protects cells from stress-induced apoptosis and supports neuronal survival and cardiac development and angiogenesis (Lochhead et al., 2012).
Identification of Genes Involved in Imatinib Response
Resistance of CML patients to imatinib therapy is an important clinical problem. The mechanisms underlying it are not fully understood but involve both BCR-ABL-dependent mechanisms and independent mechanisms, such as membrane-bound drug transporters and activation of alternative signaling pathways, that are mediated by genetic mutations and epigenetic alterations (Apperley, 2007; Balabanov et al., 2014; Trela et al., 2014). The challenge of resistance has been somewhat circumvented by the use of alternative tyrosine kinase inhibitors (TKI) or combinations of TKIs (Jabbour et al., 2015) but understanding the mechanisms underlying resistance to tyrosine kinases in general would improve treatment and clinical outcomes and understanding of the CML disease process. Previously, Carette et al. (2009) identified mutants that mediate imatinib resistance using their original KBM7 liquid-based screening approach. Though it is unclear whether they performed independent screens and how many cells survived, they expanded 11 imatinib-resistant cell lines and identified 3 mutant genes from 7 independent clones, similar to the number of mutant genes we found. The 3 genes were neurofibromin 1 (NF1) and protein tyrosine phosphatase-N1 (PTPN1), both of which had previously been shown to play an important role in the response of chronic myeloid cells to imatinib therapy (Luo et al., 2008), and PTPN12, a tyrosine phosphatase that negatively regulates c-abl activity (Cong et al., 2000). Carette et al. confirmed the absence of NFI by immunoblot in 2 independent surviving clones but did not confirm resistance to imatinib relative to wild-type cells in additional experiments. In our study, survival of 2 independent mutant clones also indicated a role for NF1 knockdown in imatinib resistance but survival was not statistically significant in our validation experiments. We did not identify PTPN1 or PTPN12 as candidates. We identified and validated a role for 2 additional mutants, CASP10 (preliminary validation), and LYRM9 (full validation), identified in 1 and 6 independent clones, respectively. It is unclear why only 1 gene overlapped between our screen and Carette’s 2009 screen but it may reflect the different screening conditions used. We treated cells in semi-solid media with 0.8, 1, 1.5, and 2 μM imatinib by adding it 1 time after cells had been plated and incubated for 24 h. In Carette’s study, a concentration of 1 μM imatinib in liquid media was used for 4 days followed by dilution to 0.3 μM for the following 2 weeks.
CASP10, together with CASP8, mediates the extrinsic death receptor-mediated pathway (Movassagh and Foo, 2008). Imatinib induces apoptosis of CML cells and the combined treatment of imatinib and other molecules that enhance imatinib-induced apoptosis has been suggested as an option for resistant patients (Trela et al., 2014). Previously, apoptosis-related gene expression profiles were shown to be associated with primary resistance to imatinib in CML patients (Ferreira et al., 2015). The function and role in resistance of LYRM9 are unknown.
In addition to the positive hits identified in the screening process, we showed that LRP5 conferred resistance to imatinib. Though a role for LRP5 in imatinib resistance was not previously known, the Wnt-β-catenin pathway has been implicated (Yuo et al., 1985; Zhang et al., 2013a).
Advantages and Limitations of Haploid Screening
Recently, we wrote a comprehensive review of functional genomic screening approaches, including yeast, haploid, RNAi, and clustered regularly interspaced short palindrome repeats-associated nuclease (CRISPR)-Cas9, and their relative advantages and disadvantages (Shen et al., 2015). Though the groundbreaking genome-editing technology CRISPR-cas9 can be applied in a variety of cell types to assess toxicity, it has not yet been applied widely for chemical screening (Shen et al., 2015). Human haploid screening has advantages over yeast screening, mainly by enabling the screening of evolutionarily nonconserved genes. It has identified host factors involved in responses to infectious agents (Carette et al., 2009, 2011a; Reiling et al., 2011; Timms et al., 2013; van den Boomen et al., 2014), drug resistance (Chen et al., 2014; Reiling et al., 2013; Winter et al., 2014), and other cellular responses (Dixon et al., 2015; Duncan et al., 2012; Lee et al., 2013). Our findings from this study, and recent data screening KBM7 cells for resistance to chlorpyrifos (Zhu et al., 2015), illustrate the potential for application of haploid screening to environmental chemicals.
As KBM7 cells are of hematopoietic origin, derived from blast-crisis CML (Kotecki et al., 1999), they are of particular relevance to FA and imatinib but some of the findings from the screening may be indicative of general toxicological effects. FA induces nasal pharyngeal carcinoma, neurotoxicity, immunotoxicity, and reproductive toxicity and may increase the risk of asthma/allergies (Tang et al., 2009; Zhang et al., 2009). KBM7 cells have been reprogrammed to induced pluripotent stem cells (iPSCs), with the potential to differentiate into all 3 germ layers (Carette et al., 2010). Use of modified variants of KBM7 cells, therefore, has the potential to extend the applicability of haploid screening to multiple cell types and toxic agents. For example, KBM7-derived iPSCs became resistant to imatinib (Carette et al., 2010) and a derivative KBM7 cell line called HAP1 became susceptible to Ebola virus (Carette et al., 2011b), allowing the identification of additional relevant genes.
Haploid screening of KBM7 cells does have limitations. First, KBM7 cells have a point mutation in TP53, a gene that encodes a major player in DNA damage response. However, apart from driver mutations other than the BCR-ABL translocation and point mutations in TP53 and NOTCH1, KBM7 cells are genetically stable and exhibit limited genetic drift (Burckstummer et al., 2013). A majority of clones (90%) remain haploid after 4–6 weeks in culture, and we have incorporated enrichment steps for haploid cells and true gene-trap mutants in our method to further ensure a stable starting population of cells and reduce potential batch effects. Second, despite our modifications, the haploid screening process described here has technical limitations including resistant colonies lacking gene-trap vector, failure of inverse PCR reactions to generate product, the generation of multiple bands requiring additional separation and purification prior to sequencing, and sequencing products that were too short or uninformative to identify host genes. False negatives appear to be an issue, apparent in our failure to confirm 2 earlier imatinib-resistant mutant genes identified by haploid screening (Carette et al., 2009). Further, LRP5 knockdown was not identified as a resistance factor in the imatinib functional screening but the LRP5 mutant conferred resistance when tested separately. Despite these technical limitations, we believe that the haploid screening methodology holds great promise as a complementary approach to identify toxicant response genes.
Third, the number of human haploid mutant clones available (3396) is limited because of the strong genomic integration bias associated with retroviruses (Burckstummer et al., 2013). Fourth, although qRT-PCR of the transcript associated with the trapped gene in a subset of representative clones in the mutant library showed that near-complete gene inactivation was achieved, not every trapped gene in KBM7 leads to full transcriptional inactivation, as a result of genomic context or alternative transcripts (Burckstummer et al., 2013). Thus, some mutants may be more or less informative, introducing bias during screening. Fifth, KBM7 cells are not completely haploid, having disomy of chromosome 8. However, a truly haploid cell line called eHAP (engineered-HAPloid) has been developed using CRISPR/Cas9-based genome engineering to excise this chromosomal fragment (Essletzbichler et al., 2014). Sixth, KBM7 is a cancer cell line and thus may not fully recapitulate the normal cellular response. However, there is strong representation of more than half of the proteins in 3-quarters of all Kyoto Encyclopedia of Genes and Genomes pathways, including diverse signaling pathways (Burckstummer et al., 2013).
CONCLUSION AND FUTURE DIRECTIONS
We have developed an improved KBM7 haploid screening protocol and identified several gene mutants with enhanced survival in the presence of FA and imatinib, some of which are novel findings. Further validation is necessary. As RNAi has limitations, we will use CRISPR to generate knockout hematopoietic cell lines for several of the FA response genes identified in this study. We will initially examine survival across a range of FA doses to evaluate dose-response. We will examine specific roles for the identified genes by functional validation in knockdown or knockout studies in cell lines in cells in vitro, or in animal models in vivo, using relevant phenotypes. We have conducted multiple validation studies for genes involved in the response to arsenic, benzene, and FA, identified through functional genomic screening (McHale et al., 2014; Zhang et al., 2014). Polymorphisms in candidate susceptibility genes will also be evaluated for their role in susceptibility to hematotoxicity and other toxicity endpoints in a human population occupationally exposed to FA (Zhang et al., 2010), using approaches similar to those applied in studies of benzene (Lan et al., 2004, 2009; Shen et al., 2006, 2011). Identification of susceptible individuals is a priority of twenty-first century risk assessment according to recent findings from the National Academy of Sciences (Mortensen and Euling, 2013).
FUNDING
Supported by National Institute of Environmental Health Science (P42ES004705 to M.T.S.) and (R01ES017452 to L.Z.).
Supplementary Material
ACKNOWLEDGMENTS
The authors are grateful to Dr Daniel P. Blanchard, Department of Molecular and Cellular Biology, University of California, Berkeley, for assistance with cloning shRNA and Western blotting and to Dr Xuefeng Ren, Department of Epidemiology and Environmental Health, University of Buffalo, New York for assistance with initiating this KBM7 study as a postdoctoral trainee in Superfund Research Program at Berkeley.
SUPPLEMENTARY DATA
Supplementary data are available online at http://toxsci.oxfordjournals.org/.
REFERENCES
- Andersen M. E., Clewell H. J., III, Bermudez E., Dodd D. E., Willson G. A., Campbell J. L., Thomas R. S. (2010). Formaldehyde: Integrating dosimetry, cytotoxicity, and genomics to understand dose-dependent transitions for an endogenous compound. Toxicol. Sci. 118, 716–731. [DOI] [PubMed] [Google Scholar]
- Apperley J. F. (2007). Part I: Mechanisms of resistance to imatinib in chronic myeloid leukaemia. Lancet Oncol. 8, 1018–1029. [DOI] [PubMed] [Google Scholar]
- Balabanov S., Braig M., Brummendorf T. H. (2014). Current aspects in resistance against tyrosine kinase inhibitors in chronic myelogenous leukemia. Drug Discov. Today Technol. 11, 89–99. [DOI] [PubMed] [Google Scholar]
- Birsoy K., Wang T., Possemato R., Yilmaz O. H., Koch C. E., Chen W. W., Hutchins A. W., Gultekin Y., Peterson T. R., Carette J. E., et al. (2013). MCT1-mediated transport of a toxic molecule is an effective strategy for targeting glycolytic tumors. Nat. Genet. 45, 104–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjorklund P., Akerstrom G., Westin G. (2007). An LRP5 receptor with internal deletion in hyperparathyroid tumors with implications for deregulated WNT/beta-catenin signaling. PLoS Med. 4, e328.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burckstummer T., Banning C., Hainzl P., Schobesberger R., Kerzendorfer C., Pauler F. M., Chen D., Them N., Schischlik F., Rebsamen M., et al. (2013). A reversible gene trap collection empowers haploid genetics in human cells. Nat. Methods 10, 965–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carette J. E., Guimaraes C. P., Varadarajan M., Park A. S., Wuethrich I., Godarova A., Kotecki M., Cochran B. H., Spooner E., Ploegh H. L., et al. (2009). Haploid genetic screens in human cells identify host factors used by pathogens. Science 326, 1231–1235. [DOI] [PubMed] [Google Scholar]
- Carette J. E., Guimaraes C. P., Wuethrich I., Blomen V. A., Varadarajan M., Sun C., Bell G., Yuan B., Muellner M. K., Nijman S. M., et al. (2011a). Global gene disruption in human cells to assign genes to phenotypes by deep sequencing. Nat. Biotechnol. 29, 542–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carette J. E., Pruszak J., Varadarajan M., Blomen V. A., Gokhale S., Camargo F. D., Wernig M., Jaenisch R., Brummelkamp T. R. (2010). Generation of iPSCs from cultured human malignant cells. Blood 115, 4039–4042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carette J. E., Raaben M., Wong A. C., Herbert A. S., Obernosterer G., Mulherkar N., Kuehne A. I., Kranzusch P. J., Griffin A. M., Ruthel G., et al. (2011b). Ebola virus entry requires the cholesterol transporter Niemann-Pick C1. Nature 477, 340–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W. W., Birsoy K., Mihaylova M. M., Snitkin H., Stasinski I., Yucel B., Bayraktar E. C., Carette J. E., Clish C. B., Brummelkamp T. R., et al. (2014). Inhibition of ATPIF1 ameliorates severe mitochondrial respiratory chain dysfunction in mammalian cells. Cell Rep. 7, 27–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong F., Spencer S., Cote J. F., Wu Y., Tremblay M. L., Lasky L. A., Goff S. P. (2000). Cytoskeletal protein PSTPIP1 directs the PEST-type protein tyrosine phosphatase to the c-Abl kinase to mediate Abl dephosphorylation. Mol. Cell 6, 1413–1423. [DOI] [PubMed] [Google Scholar]
- Costa S., Garcia-Leston J., Coelho M., Coelho P., Costa C., Silva S., Porto B., Laffon B., Teixeira J. P. (2013). Cytogenetic and immunological effects associated with occupational formaldehyde exposure. J. Toxicol. Environ. Health A 76, 217–229. [DOI] [PubMed] [Google Scholar]
- Davis R. S., Li H., Chen C. C., Wang Y. H., Cooper M. D., Burrows P. D. (2002). Definition of an Fc receptor-related gene (FcRX) expressed in human and mouse B cells. Int. Immunol. 14, 1075–1083. [DOI] [PubMed] [Google Scholar]
- de Graaf B., Clore A., McCullough A. K. (2009). Cellular pathways for DNA repair and damage tolerance of formaldehyde-induced DNA-protein crosslinks. DNA Repair 8, 1207–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon S. J., Winter G. E., Musavi L. S., Lee E. D., Snijder B., Rebsamen M., Superti-Furga G., Stockwell B. R. (2015). Human haploid cell genetics reveals roles for lipid metabolism genes in nonapoptotic cell death. ACS Chem. Biol. 10, 1604–1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan L. M., Timms R. T., Zavodszky E., Cano F., Dougan G., Randow F., Lehner P. J. (2012). Fluorescence-based phenotypic selection allows forward genetic screens in haploid human cells. PLoS One 7, e39651.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essletzbichler P., Konopka T., Santoro F., Chen D., Gapp B. V., Kralovics R., Brummelkamp T. R., Nijman S. M., Burckstummer T. (2014). Megabase-scale deletion using CRISPR/Cas9 to generate a fully haploid human cell line. Genome Res. 24, 2059–2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Facchetti F., Cella M., Festa S., Fremont D. H., Colonna M. (2002). An unusual Fc receptor-related protein expressed in human centroblasts. Proc. Natl. Acad. Sci. U.S.A. 99, 3776–3781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira A. F., de Oliveira G. L., Tognon R., Collassanti M. D., Zanichelli M. A., Hamerschlak N., de Souza A. M., Covas D. T., Kashima S., de Castro F. A. (2015). Apoptosis-related gene expression profile in chronic myeloid leukemia patients after imatinib mesylate and dasatinib therapy. Acta Haematol. 133, 354–364. [DOI] [PubMed] [Google Scholar]
- Galvan N., Lim S., Zmugg S., Smith M. T., Zhang L. (2008). Depletion of WRN enhances DNA damage in HeLa cells exposed to the benzene metabolite, hydroquinone. Mutat. Res. 649, 54–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaytan B. D., Vulpe C. D. (2014). Functional toxicology: Tools to advance the future of toxicity testing. Front. Genet. 5, 110.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman J. M., Melo J. V. (2003). Chronic myeloid leukemia–advances in biology and new approaches to treatment. N. Engl. J. Med. 349, 1451–1464. [DOI] [PubMed] [Google Scholar]
- Gramatges M. M., Bertuch A. A. (2013). Short telomeres: From dyskeratosis congenita to sporadic aplastic anemia and malignancy. Transl. Res. 162, 353–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu P., Min J. N., Wang Y., Huang C., Peng T., Chai W., Chang S. (2012). CTC1 deletion results in defective telomere replication, leading to catastrophic telomere loss and stem cell exhaustion. EMBO J. 31, 2309–2321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidel F. H., Arreba-Tutusaus P., Armstrong S. A., Fischer T. (2015). Evolutionarily conserved signaling pathways: Acting in the shadows of acute myelogenous leukemia’s genetic diversity. Clin. Cancer Res. 21, 240–248. [DOI] [PubMed] [Google Scholar]
- Hosgood H. D., III, Zhang L., Tang X., Vermeulen R., Hao Z., Shen M., Qiu C., Ge Y., Hua M., Ji Z., et al. (2013). Occupational exposure to formaldehyde and alterations in lymphocyte subsets. Am. J. Ind. Med. 56, 252–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C., Dai X., Chai W. (2012). Human Stn1 protects telomere integrity by promoting efficient lagging-strand synthesis at telomeres and mediating C-strand fill-in. Cell Res. 22, 1681–1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- IARC. ( 2006). International agency for research on cancer: Formaldehyde, 2-butoxyethanol, and 1-tert-butoxy-2propanol. http://apps.who.int/bookorders/anglais/detart1.jsp?sesslan=1&codlan=1&codcol=72&codcch=88%5Bserial online. Accessed November 1, 2015.
- Jabbour E., Kantarjian H., Cortes J. (2015). Use of second- and third-generation tyrosine kinase inhibitors in the treatment of chronic myeloid leukemia: An evolving treatment paradigm. Clin. Lymphoma Myeloma Leuk. 15, 323–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen H., Wlodarska I., Mecucci C., Hagemeijer A., Vandenberghe P., Marynen P., Cools J. (2006). Fusion of ETV6 to GOT1 in a case with myelodysplastic syndrome and t(10;12)(q24;p13). Haematologica 91, 949–951. [PubMed] [Google Scholar]
- Jia X., Jia Q., Zhang Z., Gao W., Zhang X., Niu Y., Meng T., Feng B., Duan H., Ye M., et al. (2014). Effects of formaldehyde on lymphocyte subsets and cytokines in the peripheral blood of exposed workers. PLoS One 9, e104069.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jo W. J., Loguinov A., Wintz H., Chang M., Smith A. H., Kalman D., Zhang L., Smith M. T., Vulpe C. D. (2009a). Comparative functional genomic analysis identifies distinct and overlapping sets of genes required for resistance to monomethylarsonous acid (MMAIII) and arsenite (AsIII) in yeast. Toxicol. Sci. 111, 424–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jo W. J., Ren X., Chu F., Aleshin M., Wintz H., Burlingame A., Smith M. T., Vulpe C. D., Zhang L. (2009b). Acetylated H4K16 by MYST1 protects UROtsa cells from arsenic toxicity and is decreased following chronic arsenic exposure. Toxicol. Appl. Pharmacol. 241, 294–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones C. H., Pepper C., Baird D. M. (2012). Telomere dysfunction and its role in haematological cancer. Br. J. Haematol. 156, 573–587. [DOI] [PubMed] [Google Scholar]
- Kasbek C., Wang F., Price C. M. (2013). Human TEN1 maintains telomere integrity and functions in genome-wide replication restart. J. Biol. Chem. 288, 30139–30150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy R. D., D’Andrea A. D. (2005). The Fanconi Anemia/BRCA pathway: New faces in the crowd. Genes Dev. 19, 2925–2940. [DOI] [PubMed] [Google Scholar]
- Kotecki M., Reddy P. S., Cochran B. H. (1999). Isolation and characterization of a near-haploid human cell line. Exp. Cell Res. 252, 273–280. [DOI] [PubMed] [Google Scholar]
- Lan Q., Smith M. T., Tang X., Guo W., Vermeulen R., Ji Z., Hu W., Hubbard A. E., Shen M., McHale C. M., et al. (2015). Chromosome-wide aneuploidy study of cultured circulating myeloid progenitor cells from workers occupationally exposed to formaldehyde. Carcinogenesis 36, 160–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan Q., Zhang L., Li G., Vermeulen R., Weinberg R. S., Dosemeci M., Rappaport S. M., Shen M., Alter B. P., Wu Y., et al. (2004). Hematotoxicity in workers exposed to low levels of benzene. Science 306, 1774–1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan Q., Zhang L., Shen M., Jo W. J., Vermeulen R., Li G., Vulpe C., Lim S., Ren X., Rappaport S. M., et al. (2009). Large-scale evaluation of candidate genes identifies associations between DNA repair and genomic maintenance and development of benzene hematotoxicity. Carcinogenesis 30, 50–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C. C., Carette J. E., Brummelkamp T. R., Ploegh H. L. (2013). A reporter screen in a human haploid cell line identifies CYLD as a constitutive inhibitor of NF-kappaB. PLoS One 8, e70339.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lochhead P. A., Gilley R., Cook S. J. (2012). ERK5 and its role in tumour development. Biochem. Soc. Trans. 40, 251–256. [DOI] [PubMed] [Google Scholar]
- Luo B., Cheung H. W., Subramanian A., Sharifnia T., Okamoto M., Yang X., Hinkle G., Boehm J. S., Beroukhim R., Weir B. A., et al. (2008). Highly parallel identification of essential genes in cancer cells. Proc. Natl. Acad. Sci. U.S.A. 105, 20380–20385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maltais L. J., Lovering R. C., Taranin A. V., Colonna M., Ravetch J. V., Dalla-Favera R., Burrows P. D., Cooper M. D., Davis R. S. (2006). New nomenclature for Fc receptor-like molecules. Nat. Immunol. 7, 431–432. [DOI] [PubMed] [Google Scholar]
- McHale C. M., Smith M. T., Zhang L. (2014). Application of toxicogenomic profiling to evaluate effects of benzene and formaldehyde: From yeast to human. Ann. N. Y. Acad. Sci. 1310, 74–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mechetina L. V., Najakshin A. M., Volkova O. Y., Guselnikov S. V., Faizulin R. Z., Alabyev B. Y., Chikaev N. A., Vinogradova M. S., Taranin A. V. (2002). FCRL, a novel member of the leukocyte Fc receptor family possesses unique structural features. Eur. J. Immunol. 32, 87–96. [DOI] [PubMed] [Google Scholar]
- Miyake Y., Nakamura M., Nabetani A., Shimamura S., Tamura M., Yonehara S., Saito M., Ishikawa F. (2009). RPA-like mammalian Ctc1-Stn1-Ten1 complex binds to single-stranded DNA and protects telomeres independently of the Pot1 pathway. Mol. Cell 36, 193–206. [DOI] [PubMed] [Google Scholar]
- Mortensen H. M., Euling S. Y. (2013). Integrating mechanistic and polymorphism data to characterize human genetic susceptibility for environmental chemical risk assessment in the 21st century. Toxicol. Appl. Pharmacol. 271, 395–404. [DOI] [PubMed] [Google Scholar]
- Movassagh M., Foo R. S. (2008). Simplified apoptotic cascades. Heart Fail. Rev. 13, 111–119. [DOI] [PubMed] [Google Scholar]
- North M., Romero C., Loguinov A., Smith M. T., Zhang L., Vulpe C. D. (2011a). Identification of novel biomarkers of formaldehyde toxicity in humans using functional genomics in yeast. The Toxicologist 120, 548. [Google Scholar]
- North M., Tandon V. J., Thomas R., Loguinov A., Gerlovina I., Hubbard A. E., Zhang L., Smith M. T., Vulpe C. D. (2011b). Genome-wide functional profiling reveals genes required for tolerance to benzene metabolites in yeast. PLoS One 6, e24205.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- North M., Vulpe C. D. (2010). Functional toxicogenomics: Mechanism-centered toxicology. Int. J. Mol. Sci. 11, 4796–4813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connor S. T., Lan J., North M., Loguinov A., Zhang L., Smith M. T., Gu A. Z., Vulpe C. (2012). Genome-wide functional and stress response profiling reveals toxic mechanism and genes required for tolerance to benzo[a]pyrene in S. cerevisiae. Front. Genet. 3, 316.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiling J. H., Clish C. B., Carette J. E., Varadarajan M., Brummelkamp T. R., Sabatini D. M. (2011). A haploid genetic screen identifies the major facilitator domain containing 2A (MFSD2A) transporter as a key mediator in the response to tunicamycin. Proc. Natl. Acad. Sci. U.S.A. 108, 11756–11765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiling J. H., Olive A. J., Sanyal S., Carette J. E., Brummelkamp T. R., Ploegh H. L., Starnbach M. N., Sabatini D. M. (2013). A CREB3-ARF4 signalling pathway mediates the response to Golgi stress and susceptibility to pathogens. Nat. Cell Biol. 15, 1473–1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren X., Aleshin M., Jo W. J., Dills R., Kalman D. A., Vulpe C. D., Smith M. T., Zhang L. (2011a). Involvement of N-6 adenine-specific DNA methyltransferase 1 (N6AMT1) in arsenic biomethylation and its role in arsenic-induced toxicity. Environ. Health Perspect. 119, 771–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren X., Ji Z., McHale C. M., Yuh J., Bersonda J., Tang M., Smith M. T., Zhang L. (2013). The impact of FANCD2 deficiency on formaldehyde-induced toxicity in human lymphoblastoid cell lines. Arch. Toxicol. 87, 189–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren X., Lim S., Ji Z., Yuh J., Peng V., Smith M. T., Zhang L. (2011b). Comparison of proliferation and genomic instability responses to WRN silencing in hematopoietic HL60 and TK6 cells. PLoS One 6, e14546.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren X., Lim S., Smith M. T., Zhang L. (2009). Werner syndrome protein, WRN, protects cells from DNA damage induced by the benzene metabolite hydroquinone. Toxicol. Sci. 107, 367–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reshetnikova E. S., Mechetina L. V., Volkova O. Y., Guselnikov S. V., Chikaev N. A., Kovesdi D., Alabyev B., Sarmay G., Burrows P. D., Najakshin A. M., et al. (2012). Differential expression of FCRLA in naive and activated mouse B cells. Cell Immunol. 272, 182–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridpath J. R., Nakamura A., Tano K., Luke A. M., Sonoda E., Arakawa H., Buerstedde J. M., Gillespie D. A., Sale J. E., Yamazoe M., et al. (2007). Cells deficient in the FANC/BRCA pathway are hypersensitive to plasma levels of formaldehyde. Cancer Res. 67, 11117–11122. [DOI] [PubMed] [Google Scholar]
- Santiago T., Kulemzin S. V., Reshetnikova E. S., Chikaev N. A., Volkova O. Y., Mechetina L. V., Zhao M., Davis R. S., Taranin A. V., Najakshin A. M., et al. (2011). FCRLA is a resident endoplasmic reticulum protein that associates with intracellular Igs, IgM, IgG and IgA. Int. Immunol. 23, 43–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sapmaz H. I., Sarsilmaz M., Godekmerdan A., Ogeturk M., Tas U., Kose E. (2015). Effects of formaldehyde inhalation on humoral immunity and protective effect of Nigella sativa oil: An experimental study. Toxicol. Ind. Health. DOI: 10.1177/0748233714566294. [DOI] [PubMed] [Google Scholar]
- Sarova I., Brezinova J., Zemanova Z., Gancarcikova M., Vydra J., Cermak J., Michalova K. (2011). A novel gene LRP5 on 11q13.2 is rearranged in two patients with acute myeloid leukemia. Leuk. Res. 35, e200–e202. [DOI] [PubMed] [Google Scholar]
- Seow W. J., Zhang L., Vermeulen R., Tang X., Hu W., Bassig B. A., Ji Z., Shiels M. S., Kemp T. J., Shen M., et al. (2015). Circulating immune/inflammation markers in Chinese workers occupationally exposed to formaldehyde. Carcinogenesis pii: bgv055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen H., McHale C. M., Smith M. T., Zhang L. (2015). Functional genomic screening approaches in mechanistic toxicology and potential future applications of CRISPR-Cas9. Mutat. Res. Rev. Mutat. Res. 764, 31–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen M., Lan Q., Zhang L., Chanock S., Li G., Vermeulen R., Rappaport S. M., Guo W., Hayes R. B., Linet M., et al. (2006). Polymorphisms in genes involved in DNA double-strand break repair pathway and susceptibility to benzene-induced hematotoxicity. Carcinogenesis 27, 2083–2089. [DOI] [PubMed] [Google Scholar]
- Shen M., Zhang L., Lee K. M., Vermeulen R., Hosgood H. D., Li G., Yin S., Rothman N., Chanock S., Smith M. T., et al. (2011). Polymorphisms in genes involved in innate immunity and susceptibility to benzene-induced hematotoxicity. Exp. Mol. Med. 43, 374–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song M. S., Baker G. B., Dursun S. M., Todd K. G. (2010). The antidepressant phenelzine protects neurons and astrocytes against formaldehyde-induced toxicity. J. Neurochem. 114, 1405–1413. [DOI] [PubMed] [Google Scholar]
- Stewart J. A., Wang F., Chaiken M. F., Kasbek C., Chastain P. D., II, Wright W. E., Price C. M. (2012). Human CST promotes telomere duplex replication and general replication restart after fork stalling. EMBO J. 31, 3537–3549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struski S., Mauvieux L., Gervais C., Helias C., Liu K. L., Lessard M. (2008). ETV6/GOT1 fusion in a case of t(10;12)(q24;p13)-positive myelodysplastic syndrome. Haematologica 93, 467–468. [DOI] [PubMed] [Google Scholar]
- Surovtseva Y. V., Churikov D., Boltz K. A., Song X., Lamb J. C., Warrington R., Leehy K., Heacock M., Price C. M., Shippen D. E. (2009). Conserved telomere maintenance component 1 interacts with STN1 and maintains chromosome ends in higher eukaryotes. Mol. Cell 36, 207–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang X., Bai Y., Duong A., Smith M. T., Li L., Zhang L. (2009). Formaldehyde in China: Production, consumption, exposure levels, and health effects. Environ. Int. 35, 1210–1224. [DOI] [PubMed] [Google Scholar]
- Timms R. T., Duncan L. M., Tchasovnikarova I. A., Antrobus R., Smith D. L., Dougan G., Weekes M. P., Lehner P. J. (2013). Haploid genetic screens identify an essential role for PLP2 in the downregulation of novel plasma membrane targets by viral E3 ubiquitin ligases. PLoS Pathog. 9, e1003772.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trela E., Glowacki S., Blasiak J. (2014). Therapy of chronic myeloid leukemia: Twilight of the imatinib era? ISRN Oncol. 2014, 596483.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tulpule K., Dringen R. (2013). Formaldehyde in brain: An overlooked player in neurodegeneration? J. Neurochem. 127, 7–21. [DOI] [PubMed] [Google Scholar]
- van den Boomen D. J., Timms R. T., Grice G. L., Stagg H. R., Skodt K., Dougan G., Nathan J. A., Lehner P. J. (2014). TMEM129 is a Derlin-1 associated ERAD E3 ligase essential for virus-induced degradation of MHC-I. Proc. Natl. Acad. Sci. U.S.A. 111, 11425–11430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F., Li C., Liu W., Jin Y. (2014a). Modulation of microRNA expression by volatile organic compounds in mouse lung. Environ. Toxicol. 29, 679–689. [DOI] [PubMed] [Google Scholar]
- Wang F., Stewart J., Price C. M. (2014b). Human CST abundance determines recovery from diverse forms of DNA damage and replication stress. Cell Cycle 13, 3488–3498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F., Stewart J. A., Kasbek C., Zhao Y., Wright W. E., Price C. M. (2012). Human CST has independent functions during telomere duplex replication and C-strand fill-in. Cell Rep. 2, 1096–1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson T. J., Gilfillan S., Colonna M. (2010). Fc receptor-like A associates with intracellular IgG and IgM but is dispensable for antigen-specific immune responses. J. Immunol. 185, 2960–2967. [DOI] [PubMed] [Google Scholar]
- Winter G. E., Radic B., Mayor-Ruiz C., Blomen V. A., Trefzer C., Kandasamy R. K., Huber K. V., Gridling M., Chen D., Klampfl T., et al. (2014). The solute carrier SLC35F2 enables YM155-mediated DNA damage toxicity. Nat. Chem. Biol. 10, 768–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winzeler E. A., Shoemaker D. D., Astromoff A., Liang H., Anderson K., Andre B., Bangham R., Benito R., Boeke J. D., Bussey H., et al. (1999). Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 285, 901–906. [DOI] [PubMed] [Google Scholar]
- Yamazoe M., Sonoda E., Hochegger H., Takeda S. (2004). Reverse genetic studies of the DNA damage response in the chicken B lymphocyte line DT40. DNA Repair 3, 1175–1185. [DOI] [PubMed] [Google Scholar]
- Ye X., Ji Z., Wei C., McHale C. M., Ding S., Thomas R., Yang X., Zhang L. (2013). Inhaled formaldehyde induces DNA-protein crosslinks and oxidative stress in bone marrow and other distant organs of exposed mice. Environ. Mol. Mutagen 54, 705–718. [DOI] [PubMed] [Google Scholar]
- Yuo C. Y. Ares M. JrandWeiner A. M. (1985). Sequences required for 3′ end formation of human U2 small nuclear RNA. Cell 42, 193–202. [DOI] [PubMed] [Google Scholar]
- Zhang B., Li M., McDonald T., Holyoake T. L., Moon R. T., Campana D., Shultz L., Bhatia R. (2013a). Microenvironmental protection of CML stem and progenitor cells from tyrosine kinase inhibitors through N-cadherin and Wnt-beta-catenin signaling. Blood 121, 1824–1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., McHale C. M., Greene N., Snyder R. D., Rich I. N., Aardema M. J., Roy S., Pfuhler S., Venkatactahalam S. (2014). Emerging approaches in predictive toxicology. Environ. Mol. Mutagen 55, 679–688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Steinmaus C., Eastmond D. A., Xin X. K., Smith M. T. (2009). Formaldehyde exposure and leukemia: A new meta-analysis and potential mechanisms. Mutat. Res. 681, 150–168. [DOI] [PubMed] [Google Scholar]
- Zhang L., Tang X., Rothman N., Vermeulen R., Ji Z., Shen M., Qiu C., Guo W., Liu S., Reiss B., et al. (2010). Occupational exposure to formaldehyde, hematotoxicity, and leukemia-specific chromosome changes in cultured myeloid progenitor cells. Cancer Epidemiol. Biomarkers Prev. 19, 80–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Liu X., McHale C., Li R., Zhang L., Wu Y., Ye X., Yang X., Ding S. (2013b). Bone marrow injury induced via oxidative stress in mice by inhalation exposure to formaldehyde. PLoS One 8, e74974.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J., Dubois A., Ge Y., Olson J. A., Ren X. (2015). Application of human haploid cell genetic screening model in identifying the genes required for resistance to environmental toxicants: Chlorpyrifos as a case study. J. Pharmacol. Toxicol. Methods 76, 76–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.