Abstract
Background
Intrauterine growth restriction (IUGR), which refers to reduced fetal growth in the context of placental insufficiency, is etiologically heterogeneous. IUGR is associated not only with perinatal morbidity and mortality but also with adult-onset disorders, such as cardiovascular disease and diabetes, posing a major health burden. Placental epigenetic dysregulation has been proposed as one mechanism that causes IUGR; however, the spectrum of epigenetic pathophysiological mechanisms leading to IUGR remains to be elucidated. Monozygotic monochorionic twins are particularly affected by IUGR, in the setting of severe discordant growth. Because monozygotic twins have the same genotype at conception and a shared maternal environment, they provide an ideal model system for studying epigenetic dysregulation of the placenta.
Results
We compared genome-wide placental DNA methylation patterns of severely growth-discordant twins to identify novel candidate genes for IUGR. Snap-frozen placental samples for eight severely growth-discordant monozygotic monochorionic twin pairs were obtained at delivery from each twin. A high-resolution DNA methylation array platform was used to identify methylation differences between IUGR and normal twins. Our analysis revealed differentially methylated regions in the promoters of eight genes: DECR1, ZNF300, DNAJA4, CCL28, LEPR, HSPA1A/L, GSTO1, and GNE. The largest methylation differences between the two groups were in the promoters of DECR1 and ZNF300. The significance of these group differences was independently validated by bisulfite pyrosequencing, implicating aberrations in fatty acid beta oxidation and transcriptional regulation, respectively. Further analysis of the array data identified methylation changes most prominently affecting the Wnt and cadherin pathways in the IUGR cohort.
Conclusions
Our results suggest that IUGR in monozygotic twins is associated with impairments in lipid metabolism and transcriptional regulation as well as cadherin and Wnt signaling. We show that monozygotic monochorionic twins discordant for growth provide a useful model to study one type of the epigenetic placental dysregulation that drives IUGR.
Electronic supplementary material
The online version of this article (doi:10.1186/s13148-016-0238-x) contains supplementary material, which is available to authorized users.
Keywords: Intrauterine growth restriction, Monozygotic twins, Monochorionic twins, DNA methylation, Wnt pathway, Cadherin pathway, DECR1, ZNF300, LEPR
Background
Fetal growth restriction is considered a disorder with significant etiologic heterogeneity, as there are many known fetal and maternal causes of growth restriction [1]. When growth restriction occurs in the context of placental insufficiency, it is termed intrauterine growth restriction (IUGR). IUGR, which affects 10–15 % of pregnancies [2], is associated not only with significant perinatal morbidity and mortality [2] but also with adult-onset diseases, such as cardiovascular disease and diabetes [3–5]. Thus, IUGR poses a major human health burden. The association between IUGR and adult-onset disease is thought to result from fetal programming, wherein fetal adaptive mechanisms “set” metabolic regulatory pathways in response to fetal environments, such as limited nutrition. Consequently, in a society where postnatal nutrient supply exceeds the prenatally “sensed” demands, these metabolic set points predispose the organism to disease later in life [3–5]. The molecular basis of fetal programming is likely to be mediated by epigenetic mechanisms such as DNA methylation.
Early embryonic development depends on normally functioning epigenetic modifications such as DNA methylation, to guide spatial and temporal gene activation and repression that mediate normal growth, cell differentiation and morphogenesis [6]. Likewise, normal placental growth requires appropriate epigenetic regulation of gene expression.
The spectrum of epigenetic pathophysiological mechanisms leading to IUGR remains to be elucidated. These epigenetic alterations, which can involve the fetus and/or the placenta, may affect expression of imprinted or non-imprinted genes important for normal fetal growth and development. Imprinted genes are normally expressed from a single allele in a parent of origin-specific manner [6]. This expression pattern is coordinated via CpG-rich differentially methylated regions (DMRs) in imprinted domains. Loss of imprinting, characterized by aberrant methylation of an imprinted DMR, may restrict the growth of both the placenta and the fetus, causing IUGR, as in the undergrowth disorder Russell-Silver syndrome (OMIM 180860) [6].
IUGR due to placental insufficiency typically exhibits specific diagnostic vascular and pathological placental features. It has been proposed that the intrauterine environment can affect placental development and alter placental gene expression, thereby reducing the placenta’s ability to support optimal fetal growth. Studies investigating placental epigenetic dysregulation have identified altered DNA methylation of genes involved in placental development, including trophoblast differentiation, angiogenesis, and endocrine signaling [7]. Placental epigenetic dysregulation, in particular altered DNA methylation, has been proposed as a mechanism underlying the aberrant gene expression in the pathophysiology of IUGR [7].
Monochorionic (MC) twins are monozygotic (MZ; i.e., identical) twins that share the same placenta. They provide an ideal model for studying epigenetic regulation of the placenta because of their identical genotypes and shared maternal environment. Monochorionic twins are also particularly affected by IUGR in the setting of severe discordant growth, which occurs in 20 % of MC pregnancies [8, 9]. Severe discordant growth is a major contributor to perinatal morbidity and mortality in MC twin pregnancies. Growth discordance is 2.5-fold higher in MC versus dichorionic twin pregnancies [10]. IUGR in the setting of severe growth discordance of MC twins is typically characterized by unequal placental share, where the smaller twin has a smaller share of the placenta [11]. A better understanding of the placental dysfunction underlying MC growth discordance not only holds promise for improving postnatal outcomes in this population but will also offer clues to understanding IUGR on a wider scale in singletons and the molecular basis of fetal programming relevant to increased risks for adult-onset disease.
In this study, we compared genome-wide placental DNA methylation (DNAm) patterns of severely growth-discordant MC twins. Our microarray analysis identified novel candidate differentially methylated regions for IUGR, including those overlapping the promoters of three genes DECR1, LEPR, and ZNF300. These differences were statistically significant when validated by an independent methodology, bisulfite pyrosequencing. A pathway analysis supported a role for epigenetic dysregulation in IUGR pathogenesis of the Wnt and cadherin pathways, which are known to be important in embryonic development. While a single prior study reported targeted gene expression in growth-discordant MC twins [12], the current study is the first genome-wide investigation of DNAm in MC growth-discordant twins.
Results
We sought to identify novel candidate genes for IUGR by comparing genome-wide placental DNAm using Illumina HumanMethylation450 arrays of eight severely growth-discordant MC twin pairs. The clinical characteristics of the MZ twins in this study are detailed in Additional file 1: Table S1. Growth-discordant twin pairs had birth weight differences between 21 and 59 %. All growth-restricted twins measured below the tenth centile in weight for gestational age, while their normal weight counterparts measured above the tenth centile.
A description of placental pathology for these twins is found in Additional file 1: Table S2. Pathological features typically associated with IUGR, such as velamentous cord insertion, advanced villous maturity, thrombosis, and infarction, were consistently present in the IUGR-affected twin’s placental share.
Genome-wide DNA methylation patterns
Differential DNAm analysis was performed on a total of 399,118 sites after quality control analysis of the data (see the “Methods” section). When comparing global DNAm patterns using unsupervised hierarchical clustering, twins were most similar to their own co-twins in 7/8 discordant twin pairs (Fig. 1). This suggests that IUGR is not associated with genome-wide DNAm changes, a finding previously described in other epigenome-wide association studies [13, 14].
Fig. 1.
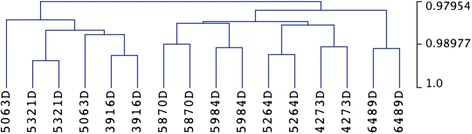
Genome-wide unsupervised hierarchical clustering. Correlation between global DNA methylation patterns involving 399,118 autosomal CpGs was highest between co-twins for 7/8 discordant twin pairs, with the twin pair 5063D being the only exception. This suggests that IUGR is not associated with genome-wide DNA methylation changes. Monozygosity was molecularly confirmed for all twin pairs. Pearson correlation with average linkage was used as the clustering metric, with the scale shown on the right. Samples 4273D, 5264D, 5063D, and 5321D overlap with samples from a previous study [12]
Differentially methylated regions associated with IUGR
To find genomic regions with DNAm differences in IUGR, we used bump hunting [15], which is known to improve the statistical power and robustness by combining methylation patterns across nearby CpG sites in smaller datasets [16]. After accounting for the potential confounding effects of the twins’ sex, gestational age, and maternal age (see the “Methods” section), our analysis revealed eight statistically significant (p < 0.05) DMRs where the magnitude of the DNAm differences between IUGR and the other twin exceeded 10 % (Table 1). These DMRs match the promoters of eight genes (listed in decreasing order of methylation difference between IUGR and controls): DECR1, ZNF300, DNAJA4, CCL28, LEPR, HSPA1A/L, GSTO1, GNE (Table 1). For six of eight genes, the IUGR twin demonstrated a gain of DNAm. Inspection of individual DMRs revealed consistent patterns of DNAm change that also corresponded well with other epigenetic features, such as histone marks, DNase hypersensitivity regions, and transcription factor binding sites (Figs. 2, 3, 4; Additional file 1: Figures S1–S2).
Table 1.
Differentially methylated regions (DMR). Shown are the DMRs with the significance level p < 0.05 as reported by the bumphunter software and spanning at least three CpGs with the average DNA methylation differences (delta beta) between IUGR and healthy twins above 10 % by magnitude in all CpGs. The DMRs are sorted by the magnitude of delta-beta differences. DMR length is measured in basepairs. Gene matches represent the overlaps between the DMR and the gene’s body or promoter region up to 1500 bp from the transcription start site. Genome coordinates are given with respect to hg19 genome assembly
| Chr | Start | End | Δβ | p value | CpGs | Length | Gene |
|---|---|---|---|---|---|---|---|
| 8 | 91013426 | 91013697 | 0.175 | 0.0027 | 6 | 272 | DECR1 |
| 5 | 150284416 | 150284796 | 0.161 | 0.0032 | 7 | 381 | ZNF300 |
| 15 | 78556834 | 78556945 | −0.152 | 0.0082 | 5 | 112 | DNAJA4 |
| 5 | 43396953 | 43397288 | −0.139 | 0.0081 | 6 | 336 | CCL28 |
| 1 | 65991176 | 65991664 | 0.133 | 0.0329 | 3 | 489 | LEPR |
| 6 | 31783322 | 31783482 | 0.130 | 0.0069 | 7 | 161 | HSPA1A/L |
| 10 | 106014615 | 106014893 | 0.126 | 0.0401 | 3 | 279 | GSTO1 |
| 9 | 36258114 | 36258885 | 0.120 | 0.0114 | 6 | 772 | GNE |
Fig. 2.
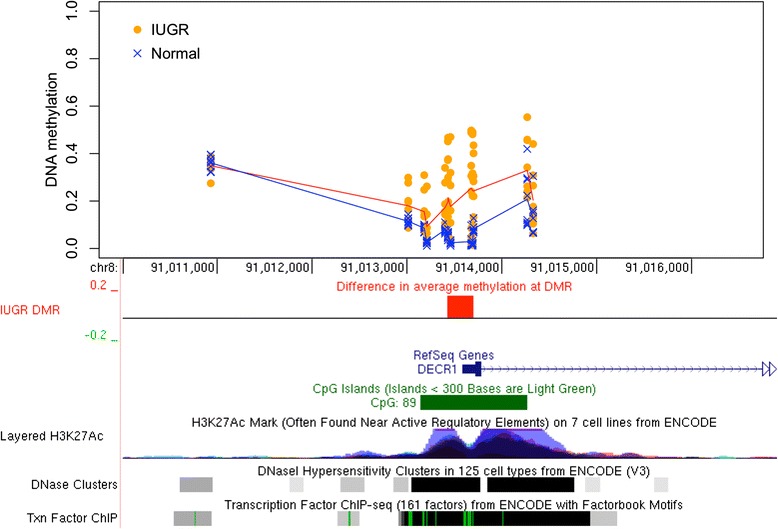
DNAm changes in IUGR twins in DECR1 promoter. Methylation levels in IUGR twins (orange circles) and unaffected co-twins (blue crosses) are shown, arranged vertically for each CpG in the region spanning the DECR1 gene promoter. The mean methylation levels are shown for the IUGR cohort (red line) and controls (blue line). The top six CpGs with methylation change above 10 %, and no more than 500 bp apart, comprise the DMR (red box), which also corresponds to the wider gap between the red and blue lines. Other genomic features, such as CpG islands, histone marks, DNase hypersensitivity clusters, and transcription factor binding sites are also shown. Visualization is provided by the UCSC Genome Browser and was inspired by DMRcate software [66]
Fig. 3.
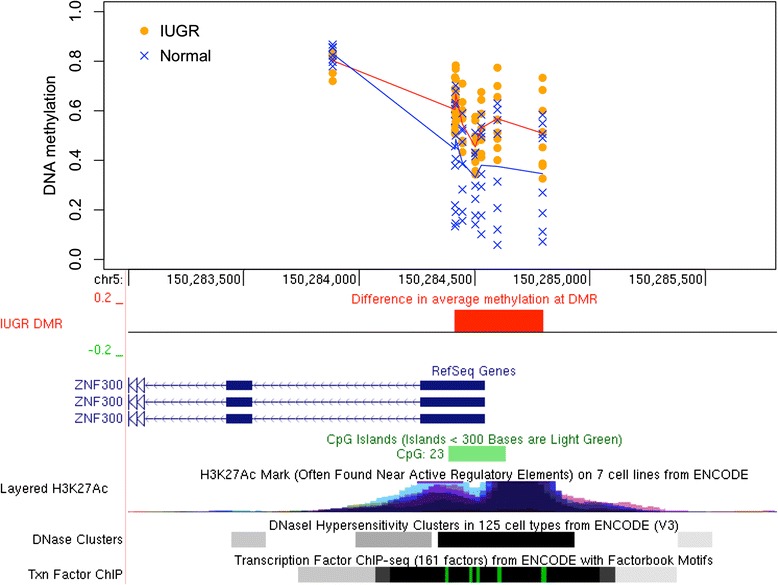
DNAm changes in IUGR twins in ZNF300 promoter. Methylation levels in IUGR twins (orange circles) and unaffected co-twins (blue crosses) are shown, arranged vertically for each CpG in the region spanning the promoter of the ZNF300 gene transcript. The mean methylation levels are shown for the IUGR cohort (red line) and controls (blue line). The top seven CpGs with methylation change above 10 % comprise the DMR (red box), which also corresponds to the wider gap between the red and blue lines
Fig. 4.
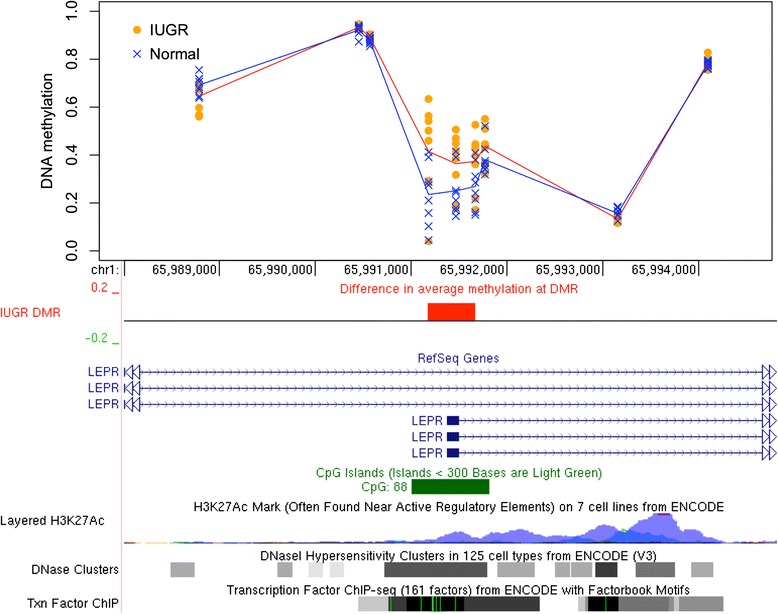
DNAm changes in IUGR twins in LEPR promoter. Methylation levels in IUGR twins (orange circles) and unaffected co-twins (blue crosses) are shown, arranged vertically for each CpG in the region spanning the promoter of one of the LEPR gene transcripts. The mean methylation levels are shown for the IUGR cohort (red line) and controls (blue line). The top three CpGs with methylation change above 10 % comprise the DMR (red box), which also corresponds to the wider gap between the red and blue lines
DECR1, ZNF300, and LEPR DNA methylation results
Three of the eight genes identified in the bump hunting analysis were chosen for validation by targeted bisulfite pyrosequencing (PyroMark Q24; Qiagen) (see the “Methods” section). DECR1 and ZNF300 were selected because they exhibited the largest methylation differences in our analysis, and LEPR was chosen because of its recent association with IUGR in a separate MZ-MC twin study [12].
The largest methylation differences were found in the promoter region of the DECR1 gene, which encodes 2,4 dienoyl-CoA reductase (DECR), a mitochondrial enzyme in the unsaturated fatty acid beta oxidation pathway vital for unsaturated fat metabolism.
Placental samples from the IUGR twins exhibited a gain of methylation in all of the CpG sites within the CpG island located in the DECR1 promoter region, compared to their normal weight counterparts (Fig. 2) (Table 1). The mean DNAm differences in IUGR (also known as delta beta or Δβ) across all twin pairs for the DECR1 was 17.5 % (bump hunting p = 0.0027), reaching as high as 22.5 and 21.4 %, respectively, for the top two differentially methylated CpG sites within the DMR. There was a very substantial gain in DNAm levels in five of eight discordant twin pairs, ranging on average between 14 and 42 % across the DMR, and a small gain in one other pair (4 %). The remaining two twin pairs showed no difference in DNAm (less than 1 % change); see Additional file 1: Figure S3 for details.
The results obtained for the two top DECR1-promoter CpGs, cg01971612 and cg18485485, were independently validated through bisulfite pyrosequencing analysis in the same set of twins (Fig. 5a). A significant difference in DNAm (mean Δβ gain of 12.6 %) was observed in the IUGR compared to the normal twin group for both CpG sites (paired t test, two-tailed, p = 0.021) (Additional file 1: Figure S4, see the “Methods” section).
Fig. 5.
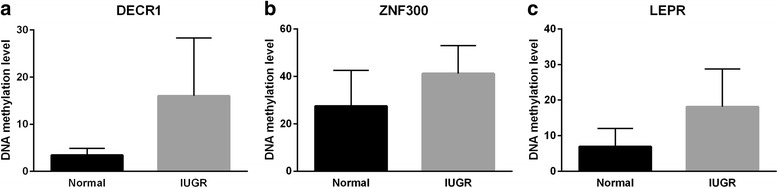
Validation of candidate IUGR regions by bisulfite pyrosequencing. Bar charts representing the DNA methylation levels at two CpG sites overlapping DECR1 (cg01971612, cg18485485) (a), three CpG sites overlapping ZNF300 (cg04675542, cg02343823, cg08580836) (b), and one CpG site overlapping LEPR (cg08234308) (c) in both IUGR (gray color) and healthy co-twins (black color)
Following DECR1, the next largest DMR showing 16 % gain of methylation in IUGR (p = 0.0032) was located in the promoter of the zinc finger protein-coding gene, ZNF300, (Fig. 3), which plays an important role in tumor growth and immune modulation. Five of the twin pairs showed a methylation gain between 18 and 32 % in IUGR at this DMR, with two more pairs showing small or negligible gains (5.4 and 1.3 %, respectively). The remaining one pair showed a small DNAm loss of 4.3 %; see Additional file 1: Figure S5 for details.
The results for three CpG sites (cg04675542, cg02343823, cg08580836) within the ZNF300 DMR were independently validated using targeted bisulfite pyrosequencing analysis (Fig. 5b). Significant differences were demonstrated for the three CpG sites in IUGR compared to the normal twin group (mean Δβ gain of 13.7 %; paired t test, two-tailed, p = 0.016) (Additional file 1: Figure S6).
In the LEPR gene, placental samples from the IUGR twins exhibited a gain of methylation at all CpG sites located near the promoter region (Fig. 4). The DMR showed an average DNAm gain of 13.3 % (p = 0.033) in IUGR twins compared to their healthy counterparts, with four of eight discordant twin pairs having very substantial gain of between 13 and 33 %. A small gain was found in three other pairs (between 3.4 and 6.4 %); the remaining one pair showed negligible DNAm difference (1.7 % change); see Additional file 1: Figure S7 for details.
DNAm levels for the top CpG site (cg08234308) within the LEPR DMR were independently validated using targeted bisulfite pyrosequencing analysis (Fig. 5c) and demonstrated a significant difference (mean Δβ gain of 11.2 %; paired t test, two-tailed, p = 0.022) between the IUGR and the normal twin groups (Additional file 1: Figure S8).
Differentially methylated CpG sites associated with IUGR
To assess the broader functional implications of the genome-wide DNAm changes in IUGR, our analysis was extended to the level of individual CpGs (see the “Methods” section). This analysis identified 296 statistically significant CpG sites (Fig. 6), which overlap 172 genes or gene-promoter regions (Additional file 2: Table S3). As an additional validation step, using these 296 CpGs to quantify the methylation profiles resulted in the IUGR cohort clustering separately from the group of eight unaffected twins (Fig. 7). This suggested that the identified collection of CpGs which overlap the list of eight genomic regions identified by bump hunting is also sufficiently representative of methylation differences in IUGR, and is therefore also likely to reflect broad functional patterns related to the disorder.
Fig. 6.
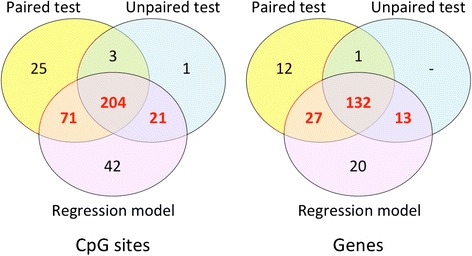
CpG sites used for functional enrichment analysis. Only the CpGs that have a DNAm difference at least 10 % in IUGR twins were considered. A set of 296 CpGs (numbers in red) was selected as the sites that were identified by limma regression analysis after accounting for confounding effects and also passed at least one of the non-parametric tests for IUGR association. The right Venn diagram shows the number of genes matching each criteria of significance, where 172 genes comprise the set tested for functional enrichment
Fig. 7.
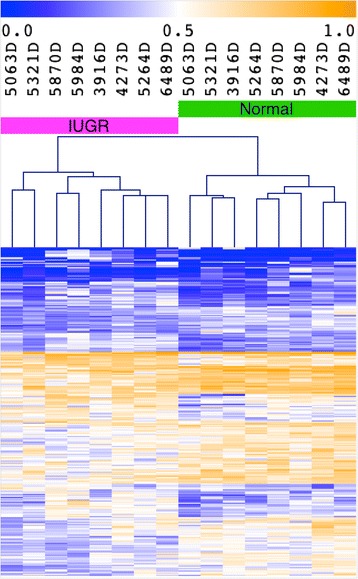
The putatively affected CpGs are able to separate the IUGR cohort from the normal cohort. Clustering heatmap shows IUGR (magenta) and normal (green) twin samples, shown in columns, forming two separate clusters in Manhattan distance metric. Rows represent the 296 affected CpGs. The color scale at the top indicates the range of the DNAm values
Functional enrichment
Enrichment analysis showed a significant over-representation of functional groups highly relevant to the IUGR phenotype, such as fetal development, intracellular signaling, and cell adhesion. For example, 55 of the 172 genes were involved in “anatomical structure development” (p = 1.0e−7 after false discovery rate (FDR) correction) and 40 genes were associated with “nervous system development” (corrected p = 2.6e−10). As many as 70 genes, or 41 % of our list, were annotated as “intrinsic to membrane” (corrected p = 0.011). Other enriched functional groups were related to cell adhesion, developmental processes, and regulation of transcription (Additional file 2: Table S4). We also analyzed the enrichment of the 296 individual CpG sites in the context of much wider genomic regions, extending up to 100,000 bp to the nearest gene, which facilitated the functional assignment of intergenic CpGs. The functional categories detected in this analysis were related to developmental processes, lipid metabolism (e.g., lipopolysaccharide metabolic process, response to cholesterol) and cellular response to various stimuli (Additional file 2: Table S5).
The enriched functional groups such as “intrinsic to membrane,” “anatomical structure development” and “nervous system development” also included most of the genes from the Wnt and cadherin pathways. Analysis of pathway enrichment confirmed that the cadherin pathway, which mediates calcium-ion-dependent cell adhesion, was prominent among the candidate genes (20 genes, p = 5.9e−10 after FDR correction), as was the related Wnt signaling pathway (23 affected genes, p = 5.0e−8 after FDR correction).
Discussion
To our knowledge, this is the first report of genome-wide DNA methylation analysis in growth-discordant MC twins. None of the top eight genes identified in our analysis is associated with a known imprinted region, implicating a mechanism of IUGR that is distinct from those impacted in imprinting disorders. Two of the eight genes differentially methylated in the IUGR group (DECR1 and GSTO1) have been previously associated with IUGR (in animals and singleton humans, respectively), suggesting (at least in part) a common pathogenic mechanism for IUGR in singletons versus that in MC growth-discordant twins [17, 18].
In general, for each DMR, the patterns of DNAm differences in IUGR (Δβ) were consistent across the majority of twin pairs, although the magnitude of Δβ across different twin pairs varied (Additional file 1: Figure S9). Overall, this heterogeneity across the twin pairs is not unusual for either MZ twin studies [14] or for IUGR studies [19].
The IUGR twins in our series exhibited heterogeneity of IUGR placental pathology (Additional file 1: Table S2), which has been shown in other larger scale studies as well [12] and may relate to both cause and/or effect of the heterogeneity in the results of our epigenetic analysis.
Prominent biological networks in IUGR
Our study demonstrates an association of IUGR with DNA methylation changes that impact the regulation of genes relating to lipid metabolism and to nervous system development. Interestingly, neonates with IUGR exhibit reduced lipid stores with significant neonatal metabolic morbidity [20] and are known to suffer neurodevelopmental difficulties compared to age-matched controls [21]. Our results implicate a role for altered DNA methylation in the cause and/or effect of these two major IUGR-related clinical impairments.
The largest methylation differences were found in the promoter region of the DECR1 gene. This gene encodes DECR, the rate-limiting mitochondrial enzyme in unsaturated fatty acid beta oxidation, which is required for growth and neurodevelopment [22] and has also been implicated in adaptive cellular stress-response mechanisms in IUGR animals [17]. An association between unsaturated fatty acids and intrauterine growth has been previously reported in that prenatal supplementation of the unsaturated fatty acid, omega-3, has been shown to increase birth weight [23], while impaired beta oxidation in fetuses with IUGR has also been suggested by several other studies [17, 24, 25]. Biochemically diagnosed DECR deficiency has been reported in two unrelated individuals, who had poor growth, hypotonia, developmental delay, and severe encephalopathy; one of these individuals died in infancy [26, 27]. Studies of Decr1−/− mice suggest that the enzyme is responsible for the induction of gluconeogenesis during fasting [28].
The next largest methylation difference identified in our analysis was a gain of methylation in the promoter of the ZNF300 gene in the IUGR group compared to controls. ZNF300 encodes a zinc finger protein, which functions as a ubiquitous transcriptional repressor [29]. Hypermethylation of this gene has been associated with decreased gene expression, modulation of inflammatory pathways, and tumor development [30]. ZNF300 knockdown studies revealed this gene’s role in cell differentiation [31]. Although ZNF300 has not previously been associated with IUGR, its strong role in cancer development supports its potential to impact fetal growth.
Another highly ranked candidate gene is the LEPR gene, which encodes the leptin receptor. LEPR deficiency in humans leads to morbid obesity and pituitary dysfunction [32–34], and polymorphisms in the LEPR gene have been associated with birth weight [35], obesity [36], and type II diabetes [37]. In animal studies, maternal diet restriction has been associated with subsequent obesity at a few months of age as well as reduced birth weight of the offspring and reduced expression of LEPR [38]. Lepr-deficient (Lepr db/db) mouse models are often used for studying type II diabetes (reviewed in [39]). To date, our study is the first to link increased LEPR methylation with IUGR in humans.
One previous study of growth-discordant MZ-MC twins assessed placental expression of 88 angiogenesis-related genes, the leptin (LEP) and LEPR genes, and also assessed methylation using an assay targeting 64 CpGs overlapping the LEP promoter [12]. They found that the smaller twins exhibited a significant increase in LEP expression along with an average gain of methylation of the LEP promoter region [12]. Four twin pairs used in that study were also used in our study (see the “Methods” section and Additional file 1: Tables S1–S2). It was difficult to compare Schrey et al.’s [12] data to our data as only six of the 63 CpG sites tested in the LEP promoter region are covered by the Illumina 450k array. Our analyses of the six CpGs for the eight MZ twin pairs and the four twin pairs overlapping the two studies did not show any significant gain of methylation in the LEP promoter region. That is, we suggest that the significant differences in LEP methylation found by Schrey and colleagues [12] were driven by CpGs not assessed in our study. The Schrey study also reported a decrease in LEPR expression in the IUGR group, though this difference was not significant [12]. A decrease in LEPR expression would be consistent with the gain of LEPR methylation found in the current study. In general, however, differences in the results of our study and those found by Schrey and colleagues [12] may largely be attributed to the use of different methodologies: genome-wide DNA methylation with limited coverage at individual promoter regions in this study, and gene expression analysis by Schrey and colleagues.
The remainder of the genes with methylation differences identified in the IUGR group (DNAJA4, CCL28, HSPA1A/L, GSTO1, and GNE) function in growth, immunity, and cell metabolism. The GSTO1 gene, which encodes an arsenic detoxification enzyme, glutathione S-transferase omega 1, is the only gene in this list to have a prior association with fetal growth restriction [18]. This enzyme also plays a role in the inflammatory response, apoptosis, and tumor development (reviewed in [40]). The mouse Gsto1 knockout model shows impaired detoxification of arsenic, a known human carcinogen [41]. DNAJA4 and HSPA1A/L encode heat-shock proteins. A variety of tumors exhibit hypermethylation of DNAJA4, which has been associated with gene silencing [42, 43]. Increased expression of DNAJA4 has been shown to suppress tumor invasion [42–44]. CCL28, which encodes the growth factor chemokine ligand 28 [45], increases placental cell apoptosis [46] and is downregulated in a variety of human malignancies [47, 48]. GNE encodes a key enzyme in sialic acid biosynthesis, a process that is often upregulated in tumor development [49]. Reduced sialylation has been shown in heterozygous Gne-deficient mice, while complete knockouts are embryonically lethal [50]. Gne-deficient mouse embryonic stem cells exhibit impaired proliferation and cell differentiation, particularly affecting muscle [51]. In humans, GNE deficiency causes Nonaka myopathy [52]. Collectively, the aberrant methylation of the genes found in our IUGR cohort appears to drive increased apoptosis, impaired cellular metabolism as well as poor growth, and invasion of the placenta.
Prominent cellular pathways in IUGR
The Wnt and cadherin pathways were significantly over-represented in the list of candidate genes generated from 296 individual CpGs representative of the IUGR epigenome. Wnt signaling is well known for its role in carcinogenesis and embryonic development, specifically body axis patterning, cell proliferation, cell migration and cell differentiation [53, 54]. Wnt signaling plays a role in placental development (trophoblast proliferation and invasion) and is altered in gestational diseases (reviewed in [55]). Our group has previously shown that the promoter region of the WNT2 gene is differentially methylated in the placentas of IUGR singletons versus controls [19]. WNT2 gene expression has also been proposed to mediate poor maternal nutrition and fetal growth restriction [56]. Our results suggest that upregulation of DNAJA4 may be an important downstream effect of impaired Wnt signaling in IUGR. Cadherins are a group of “calcium-dependent adhesion” transmembrane proteins that facilitate normal embryonic tissue organization and trophoblast invasion via cell-to-cell adhesion and cell migration (reviewed in [57]).
The precise mechanisms by which Wnt and cadherin signaling play a role in IUGR is unknown. To the best of our knowledge, this is the first report that prominently highlights these pathways in the pathogenesis of IUGR in growth-discordant MC twins.
Conclusions
This is the first report of a genome-wide DNA methylation analysis using the Infinium HumanMethylation450 BeadChip platform to investigate IUGR in MC growth-discordant twins. We identified eight candidate genes that are epigenetically altered in IUGR. Our results suggest that dysregulation of lipid metabolism and transcription, as well as impaired cadherin and Wnt signaling, are associated with IUGR. Collectively, the genes exhibiting differential regulation in growth-restricted fetuses may underlie fetal programming and may generate candidate biomarkers for IUGR and/or its related adult-onset diseases. These results should be validated by further investigation of DNA methylation and/or gene expression of the identified candidate genes in a large cohort of MZ-MC growth-discordant twins. Finally, we propose the growth-discordant MC twin model as a useful tool in the genome-wide study of IUGR.
Methods
Sample selection
This study was approved by the Mount Sinai Hospital (MSH) and the Hospital for Sick Children (SickKids) Research Ethics Boards (REB # 10-0128-E). Participants were recruited from the MSH high-risk obstetrical clinic, following diagnosis of MC discordant growth and placental insufficiency. Informed consent was obtained from each participant. Growth discordance was defined as a birth weight difference >20 % between the co-twins [58]. Birth weight was plotted as percentile for gestational age using a Canadian population-based reference chart [59]. Placental insufficiency was defined as intermittent absent or reversed end-diastolic flow. A full list of the inclusion and exclusion criteria is found in Additional file 1: Table S6.
Following these criteria, we recruited eight MC growth-discordant twin pregnancies.
Placental characteristics are detailed in Additional file 1: Table S2. Four of these pairs overlap with a previous MZ twin study of IUGR that evaluated expression of angiogenesis-related genes and targeted DNA methylation of one gene [12].
Placental sampling was performed immediately after delivery by members of the Research Center for Women’s and Infants’ Health BioBank, Toronto. At delivery, cord clamps were labeled as twin “A” or “B” to distinguish each twin’s placental share. Snap-frozen placental samples were obtained from each twin according to a previously standardized protocol (http://biobank.lunenfeld.ca/). The remainder of each placenta was sent for pathological examination. Monozygosity was molecularly confirmed for all twin pairs.
Genome-wide DNA methylation analysis
Genomic DNA from the placental samples was bisulfite treated (Qiagen). DNA methylation was then analyzed using the Infinium HumanMethylation450 BeadChip array (Illumina, San Diego, CA, USA) following the standard Infinium HD Assay Methylation Protocol Guide (Part #15019519, Illumina). The array assesses DNA methylation at single CpG sites genome-wide; however, it does not differentiate between 5-methylcytosine and 5-hydroxymethylcytosine, which is a recognized limitation of this platform [60].
All samples were run on the array together within the same batch and passed quality control. Data pre-processing such as background subtraction and normalization to internal array controls was performed in Illumina GenomeStudio v2011.1 Methylation Module 1.9 software (http://www.illumina.com).
CpG probe filtering
The 485,577 methylation probes from the HumanMethylation450 array were filtered by removing probes with missing DNA methylation values (beta values either missing or exactly zero) and with detection p values above the significance threshold 0.05. We also removed all cross-reactive probes annotated as such in [61]. To minimize possible confounding effect by ethnicity or sex, especially in the context of unpaired statistical tests between IUGR and control groups (Fig. 6), all probes for which there was a known SNP of at least 1 % minor allele frequency within 5 bp of the targeted cytosine (based on the annotation in [61]) as well as probes located on sex chromosomes were excluded, leaving 399,118 autosomal CpG sites available for analysis.
Detection of differentially methylated regions
To detect the differentially methylated regions, we used the bump hunting method of Jaffe and colleagues [15] implemented in the bumphunter Bioconductor package, setting the cutoff threshold of 10 % for the Δβ methylation difference in IUGR. To account for confounding effects, the model design matrix consisted of an intercept term, IUGR indicator variable, and twin sex, maternal age and twin’s gestational age as additional confounders. Statistical significance of the detected bumps was assessed in comparison to B = 1000 randomized bootstrap iterations. From the resulting collection of bumps, we retained only the DMRs that contained at least three CpGs with gaps no more than 500 bp, and exhibited methylation differences of 10 % or higher in IUGR while satisfying the significance level p < 0.05.
Selection of CpG sites for functional enrichment analysis
For the analysis of methylome-wide functional enrichment in IUGR, we included not only the eight DMRs from the bump hunting analysis, but also the CpG sites that were significantly different in IUGR compared to healthy co-twins. These individual CpG sites were identified using a combination of statistical criteria to account for the small sample size.
Three sets of statistical tests were performed on eight growth-discordant twin pairs at each CpG site. First, we used limma regression modeling, which is particularly well suited for small sample sizes thanks to its methodology of information borrowing across CpG sites. The model was applied to DNAm data log-transformed into M-values to improve statistical accuracy [62]. The twin’s sex, gestational age and maternal age were included in the model as covariates. CpGs were considered significant if the limma regression p values corresponding to the IUGR status satisfied the significance level p < 0.05.
Second, we used a non-parametric approach to test the between-group differences, comparing the placental methylation levels in the group of eight healthy individuals (controls) and the group of eight affected IUGR individuals. The unpaired Mann–Whitney U test was used for each CpG site.
Finally, the changes in DNA methylation level in healthy twins versus their IUGR-affected counterparts were compared using the paired Wilcoxon signed-rank test, which attempted to identify a consistent pattern of DNAm shift across all eight pairs, although not necessarily an overall difference between the IUGR and the healthy cohorts. Thus, the two non-parametric tests are complementary in finding pattern of differences across the twin pairs.
Given the large number of probes tested and a relatively small sample size, the standard methods of correction for multiple testing (such as Bonferroni or Benjamini-Hochberg FDR procedures) were too restrictive to confirm the validity of any of the individual tests. Therefore, we decided on a pragmatic strategy of using the probe-level statistical tests as guidance and complemented them with an effect-size threshold. We considered the threshold of 10 % as sufficiently stringent because relatively few CpG sites had a DNA methylation change exceeding 10 % by magnitude.
For validation purposes, we used unsupervised clustering to confirm that the resulting set of CpG sites contained a sufficiently discernible biological signal unobscured by the potential false positives or other stochastic noise in the selected loci. All DNAm profiles were first reduced to the set of probes with significant differences in IUGR according to our selection criteria. Thereafter, hierarchical clustering was applied to discern the different groups in our cohort.
Functional enrichment analysis
To identify prominent clinical mechanisms affecting and/or affected by growth restriction in our cohort, the 172 genes with evidence of differential methylation were investigated with respect to their functions and major biological roles. The analysis was performed using DAVID [63]. All p values were FDR corrected.
The 296 putatively affected CpG set was also analyzed in the context of wider genomic regions using GREAT [64]. We set the distal extension to the nearest genes as 100 kbp. The background set of probes to which the comparison was made was defined as the 399,118 autosomal CpGs used as the initial input to our analysis pipeline.
Validation analysis of DNA methylation using bisulfite pyrosequencing
Results of top candidate CpG sites (cg01971612 and cg18485485 for DECR1; cg04675542, cg02343823, and cg08580836 for ZNF300; and cg08234308 for LEPR) were validated using pyrosequencing of bisulfite converted DNA (PyroMark Q24; Qiagen). Polymerase chain reaction (PCR) and sequencing primers were designed using PyroMark Assay Design 2.0. All procedures were performed according to the manufacturer’s protocols [65]. The presence of 5-methylcytosine versus 5-hydroxymethylcytosine could not be distinguished using this method.
Abbreviations
DMR, differentially methylated region; DNAm, DNA methylation; FDR, false discovery rate; IUGR, intrauterine growth restriction; MC, monochorionic; MZ, monozygotic; PCR, polymerase chain reaction
Acknowledgements
We would like to thank Chunhua Zhao, Youliang Lou, and Yi-an Chen for their contributions to this work. This study was supported by the Mclaughlin Centre and the Canadian Institute for Health Research. Bioinformatic analyses were supported by the Canadian Centre for Computational Genomics (C3G), part of the Genome Innovation Network (GIN), funded by Genome Canada through Genome Quebec and Ontario Genomics.
Authors’ contributions
MR participated in the design of the study, carried out the sample selection, methylation array interpretation, and validation analysis, and drafted the manuscript. SC participated in the design of the study, carried out the methylation array interpretation and validation analysis, and drafted the manuscript. ALT participated in the design of the study, performed the bioinformatic analyses of the data, and drafted the manuscript. MB participated in the design of the study and oversaw bioinformatic analyses. SD, and JK participated in the design of the study. SK performed the pathological analyses on the samples. RW participated in the design, data analyses, and writing of the manuscript as corresponding senior author. All authors read and approved the final manuscript.
Competing interests
The authors declare that they have no competing interests.
Additional files
Supplementary Figures and Tables. (PDF 1827 kb)
Supplementary Tables S3–S5. (XLSX 86 kb)
Contributor Information
Maian Roifman, Email: maian.roifman@sickkids.ca.
Sanaa Choufani, Email: sanaa.choufani@sickkids.ca.
Andrei L. Turinsky, Email: turinsky@sickkids.ca
Sascha Drewlo, Email: sdrewlo@med.wayne.edu.
Sarah Keating, Email: skeating@mtsinai.on.ca.
Michael Brudno, Email: brudno@cs.toronto.edu.
John Kingdom, Email: jkingdom@mtsinai.on.ca.
Rosanna Weksberg, Email: rweksb@sickkids.ca.
References
- 1.Hendrix N, Berghella V. Non-placental causes of intrauterine growth restriction. Semin Perinatol. 2008;32(3):161–5. doi: 10.1053/j.semperi.2008.02.004. [DOI] [PubMed] [Google Scholar]
- 2.Bernstein IM, Horbar JD, Badger GJ, Ohisson A, Golan A. Morbidity and mortality among very-low-birth-weight neonates with intrauterine growth restriction. The Vermont Oxford Network. Am J Obstet Gynecol. 2000;182(1 Pt 1):198–206. doi: 10.1016/S0002-9378(00)70513-8. [DOI] [PubMed] [Google Scholar]
- 3.Barker DJ. The fetal and infant origins of adult disease. BMJ. 1990;301(6761):1111. doi: 10.1136/bmj.301.6761.1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gluckman PD, Hanson MA, Beedle AS. Early life events and their consequences for later disease: a life history and evolutionary perspective. Am J Hum Biol. 2007;19(1):1–19. doi: 10.1002/ajhb.20590. [DOI] [PubMed] [Google Scholar]
- 5.Morley R, Dwyer T, Carlin JB. Studies of twins: can they shed light on the fetal origins of adult disease hypothesis? Twin Res. 2003;6(6):520–5. doi: 10.1375/136905203322686527. [DOI] [PubMed] [Google Scholar]
- 6.Inbar-Feigenberg M, Choufani S, Butcher DT, Roifman M, Weksberg R. Basic concepts of epigenetics. Fertil Steril. 2013;99(3):607–15. doi: 10.1016/j.fertnstert.2013.01.117. [DOI] [PubMed] [Google Scholar]
- 7.Nelissen EC, van MontFoort AP, Dumoulin JC, Evers JL. Epigenetics and the placenta. Hum Reprod Update. 2011;17(3):397–41. doi: 10.1093/humupd/dmq052. [DOI] [PubMed] [Google Scholar]
- 8.Lewi L, Gucciardo L, Huber A, Jani J, Van Mieghem T, Done E, et al. Clinical outcome and placental characteristics of monochorionic diamniotic twin pairs with early- and late-onset discordant growth. Am J Obstet Gynecol. 2008;199(5):511 e1–7. doi: 10.1016/j.ajog.2008.04.022. [DOI] [PubMed] [Google Scholar]
- 9.Sebire NJ, Snijders RJ, Hughes K, Sepulveda W, Nicolaides KH. The hidden mortality of monochorionic twin pregnancies. Br J Obstet Gynaecol. 1997;104(10):1203–7. doi: 10.1111/j.1471-0528.1997.tb10948.x. [DOI] [PubMed] [Google Scholar]
- 10.Bejar R, Vigliocco G, Gramajo H, Solana C, Benirschke K, Berry C, et al. Antenatal origin of neurologic damage in newborn infants. II. Multiple gestations. Am J Obstet Gynecol. 1990;162(5):1230–6. doi: 10.1016/0002-9378(90)90024-2. [DOI] [PubMed] [Google Scholar]
- 11.Lopriore E, Pasman SA, Klumper FJ, Middeldorp JM, Walther FJ, Oepkes D. Placental characteristics in growth-discordant monochorionic twins: a matched case-control study. Placenta. 2012;33(3):171–4. doi: 10.1016/j.placenta.2011.12.004. [DOI] [PubMed] [Google Scholar]
- 12.Schrey S, Kingdom J, Baczyk D, Fitzgerald B, Keating S, Ryan G, et al. Leptin is differentially expressed and epigenetically regulated across monochorionic twin placenta with discordant fetal growth. Mol Hum Reprod. 2013;19(11):764–72. doi: 10.1093/molehr/gat048. [DOI] [PubMed] [Google Scholar]
- 13.Gordon L, Joo JE, Powell JE, Ollikainen M, Novakovic B, Li X, et al. Neonatal DNA methylation profile in human twins is specified by a complex interplay between intrauterine environmental and genetic factors, subject to tissue-specific influence. Genome Res. 2012;22(8):1395–406. doi: 10.1101/gr.136598.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wong CC, Meaburn EL, Ronald A, Price TS, Jeffries AR, Schalkwyk LC, et al. Methylomic analysis of monozygotic twins discordant for autism spectrum disorder and related behavioural traits. Mol Psychiatry. 2014;19(4):495–503. doi: 10.1038/mp.2013.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jaffe AE, Murakami P, Lee H, Leek JT, Fallin MD, Feinberg AP, et al. Bump hunting to identify differentially methylated regions in epigenetic epidemiology studies. Int J Epidemiol. 2012;41(1):200–9. doi: 10.1093/ije/dyr238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li D, Xie Z, Pape ML, Dye T. An evaluation of statistical methods for DNA methylation microarray data analysis. BMC Bioinformatics. 2015;16:217. doi: 10.1186/s12859-015-0641-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Arroyo JA, Garcia-Jones P, Graham A, Teng CC, Battaglia FC, Galan HL. Placental TonEBP/NFAT5 osmolyte regulation in an ovine model of intrauterine growth restriction. Biol Reprod. 2012;86(3):94. doi: 10.1095/biolreprod.111.094797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thomas S, Arbuckle TE, Fisher M, Fraser WD, Ettinger A, King W. Metals exposure and risk for small-for-gestational age birth in a Canadian birth cohort: the MIREC study. Environ Res. 2015;140:430–439. doi: 10.1016/j.envres.2015.04.018. [DOI] [PubMed] [Google Scholar]
- 19.Ferreira JC, Choufani S, Grafodatskaya D, Butcher DT, Zhao C, Chitayat D, et al. WNT2 promoter methylation in human placenta is associated with low birthweight percentile in the neonate. Epigenetics. 2011;6(4):440–9. doi: 10.4161/epi.6.4.14554. [DOI] [PubMed] [Google Scholar]
- 20.Longo S, Bollani L, Decembrino L, Di Comite A, Angelini M, Stronati M. Short-term and long-term sequelae in intrauterine growth retardation (IUGR) J Matern Fetal Neonatal Med. 2013;26(3):222–5. doi: 10.3109/14767058.2012.715006. [DOI] [PubMed] [Google Scholar]
- 21.Geva R, Eshel R, Leitner Y, Valevski AF, Harel S. Neuropsychological outcome of children with intrauterine growth restriction: a 9-year prospective study. Pediatrics. 2006;118(1):91–100. doi: 10.1542/peds.2005-2343. [DOI] [PubMed] [Google Scholar]
- 22.Herrera E. Lipid metabolism in pregnancy and its consequences in the fetus and newborn. Endocrine. 2002;19(1):43–55. doi: 10.1385/ENDO:19:1:43. [DOI] [PubMed] [Google Scholar]
- 23.Szajewska H, Horvath A, Koletzko B. Effect of n-3 long-chain polyunsaturated fatty acid supplementation of women with low-risk pregnancies on pregnancy outcomes and growth measures at birth: a meta-analysis of randomized controlled trials. Am J Clin Nutr. 2006;83(6):1337–44. doi: 10.1093/ajcn/83.6.1337. [DOI] [PubMed] [Google Scholar]
- 24.Cetin I, Giovannini N, Alvino G, Agostoni C, Riva E, Giovannini M, et al. Intrauterine growth restriction is associated with changes in polyunsaturated fatty acid fetal-maternal relationships. Pediatr Res. 2002;52(5):750–5. doi: 10.1203/00006450-200211000-00023. [DOI] [PubMed] [Google Scholar]
- 25.Alvino G, Cozzi V, Radaelli T, Ortega H, Herrera E, Cetin I. Maternal and fetal fatty acid profile in normal and intrauterine growth restriction pregnancies with and without preeclampsia. Pediatr Res. 2008;4(6):615–20. doi: 10.1203/PDR.0b013e31818702a2. [DOI] [PubMed] [Google Scholar]
- 26.Roe CR, Millington DS, Norwood DL, Kodo N, Sprecher H, Mohammed BS, Nada M, Schulz H, McVie R. 2,4-Dienoyl-coenzyme A reductase deficiency: a possible new disorder of fatty acid oxidation. J Clin Invest. 1990;85(5):1703–7. doi: 10.1172/JCI114624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Houten SM, Denis S, te Brinke H, Jongejan A, van Kampen AHC, Bradley EJ, Baas F, Hennekam RCM, Millington DS, Young SP, Frazier DM, Gucsavas-Calikoglu M, Wanders RJA. Mitochondrial NADP(H) deficiency due to mutation in NADK2 causes dienoyl-CoA reductase deficiency with hyperlysinemia. Hum Mol Genet. 2014;23(18):5009–16. doi: 10.1093/hmg/ddu218. [DOI] [PubMed] [Google Scholar]
- 28.Miinalainen IJ, Schmitz W, Huotari A, et al. Mitochondrial 2,4-dienoyl-CoA reductase deficiency in mice results in severe hypoglycemia with stress intolerance and unimpaired ketogenesis. PLoS Genet. 2009;5(7):e1000543. doi: 10.1371/journal.pgen.1000543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gou D, Wang J, Gao L, Sun Y, Peng X, Huang J, Li W. Identification and functional analysis of a novel human KRAB/C2H2 zinc finger gene ZNF300. Biochim Biophys Acta. 2004;1676(2):203–9. doi: 10.1016/j.bbaexp.2003.11.011. [DOI] [PubMed] [Google Scholar]
- 30.Zhao Y, Xue F, Sun J, Guo S, Zhang H, Qiu B, et al. Genome-wide methylation profiling of the different stages of hepatitis B virus-related hepatocellular carcinoma development in plasma cell-free DNA reveals potential biomarkers for early detection and high-risk monitoring of hepatocellular carcinoma. Clin Epigenetics. 2014;6(1):30. doi: 10.1186/1868-7083-6-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cai J, Gong R, Yan F, Yu C, Liu L, Wang W, et al. ZNF300 knockdown inhibits forced megakaryocytic differentiation by phorbol and erythrocytic differentiation by arabinofuranosyl cytidine in K562 cells. PLoS One. 2014;9(12):e114768. doi: 10.1371/journal.pone.0114768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Clement K, Viasse C, Lahlou N, Cabrol S, Pelloux V, Cassuto D, et al. A mutation in the human leptin receptor gene causes obesity and pituitary dysfunction. Nature. 1998;392(6674):398–401. doi: 10.1038/32911. [DOI] [PubMed] [Google Scholar]
- 33.Farooqi IS, Wangensteen T, Collins S, Kimber W, Matarese G, Keogh JM, et al. Clinical and molecular genetic spectrum of congenital deficiency of the leptin receptor. N Engl J Med. 2007;356(3):237–47. doi: 10.1056/NEJMoa063988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jiang B, Liu Y, Liu Y, Fang F, Wang X, Li B. Association of four insulin resistance genes with type 2 diabetes mellitus and hypertension in the Chinese Han population. Mol Biol Rep. 2014;41(2):925–33. doi: 10.1007/s11033-013-2937-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marginean CO, Marginean C, Voidazan S, Melit L, Crauciuc A, Duicu C, Banescu C. Correlations between leptin gene polymorphisms 223A/G, 1019 G/A, 492 G/C, 976 C/A, and anthropometrical and biochemical parameters in children with obesity: a prospective case-control study in Romanian population—the Nutrichild study. Medicine. 2016;95(12):e3115. doi: 10.1097/MD.0000000000003115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang MM, Wang J, Fan JJ, Ng TK, Sun DJ, Guo X, Teng Y, Li YB. Variations in the obesity gene “LEPR” contribute to risk of type 2 diabetes mellitus: evidence from a meta-analysis. J Diab Res. 2016;2016:5412084. doi: 10.1155/2016/5412084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Souren NY, Paulussen AD, Steyls A, Loos RJ, Stassen AP, Gielen M, et al. Common SNPs in LEP and LEPR associated with birth weight and type 2 diabetes-related metabolic risk factors in twins. Int J Obes. 2008;32:1233–1239. doi: 10.1038/ijo.2008.68. [DOI] [PubMed] [Google Scholar]
- 38.Ovilo C, Gonzalez-Bulnes A, Benitez R, Ayuso M, Barbero A, Perez-Solana ML, et al. Prenatal programming in an obese swine model: sex-related effects of maternal energy restriction on morphology, metabolism and hypothalamic gene expression. Br J Nutr. 2014;111(4):735–46. doi: 10.1017/S0007114513002948. [DOI] [PubMed] [Google Scholar]
- 39.King AJF. The use of animal models in diabetes research. Br J Pharmacol. 2012;166(3):877–894. doi: 10.1111/j.1476-5381.2012.01911.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zmorzynski S, Swiderska-Kolacz G, Koczkodaj D, Filip AA. Significance of polymorphisms and expression of enzyme-encoding genes related to glutathione in hematopoietic cancers and solid tumors. Biomed Res Int. 2015;2015:853573. doi: 10.1155/2015/853573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chowdhury UK, Zakharyan RA, Hernandez A, Avram MD, Kopplin MJ, Aposhian HV. Glutathione-S-transferase-omega [MMA(V) reductase] knockout mice: enzyme and arsenic species concentrations in tissues after arsenate administration. Toxicol Appl Pharmacol. 2006;216(3):446–57. doi: 10.1016/j.taap.2006.06.014. [DOI] [PubMed] [Google Scholar]
- 42.Alholle A, Brini AT, Gharanei S, Vaiyapuri S, Arrigoni E, Dallol A, et al. Functional epigenetic approach identifies frequently methylated genes in Ewing sarcoma. Epigenetics. 2013;8(11):1198–204. doi: 10.4161/epi.26266. [DOI] [PubMed] [Google Scholar]
- 43.Mahoney SE, Yao Z, Keyes CC, Tapscott SJ, Diede SJ. Genome-wide DNA methylation studies suggest distinct DNA methylation patterns in pediatric embryonal and alveolar rhabdomyosarcomas. Epigenetics. 2012;7(4):400–8. doi: 10.4161/epi.19463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pencheva N, Tran H, Buss C, Huh D, Drobnjak M, Busam K, Tavazoie SF. Convergent multi-miRNA targeting of ApoE drives LRP1/LRP8-dependent melanoma metastasis and angiogenesis. Cell. 2012;151(5):1068–82. doi: 10.1016/j.cell.2012.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Karlsson C, Baudet A, Miharada N, Soneji S, Gupta R, Magnusson M, et al. Identification of the chemokine CCL28 as a growth and survival factor for human hematopoietic stem and progenitor cells. Blood. 2013;121(19):3838–42. doi: 10.1182/blood-2013-02-481192. [DOI] [PubMed] [Google Scholar]
- 46.Sun C, Zhang YY, Tang CL, Wang SC, Piao HL, Toa Y, et al. Chemokine CCL28 induces apoptosis of decidual stromal cells via binding CCR3/CCR10 in human spontaneous abortion. Mol Hum Reprod. 2013;19(10):676–86. doi: 10.1093/molehr/gat038. [DOI] [PubMed] [Google Scholar]
- 47.Dimberg J, Hugander A, Wågsäter D. Protein expression of the chemokine, CCL28, in human colorectal cancer. Int J Oncol. 2006;28(2):315–9. [PubMed] [Google Scholar]
- 48.Liu GX, Lan J, Sun Y, Hu YJ, Jiang GS. Expression of the chemokine CCL28 in pleomorphic adenoma and adenolymphoma of the human salivary glands. Exp Ther Med. 2012;4(1):65–69. doi: 10.3892/etm.2012.544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kemmner W, Kessel P, Sanchez-Ruderisch H, Moller H, Hinderlich S, Schlag PM, Detjen K. Loss of UDP-N-acetylglucosamine 2-epimerase/N-acetylmannosamine kinase (GNE) induces apoptotic processes in pancreatic carcinoma cells. FASEB J. 2012;26(2):938–46. doi: 10.1096/fj.11-186700. [DOI] [PubMed] [Google Scholar]
- 50.Schwarzkopf M, Knobeloch KP, Rohde E, Hinderlich S, Wiechens N, Lucka L, Horak I, Reutter W, Horstkorte R. Sialylation is essential for early development in mice. Proc Natl Acad Sci U S A. 2002;99:5267–5270. doi: 10.1073/pnas.072066199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Krentsis IM, Sela I, Eiges R, Blanchard V, Berger M, Cohen MB, Mitrani-Rosenbaum S. GNE is involved in the early development of skeletal and cardiac muscle. PLoS One. 2011;6(6):e21389. doi: 10.1371/journal.pone.0021389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Eisenberg I, Avidan N, Potikha T, Hochner H, Chen M, Olender T, Barash M, Shemesh M, Sadeh M, Grabov-Nardini G, Shmilevich I, Friedmann A, Karpati G, Bradley WG, Baumbach L, Lancet D, Ben Asher E, Beckmann JS, Argov Z, Mitrani-Rosenbaum S. The UPD-N-acetylglucosamine 2-epimerase/N-acetylmannosamine kinase gene is mutated in recessive hereditary inclusion body myopathy. Nature Genet. 2001;29:83–87. doi: 10.1038/ng718. [DOI] [PubMed] [Google Scholar]
- 53.Clevers H. Wnt/beta-catenin signaling in development and disease. Cell. 2006;127(3):469–80. doi: 10.1016/j.cell.2006.10.018. [DOI] [PubMed] [Google Scholar]
- 54.Logan CY, Nusse R. The Wnt signaling pathway in development and disease. Annu Rev Cell Dev Biol. 2004;20:781–810. doi: 10.1146/annurev.cellbio.20.010403.113126. [DOI] [PubMed] [Google Scholar]
- 55.Knofler M, Pollheimer J. Human placental trophoblast invasion and differentiation: a particular focus on Wnt signaling. Front Genet. 2013;4:190. doi: 10.3389/fgene.2013.00190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Reamon-Buettner SM, Buschmann J, Lewin G. Identifying placental epigenetic alterations in an intrauterine growth restriction (IUGR) rat model induced by gestational protein deficiency. Reprod Toxicol. 2014;45:117–24. doi: 10.1016/j.reprotox.2014.02.009. [DOI] [PubMed] [Google Scholar]
- 57.Kokkinos MI, Murthi P, Wafai R, Thompson EW, Newgreen DF. Cadherins in the human placenta—epithelial-mesenchymal transition (EMT) and placental development. Placenta. 2010;31(9):747–55. doi: 10.1016/j.placenta.2010.06.017. [DOI] [PubMed] [Google Scholar]
- 58.Breathnach FM, McAuliffe FM, Geary M, Daly S, Higgins JR, Dornan J, et al. Prediction of safe and successful vaginal twin birth. Am J Obstet Gynecol. 2011;205(3):237 e1–7. doi: 10.1016/j.ajog.2011.05.033. [DOI] [PubMed] [Google Scholar]
- 59.Kramer MS, Platt RW, Wen SW, Joseph KS, Allen A, Abrahamowicz M, et al. A new and improved population-based Canadian reference for birth weight for gestational age. Pediatrics. 2001;108(2):E35. doi: 10.1542/peds.108.2.e35. [DOI] [PubMed] [Google Scholar]
- 60.Sandoval J, Heyn H, Moran S, Serra-Musach J, Pujana MA, Bibikova M, et al. Validation of a DNA methylation microarray for 450,000 CpG sites in the human genome. Epigenetics. 2011;6(6):692–702. doi: 10.4161/epi.6.6.16196. [DOI] [PubMed] [Google Scholar]
- 61.Chen YA, Lemire M, Choufani S, Butcher DT, Grafodatskaya D, Zanke BW, et al. Discovery of cross-reactive probes and polymorphic CpGs in the Illumina Infinium HumanMethylation450 microarray. Epigenetics. 2013;8(2):203–9. doi: 10.4161/epi.23470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Du P, Zhang X, Huang CC, Jafari N, Kibbe WA, Hou L, et al. Comparison of Beta-value and M-value methods for quantifying methylation levels by microarray analysis. BMC Bioinformatics. 2010;11:587. doi: 10.1186/1471-2105-11-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.da Huang W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 64.McLean CY, Bristor D, Hiller M, Clarke SL, Schaar BT, Lowe CB, et al. GREAT improves functional interpretation of cis-regulatory regions. Nat Biotechnol. 2010;28(5):495–01. doi: 10.1038/nbt.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Qiagen. PyroMark: PCR handbook. Sample & Assay Technologies. 2009; http://cmgm.stanford.edu/pan/doc/Pyrosequencing/PyroMark_Q24_PCR_Handbook.pdf
- 66.Peters TJ, Buckley MJ, Statham AL, Pidsley R, Samaras K, Lord VR, et al. De novo identification of differentially methylated regions in the human genome. Epigenetics Chromatin. 2015;8:6. doi: 10.1186/1756-8935-8-6. [DOI] [PMC free article] [PubMed] [Google Scholar]


