Abstract
Alanine, serine, cysteine-preferring transporter 2 (ASCT2; SLC1A5) mediates uptake of glutamine, a conditionally essential amino acid in rapidly proliferating tumour cells. Uptake of glutamine and subsequent glutaminolysis is critical for activation of the mTORC1 nutrient-sensing pathway, which regulates cell growth and protein translation in cancer cells. This is of particular interest in breast cancer, as glutamine dependence is increased in high-risk breast cancer subtypes. Pharmacological inhibitors of ASCT2-mediated transport significantly reduced glutamine uptake in human breast cancer cell lines, leading to the suppression of mTORC1 signalling, cell growth and cell cycle progression. Notably, these effects were subtype-dependent, with ASCT2 transport critical only for triple-negative (TN) basal-like breast cancer cell growth compared with minimal effects in luminal breast cancer cells. Both stable and inducible shRNA-mediated ASCT2 knockdown confirmed that inhibiting ASCT2 function was sufficient to prevent cellular proliferation and induce rapid cell death in TN basal-like breast cancer cells, but not in luminal cells. Using a bioluminescent orthotopic xenograft mouse model, ASCT2 expression was then shown to be necessary for both successful engraftment and growth of HCC1806 TN breast cancer cells in vivo. Lower tumoral expression of ASCT2 conferred a significant survival advantage in xenografted mice. These responses remained intact in primary breast cancers, where gene expression analysis showed high expression of ASCT2 and glutamine metabolism-related genes, including GLUL and GLS, in a cohort of 90 TN breast cancer patients, as well as correlations with the transcriptional regulators, MYC and ATF4. This study provides preclinical evidence for the feasibility of novel therapies exploiting ASCT2 transporter activity in breast cancer, particularly in the high-risk basal-like subgroup of TN breast cancer where there is not only high expression of ASCT2, but also a marked reliance on its activity for sustained cellular proliferation.
Introduction
Despite being a highly heterogenous disease, the breast cancer therapeutic strategy is dictated largely by tumoral expression of only three cell surface markers: oestrogen receptor (ER), progesterone receptor and human epidermal growth factor receptor 2 (HER2). Tumours that lack clinically detectable expression of all three receptors are classified as triple-negative (TN) and, in the absence of any targeted therapies, rely largely on surgery, radiotherapy and cytotoxic chemotherapy for their management.1 TN breast cancers generally display distinct features on gene expression analysis, with the PAM50 classification2, 3 and subsequent molecular profiling4 showing enrichment of genes associated with proliferation, hypoxia and stem-like features. As a result, TN breast cancers have poorer prognosis overall,5, 6, 7 and are usually associated with increased recurrence and death compared with other subtypes.8 Attempts to stratify this generally poor-prognosis subtype into clinically useful subgroups have revealed at least seven different molecular subtypes of TN breast cancer,9 highlighting the need for novel and effective targeted therapies for these patients. Because glutamine dependence is increased in proliferative subtypes of breast cancer,10, 11 we hypothesized that highly proliferative TN tumours may be uniquely sensitive to inhibitors of glutamine uptake.
Alanine, serine, cysteine-preferring transporter 2 (ASCT2; SLC1A5) is a cell surface solute-carrying transporter that mediates uptake of neutral amino acids including glutamine.12, 13 Although a non-essential amino acid in normal cells, the demand for glutamine is dramatically increased throughout malignant transformation to support increased metabolic demands; namely, provision of catabolic substrates for ATP production and anabolic substrates for macromolecule biosynthesis.14 The intracellular glutamine pool is also critical for sustained activation of mTORC1 signalling,15 a master regulator of cell growth and protein translation,16 as well as prevention of apoptosis (reviewed in ref. 17). Blocking ASCT2 to prevent glutamine uptake has been shown to successfully prevent tumour cell proliferation in melanoma,18 non-small cell lung cancer,19, 20 prostate cancer 21 and acute myeloid leukaemia.22
In this study, we aimed to investigate the role of ASCT2 in breast cancer using a combination of in vitro and in vivo approaches combined with gene expression analysis of clinical TN breast cancer patient samples. We show that although ASCT2 is highly expressed in most breast cancer subtypes, only basal-like TN breast cancer cells require ASCT2-mediated uptake of glutamine to sustain mTORC1 signalling, cell growth and cell cycle progression. Targeted knockdown of ASCT2 showed that loss of ASCT2 alone was sufficient to cause rapid cell death in vitro and reduce engraftment and subsequent growth of xenografted cells in vivo. These effects were accompanied by significantly altered gene expression, which suggested dynamic regulation of ASCT2 and other glutamine metabolism-related genes (GLS, GLUL) by a coordinated MYC- and ATF4-driven transcriptional programme in TN breast cancer. Absolute mRNA measurements of these genes in TN breast cancer samples showed that these genes were highly expressed across all samples and were significantly correlated in a subgroup of patients. Our data provide the first conclusive evidence that ASCT2 represents a novel therapeutic axis in a challenging subset of breast cancers.
Results and Discussion
Basal-like TN breast cancer cell growth is dependent on ASCT2 activity
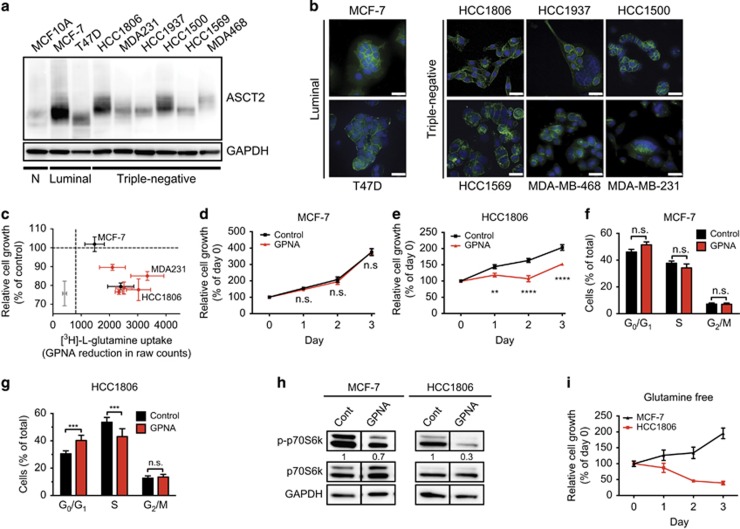
We examined ASCT2 protein expression and cell surface localization in a normal basal mammary cell line (MCF10A) and a variety of breast cancer cell lines representing the luminal (ER+, HER2− MCF-7 and T47D), basal-like (triple-negative, TN; HCC1806, HCC1937, HCC1500, HCC1569, MDA-MB-468/MDA468) and claudin-low (CL; MDA-MB-231/MDA231) molecular subtypes of breast cancer (Figures 1a and b). ASCT2 protein was expressed and present on the plasma membrane for all the cell lines (shown in green, Figure 1b), although mainly cytoplasmic staining was seen in the claudin-low4 glutamine-sensitive11 cell line, MDA-MB-231 (Figure 1b). To determine which cells require ASCT2 function, we examined radiolabelled glutamine uptake in the presence of the ASCT2 inhibitor l-γ-glutamyl-p-nitroanilide (GPNA23). GPNA significantly decreased ASCT2 function in normal (grey), luminal (black) and basal-like (red) breast cell lines, but showed variable efficacy in inhibiting cell growth (Figure 1c and Supplementary Figures 1A–E). As previously reported, basal-like cells, but not luminal cells, were uniquely sensitive to glutamine availability,24, 25 and had significantly lower growth rates when treated with GPNA (Supplementary Figures 1B–E) despite maintaining relatively high levels of glutamine transport (Supplementary Figure 1A). Both MCF-7 and T47D are believed to be glutamine independent owing to increased levels of glutamine synthetase expression, which allows them to synthesize glutamine from glutamate.26 Interestingly, although we confirmed that luminal MCF-7 cells were insensitive to ASCT2 inhibition and subsequent glutamine deprivation, T47D cells were not. This suggests that other amino acids transported by ASCT2 may be important for T47D proliferation. We therefore chose two cell lines with distinct clinical backgrounds and biological behaviours for further analysis: MCF-7 (luminal) and HCC1806 (basal-like TN). These two cell lines exhibited high cell surface ASCT2 expression (Figures 1a and b) but differential responses to GPNA.
Figure 1.
Inhibiting glutamine uptake represses cell growth in HCC1806 basal-like breast cancer cells but not luminal MCF-7 cells. (a) ASCT2 protein (60–75 kDa; Cell Signaling, Beverly, MA, USA; Cat. No. 8057 S) expression was measured by western blotting in normal mammary cells (MCF10A), two luminal (MCF-7, T47D) and six triple-negative breast cancer cell lines including basal-like (HCC1806, HCC1937, MDA-MB-468/MDA468) and claudin-low (CL; MDA-MB-231/MDA231) subsets, with GAPDH (37 kDa; Abcam, Cambridge, UK; Cat. No. ab8245) used as loading control. MCF-7 and MCF10A cells were a kind gift from the Vascular Biology Laboratory, Centenary Institute, Sydney, New South Wales, Australia, and identity confirmed using STR testing. T47D, HCC1806, HCC1937, MDA231 and MDA468 cells were obtained from ATCC, Manassas, VA, USA. HCC1500 and HCC1569 cells were a kind gift from the Tumour Progression Laboratory, Garvan Institute of Medical Research, Sydney, New South Wales, Australia. All cell lines were routinely confirmed to be mycoplasma-free using PCR testing. (b) ASCT2 localization was determined in breast cancer cell lines by immunofluorescence staining with primary antibody as in a, goat anti-rabbit AlexaFluor488 (Life Technologies, Carlsbad, CA, USA; Cat. No. A11034; green) as secondary antibody, and nuclei visualized using 4',6-diamidino-2-phenylindole (DAPI; blue). Scale bar, 30 μm. (c) Normal mammary cells (MCF10A; grey), luminal breast cancer cell lines (MCF-7, T47D; black), and triple-negative breast cancer cell lines (HCC1806, HCC1937, HCC1500, HCC1569, MDA231; red) were treated with l-γ-glutamyl-p-nitroanilide (GPNA; 1 mM; Sigma-Aldrich, St Louis, MO, USA) along with 0.3 mCi [3H]-l-glutamine (PerkinElmer, Waltham, MA, USA) for 15 min to assess uptake as previously described.21 Uptake data are presented as reduction in raw scintillation counts by GPNA compared with control, whereas MTT data are expressed as % inhibition compared with control. MCF-7 (d) and HCC1806 (e) cells were cultured in 96-well plates (MCF-7: 3 × 103 cells/well; HCC1806: 1 × 103 cells/well) for 3 days with or without 1 mM GPNA. MTT assays were conducted every 24 h to assess cell growth according to manufacturer's instructions (MTT Cell Growth Assay Kit; Merck-Millipore, Billerica, MA, USA). Cell cycle progression for MCF-7 (f) and HCC1806 (g) cells was analysed using APC-BrdU Flow Kit according to manufacturer's instructions (BD Pharmingen, San Jose, CA, USA) after 24 h culture with or without 1 mM GPNA. (h) MCF-7 and HCC1806 cells were incubated in GPNA or control media for 6 h. Total and phosphorylated (p-) p70S6K signalling protein (70, 85 kDa; both from Cell Signaling, Beverly, MA, USA; Cat. No. 9205S/9202S) was then detected by western blotting, with GAPDH (37 kDa) as loading control. (i) MCF-7 and HCC1806 cells were cultured in 96-well plates (MCF-7: 3 × 103 cells/well; HCC1806: 1 × 103 cells/well) in glutamine-free media (containing 10% dialysed FBS) for 3 days with MTT assays conducted every 24 h to assess cell growth according to manufacturer's instructions (MTT Cell Growth Assay Kit; Merck-Millipore, Billerica, MA, USA). Asterisks denote P-values as follows: *P⩽0.05; **P⩽0.01; ***P⩽0.001; ****P⩽0.0001, NS, not significant; two-way analysis of variance. Data in c–g and i represent mean±s.e.m., n⩾3 experiments. Data in a, b and h are representative of three independent experiments, with blots in H cropped to exclude extraneous lanes. Full blots are in Supplementary Figure 1.
We first confirmed that despite significant inhibition of glutamine uptake in both cell lines, GPNA treatment repressed cell growth (Figures 1d and e) and cell cycle progression (Figures 1f and g) in HCC1806 cells only. We next showed that inhibition of ASCT2-mediated glutamine uptake reduced the mTORC1 signalling axis in HCC1806 cells (Figure 1h; Supplementary Figure 1F). As mTORC1 inhibition mediates G1 blockade,26, 27 this is a likely mechanism of action, as previously published.15, 28 As ASCT2 inhibition has also been shown to activate mTORC2 activity upstream of apoptotic signalling,29 downstream phosphorylation levels of mTORC2 substrate, Akt (p-Ser473) were measured after 6 h GPNA treatment and showed a modest increase in HCC1806 cells (Supplementary Figure 1G). This change was reflected in a significant increase in the number of apoptotic cells in HCC1806 cells, but no change in viability in MCF-7 cells (Supplementary Figures 1H and I). Thus, in contrast to the basal-like cell line, the luminal cell line was largely insensitive to glutamine deprivation alone. We confirmed that MCF-7 cells were able to sustain proliferation in the absence of exogenous glutamine, compared with the dramatic loss of cell viability in the basal-like HCC1806 cells (Figure 1i). Together, these data indicated a differential reliance on ASCT2 activity across breast cancer cell lines, whereby only basal-like cells required ASCT2-mediated glutamine uptake for cell growth.
ASCT2 expression is required for basal-like breast cancer cell growth in vitro
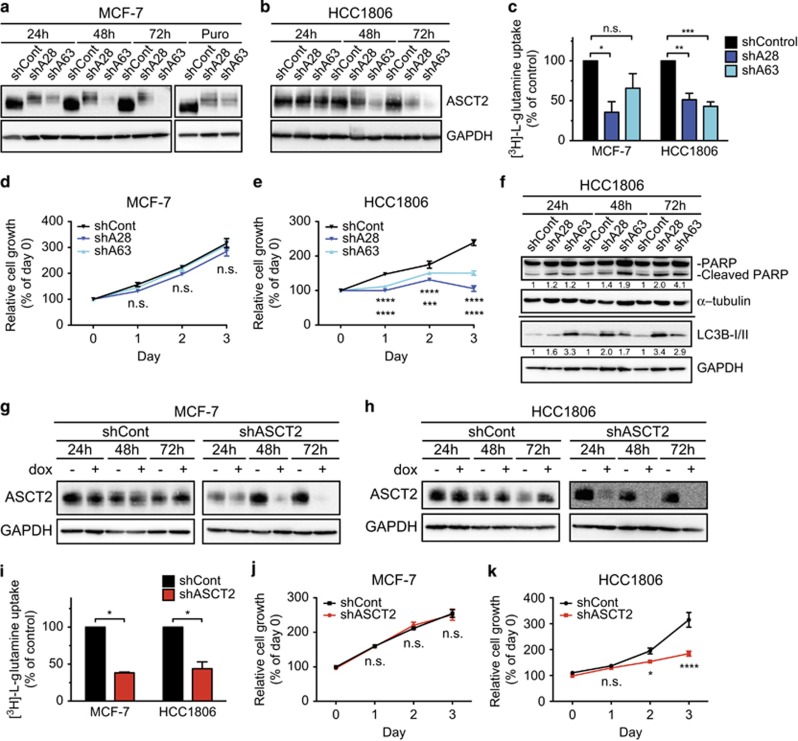
We next performed targeted knockdown of ASCT2 to determine whether ASCT2 was directly responsible for the observed glutamine-dependent effects on basal-like breast cancer cell growth. This was achieved by lentiviral transduction of a control shRNA (shCont; plant miRNA ath-mir159a, sequence and specificity detailed previously30), or one of two different shRNAs against ASCT2 (shA28, sequence in Figure 2 legend; or shA63 (ref. 21)). Protein knockdown was confirmed in MCF-7 and HCC1806 cells (Figures 2a and b) by western blotting. Glutamine uptake was reduced in both MCF-7 and HCC1806 cells transduced with shA28 and shA63, as compared with cells transduced with shCont (Figure 2c). ASCT2 knockdown had no effect on MCF-7 cell growth (Figure 2d), whereas expression of either shRNA against ASCT2 significantly reduced HCC1806 cell growth in the 72 h following transduction (Figure 2e), and caused an increase in cleaved PARP protein levels and LC3B-II accumulation (Figure 2f), as well as significantly increased levels of cleaved-caspase 3 as detected by immunofluorescence microscopy (Supplementary Figures 2A and B). Furthermore, the analysis of CyQUANT/PI staining showed a significant decrease in live cell numbers coupled with a significant increase in dead cells (PI+) after 72 h (Supplementary Figures 2C–E).
Figure 2.
ASCT2 expression is required for HCC1806 cell growth. MCF-7 (a) and HCC1806 (b) cells were transduced with a lentiviral vector (pLKO.1) containing control shRNA (shCont; shRNA sequence and specificity detailed previously21) or one of two shRNAs against ASCT2 (shA28: 5′CCGGGCTGCTTATCCGCTTCTTCAACTCGAGTTGAAGAAGCGGATAAGCAGCTTTTTG-3′ or shA63: 5′-CCGGCTGGATTATGAGGAATGGATACTCGAGTATCCATTCCTCATAATCCAGTTTTTG-3′ both from Sigma-Aldrich, St Louis, MO, USA). ASCT2 expression (60–75 kDa) was measured by western blotting at 24, 48 and 72 h post transduction and after long-term puromycin (Puro) selection (a only). (c), MCF-7 cells (Puro) and HCC1806 cells (72 h) were incubated with [3H]-l-glutamine for 15 min as previously described21 to assess uptake after ASCT2 knockdown. Transduced MCF-7 (d) and HCC1806 (e) cells were cultured for 3 days post selection or post transduction, respectively. MTT assays were conducted every 24 h to assess the cell growth according to manufacturer's instructions (MTT Cell Growth Assay Kit; Merck-Millipore, Billerica, MA, USA). (f), Levels of full-length and cleaved poly ADP ribose polymerase (PARP) protein (116 and 89 kDa; Cell Signaling, Beverly, MA, USA; Cat. No. 9541 S) and LC3B-I/II protein forms (14, 16 kDa; Cell Signaling, Beverly, MA, USA; Cat. No. 2775 S) with GAPDH as loading control (37 kDa) were measured in HCC1806 cells by western blotting at 24, 48 and 72 h post transduction. MCF-7 (g) and HCC1806 (h) cells were transduced with lentiviral vector (pFH1tUTG) that encoded doxycycline (dox)-inducible expression of the control shRNA (shCont; sequence and specificity detailed previously21) or the shA63 shRNA (shASCT2; shRNA sequence as above) also encoding an eGFP reporter tag.34 Transduced cells were sorted for GFP-positive cells using fluorescence-activated cell sorting and then shRNA expression was induced by treatment with 1 μg/ml dox. ASCT2 expression (60–75 kDa) was measured by western blotting at 24, 48 and 72 h ±dox with GAPDH as loading control (37 kDa), (i), Transduced MCF-7 and HCC1806 cells were incubated with 0.3 mCi [3H]-l-glutamine to assess uptake after 72 h dox treatment, as previously described.21 Transduced MCF-7 (j) and HCC1806 (k) cells were cultured with dox for 3 days and MTT assays were conducted every 24 h to assess the cell growth according to manufacturer's instructions (MTT Cell Growth Assay Kit; Merck-Millipore, Billerica, MA, USA). Asterisks denote P-values as follows: *P⩽0.05; **P⩽0.01; ***P⩽0.001; ****P⩽0.0001, NS, not significant; two-way analysis of variance. Data in c–e, i–k represent mean±s.e.m., n⩾3 experiments. Data in a, b, f–h are representative of three independent experiments. For MTT assays (e), stars under growth curves refer to shCont vs shA28 (top) and shCont vs shA63 (bottom).
As ASCT2 activity and glutamine availability affects several intracellular pathways, including mTORC1 lysosomal translocation,31, 32 caspase activation, PARP cleavage33 and autophagy,22, 28 these are the likely mechanisms of action for the observed growth inhibition and induction of apoptosis. Furthermore, in addition to glutamine, ASCT2 transports other amino acids including alanine, serine, cysteine, threonine and asparagine.13 It is possible that depletion of these amino acids also has an important role in the control of cell growth and apoptosis in TNBC.
Stable antibiotic selection of shASCT2-expressing cells was only successful in MCF-7 cells as HCC1806 cells died rapidly after transduction, further suggesting induction of apoptosis and confounding the results for uptake and cell growth assays. We therefore generated an inducible ASCT2 shRNA (shA63) using a doxycycline-inducible lentiviral vector.34 ASCT2 knockdown was confirmed in MCF-7 (Figure 2g) and HCC1806 cells (Figure 2h) in the presence of doxycycline at 24, 48 and 72 h. A significant reduction in glutamine uptake was observed in both MCF-7 and HCC1806 shASCT2 cells as compared with shCont after 72 h doxycycline treatment (Figure 2i); however, MCF-7 cell growth was again unaffected by ASCT2 knockdown (Figure 2j) whereas HCC1806 cells showed a significant reduction in proliferation when cultured in doxycycline (Figure 2k). As MTT assay results may be confounded by changes in cellular metabolism, we confirmed the significant inhibitory effect on cell growth by analysing uptake of CyQUANT live cell stain (Supplementary Figure 3A). These are the first data to conclusively show that ASCT2 loss is sufficient to significantly reduce basal-like breast cancer cell growth in vitro.
ASCT2 knockdown in vivo represses basal-like tumour growth and improves xenografted mouse survival
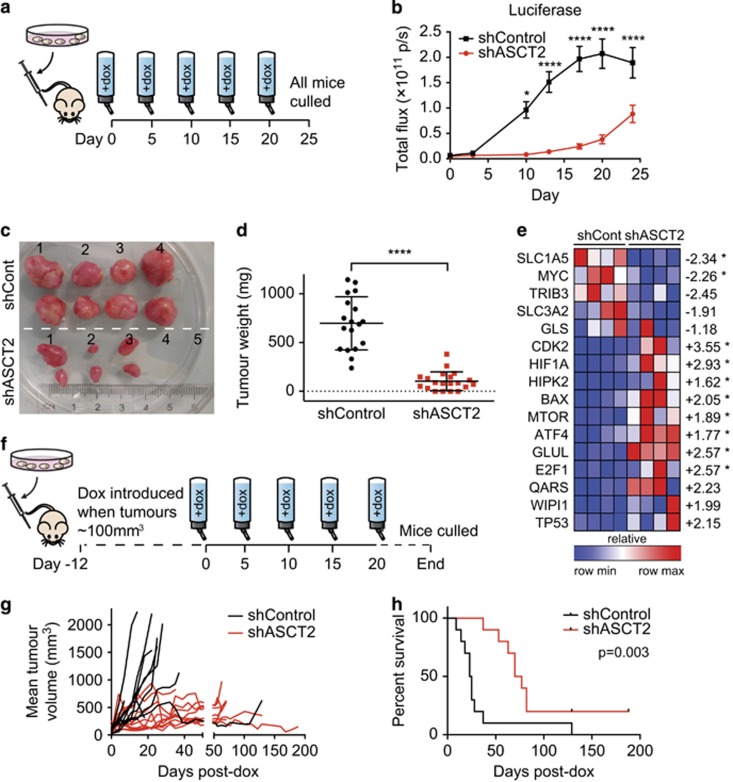
To enable analysis of ASCT2 knockdown on in vivo cell growth, HCC1806 cells were transduced with an additional lentivirus engineered to enable stoichiometric expression of both mCherry and firefly luciferase reporters. Sorted HCC1806-mCherryhi GFP+ cells (shCont and shASCT2) were then orthotopically injected into the mammary fat pad of athymic nu/nu mice, after confirming doxycycline-inducible ASCT2 protein knockdown (Supplementary Figure 3B) and repression of cell growth in vitro (Supplementary Figure 3A). Mice were then imaged twice weekly while doxycycline was delivered in the drinking water from Day 0 (as per Figure 3a). Knockdown of ASCT2 caused a reduction in tumour take-rate, with only 80% of shASCT2 tumours engrafting successfully compared with 100% of shCont tumours, as well as statistically significant decreases in tumour growth (Figure 3b, Supplementary Figure 3C), size (Figure 3c) and weight (Figure 3d) at end point. Relative quantification of mRNA in tumours collected at end point also showed significantly altered gene expression as a result of ASCT2 knockdown throughout tumour establishment and growth (Figure 3e). As expected, ASCT2 (SLC1A5) expression was significantly decreased (2.34-fold, P=0.0286). Expression of several master regulators of tumour metabolism was altered, including a significant reduction (2.26-fold, P=0.0286) in oncogenic MYC (amplified in HCC1806 cells35) and upregulation of transcription factors, HIF1a (2.93-fold, P=0.0286) and ATF4 (1.77-fold, P=0.0286). Together, these changes indicate a shift to glycolytic metabolism as a cellular survival mechanism when ASCT2 expression is lost. They were accompanied by increased expression of glutaminogenic genes (glutamine–ammonia ligase, GLUL, 2.57-fold, P=0.0286; glutamyl-tRNA synthetase, QARS, 2.23-fold, NS (not significant)). Also significantly increased were several genes with known pro-apoptotic roles in cellular stress (HIPK2, BAX36, 37), suggesting initiation of a dynamic transcriptional programme in response to loss of ASCT2 transport.
Figure 3.
ASCT2 knockdown in vivo represses tumour growth and improves survival. (a) HCC1806 cells expressing an additional mCherry-luciferase reporter construct (pHIV1SDm-CMV-mCherry-P2A-luc47) and doxycycline-inducible shRNAs (see Figure 2h legend), shControl or shASCT2 (pFH1tUTG34) were sorted for mCherryhi/GFP+ cells using fluorescence-activated cell sorting and then 4 × 106 cells suspended in a solution of 50% Matrigel in RPMI (100 μl) were implanted orthotopically into the left and right abdominal mammary glands of 8–12-week-old female athymic nu/nu mice (Animal Resource Centre, Perth, Western Australia, Australia), five mice per group from two separate experiments (n=10). Sample size was estimated on the basis of previous publications21 with no randomization or blinding. Doxycycline (200 μg/ml) was administered ad libitum in drinking water from Day 0 and tumour growth was measured twice weekly until individual shControl tumours reached ~1000 mm3, when all the mice were humanely euthanized in accordance with ethics approval protocol 2013/030 A from Sydney Local Health District Animal Welfare Committee. (b) Bioluminescence was detected twice weekly using a Xenogen IVIS Lumina II following intraperitoneal injection of 150 mg/kg d-luciferin. At end point, all the mice were euthanized and tumours collected for the measurement of size (c) and weight (d; error bars represent mean±s.d.). (e) Tumour RNA was extracted from shCont and shASCT2 (n=4 each) tumours using TRI Reagent, then reverse transcribed using SuperScript III (Life Technologies, Carlsbad, CA, USA) and random hexamers. Relative gene expression (GAPDH/ACTB) was measured by RT-qPCR using a custom Taqman Low-Density Array (Taqman, Life Technologies) run on a QuantStudio 12 K Flex Real-Time PCR System. Heat map of relative expression (compared with the first shControl lane) for each gene was generated in Gene-E (Broad Institute). Average fold-change between the shControl and shASCT2 groups are indicated to the right of the heat-map, asterisks indicate significant difference (P<0.05, n=4; Mann–Whitney U-test). (f), HCC1806-mCherryhi/GFP+ cells (2 × 106) were injected orthotopically as in a and allowed to establish (tumour size ~100 mm3) before the administration of doxycycline (200 μg/ml). Tumour size was measured twice weekly using digital calipers to calculate mean tumour volume per mouse (g) and individual mice were euthanized when they reached ethical end point (individual tumour size >1000 mm3; h). Data in b represents mean±s.e.m., n=9 (shCont; one mouse was euthanized owing to ulceration at tumour site and is not included in the analyses) or n=10 (shASCT2) mice (with two tumours per mouse), five mice per group in two independent experiments. Data in g represents mean, n=10 (shCont) or n=10 (shASCT2) mice (with two tumours per mouse), five mice per group in two independent experiments. Asterisks denote P-values as follows: *P⩽0.05; **P⩽0.01; ***P⩽0.001; ****P⩽0.0001, NS, not significant; two-way analysis of variance (b), Mann–Whitney U-test (d and e), or log-rank Mantel–Cox test (h). Data in c are representative of two independent experiments.
To ensure that a reduced tumour take-rate was not masking the effects of ASCT2 knockdown, we next allowed tumours to establish before introduction of doxycycline (as per Figure 3f). In this setting, ASCT2 knockdown continued to significantly reduce tumour volume (Figure 3g) and also more than tripled the median survival time from 24 to 73.5 days post doxycycline treatment. This indicated that lower tumoral ASCT2 expression conferred a significant survival advantage even in established tumours (Figure 3h; log-rank HR=3.405 (1.951–15.69), P=0.003, Mantel–Cox test). These data indicate that ASCT2 expression is required for successful basal-like tumour engraftment and tumour growth in vivo, and suggest that the presence of ASCT2 expression decreases time to ethical end point in mice with basal-like breast cancer xenografts. This is consistent with recent findings showing that low ASCT2 expression correlated with increased disease-free survival in a large cohort of human breast cancer patient samples, and showed a strong association with other clinicopathological predictors of poor prognosis, such as positive node status and absence of ER expression.38 Together these data indicated that ASCT2 expression promotes basal-like breast tumour growth in vivo, and is similarly associated with aggressive biological behaviour in clinical disease.
Triple-negative breast cancers globally increase ASCT2 expression and glutamine metabolism
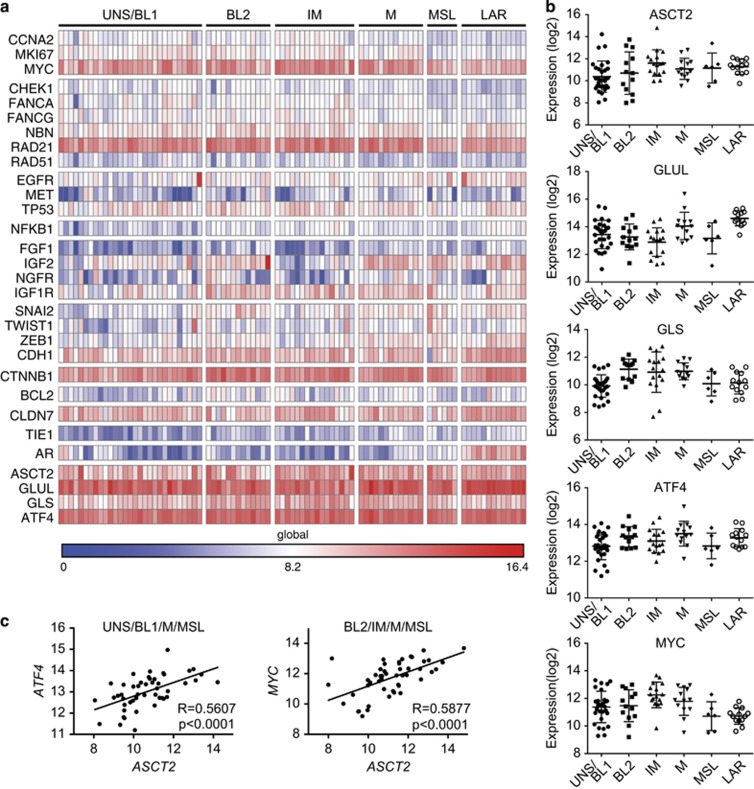
Given that basal-like tumours make up at least half of the broader clinical TN subtype,39 we hypothesized that ASCT2 may be a critical driver of cell growth in these breast cancers and thus present a possible axis for targeted therapeutic intervention. We therefore designed a custom NanoString nCounter DX code set to quantify absolute RNA expression of a subset of glutamine metabolism genes in a cohort of 90 TN breast cancer patients. Using a previously defined TN breast cancer gene expression signature,9 samples were manually clustered into six TN subgroups based on 26 genes (Figure 4a). Global gene expression for this subset (as opposed to relative gene expression), revealed high expression of a number of genes associated with TN breast cancer, including MYC, RAD21 and CTNNB1, with widespread downregulation of hormone-receptor signalling genes (AR) outside the LAR subtype. Strikingly, however, expression of ASCT2 and glutamine metabolism-related genes (GLS, GLUL) was uniformly high across all TN subgroups (Figures 4a and b), suggesting a broad reliance on glutamine availability in both basal-like and non-basal-like TN breast cancers and reinforcing a link between increased glutamine metabolism and clinically aggressive breast cancers.40 Furthermore, the stress-responsive transcription factor, ATF4, was also highly expressed in all TN subsets (Figures 4a and b). ATF4 expression was also significantly increased after ASCT2 knockdown in vivo (Figure 3e), supporting previous data in PC-3 prostate cancer cells in vitro.21 In that study, GPNA was shown to increase not only ATF4 expression, but also led to concomitant increases in a range of amino acid transporters including ASCT2, SLC1A4, SLC3A2, SLC7A11, SLC7A1 and SLC38A1.21 Although it has been shown that ATF4 directly regulates many of these transporters,30, 41 in our study, increased ATF4 expression after ASCT2 knockdown in vivo was not able to increase either ASCT2 or SLC3A2 expression (Figure 3e). Despite this, in patient samples, our data show that ASCT2 expression correlates with ATF4 expression in UNS/BL1, M and MSL subtypes (P<0.001, r=5607; Figure 4c and Supplementary Table 1), suggesting direct transcriptional regulation. Other recent data has shown that ASCT2 is not only regulated by ATF4, but also by N-Myc,42, 43 adding to the well described role of c-Myc in transcriptionally regulating glutamine metabolism.44, 45 Similar to ATF4, ASCT2 expression correlated with MYC in BL2, IM, M and MSL subsets (P<0.0001, r=0.5877; Figure 4c and Supplementary Table 1). It is therefore possible that while amino acid transporters are increased in a variety of cancers, this may merely reflect Myc- and ATF4-mediated transcriptional programmes, rather than the reliance of a particular cancer on the relevant substrate amino acid(s). It is therefore crucial to determine which cancer, and subsets, not only express the relevant transporters and enzymes, but importantly rely on them for critical metabolic functions. It is clear that the ER+ breast cancer cell line MCF-7, despite high ASCT2 expression, is not sensitive to glutamine deprivation (Figure 1). However, our data for five different basal-like TN breast cancer cell lines, together with a recent report showing the importance of ASCT2 and SNAT2 in regulating glutamine uptake and growth in the claudin-low TN cell line, MDA-MB-231,38 suggests that the majority of TN molecular subsets may be susceptible to therapies targeting glutamine uptake or glutamine metabolic pathways. Therefore, further preclinical development and testing of therapeutics targeting ASCT2, SNAT2 or downstream metabolic pathways such as glutaminase24, 46 should therefore be undertaken across a range of TN breast cancer subtypes.
Figure 4.
ASCT2 is highly expressed in human triple-negative breast cancer patient samples. RNA was extracted from 96 formalin-fixed paraffin-embedded triple-negative (TN) patient sample cores (AllPrep DNA/RNA FFPE Kit, Qiagen, Melbourne, Victoria, Australia) obtained from Royal Prince Alfred Hospital and Concord Repatriation General Hospital, Australia, in accordance with ethics approval from Sydney Local Health District Ethics Board with informed patient consent. Unamplified mRNA levels were measured directly using a custom probe set on the NanoString nCounter DX platform according to manufacturer's instructions and normalized to mean expression of six housekeeper genes (CLTC, GAPDH, GUSB, HPRT1, PGK1, TUBB). Six patient samples failed quality control checks and were excluded from subsequent analysis. Heat map (a) was generated in Gene-E (Broad Institute) and shows the global absolute gene expression (log2, global min and max values shown) after manual clustering into TN breast cancer subtypes according to gene expression patterns previously described9 (UNS/BL1, unstable/basal-like 1; BL2, basal-like 2; IM, immunomodulatory; M, mesenchymal; MSL, mesenchymal stem-like; LAR, luminal androgen receptor). Log2 gene expression of ASCT2/SLC1A5, glutamine metabolism-related genes (GLUL, GLS) and transcription factors, MYC and ATF4 (b) across TN subtypes in our patient cohort as defined in a. Correlation between ASCT2/SLC1A5 and transcriptional regulators, MYC and ATF4, was analysed for TN patient subgroups in our cohort (Supplementary Table 1), and subgroups that showed significant correlations (r>0.4, P<0.05; Pearson's correlation statistic; exact P-values shown inset) were grouped and the correlation shown in c. Black line shows the best-fit linear regression (SLC1A5 vs ATF4: r2=0.314; SLC1A5 vs MYC: r2=0.345).
Acknowledgments
This work was supported by grants from the National Breast Cancer Foundation (ECF-12-05 to JH); Movember through the Prostate Cancer Foundation of Australia (YI0813 to QW; YI0707 to JH); Cancer Institute NSW (WR and MvG); Cancer Council NSW (APP1080503 to JH); University of Sydney (AM); Cure the Future and an anonymous foundation (JEJR); Tour de Cure Fellowship (CGB); Sydney Breast Cancer Foundation and the RT Hall Foundation (SO'T). We thank Michael P Kuligowski from Centenary Imaging Facility for assistance with Volocity image analysis and staff at the Advanced Cytometry Facility, Sydney for FACS assistance.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on the Oncogene website (http://www.nature.com/onc)
Supplementary Material
References
- Gluz O, Liedtke C, Gottschalk N, Pusztai L, Nitz U, Harbeck N. Triple-negative breast cancer—current status and future directions. Ann Oncol 2009; 20: 1913–1927. [DOI] [PubMed] [Google Scholar]
- Sorlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci USA 2001; 98: 10869–10874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA et al. Molecular portraits of human breast tumours. Nature 2000; 406: 747–752. [DOI] [PubMed] [Google Scholar]
- Prat A, Parker JS, Karginova O, Fan C, Livasy C, Herschkowitz JI et al. Phenotypic and molecular characterization of the claudin-low intrinsic subtype of breast cancer. Breast Cancer Res 2010; 12: R68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dent R, Trudeau M, Pritchard KI, Hanna WM, Kahn HK, Sawka CA et al. Triple-negative breast cancer: clinical features and patterns of recurrence. Clin Cancer Res 2007; 13: 4429–4434. [DOI] [PubMed] [Google Scholar]
- Haffty BG, Yang Q, Reiss M, Kearney T, Higgins SA, Weidhaas J et al. Locoregional relapse and distant metastasis in conservatively managed triple negative early-stage breast cancer. J Clin Oncol 2006; 24: 5652–5657. [DOI] [PubMed] [Google Scholar]
- Rakha EA, El-Sayed ME, Green AR, Lee AH, Robertson JF, Ellis IO. Prognostic markers in triple-negative breast cancer. Cancer 2007; 109: 25–32. [DOI] [PubMed] [Google Scholar]
- Chen XS, Ma CD, Wu JY, Yang WT, Lu HF, Wu J et al. Molecular subtype approximated by quantitative estrogen receptor, progesterone receptor and Her2 can predict the prognosis of breast cancer. Tumori 2010; 96: 103–110. [DOI] [PubMed] [Google Scholar]
- Lehmann BD, Bauer JA, Chen X, Sanders ME, Chakravarthy AB, Shyr Y et al. Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J Clin Invest 2011; 121: 2750–2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Kim do H, Jung WH, Koo JS. Expression of glutamine metabolism-related proteins according to molecular subtype of breast cancer. Endocr Relat Cancer 2013; 20: 339–348. [DOI] [PubMed] [Google Scholar]
- Timmerman LA, Holton T, Yuneva M, Louie RJ, Padro M, Daemen A et al. Glutamine sensitivity analysis identifies the xCT antiporter as a common triple-negative breast tumor therapeutic target. Cancer Cell 2013; 24: 450–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanai Y, Hediger MA. The glutamate/neutral amino acid transporter family SLC1: molecular, physiological and pharmacological aspects. Pflugers Arch 2004; 447: 469–479. [DOI] [PubMed] [Google Scholar]
- Kekuda R, Prasad PD, Fei YJ, Torres-Zamorano V, Sinha S, Yang-Feng TL et al. Cloning of the sodium-dependent, broad-scope, neutral amino acid transporter Bo from a human placental choriocarcinoma cell line. J Biol Chem 1996; 271: 18657–18661. [DOI] [PubMed] [Google Scholar]
- DeBerardinis RJ, Mancuso A, Daikhin E, Nissim I, Yudkoff M, Wehrli S et al. Beyond aerobic glycolysis: transformed cells can engage in glutamine metabolism that exceeds the requirement for protein and nucleotide synthesis. Proc Natl Acad Sci USA 2007; 104: 19345–19350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duran RV, Oppliger W, Robitaille AM, Heiserich L, Skendaj R, Gottlieb E et al. Glutaminolysis activates Rag-mTORC1 signaling. Mol Cell 2012; 47: 349–358. [DOI] [PubMed] [Google Scholar]
- Wullschleger S, Loewith R, Hall MN. TOR signaling in growth and metabolism. Cell 2006; 124: 471–484. [DOI] [PubMed] [Google Scholar]
- Green DR, Galluzzi L, Kroemer G. Cell biology. Metabolic control of cell death. Science 2014; 345: 1250256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Beaumont KA, Otte NJ, Font J, Bailey CG, van Geldermalsen M et al. Targeting glutamine transport to suppress melanoma cell growth. Int J Cancer 2014; 135: 1060–1071. [DOI] [PubMed] [Google Scholar]
- Hassanein M, Hoeksema MD, Shiota M, Qian J, Harris BK, Chen H et al. SLC1A5 mediates glutamine transport required for lung cancer cell growth and survival. Clin Cancer Res 2013; 19: 560–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassanein M, Qian J, Hoeksema MD, Wang J, Jacobovitz M, Ji X et al. Targeting SLC1A5-mediated glutamine dependence in non-small cell lung cancer. Int J Cancer 2015; 137: 1587–1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Hardie RA, Hoy AJ, van Geldermalsen M, Gao D, Fazli L et al. Targeting ASCT2-mediated glutamine uptake blocks prostate cancer growth and tumour development. J Pathol 2015; 236: 278–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willems L, Jacque N, Jacquel A, Neveux N, Maciel TT, Lambert M et al. Inhibiting glutamine uptake represents an attractive new strategy for treating acute myeloid leukemia. Blood 2013; 122: 3521–3532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esslinger CS, Cybulski KA, Rhoderick JF. Ngamma-aryl glutamine analogues as probes of the ASCT2 neutral amino acid transporter binding site. Bioorg Med Chem 2005; 13: 1111–1118. [DOI] [PubMed] [Google Scholar]
- Gross MI, Demo SD, Dennison JB, Chen L, Chernov-Rogan T, Goyal B et al. Antitumor activity of the glutaminase inhibitor CB-839 in triple-negative breast cancer. Mol Cancer Ther 2014; 13: 890–901. [DOI] [PubMed] [Google Scholar]
- Kung HN, Marks JR, Chi JT. Glutamine synthetase is a genetic determinant of cell type-specific glutamine independence in breast epithelia. PLoS Genet 2011; 7: e1002229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fingar DC, Blenis J. Target of rapamycin (TOR): an integrator of nutrient and growth factor signals and coordinator of cell growth and cell cycle progression. Oncogene 2004; 23: 3151–3171. [DOI] [PubMed] [Google Scholar]
- Saqcena M, Menon D, Patel D, Mukhopadhyay S, Chow V, Foster DA. Amino acids and mTOR mediate distinct metabolic checkpoints in mammalian G1 cell cycle. PLoS One 2013; 8: e74157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicklin P, Bergman P, Zhang B, Triantafellow E, Wang H, Nyfeler B et al. Bidirectional transport of amino acids regulates mTOR and autophagy. Cell 2009; 136: 521–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs BC, Finger RE, Onan MC, Bode BP. ASCT2 silencing regulates mammalian target-of-rapamycin growth and survival signaling in human hepatoma cells. Am J Physiol Cell Physiol 2007; 293: C55–C63. [DOI] [PubMed] [Google Scholar]
- Wang Q, Bailey CG, Ng C, Tiffen J, Thoeng A, Minhas V et al. Androgen receptor and nutrient signaling pathways coordinate the demand for increased amino acid transport during prostate cancer progression. Cancer Res 2011; 71: 7525–7536. [DOI] [PubMed] [Google Scholar]
- Sancak Y, Bar-Peled L, Zoncu R, Markhard AL, Nada S, Sabatini DM. Ragulator-Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell 2010; 141: 290–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancak Y, Peterson TR, Shaul YD, Lindquist RA, Thoreen CC, Bar-Peled L et al. The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science 2008; 320: 1496–1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs BC, Perez JC, Suetterlin JE, Chaudhry SB, Bode BP. Inducible antisense RNA targeting amino acid transporter ATB0/ASCT2 elicits apoptosis in human hepatoma cells. Am J Physiol Gastrointest Liver Physiol 2004; 286: G467–G478. [DOI] [PubMed] [Google Scholar]
- Herold MJ, van den Brandt J, Seibler J, Reichardt HM. Inducible and reversible gene silencing by stable integration of an shRNA-encoding lentivirus in transgenic rats. Proc Natl Acad Sci USA 2008; 105: 18507–18512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin AA, Kim S et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature 2012; 483: 603–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puca R, Nardinocchi L, Givol D, D'Orazi G. Regulation of p53 activity by HIPK2: molecular mechanisms and therapeutical implications in human cancer cells. Oncogene 2010; 29: 4378–4387. [DOI] [PubMed] [Google Scholar]
- Yee KS, Wilkinson S, James J, Ryan KM, Vousden KH. PUMA- and Bax-induced autophagy contributes to apoptosis. Cell Death Differ 2009; 16: 1135–1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon YJ, Khelifa S, Ratnikov B, Scott DA, Feng Y, Parisi F et al. Regulation of glutamine carrier proteins by RNF5 determines breast cancer response to ER stress-inducing chemotherapies. Cancer Cell 2015; 27: 354–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perou CM. Molecular stratification of triple-negative breast cancers. Oncologist 2010; 15: 39–48. [DOI] [PubMed] [Google Scholar]
- Terunuma A, Putluri N, Mishra P, Mathe EA, Dorsey TH, Yi M et al. MYC-driven accumulation of 2-hydroxyglutarate is associated with breast cancer prognosis. J Clin Invest 2014; 124: 398–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Tiffen J, Bailey CG, Lehman ML, Ritchie W, Fazli L et al. Targeting amino acid transport in metastatic castration-resistant prostate cancer: effects on cell cycle, cell growth, and tumor development. J Natl Cancer Inst 2013; 105: 1463–1473. [DOI] [PubMed] [Google Scholar]
- Ren P, Yue M, Xiao D, Xiu R, Gan L, Liu H et al. ATF4 and N-Myc coordinate glutamine metabolism in MYCN-amplified neuroblastoma cells through ASCT2 activation. J Pathol 2015; 235: 90–100. [DOI] [PubMed] [Google Scholar]
- Qing G, Li B, Vu A, Skuli N, Walton ZE, Liu X et al. ATF4 regulates MYC-mediated neuroblastoma cell death upon glutamine deprivation. Cancer Cell 2012; 22: 631–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao P, Tchernyshyov I, Chang TC, Lee YS, Kita K, Ochi T et al. c-Myc suppression of miR-23a/b enhances mitochondrial glutaminase expression and glutamine metabolism. Nature 2009; 458: 762–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wise DR, DeBerardinis RJ, Mancuso A, Sayed N, Zhang XY, Pfeiffer HK et al. Myc regulates a transcriptional program that stimulates mitochondrial glutaminolysis and leads to glutamine addiction. Proc Natl Acad Sci USA 2008; 105: 18782–18787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacque N, Ronchetti AM, Larrue C, Meunier G, Birsen R, Willems L et al. Targeting glutaminolysis has anti-leukemic activity in acute myeloid leukemia and synergizes with BCL-2 inhibition. Blood 2015; 126: 1346–1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiffen JC, Bailey CG, Ng C, Rasko JE, Holst J. Luciferase expression and bioluminescence does not affect tumor cell growth in vitro or in vivo. Mol Cancer 2010; 9: 299. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.