Abstract
Oil palm (Elaeis guineensis) is the most productive oil bearing crop worldwide. It has three fruit forms, namely dura (thick-shelled), pisifera (shell-less) and tenera (thin-shelled), which are controlled by the SHELL gene. The fruit forms exhibit monogenic co-dominant inheritance, where tenera is a hybrid obtained by crossing maternal dura and paternal pisifera palms. Commercial palm oil production is based on planting thin-shelled tenera palms, which typically yield 30% more oil than dura palms, while pisifera palms are female-sterile and have little to no palm oil yield. It is clear that tenera hybrids produce more oil than either parent due to single gene heterosis. The unintentional planting of dura or pisifera palms reduces overall yield and impacts land utilization that would otherwise be devoted to more productive tenera palms. Here, we identify three additional novel mutant alleles of the SHELL gene, which encode a type II MADS-box transcription factor, and determine oil yield via control of shell fruit form phenotype in a manner similar to two previously identified mutant SHELL alleles. Assays encompassing all five mutations account for all dura and pisifera palms analyzed. By assaying for these variants in 10,224 mature palms or seedlings, we report the first large scale accurate genotype-based determination of the fruit forms in independent oil palm planting sites and in the nurseries that supply them throughout Malaysia. The measured non-tenera contamination rate (10.9% overall on a weighted average basis) underscores the importance of SHELL genetic testing of seedlings prior to planting in production fields. By eliminating non-tenera contamination, comprehensive SHELL genetic testing can improve sustainability by increasing yield on existing planted lands. In addition, economic modeling demonstrates that SHELL gene testing will confer substantial annual economic gains to the oil palm industry, to Malaysian gross national income and to Malaysian government tax receipts.
Keywords: oil palm, tenera, fruit form, shell, genetic testing
Introduction
The Elaeis genus comprises two interfertile species, E. guineensis of West African origin and E. oleifera of Central and South American origin (Zeven, 1965; Cochard et al., 2005). Due to higher oil yield, E. guineensis is the predominant species in commercial planting sites, and oil palm cultivation accounts for 45% of edible oil production worldwide. Oil palm has three naturally occurring fruit forms (dura, pisifera, and tenera) which vary in oil yield depending on the presence and thickness of the distinct lignified shell surrounding the fruit kernel. Dura fruits have a thick shell (2–8 mm) and produce approximately 5.3 tons of oil per hectare per year. Pisifera fruits have no shell, are typically female sterile, and bunches prematurely rot prior to oil production. Crosses of dura and pisifera palms produce hybrids having tenera fruit with a relatively thin shell (0.5–3 mm) surrounded by a distinct fiber ring and can produce from 7.4 tons (Hartley, 1988) to as high as 13.6 tons of oil per hectare per year, as seen in elite individuals (Sharma and Tan, 1999). Therefore, tenera hybrids produced by selection of maternal (dura) and paternal (pisifera) pools are the basis for commercial palm oil production throughout southeast Asia (Rajanaidu et al., 2000).
The shell fruit form phenotype displays co-dominant monogenic inheritance (Beirnaert and Vanderweyen, 1941). Our previous identification of the SHELL gene and two mutations responsible for tenera and pisifera fruit forms explained the single gene heterosis exhibited in tenera palms (Singh et al., 2013a). The discovery was recently independently verified and converted to a PCR based assay (Ritter et al., 2015). SHELL is a type II MADS-box transcription factor homologous to Arabidopsis SEEDSTICK (STK) and rice OsMADS13, which are members of transcription factor networks controlling differentiation of the ovule, seed and lignified endocarp in Arabidopsis (Favaro et al., 2003; Pinyopich et al., 2003; Dinneny and Yanofsky, 2005) and ovule differentiation and female fertility in rice (Dreni et al., 2007). MADS-box proteins function through heterodimerization with other MADS-box family members, and as predicted by homology, wild-type SHELL heterodimerizes with the rice SEPALLATA MADS-box protein, OsMADS24, in yeast two-hybrid assays (Singh et al., 2013a). The shMPOB mutant allele, identified among descendants of the Nigerian tenera accession T128, is a T-to-C polymorphism that results in a leucine-to-proline amino acid change within the highly conserved MADS-box domain. The shAVROS mutant allele, identified within a multigenerational pedigree spanning five-decades segregating for the Congo-derived AVROS pisifera allele, is an A-to-T polymorphism resulting in a lysine-to-asparagine substitution, which is two amino acids carboxy-terminal to the shMPOB amino acid change (Singh et al., 2013a). Both mutations occur within the α-helical structure, characteristic of all MADS-box domains, which is involved in both heterodimerization and DNA binding. Palms producing thick-shelled dura fruits are homozygous for the wild-type (ShDeliDura) nucleotide at each of the two variant nucleotide positions. Heterozygosity for either the shMPOB or shAVROS alleles (ShDeliDura/shMPOB or ShDeliDura/shAVROS) results in palms producing thin-shelled tenera fruits. Palms homozygous for either mutation or heteroallelic for both mutations (a shAVROS mutation on one chromosome and a shMPOB mutation on the other) produce shell-less pisifera fruits. Cases of heteroallelic pisifera palms indicated that shAVROS and shMPOB alleles do not complement each other, thus confirming the identity of the SHELL gene (Singh et al., 2013a).
The intention of the oil palm industry is to plant only high-yielding tenera hybrids in production fields. However, non-tenera contamination can arise due to several reasons, including unintentional use of pollen from a non-pisifera palm, self-pollination of dura parental palms, open pollination of dura parental palms by surrounding dura palms, and imprecise selection of seeds or seedlings (Corley, 2005). The phenotypic identification of non-tenera contaminant palms is based on the observation of a cross-section of mature fruit. Since fruiting first occurs 3–4 years after field planting, the replacement of contaminants is not economical due to the large size of palms at this stage. With the discovery of SHELL and the mutations responsible for fruit form, DNA-based fruit form prediction is possible in nurseries prior to field planting.
We report the application of SHELL genetic testing in the assessment of non-tenera contamination rates in planting and nursery sites throughout Malaysia. Contamination rates are substantially higher than the <1% theoretically achievable through careful control of pollination (Corley, 2005). DNA sequencing identified three novel mutant alleles of SHELL, each resulting in the tenera fruit form when heterozygous. Like the shAVROS and shMPOB mutations, each mutant allele is a missense mutation resulting in substitution of a conserved amino acid within the SHELL MADS-box domain, indicating that all five SHELL mutations share a common or closely related molecular mechanism impacting oil palm fruit form.
We also constructed a 48-parameter, 4-stage economic model to determine the losses incurred by the inadvertent cultivation of non-tenera palms in Malaysia. The findings suggest that comprehensive DNA-based screening and removal of non-tenera contaminants at the nursery will increase yield and result in substantial economic gains while optimizing use of existing planted area – a positive step toward sustainability.
Materials and Methods
Sampling of Independent Planting Sites and Independent Nurseries
Non-tenera contamination rates were assessed in representative independent planting sites surrounding the Malaysian Palm Oil Board’s (MPOB’s) 6 research stations, located in Peninsular Malaysia (Teluk Intan, Kluang, Keratong, Hulu Paka), Sarawak (Sessang) and Sabah (Lahad Datu). Non-tenera contamination rates were also assessed in three independent nursery sites in each of seven sampling sub-regions encompassing the six geographical locations selected for the independent planters. The seven sampling sub-regions include Perak, Selangor, Negeri Sembilan, Melaka, Johor, and Sarawak. In total, approximately 200 palms from each of 36 independent planting sites (n = 6,272) and from each of 21 nursery sites (n = 3,952) were randomly selected and genotyped by allele-specific PCR.
PCR Based Genetic Testing
In a total of 10,224 samples, genotypes at the shAVROS and shMPOB variant nucleotide positions were determined by SureSawitTM SHELL Kit (Orion Biosains1). Approximately 20 ng of genomic DNA were used as template for each of four independent allele-specific PCR reactions: (i) exon 1 specific primers in which one primer overlapped the shMPOB variant nucleotide with the wild-type base and amplified only the wild-type allele at the variant nucleotide position, (ii) exon 1 specific primers in which one primer overlapped the shMPOB variant nucleotide with the shMPOB base and amplified only the shMPOB allele at the variant nucleotide position, (iii) exon 1 specific primers in which one primer overlapped the shAVROS variant nucleotide with the wild-type base and amplified only the wild type allele at the variant nucleotide position, and (iv) exon 1 specific primers in which one primer overlapped the shAVROS variant nucleotide with the mutant base and amplified only the shAVROS allele at the variant nucleotide position. PCR wells were scored for amplification at end point by SYBR Green emission. Genotypes were scored based on the amplification pattern across the four reactions. Samples that are wild-type at both variant positions (ShDeliDura/ShDeliDura) amplify in reactions (i) and (iii) only. ShDeliDura/shMPOB samples amplify in reactions (i), (ii), and (iii) only. ShDeliDura/shAVROS samples amplify in reactions (i), (iii), and (iv) only. shAVROS/shAVROS samples amplify in reactions (i) and (iv) only. shMPOB/shMPOB samples amplify in reactions (ii) and (iii) only. Finally, heteroallelic shAVROS/shMPOB samples amplify in all four reactions. Primer sequences and PCR conditions are provided in Supplementary Table S1.
DNA Sequencing
A total of 1,132 palm or seedling samples were genotyped by allele-specific PCR as wild-type at both the shAVROS and shMPOB variant nucleotide positions. Although, these palms or seedlings would be predicted to be dura phenotype based on the genotypes at the shAVROS and shMPOB positions alone, they could, in fact, be tenera phenotype due to SHELL mutations independent of the shAVROS and shMPOB mutations. Therefore, exon 1 of SHELL was PCR amplified using flanking intronic primers (Singh et al., 2013a) and Sanger sequenced. Primer sequences were confirmed to be unique in the reference pisifera genome and free of polymorphic nucleotides (Singh et al., 2013b). SHELL exon 1 was amplified from 20 ng of purified genomic DNA under standard PCR amplification conditions (Singh et al., 2013a). Amplicons were treated with exonuclease I and shrimp alkaline phosphatase to remove unincorporated primers and deoxynucleotides. An aliquot of each amplicon was Sanger sequenced (ABI 3730) using an amplicon specific primer. All sequencing data was aligned to the reference pisifera genome sequence, and data was analyzed to determine the genotype at each exon 1 base. SHELL exons 2–7 were similarly Sanger sequenced in a subset of samples. Exons 3–6 were amplified using an intron 2 forward primer and an intron 6 reverse primer. Exons 2 and 7 were separately amplified using intronic primers flanking each exon. All primer sequences and PCR conditions are provided in Supplementary Table S1.
Genotype/Phenotype Comparisons
To determine concordance of fruit form phenotype with SHELL genotype, 512 previously unanalyzed samplings were phenotyped by visual inspection of shell thickness in cross-sectioned ripe fruits and genotyped by Sanger sequencing of SHELL exon 1, as described above. Phenotyping and genotyping were performed by different individuals. Individuals involved in phenotyping were blinded to genotyping calls, and individuals involved in genotyping were blinded to phenotyping calls. Genotypes and phenotypes were compared only after each set of calls was finalized. To address initial genotype/phenotype discordances, 15 discordant palms, as well as 15 palms that were concordant in the initial analysis, were sampled a second time for independent blinded phenotyping and genotyping as described above.
Economic Impact Modeling of Comprehensive SHELL Genetic Testing
To assess the economic impact of SHELL genetic testing on the Malaysian oil palm industry, Malaysian gross national income (GNI) and tax revenues of the Malaysian government, a 48-parameter, four-stage economic model that involves breeders, nurseries, planting sites and mills was constructed (Table 4). In the model, two scenarios were compared – a baseline scenario representing the current state of the independent planters and a SHELL gene testing scenario, which forecasts changes in the production of crude palm oil (CPO), palm kernel oil (PKO), and palm kernel cake (PKC) in the deployment of comprehensive DNA testing and culling at the nursery stage. Contamination rates utilized for economic modeling were based on weighted averages of observed contamination in the surveyed independent planting sites, as these values represent the closest approximation of contamination currently in production fields. Observed contamination rates within each planting region were weighted according to the hectare size of planting area for each region, resulting in the pisifera (2.8%) and dura (8.1%) contamination rates utilized in the baseline scenario. Note that non-weighted contamination rates are reported in Table 1. Assuming that comprehensive SHELL gene testing would enable the molecular based identification and subsequent culling of all contaminant pisifera and dura palms at the nursery stage, we modeled 100% tenera rates in the selected sites in the SHELL gene testing scenario.
Table 4.
Parameters for Economic Impact Model.
| Definition | Value | |||
|---|---|---|---|---|
| Contamination rates: | ||||
| Initial fraction of Tenera | 89.10% | |||
| Initial fraction of Dura | 8.10% | |||
| Initial fraction of Pisifera | 2.80% | |||
| Final fraction of Tenera | 100.00% | |||
| Final fraction of Dura | 0.00% | |||
| Final fraction of Pisifera | 0.00% | |||
| Plantation: | ||||
| Number of trees per hectare | 143 | |||
| Total planted area (in HA) | 5,392,235 | |||
| Fraction cultivated by Ind. smallholders | 15.00% | |||
| Total planted area of Ind. smallholders (in HA) | 808,835 | |||
| Age structure and productivity: | ||||
| Initial fraction of immature palms (age < 4) | 13.00% | |||
| Initial fraction of young palms (age 5–8) | 17.00% | |||
| Initial fraction of prime palms (age 9–18) | 35.00% | |||
| Initial fraction of aging palms (age 19–24) | 20.00% | |||
| Initial fraction of old palms (age 25–30) | 15.00% | |||
| Productivity – young palms (MT FFB/HA/YR)(D&T) | 15 | |||
| Productivity – prime palms (MT FFB/HA/YR)(D&T) | 23 | |||
| Productivity – aging palms (MT FFB/HA/YR)(D&T) | 17 | |||
| Productivity – old palms (MT FFB/HA/YR)(D&T) | 15 | |||
| Definition | Parameter | Value | ||
| Milling: | ||||
| CPO mill extraction efficiency | ECPO | 86.00% | ||
| PKO mill extraction efficiency | EPKO | 97.00% | ||
| Valuation: | ||||
| Initial CPO price (in RM) | PCPO | 2,869 | ||
| Initial PKO price (in RM) | PPKO | 3,781 | ||
| Initial PKC price (in RM) | PPKC | 370 | ||
| CPO price inflation rate | RI | 4.79% | ||
| Bond rate for Malaysia | RB | 3.55% | ||
| MY corporate income tax rate | Tax | 25.00% | ||
| MY econ multiplier (oil palm primary products) | M | 1.511 | ||
| Fruit composition (mass ratios): | ||||
| Tenera: fruit to bunch (Average) | PF/FFB(T)(Average) | 0.649 | ||
| Tenera: fruit to bunch (SD) | PF/FFB(T)(SD) | 0.041 | ||
| Dura: fruit to bunch (Average) | PF/FFB(D)(Average) | 0.612 | ||
| Dura: fruit to bunch (SD) | PF/FFB(D)(SD) | 0.038 | ||
| Tenera: mesocarp to fruit (Average) | Msc/PF(T)(Average) | 0.865 | ||
| Tenera: mesocarp to fruit (SD) | Msc/PF(T)(SD) | 0.027 | ||
| Dura: mesocarp to fruit (Average) | Msc/PF(D)(Average) | 0.612 | ||
| Dura: mesocarp to fruit (SD) | Msc/PF(D)(SD) | 0.026 | ||
| Tenera: shell to fruit (Average) | PKS/PF(T)(Average) | 0.07 | ||
| Tenera: shell to fruit (SD) | PKS/PF(T)(SD) | 0.008 | ||
| Dura: shell to fruit (Average) | PKS/PF(D)(Average) | 0.306 | ||
| Dura: shell to fruit (SD) | PKS/PF(D)(SD) | 0.026 | ||
| Tenera: kernel to fruit (Average) | PK/PF(T)(Average) | 0.065 | ||
| Tenera: kernel to fruit (SD) | PK/PF(T)(SD) | 0.005 | ||
| Dura: kernel to fruit (Average) | PK/PF(D)(Average) | 0.082 | ||
| Dura: kernel to fruit (SD) | PK/PF(D)(SD) | 0.01 | ||
| Tenera: CPO content of mesocarp (Average) | CPO/Msc(T)(Average) | 0.55 | ||
| Dura: CPO content of mesocarp (Average) | CPO/Msc(D)(Average) | 0.55 | ||
| Tenera: PKO content of kernel (Average) | PKO/PK(T)(Average) | 0.47 | ||
| Dura: PKO content of kernel (Average) | PKO/PK(D)(Average) | 0.47 | ||
MT, metric tons; FFB, fresh fruit bunch; HA, hectare; T, tenera; D, dura; CPO, crude palm oil; PKO, palm kernel oil; MY, Malaysia; PF, palm fruit; Msc, mesocarp; PKS, palm kernel shell; PK, palm kernel; PKC, palm kernel cake; RM, Malaysian Ringgit.
Table 1.
Non-tenera contamination by region.
| Regiona | No. sites surveyed | Palms genotyped | tenerab (%) | durac (%) | pisiferad (%) | Unadjusted non-tenera contaminatione (%) |
|---|---|---|---|---|---|---|
| 1 | 6 | 1,132 | 88.9 | 7.4 | 3.7 | 11.1 |
| 2 | 6 | 1,133 | 89.1 | 9.7 | 1.1 | 10.9 |
| 3 | 6 | 882 | 96.4 | 2.8 | 0.8 | 3.6 |
| 4 | 6 | 1,092 | 97.5 | 2.3 | 0.2 | 2.5 |
| 5 | 3 | 539 | 92.8 | 7.1 | 0.2 | 7.2 |
| 6 | 3 | 571 | 79.7 | 20.1 | 0.2 | 20.3 |
| 7 | 3 | 576 | 76.9 | 22.7 | 0.3 | 23.1 |
| 8 | 3 | 571 | 84.2 | 15.6 | 0.2 | 15.8 |
| 9 | 3 | 550 | 95.8 | 3.8 | 0.4 | 4.2 |
| 10 | 6 | 921 | 91.6 | 8.1 | 0.2 | 8.4 |
| 11 | 6 | 1,112 | 81.0 | 11.8 | 7.2 | 19.0 |
| 12 | 3 | 572 | 88.5 | 11.4 | 0.2 | 11.5 |
| 13 | 3 | 573 | 94.1 | 5.6 | 0.3 | 5.9 |
| Total | 57 | 10,224 | 89.3 | 9.2 | 1.5 | 10.7 |
aRegions within Peninsular Malaysia (1–9) and Sabah and Sarawak (10–13) were surveyed.
bTenera genotypes include ShDeliDura/shAVROS, ShDeliDura/shMPOB, ShDeliDura/shMPOB2, ShDeliDura/shMPOB3, or ShDeliDura/shMPOB4.
cDura genotypes are wild-type at all five variant nucleotide positions.
dPisifera genotypes include shAVROS/shAVROS, shMPOB/shMPOB, or shAVROS/shMPOB2.
eNote that national weighted average contamination rate adjusted for the regional planting area is 10.9%.
Monte Carlo simulation was used to mimic variation around four key fruit composition parameters including fruit to bunch, mesocarp to fruit, shell to fruit, and kernel to fruit for tenera and dura palms, where 1,000 values were selected at random from normal distributions of each parameter for each fruit form for each year in the 60-years simulation (Table 4). For pisifera palms in the baseline scenario, we assumed these trees produced no fruit and were a complete loss.
We held planted area managed by independent planters at reported 2015 levels throughout the simulation (0.809 M HA, representing 15% of the 5.39 M HA of total planted area in Malaysia), and assumed an average planting density of 143 palms per hectare. Assumptions surrounding the age structure and productivity of palms in year 0 of the simulation are summarized in Table 4, as are mill extraction efficiencies and other parameters.
The mass of CPO, PKO, and PKC for the baseline and SHELL testing scenarios were computed from the following mass balance equations:
-
(i)
FFB (fresh fruit bunch) = EFB (empty fruit bunch) + PF (palm fruit);
-
(ii)
PF (palm fruit) = Msc (mesocarp) + PK (kernel) + PKS (palm kernel shell);
-
(iii)
Msc (mesocarp) = CPO (crude palm oil) + MF (mesocarp fruit fiber) + POMEMSC (fraction of palm oil mill effluent from mesocarp);
-
(iv)
PK (kernel) = PKO (palm kernel oil) + PKC (palm kernel cake) + WWPK (fraction of palm kernel mill waste water from kernel).
Substituting terms of equation (i) with equations (ii), (iii), and (iv) yields equation (v):
-
(v)
FFB = EFB + CPO + MF + POMEMSC + PKO + PKC + WWPK + PKS.
To determine the selling price (P) of CPO and PKO at a future date (i.e., future month A), a regression of monthly closing prices denominated in Malaysian Ringgit from January 1995 through April 2015 for CPO2 and from January 1996 through April 2015 for PKO3, was computed and the following exponential trend equations and effective inflation rates were determined and used in the model (historic prices for PKO denominated in Malaysian Ringgit were not available in 1995):
PCPO(month A) = 1107.9 × e(0.0039 × B); an effective inflation rate of 4.79%
PPKO(month A) = 1602.5 × e(0.0037 × B); an effective inflation rate of 4.54%
where B equals the number of months from January 1995 to Month A.
A price inflation rate of 4.79% (based on the effective inflation rate of CPO) and a bond rate of 3.55% were used for time value of money corrections, an economic multiplier of 1.511 (as determined for oil palm primary products in Malaysia; Bekhet, 2011) was used to convert industry economic gains into increases in GNI, and a weighted average tax rate of 25% (the standard Malaysian corporate income tax rate since 2009) applied to increases in GNI was used to compute increases in Malaysian government tax receipts. The costs of sampling and culling labor at the nursery, shipping sampled material to a centralized laboratory and the costs of SHELL allele testing were not included in the model.
Results
Non-tenera Contamination Trial Design
Adult palms and seedlings were selected among six planting sites and three nearby independent nurseries within multiple geographical regions throughout Peninsular Malaysia, Sabah and Sarawak (Table 1). The number of sampled palms per region ranged from 539 to 1,133, for a total of 10,224 independent palms. To ensure that the palms sampled from each site were representative, plants were randomly selected from several operational areas within each site. SHELL genotypes for two previously described mutations (shAVROS and shMPOB) were determined by allele-specific PCR assays designed to amplify either the dura or pisifera allele (see Materials and Methods). In addition to shAVROS and shMPOB mutant alleles, Table 1 includes three novel SHELL mutations described below.
Non-tenera Contamination
Palms were classified as genetically non-tenera if the genotype was either homozygous wild type at both variant nucleotide positions and therefore dura fruit form, or homozygous or heteroallelic for shAVROS and/or shMPOB mutant alleles (shAVROS/shAVROS, shMPOB/shMPOB, or shAVROS/shMPOB) and therefore pisifera fruit form. Palms that were homozygous or heteroallelic for any of three novel SHELL gene mutations, described below, were also classified as non-tenera. Palms identified as genetically dura were confirmed by DNA sequencing to have no other SHELL gene mutations that could have caused the palm to be a tenera phenotype palm (see Materials and Methods). The unadjusted non-tenera contamination across all palms sampled was 10.7% [95% confidence interval (CI) 10.1–11.9%; Table 1]. The average contamination rates from each region were then weighted by each region’s respective total oil palm planted area resulting in a national weighted average contamination rate in independent planting sites of 10.9% (2.8% pisifera and 8.1% dura).
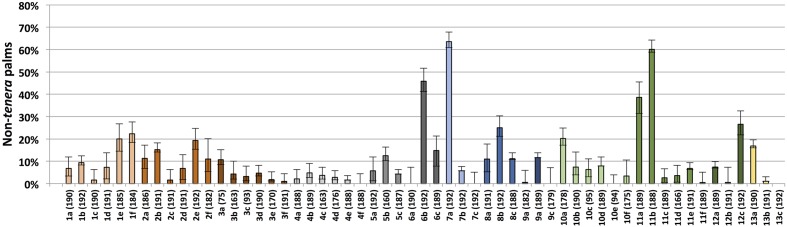
Both dura and pisifera contamination were observed in each region with dura contamination exceeding that of pisifera in each case (Table 1). This finding indicates that contamination is not completely explained by self or open pollination of dura palms by dura pollen (which would yield only dura seeds) or by unintentional pollination of dura palms by tenera pollen (which would yield dura or tenera seeds), as pisifera seeds could only be produced from tenera × pisifera or tenera × tenera crosses. Regional unweighted non-tenera contamination ranged from 2.5% (Region 4) to 23.1% (Region 7). There was significant variation in non-tenera contamination between different sites within a given region, with the exception of the two lowest overall contamination sites (Regions 3 and 4; Figure 1). Furthermore, individual sites with very high contamination rates were identified, for example, site 7a with 63.5% (95% CI 56.5–70.1%) non-tenera palms and site 11b with 60.1% (95% CI 52.9–67.0%) non-tenera palms. Only 6 of 57 sites had no detectable non-tenera palms (Figure 1). Palms of all age groups sampled (from less than 5 years to greater than 10 years after planting) showed non-tenera contamination, indicating that current controlled pollination practices have not overcome the problem.
FIGURE 1.
Non-tenera contamination rates within each individual sampled site. Palms and nursery seedlings genotyped as either wild-type (dura), homozygous mutant for any of the five identified SHELL gene MADS box mutations (pisifera) or heteroallelic for any two of the five mutations (pisifera) were classified as non-tenera. On the x-axis, numbers 1–13 indicate sampled regions according to Table 1. Letters represent each independent site sampled within the indicated region. Numbers in parentheses indicate the number of palms or seedlings genotyped per region/site. Error bars indicate 95% confidence intervals.
Identification of Novel Mutant Alleles of SHELL
We previously identified two independent but closely related mutations within the SHELL MADS-box domain (shAVROS and shMPOB) that each results in the tenera fruit form phenotype when heterozygous and the pisifera fruit form phenotype when homozygous or heteroallelic with each other (Singh et al., 2013a). However, recognizing that additional mutant alleles of SHELL could exist in commercial material, exon 1 of the SHELL gene (encoding the entire MADS-box domain) was sequenced in each of the DNA samples with a wild-type genotype at both the shAVROS and shMPOB nucleotide variant positions (see Materials and Methods).
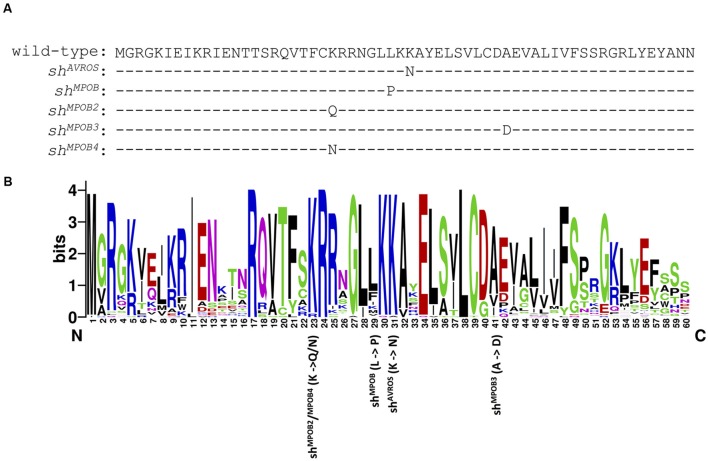
Among 1,132 palms genotyped as homozygous wild-type at the shAVROS and shMPOB nucleotide positions, 235 (20.8%) were heterozygous for one of three novel non-synonymous nucleotide variants within the MADS-box coding region (Table 2; Figure 2A), and no palms were found to be homozygous for these variants. Two variants [shMPOB2 (lysine-to-glutamine) and shMPOB4 (lysine-to-asparagine)] result in the substitution of the same conserved lysine six residues amino-terminal to the shMPOB leucine-to-proline position. The shMPOB3 variant results in an alanine-to-aspartate substitution 10 residues carboxy-terminal to the shAVROS lysine-to-asparagine position (Figure 2). The three novel mutant SHELL alleles varied in frequency. The shMPOB2 variant was detected in 1.4% of the 10,224 palms analyzed (Table 2), and was present in 7 of the 13 geographic regions at rates ranging from 0.1% (Region 1) to 6.4% (Region 10). The shMPOB3 variant was detected in 7 of 13 regions, at a rate ranging from 0.3% (Region 7) to 2.1% (Region 1), representing 0.8% of all palms analyzed. Finally, the shMPOB4 variant was detected in only two palms (in Region 1 and Region 3), representing 0.02% of all palms analyzed. Given the conservation of these residues within the MADS-box domain motif (Figure 2B) and the proximity to the mutations previously shown to determine fruit form phenotype (Singh et al., 2013a), we hypothesized that these variants represent novel mutant alleles of SHELL that also control oil palm shell fruit form. Note that the non-tenera contamination rates described in Table 1 and Figure 1 were calculated based on all five SHELL gene variants. All seven exons of SHELL were sequenced in a panel of 112 palms that were homozygous wild-type at all five variant nucleotide positions. While three synonymous single nucleotide polymorphisms (SNPs) were identified within exon 7, these would not be predicted to impact fruit form as they code for the wild-type amino acid. One non-synonymous SNP resulting in a conservative glutamate-to-aspartate substitution 12 amino acids from the SHELL carboxy terminus was identified.
Table 2.
Novel SHELL alleles.
| Potential dura contaminantsa | shMPOB2 | shMPOB3 | shMPOB4 | |
|---|---|---|---|---|
| Region 1 | 110 | 1 (0.1%)b | 24 (2.1%) | 1 (0.1%) |
| Region 2 | 126 | – | 16 (1.4%) | – |
| Region 3 | 32 | – | 5 (0.6%) | 1 (0.1%) |
| Region 4 | 36 | – | 11 (1.0%) | – |
| Region 5 | 37 | 1 (0.2%) | – | – |
| Region 6 | 114 | 18 (3.2%) | – | – |
| Region 7 | 129 | 12 (2.1%) | 2 (0.3%) | – |
| Region 8 | 89 | 5 (0.9%) | – | – |
| Region 9 | 20 | – | – | – |
| Region 10 | 145 | 59 (6.4%) | 11 (1.2%) | – |
| Region 11 | 199 | 52 (4.7%) | 16 (1.4%) | – |
| Region 12 | 65 | – | – | – |
| Region 13 | 30 | – | – | – |
| Total | 1,132 | 148 (1.4%) | 85 (0.8%) | 2 (0.02%) |
aNumber of palms genotyped as wild-type at both the shAVROS and shMPOB variant nucleotide positions.
bInteger value represents number of palms per site heterozygous for the indicated nucleotide variant, and parentheses denote the percentage of the indicated heterozygous palms among all palms analyzed per site.
FIGURE 2.
SHELL MADS box domain mutations associated with fruit form phenotype. (A) The amino acid sequence of the wild-type (dura) SHELL MADS box domain is shown at the top. Amino acid substitutions encoded by the shAVROS, shMPOB, shMPOB2, shMPOB3, and shMPOB4 mutations are each indicated below the wild-type amino acid sequence. (B) Logo plot of amino acid conservation among 51 independent oil palm MADS box gene predictions within the oil palm reference genome sequence. The positions of substituted amino acids encoded by the five SHELL mutations are indicated below the plot. The shMPOB2 (K-to-Q) and shMPOB4 (K-to-N) mutations alter the same amino acid (position 23 in the Logo plot). The shMPOB, shAVROS, and shMPOB3 mutations alter amino acid positions 29, 31, and 41 in the Logo plot, respectively. Each mutation results in alteration of the most prevalent amino acid to an amino acid not utilized in any oil palm MADS box gene prediction at that position.
To demonstrate that the three novel mutant SHELL alleles conferred the shell fruit form trait, adult palms from populations in which the alleles were detected were sampled and scored for fruit form. In total, 512 sampled palms with mature fruit bunches were visually phenotyped and DNA sequenced (see Materials and Methods). In an initial genotype-by-phenotype comparison, 97.1% of genotypes matched the predicted phenotype, assuming that all five mutant alleles result in tenera phenotype when heterozygous and pisifera phenotype when homozygous. A 2.9% discordance rate is within the accuracy norms of visual phenotyping in plantation settings (Singh et al., 2013a). The 15 discordant palms included nine ShDeliDura/shAVROS palms and one ShDeliDura/shMPOB2 palm phenotyped as dura, two shAVROS/shAVROS palms and one ShDeliDura/ ShDeliDura palm phenotyped as tenera and 2 ShDeliDura/shAVROS palms phenotyped as pisifera. There were no discordances in which a genotypically pisifera palm was phenotyped as dura, or vice versa, as these phenotypes are the most easily discriminated by visual inspection of shell thickness. However, in addition to a low rate of visual phenotyping inaccuracy, discordances could arise due to inaccuracies of genotyping as well as due to potential sample collection and processing errors that may arise, especially under conditions where 100–1000s of samples are collected in field settings. To address this, the 15 discordant palms, as well as 15 palms that were concordant in the initial analysis, were sampled a second time for independent blinded phenotyping and genotyping (see Materials and Methods). In this independent round of sampling, no phenotype or genotype changes occurred among the 15 originally concordant palms, and therefore these remained genotype-by-phenotype concordant. However, among the 15 originally discordant palms, all 15 were either scored as a different phenotype than in the first sampling (six palms) or genotyping of the repeated sampling yielded a different genotype than the first sampling (nine palms), and these changes resolved each of the 15 discordances. These results suggest that the very low rate of discordances was due to a combination of phenotyping and sample processing inaccuracies. First, all genotyping in this genotype-by-phenotype comparison was performed by Sanger DNA sequencing, which has a very low error rate. Second, if genotyping inaccuracies were the cause of discordances, then genotype calls should have changed at a similar rate among the initially concordant and discordant palm sets. However, 60% of initial discordant palms changed genotype upon second sampling compared to no palms changing genotype within the initially concordant set (p < 0.0003, chi squared test). To further support this conclusion, we performed three independent PCR amplification and Sanger sequencing assays on each of the 30 original sampling DNA preps, as well as each of the 30 resampled DNA preps. In every case (180 independent assays), genotype calls were consistent between replicated assays of the same sample and agreed with the original genotype call of the same prep, thus demonstrating the accuracy of genotype calls.
After resolution of rare discordances, the five SHELL MADS box domain mutations accounted for 100% of observed tenera and pisifera phenotypes (Table 3; Figure 3). Among the 422 phenotypically tenera palms, 385 were heterozygous shAVROS/ShDeliDura, two were heterozygous shMPOB/ShDeliDura, two were heterozygous shMPOB2/ShDeliDura, 31 were heterozygous shMPOB3/ShDeliDura, and two were heterozygous shMPOB4/ShDeliDura. All 56 phenotypically dura palms were wild-type at each of the five variant positions. Finally, among 34 phenotypically pisifera palms, 33 were homozygous shAVROS/shAVROS and one was heteroallelic with the shMPOB2 allele on one chromosome and the shAVROS allele on the other. This heteroallelic pisifera palm suggests that shMPOB2 does not complement the shAVROS allele, thus confirming that the shMPOB2 mutation (and, likely, the shMPOB4 mutation, as it involves substitution of the same amino acid) functionally determines oil palm shell fruit form. Furthermore, the 100% concordance between heterozygosity for shMPOB3 and tenera phenotype strongly supports the conclusion that this allele is functionally equivalent to the other four mutant alleles. The finding that all tenera (n = 442) and all pisifera (n = 34) phenotype palms could be explained by heterozygous (tenera) or homozygous/heteroallelic (pisifera) mutations involving the five SHELL MADS box domain mutations indicates that the single conservative missense amino acid substitution detected in exon 7 likely does not impact shell fruit form phenotype. While additional mutant alleles impacting fruit form may be present in other oil palm populations, the data confirm that the three novel SHELL alleles affect fruit form and suggest that these five mutations are responsible for at least the vast majority of tenera and pisifera phenotypes within this wide sampling of planting sites and nurseries.
Table 3.
Concordance of SHELL genotype and fruit form phenotype.
| Observed phenotypesb |
||||
|---|---|---|---|---|
| Predicted phenotypesa | Genotypes | tenera | pisifera | dura |
| tenera | ShDeliDura/shAVROS | 385 | – | – |
| ShDeliDura/shMPOB | 2 | – | – | |
| ShDeliDura/shMPOB2 | 2 | – | – | |
| ShDeliDura/shMPOB3 | 31 | – | – | |
| ShDeliDura/shMPOB4 | 2 | – | – | |
| pisifera | shAVROS/shAVROS | – | 33 | – |
| shAVROS/shMPOB2 | – | 1 | – | |
| dura | ShDeliDura/ShDeliDura | – | - | 56 |
aFruit form phenotypes predicted by the indicated genotype.
bFruit form phenotypes determined by visual inspection.
FIGURE 3.
Fruit form phenotypes. Photographs of three whole fruits (top) and three cross-sectioned fruits (bottom) from a wild-type dura palm (A), and from ShDeliDura/shAVROS (B), ShDeliDura/shMPOB (C), ShDeliDura/shMPOB2 (D), ShDeliDura/shMPOB3 (E), or ShDeliDura/shMPOB4 (F) tenera palms are shown. Arrows indicate the thick shell of dura fruit (A) and the thin shell of tenera fruits from palms with different heterozygous mutant alleles (B–F).
Economic Impact Modeling of Comprehensive SHELL Genetic Testing
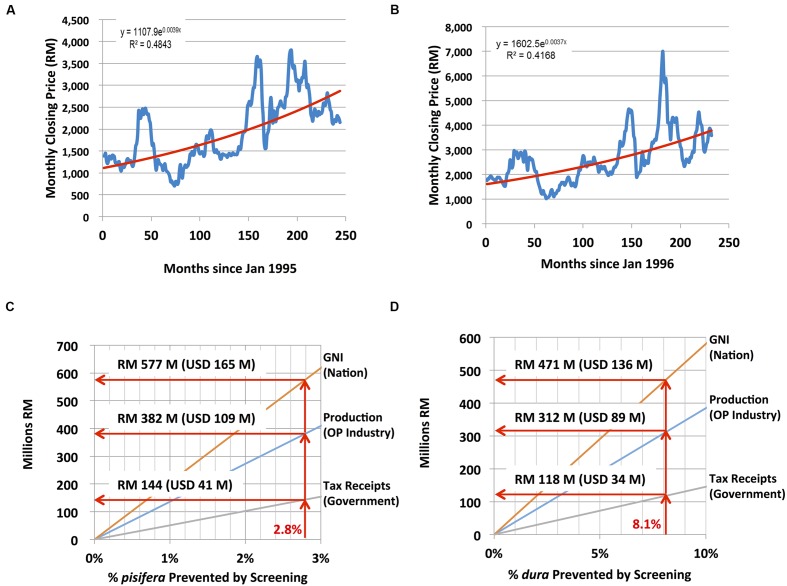
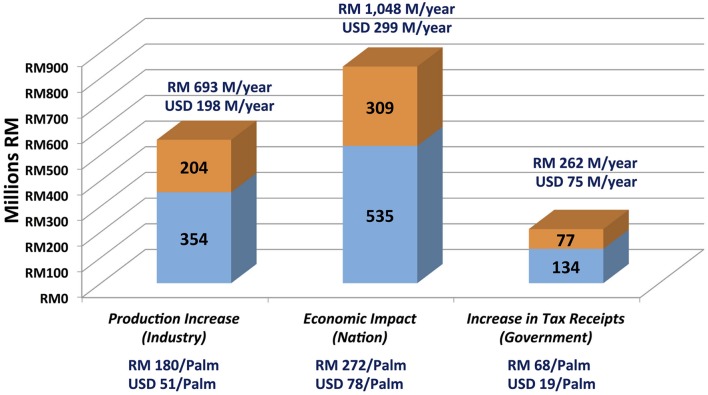
Nursery stage DNA testing of the estimated 3.9 million palms planted at independent planting sites each year would enable nursery operators to cull contaminant palms before they are field planted. To assess the economic impact of SHELL genetic testing in the independent sector, a 48-parameter, four-stage economic model that involves breeders, nurseries, planting sites and mills was constructed (Table 4, see Materials and Methods). The monthly closing prices for CPO and PKO and the fit of the regression curves for CPO and PKO are plotted in Figures 4A,B, respectively, where the regression equations were able to explain 48.43 and 41.68% of the historical pricing data, and regression predictions were used for future CPO and PKO pricing in the model (see Materials and Methods). The economic model determined two curves, which compute the annual gains at steady state excluding sampling, testing and culling costs (i) to the oil palm industry, (ii) to Malaysian GNI, and (iii) to the Malaysian government in the form of increased tax revenues. The gains are determined as a function of the weighted average national independent planting site pisifera (Figure 4C) and dura (Figure 4D) contamination prevented in the SHELL gene screening scenario (see Materials and Methods). The SHELL gene screening scenario would increase GNI, industry income, and government tax income as low yielding contaminant palms are replaced by high yielding tenera palms. At steady state, SHELL gene DNA testing in the independent sector alone would add Malaysian Ringgit (RM) 1.05 billion to Malaysian GNI annually by comprehensive screening (or RM 272 in gains per screened palm - including all tenera and non-tenera palms tested), RM 0.693 BN of increased production annually to oil palm industry members (or RM 180 per screened palm) and RM 0.26 BN of increased tax receipts annually (or RM 68 in new taxes per screened palm; Figure 5). Using the average RM to USD exchange rate over the past 20 years of 3.5, SHELL gene DNA testing would add ∼$300M USD to Malaysian GNI annually (or $77.71 USD per screened palm), $198M USD of increased production annually to oil palm industry members (or $51.42 USD per screened palm), and ∼$75M USD of increased tax receipts annually (or $19.43 USD per screened palm; Figure 5).
FIGURE 4.
Economic impact modeling of comprehensive SHELL genetic testing. To determine selling price of CPO (A) and PKO (B) at a future date, monthly closing prices (in Malaysian Ringgit, RM) are plotted from January 1995 through April 2015 (A) and from January 1996 through April 2015 (historic prices for PKO denominated in Malaysian Ringgit were not available in 1995); (B). A regression of monthly closing prices was computed as described in section “Materials and Methods.” Annual economic impact of preventing pisifera (C) and dura (D) contamination in independent planting sites at steady state. RM to U.S. dollar (USD) conversions are based on an average exchange rate over the past 20 years of 3.5RM/USD.
FIGURE 5.
Annual economic impact of SHELL genetic testing on the oil palm industry, gross national income and the Malaysian government. Calculated annual increases (Malaysian Ringgit, RM) are plotted as proportions obtained by genetic screening based prevention of dura (blue) and pisifera (orange) contamination among independent planting sites. Gains per individual palm screened prior to commercial planting including tenera and non-tenera palms are indicated below each column. RM to U.S. dollar (USD) conversions are based on an average exchange rate over the past 20 years of 3.5RM/USD.
Discussion
Since the publication of the E. guineensis and E. oleifera reference genome sequences in Singh et al. (2013b), several critical milestones toward oil palm industry sustainability have been achieved. These advances include the identifications of the SHELL gene (Singh et al., 2013a), the VIRESCENS gene and mutations responsible for oil palm fruit color (Singh et al., 2014), as well as the MANTLED gene and the epigenetic abnormality responsible for the somaclonal mantling phenotype (Ong-Abdullah et al., 2015). These discoveries have introduced strategies for genetic or epigenetic testing impacting sustainability by ensuring that land devoted to oil palm production is optimally utilized. The present study will enhance the sensitivity of DNA-based screening for non-tenera contamination through the addition of three novel mutant alleles of SHELL to the two previously identified alleles (Singh et al., 2013a). These five mutations, clustering within a highly conserved 19 amino acid span of the MADS box domain, were sufficient to account for all tenera and pisifera phenotype palms within the diverse geography surveyed.
These findings, for the first time, also enabled a direct assessment of non-tenera contamination in independent planting sites and nurseries across Malaysia. Within these populations, contamination rates were substantially higher than the <1% contamination rate theoretically achievable through careful control of pollination (Corley, 2005). Although, contamination rates varied significantly both between surveyed regions (ranging from 2.5 to 30.0%) and between specific sites within those regions, no region was found to be free of contamination. Overall, the measured non-tenera contamination rate across all regions/sites on a weighted average basis was 10.9% (8.1% dura and 2.8% pisifera).
Given directly measured contamination rates among independent planting sites and nurseries, it is possible to model the real-world economic impact of the implementation of comprehensive SHELL genetic testing at the nursery stage in this sector, which would effectively eliminate the inadvertent planting of non-tenera materials. Economic modeling predicts that the comprehensive genetic testing to prevent cultivation of non-tenera palms in the independent sector alone would add ∼RM 1.05 BN ($300M USD) to Malaysian GNI annually, which represents RM 272 ($77.71 USD) in gains per screened palm. Screening would contribute RM 0.693 BN ($198M USD) to oil palm industry members annually representing RM 180 ($51.42 USD) per screened palm. Finally, SHELL gene testing would increase the Malaysian tax revenues annually by RM 0.26 BN (∼$75M USD) or RM 68 ($19.43 USD) for each palm screened. Therefore, SHELL genetic testing would have a major positive economic impact while improving oil palm sustainability by optimizing the utilization of existing planted area.
Although, over 10,000 palms and seedlings derived from multiple sites within each of 13 geographic regions were analyzed in the present study, it is noted that this survey of 36 independent planting sites and 21 nurseries represents only a small sampling of the over 200,000 independent planting sites throughout Malaysia. The findings reveal that non-tenera contamination is likely a concern throughout the entire palm industry, and they demonstrate that comprehensive genetic testing will have major positive environmental and economic impacts. However, substantially larger surveys of contamination, utilizing now available SHELL genetic testing, will be a critical area of future oil palm research.
Author Contributions
E-TL, MA, RN, NL, JO, Ravigadevi Sambanthamurthi, and Rajinder Singh conceptualized the research program. LO, E-TL, MA, RN, MM, WO, AM, NL, JO, Y-MC, Ravigadevi Sambanthamurthi, and Rajinder Singh designed the experiments and coordinated the project. LO, NT, MAB, AVB, MB, ML, Y-MC, and Rajinder Singh conducted laboratory experiments. LO, E-TL, MA, NL, JO, MAB, AVB, NJ, MB, ML, SWS, Ravigadevi Sambanthamurthi, and Rajinder Singh assisted in data analysis. E-TL, JN, K-LC, MH, NA, and SWS performed bioinformatics analysis. LO, AF, AK, and SB coordinated collection of samples. RN, NL, AH-O, AN, HC, and SAS participated in the economic analysis. LO, E-TL, MA, RN, NL, JO, Ravigadevi Sambanthamurthi, and Rajinder Singh prepared and revised the manuscript. All authors approved the final manuscript.
Conflict of Interest Statement
AK and SB are employees of Orion Biosains, a biotechnology company located in Puchong, Selangor, Malaysia that offers SHELL genetic testing to the oil palm industry under a royalty bearing license from the Malaysian Palm Oil Board. Orion Biosains is an affiliate of Orion Genomics, LLC based in St. Louis, MO. NL, JO, and AF are directors of Orion Biosains. NL, JO, AF, MAB, AVB, MB, ML, NJ, SWS, and CB are employees and stock holders of Orion Genomics, LLC. LO, E-TL, MA, RN, NT, JN, MM, K-LC, MH, NA, WO, AM, Y-MC, Ravigadevi Sambanthamurthi, and Rajinder Singh are employees of the Malaysian Palm Oil Board. LO, E-TL, MA, RN, SWS, NL, JO, AVB, MAB, Ravigadevi Sambanthamurthi, and Rajinder Singh are co-inventors on pending patents covering SHELL genetic testing.
Acknowledgments
The authors would like to thank MPOB for permission to publish this manuscript. We would also like to thank the staff of MPOB’s Breeding and Tissue Culture Unit at the various research stations for their assistance in the sampling process. We also extend our appreciation to the Integration Research & Extension Division as well as the Licensing & Enforcement Division, especially Ms. Nur Hanani Mansor, Mr. Moktarudin Hj. Ahmad, and Mr. Azhar Ahmad for their assistance in conducting this study. We thank Rob Martienssen for critical advice and guidance and for helpful comments on preparation of the manuscript.
Funding. This work was funded by the Malaysian Palm Oil Board and Orion Genomics, LLC.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2016.00771
References
- Beirnaert A., Vanderweyen R. (1941). Contribution a l’etude genetique et biometrique des varieties d’Elaeis guineensis Jacquin. Publ. Inst. Nat. Etude agron. Congo Belge Ser. Sci. 27 1–101. [Google Scholar]
- Bekhet H. A. (2011). Output, income and employment multipliers in Malaysian Economy: input-output approach. Int. Bus. Res. 4 208–223. [Google Scholar]
- Cochard B., Amblard P., Durand-Gasselin T. (2005). Oil palm genetic improvement and sustainable development. Oleagineux Corps Gras Lipides 12 141–147. 10.1051/ocl.2005.0141 [DOI] [Google Scholar]
- Corley R. H. V. (2005). Illegitimacy in oil palm breeding – A review. J. Oil Palm Res. 17 64–69. [Google Scholar]
- Dinneny J. R., Yanofsky M. F. (2005). Drawing lines and borders: how the dehiscent fruit of Arabidopsis is patterned. Bioessays 27 42–49. 10.1002/bies.20165 [DOI] [PubMed] [Google Scholar]
- Dreni L., Jacchia S., Fornara F., Fornari M., Ouwerkerk P. B., An G., et al. (2007). The D-lineage MADS-box gene OsMADS13 controls ovule identity in rice. Plant J. 52 690–699. 10.1111/j.1365-313X.2007.03272.x [DOI] [PubMed] [Google Scholar]
- Favaro R., Pinyopich A., Battaglia R., Kooiker M., Borghi L., Ditta G., et al. (2003). MADS-box protein complexes control carpel and ovule development in Arabidopsis. Plant Cell 15 2603–2611. 10.1105/tpc.015123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartley C. (1988). The Oil Palm. London: Longman. [Google Scholar]
- Ong-Abdullah M., Ordway J. M., Jiang N., Ooi S. E., Kok S. Y., Sarpan N., et al. (2015). Loss of Karma transposon methylation underlies the mantled somaclonal variant of oil palm. Nature 525 533–537. 10.1038/nature15365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinyopich A., Ditta G. S., Savidge B., Liljegren S. J., Baumann E., Wisman E., et al. (2003). Assessing the redundancy of MADS-box genes during carpel and ovule development. Nature 424 85–88. 10.1038/nature01741 [DOI] [PubMed] [Google Scholar]
- Rajanaidu N., Kushairi A., Rafii M., Mohd Din A., Maizura I., Jalani B. S. (2000). Advances In Oil Palm Research. Bangi, Selangor: Malaysian Palm Oil Board. [Google Scholar]
- Ritter E., Lopez de Armentia E., Erika P., Herrero J., Niggrum Y. P., Santika B., et al. (2015). Development of a molecular marker system to distinguish shell thinkness in oil palm genotypes. Euphytica 207 367–376. 10.1007/s10681-015-1553-6 [DOI] [Google Scholar]
- Sharma M., Tan Y. P. (1999). “Oil palm breeding programmes and the performance of DxP planting materials at United Plantations Berhad,” in Proceedings of Seminar on Sourcing of Oil Palm Planting Materials for Local and Overseas Joint Venture, (Malaysia: Palm Oil Research Institute; ), 118–135. [Google Scholar]
- Singh R., Low E. T., Ooi L. C., Ong-Abdullah M., Nookiah R., Ting N. C., et al. (2014). The oil palm VIRESCENS gene controls fruit colour and encodes a R2R3-MYB. Nat. Commun. 5 4106 10.1038/ncomms5106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R., Low E. T., Ooi L. C., Ong-Abdullah M., Ting N. C., Nagappan J., et al. (2013a). The oil palm SHELL gene controls oil yield and encodes a homologue of SEEDSTICK. Nature 500 340–344. 10.1038/nature12356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R., Ong-Abdullah M., Low E. T., Manaf M. A., Rosli R., Nookiah R., et al. (2013b). Oil palm genome sequence reveals divergence of interfertile species in Old and New worlds. Nature 500 335–339. 10.1038/nature12309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeven A. C. (1965). The origin of the oil palm. J. Niger. Inst. Oil Palm Res. 4 218–225. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.