Abstract
Olfaction poses one of the most complex ligand-receptor matching problems in biology due to the unparalleled multitude of odor molecules facing a large number of cognate olfactory receptors. We have recently deorphanized an olfactory receptor, TAAR13c, as a specific receptor for the death-associated odor cadaverine. Here we have modeled the cadaverine/TAAR13c interaction, exchanged predicted binding residues by site-directed mutagenesis, and measured the activity of the mutant receptors. Unexpectedly we observed a binding site for cadaverine at the external surface of the receptor, in addition to an internal binding site, whose mutation resulted in complete loss of activity. In stark contrast, elimination of the external binding site generated supersensitive receptors. Modeling suggests this site to act as a gate, limiting access of the ligand to the internal binding site and thereby downregulating the affinity of the native receptor. This constitutes a novel mechanism to fine-tune physiological sensitivity to socially relevant odors.
The interaction of odors with their cognate receptors constitutes one of the most complex ligand/receptor binding problems in biology due to the sheer quantity of potential odor molecules facing a limited albeit huge number of different olfactory receptors which in some species comprise close to 10% of all proteins1,2. The tuning width of these receptors is extremely variable, with odor spectra ranging from exceedingly broad3,4 to monospecific5. In some cases a single functional group of the ligand dominates the specificity of the ligand/receptor interaction, in other cases an ensemble of chemical features is recognized6,7,8. However, only in very few cases do we possess a molecular understanding of the binding interaction between an odorant and its receptor9. So far crystal structures are not available for any olfactory receptor, and thus prediction of olfactory receptor structures has relied on modeling studies using established templates such as the beta-adrenergic receptor (β-AR)10 and rhodopsin11, further supported by site-directed mutagenesis and subsequent functional analysis of mutant receptors12.
We have used a similar approach to unravel the ligand interaction of a zebrafish olfactory receptor specific for aliphatic diamines, TAAR13c6. The trace amine associated receptor (TAAR) family is the only olfactory receptor family that is much larger in teleost fish compared to tetrapods, suggesting an essential role for TAARs in fish13. Zebrafish possess 112 taar genes, compared to only 15 in mouse and even less in the amphibian and avian lineages6. Since zebrafish serve as a model system for vertebrates, and their olfactory system is qualitatively similar to that of vertebrates including mammals14, zebrafish are well suited to gain deeper insight into vertebrate olfactory receptor properties.
We have recently shown TAAR13c to be a highly sensitive and specific receptor for the death-associated odor cadaverine6, which emanates from carrion via bacterial decarboxylation of lysine. Cadaverine is strongly repulsive for humans and, interestingly, it also elicits strong innate aversive behavior in zebrafish6. At low concentrations of cadaverine mostly TAAR13c-expressing neurons get activated suggesting that a single olfactory receptor might suffice to generate a powerful odor-driven behavior6. Here we aimed to understand the molecular basis at the very beginning of this neural circuit, i.e. the interaction of TAAR13c with its native ligand cadaverine.
We have performed thorough modeling of the TAAR13c receptor to identify potential binding site residues, and found all of them clustering in the upper third of the transmembrane domains of TAAR13c. We mutated several of these candidate residues and compared the activation of mutant to wildtype receptors in a heterologous cell expression system. Two aspartates, Asp1123.32 and Asp2025.42, buried in the plane of the membrane, were identified as essential components of an internal binding site for cadaverine. Another aspartate, Asp2796.58, was found to constitute an essential residue of a second binding site located at the extracellular surface of the receptor. Both conservative and non-conservative substitutions of Asp2796.58 generated supersensitive receptors. Based on our modeling data we suggest the external binding site to act as a gate, which cadaverine has to pass on its way to the internal binding site. As long as the external binding site is occupied, the gate is closed, and thus limits the free access of cadaverine to the internal binding site. This constitutes a novel molecular mechanism for regulating ligand access to the activating binding site. To the best of our knowledge, such a gating mechanism has not been suggested for any olfactory receptor of any species so far.
Results
Modeling of TAAR13c predicts the expected 7TM structure and two additional small helices in ECL2 and C-terminus
We have modeled the TAAR13c structure as a prerequisite to gain structural insights into the molecular architecture and functional constraints of its binding pocket. The homology model of TAAR13c was based on X-ray crystal structures of six templates (see Mat. & Meth and Supplementary Data 2). The TAAR13c primary structure shares a maximal identity of 33% to its closest homolog, the β1 adrenergic receptor (β1AR; Protein Data Bank Entry 4AMJ)15.
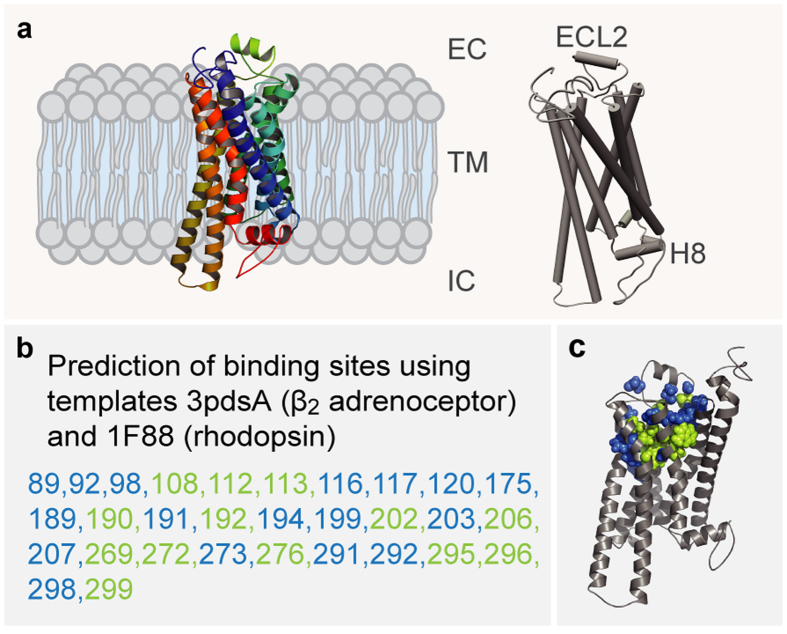
The homology model of TAAR13c (Fig. 1a) revealed the canonical bundle of seven transmembrane (TM) α-helices followed by an eighth intracellular helix (H8) running parallel to the membrane axis. In rhodopsin, an interaction between H8 and TM7 keeps the receptor in a prereceptive state16, but no such interaction was seen in the TAAR13c model. Interestingly, in the model based on β1AR we also observed a short α-helix in extracellular loop 2 (ECL2) of TAAR13c, which is absent in most class A GPCRs, but present in the β-adrenergic receptors17. Furthermore, a disulfide bridge is predicted between Cys1053.25 at the extracellular end of TM3 and Cys190 in ECL2. This disulphide bond provides conformational restraint and is important for effectively tethering ECL2 to the helical bundle18. The highly conserved landmark motif DRH/Y, here DRH, is located at the cytoplasmic end of TM3 as expected17. This motif stabilizes the inactive state in some receptors, and governs G-protein coupling in other receptors19. We also observed the ‘ionic lock’ between the DRH motif and a glutamate residue at the cytosolic surface of TM6. Due to a salt bridge that is formed between Arg1303.50 of the DRH motif and Glu2516.34 in TM6, the third and sixth TM helices are connected, a feature which is conserved among all family A GPCRs17.
Figure 1. Homology modeling of TAAR13c predicts 7 TM, two additional short helices, and potential binding residues clustered in the upper third of the TM region.
(a) Cartoon representation of the TAAR13c model based on comparison with six crystal structures shows the expected seven transmembrane domains and two short extra helices. The planks representation to the right shows a short α-helix to be located in ECL2 and an intracellular eighth helix, H8 located parallel to the membrane plane. (b) Ligand binding residues (given as residue number in TAAR13c) as predicted by sequence profile comparison with binding sites of PDB templates 3pdsA (FAUC-50-β2 adrenoceptor complex) and 1F88 (bovine rhodopsin); blue, residues only reported in one of the models; green, residues predicted in both models. (c) Predicted binding residues listed in panel (b) shown as spheres in the TAAR13c structure, color code as before. Note the presence of two ‘green’ columns.
Docking of cadaverine validates a binding pocket in the outer-third of the TM domain
We then used COACH20 to predict putative cadaverine binding residues by sequence profile comparison of TAAR13c with binding sites of several PDB structures, with best fits found for 3pdsA (FAUC50/β2 adrenoceptor complex) and 1F88 (bovine rhodopsin) in the TAAR13c homology model (Fig. 1b). COACH is a meta-server approach to generate complementary ligand binding site predictions using comparative methods, which recognize ligand-binding templates from BioLiP protein function database by binding-specific substructure and sequence profile comparisons21. Initially 30 such residues were found. They all clustered in the upper third of the TM domain suggesting that the putative binding pocket is located within this region (Fig. 1c). The classical amine-binding motif of aminergic receptors consists of Asp1123.32 and Trp2967.40, both of which were also predicted as binding partners of cadaverine22. In close proximity to these residues and at the same plane of the membrane another aspartate residue, Asp2025.42, was predicted as a binding partner. We hypothesized that this residue might be involved in binding to the second amino group of cadaverine and examined the region surrounding Asp1123.32, Asp2025.42 and Trp2967.40 (i.e. the upper one-third of TM3, 5, and 6) by computational docking of cadaverine.
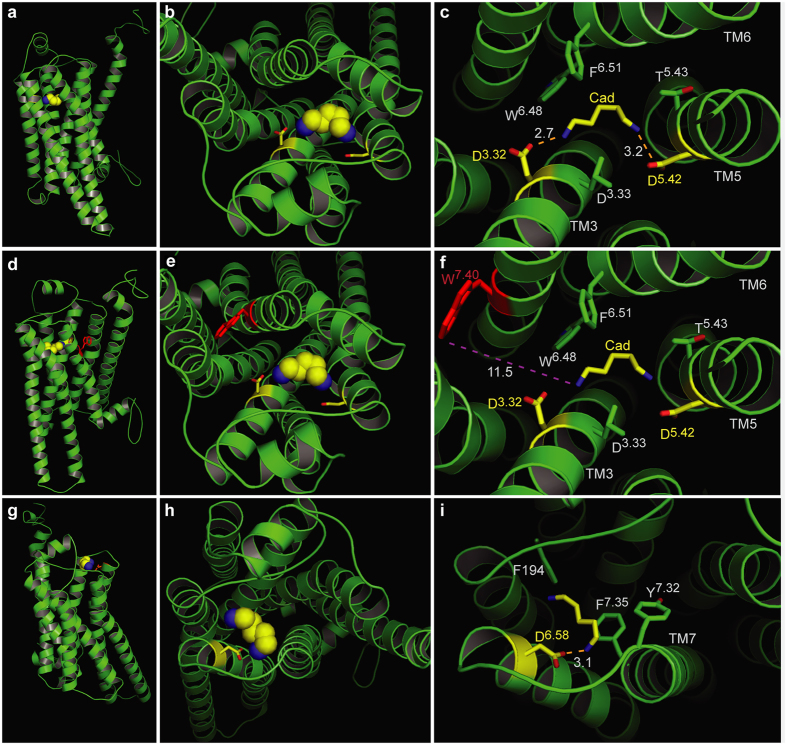
The docking results confirmed the involvement of Asp1123.32 and Asp2025.42 in ligand binding. Our results suggest that one amino group of cadaverine (protonated at physiological pH) forms a salt bridge with Asp1123.32 at a distance of 2.7 Å (Fig. 2c, Table 1), well within the range given for salt bridges 1.75–4.0 Å23. This Asp1123.32 is stabilized by a hydrogen bond to the hydroxyl group of Tyr299. A similar salt bridge has been described for β1- and β2ARs24. The second protonated amino group of cadaverine was docked 3.2 Å away from Asp2025.42 allowing formation of another salt bridge (Fig. 2c). This particular residue is known to undergo binding interactions with ligands in many other GPCRs, forming hydrogen bonds, van der Waals interactions and salt bridges25. Further binding residues validated by docking were Leu1133.33, Thr2035.43, Trp2696.48, and Phe2726.51, all situated within 5 Å distance from cadaverine and thus well within the range of van der Waals interactions (3–6 Å26) (Supplementary Table 1). Thus, these residues are likely candidates for stabilizing the hydrophobic backbone of cadaverine. In addition to cadaverine, diaminoheptane, which has a very similar affinity to TAAR13c6, forms the same two salt bridges as cadaverine (2.9 Å distance to Asp1123.32 and 2.4 Å to Asp2025.42).
Figure 2. Two binding sites for cadaverine predicted by docking to wildtype TAAR13c.
TAAR13c structure (green) is shown with cadaverine (cad; yellow, backbone; blue, amino groups) and coordinating aspartate residues (yellow, backbone; red, carboxyl group). (a) Sideview of TAAR13c shows spatial position of cadaverine docked to the internal binding site, located in the external third of the TM region. (b) Enlarged view from the extracellular surface onto the same binding site shown in panel (a). (c) Enlargement from panel (b) (same view) showing cadaverine, the major interacting residues and the distances in Å from the carboxyl groups of aspartates D3.32 and D5.42 to the amino groups of cadaverine. Salt bridges are visualized as orange dashed lines. (d) Side view (turned about 90° compared to panel (a) shows the side chain of W7.40 located away from the predicted binding site. (e) View from the extracellular surface, enlarged, same orientation as panel (b). W7.40 is positioned on the distal side of its TM relative to the binding site. (f) Enlargement from panel (e) (same view) showing cadaverine, the two interacting aspartate residues and the large distance to the side chain of W7.40 (purple dashed line), which is thus unlikely to participate in binding interactions. (g) Side view (turned about 90° compared to panel (a) showing cadaverine bound at an additional binding site on the external surface. (h) View onto the extracellular surface, enlarged, orientation turned 180° relative to panel (b). A single aspartate (D6.58) coordinates cadaverine. (i) Enlargement from panel (h) (similar view) showing cadaverine, the major interacting residues and the distance from the carboxyl group of D6.58 to the amino group of cadaverine. The salt bridge is visualized as orange dashed line.
Table 1. EC50 and contact distances for cadaverine and putrescine amino groups with Asp1123.32 and Asp2025.42 for mutant and wildtype TAAR13c.
| Wildtype/Mutant | EC50 (μM, mean+/−SEM) |
Distance to Cad-NH2 |
Distance to Put-NH2 |
|||
|---|---|---|---|---|---|---|
| cad | put | pos112 | pos202 | pos112 | pos202 | |
| TAAR13c | 15.2 ± 2.2 | >1000 | 2.7 | 3.2 | 3.0 | 5.5 |
| D112A | loss | loss | 2.4 | 10.8 | 10.7 | 4.9 |
| D112E | ≫1000 | loss | 9.2 | 3.2 | 9.8 | 5.0 |
| D112N | loss | loss | 12.0 | 3.1 | 12.0 | 3.1 |
| D202A | loss | loss | 2.4 | 10.8 | 10.7 | 4.9 |
| D202E | loss | loss | 9.2 | 3.2 | 9.8 | 5.0 |
| D202N | loss | loss | 3.0 | 9.5 | 2.5 | 10.6 |
| D279A | 2.1 ± 1.0 | 31 ± 13 | 3.0 | 3.7 | 3.0 | 6.4 |
| D279E | 0.72 ± 0.24 | 10 ± 4 | 2.9 | 3.5 | 3.0 | 5.6 |
| D279N | 1.1 ± 0.8 | 7.3 ± 3.6 | 2.9 | 3.5 | 3.0 | 5.6 |
| W296F | 42 ± 6 | >1000 | – | – | – | – |
| W296G | 55 ± 48 | loss | – | – | – | – |
| W296Y | 28 ± 8 | >1000 | – | – | – | – |
| D78E | 64 ± 28 | >1000 | – | – | – | – |
Docking did not confirm Trp2967.40 as a binding residue in TAAR13c. Notably, Trp2967.40 is 11.5 Å away from the docked cadaverine, and furthermore the residue is located on the distal side of TM7 relative to cadaverine, excluding a van der Waals interaction (Fig. 2f). This was unexpected, because in class A GPCRs this tryptophan is highly conserved, and serves to stabilize the hydrophobic backbone of amines as part of the amine-binding motif22.
In addition to cadaverine, TAAR13c is activated by putrescine, a smaller diamine, albeit with much lower affinity6. Docking of putrescine into the TAAR13c model revealed the same salt bridge with Asp1123.32 as for cadaverine, albeit at a slightly larger distance of 3.0 Å (Table 1). However, Asp2025.42 is not able to form the second salt bridge, because the distance of 5.5 Å between the amino group of putrescine and the carboxylic group of Asp2025.42 (Table 1) is too large for a typical salt bridge and only allows a rather weak binding interaction27. This finding would explain the decrease in affinity of TAAR13c for putrescine compared to cadaverine. Nevertheless the weak interaction between Asp2025.42 and the second amino group of putrescine seems to be relevant because the corresponding monoamine (butylamine) is not able to activate TAAR13c at all6.
Taken together, these data strongly suggest that TAAR13c activation relies on the interaction of two amino groups provided by the ligand with two negatively charged residues in the binding cavity of the receptor, and that stabilization of the ligand backbone is not achieved by the canonical tryptophan, here Trp2967.40, in TM7 of the DW motif.
Docking of cadaverine suggests a second binding site for diamines
In addition to the above-mentioned residues, we identified another docking site on the extracellular surface of the receptor (Fig. 2g,h). The main binding residue of this second site is Asp2796.58, which forms a salt bridge with one amino group of cadaverine (Fig. 2i) and putrescine, each at a distance of 3.1 Å (Table 1). Surrounding apolar residues Phe194 in ECL2 and Phe2917.35 in TM7 are also predicted as interaction partners, and presumably serve to stabilize the ligand’s apolar backbone (Supplementary Table 1). No residue coordinating the second amino group was detected in this docking site, suggesting that it might not discriminate for chain length. Thus, this site differs in two properties from the internal docking site: (i) it does not noticeably distinguish between cadaverine and putrescine, and (ii) it does not require a second amino group to bind the ligand. Since TAAR13c activation strongly depends on the ligand’s chain length and absolutely requires the second positive charge of the ligand6, this second docking site containing Asp2796.58 is rather unlikely to serve as the ligand binding site that activates the receptor. In order to provide independent experimental proof for the predicted docking residues, we generated a series of receptor mutants by site-directed mutagenesis and studied receptor activity after heterologous expression in mammalian cells.
Mutation of the aminergic DW motif shows only the aspartate as required for ligand binding
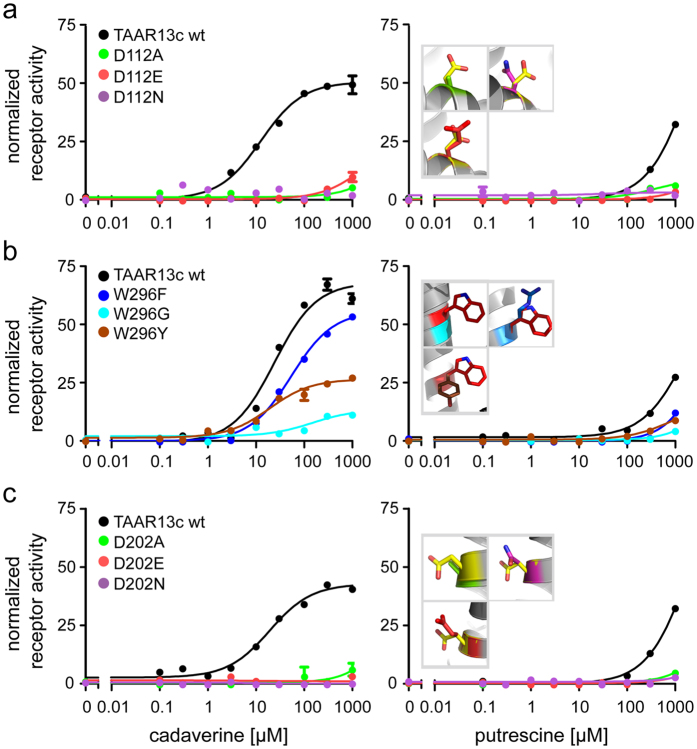
As described above, docking predicted only the Asp1123.32 but not the Trp2967.40 residue of the conserved DW motif to interact with cadaverine (Fig. 2) and putrescine. A series of substitutions were generated for both residues to examine their effects on receptor activity. For the aspartate we chose D112E as a conservative exchange, since the charge is kept and only its position is slightly changed due to the longer side chain in glutamate. Other mutations employed were D112N, which eliminates the charge, but keeps the polarity, and finally D112A, which removes both the charge and polarity. Mutant receptors were stably transfected into HEK293 cells that constitutively express the A2 subunit of an olfactory cyclic nucleotide-gated (CNG) ion channel (see Mat. & Meth.). Activation of TAAR receptors and subsequent cAMP production could be monitored in these cell lines as elevated Ca2+ levels due to Ca2+ influx through cAMP-dependent opening of the CNG channels. We observed that even the most conservative exchange, D112E, reduced TAAR13c activation drastically, and shifted the dose response curve for cadaverine more than two orders of magnitude to higher concentrations (Fig. 3a, Table 1). Mutation to either asparagine or alanine completely abolished cadaverine-evoked activity. Consistent with these experimental results, docking simulations with cadaverine showed the absence of the wildtype salt bridge in all three mutants (Table 1). Taken together we conclude that Asp1123.32 is a pivotal part of the binding site leading to activation of the TAAR13c receptor by cadaverine.
Figure 3.
Concentration-response curves of TAAR13c mutated in the internal binding site and Trp2967.40. HEK293 cell lines constitutively expressing CNG channels and either wildtype or TAAR13c mutants were incubated with concentration series of cadaverine or putrescine (10 nM to 1 mM). Changes in intracellular Ca2+ were detected by Fluo-4 and calculated as ∆F/F. Values were normalized to the fluorescence ratio obtained with NKH477, an agonist of membrane-bound adenylyl cyclases. Representative binding curves are shown color-coded for mutation (green, A; red, E; magenta, N; blue, F; cyan, G; brown, Y), error bars (SD, n = 4) are shown if exceeding symbol size. Left column, responses to cadaverine; right column, responses to putrescine. Insets in right panels show the positions of the mutant side chains (same color as for the respective binding curves) overlayed over the wildtype residue (D, yellow and W, red, respectively). (a) Mutation of the D112 residue to D112E, D112N and D112A results in almost complete loss of activation by cadaverine, with small residual activity only at the highest concentration tested, 1mM. Activation by putrescine is abolished in all D112 mutants. (b) Mutants W296F, W296Y and even W296G were activated by both cadaverine and putrescine, at similar EC50’s as wildtype TAAR13c. Efficacy for W296F is very similar to wildtype TAAR13c but reduced for W296Y and W296G. (c) Mutation of the D202 residue to D202E, D202N, and D202A results in complete loss of activation by cadaverine and putrescine.
Similar effects were observed for the mutant receptors when putrescine was applied as a ligand (Fig. 3a, Table 1), and again, even the D112E variant displayed a drastic reduction of the putrescine response. These results confirm the involvement of Asp1123.32 also for the predicted interaction with putrescine. We were intrigued that binding to putrescine, which is one methylene group shorter than cadaverine, could not be improved by the longer side chain of glutamate28, and therefore performed docking simulations with the D112E mutant and putrescine. Indeed, the distance between the charged glutamate side chain and the amino group of putrescine was predicted as 9.8 Å (Table 1), well beyond the range of a salt bridge27, and thus consistent with the drastic reduction in affinity observed for the D112E receptor mutant. In fact, docking of putrescine showed for all three mutants that at most one amino group is able to form a typical salt bridge, whereas the other amino group is too far away for a binding interaction (Table 1). Taken together, experimental and modeling results obtained with Asp1123.32 mutants and putrescine again confirmed the involvement of Asp1123.32 in ligand binding as predicted by modeling and docking the wildtype receptor (see above).
The tryptophan residue Trp2967.40 of the DW motif13 was not predicted to be part of the binding site (Fig. 2f). We therefore examined whether replacement of this residue might affect receptor activity. Three mutants were generated, either exchanging the tryptophan for phenylalanine (less bulky), tyrosine (switch to polar residue) or glycine (no side chain interaction possible). Stably transfected cell lines were established with all mutants, and receptor activity was examined with a series of cadaverine and putrescine concentrations (Fig. 3b).
Cadaverine and putrescine were able to activate all three mutants, with similar affinity as wildtype, even for the drastic W296G exchange (Fig. 3b, Table 1), in sharp contrast to the almost complete loss of activity in all Asp1123.32 mutants. Furthermore, the efficacy of W296F was similar to wildtype TAAR13c. A slight reduction of efficacy was observed for the W296Y mutant, which might be caused by a different interaction of the more hydrophilic tyrosine with neighboring side chains compared to the tryptophan in wildtype TAAR13c. Efficacy was strongly reduced in the W296G mutant, conceivably due to a loss of structural stability by insertion of the highly flexible glycine into a transmembrane domain. Taken together, binding experiments for all three mutants, W296F, W296Y, and W296G, show little loss in affinity and (with one exception) efficacy, confirming that Trp2967.40 is irrelevant for the binding interaction to cadaverine and putrescine, as predicted by the theoretical model.
A second aspartate, Asp2025.42 in TM5 is required for activation of TAAR13c by cadaverine and putrescine
Docking studies suggested an interaction of Asp2025.42 with the second amino group of cadaverine (Fig. 2c). We followed a similar strategy as before and generated cell lines expressing the following receptor mutants: D202E, D202N, and D202A. Functional testing with cadaverine revealed that even the most subtle replacement, the D202E substitution, eliminated the activity of the receptor completely (Fig. 3c). The same results were obtained for the other substitutions of D202 to either asparagine or alanine (Fig. 3c). Docking simulations for cadaverine with the mutants showed a large distance of about 10 Å between the mutated residues and the second amino group of cadaverine, consistent with a loss of the second salt bridge present in the wildtype receptor (Table 1). Hence we conclude that Asp2025.42 is another pivotal residue in the cadaverine binding site and participates in the activation of the TAAR13c receptor.
As observed for cadaverine, all three receptor variants remained quiescent when cell lines were treated with putrescine (Fig. 3, Table 1). Docking data showed that the distance between the glutamate and the second amino group of putrescine was about twice as large as in the wildtype receptor (Table 1), consistent with the inability of putrescine to activate the D202E mutant receptor. A similar result was obtained for the other two mutations (Table 1). Thus, all experiments performed with Asp2025.42 mutants are consistent with docking results for these mutants, and confirm the prediction of Asp2025.42 as an essential component of the diamine binding site that can activate TAAR13c.
Elimination of the external binding site for diamines generates a supersensitive receptor
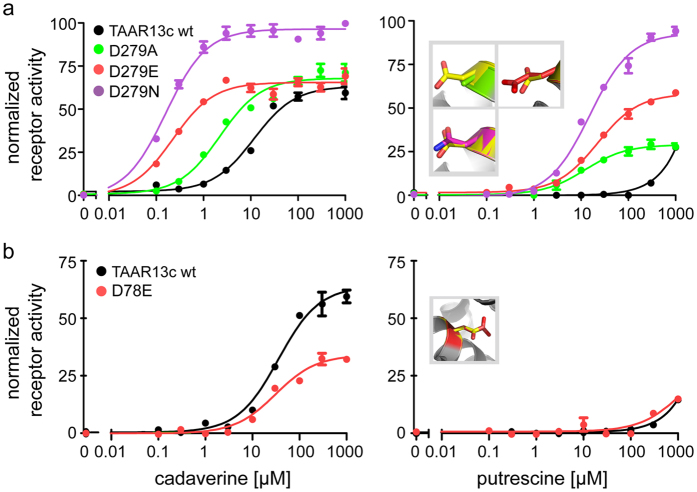
To examine the relevance of the potential binding site on the extracellular face of TAAR13c predicted by docking, we generated three variants of Asp2796.58, i.e. D279E, D279N, and D279A, and expressed them stably in cell lines. All three receptor variants displayed a massive increase in affinity and efficacy (Fig. 4a). The most pronounced effect was observed for D279E, whose apparent affinity (EC50) to cadaverine increased over twentyfold (Table 1). For the D279N and the D279A mutant receptors we observed a 14fold and a 7fold increase of EC50, respectively (Table 1). The efficacy for all three mutants increased as well, with the most pronounced effect seen for the D279N mutant - about twice the value of wildtype TAAR13c (Fig. 4a). Hence modifications of Asp2796.58 result in supersensitive receptors. A similar increase in affinity and efficacy of these three receptor variants was measured when cell lines were treated with putrescine (Fig. 4a, Table 1). Notably, EC50’s of the mutants for putrescine reached values of 7–30 μM, very similar to those determined for cadaverine on wildtype TAAR13c.
Figure 4. Concentration-response curves of TAAR13c mutated in the external binding site and a control site.
HEK293 cell lines constitutively expressing CNG channels and either wildtype or TAAR13c mutants were incubated with concentration series of cadaverine or putrescine (10 nM to 1 mM). Changes in intracellular Ca2+ were detected by Fluo-4 and calculated as ∆F/F. Values were normalized to the fluorescence ratio obtained with NKH477, an agonist of membrane-bound adenylyl cyclases. Representative binding curves are shown color-coded for mutation (green, A; red, E; magenta, N; blue, F; cyan, G; brown, Y), error bars (SD, n = 4) are shown if exceeding symbol size. Left column, responses to cadaverine; right column, responses to putrescine. Insets in right panels show the positions of the mutant side chains (same color as for the respective binding curves) overlayed over the wildtype residue (D, yellow and W, red, respectively). (a) For mutant D279E the apparent affinity to cadaverine and putrescine as estimated by EC50 increased about twenty and hundredfold, respectively, compared to wildtype. EC50 values for binding of cadaverine to D279N and D279A mutants increased 14fold and 7fold, respectively. Notably, the EC50 values for binding of putrescine to D279N and D279A mutants were as low as those observed for wildtype TAAR13c binding to cadaverine. (b) Mutant D78E bound cadaverine and putrescine similarly well compared to wildtype. Since this aspartate residue is located in the TM region, but outside of the ligand binding site, this result shows that an exchange of aspartate to glutamate in the TM region per se does not impair receptor activity.
When we performed docking simulations, no external binding site was predicted for all three mutants, i.e. the modification of Asp2796.58 eliminated this binding site. The internal binding site, however, remained intact, with distances to the ligand amino groups in the range of those observed for wildtype TAAR13c (Table 1), and similar subsets of additional contact sites (Supplementary Table 1).
These observations are consistent with the hypothesis that a ligand-occupied external binding site blocks access to the internal binding site. Elimination of the external binding site relieves this block, leading to an increase in effective ligand concentration in the vicinity of the internal binding site, thus generating a supersensitive receptor.
Discussion
It is a complex but fascinating challenge to gain deeper understanding how the binding of a multitude of different odor molecules to a large cohort of olfactory receptors evokes specific cellular responses. In recent years ligands have been reported for many olfactory receptors4,6,29,30. However, only in very few cases do we possess a molecular understanding of the binding interaction between an odorant and its receptor9. So far crystal structures are not available for any olfactory receptor, and thus prediction of olfactory receptor structures has relied on modeling studies. Here we used modeling and docking to predict binding interactions of a receptor linked to a robust innate avoidance behavior. This receptor is a member of the TAAR family of olfactory receptors, TAAR13c, which is activated by the death-associated odor cadaverine in the micromolar range, and, much less efficiently, by the closely related diamine putrescine6.
We applied a two-pronged approach to elucidate the mechanism of TAAR13c activation: (i) receptor modeling and docking to uncover residues participating in ligand binding and receptor activation; (ii) site-directed mutagenesis of predicted binding residues followed by functional analysis in an heterologous expression system. For wildtype TAAR13c and all mutants we performed docking simulations with cadaverine and putrescine, and determined bond length between pivotal binding residues of the receptor and ligand amino groups as an estimate of binding strength. In all cases predicted alterations in bond lengths of the mutant receptors were consistent with the experimentally observed changes in activity profiles.
Our results identified Asp1123.32 and Asp2025.42, but not Trp2967.40 as essential elements of a binding site for cadaverine and putrescine in the upper third of the TM region (internal binding site). The results for the mutation of the two aspartates to alanine reaffirm a recent independent study31. Our broader approach to the mutational analyses including the conservative exchanges from aspartate to glutamate and asparagine allows us to confidently state that the observed loss of receptor activation is not due to general conformational changes caused by the rather drastic aspartate to alanine exchange. Furthermore, we also examined whether an aspartate to glutamate transition as such could impair receptor function. We mutated an aspartate residue within the TM region of the receptor, but outside of the predicted binding site (D782.50) to glutamate. This mutation did not impair the receptor’s properties, strongly suggesting that the exchange of an aspartate to glutamate in the TM region as such is not sufficient to generate coarse conformational changes resulting in a loss of receptor function (Fig. 4b). Similarly, mutations of Trp2967.40 did not alter the receptor’s activation profile by cadaverine and putrescine. Thus, the drastic changes in TAAR13c affinity seen even for the conservative D112E and D202E exchanges most likely are due to the structural change within the binding pocket. The requirement for binding residues for two functional groups we reported here is fully consistent with the ligand profile of TAAR13c6.
Asp1123.32 and Trp2967.40 together constitute the so-called DW motif, which is highly conserved in aminergic receptors25,32. Asp1123.32 is assumed to coordinate the amino group of amine-containing ligands, and Trp2967.40 is supposed to interact with the apolar or aromatic backbone. However, the orientation of the diamines cadaverine and putrescine in the binding pocket necessitated by coordination of the second amino group to Asp2025.42 precludes coordination of the backbone by Trp2967.40, in accordance with the experimental results.
Interestingly, a second binding site for cadaverine and putrescine was predicted at the external surface of TAAR13c, right above the internal binding site discussed in the preceding paragraph. Mutation of the pivotal element of this binding site, Asp2796.58, resulted in a supersensitive receptor, which displayed an increase in apparent affinity (EC50) for cadaverine and putrescine of up to two orders of magnitude. This rather unexpected result prompted us to examine the predicted structure of TAAR13c for possible mechanistic explanations. Both the internal and the external binding site were predicted by modeling and confirmed by mutational analysis as discussed above. However, whereas the internal binding site (Asp1123.32, Asp2025.42) is necessary for receptor activation, the external binding site (Asp2796.58) is not, and in fact impairs receptor activation.
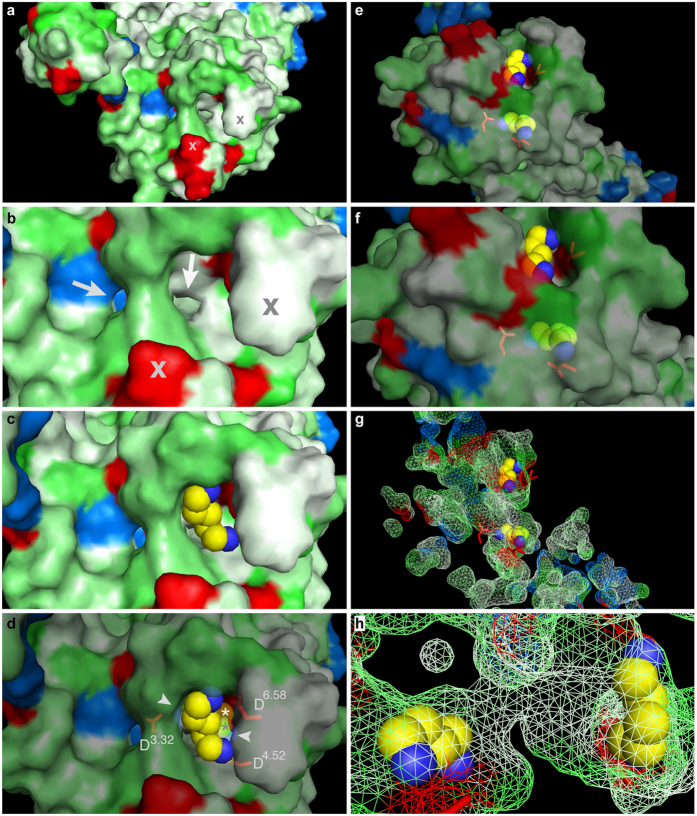
We hypothesized that the external binding site, when occupied, might block access of the ligand to the internal binding site. There are only two conceivable access points to the internal binding site, highlighted in the surface view of the docking model (Fig. 5a,b). One of these sites is flanked by a positively charged residue, Arg922.64. This constraint makes it unlikely to allow passage of positively charged diamine compounds into the receptor’s internal binding pocket.
Figure 5. Occupied external binding site blocks access of cadaverine to the interior binding and activation site.
Surface view (panels a–f) and mesh view (panels g,h) of the TAAR13c model docked with cadaverine visualizes a potential access path for cadaverine. (a) View from the extracellular surface; red, acidic residues; blue, basic residues; green, polar residues; white, hydrophobic residues; cross, same protrusions as marked in panel (b). (b) Enlarged detail of panel a; arrows point to two potential access paths for passage of cadaverine to the internal binding site. Note that the positively charged Arg922.64 precludes access via left route. (c) Cadaverine docked into the external binding site, same view as in panel b. Note that the access to the internal binding site is blocked. (d) Same view as panel (b,c), but partly transparent; aspartates of external (D6.58) and internal (D3.32 and D4.52) binding sites are shown in stick mode; asterisks mark cadaverine bound to the internal binding site; arrowheads point to the amino groups of cadaverine docked into the internal binding site, which lies right below the external binding site shown in top view. (e) Slightly tilted view onto the external surface to display both binding sites at once, external binding site is up. Partial transparency visualizes cadaverine docked into the internal binding site, down; aspartates of external and internal binding sites are shown in stick mode, red. (f) Enlargement of panel (e). (g) Same orientation and magnification as in panel e; mesh view of all predicted cavities, with cadaverine docked into the external and internal binding site. (h) Enlarged view turned 90° counter clockwise and 90° front-to-back compared to panel (g). Note the tunnel connecting the external (left) and internal (right) binding site.
The second access point has no such restrictions and thus appears suitable to allow access of the ligand to the internal binding site (Fig. 5b,h). This access point is located directly below the external binding site and thus can be expected to be blocked as long as the external binding site is occupied by ligand (Fig. 5b,c). Only after dissociation of the ligand from the external binding site there is a chance for it to travel towards the internal binding site (Fig. 5h). Thus the external binding site acts as a gate that may be closed (binding site occupied by ligand) or open (binding site not occupied). Elimination of the external binding site by exchanging Asp2796.58 destroys the gate and ligand access to the internal binding site is no longer impeded by the intermediary step of binding to the external site. Thus, a supersensitive receptor is generated. In the wildtype TAAR13c, the external binding site serves to downregulate the affinity to diamines, conceivably to adjust it to a physiologically meaningful range. This phenomenon constitutes a novel mechanism in olfactory receptor function.
Some recent reports in the literature describe increases in affinity for mutations of the ligand binding site that leads to receptor activation33,34. These effects are generally small (often two fold, in rare cases up to tenfold). In contrast, here we report a considerably stronger effect on receptor affinity - up to two orders of magnitude - by introducing mutations far away from the activating binding site, and also far away from the intracellular signal transducing regions. Downregulation of a receptor’s activity by occupation of an external ligand binding site constitutes a distinct, novel mechanism, by which odorant access to the internal ligand binding site that leads to receptor activation is impaired. Future studies will be required to elucidate, whether such a mechanism might occur in perception of other socially relevant odors, and how widespread it might be in olfactory receptor activation.
During evolution, the affinity of olfactory receptors had to be constantly tuned to physiologically relevant odor concentrations. Sometimes this may have amounted to downtuning the affinity, either directly at the activating binding site, or indirectly via a ligand-regulated gating mechanism such as described here for TAAR13c. Gating mechanisms to regulate open/closed states of ion channels and transmitter receptors are well known. Here we show for the first time that gating also plays a role in olfactory perception.
Materials and Methods
Heterologous expression of TAAR13c receptor mutants
Cell lines that constitutively expressed either the wildtype TAAR13c receptor or a receptor mutant were generated using a previously established protocol35. We used a cell line that had been stably transfected with a gene encoding a variant of the A2-subunit of the olfactory cyclic nucleotide-gated (CNG) ion channel36,37. Approximately 8 μg of the respective TAAR13c expression vectors were introduced into ~4 × 105 cells by a modified calcium-phosphate method38. Stably transfected cells were selected in the presence of the antibiotic G418 (0.8 mg/ml). Expression of TAAR13c was monitored by Western blotting (Supplementary Data 1) with specific anti-TAAR13c antibodies6 and anti-Rhodopsin antibodies (Sigma Aldrich, Taufkirchen, Germany).
Monitoring functional TAAR13c receptor activity in cell lines
Activation of TAAR13c by cadaverine and putrescine evoked a rise in intracellular cAMP concentration that activates the CNG channels35 and thereby causes an influx of Ca2+ through the open channels. Changes in [Ca2+]i were monitored with the Ca2+-sensitive fluorescent dye Fluo-4. Cells were grown in 96-well dishes to a density of approximately 2 × 104 cells per well. Cells were loaded at room temperature with Fluo-4 AM as described previously37. After 90 min, the loading solution was substituted for dye-free ECS (extracellular solution; 120 mM NaCl, 5 mM KCl, 2 mM MgCl2, 2 mM CaCl2, 10 mM HEPES, and 10 mM glucose, pH 7.4 [NaOH]) containing 100 μM IBMX. The plate was transferred into a fluorescence reader (FLUOstar Omega, BMG Labtech, Offenburg, Germany) to monitor Fluo-4 fluorescence. The excitation wavelength was 485 nm. Fluorescence emission was detected at 520 nm. A concentration series of cadaverine or putrescine (10−7 to 10−3 M) as well as 10−5 M NKH477 (positive control) was added once Fluo-4 fluorescence had reached a stable value in each well. The changes in Fluo-4 fluorescence were recorded automatically. The fluorescence signal generated by the adenylyl cyclase activator NKH477 was set to 100% as an internal standard. Concentration–response curves were established from at least three independent experiments with quadruplicate measurements in each experiment.
Data were analyzed and displayed using Prism 5.04 software (GraphPad, San Diego, CA, USA).
Introduction of mutations into TAAR13c
As starting point for mutagenesis we used a full length TAAR13c cDNA construct that harbored an N-terminal extension encoding the first 20 amino acids of bovine rhodopsin in pcDNA3.1 (−) expression vector6. Point mutations were introduced using the QuikChange Site-directed mutagenesis kit (Agilent Technologies, Santa Clara, USA). In brief, PCR reactions were performed using PfuUltra High-Fidelity DNA Polymerase with the above described plasmid as a template, and mutagenic primers listed in Table 2. Parental strands, which are methylated in contrast to the PCR products, were selectively digested with Dpn1 enzyme and the resulting product was transformed into XL-1 blue supercompetent E.coli cells by electroporation. Screening of recombinants was done by colony PCR using wild type TAAR13c primers. Colonies positive for the desired mutation were grown under standard conditions in LB broth and the plasmids were isolated using a plasmid DNA purification kit from Zymo research (California, USA). All mutations were verified by DNA sequencing.
Table 2. Sequences of mutagenic primers used.
| Wildtype/Mutant | Forward Primer, 5′-3′orientation | Reverse Primer, 5′-3′ orientation |
|---|---|---|
| TAAR13c | atggatttatcatcacaagaa | tcaaaccgtaaataaattgat |
| D112A | ccggttttgccctgtttctcac | gtgagaaacagggcaaaaccgg |
| D112N | acaccggttttaacctgtttctcac | gtgagaaacaggttaaaaccggtgt |
| D112E | ccggttttgaactgtttctcac | gtgagaaacagttcaaaaccgg |
| D202A | tggtcagttttagccacattacta | tagtaatgtggctaaaactgacca |
| D202N | tggtcagttttaaacacattacta | tagtaatgtgtttaaaactgacca |
| D202E | tggtcagttttagagacattacta | tagtaatgtctctaaaactgacca |
| D279A | actctctggtggctccctacattaac | gttaatgtagggagccaccagagagt |
| D279N | actctctggtgaatccctacattaac | gttaatgtagggattcaccagagagt |
| D279E | actctctggtggagccctacattaac | gttaatgtagggctccaccagagagt |
| W296G | tgatgcatttggtgggttaggctacac | gtgtagcctaacccaccaaatgcatca |
| W296Y | tgatgcatttggttacttaggctacac | gtgtagcctaagtaaccaaatgcatca |
| W296F | tgatgcatttggtttcttaggctacac | gtgtagcctaagaaaccaaatgcatca |
| D78E | ctggctctggcggaactgctgg | ccagcagttccgccagagccag |
Homology modeling and ligand docking of TAAR13c
Homology models of TAAR13c were generated using GPCR-I-TASSER39 based on the crystal structure of six homologous templates and sequence alignments of these templates with TAAR13c (Supplementary Data 2). The sequence alignments were verified by inspection for proper aligning of conserved motifs and disulfide bridges. The model with the highest C-score (−0.36), which is well within the confidence range40, was chosen as the final structure. In initial experiments the mutant structure was generated with the above protocol and compared to a mutant model generated using side chain substitution in the wildtype structure using Chimera41. We observed no detectable RMSD difference (<0.05) between models generated by these two methods. Thus, subsequently homology models for mutants were generated by side chain substitution in the wildtype homology model. The receptor model was prepared for docking by adding protons and charges were assigned to ionizable side chains using the DockPrep function in Chimera. The model was then subjected to minimization using ModRefiner42. The final model backbone conformation was inspected by Ramachandran plot using Rampage43 (Supplementary Fig. 1). The ligand was used with flexible, rotatable bonds and was docked into the binding site using Autodock Vina44 with default parameter setting (iterated local search global optimizer)44. The resulting conformations were analyzed using AutoDock Tools45. PyMol46 was used for visualization of various ligand conformations and for preparing figures.
Additional Information
How to cite this article: Sharma, K. et al. Elimination of a ligand gating site generates a supersensitive olfactory receptor. Sci. Rep. 6, 28359; doi: 10.1038/srep28359 (2016).
Supplementary Material
Acknowledgments
We thank Lena Sachs, who was involved in an early stage of the project. We gratefully acknowledge financial support from the German Science foundation (grants KO 1046/7-1 and RTG 1960 to S.I.K.), and the International Graduate Schools IGSDHD (K.S., G.A.) and IGS-GFG (A.H.).
Footnotes
The authors declare no competing financial interests.
Author Contributions The experiments were conceived by S.I.K. and designed by K.S., G.A., S.I.K. and A.B. Experiments were performed by S.B., K.S., G.A. and A.H. Illustrations were drafted by K.S., S.B. and S.I.K. Data analysis was done by K.S. and S.B. K.S., A.B. and S.I.K. wrote the paper.
References
- Strotmann J., Levai O., Fleischer J., Schwarzenbacher K. & Breer H. Olfactory receptor proteins in axonal processes of chemosensory neurons. The Journal of neuroscience: the official journal of the Society for Neuroscience 24, 7754–7761, doi: 10.1523/JNEUROSCI.2588-04.2004 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young J. M. & Trask B. J. The sense of smell: genomics of vertebrate odorant receptors. Hum Mol Genet 11, 1153–1160 (2002). [DOI] [PubMed] [Google Scholar]
- Grosmaitre X. et al. SR1, a mouse odorant receptor with an unusually broad response profile. The Journal of neuroscience: the official journal of the Society for Neuroscience 29, 14545–14552, doi: 10.1523/JNEUROSCI.2752-09.2009 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito H., Chi Q., Zhuang H. & Matsunami H. & Mainland, J. D. Odor coding by a Mammalian receptor repertoire. Sci Signal 2, ra9, doi: 10.1126/scisignal.2000016 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J. & Webb D. M. Evolutionary deterioration of the vomeronasal pheromone transduction pathway in catarrhine primates. Proc Natl Acad Sci USA 100, 8337–8341, doi: 10.1073/pnas.1331721100 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain A. et al. High-affinity olfactory receptor for the death-associated odor cadaverine. Proc Natl Acad Sci USA 110, 19579–19584, doi: 10.1073/pnas.1318596110 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Pacifico R., Cawley D., Feinstein P. & Bozza T. Ultrasensitive detection of amines by a trace amine-associated receptor. The Journal of neuroscience: the official journal of the Society for Neuroscience 33, 3228–3239, doi: 10.1523/JNEUROSCI.4299-12.2013 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachse S., Rappert A. & Galizia C. G. The spatial representation of chemical structures in the antennal lobe of honeybees: steps towards the olfactory code. The European journal of neuroscience 11, 3970–3982 (1999). [DOI] [PubMed] [Google Scholar]
- Araneda R. C., Kini A. D. & Firestein S. The molecular receptive range of an odorant receptor. Nat Neurosci 3, 1248–1255, doi: 10.1038/81774 (2000). [DOI] [PubMed] [Google Scholar]
- Cherezov V. et al. High-Resolution Crystal Structure of an Engineered Human β2-Adrenergic G Protein–Coupled Receptor. Science 318, 1258–1265, doi: 10.1126/science.1150577 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palczewski K. et al. Crystal structure of rhodopsin: A G protein-coupled receptor. Science 289, 739–745 (2000). [DOI] [PubMed] [Google Scholar]
- Kobilka B. K. et al. Chimeric alpha 2-,beta 2-adrenergic receptors: delineation of domains involved in effector coupling and ligand binding specificity. Science 240, 1310–1316 (1988). [DOI] [PubMed] [Google Scholar]
- Hussain A., Saraiva L. R. & Korsching S. I. Positive Darwinian selection and the birth of an olfactory receptor clade in teleosts. Proc Natl Acad Sci USA 106, 4313–4318, doi: 10.1073/pnas.0803229106 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain A. The Olfactory Nervous System Of Terrestrial And Aquatic Vertebrates. Nature precedings hdl:10101/npre.2011.6642.1 (2011). [Google Scholar]
- Warne T. et al. Structure of a beta1-adrenergic G-protein-coupled receptor. Nature 454, 486–491, doi: 10.1038/nature07101 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchberg K. et al. Conformational dynamics of helix 8 in the GPCR rhodopsin controls arrestin activation in the desensitization process. Proc Natl Acad Sci USA 108, 18690–18695, doi: 10.1073/pnas.1015461108 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbaum D. M., Rasmussen S. G. & Kobilka B. K. The structure and function of G-protein-coupled receptors. Nature 459, 356–363, doi: 10.1038/nature08144 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheatley M. et al. Lifting the lid on GPCRs: the role of extracellular loops. Br J Pharmacol 165, 1688–1703, doi: 10.1111/j.1476-5381.2011.01629.x (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rovati G. E., Capra V. & Neubig R. R. The highly conserved DRY motif of class A G protein-coupled receptors: beyond the ground state. Mol Pharmacol 71, 959–964, doi: 10.1124/mol.106.029470 (2007). [DOI] [PubMed] [Google Scholar]
- Yang J., Roy A. & Zhang Y. Protein-ligand binding site recognition using complementary binding-specific substructure comparison and sequence profile alignment. Bioinformatics 29, 2588–2595, doi: 10.1093/bioinformatics/btt447 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Roy A. & Zhang Y. BioLiP: a semi-manually curated database for biologically relevant ligand-protein interactions. Nucleic acids research 41, D1096–1103, doi: 10.1093/nar/gks966 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang E. S. Construction of a sequence motif characteristic of aminergic G protein-coupled receptors. Protein science: a publication of the Protein Society 12, 1360–1367, doi: 10.1110/ps.0305603 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blundell T., Barlow D., Borkakoti N. & Thornton J. Solvent-induced distortions and the curvature of alpha-helices. Nature 306, 281–283 (1983). [DOI] [PubMed] [Google Scholar]
- Rosenbaum D. M. et al. GPCR engineering yields high-resolution structural insights into beta2-adrenergic receptor function. Science 318, 1266–1273, doi: 10.1126/science.1150609 (2007). [DOI] [PubMed] [Google Scholar]
- Surgand J. S., Rodrigo J., Kellenberger E. & Rognan D. A chemogenomic analysis of the transmembrane binding cavity of human G-protein-coupled receptors. Proteins 62, 509–538, doi: 10.1002/prot.20768 (2006). [DOI] [PubMed] [Google Scholar]
- Rowland R. S. & Taylor R. Intermolecular Nonbonded Contact Distances in Organic Crystal Structures: Comparison with Distances Expected from van der Waals Radii. The Journal of Physical Chemistry 100, 7384–7391, doi: 10.1021/jp953141+ (1996). [DOI] [Google Scholar]
- Barlow D. J. & Thornton J. M. Ion-pairs in proteins. J Mol Biol 168, 867–885 (1983). [DOI] [PubMed] [Google Scholar]
- Hall S. E., Floriano W. B., Vaidehi N. & Goddard W. A. 3rd. Predicted 3-D structures for mouse I7 and rat I7 olfactory receptors and comparison of predicted odor recognition profiles with experiment. Chemical senses 29, 595–616, doi: 10.1093/chemse/bjh063 (2004). [DOI] [PubMed] [Google Scholar]
- Krautwurst D., Yau K. W. & Reed R. R. Identification of ligands for olfactory receptors by functional expression of a receptor library. Cell 95, 917–926 (1998). [DOI] [PubMed] [Google Scholar]
- Li J., Haddad R., Chen S., Santos V. & Luetje C. W. A broadly tuned mouse odorant receptor that detects nitrotoluenes. Journal of neurochemistry 121, 881–890, doi: 10.1111/j.1471-4159.2012.07740.x (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q. et al. Non-classical amine recognition evolved in a large clade of olfactory receptors. Elife 4, doi: 10.7554/eLife.10441 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katritch V., Cherezov V. & Stevens R. C. Structure-function of the G protein-coupled receptor superfamily. Annu Rev Pharmacol Toxicol 53, 531–556, doi: 10.1146/annurev-pharmtox-032112-135923 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katada S., Hirokawa T., Oka Y., Suwa M. & Touhara K. Structural basis for a broad but selective ligand spectrum of a mouse olfactory receptor: mapping the odorant-binding site. The Journal of neuroscience: the official journal of the Society for Neuroscience 25, 1806–1815, doi: 10.1523/jneurosci.4723-04.2005 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato A., Katada S. & Touhara K. Amino acids involved in conformational dynamics and G protein coupling of an odorant receptor: targeting gain-of-function mutation. Journal of neurochemistry 107, 1261–1270, doi: 10.1111/j.1471-4159.2008.05693.x (2008). [DOI] [PubMed] [Google Scholar]
- Wachten S., Schlenstedt J., Gauss R. & Baumann A. Molecular identification and functional characterization of an adenylyl cyclase from the honeybee. Journal of neurochemistry 96, 1580–1590, doi: 10.1111/j.1471-4159.2006.03666.x (2006). [DOI] [PubMed] [Google Scholar]
- Ludwig J., Margalit T., Eismann E., Lancet D. & Kaupp U. B. Primary structure of cAMP-gated channel from bovine olfactory epithelium. FEBS Lett 270, 24–29 (1990). [DOI] [PubMed] [Google Scholar]
- Balfanz S. et al. Functional characterization of transmembrane adenylyl cyclases from the honeybee brain. Insect biochemistry and molecular biology 42, 435–445, doi: 10.1016/j.ibmb.2012.02.005 (2012). [DOI] [PubMed] [Google Scholar]
- Chen C. & Okayama, H. High-efficiency transformation of mammalian cells by plasmid DNA. Mol Cell Biol 7, 2745–2752 (1987). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Yang J., Jang R. & Zhang Y. GPCR-I-TASSER: A Hybrid Approach to G Protein-Coupled Receptor Structure Modeling and the Application to the Human Genome. Structure 23, 1538–1549, doi: 10.1016/j.str.2015.06.007 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge L. et al. Differential proteomic analysis of the anti-depressive effects of oleamide in a rat chronic mild stress model of depression. Pharmacol Biochem Behav 131, 77–86, doi: 10.1016/j.pbb.2015.01.017 (2015). [DOI] [PubMed] [Google Scholar]
- Pettersen E. F. et al. UCSF Chimera–a visualization system for exploratory research and analysis. Journal of computational chemistry 25, 1605–1612, doi: 10.1002/jcc.20084 (2004). [DOI] [PubMed] [Google Scholar]
- Xu D. & Zhang Y. Improving the physical realism and structural accuracy of protein models by a two-step atomic-level energy minimization. Biophys J 101, 2525–2534, doi: 10.1016/j.bpj.2011.10.024 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovell S. C. et al. Structure validation by Calpha geometry: phi,psi and Cbeta deviation. Proteins 50, 437–450, doi: 10.1002/prot.10286 (2003). [DOI] [PubMed] [Google Scholar]
- Trott O. & Olson A. J. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. Journal of computational chemistry 31, 455–461, doi: 10.1002/jcc.21334 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris G. M. et al. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. Journal of computational chemistry 30, 2785–2791, doi: 10.1002/jcc.21256 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrödinger L. L. C. The PyMOL Molecular Graphics System. Version 1.8 (2015). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.