Abstract
Background
In cereal crops such as wheat, an optimal timing of developmental transitions is required to maximize grain yield. Many of these developmental changes are precisely regulated by changes in the duration, intensity or quality of light. Phytochromes are dimeric photoreceptors that absorb light maximally in the red and far-red wavelengths and induce large-scale transcriptional changes in response to variation in light quality. In wheat, PHYC is required for early flowering under long days. However, it is currently unknown whether this function requires the presence of PHYB. In this study, we characterized the role of PHYB in wheat development and used RNA-seq to analyze and compare the transcriptomes of phyB-null and phyC-null TILLING mutants.
Results
Under long-day photoperiods, phyB-null plants exhibit a severe delay in flowering comparable to the delay observed in phyC-null plants. These results demonstrate that both genes are required for the induction of wheat flowering under long days. Using replicated RNA-seq studies we identified 82 genes that are significantly up or down regulated in both the phyB-null and phyC-null mutant relative to their respective wild-type controls. Among these genes are several well-characterized positive regulators of flowering, including PPD1, FT1 and VRN1. Eight-fold more genes were differentially regulated only in the phyB-null mutant (2202) than only in the phyC-null mutant (261). The PHYB-regulated genes were enriched in components of the auxin, gibberellin and brassinosteroid biosynthesis and signaling pathways, and in transcription factors with putative roles in regulating vegetative development and shade-avoidance responses. Several genes involved in abiotic stress tolerance pathways were also found to be regulated by PHYB.
Conclusions
PHYB and PHYC are both required for the photoperiodic induction of wheat flowering, whereas PHYB alone regulates a large number of genes involved in hormone biosynthesis and signaling, shade-avoidance response, and abiotic stress tolerance. Our analysis provides a comprehensive overview of the PHYB- and PHYC-mediated transcriptional changes during light signaling, and an initial step towards the dissection of this regulatory gene network in wheat. This further dissection will be required to explore the individual phytochrome-mediated developmental responses and to evaluate their potential to improve wheat adaptation to changing environments.
Electronic supplementary material
The online version of this article (doi:10.1186/s12870-016-0831-3) contains supplementary material, which is available to authorized users.
Keywords: Wheat, PHYB, PHYC, Photomorphogenesis, RNA-seq, Shade-avoidance, Flowering
Background
Plants utilize external cues to precisely coordinate their growth and development with environmental conditions that maximize reproductive success. In cereal crops such as wheat, this has a direct impact on grain production, so understanding the regulatory mechanisms underlying these responses has important practical implications.
Plants are exquisitely adapted to detect variation in the quality, intensity and duration of light signals, and in response undergo rapid and dynamic transcriptional changes. These responses are mediated by several classes of photoreceptors, which absorb light of different wavelengths [1]. Among the photoreceptor families, the phytochromes absorb light maximally in the red (R) and far-red (FR) spectrum and modulate several important biological processes, including seed germination, flowering development, circadian rhythms and shade-avoidance [2, 3].
The phytochrome protein consists of two modules, both of which are essential for light signaling. The N-terminal photosensory core module is required for chromophore binding and photoconvertibility and is comprised of three domains: PAS (named from homology to PERIOD, ARYL HYDROCARBON RECEPTOR NUCLEAR TRANSPORTER and SINGLE MINDED), GAF (cGMP phosphodiesterase/adenylate cyclase/FhlA) and PHY (phytochrome-specific domains) [3]. The C-terminal module is comprised of two tandem PAS domains and a histidine kinase-like domain and is required for downstream regulatory function [4].
Phytochrome proteins can have one of two interchangeable isomeric forms; the biologically inactive, R light-absorbing Pr form and the biologically active, FR light-absorbing Pfr form [3]. Phytochromes are synthesized in the Pr form in the cytosol and, upon absorption of R light, undergo rapid conformational change to the active Pfr form, which results in their import into the nucleus [5]. In darkness or upon absorption of FR light, phytochromes in the Pfr state revert to the inactive Pr state [3]. In the nucleus, phytochrome dimers interact with a small subset of basic helix-loop-helix (bHLH) transcription factors known as PHYTOCHROME INTERACTING FACTORS (PIFs) [6]. The PIFs directly regulate a set of downstream targets by binding to conserved ‘G-box’ elements in their promoters [7]. The PIF primary targets then activate an array of secondary responses, including other transcription factors and regulators of growth and development [8]. The interaction between phytochromes and PIFs triggers the rapid multi-site phosphorylation of the latter, tagging them for degradation by the 26S proteasome machinery [9]. Phytochromes in the Pfr state inhibit the regulatory activity of PIF proteins by releasing them from their DNA targets [10, 11]. The PHY-PIF interaction also induces the degradation of the PHY protein, as part of a feedback regulatory mechanism to control active PHY levels [12]. This flexible mechanism allows for the precise adjustment of plant growth and development to subtle variations in light quality.
The phytochromes are encoded by three main clades of genes; PHYA, PHYB and PHYC [13]. While in the dicot lineage, gene duplication events within the PHYB clade have given rise to the PHYD and PHYE genes, the genomes of most monocot species, including wheat and barley, contain a single copy of each of the three phytochrome genes [13]. In Arabidopsis, a series of phy null mutants have been used to characterize the distinct and overlapping roles played by each phytochrome during development [2]. PHYA is the predominant phytochrome in seedling photomorphogenesis and regulates hypocotyl elongation during de-etiolation and the response to low fluence light [14]. PHYB, partially redundantly with the related PHYD and PHYE genes, regulates vegetative development, including the shade-avoidance syndrome, a response characterized by changes in plant architecture and growth under low ratios of R light to FR light (R/FR) to avoid shading by surrounding vegetation [15]. PHYC plays a more limited role and regulates a variety of photomorphogenesis responses throughout development in combination with other phytochromes. In both Arabidopsis and rice, PHYC activity is dependent on a functional PHYB protein [16–18]. However, in wheat, the PHYC protein is stable in the absence of other phytochromes and is sufficient to induce photomorphogenic changes when introduced into an Arabidopsis plant lacking functional endogenous phytochromes [19].
In the long-day (LD) grasses, such as wheat, barley and Brachypodium, PHYC plays a critical role in the acceleration of flowering under inductive LD conditions [19–21]. In these species PHYC is essential for the light activation of the PHOTOPERIOD 1 gene (PPD1 = PSEUDO RESPONSE REGULATOR37, PRR37) [19–21], which is responsible for most of the natural variation in photoperiodic response in the temperate cereals [22–25]. Under LD, PPD1 upregulates FLOWERING LOCUS T1 (FT1), which encodes a mobile protein with homology to Phosphatidylethanolamine-Binding Proteins (PEBPs) domain [26]. The FT1 protein is transported through the phloem from the leaves to the shoot apical meristem, where it is assembled into a hexameric protein complex that directly activates the expression of the meristem identity gene VRN1 [27, 28].
Wheat phyC-null mutant plants flower more than 100 days later than the wild-type control and this delay is associated with the downregulation of both PPD1 and FT1 [19]. The delay in flowering in the phyC-null mutant is more severe than the effect observed in either ppd1-null [29] or ft1-null [30] mutants, suggesting that in addition to the PPD1-FT1-mediated effect on flowering, PHYC also regulates other floral activation pathways. These additional effects on flowering time might be associated with the transcriptional changes observed in several components of the circadian clock in the phyC-null mutant [19].
Protein interaction studies demonstrated that the wheat PHYC protein can form both PHYC-PHYC homodimers and PHYB-PHYC heterodimers [19], but it is currently unknown whether PHYB is also necessary for the LD induction of flowering in wheat. In this study we show that the phyB-null mutant flowers even later than the phyC-null mutant, suggesting that both phytochromes are critical for flowering induction in wheat. We also describe morphological differences in the vegetative phenotype of the two mutants and characterize the subsets of genes regulated by each phytochrome using replicated RNA-seq studies. We show that both PHYB and PHYC are required for the induction of several flowering genes, and that more genes are differentially regulated only in the phyB-null mutant than only in the phyC-null mutant. PHYB-regulated targets include multiple genes involved in vegetative development, hormone biosynthesis and signaling, the shade-avoidance response, and abiotic stress tolerance. Our analysis provides insight into the downstream regulatory networks controlled by wheat PHYB and PHYC and identifies additional targets to further dissect light-mediated developmental signals in wheat.
Results
Characterization of the phyB-null mutant
Using a Targeting Induced Local Lesions in Genomes (TILLING) population of the tetraploid wheat variety Kronos we identified 206 mutations in the coding regions of the A and B genome copies of PHYB (henceforth PHYB-A and PHYB-B, respectively). Among these mutations, we selected line T4-2711 carrying a C to T change at nucleotide 1756 in PHYB-A. This phyB-A mutation generates a premature stop codon at position 586 (R586*) and a deletion of the last 641 amino acids including the entire regulatory module (Fig. 1a). For the PHYB-B gene, we selected line T4-2078 carrying a C to T change at nucleotide 3079. This phyB-B mutation generates a premature stop codon at position 1027 (Q1027*) that results in a C-terminally truncated protein lacking the distal 140 amino acids, including the histidine kinase domain (Fig. 1a). Since these C-terminal domains are required for phytochrome signaling [4], there is a high probability that both selected mutant lines encode non-functional PHYB proteins, and are thus loss-of-function mutations. Both mutants were backcrossed twice to wild-type Kronos to reduce the background mutation load, and were then intercrossed to select a plant homozygous for both mutations (phyB-A/phyB-B), hereafter referred to as phyB-null. We confirmed the presence of these mutations in the cDNA of each mutant line using RT-PCR and Sanger sequencing.
Fig. 1.
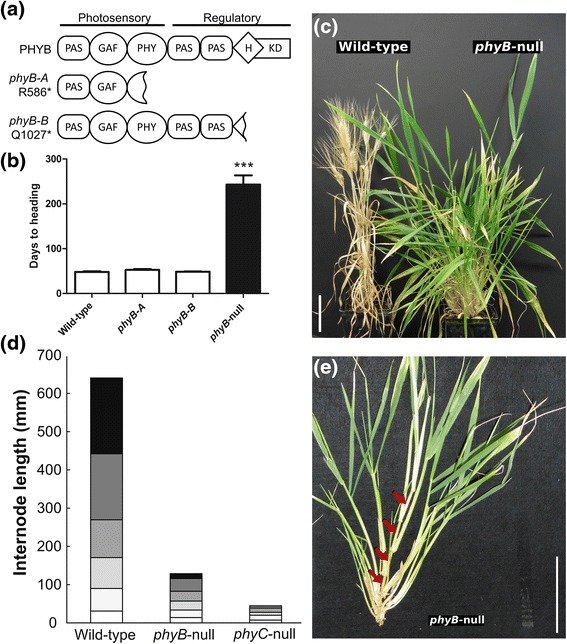
Characterization of the phyB-null mutant. a Schematic representation of the conserved functional domains of the PHYB protein and selected phyB-A and phyB-B TILLING mutants. b Flowering time of phyB-null mutant plants under LD photoperiod. Data represent the mean of at least five plants (*** P < 0.001). c Wild type and phyB-null mutant plants 60-days after sowing. d Stem length at 60 days. Individual internodes are indicated by different shades. White bars indicate the oldest internode and black bars the youngest internode. e Detail of a tiller from a phyB-null mutant plant 60-days after sowing showing elongated internodes (red arrows). Bar = 10 cm
Under LD conditions, neither the phyB-A (4.7 days delay, P >0.05) nor phyB-B (0.3 days delay, P >0.05) single mutants exhibited significant changes in flowering time when compared to wild-type sister lines (Fig. 1b). However, the phyB-null mutant (lacking any functional PHYB gene) exhibited a severe delay in flowering of 195 days (P <0.001, Fig. 1b). Furthermore, the emerged spikes did not set seeds, despite the formation of all constituent parts of the spikelet and floral organs (Additional file 1: Figure S1). This significant delay in flowering is even more severe than the large delay previously observed in the phyC-null mutant (108-days delay in flowering [19]) and demonstrates that both PHYB and PHYC genes are required for the induction of wheat flowering under LD.
The late flowering phenotype of the phyB-null mutant was associated with an extended vegetative developmental phase and other differences in the plant’s morphology (Fig. 1c). The rate of leaf emergence in phyB-null mutants was significantly faster (average 3.3 days per leaf) than in either the wild-type control plants (3.7 days per leaf, P <0.001) or the phyC-null mutant (3.5 days per leaf, P <0.05, Additional file 1: Figure S2a). We measured the size of the most recently expanded leaf at three different timepoints (36, 43 and 50 days). At each stage, leaves of the phyB-null mutant plant were significantly longer (P <0.05) than the wild-type control (Additional file 1: Figure S2b). Leaves in the phyB-null plant were also wider, on average, than the wild-type control, although these differences were significant only at 36 days (P <0.05, Additional file 1: Figure S2c). By comparison, the phyC-null mutant showed significantly longer (P <0.001, Additional file 1: Figure S2b) and narrower leaves (P <0.05, Additional file 1: Figure S2c) than the wild-type at all three timepoints. We also measured stem length at 60 days of age, at which stage the wild-type plant had initiated flowering development, associated with rapid stem elongation (Fig. 1d). As expected for plants in the vegetative stage, both phyB-null and phyC-null mutants exhibited greatly reduced stem elongation when compared to the wild-type, although the total number of internodes was unaffected in both mutants. Interestingly, the internodes in the phyB-null mutant were significantly (P <0.01) longer than the phyC-null mutant (Fig. 1d and e, Additional file 1: Figure S1e). Finally, we compared germination rates between phyB-null and wild-type controls and found no significant differences.
Taken together, these observations suggest that while both PHYB and PHYC genes are important during the regulation of flowering development, PHYB also appears to play an important role during vegetative development, influencing the rate of leaf production and cellular elongation in both stems and leaves.
Plants with mutations in only one PHYB homoeologue (phyB-A or phyB-B) showed no phenotypic differences with the wild-type plants, which is consistent with the lack of differences in flowering time. This last result demonstrated that the PHYB-A and PHYB-B homoeologues exhibit a large degree of functional redundancy.
Identification of high-confidence differentially expressed genes
To analyze and compare genes regulated by PHYB and PHYC we performed replicated RNA-seq studies. Leaf tissue was harvested in the morning from LD-grown four-week-old phyB-null and phyC-null mutant plants, and from their respective wild-type sister lines, which were used as controls (Additional file 1: Figure S3). To reduce the incidence of false positives (genes incorrectly defined as differentially expressed between genotypes), we performed the complete experiment twice, using four biological replicates per genotype in each experiment. We sequenced a total of 32 RNA-seq libraries (four biological replications * four genotypes * two experiments), generating an average of 49.1 million 50 bp single-end reads per sample (Additional file 1: Table S1). We mapped 95.3 % of these reads to gene-coding regions identified within the draft assembly of the wheat genome from the International Wheat Genome Sequencing Consortium (v2.2, see Methods) [31]. All subsequent analyses were performed using only those sequencing reads that mapped uniquely to one transcribed locus (average 58.5 % of reads, Additional file 1: Table S1).
A Principal Component Analysis including the expression results for phyB-null, phyC-null and their respective controls showed a good separation among genotypes (Additional file 1: Figure S4a). The first principal component separated the phyC-null samples from those obtained from the wild-type sister plants. The second principal component separated the phyB-null samples from those corresponding to the wild-type sister plants. Limited differences were detected in these two components between the two experimental replications (Additional file 1: Figure S4a).
When each phytochrome was analyzed separately, the two PHYB experimental replications clustered together (Additional file 1: Figure S4b), but the two PHYC experimental replications clustered in two groups separated by the second principal component (Additional file 1: Figure S4c). These two PHYC experimental replications were performed in growth chambers of the same brand and model, and with identical halide light configurations. However, after we saw the differences in gene expression we reexamined the chambers and realized that they differed in their ballast systems, which may have caused some of the observed variation between experimental replicates. Although light intensity at R (~660 nm) and FR (~730 nm) wavelengths varied in these two chambers, the R/FR ratio was similar (2.61 in the first chamber and 2.83 in the second). Despite this variability, the first component clearly differentiated the phyC-null samples from the wild-type sister plants (Additional file 1: Figure S4c).
Defining PHYB and PHYC regulated genes in wheat
Pair-wise comparisons of gene expression values identified 2306 high-confidence PHYB-regulated genes which were differentially expressed between the phyB-null mutant and wild-type sister lines in the same direction in both experimental replications (4315 PHYB-regulated genes in experiment one and 5955 in experiment two at False Discovery Rate (FDR)-adj P <0.01, Fig. 2a). Only one gene was differentially expressed in both experimental replicates in opposite directions, and was excluded from the analysis. Because the experimental replications were performed independently, these high-confidence genes have an expected FDR < 0.0001 (<0.01 * <0.01). We also identified a further 5656 genes which were differentially expressed in just one of the two experimental replications, which are hereafter referred to as FDR-adj P < 0.01 PHYB-regulated genes. A similar comparison between the phyC-null mutant and wild-type identified 365 high-confidence PHYC-regulated genes regulated in the same direction (1302 PHYC-regulated genes in experiment one and 801 in experiment two at FDR-adj P <0.01, Fig. 2b) and 1373 FDR-adj P < 0.01 PHYC-regulated genes. Normalized expression values and FDR-adjusted P values for all genes are provided in Additional file 2 to facilitate the re-analysis of this data using different levels of stringency.
Fig. 2.
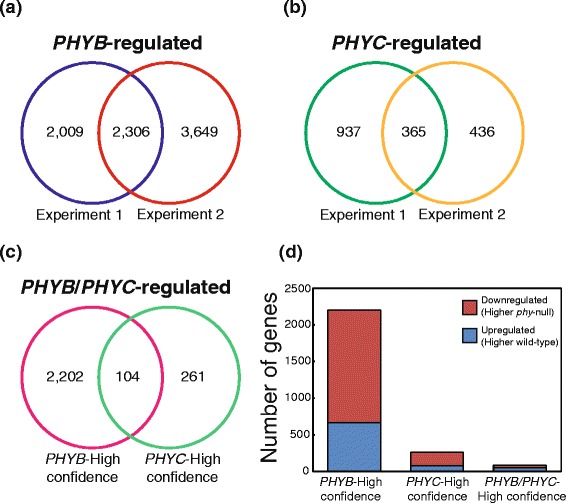
Numbers of differentially expressed genes in each experimental replicate regulated by (a) PHYB, (b) PHYC and (c) By both PHYB and PHYC. d Proportion of up- and down-regulated genes in each class
To limit the impact of variation between experimental replicates, and to reduce the incidence of false positives, we focused all subsequent analyses only on high-confidence PHYB and PHYC differentially-expressed genes (Additional file 3). Full details of FDR-adj P < 0.01 PHYB- and PHYC-regulated genes are provided in Additional file 4. Furthermore, all expression data is publicly available through “WheatExp”, an online wheat expression database and visualization tool [32].
A comparison of the 2567 high-confidence genes differentially regulated by PHYB and/or PHYC revealed 104 that were commonly regulated by both genes (Fig. 2c). Of these 104 common genes, 82 were differentially regulated in a concerted manner in the same direction (i.e., induced by both PHYB and PHYC). The remaining 22 genes were regulated in the opposite directions (i.e., induced by PHYB but suppressed by PHYC or vice versa). The number of high-confidence genes that were detected only in the phyB-null mutant (2202) was 8.5-fold higher than the number differentially expressed only in the phyC-null mutant (261, Fig. 2c). Similar proportions were identified among the 6693 FDR-adj P <0.01 PHY-regulated genes; 5320 were differentially expressed only in the phyB-null mutant, 1037 were detected only in the phyC-null mutant and 336 were commonly regulated in both mutants (Additional file 4). A majority of the high-confidence PHYB-regulated (69.9 %) and PHYC-regulated (70.9 %) genes exhibited higher expression levels in the respective phy-null mutant, suggesting that they are downregulated by the active PHY protein (Fig. 2d). In contrast, among the 82 genes regulated by both PHYB and PHYC, the majority (59.8 %) exhibited lower expression levels in the respective phy-null mutant, suggesting that they are upregulated by the functional PHY proteins (Fig. 2d).
Genes with similar effects in the phyB-null and phyC-null mutants include multiple flowering regulation genes
Functional analysis of the 82 genes similarly regulated in the phyB-null and phyC-null mutants showed significant enrichment for genes with roles in transcriptional regulation (Additional file 1: Table S2). Consistent with the late-flowering phenotypes of phyB-null and phyC-null plants, both mutants showed greatly reduced transcript levels of a number of well-characterized positive regulators of flowering, including PPD-B1, VERNALIZATION1, FRUITFULL2 and FT1 (Table 1). The A-genome copy of PPD1 is also downregulated in both phyB-null and phyC-null plants but is not among the high-confidence genes (Traes_2AS_2FCD59730, Additional file 2). The PPD-A1b allele in Kronos carries a deletion in the promoter region that causes altered expression levels independently of the photoperiod [24]. Our results demonstrate that this allele is expressed even in the absence of functional PHYB and PHYC phytochromes. Several other genes involved in the regulation of flowering time were also downregulated in both mutants, including GIGANTEA (GI), a circadian clock output gene, CONSTANS9 (CO9), a regulator of barley flowering under SD [33], and two flowering activators belonging to the GATA-domain family of transcriptional regulators [34, 35] (Table 1).
Table 1.
Selected flowering time genes regulated in concert by both PHYB and PHYC
| Avg. normalized counts | ||||||
|---|---|---|---|---|---|---|
| Gene | Ensembl ID | PHYB | PHYC | |||
| Wild-type | phyB-null | Wild-type | phyC-null | Putative function | ||
| Downregulated in phyB-null and phyC-null mutants | ||||||
| PPD-B1 | Traes_2BS_8BED816B1.1 | 4,505 | 37 | 2,285 | 79 | Flowering induction |
| VRN-A1 | Traes_5AL_13E2DEC48.2 | 5,605 | 306 | 2,647 | 272 | Meristem identity |
| VRN-B1 | Traes_5BL_89636D032.1 | 1,573 | 73 | 884 | 89 | Meristem identity |
| FUL-A2 | Traes_2AL_20C2D79E1.2 | 820 | 2 | 279 | 3 | Meristem identity |
| GI-B | contig105156 | 739 | 233 | 642 | 169 | Circadian clock output |
| FT-A1 | Traes_7AS_EBD5F1F54.1 | 590 | 2 | 331 | 1 | Flowering induction |
| FT-B1 | Traes_7BS_581AA844D.1 | 797 | 1 | 267 | 2 | Flowering induction |
| GATA-B1 | contig67606 | 69 | 1 | 77 | 1 | Flowering induction |
| GATA-A7 | td-k51_contig_84804 | 194 | 2 | 225 | 2 | Flowering induction |
| CO-B9 | Traes_1BL_688EF6A1A.1 | 594 | 197 | 410 | 158 | Flowering induction |
| Upregulated in phyB-null and phyC-null mutants | ||||||
| TaAGL33-A | Traes_5AL_6FF34F1C3.2 | 43 | 300 | 51 | 248 | Flowering induction |
| TaAGL33-B | Traes_4BL_5AF7ACF03.2 | 5 | 132 | 6 | 62 | Flowering induction |
| CO-A1 | Traes_7AS_F46AC277B.1 | 19 | 219 | 14 | 126 | Flowering induction |
We also identified several flowering time genes which showed the opposite response, with significantly elevated expression levels in both the phyB-null and phyC-null mutant (Table 1). These included CONSTANS1 (CO1), a gene central to flowering induction in Arabidopsis [36] and both A and B homoeologues of AGAMOUS-LIKE 33 (TaAGL33) a gene homologous to HvODDSOC2, which delays flowering when overexpressed in barley [37] (Table 1).
To further explore the role of PHYB in the regulation of flowering, we selected six flowering time genes for validation by qRT-PCR and assayed their transcript levels in wild-type and phyB-null plants at three timepoints (two-week, four-week and six-week old plants). Four genes (PPD1, FT1, VRN1 and CO1) were chosen from the high-confidence genes regulated by both PHYB and PHYC. VRN1 and FT1 expression was significantly higher in the wild-type than the phyB-null mutant at all three timepoints, PPD1 expression was higher at the four-week and six-week timepoints and CO1 expression was significantly reduced in wild-type plants at the six-week timepoint, confirming the role of PHYB in the regulation of these genes (Additional file 1: Figure S5). These four genes were shown to be regulated by PHYC in a previous study [19]. Two additional genes from the FDR-adj P < 0.01 PHYB-regulated genes were also selected (FT2 and VRN2, Additional file 4). There were no significant differences in the expression of either of these genes between wild-type and the phyB-null mutant at the four-week timepoint, consistent with the RNA-seq data. However, the expression of both genes was significantly different between genotypes at six-weeks, demonstrating that PHYB regulates the expression of these genes at a later stage of development (Additional file 1: Figure S5).
High-confidence genes differentially regulated only in the phyC-null mutant
We next examined the 261 high-confidence genes that showed a significant response in the phyC-null mutant, but showed no significant differential expression in the phyB-null mutant. Functional enrichment data showed an over-representation of genes with roles in photoperiodism and flowering time (Additional file 1: Table S2). These included FT3, a member of the PEBP gene family that is expressed under SD at higher levels than FT1 and FT2 [30] and the circadian clock gene PRR95, which are both downregulated in the phyC-null mutant (Table 2). Also included in this list was TaAGL41, a MADS-box gene which is a close homolog of TaAGL33/ODDSOC2, which was upregulated in the phyC-null mutant (Table 2).
Table 2.
Selected PHYC-specific regulated genes
| Gene | Ensembl ID | Avg. normalized counts | Putative function | |
|---|---|---|---|---|
| Wild-type | phyC-null | |||
| Downregulated in phyC-null mutants | ||||
| FT-B3 | Traes_1BL_2C43B822A.1 | 59 | 7 | Flowering regulation |
| PRR95-A | Traes_5AL_852A1474C.1 | 1,423 | 456 | Circadian clock |
| Upregulated in phyC-null mutants | ||||
| TaAGL41-B | TRAES3BF009000010CFD_t1 | 45 | 121 | Flowering regulation |
High-confidence genes differentially regulated only in the phyB-null mutant
The genes differentially regulated only in the phyB-null mutant included those with putative roles in flowering, hormone biosynthesis and signaling, shade-avoidance, and abiotic stress tolerance (Table 3).
Table 3.
Selected PHYB-specific regulated genes
| Gene | Ensembl ID | Avg. normalized counts | ||
|---|---|---|---|---|
| Wild-type | phyB-null | Putative function | ||
| Flowering time | ||||
| CCA-B1 | Traes_7BL_998EC9F74.2 | 29,787 | 13,084 | Circadian clock output |
| LHY-B | Traes_6BL_2F2381640.1 | 126 | 43 | Circadian clock output |
| FT-like_chr3-A | Traes_3AS_4310A2281.1 | 4 | 44 | PEBP-family |
| FT-like_chr3-B | TRAES3BF053100340CFD_t1 | 2 | 303 | PEBP-family |
| FT-like_chr6-A | Traes_6AL_66B24F155.1 | 21 | 138 | PEBP-family |
| FT-like_chr2-A | Traes_2AL_2F198B97C.1 | 0 | 10 | PEBP-family |
| FT-like_chr5-1-A | Traes_5AL_96274800D.1 | 1 | 95 | PEBP-family |
| FT-like_chr5-2-A | Traes_5AL_EFB6E50C9.2 | 179 | 27 | PEBP-family |
| FPF-like-A1 | Traes_2AS_329CCD131.1 | 63 | 7,688 | Flowering promotion |
| FPF-like-B1 | Traes_2BS_EA4D55C79.1 | 35 | 3,503 | Flowering promotion |
| FPF-like-B2 | Traes_2BL_600226046.1 | 140 | 1,538 | Flowering promotion |
| FPF-like-A3 | Traes_6AS_447C71E5B.1 | 13 | 70 | Flowering promotion |
| FPF-like-A4 | Traes_5AL_6FD9AE7EB.1 | 91 | 7 | Flowering promotion |
| VIL-A2 | Traes_6AS_DF6C22BF3.1 | 1,315 | 539 | Vernalization |
| VIL-B2 | Traes_6BS_2B746261B.1 | 1,030 | 337 | Vernalization |
| Hormone biosynthesis and signaling | ||||
| TAR-A2 | Traes_3AS_FF5C06A87.1 | 26 | 94 | Auxin biosynthesis |
| TAR-B2 | td-k55_contig_11729 | 559 | 1609 | Auxin biosynthesis |
| IAA-A12 | td-k61_contig_15261 | 1 | 20 | AUX/IAA |
| IAA-B12 | Traes_5BL_9301BD154.1 | 41 | 143 | AUX/IAA |
| IAA-B15 | Traes_1BS_8A19C460B.1 | 191 | 440 | AUX/IAA |
| IAA-A16 | Traes_3AS_771897131.2 | 71 | 163 | AUX/IAA |
| IAA-B17 | Traes_7BL_74071485F.2 | 1,041 | 1,859 | AUX/IAA |
| IAA-A19 | Traes_1AL_859346448.1 | 380 | 689 | AUX/IAA |
| ARF-B6 | Traes_6BS_BD894AD26.1 | 335 | 88 | ARF |
| ARF-B9 | contig33661 | 16 | 0 | ARF |
| ARF-B17 | Traes_7BL_66296695F.1 | 2,071 | 493 | ARF |
| WAT1-like-A1 | td-k25_contig_69900 | 3 | 16 | Auxin transport |
| WAT1-like-B2 | Traes_5BL_CBB9D7E7F.1 | 552 | 114 | Auxin transport |
| PIN3-like-B | Traes_6BL_F93C57B09.1 | 69 | 152 | Auxin transport |
| GA20ox-A1 | Traes_4AL_FABDF4EDA.1 | 77 | 423 | GA biosynthesis |
| GA20ox-B2 | td-k51_contig_72276 | 20 | 2,482 | GA biosynthesis |
| GA20ox-A4 | Traes_1AL_3A716350F.2 | 4 | 229 | GA biosynthesis |
| GA20ox-B4 | Traes_1BL_32506F819.1 | 8 | 219 | GA biosynthesis |
| GA2ox-A6 | Traes_2AL_BA387175F.1 | 2 | 37 | GA catabolism |
| GA2ox-A11 | Traes_4AS_7DC625FF5.1 | 3 | 47 | GA catabolism |
| GA2ox-B11 | Traes_4BL_63EFE8C91.1 | 1 | 36 | GA catabolism |
| ACC synthase-A | Traes_4AL_A5B9F7B36.1 | 2 | 28 | Ethylene biosynthesis |
| ACC oxidase-B | Traes_5BL_6AAC89B49.1 | 164 | 857 | Ethylene biosynthesis |
| BZR1-like-A | Traes_3AS_5A6A80EA5.1 | 153 | 916 | BR signaling |
| BZR1-like-B | td-k21_contig_21617 | 113 | 681 | BR signaling |
| PYL4-like-A1 | Traes_4AS_72BEF89AC.1 | 115 | 19 | ABA signaling |
| PYL4-like-B1 | Traes_4BL_E43C1BB11.1 | 322 | 80 | ABA signaling |
| PYL4-like-B2 | Traes_2BS_6428AA6CC.1 | 323 | 146 | ABA signaling |
| Transcription factors | ||||
| PIF-B3 | Traes_1BS_D1FCBFBE8.1 | 35 | 3,065 | bHLH |
| bHLH47-like1-A | Traes_2AS_9AEA9BDEA.1 | 87 | 5,046 | bHLH |
| bHLH47-like2-A | Traes_2AS_C4568AE60.1 | 606 | 1,740 | bHLH |
| PIL1-like-A | Traes_5AL_85F3BE385.2 | 323 | 698 | bHLH |
| BIM-A2 | Traes_5AL_0D4BDDDCD.1 | 93 | 248 | bHLH |
| BIM-B2 | Traes_5BL_AAC9C7238.2 | 615 | 1,453 | Bhlh |
| ATHB2-A | Traes_2AL_EF9549D16.1 | 24 | 640 | Homeobox domain |
| ATHB2-B | Traes_2BL_02479C76A.1 | 4 | 332 | Homebox domain |
| SPL14-like-A | Traes_7AS_FB2A769B5.1 | 93 | 182 | SPL |
| SPL14-like-B | Traes_7BS_7ACA0B10A.2 | 36 | 128 | SPL |
| Growth and cell elongation | ||||
| XTH-like-B | Traes_4BS_64FB912EE.1 | 18 | 103 | Cell elongation |
| LNG-1-like-A | Traes_2AS_2C93BAE62.1 | 204 | 1,102 | Cell elongation |
| LNG-2-like-A | td-k25_contig_4862 | 0 | 23 | Cell elongation |
| PROG –A1 | td-k41_contig_81185 | 90 | 502 | Vegetative growth |
| PROG –B1 | td-k41_contig_81185 | 16 | 173 | Vegetative growth |
| BSH-B | Traes_1BS_E09101AE8.1 | 296 | 640 | Vegetative growth |
| GT-A1 | Traes_4AS_1EA23DE08.1 | 11 | 92 | Vegetative growth |
| TZP-A | Traes_3AL_5006FA990.1 | 1,766 | 694 | Architecture |
| TZP-B | isotig04736 | 7,439 | 2,774 | Architecture |
| Cold tolerance | ||||
| ICE-B41 | td-k35_contig_73009 | 103 | 12 | CBF induction |
| WCOR413-A | Traes_5AL_F7649C79D.1 | 31 | 125 | Cold response |
| WCS19a-B | Traes_2BL_6382E3EFF.1 | 1 | 23 | Cold response |
| WCOR14-B | td-k25_contig_47404 | 5 | 208 | Cold response |
| WCOR15-A | Traes_2AL_079988C38.1 | 18 | 1,025 | Cold response |
At the morning time point sampled in this study, the transcript levels of LATE ELONGATED HYPOCOTYL (LHY) and CIRCADIAN CLOCK ASSOCIATED1 (CCA1) were significantly higher in the wild-type than in the phyB-null mutant.
In addition to its shared role with PHYC in activating FT1 expression, PHYB also regulates six other uncharacterized members of the PEBP gene family independently of PHYC (Table 3). These six genes are distinct from the FT-like genes described previously in wheat [30]. Interestingly, only one of these putative FT-like genes (the orthologue of HvMOTHER OF FT1 = FT-like_chr5-2, Table 3) showed the same transcriptional response as FT1 (downregulation in the phyB-null mutant), with the remainder showing significantly higher expression in the phyB-null mutant. PHYB also specifically regulates five FLOWERING-PROMOTING FACTOR-like (FPF-like) genes, four of which are significantly upregulated in the late-flowering phyB-null mutant (Table 3).
The wheat VERNALIZATION INSENSITVE 3-LIKE 2 gene (VIL2) [38] encodes a plant homeodomain (PHD)-finger protein and was significantly downregulated in the phyB-null mutant (Table 3). This gene is related to the Arabidopsis VERNALIZATION INSENSITIVE 3 gene, which plays an important role in the vernalization response in Arabidopsis (Table 3). However, the function of this gene in wheat has yet to be established [38].
PHYB-regulated targets exhibit an enrichment of genes with roles in the biosynthesis and signaling of several classes of hormones (Additional file 1: Table S2). Within the auxin pathway, the phyB-null mutant exhibited an upregulation of the auxin biosynthesis gene TRYPTOPHAN AMINOTRANSFERASE RELATED 2 (TAR2) and five AUX/IAA genes, and a downregulation of three AUXIN RESPONSE FACTOR (ARF) genes (Table 3). Auxin signaling is modulated by the hormone’s cellular location, which is controlled by the activity of auxin transporters [39]. We identified three differentially expressed genes encoding auxin transporters, two homologous to the vacuole-localized WALLS ARE THIN 1 (WAT1) and another to the plasma-membrane localized PIN-FORMED3 (PIN3) (Table 3, Fig. 5).
Fig. 5.
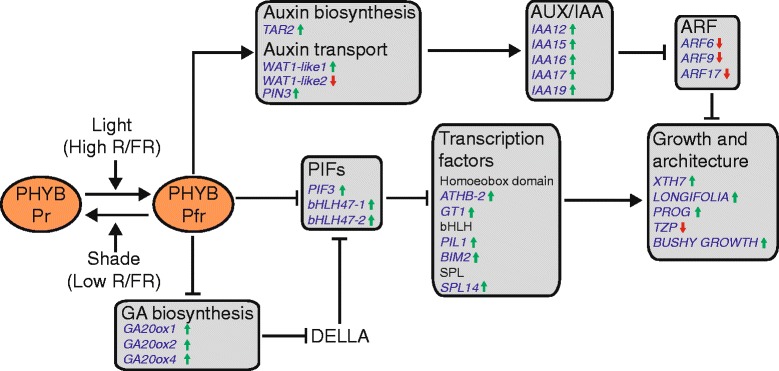
Putative members of the shade-avoidance response pathway in wheat. Genes significantly up (⬆) or down (⬇) regulated in the phyB-null mutant are displayed. Gene names are in blue
The phenotypic changes in the phyB-null mutant were consistent with an increased rate of gibberellin (GA) biosynthesis, a plant hormone closely linked to phytochrome-mediated growth promotion [40] and to wheat spike development [41]. Three members of the GA20oxidase family, which encode enzymes catalyzing the rate-limiting reaction in GA biosynthesis, were upregulated, as were two genes encoding GA-deactivating enzymes, GA2ox6 and GA2ox11 (Table 3). The expression of both A and B homoeologues of BRASSINAZOLE RESISTANT 1 (BZR1), a brassinosteroid-responsive transcription factor which positively regulates cell growth, were upregulated in the phyB-null mutant (Table 3), as were two ethylene biosynthesis genes, one encoding an ACC-synthase and another encoding an ACC-oxidase (Table 3). We also found that phyB-null mutant exhibited reduced expression of two putative abscisic acid (ABA) receptors of the PYRABACTIN-RESISTANCE-LIKE (PYL) gene family, indicative of a reduced sensitivity to this hormone (Table 3). Taken together, these findings indicate that PHYB plays a prominent role in regulating GA, BR, auxin, ABA and ethylene biosynthesis, transport and signaling pathways.
In Arabidopsis, a subset of genes has been identified which are differentially expressed in response to low R/FR and that are required for the shade-avoidance response. These genes include different classes of transcription factors, particularly members of the bHLH and homeobox leucine zipper family. We identified three bHLH genes which were significantly upregulated in the phyB-null mutant with high homology to characterized PIF genes (Table 3). Other bHLH genes upregulated in the phyB-null mutant included the wheat orthologue of PHYTOCHROME INTERACTING LIKE PROTEIN1 (PIL1), which regulates cell wall expansion genes during rice stem elongation [42] and BES1-INTERACTING MYC-LIKE PROTEIN 2 (BIM2), a bHLH transcription factor with a role in mediating brassinosteroid signaling during shade-avoidance [43] (Table 3). We also found a significant upregulation in the phyB-null mutant of two homeobox leucine zipper transcription factors. The first, ATHB2, is a direct PIF target which regulates transcriptional responses to light quality during shade-avoidance [44], while the second, GRASSY TILLERS 1 (GT1), regulates tillering and bud outgrowth in the grasses [45]. Finally, one member of the SQUAMOSA PROMOTER BINDING-LIKE (SPL) family of transcription factors with highest similarity to Arabidopsis SPL14 was upregulated in the phyB-null mutant. Mutations in this gene result in plants with elongated petioles, a characteristic trait of the shade-avoidance response [46].
Several regulators of cellular growth and plant architecture were significantly upregulated in the phyB-null mutant (Table 3). These included a putative XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE (XTH) gene, which encodes an enzyme that facilitates cell wall elongation to enhance cellular growth and expansion [47], PROSTRATE GROWTH 1 (PROG1), a Cys2-His2 zinc-finger protein regulating plant architecture and panicle angle [48, 49], two members of the LONGIFOLIA-like (LNG-like)gene family, which promote longitudinal cell elongation in Arabidopsis [50] and BUSHYGROWTH (BSH), a member of the SUCROSE NON FERMENTING (SNF) gene family which has a putative role in regulating plant architecture and seed set [51].
In Arabidopsis, PHYB modulates freezing tolerance in response to changes in light quality [52]. A connection between these two traits may also exist in wheat, since we found that the wheat phyB-null mutant exhibited a significant upregulation of four COLD REGULATED (COR) genes and downregulation of one member of the INDUCER of CBF EXPRESSION (ICE) gene family [53] (Table 3).
In addition to protein-coding genes, changes in development can be induced by non-coding parts of the genome, such as miRNAs [54]. We identified loci encoding pri-miRNAs that were specifically upregulated in either the phyB-null or phyC-null mutants (Table 4). In the phyB-null mutant, two homoeologous loci encoding pri-miR530 were the most significant differentially expressed genes in our experiment. The mature miR530 is predicted to target a transcription factor with high similarity to TANDEM ZINC KNUCKLE/PLU3 (TZP), which has a role in regulating plant growth and architecture downstream of the circadian clock and light signaling pathways [55]. This gene exhibits the expected inverse transcriptional response to pri-miR530, with significantly reduced expression in the phyB-null mutant (Table 4). We also found a significant upregulation in the phyB-null mutant of one locus encoding pri-miR393 and another encoding pri-miR156g (Table 4). miR393 is predicted to target two TIR1-like members of auxin receptors [56–59], but the expression of these two targets was not significantly reduced in the phyB-null mutant (Table 4). Members of the miR156 family target multiple SPL genes during development [60], but we did not observe significant changes in the expression of any SPL genes carrying the miR156 target site at this developmental stage (Table 4).
Table 4.
PHY-regulated pri-miRNAs and the predicted targets of their mature miRNA in wheat
| Pri-miRNA counts | Target counts | |||||
|---|---|---|---|---|---|---|
| Pri-miRNA | Ensembl ID | Wild-type | phyB-null | Putative target | Wild-type | phyB-null |
| PHYB targets | ||||||
| miR530-A | isotig11608 | 16 | 7,627 | TZP | 4,603 | 1,734 |
| miR530-B | isotig11608 | 12 | 3,518 | |||
| miR393b-A | td-k31_contig_70403 | 11 | 45 | TIR1-like1 | 269 | 270 |
| TIR1-like2 | 469 | 391 | ||||
| miR156g-A | td-k31_contig_75281 | 34 | 123 | SPL family | Multiple | Multiple |
| PHYC targets | Wild-type | phyC-null | Putative target | Wild-type | phyC-null | |
| miR5200-A | td-k35_contig_61886 | 72 | 955 | FT1 | 342 | 2 |
| miR5200-B | td-k21_contig_55327 | 2,613 | 12,198 | |||
In the phyC-null mutant, we identified a significant upregulation in the expression of both homoeologues of the precursors of miR5200 (Table 4), a miRNA which represses FT1 expression in SD in Brachypodium (Fig. 4b) [61]. Increased levels of miR5200 in the phyC-null mutant may contribute to the very low expression of FT1 in these plants.
Fig. 4.
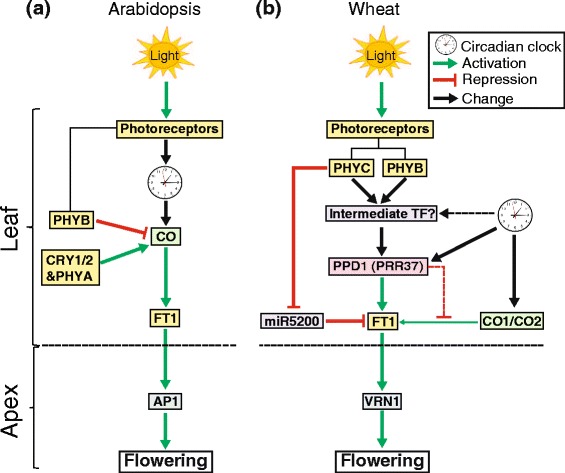
Simplified models of photoperiodic regulation of flowering in (a) Arabidopsis (LD dicot species) based on Valverde et al. [65] and (b) wheat (LD monocot species) based on Chen and Dubcovsky 2012 [87] and Chen et al. 2014 [19]. In Arabidopsis the photoperiodic response is regulated by CONSTANS (CO). In the absence of PHYB, CO proteins accumulate, inducing flowering. In wheat, PPD1/PRR37 is the central regulator of the photoperiodic response. The effects of CO in monocots are observed only in the absence of PPD1/PRR37 [88, 89]. In wheat, PHYB and PHYC are required for the light activation of PPD1/PRR37 so both the phyB-null and phyC-null mutants exhibit a late-flowering phenotype
Finally, we identified 23 transcripts annotated as repetitive elements among the high-confidence PHYB-regulated genes and 11 repetitive elements among the PHYC-regulated genes (Additional file 3). Induced expression of wheat repetitive elements has been observed under stressful conditions [62], but the effect of these changes is currently unknown. Differentially regulated repetitive elements in this study include both DNA transposons and retrotransposons, and the majority were upregulated in the phy-null mutant relative to the wild-type (65 and 91 % were upregulated in the phyB-null mutant and phyC-null mutant, respectively). No repetitive elements were identified among the 82 differentially expressed genes regulated in a concerted manner by both PHYB and PHYC proteins.
Discussion
Phytochromes are ubiquitous among photosynthetic eukaryotes and, within flowering plants, share a highly conserved protein structure [13]. However, despite their similarities at the protein level, differences in phytochrome function have been reported among different species of flowering plants. For example, in both rice and Arabidopsis the PHYC protein is unstable in the absence of other phytochromes, whereas the wheat PHYC protein is stable and functional when transformed into an Arabidopsis mutant lacking all phytochromes [19]. This suggests that in wheat, the PHYB and PHYC proteins can act both independently and in concert to control downstream regulatory pathways.
To characterize genes regulated by PHYB and PHYC in wheat, we applied a highly stringent approach using replicated RNA-seq studies. A FDR-adj P <0.01 was used for each experiment and only those genes that were significant in both experiments were defined as high-confidence genes. Since the two experiments were independent, the high-confidence genes have an FDR < 0.0001, resulting in less than one expected false positive in each selected set (PHYB = 0.04 and PHYB = 0.23). This stringent criterion is likely to exclude many genuine PHY-regulated genes (false negatives), so users of this data are encouraged to analyze the data using different levels of stringency (Additional file 2). The subset of FDR-adj P <0.01 PHYB- and PHYC-regulated genes significant in just one experiment (Additional file 4) likely includes additional true positives which may provide valuable insight into the PHY-mediated regulation of light signaling. Currently, many of the differentially expressed genes identified in this study lack annotation, indicating that additional research will be required to determine their function and their role in the transduction of light signals in wheat.
Potential mechanisms for the concerted action of PHYB and PHYC
We identified 104 genes that were differentially regulated in both phyB-null and phyC-null mutants, suggesting that PHYB and PHYC can act in a concerted manner to regulate a small subset of target genes. Figure 3 presents alternative mechanisms to explain this concerted action. If a gene/protein is regulated by the PHYB-PHYC heterodimer, loss-of-function mutations in either of these phytochromes will result in similar changes in the expression (either induction or repression) of the target gene and all its downstream targets (Fig. 3a). A similar concerted effect will also be observed if PHYB-PHYB and PHYC-PHYC homodimers each regulate the same transcription factor independently, or alternatively, regulate separate transcription factors that have similar effects on the regulation of a third gene (Fig. 3b). If the PHYB-PHYB and PHYC-PHYC homodimers regulate genes that have opposite effects on the regulation of the target gene, the phyB-null and phyC-null mutants will exhibit changes in the expression of this gene in opposing directions (Fig. 3c). This last mechanism can explain the 22 high-confidence genes that were differentially regulated in the phyB-null and phyC-null mutants in the opposite direction (Additional file 3). These regulatory pathways are likely to be more complex than the simplified mechanisms presented in Fig. 3, as some genes may be regulated by both homo- and hetero-PHY dimers. This model is presented only to show that multiple mechanisms can generate similar concerted effects.
Fig. 3.
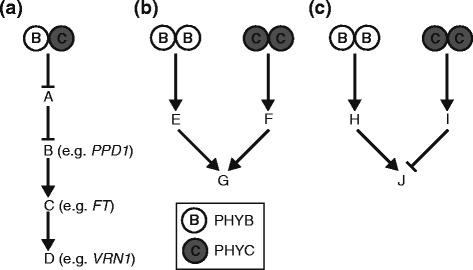
Potential mechanisms of concerted regulation of downstream genes by PHYB and PHYC. a Heterodimer regulation: regulation by a PHYB/PHYC heterodimer results always in changes in the same direction in phyB-null and phyC-null mutants. b-c Homodimer regulation: independent regulation by PHYB/PHYB and PHYC/PHYC homodimers can result in changes in the same or opposite direction. b Gene “G” is differentially regulated in the same direction in both phyB-null and phyC-null mutants. c Gene “J’ is upregulated in the phyB-null and downregulated in phyC-null mutant
In this transcriptome analysis more than 95 % of the high-confidence genes showed significant differences in expression either in the phyB-null or phyC-null mutants alone (Fig. 2c). Genes regulated by PHYB-PHYB or PHYB-PHYA dimers are likely to be included in the set of genes differentially expressed only in the phyB-null mutant, whereas those regulated by PHYC-PHYC or PHYC-PHYA dimers are likely to be included in the set of genes differentially expressed only in the phyC-null mutant. However, these two sets also include genes that are regulated in the same direction in both mutants, but that are highly-significant (significant in both experiments) only for one of the phytochrome mutants.
Differences in phytochrome function among flowering plants
Our study also revealed variable functions of the phytochromes in the regulation of flowering between monocot and dicot flowering plants. Whereas loss-of-function mutations in PHYB are associated with early flowering in Arabidopsis [63], in wheat they are associated with an extreme delay in flowering (Fig. 1b), despite both species exhibiting accelerated flowering in a LD photoperiod. In addition, loss-of-function mutations in PHYC have a limited effect on Arabidopsis flowering under LD [17, 64], but result in extreme late flowering in wheat [19], barley [20] and Brachypodium [21].
These contrasting effects are more likely due to changes downstream of the phytochrome signaling pathway that occurred during the divergence of monocots and dicots than to changes in the phytochrome proteins. In Arabidopsis, LD acceleration of flowering is mediated by CONSTANS (CO). Since PHYB reduces CO abundance during the morning [65] (Fig. 4a), phyB loss-of-function mutations in Arabidopsis result in the accumulation of CO and the subsequent activation of FT and accelerated flowering (Fig. 4a). By contrast, in the temperate grasses the acceleration of flowering under LD is mainly mediated by PPD1/PRR37, a Pseudo Response Regulator that originated from a gene duplication in the grass lineage that originated PRR37 and PRR73, and that was independent of the event that originated the PRR3 and PRR7 genes in Arabidopsis [66] (Fig. 4b). The sub-functionalization of the duplicated PPD1/PRR37 as a photoperiod gene in the grass lineage is not observed in the corresponding Arabidopsis PRR3 or PRR7 genes. In wheat, because both PHYB (Fig. 1b) and PHYC [19] are required for the light activation of PPD1, and its downstream target FT1, loss-of-function mutations in either phytochrome result in extreme delays in flowering under LD, despite the upregulation of CO1 in these mutants [19] (Fig. 4b).
Additional effects of PHYB and PHYC on flowering induction in wheat
Although the PPD1 gene plays a central role in the regulation of the photoperiod response in wheat, the strong downregulation of PPD1 in the phyB-null (120-fold reduction) and phyC-null mutants (30-fold reduction) is not sufficient to explain the drastic delay in flowering time observed in these mutants in LD photoperiods (Fig. 1b, [19]). The combined loss-of-function of the three homoeologues of PPD1 in the hexaploid wheat variety Paragon delays flowering by only one month, whereas the phyB-null and phyC-null mutants exhibit a flowering delay of more than three months. This suggests that PHYB and PHYC also regulate other flowering genes. Additional genes associated with flowering time identified in our study include positive (e.g., GATA transcription factors, VIN3) and negative (e.g., TaAGL33) regulators of flowering. Of the six uncharacterized members of the PEBP family found in our study, five were more highly expressed in the late-flowering phyB-null mutant than in the wild type (Table 3). Some PEBP proteins have been shown to act as floral repressors in other species [67], so it is possible that these upregulated FT-like genes contribute to the late flowering phenotype of the phyB-null mutant. Alternatively, they may be part of a feedback loop that compensates for the lack of FT1 expression and are actually favoring the eventual late flowering of the phyB-null mutant. It would be interesting to characterize loss-of-function mutants for these genes to understand their specific role in regulating flowering time in wheat. We also identified several differentially expressed genes which encode components of the circadian clock (e.g., LHY, CCA1, GI and PRR95). In Arabidopsis, approximately one-third of all genes are transcriptionally regulated by the circadian clock [68], suggesting that the changes generated in the core clock genes in the phyB-null (Table 3) and the phyC-null mutant [19] may result in large-scale changes in gene expression that contribute to the large delay in flowering time observed in these mutants. In the phyC-null mutant, the upregulation of miR5200 and the post-transcriptional downregulation of FT1 [61] may also contribute to the late flowering phenotype of this mutant (Fig. 4b).
PHYB and the shade-avoidance response in wheat
Observations from our phenotypic and transcriptomic studies suggest that PHYC likely plays a narrower role than PHYB in wheat development. Although characterized by delayed flowering and changes in spikelet and floret morphology (e.g., a reduced number of florets and elongated rachillas, glumes, and awns), the phyC-null mutant still produces normal flowers and seeds [19]. The strong effect on flowering time and spike morphology, together with the known role in flowering regulation of many of the genes differentially regulated in the phyC-null mutant suggest that a central role of PHYC is to regulate flowering time and spike development in wheat. However, the longer and narrower leaves found in the phyC-null mutant indicate that this gene also affects other developmental processes.
The phyB-null mutant plants flower later, are sterile and exhibit altered vegetative development (e.g., elongation of the internodes between tillers, longer and wider leaves and an increased rate of leaf production) (Fig. 1c, Additional file 1: Figure S2). In agreement with the phenotypic differences between mutants we found that the phyB-null mutant exhibited differential expression of a larger and more diverse set of genes than the phyC-null mutant. For example, a number of auxin, gibberellin, brassinosteroid and ethylene biosynthesis and signaling genes are differentially expressed in the phyB-null mutant, all hormones which are important regulatory components of the shade-avoidance response [39, 69–71] (Table 3 and Fig. 5). In addition, the phyB-null mutant showed expression changes in the wheat orthologues of several shade-avoidance regulatory genes and in genes with putative roles in regulating cell elongation and architecture (Table 3 and Fig. 5) [48–50, 55].
Experiments controlling R/FR ratios to simulate shaded conditions in dense stands found that wheat yields, even in modern varieties, were reduced in light conditions of low R/FR ratios [72]. Since the low R/FR ratios used in these experiments did not promote stem or leaf sheath growth, the negative effects on yield were attributed to delayed spike development and reduced floret number [72]. Low R/FR ratios favors formation of the inactive Pr form of the phytochromes, so the delayed flowering under these conditions is consistent with the observed delayed flowering in the loss-of function phyB-null and phyC-null mutants. By contrast, in Arabidopsis and other dicot species, low R/FR light ratios (or phyB mutations) result in accelerated reproductive development, a characteristic of the shade-avoidance response [15]. To better characterize the impact of the shade avoidance response in wheat, it will be necessary to separate the effects of low R/FR light on flowering from the effects it has on vegetative development by dissecting the complex transcriptional networks controlled by PHYB.
Further studies will also be required to investigate the links between PHYB-mediated light signaling and tolerance to abiotic stresses such as cold and drought. The wheat phyB-null mutant exhibits a significant reduction in the expression of two ABA receptors, a transcriptional profile which, in Arabidopsis, is indicative of reduced ABA-sensitivity and lower drought tolerance through effects on stomatal conductance [73]. PHYB also activates members of the cold acclimation pathway in response to low R/FR light in Arabidopsis [52]. The principal targets during this response are members of the C-REPEAT/DRE BINDING FACTOR (CBF) family of transcriptional regulators. CBF genes activate downstream COR genes which play a functional role protecting the cell against frost and desiccation damage [52]. While we did not detect any differentially-expressed CBF genes within our dataset, four wheat COR genes were upregulated in the phyB-null mutant and one member of the INDUCER of CBF EXPRESSION (ICE) gene family [53] was downregulated (Table 3), suggesting that the role of PHYB in the light-mediated activation of the cold acclimation pathway observed in Arabidopsis [52] may be conserved in wheat.
Effect of PHYB and PHYC in the regulation of repetitive elements and miRNAs
In addition to their effect on the expression of hundreds of protein-coding genes, both the phyB-null and phyC-null mutants exhibit differential expression of several repetitive elements. This suggests a possible role of the phytochromes (or more likely of its downstream targets) in the transcriptional regulation of such elements.
In addition to directly affecting the rate of transcription, gene activity may also be impacted post-transcriptionally through the activity of miRNAs, a class of small ncRNAs which target specific mRNA transcripts for cleavage and thus inactivation [74]. Both phy-null mutants showed high-confidence differentially expressed miRNAs. The upregulation of pri-miR5200 in the phyC-null mutant relative to the wild-type is consistent with the increased levels of miR5200 observed under SD than under LD in Brachypodium [61]. Since phyC-null plants cannot perceive the LD signal [19], miR5200 levels are maintained at high levels even under LD, whereas they are greatly reduced in these conditions in wild-type wheat and Brachypodium plants [61]. The post-transcriptional downregulation of the central wheat flowering promoter FT1 by miR5200 may contribute to the extremely late flowering of the phyC-null mutant. The phyB-null mutant exhibited increased expression of pri-miR156 (Table 4), which has been associated with a prolonged vegetative state in maize [75] and switchgrass [76], consistent with the phenotype of the wheat phyB-null mutant.
Conclusion and future directions
This study demonstrates that both PHYB and PHYC are required for the acceleration of wheat flowering under LD, an effect which is partially mediated by the transcriptional activation of PPD1. Both phyB-null and phyC-null mutants exhibit changes in the expression of circadian clock genes, and their disruption may contribute to the dramatic flowering delay observed in these plants. In addition, this study revealed that PHYB and PHYC also specifically regulate certain pathways. PHYC activity, but not PHYB is required for the downregulation of miR5200, a post-transcriptional repressor of the flowering promoter FT1. PHYB is actively involved in the regulation of the shade-avoidance response. In contrast with Arabidopsis and the SD grasses, phyB-null mutations in wheat (as well as reduced R/FR ratios) result in delayed flowering. Therefore, a modification of wheat responses to growth in dense stands will require the separation of the effects of PHYB on flowering from its effects on other components of the shade-avoidance response. The PIFs are excellent candidates to initiate the dissection of these pathways since they interact directly with the phytochromes and are critical hubs that integrate temperature, light and hormonal signals to regulate development [6, 77]. RNA-seq experiments using different combinations of wheat pif-null mutants may help to dissect the complex PHYB/PHYC effects described in this study.
Methods
Plant materials
We identified phyB-null mutant lines by screening a TILLING population in the tetraploid Triticum turgidum L. subsp. durum (Desf.) var. Kronos using a protocol described previously [78]. Full-length genomic sequences of the PHYB-A and PHYB-B homoeologues were identified from the draft genome assemblies of Triticum urartu [79] and Aegilops speltoides, respectively, and used to design homoeologue-specific primers to amplify fragments of each gene in ‘Kronos’. PCR-amplification of specific fragments were performed using the following conditions – 95 °C for 5 m, 40 cycles of: 95 °C for 30 s, 62/66 °C for 30 s, 72 °C 1 min/kb; 72 °C 7 min (annealing temperatures were 62 °C and 66 °C for the PHYB-A and PHYB-B TILLING fragments, respectively). Mutations in PHYB-A were detected by CelΙ digestion of a 1875 bp PCR product amplified using the primers PHYB-A-F1 (5’-CTCTCCATCGCTGACGCAGTT-3’) and PHYB-A-R1 (5’-GATTGCTCTGACCCAAATGTCTTC-3’), and mutations in PHYB-B were identified by digesting a 1007 bp PCR fragment amplified with primers PHYB-B-F1 (5’-CCATGTTTGCAGATGTTGCAG-3’) and PHYB-B-R2 (5’-AGGTGTACATCCAGTCAGGTTGCA-3’). A CAPS marker was developed to genotype the phyB-B mutation by digesting the amplified TILLING product with the restriction enzyme HpyCh4V and running products on a 3 % polyacrylamide gel stained with ethidium bromide. After digestion with this enzyme the mutant allele shows a 407 bp band and the wild-type allele shows two fragments of 224 bp and 183 bp. The phyB-A mutation was detected by Sanger sequencing using the sequencing primer 5’-ATATCATCGAGTGGTTGACG-3’. The selected M3 lines carrying phyB-A and phyB-B null mutations were each backcrossed twice to wild-type ‘Kronos’ to reduce the impact of background mutations, before combining them to generate a phyB-null mutant line. Wild-type BC2F2 sister lines from these crosses were used as control plants in each experiment. Because phyB-null plants were sterile, backcrosses and F2 seed production were performed maintaining one of the mutations in a heterozygous state. Null mutations for PHYC were described previously [19]. The source of all plant materials was UC Davis.
Growth conditions
All plants were grown in PGR15 growth chambers (Conviron, Manitoba, Canada) under LD conditions (16 h light/8 h dark) at 20 °C day/18 °C night temperatures and a light intensity of ~260 μM m−2 s−1. All chambers used similar halide light configurations and were located in the same room. PHYB and PHYC experiments were run separately. The two replications of the PHYB experiment were performed one after the other in the same chamber, but the two replications of the PHYC experiment were performed in separate chambers that we later realized had different ballast systems. The light intensity in each growth chamber at R (655-665 nm) and FR (725-735 nm) wavelengths was measured using a FieldSpec® HandHeld 2 visible near-infra-red Spectroradiometer (ASN Inc., Boulder Colorado). All statistical comparisons between mutant and wild-type controls are made within the same chamber so they are unaffected by variation between chambers.
RNA-seq library construction and sequencing
The fully extended 3rd leaf of four-week-old plants was harvested 4 h after the beginning of a 16 h light period (LD) for each genotype and stored immediately in liquid nitrogen. For each experiment, four biological replicates were used. Leaf tissues were ground into a fine powder in liquid nitrogen and total RNA was extracted using the Spectrum™ Plant Total RNA kit (Sigma-Aldrich, St. Louis, MO). Sequencing libraries were produced using the TruSeq RNA Sample Preparation kit v2 (Illumina, San Diego, CA), according to the manufacturer’s instructions. Library quality was determined using a high-sensitivity DNA chip run on a 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA). Libraries were barcoded to allow multiplexing within a single lane and were sequenced using the 50 bp SE module on a HiSeq2000 sequencer at the UC Davis Genome Center.
Raw reads were processed using a pipeline incorporating “Scythe” (https://github.com/vsbuffalo/scythe) to remove Illumina adapter contamination (Default options) and “Sickle” (https://github.com/najoshi/sickle) to remove low-quality reads (Default options except –l 25 –q 25). Trimmed reads were mapped to the A and B chromosome arms from the latest version of the draft wheat genome assembly in the hexaploid variety Chinese Spring (v2.2) from the IWGSC [31]. RNA-seq reads were mapped using GSNAPl, a splicing-aware aligner (version 05-09-2013, default parameters except -m 2 -n 1 -N 1 -A sam [80]) to generate Sequence Alignment/Map (SAM) files for each sample.
To define regions corresponding to transcribed gene-coding regions within this reference assembly, we performed a similar analysis as described previously [81]. Briefly, a non-redundant set of wheat transcripts from several transcriptomes were mapped to the A and B chromosome arm assemblies separately using GMAP (Version 05-09-2013), default parameters except -n 1 --nofails --cross-species -f samse -x 0 [82]. Bedtools cluster (-d 0) was then used to merge overlapping aligned regions, followed by bedtools merge to merge overlapping regions into a single putative transcribed region. The resulting General Feature Format (GFF) file consisted of 150,754 genomic ranges, each representing the genomic contig identifier and the start and end coordinate of the putative transcribed region.
Raw count values were generated using ht-seq count (-m union) using the generated GFF file and individual Sequence Alignment/Mapping (SAM) files for each sample. All reads with a mapping quality (MAPQ) value less than 40 from the SAM file (signifying an ambiguous, non-unique mapping position), were discarded at this stage, ensuring that expression values were generated using only uniquely-mapped reads. This approach generates homoeologue-specific expression profiles [32]. The percent of uniquely-mapped reads in each sample is described in Additional file 1: Table S1.
We used a custom ‘R’ package ‘noleaven’ (https://github.com/topherconley/noleaven) to remove contigs which had zero or very low numbers of counts. For each experimental replication, contigs which had less than three reads mapping to at least two biological replicates in the experiment were removed.
Raw counts were normalized using DESeq (Version 1.12.1 [83], R Version 2.14.2). After normalization, we applied the statistical tests implemented in both DESeq and edgeR [84] to classify differentially expressed genes in pair-wise comparisons. The P-values generated by both analyses were adjusted for FDR, using the procedure of Benjamini and Hochberg [85] and we selected a stringent cutoff of adjusted P ≤0.01 for significance for both tests within each experimental replication. Throughout the paper, both DESeq and edgeR results are presented as FDR-adjusted P values. We then selected for further analyses those genes that were significant in both replications under the criteria outlined above, and designated those genes as “high-confidence” differentially expressed genes (FDR < 0.0001).
Functional annotation
For functional annotation, we identified the longest transcribed contig mapping to each genomic locus and performed a BLASTX against the nr protein database (nr.28, Apr 24, 2015 release, NCBI) and a BLASTP using the translated ORF against the Pfam database version 27.0 with InterProScan version 5.13 to identify conserved protein domains. The output was used to infer GO terms associated with each genomic locus using BLAST2GO version 2.6.5 and used the ‘R’ package TopGO version 2.14.0 to perform an enrichment analysis among the differentially regulated gene sets. “Biological Process” terms were obtained and significance values for enrichment were calculated using ‘classic’ Fishers’ exact test, as implemented in TopGO. Wheat miRNAs were annotated based on the closest rice homologue identified from the miRNA database “miRBase” (Release 21, [86]), except for miR5200, which was identified and annotated using the Brachypodium homologues [61].
qRT-PCR validation of flowering time genes
The most recently-emerged leaf from wild-type and phyB-null plants were collected four hours after the beginning of a 16-h photoperiod from 2-week, 4-week and 6-week old plants. Harvested tissue was ground to a fine powder in liquid nitrogen and RNA was extracted as described above for RNA-seq library preparation. cDNA was synthesized using the High Capacity Reverse Transcription Kit (Applied Biosystems, Foster City, CA) according to the manufacturer’s instructions. Quantitative RT-PCR was performed using SYBR Green and a 7500 Fast Real-Time PCR system (Applied Biosystems, Foster City, CA). Primers for the target genes PPD1, CO1, FT1, FT2, VRN1, VRN2 and the control gene ACTIN were described previously [19, 27]. Expression data are presented as fold-ACTIN levels.
Abbreviations
ABA, Abscisic Acid; bHLH, basic Helix Loop Helix; BR, Brassinosteroid; FDR, False Discovery Rate; FR, Far red; GA, Gibberellin; GAF, cGMP phosphodiesterase/adenylate cyclase/FhlA; GFF, General Feature Format; LD, Long Day; MAPQ, Mapping Quality; PAS, period, aryl hydrocarbon receptor nuclear transporter and single minded; PEBP, Phosphatidylethanolamine-Binding Proteins; PHY, Phytochrome; PIF, Phytochrome Interacting Factor; R, Red; SAM, Sequence Alignment/Map; SD, Short Day; TILLING, Targeting Induced Local Lesions IN Genomes
Acknowledgements
We thank Dr. Neal Willits (Dept. Statistics, UC Davis) for valuable discussions regarding statistical significance, Tyson Howell for help in interpreting statistical data and Mei Yee Lau for excellent technical support.
Funding
This project was supported by NRI grants number 2011-67013-30077 and 2016-67013-24617 from the USDA National Institute of Food and Agriculture and by the Howard Hughes Medical Institute and the Gordon and Betty Moore Foundation grant number GBMF3031. Support for publication costs was provided by the Colorado State University Libraries Open Access Research and Scholarship Fund.
Availability of data and materials
RNA-seq reads, mapping reference and annotation files for this study were deposited in the National Center for Biotechnology Information’s Gene Expression Omnibus under accession number GSE79049 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE79049).
Authors' contributions
SP and JD designed the experiment. AC and NK generated and characterized the PHYB and PHYC mutants. SP performed and analyzed the RNA-seq experiment and generated the first and last draft of the manuscript. JMD performed the miRNA analyses. JD contributed to the statistical analysis and to the revision of the manuscript. All authors have read and approved the manuscript.
Competing interests
The authors declare that they have no competing interests.
Additional files
Summary of RNA-seq reads and mapping. Table S2: TopGO analysis for functional enrichment for genes regulated by PHYB, PHYC and in concert by both PHYB and PHYC. Figure S1. Spike and floral organ phenotype of the phyB-null mutant. (a) Whole spike, (b) Single spikelet (c) Separated spikelet and (d) stamen and stigma. (e) Comparison of internode length between 83-day-old phyB-null, phyC-null and wild-type Kronos plants. Leaves have been removed to facilitate visualization of internodes. Nodes are indicated by purple arrows. Bar = 10 cm. Figure S2. Vegetative phenotype of wild-type control, phyB-null and phyC-null plants. (a) Leaf emergence rate, (b) leaf length and (c) width at three different timepoints. * P <0.05; ** P <0.01. Figure S3. Phenotype of four-week old PHYB wild-type, phyB-null, PHYC wild-type and phyC-null plants at the stage of harvest for RNA-seq analysis. Figure S4. Principal Component Analysis of normalized expression values of all genes. (a) All libraries, (b) PHYB libraries and (c) PHYC libraries. Figure S5. Relative transcript levels of six flowering time genes determined by qRT-PCR in wild-type and phyB-null mutants at three stages of development (Leaves from two-week, four-week and six-week-old plants). Expression levels are presented as fold-ACTIN. * P < 0.05, ** P < 0.01, *** P < 0.001. (PDF 537 kb)
All RNA-seq data. Columns describe the genomic locus, the associated longest transcript which mapped to this locus, top BLAST hit and description and e-value, normalized counts from each library, fold-change (wild-type/phy-null mutant, genes highlighted green are upregulated in the phy-null mutant, genes highlighted red are downregulated in the phy-null mutant) and FDR-adjusted P values from DESeq and EdgeR (Padj <0.01 highlighted in yellow). (XLSX 60849 kb)
High-confidence PHYB/PHYC-regulated genes. In different tabs, 2202 PHYB-regulated genes, 261 PHYC-regulated genes, 82 genes showing concerted regulation by PHYB and PHYC in the same direction and 22 genes showing concerted regulation by PHYB and PHYC in the opposite direction. Columns describe the genomic locus, the associated longest transcript which mapped to this locus, top BLAST hit and description and e-value, normalized counts from each library, fold-change (wild-type/phy-null mutant, genes highlighted green are upregulated in the phy-null mutant, genes highlighted red are downregulated in the phy-null mutant) and FDR-adjusted P values from DESeq and EdgeR (Padj <0.01 highlighted in yellow). (XLSX 1200 kb)
FDR-adj P <0.01 PHYB/PHYC-regulated genes significant in just one experiment. In different tabs, 5320, PHYB-regulated genes, 1037 PHYC-regulated genes and 336 genes showing concerted regulation by PHYB and PHYC. Columns describe the genomic locus, the associated longest transcript which mapped to this locus, top BLAST hit and description and e-value, normalized counts from each library, fold-change (wild-type/phy-null mutant, genes highlighted green are upregulated in the phy-null mutant, genes highlighted red are downregulated in the phy-null mutant) and FDR-adjusted P values from DESeq and EdgeR (Padj <0.01 highlighted in yellow). (XLSX 3100 kb)
Contributor Information
Stephen Pearce, Email: stephen.pearce@colostate.edu.
Nestor Kippes, Email: nfkippes@ucdavis.edu.
Andrew Chen, Email: a.chen2@uq.edu.au.
Juan Manuel Debernardi, Email: jmdebernardi@ucdavis.edu.
Jorge Dubcovsky, Email: jdubcovsky@ucdavis.edu.
References
- 1.Möglich A, Yang X, Ayers RA, Moffat K. Structure and function of plant photoreceptors. Annu Rev Plant Biol. 2010;61:21–47. doi: 10.1146/annurev-arplant-042809-112259. [DOI] [PubMed] [Google Scholar]
- 2.Franklin KA, Quail PH. Phytochrome functions in Arabidopsis development. J Exp Bot. 2010;61:11–24. doi: 10.1093/jxb/erp304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rockwell NC, Su YS, Lagarias JC. Phytochrome structure and signaling mechanisms. Annu Rev Plant Biol. 2006;57:837–58. doi: 10.1146/annurev.arplant.56.032604.144208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Müller R, Fernández AP, Hiltbrunner A, Schäfer E, Kretsch T. The histidine kinase-related domain of Arabidopsis phytochrome A controls the spectral sensitivity and the subcellular distribution of the photoreceptor. Plant Physiol. 2009;150:1297–309. doi: 10.1104/pp.109.135988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nagatani A. Light-regulated nuclear localization of phytochromes. Curr Opin Plant Biol. 2004;7:708–11. doi: 10.1016/j.pbi.2004.09.010. [DOI] [PubMed] [Google Scholar]
- 6.Leivar P, Monte E. PIFs: systems integrators in plant development. Plant Cell. 2014;26:56–78. doi: 10.1105/tpc.113.120857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Leivar P, Tepperman JM, Cohn MM, Monte E, Al-Sady B, Erickson E, Quail PH. Dynamic antagonism between phytochromes and PIF family basic helix-loop-helix factors induces selective reciprocal responses to light and shade in a rapidly responsive transcriptional network in Arabidopsis. Plant Cell. 2012;24:1398–419. doi: 10.1105/tpc.112.095711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hornitschek P, Kohnen MV, Lorrain S, Rougemont J, Ljung K, López-Vidriero I, Franco-Zorrilla JM, Solano R, Trevisan M, Pradervand S, et al. Phytochrome interacting factors 4 and 5 control seedling growth in changing light conditions by directly controlling auxin signaling. Plant J. 2012;71:699–711. doi: 10.1111/j.1365-313X.2012.05033.x. [DOI] [PubMed] [Google Scholar]
- 9.Al-Sady B, Ni W, Kircher S, Schäfer E, Quail PH. Photoactivated phytochrome induces rapid PIF3 phosphorylation prior to proteasome-mediated degradation. Mol Cell. 2006;23:439–46. doi: 10.1016/j.molcel.2006.06.011. [DOI] [PubMed] [Google Scholar]
- 10.Park E, Park J, Kim J, Nagatani A, Lagarias JC, Choi G. Phytochrome B inhibits binding of phytochrome-interacting factors to their target promoters. Plant J. 2012;72:537–46. doi: 10.1111/j.1365-313X.2012.05114.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sakuraba Y, Jeong J, Kang MY, Kim J, Paek NC, Choi G. Phytochrome-interacting transcription factors PIF4 and PIF5 induce leaf senescence in Arabidopsis. Nat Commun. 2014;5:4636. doi: 10.1038/ncomms5636. [DOI] [PubMed] [Google Scholar]
- 12.Ni W, Xu SL, Tepperman JM, Stanley DJ, Maltby DA, Gross JD, Burlingame AL, Wang ZY, Quail PH. A mutually assured destruction mechanism attenuates light signaling in Arabidopsis. Science. 2014;344:1160–4. doi: 10.1126/science.1250778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li FW, Melkonian M, Rothfels CJ, Villarreal JC, Stevenson DW, Graham SW, Wong GK, Pryer KM, Mathews S. Phytochrome diversity in green plants and the origin of canonical plant phytochromes. Nat Commun. 2015;6:7852. doi: 10.1038/ncomms8852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Casal JJ, Candia AN, Sellaro R. Light perception and signalling by phytochrome A. J Exp Bot. 2014;65:2835–45. doi: 10.1093/jxb/ert379. [DOI] [PubMed] [Google Scholar]
- 15.Casal JJ. Photoreceptor signaling networks in plant responses to shade. Annu Rev Plant Biol. 2013;64:403–27. doi: 10.1146/annurev-arplant-050312-120221. [DOI] [PubMed] [Google Scholar]
- 16.Franklin KA, Davis SJ, Stoddart WM, Vierstra RD, Whitelam GC. Mutant analyses define multiple roles for phytochrome C in Arabidopsis photomorphogenesis. Plant Cell. 2003;15:1981–9. doi: 10.1105/tpc.015164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Monte E, Alonso JM, Ecker JR, Zhang Y, Li X, Young J, Austin-Phillips S, Quail PH. Isolation and characterization of phyC mutants in Arabidopsis reveals complex crosstalk between phytochrome signaling pathways. Plant Cell. 2003;15:1962–80. doi: 10.1105/tpc.012971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Takano M, Inagaki N, Xie X, Yuzurihara N, Hihara F, Ishizuka T, Yano M, Nishimura M, Miyao A, Hirochika H, et al. Distinct and cooperative functions of phytochromes A, B, and C in the control of deetiolation and flowering in rice. Plant Cell. 2005;17:3311–25. doi: 10.1105/tpc.105.035899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen A, Li C, Hu W, Lau MY, Lin H, Rockwell NC, Martin SS, Jernstedt JA, Lagarias JC, Dubcovsky J. PHYTOCHROME C plays a major role in the acceleration of wheat flowering under long-day photoperiod. Proc Natl Acad Sci U S A. 2014;111:10037–44. doi: 10.1073/pnas.1409795111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nishida H, Ishihara D, Ishii M, Kaneko T, Kawahigashi H, Akashi Y, Saisho D, Tanaka K, Handa H, Takeda K, et al. Phytochrome C is a key factor controlling long-day flowering in barley. Plant Physiol. 2013;163:804–14. doi: 10.1104/pp.113.222570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Woods DP, Ream TS, Minevich G, Hobert O, Amasino RM. PHYTOCHROME C is an essential light receptor for photoperiodic flowering in the temperate grass, Brachypodium distachyon. Genetics. 2014;198:397–408. doi: 10.1534/genetics.114.166785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Beales J, Turner A, Griffiths S, Snape JW, Laurie DA. A Pseudo-Response Regulator is misexpressed in the photoperiod insensitive Ppd-D1a mutant of wheat (Triticum aestivum L.) Theor Appl Genet. 2007;115:721–33. doi: 10.1007/s00122-007-0603-4. [DOI] [PubMed] [Google Scholar]
- 23.Turner A, Beales J, Faure S, Dunford RP, Laurie DA. The pseudo-response regulator Ppd-H1 provides adaptation to photoperiod in barley. Science. 2005;310:1031–4. doi: 10.1126/science.1117619. [DOI] [PubMed] [Google Scholar]
- 24.Wilhelm EP, Turner AS, Laurie DA. Photoperiod insensitive Ppd-A1a mutations in tetraploid wheat (Triticum durum Desf.) Theor Appl Genet. 2009;118:285–94. doi: 10.1007/s00122-008-0898-9. [DOI] [PubMed] [Google Scholar]
- 25.Worland A. The influence of flowering time genes on environmental adaptability in European wheats. Euphytica. 1996;89:49–57. doi: 10.1007/BF00015718. [DOI] [Google Scholar]
- 26.Yan L, Fu D, Li C, Blechl A, Tranquilli G, Bonafede M, Sanchez A, Valarik M, Yasuda S, Dubcovsky J. The wheat and barley vernalization gene VRN3 is an orthologue of FT. Proc Natl Acad Sci U S A. 2006;103:19581–6. doi: 10.1073/pnas.0607142103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li C, Dubcovsky J. Wheat FT protein regulates VRN1 transcription through interactions with FDL2. Plant J. 2008;55:543–54. doi: 10.1111/j.1365-313X.2008.03526.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li C, Lin H, Dubcovsky J. Factorial combinations of protein interactions generate a multiplicity of florigen activation complexes in wheat and barley. Plant J. 2015;84:70–82. doi: 10.1111/tpj.12960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shaw LM, Turner AS, Herry L, Griffiths S, Laurie DA. Mutant alleles of Photoperiod-1 in wheat (Triticum aestivum L.) that confer a late flowering phenotype in long days. PLoS One. 2013;8:e79459. doi: 10.1371/journal.pone.0079459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lv B, Nitcher R, Han X, Wang S, Ni F, Li K, et al. Characterization of FLOWERING LOCUS T1 (FT1) gene in Brachypodium and wheat. PLoS One. 2014;9:e94171. [DOI] [PMC free article] [PubMed]
- 31.IWGSC A chromosome-based draft sequence of the hexaploid bread wheat (Triticum aestivum) genome. Science. 2014;345:1251788. doi: 10.1126/science.1251788. [DOI] [PubMed] [Google Scholar]
- 32.Pearce S, Vazquez-Gross H, Herin SY, Hane D, Wang Y, Gu YQ, Dubcovsky J. WheatExp: an RNA-seq expression database for polyploid wheat. BMC Plant Biol. 2015;15:299. doi: 10.1186/s12870-015-0692-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kikuchi R, Kawahigashi H, Oshima M, Ando T, Handa H. The differential expression of HvCO9, a member of the CONSTANS-like gene family, contributes to the control of flowering under short-day conditions in barley. J Exp Bot. 2012;63:773–84. doi: 10.1093/jxb/err299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Richter R, Behringer C, Müller IK, Schwechheimer C. The GATA-type transcription factors GNC and GNL/CGA1 repress gibberellin signaling downstream from DELLA proteins and PHYTOCHROME-INTERACTING FACTORS. Genes Dev. 2010;24:2093–104. doi: 10.1101/gad.594910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Richter R, Behringer C, Zourelidou M, Schwechheimer C. Convergence of auxin and gibberellin signaling on the regulation of the GATA transcription factors GNC and GNL in Arabidopsis thaliana. Proc Natl Acad Sci U S A. 2013;110:13192–7. doi: 10.1073/pnas.1304250110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Suárez-López P, Wheatley K, Robson F, Onouchi H, Valverde F, Coupland G. CONSTANS mediates between the circadian clock and the control of flowering in Arabidopsis. Nature. 2001;410:1116–20. doi: 10.1038/35074138. [DOI] [PubMed] [Google Scholar]
- 37.Greenup AG, Sasani S, Oliver SN, Talbot MJ, Dennis ES, Hemming MN, Trevaskis B. ODDSOC2 is a MADS box floral repressor that is down-regulated by vernalization in temperate cereals. Plant Physiol. 2010;153:1062–73. doi: 10.1104/pp.109.152488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fu D, Dunbar M, Dubcovsky J. Wheat VIN3-like PHD finger genes are up-regulated by vernalization. Mol Genet Genomics. 2007;277:301–13. doi: 10.1007/s00438-006-0189-6. [DOI] [PubMed] [Google Scholar]
- 39.Keuskamp DH, Pollmann S, Voesenek LA, Peeters AJ, Pierik R. Auxin transport through PIN-FORMED 3 (PIN3) controls shade avoidance and fitness during competition. Proc Natl Acad Sci U S A. 2010;107:22740–4. doi: 10.1073/pnas.1013457108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hisamatsu T, King RW, Helliwell CA, Koshioka M. The involvement of gibberellin 20-oxidase genes in phytochrome-regulated petiole elongation of Arabidopsis. Plant Physiol. 2005;138:1106–16. doi: 10.1104/pp.104.059055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pearce S, Vanzetti LS, Dubcovsky J. Exogenous gibberellins induce wheat spike development under short days only in the presence of VERNALIZATION1. Plant Physiol. 2013;163:1433–45. doi: 10.1104/pp.113.225854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Todaka D, Nakashima K, Maruyama K, Kidokoro S, Osakabe Y, Ito Y, Matsukura S, Fujita Y, Yoshiwara K, Ohme-Takagi M, et al. Rice phytochrome-interacting factor-like protein OsPIL1 functions as a key regulator of internode elongation and induces a morphological response to drought stress. Proc Natl Acad Sci U S A. 2012;109:15947–52. doi: 10.1073/pnas.1207324109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cifuentes-Esquivel N, Bou-Torrent J, Galstyan A, Gallemí M, Sessa G, Salla Martret M, Roig-Villanova I, Ruberti I, Martinez-García JF. The bHLH proteins BEE and BIM positively modulate the shade avoidance syndrome in Arabidopsis seedlings. Plant J. 2013;75:989–1002. doi: 10.1111/tpj.12264. [DOI] [PubMed] [Google Scholar]
- 44.Steindler C, Matteucci A, Sessa G, Weimar T, Ohgishi M, Aoyama T, Morelli G, Ruberti I. Shade avoidance responses are mediated by the ATHB-2 HD-zip protein, a negative regulator of gene expression. Development. 1999;126:4235–45. doi: 10.1242/dev.126.19.4235. [DOI] [PubMed] [Google Scholar]
- 45.Whipple CJ, Kebrom TH, Weber AL, Yang F, Hall D, Meeley R, Schmidt R, Doebley J, Brutnell TP, Jackson DP. grassy tillers1 promotes apical dominance in maize and responds to shade signals in the grasses. Proc Natl Acad Sci U S A. 2011;108:E506–12. doi: 10.1073/pnas.1102819108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stone JM, Liang X, Nekl ER, Stiers JJ. Arabidopsis AtSPL14, a plant-specific SBP-domain transcription factor, participates in plant development and sensitivity to fumonisin B1. Plant J. 2005;41:744–54. doi: 10.1111/j.1365-313X.2005.02334.x. [DOI] [PubMed] [Google Scholar]
- 47.Rose JK, Braam J, Fry SC, Nishitani K. The XTH family of enzymes involved in xyloglucan endotransglucosylation and endohydrolysis: current perspectives and a new unifying nomenclature. Plant Cell Physiol. 2002;43:1421–35. doi: 10.1093/pcp/pcf171. [DOI] [PubMed] [Google Scholar]
- 48.Jin J, Huang W, Gao JP, Yang J, Shi M, Zhu MZ, Luo D, Lin HX. Genetic control of rice plant architecture under domestication. Nat Genet. 2008;40:1365–9. doi: 10.1038/ng.247. [DOI] [PubMed] [Google Scholar]
- 49.Tan L, Li X, Liu F, Sun X, Li C, Zhu Z, Fu Y, Cai H, Wang X, Xie D, et al. Control of a key transition from prostrate to erect growth in rice domestication. Nat Genet. 2008;40:1360–4. doi: 10.1038/ng.197. [DOI] [PubMed] [Google Scholar]
- 50.Lee YK, Kim GT, Kim IJ, Park J, Kwak SS, Choi G, et al. LONGIFOLIA1 and LONGIFOLIA2, two homologous genes, regulate longitudinal cell elongation in Arabidopsis. Development. 2006;133:4305–14. [DOI] [PubMed]
- 51.Brzeski J, Podstolski W, Olczak K, Jerzmanowski A. Identification and analysis of the Arabidopsis thaliana BSH gene, a member of the SNF5 gene family. Nucleic Acids Res. 1999;27:2393–9. doi: 10.1093/nar/27.11.2393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Franklin KA, Whitelam GC. Light-quality regulation of freezing tolerance in Arabidopsis thaliana. Nat Genet. 2007;39:1410–3. doi: 10.1038/ng.2007.3. [DOI] [PubMed] [Google Scholar]
- 53.Badawi M, Reddy YV, Agharbaoui Z, Tominaga Y, Danyluk J, Sarhan F, Houde M. Structure and functional analysis of wheat ICE (inducer of CBF expression) genes. Plant Cell Physiol. 2008;49:1237–49. doi: 10.1093/pcp/pcn100. [DOI] [PubMed] [Google Scholar]
- 54.Jones-Rhoades MW, Bartel DP, Bartel B. MicroRNAS and their regulatory roles in plants. Annu Rev Plant Biol. 2006;57:19–53. doi: 10.1146/annurev.arplant.57.032905.105218. [DOI] [PubMed] [Google Scholar]
- 55.Loudet O, Michael TP, Burger BT, Le Mette C, Mockler TC, Weigel D, Chory J. A zinc knuckle protein that negatively controls morning-specific growth in Arabidopsis thaliana. Proc Natl Acad Sci U S A. 2008;105:17193–8. doi: 10.1073/pnas.0807264105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li YF, Zheng Y, Jagadeeswaran G, Sunkar R. Characterization of small RNAs and their target genes in wheat seedlings using sequencing-based approaches. Plant Sci. 2013;203-204:17–24. doi: 10.1016/j.plantsci.2012.12.014. [DOI] [PubMed] [Google Scholar]
- 57.Sun F, Guo G, Du J, Guo W, Peng H, Ni Z, et al. Whole-genome discovery of miRNAs and their targets in wheat (Triticum aestivum L.). BMC Plant Biol. 2014;14:142. [DOI] [PMC free article] [PubMed]
- 58.Sun W, Xu XH, Wu X, Wang Y, Lu X, Sun H, et al. Genome-wide identification of microRNAs and their targets in wild type and phyB mutant provides a key link between microRNAs and the phyB-mediated light signaling pathway in rice. Front Plant Sci. 2015;6:372. [DOI] [PMC free article] [PubMed]
- 59.Tang Z, Zhang L, Xu C, Yuan S, Zhang F, Zheng Y, et al. Uncovering small RNA-mediated responses to cold stress in a wheat thermosensitive genic male-sterile line by deep sequencing. Plant Physiol. 2012;159:721–38. [DOI] [PMC free article] [PubMed]
- 60.Wang JW, Czech B, Weigel D. miR156-regulated SPL transcription factors define an endogenous flowering pathway in Arabidopsis thaliana. Cell. 2009;138:738–49. doi: 10.1016/j.cell.2009.06.014. [DOI] [PubMed] [Google Scholar]
- 61.Wu L, Liu D, Wu J, Zhang R, Qin Z, Liu D, Li A, Fu D, Zhai W, Mao L. Regulation of FLOWERING LOCUS T by a microRNA in Brachypodium distachyon. Plant Cell. 2013;25:4363–77. doi: 10.1105/tpc.113.118620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Echenique V, Stamova B, Wolters P, Lazo G, Carollo L, Dubcovsky J. Frequencies of Ty1-copia and Ty3-gypsy retroelements within the Triticeae EST databases. Theor Appl Genet. 2002;104:840–4. doi: 10.1007/s00122-001-0849-1. [DOI] [PubMed] [Google Scholar]
- 63.Reed JW, Nagpal P, Poole DS, Furuya M, Chory J. Mutations in the gene for the red/far-red light receptor phytochrome B alter cell elongation and physiological responses throughout Arabidopsis development. Plant Cell. 1993;5:147–57. doi: 10.1105/tpc.5.2.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hu W, Franklin KA, Sharrock RA, Jones MA, Harmer SL, Lagarias JC. Unanticipated regulatory roles for Arabidopsis phytochromes revealed by null mutant analysis. Proc Natl Acad Sci U S A. 2013;110:1542–7. doi: 10.1073/pnas.1221738110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Valverde F, Mouradov A, Soppe W, Ravenscroft D, Samach A, Coupland G. Photoreceptor regulation of CONSTANS protein in photoperiodic flowering. Science. 2004;303:1003–6. doi: 10.1126/science.1091761. [DOI] [PubMed] [Google Scholar]
- 66.Farré EM, Liu T. The PRR family of transcriptional regulators reflects the complexity and evolution of plant circadian clocks. Curr Opin Plant Biol. 2013;16:621–9. doi: 10.1016/j.pbi.2013.06.015. [DOI] [PubMed] [Google Scholar]
- 67.Wickland DP, Hanzawa Y. The FLOWERING LOCUS T/TERMINAL FLOWER 1 gene family: functional evolution and molecular mechanisms. Mol Plant. 2015;8:983–97. doi: 10.1016/j.molp.2015.01.007. [DOI] [PubMed] [Google Scholar]
- 68.Covington MF, Maloof JN, Straume M, Kay SA, Harmer SL. Global transcriptome analysis reveals circadian regulation of key pathways in plant growth and development. Genome Biol. 2008;9:R130. doi: 10.1186/gb-2008-9-8-r130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Djakovic-Petrovic T, de Wit M, Voesenek LA, Pierik R. DELLA protein function in growth responses to canopy signals. Plant J. 2007;51:117–26. doi: 10.1111/j.1365-313X.2007.03122.x. [DOI] [PubMed] [Google Scholar]
- 70.Pierik R, Djakovic-Petrovic T, Keuskamp DH, de Wit M, Voesenek LA. Auxin and ethylene regulate elongation responses to neighbor proximity signals independent of gibberellin and della proteins in Arabidopsis. Plant Physiol. 2009;149:1701–12. doi: 10.1104/pp.108.133496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tao Y, Ferrer JL, Ljung K, Pojer F, Hong F, Long JA, Li L, Moreno JE, Bowman ME, Ivans LJ, et al. Rapid synthesis of auxin via a new tryptophan-dependent pathway is required for shade avoidance in plants. Cell. 2008;133:164–76. doi: 10.1016/j.cell.2008.01.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ugarte CC, Trupkin SA, Ghiglione H, Slafer G, Casal JJ. Low red/far-red ratios delay spike and stem growth in wheat. J Exp Bot. 2010;61:3151–62. doi: 10.1093/jxb/erq140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.González CV, Ibarra SE, Piccoli PN, Botto JF, Boccalandro HE. Phytochrome B increases drought tolerance by enhancing ABA sensitivity in Arabidopsis thaliana. Plant Cell Environ. 2012;35:1958–68. doi: 10.1111/j.1365-3040.2012.02529.x. [DOI] [PubMed] [Google Scholar]
- 74.Bologna NG, Voinnet O. The diversity, biogenesis, and activities of endogenous silencing small RNAs in Arabidopsis. Annu Rev Plant Biol. 2014;65:473–503. doi: 10.1146/annurev-arplant-050213-035728. [DOI] [PubMed] [Google Scholar]
- 75.Chuck G, Cigan AM, Saeteurn K, Hake S. The heterochronic maize mutant Corngrass1 results from overexpression of a tandem microRNA. Nat Genet. 2007;39:544–9. doi: 10.1038/ng2001. [DOI] [PubMed] [Google Scholar]
- 76.Chuck GS, Tobias C, Sun L, Kraemer F, Li C, Dibble D, Arora R, Bragg JN, Vogel JP, Singh S, et al. Overexpression of the maize Corngrass1 microRNA prevents flowering, improves digestibility, and increases starch content of switchgrass. Proc Natl Acad Sci U S A. 2011;108:17550–5. doi: 10.1073/pnas.1113971108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.de Lucas M, Prat S. PIFs get BRright: PHYTOCHROME INTERACTING FACTORs as integrators of light and hormonal signals. New Phytol. 2014;202:1126–41. doi: 10.1111/nph.12725. [DOI] [PubMed] [Google Scholar]
- 78.Uauy C, Paraiso F, Colasuonno P, Tran RK, Tsai H, Berardi S, Comai L, Dubcovsky J. A modified TILLING approach to detect induced mutations in tetraploid and hexaploid wheat. BMC Plant Biol. 2009;9:115. doi: 10.1186/1471-2229-9-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ling HQ, Zhao S, Liu D, Wang J, Sun H, Zhang C, Fan H, Li D, Dong L, Tao Y, et al. Draft genome of the wheat A-genome progenitor Triticum urartu. Nature. 2013;496:87–90. doi: 10.1038/nature11997. [DOI] [PubMed] [Google Scholar]
- 80.Wu TD, Nacu S. Fast and SNP-tolerant detection of complex variants and splicing in short reads. Bioinformatics. 2010;26:873–81. doi: 10.1093/bioinformatics/btq057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Pearce S, Tabbita F, Cantu D, Buffalo V, Avni R, Vazquez-Gross H, Zhao R, Conley CJ, Distelfeld A, Dubcovksy J. Regulation of Zn and Fe transporters by the GPC1 gene during early wheat monocarpic senescence. BMC Plant Biol. 2014;14:368. doi: 10.1186/s12870-014-0368-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wu TD, Watanabe CK. GMAP: a genomic mapping and alignment program for mRNA and EST sequences. Bioinformatics. 2005;21:1859–75. doi: 10.1093/bioinformatics/bti310. [DOI] [PubMed] [Google Scholar]
- 83.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–40. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Benjamini Y, Hochberg Y. Controlling the false discovery rate - a practical and powerful approach to multiple testing. J R Stat Soc Series B Stat Methodol. 1995;57:289–300. [Google Scholar]
- 86.Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34:D140–4. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Chen A, Dubcovsky J. Wheat TILLING mutants show that the vernalization gene VRN1 down-regulates the flowering repressor VRN2 in leaves but is not essential for flowering. PLoS Genet. 2012;8:e1003134. doi: 10.1371/journal.pgen.1003134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Alqudah AM, Sharma R, Pasam RK, Graner A, Kilian B, Schnurbusch T. Genetic dissection of photoperiod response based on GWAS of pre-anthesis phase duration in spring barley. PLoS One. 2014;9:e113120. doi: 10.1371/journal.pone.0113120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Yang S, Murphy RL, Morishige DT, Klein PE, Rooney WL, Mullet JE. Sorghum phytochrome B inhibits flowering in long days by activating expression of SbPRR37 and SbGHD7, repressors of SbEHD1, SbCN8 and SbCN12. PLoS One. 2014;9:e105352. doi: 10.1371/journal.pone.0105352. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
RNA-seq reads, mapping reference and annotation files for this study were deposited in the National Center for Biotechnology Information’s Gene Expression Omnibus under accession number GSE79049 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE79049).


