Abstract
Age-related macular degeneration (AMD) is an ocular neurodegenerative disorder and is the leading cause of legal blindness in Western societies, with a prevalence of up to 8 % over the age of 60, which continues to increase with age. AMD is characterized by the progressive breakdown of the macula (the central region of the retina), resulting in the loss of central vision including visual acuity. While its molecular etiology remains unclear, advances in genetics and genomics have illuminated the genetic architecture of the disease and have generated attractive pathomechanistic hypotheses. Here, we review the genetic architecture of AMD, considering the contribution of both common and rare alleles to susceptibility, and we explore the possible mechanistic links between photoreceptor degeneration and the alternative complement pathway, a cascade that has emerged as the most potent genetic driver of this disorder.
Background
Age-related macular degeneration is the third leading cause of vision loss worldwide. It is a late-onset disease with a complex etiology. Major risk factors contributing to susceptibility include age, family history, and smoking [1–3]. The earliest clinical manifestations of age-related macular degeneration (AMD) are focal deposits of debris, termed drusen, which are also considered to be a normal part of aging, present almost ubiquitously in the eyes of healthy individuals over the age of 50. Progression into the spectrum of pathological consequences begins with excessive accumulation of drusen in the central retina during the early/intermediate stages of AMD, followed by localized inflammation, and ultimately neurodegeneration in the macula that characterizes advanced stages of AMD.
Combining epidemiological and genetic approaches has enabled the identification of environmental and genetic contributors to AMD, both of which have tracked with technological advances in conceptual and practical statistical and genomic tools. Linkage of an AMD locus to 1q32 [4, 5], and the genome-wide association at the complement factor H (CFH) locus [6] led to the identification of the first common genome-wide significant risk variant, Y402H, that has upon sequencing in ethnic stratified cohorts revealed differential frequencies ranging from 5 % in East Asian populations to 35 % in European populations [7, 8]. This discovery propagated numerous genetics and genomic studies that have contributed to our understanding of the pathomechanisms contributing to AMD. Notably, the subsequent association of common and rare alleles at or near several additional complement genes (CFH, C2/CFB, C3, CFI, and C9) has had a significant impact of the formation of pathomechanistic hypotheses, with the cumulative evidence both from human genetics but also from histopathological studies highlighting a major role of the alternative complement pathway as a driver of AMD [9–21]. Here, we synthesize genetic evidence from rare- and common-allele studies in AMD, and we discuss the emergent picture of the genetic architecture of this atypical complex trait. Moreover, benefiting from the discovery of likely potent coding mutations in genes encoding complement components, we explore how these mutations might impede specific functions and discuss the potential contribution of aspects of this pathway to AMD pathogenesis.
AMD histopathology
The retina is composed of five major layers: the neurosensory retina; the retinal pigment epithelium (RPE); Bruch’s membrane (BM); the choriocapillaris; and the choroid (Fig. 1). The sensory cells of the neural retina are the photoreceptors (rods and cones), which through phototransduction convert light into electrical signals. Adjacent to the photoreceptors is the RPE, a part of the blood-ocular barrier and has numerous functions, including photoreceptor outer segment phagocytosis; regulation of the transportation nutrients; and cytokine secretion [22]. Lining the basal side of the RPE is BM, which is a pentalaminar extracellular matrix (ECM) consisting of elastin semipermeable barrier between inner and outer collagenous zones that provides connective tissue-based structural support and transport of waste from photoreceptors and RPE to the choroid and nutrients from the choroid to the RPE. The outermost layer of BM is composed of the basement membrane of the choriocapillaris, a fenestrated capillary bed that together with the choroid, a highly vascularized, pigmented tissue, are responsible for supplying the high metabolic demands of oxygen and nutrients to the outer retina [23].
Fig. 1.
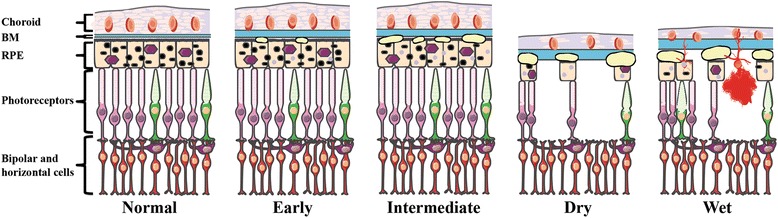
Illustration of the anatomical retinal pathology associated with the various AMD subtypes. Diagram of the outer layers of the human central retina in normal and in AMD. As the disease progresses, Bruch’s membrane (BM) increases in thickness. Early AMD is associated with small drusen and retinal pigmented epithelium (RPE) pigment abnormalities. As the disease progresses to the intermediate form, additional drusen are observed. In the two late forms of AMD (Dry and Wet), there is extensive drusen and photoreceptor cell death, with atrophy of the RPE and choroid in the Dry form and choroidal neovascularization (CNV), hemorrhaging, and RPE detachment in the Wet form. In all forms, the underlying circuitry including the horizontal and bipolar cells remains intact initially. This figure was prepared using Servier Medical Art (http://www.servier.com/Powerpoint-image-bank)
During the natural process of aging, the human eye undergoes physiological changes that include changes in the distribution of photoreceptors (30 % loss of rod photoreceptors [24]); thickening of BM [25]; and accumulation of sub-RPE debris, including drusen [22, 26–29]. Drusen are composed of esterified cholesterol, phospholipids, lipofuscin, inflammatory components (e.g., complement), and other intra- and extraocular degenerative materials [30, 31]. They are classified according to their appearance, with inert, “hard” drusen being small with well-demarcated borders in contrast to the generally pathogenic “soft” drusen that lack distinct borders and can range in size from <63 to >124 μm in diameter, the larger diameters coinciding with disease progression [32].
The clinical pathology of AMD has been described extensively [3, 33]. It involves the progressive degeneration of the macula, a small, pigmented area at the center of the retina that has the densest concentration of photoreceptors and is responsible for visual acuity. Damage to the macula results in loss of central vision. Initial indications of AMD are seen as focal hyperpigmentation of the RPE and accumulation of sub-RPE deposits, including drusen, both between BM and the RPE and within the RPE itself [34, 35].
Even though the degeneration of rods and cones does define the end-stage of the disorder, AMD is not a primary disease of the photoreceptor; most of the candidate processes, biochemical pathways, and molecules exert their effects primarily on the RPE-choroid complex [36].
AMD subtypes
The anatomical histopathology and clinical progression contribute to the clinical definition of four major AMD subtypes as categorized by the Age-Related Eye Disease Study (AREDS) severity scale grading system [37]: 1. Early AMD; 2. Intermediate AMD; 3. Advanced non-neovascular (“Dry” or geographic atrophy) AMD; and 4. Advanced neovascular (“Wet” or exudative) AMD [32, 38, 39]. Early AMD features few small (<63 μm) or medium-sized drusen (63–124 μm) and pigmentary abnormalities in the RPE, resulting in either mild visual impairment (blurred vision or decreased contrast sensitivity) or can be asymptomatic. The progression from early to intermediate AMD is hallmarked by the appearance of at least one large druse (>125 μm), along with numerous medium-size drusen. This pathology can progress to one of the two advanced forms of AMD: Dry AMD (non-neovascular), characterized by presence of drusen and atrophy of the RPE and choroid; or Wet AMD (neovascular), defined by newly formed vessels (choroidal neovascular membranes (CNV)) and RPE detachment. While occurring gradually in the Dry form versus suddenly/profoundly in the Wet form, the end result of both of these processes leads to photoreceptor cell death and vision loss (Fig. 1, Table 1).
Table 1.
Characteristics of AMD Clinical Subtypes (based on AREDS)
| AMD Subtype | |||||
|---|---|---|---|---|---|
| Early | Intermediate | Advanced Dry/Geographic Atrophy | Advanced Neovascular/Exudative/Wet | ||
| Clinical Features | Drusen | Few small (<63 μm) or medium sized (63–124 μm) | Numerous medium (63–124 μm) + at least 1 large (>125 μm) sized | Extensive medium (63–124 μm) + large (>125 μm) sized | Extensive medium (63–124 μm) + large (>125 μm) sized |
| Bruch’s membrane | Thickening | Thickening | Thickening | Thickening | |
| Retinal pigment epithelium | Pigmentary abnormalities | Pigmentary abnormalities; atrophy excluding fovea | Atrophy including fovea/macula | Detachment | |
| Choroid/choriocapillaris | Decreased vascular density | Decreased vascular density | Atrophy | Choroidal neovascularization; hemorrhage; leak fluid | |
| Neural retina | None | Photoreceptor thinning above drusen | Photoreceptor cell death | Photoreceptor detachment and cell death | |
| Impairment/outcome | Mild visual impairment (blurred vision or decreased contrast sensitivity) or asymptomatic | Mild visual impairment (blurred spots in central vision or decreased contrast sensitivity) or asymptomatic | Blurred central vision; gradual vision loss | Sudden/profound vision loss | |
Genetics of AMD: common variants versus rare variants
AMD exemplifies complex disorders: natural history and epidemiological studies have highlighted a prominent environmental role in disease risk with factors such as smoking resulting in a relative risk (RR) of >2, diet and obesity (RR > 2), nutritional supplements (odds ratio (OR) ~0.6) all contributing to pathogenicity [40]. In addition, family- and population-based studies have also highlighted that AMD carries a significant genetic burden, demonstrating >45 % concordance found in monozygotic and dizygotic twins as well as a recurrence-risk ratio between three to six times greater in siblings than in the general population [41–46].
In many ways, the dissection of the genetic basis of AMD has been an early beneficiary of advances in genetic and genomic technologies, wherein the development of both analytic platforms (genotyping, sequencing) as well as statistical methods, have found early application in this field.
Before our ability to query the genome in toto, complex trait studies focused on more traditional tools such as linkage and candidate gene association studies. Utilizing the latter, AMD was shown to be associated with two genes, ABCA4 (ATP-binding cassette subfamily A member 4) and APOE (apolipoprotein E), both of which only confer a minor risk contribution to AMD. ABCA4 is a causal gene for Stargardt disease, a retinopathy whose clinical symptoms overlap with AMD [47, 48]; APOE is a gene associated with Alzheimer’s disease (AD), a neurological degenerative disorder, whose pathology overlaps with AMD [49]. Nonetheless, and despite this initial discovery, the candidate gene approach was largely unsuccessful in identifying genetic contributors to AMD. This was due in part to limited cohort sizes, imperfect statistical methodologies, and unclear pathomechanisms hindering the ability to define candidate genes and in part due to our lack of appreciation of the extensive rare and common variation carried in human genomes [50–52].
In the era following the completion of the human genome project and the International HapMap Project, traditional family-based studies retained utility, as exemplified by the family-based linkage mapping of an AMD locus on 1q25-q31 and on 10q26 [4, 53]. However, transformative genetic progress in AMD was driven by the common-allele hypothesis explored through genome-wide association studies (GWAS) [6, 54]. Remarkably, modest GWAS analysis on 96 cases and 50 controls yielded a genome-wide significant signal (P < 10−7) on 1q32 of a common single nucleotide polymorphism (SNP) on a gene encoding CFH that revealed a polymorphism in linkage disequilibrium, Y402H, that has proven to be a common, predisposing allele that confers a two- to sevenfold increased risk for developing AMD [6, 9, 55, 56].
Soon after the identification of the CFH locus, another modestly sized GWAS performed on 96 cases and 130 controls yielded a genome-wide significant signal on 10q26, another primary locus for AMD that contains the candidate genes ARMS2/HTRA1 (age-related maculopathy susceptibility 2/HtrA serine peptidase 1) coding for a protein of unknown function and a serine protease, respectively [54, 57]. Thereafter, additional GWAS studies with progressively increasing cohort size led to the identification of more associated loci but generally resulted in diminishing returns with many of the loci having a minor contribution to AMD (Table 2).
Table 2.
Past genome-wide association studies of AMD
| Discovered gene/locus | Method | # Cases/Controls | OR | Reference |
|---|---|---|---|---|
| CFH | Affymetrix GeneChip Mapping 100K Set of microarrays | 96/50 | 4.6–7.4 | [6] |
| HTRA1/LOC387715 | Affymetrix GeneChip Mapping 100K Set of microarrays | 99/131 | 1.66–11.14 | [54] |
| LIPC | Affymetrix SNP 6.0 GeneChip and Sequenom | 979/1709 | 0.82 | [124] |
| CETP | Affymetrix SNP 6.0 GeneChip and Sequenom | 979/1709 | 1.15 | [124] |
| TIMP3 | Illumina Human370 Bead Chips and Illumina Infinium II assay | 2157/1150 | 0.63 | [125] |
| SKIV2L | Affymetrix SNP 5.0 GeneChip | 1896/1866 | 0.54 | [126] |
| MYRIP | Affymetrix SNP 5.0 GeneChip | 1896/1866 | 0.86 | [126] |
| TNFRSF10A/LOC389641 | Illumina Human610-Quad BeadChip and Illumina HumanHap550v3 BeadChip | 1536/18894 | 0.73 | [127] |
| FRK/COL10A1 | Affymetrix SNP 6.0 GeneChip and Illumina HumanCNV370v1 Bead Array | 2594/4134 | 0.87 | [128] |
| VEFGA | Affymetrix SNP 6.0 GeneChip and Illumina HumanCNV370v1 Bead Array | 2594/4134 | 1.15 | [128] |
| COL8A1/FILIP1L | Meta-analysis of GWAS | >17000/>60000 | 1.23 | [129] |
| APOE | Meta-analysis of GWAS | >17000/>60000 | [129] | |
| IER3/DDR1 | Meta-analysis of GWAS | >17000/>60000 | 1.16 | [129] |
| SLC16A8 | Meta-analysis of GWAS | >17000/>60000 | 1.15 | [129] |
| TGFBR1 | Meta-analysis of GWAS | >17000/>60000 | 1.13 | [129] |
| RAD51B | Meta-analysis of GWAS | >17000/>60000 | 1.11 | [129] |
| ADAMTS9/MIR548A2 | Meta-analysis of GWAS | >17000/>60000 | 1.1 | [129] |
| B3GALTL | Meta-analysis of GWAS | >17000/>60000 | 1.1 | [129] |
Meta-analysis genotyping method/platforms include Illumina HumanHap300, Human610-Quad, HumanHapCNV370, or HumanCNV370v1 BeadChips, Affymetrix 250K Nspl, Illumina Human670-QuadCustom chip, Illumina 660-Quadv1A, Illumina 610-Quad, Illumina Infinium HumanHap300K, HumanHap550v1, or HumanHap550v3 BeadChip, Illumina Infinium II HumanHap550, Affymetrix GeneChip Human Mapping 250k Styl Array, Affymetrix 6.0 or 1M
The associated loci discovered also germinated targeted sequencing strategies with a goal of identifying rare variants that might (a) provide direct causal evidence for the gene(s) in associated regions; (b) improve our measure of the overall risk of such genes to AMD pathogenesis; and (c) inform the direction of effect. Targeted high-throughput sequencing in combination with genotyping of the CFH locus led to the identification of a highly penetrant, rare variant R1210C which was shown to be associated with advanced AMD and to potentially result in disease onset 6 years earlier [58]. Similarly, in a smaller discovery cohort of 84 unrelated AMD patients, another penetrant, rare missense mutation in complement factor I (CFI), was unveiled, encoding G119R. This allele confers high risk to AMD (OR = 22.2), with patients with the heterozygous mutation having lower FI serum concentration [19].
With a goal of identifying elusive rare variants, a larger targeted exon sequencing study was undertaken in 2013, in which all exons of the 681 genes mapping at or close to all the AMD reported loci were sequenced in 1676 cases, 745 controls, and 36 siblings with discordant disease status [59]. This study combined sequencing-based genotypes, with the exome chip genome-wide genotyping data, with SNP genotyping to uncover K155Q (OR = 2.8) in the complement component C3 gene (C3) and P167S (OR = 2.2) in C9 as contributors to AMD disease pathophysiology. Importantly, the study also showed an overall increase in the burden of rare variants in cases compared to controls. Much of that signal originated from specific genes, such as CFI (7.8 % in cases compared to 2.3 % in controls). However, the data suggested that there were variants in additional genes that were likely involved but could not be powered sufficiently to show association for specific alleles [59]. More recent studies have involved a targeted capture enriched for complement components that led to discovery of enrichment of rare variants in CFH in AMD patients [60] as well as the largest AMD GWAS study to date that examined >12 million variants in 16,144 patients and 17,832 controls [61]. This study leads to the identification of 52 common and rare variants across 34 loci, 16 of which reached genome-wide significance for the first time [61]. Taken together, these studies highlight two key points. First, they showed that, for AMD, a blend of rare and common alleles can contribute substantially to the disease burden and, as accepted by evolutionary theory, coding alleles of major functional effect can have a much larger contribution to susceptibility. Second, however, was the more sobering observation that AMD is quite different from most other common traits studied to date. A few exceptions notwithstanding, the genetic architecture of AMD has intimated the presence of a few genes in which common, modestly penetrant alleles account for a significant fraction of the genetic burden, with rare coding variants in the same genes providing both causal evidence and adding further to the population burden [6, 58, 62, 63]. When juxtaposed with the >1000 GWAS executed for a variety of traits, this landscape has, for the most part, been confined to this disorder. The reason for this is unclear. These observations might underscore a “winner’s curse” of the first major complex trait’s GWAS successes. More importantly, they raise the possibility that the biochemical underpinnings of AMD might be fundamentally different from other complex traits and that understanding the reasons for such differences might inform our approach to both genetic discovery and therapeutic design.
Also contrasting other GWAS approaches in other diseases, a significant fraction of the susceptibility signal in AMD has mapped to, or near, genes encoding components of the complement cascade, although this is not the sole pathway implicated. Overall, the genes and alleles thought to confer significant susceptibility to AMD pathology can be clustered broadly into five major pathways (Table 3): (a) the inflammation and immune response, (b) lipid metabolism and transport, (c) extracellular matrix and cell adhesion, (d) angiogenesis, and (e) cellular stress responses. Among these, risk assessment analysis for either common or rare alleles has highlighted in genes encoding complement pathway components. The common variants near six complement genes, CFH, C2/CFB, C3, CFI, and C9 together, account for almost 60 % of the AMD genetic risk [36]. Notably, for the rare alleles found in sufficient recurrent rates to empower meaningful studies, the individual risk to AMD is sharply higher and appears as if they are almost Mendelian, as observed in the case of CFI [19].
Table 3.
Genes associated with AMD that cluster into five major pathways
| Inflammation and immune response | Cell stress response | ||||||
| C2 | Complement component 2 | HLA-C | Major histocompatibility complex, class I, C | ABCA4 | ATP-binding cassette subfamily A member 4 | HTRA1 | HtrA serine peptidase 1 |
| C3 | Complement component 3 | IL8 | Interleukin B | ACE | Angiotensin I converting enzyme 1 | RORA | RAR-related orphan receptor alpha |
| CFB | Complement factor B | MMP9 | Matrix metallopeptidase 9 | APOE | Apoliporotein E | SOD2 | Superoxide dismutase 2, mitochondrial |
| CFH | Complement factor H | PLEKHA1 | Pleckstrin homology domain containing, family A member 1 | ARMS2 | Age-related maculopathy susceptibility 2 | TF | Transferrin |
| CFD | Complement factor D | RORA | RAR-related orphan receptor alpha | CST3 | Cystatin C | TLR3 | Toll-like receptor 3 |
| CFHR1-5 | Complement factor H-related 1-5 | SERPING1 | Serpin peptidase inhibitor, clade G, member 1 | CX3CR1 | Chemokine receptor 1 | TLR4 | Toll-like receptor 4 |
| CFI | Complement factor I | TLR3 | Toll-like receptor 3 | CYP24A1 | Cytochrome P450, family 24, subfamily A peptide 1 | VLDLR | Very low-density lipoprotein receptor |
| C9 | Complement component 9 | TLR4 | Toll-like receptor 4 | GSTM1 | Glutathione S-transferase mu 1 | TNFRSF10A/LOC389641 | Tumor necrosis factor receptor superfamily, member 10a |
| CX3CR1 | Chemokine receptor 1 | VLDLR | Very low-density lipoprotein receptor | GSTP1 | Glutathione S-transferase pi 1 | IER3 | Immediate early response 3 |
| F13B | Coagulation factor XIII, B polypeptide | VTN | Vitronectin | GSTT1 | Glutathione S-transferase tau 1 | TGFBR1 | Transforming growth factor, beta receptor 1 |
| Lipid metabolism and transport | Extracellular matrix and cell adhesion | ||||||
| ABCA1 | ATP-binding cassette, subfamily A, member 1 | FADS1-3 | Fatty acid desaturases 1-3 | ACE | Angiotensin 1 converting enzyme 1 | ROBO1 | Roundabout, axon guidance receptor, homolog 1 |
| ABCA4 | ATP-binding cassette, subfamily A, member 4 | LIPC | Hepatic lipase | ARMS2 | Age-related maculopathy susceptibility 2 | TIMP3 | Tissue inhibitor of metalloproteinase 3 |
| APOE | Apolipoprotein E | LPL | Lipoprotein lipases | ADAMTS9 | ADAM metallopeptidase with trhombospondin type 1 motif, 9 | MMP19 | Matrix metallopeptidase 19 |
| CETP | Cholesteryl ester transfer protein, plasma | LRP6 | Low-density lipoprotein receptor-related protein 6 | COL8A1 | Collagen, type VIII, alpha 1 | PCOLCE | Procollagen c-endopeptidase enhancer |
| CFHR1-5 | Complement factor H-related 1-5 | RORA | RAR-related orphan receptor alpha | COL10A1 | Collagen, type X, alpha 1 | VTN | Vitronectin |
| CYP24A1 | Cytochrome P450, family 24, subfamily A peptide 1 | VLDLR | Very low-density lipoprotein receptor | CST3 | Cystatin C | ABCA7 | ATP-binding cassette, cubfamily A, member 7 |
| ELOVL4 | ELVL fatty acid elongase 4 | PLTP | Phospholipid transfer protein | CX3CR1 | Chemokine receptor 1 | ACTG1 | Actin gamma 1 |
| Angiogenesis | F13B | Coagulation factor XIII, B polypeptide | BCAR1 | Breast cancer anti-estrogen resistance 1 | |||
| ACE | Angiotensin I converting enzyme 1 | LRP6 | Low-density lipoprotein receptor-related protein 6 | FBLN5 | Fibulin 5 | COL4A4 | Collagen, type IV, alpha 4 |
| COL10A1 | Collagen, type X, alpha 1 | MMP9 | Matrix metallopeptidase 9 | HMCN1 | Hemicentin | ITGA7 | Integrin, alpha 7 |
| COL8A1 | Collagen, type VIII, alpha 1 | RORA | RAR-related orphan receptor alpha | HTRA1 | HtrA serine peptidase 1 | MYL2 | Myosin, light chain 2, regulatory, cardiac, slow |
| CST3 | Cystatin C | SERPINF1 | Serpin peptidase inhibitor, clade F | MMP9 | Matrix metallopeptidase 1 | ||
| FBLN5 | Fibulin 5 | TIMP3 | Tissue inhibitor of metalloproteinase 3 | ||||
| GDF6 | Growth differentiation factor 6 | VEGFA | Vascular endothelial growth factor A | ||||
| HTRA1 | HtrA serine peptidase 1 | VLDLR | Very low-density lipoprotein receptor | ||||
| IL8 | Interleukin 8 | ||||||
A pathogenic route to AMD: alternative complement pathway
One of the leading candidates for predisposition to AMD is the inflammatory pathogenesis theory, which hypothesizes dysregulation of the immune response, specifically complement system [6, 9, 12, 14, 15, 20, 55, 56, 64–68]. Reports dating back to 1875 have hypothesized that macular lesions were due to inflammation and that disciform degeneration were associated with choroidal inflammation [69]. Since then multiple components of the complement pathway have been linked to AMD and its pathological consequences [15, 16, 30, 64, 70, 71]. The complement system is a specialized part of innate immunity that can respond to antigen-antibody complexes (classical pathway) or bacterial mannose groups (lectin pathway) and can also be active in a low-level continuous state (alternative pathway (AP)), to allow for an immediate amplified response [72]. All complement pathways culminate in the creation of the membrane attack complex (MAC) for cell lysis and organismal defense.
AMD is not the first human genetic disorder associated with AP complement dysfunction. A number of disorders are thought to be the result of excessive AP activation, including membranoproliferative glomerulonephritis type II (MPGN type II); atypical hemolytic uremic syndrome (aHUS); and paroxysmal nocturnal hemoglobinuria (PNH, a rare form of hemolytic anemia). Of note, MPGN type II, which is characterized by renal disease and low serum C3 levels, is also associated with complete FH deficiency, arguing for a mechanistic similarity between MPGN and AMD [55, 73]. Similarly, aHUS has been associated with low FH levels in addition to low C3 levels and has mutations in several AP components, however, at a unique haplotype compared to AMD and MPGN [73]. PNH is caused by mutations in phosphatidylinositol glycan-complementation class A (PIGA) that is essential in the establishment of glycophosphatidylinositol anchors. Two known inhibitors of AP activation are glycolipid-anchored proteins that are required for the regulation at the C3 convertase step and MAC assembly [74].
Dysregulation of the AP specifically is currently thought to underlie AMD. Several lines of supporting evidence include (1) the presence of complement components in the choriocapillaris and the retina, especially in drusen [14–16, 75]; (2) increased MAC in the choriocapillaris of AMD patients [18]; and (3) the genetic association of C2, C3, CFB, CFI, and the regulator of complement activation (RCA) gene cluster on 1q32 (which includes CFH and CFHR1-5) with AMD [9–13, 17, 19–21]. However, the effects of variation within the components of the AP on AMD pathogenicity is largely unknown.
AMD is associated with activators of alternative pathway components
Low levels of constitutive complement activation via the AP allows for immediate immune response. It involves the central molecule of all three arms of the complement pathway, C3. In the AP, C3, is activated in two ways (Fig. 2): (1) it is either cleaved by convertases/plasma proteases to generate C3b or (2) a small portion is hydrolyzed spontaneously to C3H2O establishing a continuous “tick-over” ready for immediate C3b deposition on pathogens for target opsonization [76–79]. Factor B (FB) binds to C3H2O or C3b and the complex is cleaved by the plasma protease factor D (FD) forming the essential C3 convertase (C3bBb or C3H2OBb), leading to an amplification loop that cleaves and assembles C3 to C3b to C3bBb continuously. The accumulation of C3b leads to C3b binding to C3bBb, thereby creating a new enzyme, the C5 convertase (C3bBbC3b) that cleaves C5 to C5a (an anaphylatoxin and chemoattractant) and C5b, a component of the lytic pore which combines with C6–C9 to form the membrane attack complex (MAC) resulting in cell lysis [72].
Fig. 2.
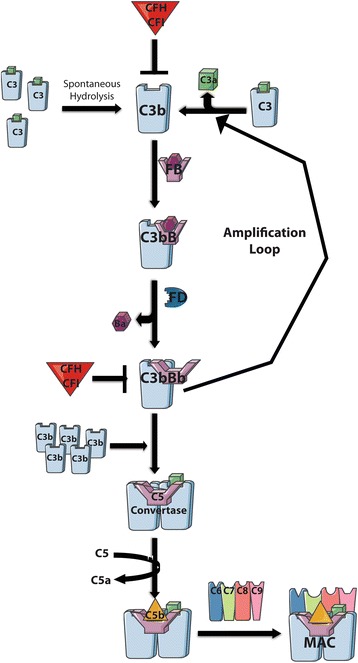
The alternative complement pathway and the formation of the C3 convertase. In the AP, the generation of C3b can occur by either spontaneous hydrolysis of C3 (“tick-over” allowing for continuous low-level activation) or by plasma proteolytic cleavage all allowing for immediate C3b deposition. C3b forms the C3 convertase upon binding to FB and cleavage by FD resulting in an amplification loop producing additional C3b to stimulate a large immune response. C3b additionally binds to the C3 convertase leading to the formation of the C5 convertase initiating the terminal pathway and the establishment of the MAC. This figure was prepared using Servier Medical Art (http://www.servier.com/Powerpoint-image-bank)
The AP has the unique characteristic of being able to be activated spontaneously, allowing for an immediate immune response, which must be tightly regulated to prevent excessive activation. Intrinsically, the AP has two major negative regulators encoded by CFH and CFI. CFH is the soluble inhibitor of the complement cascade and encodes a secreted glycoprotein, FH that acts as cofactor for FI mediated cleavage of C3b as well as accelerating decay of the C3bBb convertase.
Many cells also express membrane-associated AP regulators that hinder the C3 amplification loop and/or prevent the deposition/accumulation of C3b on self-tissue. These membrane-associated proteins include membrane cofactor protein (MCP), decay-accelerating factor (DAF), and complement receptor 1 (CR1) all of which are encoded by genes located within the RCA gene cluster on 1q32 [80]. MCP and CR1 have cofactor activity for CFI, while CR1 along with DAF have decay acceleration activity. Differential expression of MCP has been linked to AMD and upon the addition of an environmental stimulus such as smoking, a risk factor of AMD, both MCP and DAF are down regulated [81, 82]. Both activating and inhibiting components have been reported to harbor variants that potentially have functional impacts on the AP (Table 4), with an increase in overall mutational burden being reported in CFH and CFI in AMD patients [60, 83]. These perturbations of the components themselves or of their ability to interact with other AP components typically result in uncontrolled AP activation.
Table 4.
AP complement variants
| Gene | Variant | Position | OR | Common/Rare | Domain | Variant domain effect | Effect on AP | Ref |
|---|---|---|---|---|---|---|---|---|
| C3 | R102G | 19:6718387 | 1.7–2.6 | Common | Macroglobulin 1 | Decreased binding efficiency of C3b to its inhibitor CFH decreasing CFH’s cofactor activity | Increased AP | [20, 65] |
| C3 | P314L | 19:6713262 | 1.5 | Common | Macroglobulin 3 | Decreased binding efficiency of C3b to its inhibitor CFH decreasing CFH’s cofactor activity | Increased AP | [65] |
| C3 | K155Q | 19:6718146 | 2.8–3.8 | Rare | Macroglobulin 2 | Reduces binding to FH and C3b proteolytic cleavage by CFI | Increase C3 convertase production | [59, 65] |
| CFB | R32Q | 6:31914180 | 0.32 | Common | Ba | Reduced affinity for C3b and reduced hemolytic activity | Decrease formation of C3 convertase; reduced AP | [130] |
| CFB | L9H | 6:31914024 | 0.37 | Common | Signal peptide | Affects secretion of CFB | Decrease formation of C3 convertase; reduced AP | [12] |
| CFH | Y402H | 1:196659291 | 2.45–5.57 | Common | Complement control protein 7 | Affects binding to GAG heparin sulfate, sialic acid, and C-reactive protein | Reduces ability of CFH to degrade C3: increased AP | [131] |
| CFH | R1210C | 1:196716375 | 23.11 | Rare | Complement control protein 20 | Defective binding to C3b, C3d, and heparin | Impaired CFH attachment to host surfaces; reduces AP regulation | [58] |
| CFH | R53C | 1:196642206 | Rare | Complement control protein 1 | Reduced accelerating activity for AP C3 convertase | Altered CFH-mediated cofactor activity or decay-accelerating activity; increased AP | [63] | |
| CFH | D90G | 1:196643011 | Rare | Complement control protein 2 | Alters CFI cofactor activity | Decreased cofactor-mediated inactivation; increased AP | [63] | |
| CFH | I62V | 1:196642233 | 1.95–2.79 | Common | Complement control protein 2 | Binds more efficiently to C3b | Enhanced cofactor activity and increased formation of iC3b; reduced AP | [63] |
| CFH | N1050Y | 1:196712596 | 0.4 | Common | Complement control protein 18 | Possibly affects GAG and sialic acid binding | Increased AP | [62] |
| CFHR1/3 | CFHR1/3del | 0.29 | Common | n/a | n/a | Reduces cofactor activity for CFI and inhibits C5 convertase; reduced AP | [13] [132–135] | |
| CFI | G119R | 4:110685820 | 22.2 | Rare | Scavenger receptor cysteine-rich | Perturbs interdomain packing and stability of CFI | Diminished ability to degrade C3b; increased formation of C5 convertase; increased AP | [19] |
| CFI | G188A | 4:110682768 | Rare | Scavenger receptor cysteine-rich | Perturbs interdomain packing and stability of CFI | Diminished ability to degrade C3b; increased formation of C5 convertase; increased AP | [19] | |
| C9 | P167S | 5:39331894 | 2.2 | Rare | Membrane attack complex/perforin | Alters oligomerization and possibly inhibits C9's lytic activity | Affects binding with CD59; alters pore formation | [59] |
Pathogenic outcomes of the AP
Advanced age is the only risk factor common to all AMD patients. During the aging process, the physiological changes include redistribution of the photoreceptors, thickening of BM, and accumulation of debris in the eye in conjunction with environmental factors and genetic variation contribute to cascading dysfunction of physiological pathways such as lipid transport, angiogenesis, stress response, and ECM remodeling [24–26]. In turn, disruption of each of these pathways can lead to an inflammatory and an AP immune response ultimately culminating in cell death [72]. Each of these pathways has been hypothesized to alter components of the retina [36, 84] affecting the interdependence of the photoreceptors, RPE, BM, and choriocapillaris making it challenging to understand the model for AMD pathogenesis.
During the early stages of AMD, sub-RPE deposit formation (drusen and soft basal linear deposits) between the RPE and BM occurs and is also thought to be the main site of immune complex formation in AMD. While containing over 40 % lipids, other components of sub-RPE deposits include TIMP3 (TIMP metallopeptidase inhibitor 3), which plays a role in ECM maintenance and remodeling [85, 86]; amyloid beta (Aβ) that is produced either systemically or from the RPE and is proangiogenic and a known activator of complement [87]; apolipoproteins, which are also generated systemically and by the RPE [88]); and CFH and other complement components [30].
Lipid accumulation, similar to that seen in atherosclerosis, can increase choroidal vessel resistance preventing the choriocapillaris from properly clearing additional lipoproteins from the RPE and BM [89]. The accumulation of lipoproteins along with Aβ lead to the formation of a lipid “wall” external to the RPE [90, 91] that is the precursor to basal linear deposits that forms between the RPE basement membrane and the inner collagenous zone of BM [92]. The accumulated, peroxidizable lipoproteins are oxidized contributing to RPE damage [93]. Additionally, one byproduct of lipid peroxidation product is malondialdehyde (MDA), a marker for oxidative stress that contributes to RPE dysfunction [94] and a binding partner to CFH, allowing for an endogenous anti-inflammatory mediated response to this pro-inflammatory byproduct [95]. Similar to MDA, another cholesterol oxidation product having pro-inflammatory effects is 7-ketocholesterol (7KCh), which has been associated with cytotoxicity and has been shown to interact with retinal microglia, possibly promoting choroidal neovascularization [96] [97]. Further, these oxidized low-density lipoproteins can bind C-reactive protein initiating an inflammatory response leading to complement activation [98, 99]. Supplementing evidence has also been reported that complement activation leads to the recruitment of mononuclear phagocytes that contribute to RPE pathology in AMD [100]. All points that support immune system activation as a consequence of pathologic lipid accumulation.
The majority of complement is synthesized primarily by the liver and is delivered through circulation; however, some local tissues are also able to synthesize complement components, specifically the RPE and choroid [55]. Upon factors such as aging, oxidative stress (including cigarette smoke which that has been reported in vitro to activate C3 [101]), and lipid accumulation, increased AP activation in the RPE, which is thought to be the site of primary dysfunction in AMD, is primed for genetic predispositions to onset disease progression [14, 75, 101]. Upon AP activation, initiation of the terminal pathway ensues, forming MAC in BM and the choriocapillaris contributing to compromised function of the tight interaction of the RPE-BM-choroid complex [18].
Additional clues and emerging thoughts on pathogenic contributions to complex disease
There have been multiple insights implicating complement components in the pathobiology of AMD including genetic associations and accumulation of complement components in sub-RPE deposits of patient retinas [30, 102]. With common variants accounting for 65 % of the heritability of AMD, the search for rare variant contributors has only recently been undertaken through the advances of next generation sequencing [40]. Rare variants of C3, CFH, CFI, and C9 that have thus far been associated with AMD have been shown to play a role in either the complement pathway, to have an impact on the mutational load, or to hold the promise of putative future therapeutic targets [19, 58, 59, 63, 65]. However, a leading limitation in understanding disease outcome has been the lack of comprehension of the functional impact of the genetic contributors themselves. By combining the common variant common disease and rare variant common disease hypotheses [103–105], the idea of mutational burden emerges as a prognostic/diagnostic alternative for AMD, as exemplified by both common and rare variants in AP inhibitors leading to pathogenic dysregulated AP activity (Table 4). The latter highlights the necessity to functionally assess the role of a variant(s) in a gene(s) in order to more explicitly understand AMD pathogenesis. This highlights the need for development of animal models which are currently in development, for example, a transgenic mouse model expressing the human normal (Y402) and AMD-risk associated (Y402H) variants of FH has recently been described [106].
Familial aggregation is observed in most complex disease since there is greater likelihood of sharing disease-predisposing genotypes; however, non-genetic factors can contribute to discordant phenotypes [40]. In an effort to understand disease phenotypic outcomes, most studies have started by trying to identify causal variants. The identification of susceptibility variants is hindered by numerous confounding factors that have to be acknowledged to gain insight into disease mechanism. (1) Excluding a few cases such as AD and AMD, the vast majority of common variants exhibit a modest effect therefore large cohorts are required to power the findings [107, 108]. (2) Rare, disease-causing mutations, while numerous, are rarely observed in the general population [60, 83, 109, 110]. (3) SNP association of a locus does not imply that the causal variant itself is a SNP, as observed in Crohn’s disease where the causal mutations is a deletion upstream of the promoter [111]. (4) Multiple distinct alleles either common, rare, or both, can be present at a single locus [58, 112]. 5) Allele frequencies vary among ethnic groups, as not only observed in AMD but also described in Hirschsprung’s disease in which the causal variant has a minor allele frequency (MAF) ranging from 0.01 to 0.45 depending on the population [113]. (6) Causality due to non-coding variants is difficult to establish, similar to what was observed in coronary artery disease [114–116]. (7) Similar to pathways functioning in multiple processes, variants can also be pleiotropic, with their effect being dependent on the genetic content in which they are identified [117]. 8) Genes with causal variants identified need to be further studied for additional variation that could contribute to disease, as was seen in the case when delving deeper into CFH and CFI in AMD [19, 58]. (9) Structural variation cannot be ignored, as was observed in the case of Bardet-Biedl Syndrome in which copy number variations lead to disease progression (unpublished data). (10) Environmental factors can modify genetic effects and phenotypic outcomes [2, 101, 118].
Conclusions
Genetics has aided in the understanding of the complex, multifactorial nature of AMD. However, with modest signals differentiating the Dry versus Wet advanced forms of AMD, genetics has not revealed a strong predictive value on phenotypic severity or progression. Despite the long road of discovery that lies ahead, various lines of evidence linking the AP to AMD disease pathogenesis including (a) clinical phenotypes that associate with deficiencies of AP regulators [119]; (b) linkage of SNPs in AP components to disease risk [11, 120]; and (c) the functional analysis of individual AP components in the establishment of clinical phenotypes observed in both mouse and zebrafish models [74]. With numerous mouse models recapitulating at least 10 distinct human disorders, such as rheumatoid arthritis, traumatic brain injury, and AMD, the effects of AP on development and homeostasis have been established, making it clear that dysregulation/dysfunction of the process of innate immunity plays a contributory role in disease outcome [74]. Predominant functional assays for measuring the impact of variation on the complement pathway have been hemolytic and enzyme immunoassays [59, 121–123]; focus should and has begun to be placed on additional in vivo model systems to further understand the roles of AP disease contributors in a more systemic context [100, 106].
Acknowledgements
We thank Allison Ashley-Koch, Maria Kousi, and Erik Madsen for their thoughtful comments on the manuscript.
Authors’ contributions
The manuscript was written by PLT. All authors critically revised the manuscript. All authors read and approved the final manuscript.
Competing interests
The authors declare that they have no competing interests.
Abbreviations
- 7KCh
7-ketocholesterol
- ABCA4
ATP-binding cassette subfamily A member 4
- AD
Alzheimer’s disease
- aHUS
atypical hemolytic uremic syndrome
- AMD
age-related macular degeneration
- AP
alternative pathway
- APOE
apolipoprotein E
- AREDS
Age-Related Eye Disease Study
- ARMS2
age-related maculopathy susceptibility 2
- Aβ
amyloid beta
- BM
Bruch’s membrane
- C2
complement component 2
- C3
complement component 3
- C9
complement component 9
- CFB or FB
complement factor B
- CFH or FH
complement factor H
- CFI or FI
complement factor I
- CNV
choroidal neovascular membranes
- CR1
complement receptor 1
- DAF
decay-accelerating factor
- ECM
extracellular matrix
- FD
complement factor D
- GWAS
genome-wide association studies
- HTRA1
HtrA serine peptidase 1
- MAC
membrane attack complex
- MAF
minor allele frequency
- MCP
membrane cofactor protein
- MDA
malondialdehyde
- MPGN
membranoproliferative glomerulonephritis
- OR
odds ratio
- PIGA
phosphatidylinositol glycan-complement class A
- PNH
paroxysmal nocturnal Hemoglobinuria
- RCA
regulator of complement activation
- RPE
retinal pigment epithelium
- RR
relative risk
- SNP
single nucleotide polymorphism
- TIMP3
TIMP metallopeptidase inhibitor 3
References
- 1.Klein R, Peto T, Bird A, Vannewkirk MR. The epidemiology of age-related macular degeneration. Am J Ophthalmol. 2004;137:486–95. doi: 10.1016/j.ajo.2003.11.069. [DOI] [PubMed] [Google Scholar]
- 2.Thornton J, et al. Smoking and age-related macular degeneration: a review of association. Eye (Lond) 2005;19:935–44. doi: 10.1038/sj.eye.6701978. [DOI] [PubMed] [Google Scholar]
- 3.Young RW. Pathophysiology of age-related macular degeneration. Surv Ophthalmol. 1987;31:291–306. doi: 10.1016/0039-6257(87)90115-9. [DOI] [PubMed] [Google Scholar]
- 4.Weeks DE, et al. Age-related maculopathy: a genomewide scan with continued evidence of susceptibility loci within the 1q31, 10q26, and 17q25 regions. Am J Hum Genet. 2004;75:174–89. doi: 10.1086/422476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Weeks DE, et al. Age-related maculopathy: an expanded genome-wide scan with evidence of susceptibility loci within the 1q31 and 17q25 regions. Am J Ophthalmol. 2001;132:682–92. doi: 10.1016/S0002-9394(01)01214-4. [DOI] [PubMed] [Google Scholar]
- 6.Klein RJ, et al. Complement factor H polymorphism in age-related macular degeneration. Science (New York, NY) 2005;308:385–9. doi: 10.1126/science.1109557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kondo N, Bessho H, Honda S, Negi A. Complement factor H Y402H variant and risk of age-related macular degeneration in Asians: a systematic review and meta-analysis. Ophthalmology. 2011;118:339–44. doi: 10.1016/j.ophtha.2010.06.040. [DOI] [PubMed] [Google Scholar]
- 8.Thakkinstian A, et al. Systematic review and meta-analysis of the association between complement factor H Y402H polymorphisms and age-related macular degeneration. Hum Mol Genet. 2006;15:2784–90. doi: 10.1093/hmg/ddl220. [DOI] [PubMed] [Google Scholar]
- 9.Edwards AO, et al. Complement factor H polymorphism and age-related macular degeneration. Science (New York, NY) 2005;308:421–4. doi: 10.1126/science.1110189. [DOI] [PubMed] [Google Scholar]
- 10.Ennis S, Gibson J, Cree AJ, Collins A, Lotery AJ. Support for the involvement of complement factor I in age-related macular degeneration. Eur J Hum Genet. 2010;18:15–6. doi: 10.1038/ejhg.2009.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fagerness JA, et al. Variation near complement factor I is associated with risk of advanced AMD. Eur J Hum Genet. 2009;17:100–4. doi: 10.1038/ejhg.2008.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gold B, et al. Variation in factor B (BF) and complement component 2 (C2) genes is associated with age-related macular degeneration. Nat Genet. 2006;38:458–62. doi: 10.1038/ng1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hageman GS, et al. Extended haplotypes in the complement factor H (CFH) and CFH-related (CFHR) family of genes protect against age-related macular degeneration: characterization, ethnic distribution and evolutionary implications. Ann Med. 2006;38:592–604. doi: 10.1080/07853890601097030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hageman GS, et al. An integrated hypothesis that considers drusen as biomarkers of immune-mediated processes at the RPE-Bruch’s membrane interface in aging and age-related macular degeneration. Prog Retin Eye Res. 2001;20:705–32. doi: 10.1016/S1350-9462(01)00010-6. [DOI] [PubMed] [Google Scholar]
- 15.Johnson LV, Leitner WP, Staples MK, Anderson DH. Complement activation and inflammatory processes in Drusen formation and age related macular degeneration. Exp Eye Res. 2001;73:887–96. doi: 10.1006/exer.2001.1094. [DOI] [PubMed] [Google Scholar]
- 16.Johnson LV, Ozaki S, Staples MK, Erickson PA, Anderson DH. A potential role for immune complex pathogenesis in drusen formation. Exp Eye Res. 2000;70:441–9. doi: 10.1006/exer.1999.0798. [DOI] [PubMed] [Google Scholar]
- 17.Li M, et al. CFH haplotypes without the Y402H coding variant show strong association with susceptibility to age-related macular degeneration. Nat Genet. 2006;38:1049–54. doi: 10.1038/ng1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mullins RF, et al. The membrane attack complex in aging human choriocapillaris: relationship to macular degeneration and choroidal thinning. Am J Pathol. 2014;184:3142–53. doi: 10.1016/j.ajpath.2014.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van de Ven JP, et al. A functional variant in the CFI gene confers a high risk of age-related macular degeneration. Nat Genet. 2013;45:813–7. doi: 10.1038/ng.2640. [DOI] [PubMed] [Google Scholar]
- 20.Yates JR, et al. Complement C3 variant and the risk of age-related macular degeneration. N Engl J Med. 2007;357:553–61. doi: 10.1056/NEJMoa072618. [DOI] [PubMed] [Google Scholar]
- 21.Zhan X, et al. Identification of a rare coding variant in complement 3 associated with age-related macular degeneration. Nat Genet. 2013;45:1375–9. doi: 10.1038/ng.2758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Strauss O. The retinal pigment epithelium in visual function. Physiol Rev. 2005;85:845–81. doi: 10.1152/physrev.00021.2004. [DOI] [PubMed] [Google Scholar]
- 23.Lutty GA, et al. Development of the human choriocapillaris. Eye (Lond) 2010;24:408–15. doi: 10.1038/eye.2009.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Curcio CA, Owsley C, Jackson GR. Spare the rods, save the cones in aging and age-related maculopathy. Invest Ophthalmol Vis Sci. 2000;41:2015–8. [PubMed] [Google Scholar]
- 25.Feeney-Burns L, Ellersieck MR. Age-related changes in the ultrastructure of Bruch’s membrane. Am J Ophthalmol. 1985;100:686–97. doi: 10.1016/0002-9394(85)90625-7. [DOI] [PubMed] [Google Scholar]
- 26.Curcio CA, Millican CL, Bailey T, Kruth HS. Accumulation of cholesterol with age in human Bruch’s membrane. Invest Ophthalmol Vis Sci. 2001;42:265–74. [PubMed] [Google Scholar]
- 27.Delori FC, Goger DG, Dorey CK. Age-related accumulation and spatial distribution of lipofuscin in RPE of normal subjects. Invest Ophthalmol Vis Sci. 2001;42:1855–66. [PubMed] [Google Scholar]
- 28.Eldred GE, Miller GV, Stark WS, Feeney-Burns L. Lipofuscin: resolution of discrepant fluorescence data. Science (New York, NY) 1982;216:757–9. doi: 10.1126/science.7079738. [DOI] [PubMed] [Google Scholar]
- 29.Moreira EF, Larrayoz IM, Lee JW, Rodriguez IR. 7-Ketocholesterol is present in lipid deposits in the primate retina: potential implication in the induction of VEGF and CNV formation. Invest Ophthalmol Vis Sci. 2009;50:523–32. doi: 10.1167/iovs.08-2373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mullins RF, Russell SR, Anderson DH, Hageman GS. Drusen associated with aging and age-related macular degeneration contain proteins common to extracellular deposits associated with atherosclerosis, elastosis, amyloidosis, and dense deposit disease. Faseb J. 2000;14:835–46. [PubMed] [Google Scholar]
- 31.Russell SR, Mullins RF, Schneider BL, Hageman GS. Location, substructure, and composition of basal laminar drusen compared with drusen associated with aging and age-related macular degeneration. Am J Ophthalmol. 2000;129:205–14. doi: 10.1016/S0002-9394(99)00345-1. [DOI] [PubMed] [Google Scholar]
- 32.Jager RD, Mieler WF, Miller JW. Age-related macular degeneration. N Engl J Med. 2008;358:2606–17. doi: 10.1056/NEJMra0801537. [DOI] [PubMed] [Google Scholar]
- 33.Sarks SH. Ageing and degeneration in the macular region: a clinico-pathological study. Br J Ophthalmol. 1976;60:324–41. doi: 10.1136/bjo.60.5.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sarks SH, Arnold JJ, Killingsworth MC, Sarks JP. Early drusen formation in the normal and aging eye and their relation to age related maculopathy: a clinicopathological study. Br J Ophthalmol. 1999;83:358–68. doi: 10.1136/bjo.83.3.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Spraul CW, Grossniklaus HE. Characteristics of Drusen and Bruch’s membrane in postmortem eyes with age-related macular degeneration. Arch Ophthal. 1997;115:267–73. doi: 10.1001/archopht.1997.01100150269022. [DOI] [PubMed] [Google Scholar]
- 36.Fritsche LG, et al. Age-related macular degeneration: genetics and biology coming together. Annu Rev Genomics Hum Genet. 2014;15:151–71. doi: 10.1146/annurev-genom-090413-025610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Group, A.-R. E. D. S. R Risk factors associated with age-related macular degeneration. A case-control study in the Age-Related Eye Disease Study: Age-Related Eye Disease Study report number 3. Ophthalmology. 2000;107:2224–32. doi: 10.1016/S0161-6420(00)00409-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Davis MD, et al. The Age-Related Eye Disease Study severity scale for age-related macular degeneration: AREDS report no. 17. Arch Ophthal. 2005;123:1484–98. doi: 10.1001/archopht.123.11.1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ferris FL, et al. A simplified severity scale for age-related macular degeneration: AREDS report no. 18. Arch Ophthal. 2005;123:1570–4. doi: 10.1001/archopht.123.11.1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sobrin L, Seddon JM. Nature and nurture- genes and environment- predict onset and progression of macular degeneration. Prog Retin Eye Res. 2014;40:1–15. doi: 10.1016/j.preteyeres.2013.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hammond CJ, et al. Genetic influence on early age-related maculopathy: a twin study. Ophthalmology. 2002;109:730–6. doi: 10.1016/S0161-6420(01)01049-1. [DOI] [PubMed] [Google Scholar]
- 42.Klaver CC, et al. Genetic risk of age-related maculopathy. Population-based familial aggregation study. Arch Ophthal. 1998;116:1646–51. doi: 10.1001/archopht.116.12.1646. [DOI] [PubMed] [Google Scholar]
- 43.Klein ML, Mauldin WM, Stoumbos VD. Heredity and age-related macular degeneration. Observations in monozygotic twins. Arch Ophthal. 1994;112:932–7. doi: 10.1001/archopht.1994.01090190080025. [DOI] [PubMed] [Google Scholar]
- 44.Meyers SM, Greene T, Gutman FA. A twin study of age-related macular degeneration. Am J Ophthalmol. 1995;120:757–66. doi: 10.1016/S0002-9394(14)72729-1. [DOI] [PubMed] [Google Scholar]
- 45.Seddon JM, Ajani UA, Mitchell BD. Familial aggregation of age-related maculopathy. Am J Ophthalmol. 1997;123:199–206. doi: 10.1016/S0002-9394(14)71036-0. [DOI] [PubMed] [Google Scholar]
- 46.Seddon JM, Cote J, Page WF, Aggen SH, Neale MC. The US twin study of age-related macular degeneration: relative roles of genetic and environmental influences. Arch Ophthal. 2005;123:321–7. doi: 10.1001/archopht.123.3.321. [DOI] [PubMed] [Google Scholar]
- 47.Allikmets R, et al. Mutation of the Stargardt disease gene (ABCR) in age-related macular degeneration. Science (New York, NY) 1997;277:1805–7. doi: 10.1126/science.277.5333.1805. [DOI] [PubMed] [Google Scholar]
- 48.Zhang K, et al. The ABCR gene in recessive and dominant Stargardt diseases: a genetic pathway in macular degeneration. Genomics. 1999;60:234–7. doi: 10.1006/geno.1999.5896. [DOI] [PubMed] [Google Scholar]
- 49.Klaver CC, et al. Genetic association of apolipoprotein E with age-related macular degeneration. Am J Hum Genet. 1998;63:200–6. doi: 10.1086/301901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Flint J, Mackay TF. Genetic architecture of quantitative traits in mice, flies, and humans. Genome Res. 2009;19:723–33. doi: 10.1101/gr.086660.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Risch N, Merikangas K. The future of genetic studies of complex human diseases. Science (New York, NY) 1996;273:1516–7. doi: 10.1126/science.273.5281.1516. [DOI] [PubMed] [Google Scholar]
- 52.Peter I, Seddon JM. Genetic epidemiology: successes and challenges of genome-wide association studies using the example of age-related macular degeneration. Am J Ophthalmol. 2010;150:450–452.e452. doi: 10.1016/j.ajo.2010.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Majewski J, et al. Age-related macular degeneration—a genome scan in extended families. Am J Hum Genet. 2003;73:540–50. doi: 10.1086/377701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dewan A, et al. HTRA1 promoter polymorphism in wet age-related macular degeneration. Science (New York, NY) 2006;314:989–92. doi: 10.1126/science.1133807. [DOI] [PubMed] [Google Scholar]
- 55.Hageman GS, et al. A common haplotype in the complement regulatory gene factor H (HF1/CFH) predisposes individuals to age-related macular degeneration. Proc Natl Acad Sci U S A. 2005;102:7227–32. doi: 10.1073/pnas.0501536102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Haines JL, et al. Complement factor H variant increases the risk of age-related macular degeneration. Science (New York, NY) 2005;308:419–21. doi: 10.1126/science.1110359. [DOI] [PubMed] [Google Scholar]
- 57.Rivera A, et al. Hypothetical LOC387715 is a second major susceptibility gene for age-related macular degeneration, contributing independently of complement factor H to disease risk. Hum Mol Genet. 2005;14:3227–36. doi: 10.1093/hmg/ddi353. [DOI] [PubMed] [Google Scholar]
- 58.Raychaudhuri S, et al. A rare penetrant mutation in CFH confers high risk of age-related macular degeneration. Nat Genet. 2011;43:1232–6. doi: 10.1038/ng.976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Seddon JM, et al. Rare variants in CFI, C3 and C9 are associated with high risk of advanced age-related macular degeneration. Nat Genet. 2013;45:1366–70. doi: 10.1038/ng.2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Triebwasser MP, et al. Rare variants in the functional domains of complement factor H are associated with age-related macular degeneration. Invest Ophthalmol Vis Sci. 2015;56:6873–8. doi: 10.1167/iovs.15-17432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fritsche LG, et al. A large genome-wide association study of age-related macular degeneration highlights contributions of rare and common variants. Nat Genet. 2016;48:134–43. doi: 10.1038/ng.3448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Boon CJ, et al. Basal laminar drusen caused by compound heterozygous variants in the CFH gene. Am J Hum Genet. 2008;82:516–23. doi: 10.1016/j.ajhg.2007.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yu Y, et al. Whole-exome sequencing identifies rare, functional CFH variants in families with macular degeneration. Hum Mol Genet. 2014;23:5283–93. doi: 10.1093/hmg/ddu226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Anderson DH, Mullins RF, Hageman GS, Johnson LV. A role for local inflammation in the formation of drusen in the aging eye. Am J Ophthalmol. 2002;134:411–31. doi: 10.1016/S0002-9394(02)01624-0. [DOI] [PubMed] [Google Scholar]
- 65.Despriet DD, et al. Complement component C3 and risk of age-related macular degeneration. Ophthalmology. 2009;116:474–480.e472. doi: 10.1016/j.ophtha.2008.09.055. [DOI] [PubMed] [Google Scholar]
- 66.Maller JB, et al. Variation in complement factor 3 is associated with risk of age-related macular degeneration. Nat Genet. 2007;39:1200–1. doi: 10.1038/ng2131. [DOI] [PubMed] [Google Scholar]
- 67.Jakobsdottir J, Conley YP, Weeks DE, Ferrell RE, Gorin MB. C2 and CFB genes in age-related maculopathy and joint action with CFH and LOC387715 genes. PLoS One. 2008;3:e2199. doi: 10.1371/journal.pone.0002199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Park KH, Fridley BL, Ryu E, Tosakulwong N, Edwards AO. Complement component 3 (C3) haplotypes and risk of advanced age-related macular degeneration. Invest Ophthalmol Vis Sci. 2009;50:3386–93. doi: 10.1167/iovs.08-3231. [DOI] [PubMed] [Google Scholar]
- 69.Pagenstecher, H. a. G., C. Atlas der Pathologischen Anatomie des Augapfels. CW Kreidel, Wiesbaden (1875).
- 70.Hanssen R. Zur anatomie der scheibenformigen degeneration der netzhaumitte. Z. Augenh. 1930. 360-368.
- 71.Penfold PL, Killingsworth MC, Sarks SH. Senile macular degeneration: the involvement of immunocompetent cells. Graefes Arch Clin Exp Ophthalmol. 1985;223:69–76. doi: 10.1007/BF02150948. [DOI] [PubMed] [Google Scholar]
- 72.Ricklin D, Hajishengallis G, Yang K, Lambris JD. Complement: a key system for immune surveillance and homeostasis. Nat Immunol. 2010;11:785–97. doi: 10.1038/ni.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Pickering MC, et al. Spontaneous hemolytic uremic syndrome triggered by complement factor H lacking surface recognition domains. J Exp Med. 2007;204:1249–56. doi: 10.1084/jem.20070301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Holers VM. The spectrum of complement alternative pathway-mediated diseases. Immunol Rev. 2008;223:300–16. doi: 10.1111/j.1600-065X.2008.00641.x. [DOI] [PubMed] [Google Scholar]
- 75.Anderson DH, et al. The pivotal role of the complement system in aging and age-related macular degeneration: hypothesis re-visited. Prog Retin Eye Res. 2010;29:95–112. doi: 10.1016/j.preteyeres.2009.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Pangburn MK, Schreiber RD, Muller-Eberhard HJ. Formation of the initial C3 convertase of the alternative complement pathway. Acquisition of C3b-like activities by spontaneous hydrolysis of the putative thioester in native C3. J Exp Med. 1981;154:856–67. doi: 10.1084/jem.154.3.856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Pangburn MK, Muller-Eberhard HJ. Initiation of the alternative complement pathway due to spontaneous hydrolysis of the thioester of C3. Ann N Y Acad Sci. 1983;421:291–8. doi: 10.1111/j.1749-6632.1983.tb18116.x. [DOI] [PubMed] [Google Scholar]
- 78.Bexborn F, Andersson PO, Chen H, Nilsson B, Ekdahl KN. The tick-over theory revisited: formation and regulation of the soluble alternative complement C3 convertase (C3(H2O)Bb) Mol Immunol. 2008;45:2370–9. doi: 10.1016/j.molimm.2007.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lachmann PJ, Halbwachs L. The influence of C3b inactivator (KAF) concentration on the ability of serum to support complement activation. Clin Exp Immunol. 1975;21:109–14. [PMC free article] [PubMed] [Google Scholar]
- 80.Pardo-Manuel de Villena F, Heine-Suner D, Rodriguez de Cordoba S. Ordering of the human regulator of complement activation gene cluster on 1q32 by two-colour FISH. Cytogenet Cell Genet. 1996;72:339–41. doi: 10.1159/000134217. [DOI] [PubMed] [Google Scholar]
- 81.Fett AL, Hermann MM, Muether PS, Kirchhof B, Fauser S. Immunohistochemical localization of complement regulatory proteins in the human retina. Histol Histopathol. 2012;27:357–64. doi: 10.14670/HH-27.357. [DOI] [PubMed] [Google Scholar]
- 82.Wang L, et al. Nrf2 signaling modulates cigarette smoke-induced complement activation in retinal pigmented epithelial cells. Free Radic Biol Med. 2014;70:155–66. doi: 10.1016/j.freeradbiomed.2014.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kavanagh, D. et al. Rare genetic variants in the CFI gene are associated with advanced age-related macular degeneration and commonly result in reduced serum factor I levels. Hum Mol Genet. 2015. doi:10.1093/hmg/ddv091. [DOI] [PMC free article] [PubMed]
- 84.Miller JW. Age-related macular degeneration revisited—piecing the puzzle: the LXIX Edward Jackson memorial lecture. Am J Ophthalmol. 2013;155:1–35 e13. doi: 10.1016/j.ajo.2012.10.018. [DOI] [PubMed] [Google Scholar]
- 85.Fariss RN, Apte SS, Olsen BR, Iwata K, Milam AH. Tissue inhibitor of metalloproteinases-3 is a component of Bruch’s membrane of the eye. Am J Pathol. 1997;150:323–8. [PMC free article] [PubMed] [Google Scholar]
- 86.Kamei M, Hollyfield JG. TIMP-3 in Bruch’s membrane: changes during aging and in age-related macular degeneration. Invest Ophthalmol Vis Sci. 1999;40:2367–75. [PubMed] [Google Scholar]
- 87.Yoshida T, et al. The potential role of amyloid beta in the pathogenesis of age-related macular degeneration. J Clin Invest. 2005;115:2793–800. doi: 10.1172/JCI24635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Anderson DH, et al. Local cellular sources of apolipoprotein E in the human retina and retinal pigmented epithelium: implications for the process of drusen formation. Am J Ophthalmol. 2001;131:767–81. doi: 10.1016/S0002-9394(00)00961-2. [DOI] [PubMed] [Google Scholar]
- 89.Friedman E. A hemodynamic model of the pathogenesis of age-related macular degeneration. Am J Ophthalmol. 1997;124:677–82. doi: 10.1016/S0002-9394(14)70906-7. [DOI] [PubMed] [Google Scholar]
- 90.Ruberti JW, et al. Quick-freeze/deep-etch visualization of age-related lipid accumulation in Bruch’s membrane. Invest Ophthalmol Vis Sci. 2003;44:1753–9. doi: 10.1167/iovs.02-0496. [DOI] [PubMed] [Google Scholar]
- 91.Curcio CA, Johnson M, Rudolf M, Huang JD. The oil spill in ageing Bruch membrane. Br J Ophthalmol. 2011;95:1638–45. doi: 10.1136/bjophthalmol-2011-300344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Curcio CA, Johnson M, Huang JD, Rudolf M. Aging, age-related macular degeneration, and the response-to-retention of apolipoprotein B-containing lipoproteins. Prog Retin Eye Res. 2009;28:393–422. doi: 10.1016/j.preteyeres.2009.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Gordiyenko N, et al. RPE cells internalize low-density lipoprotein (LDL) and oxidized LDL (oxLDL) in large quantities in vitro and in vivo. Invest Ophthalmol Vis Sci. 2004;45:2822–9. doi: 10.1167/iovs.04-0074. [DOI] [PubMed] [Google Scholar]
- 94.Kaemmerer E, Schutt F, Krohne TU, Holz FG, Kopitz J. Effects of lipid peroxidation-related protein modifications on RPE lysosomal functions and POS phagocytosis. Invest Ophthalmol Vis Sci. 2007;48:1342–7. doi: 10.1167/iovs.06-0549. [DOI] [PubMed] [Google Scholar]
- 95.Weismann D, et al. Complement factor H binds malondialdehyde epitopes and protects from oxidative stress. Nature. 2011;478:76–81. doi: 10.1038/nature10449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Indaram M, et al. 7-Ketocholesterol increases retinal microglial migration, activation, and angiogenicity: a potential pathogenic mechanism underlying age-related macular degeneration. Sci Rep. 2015;5:9144. doi: 10.1038/srep09144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Rodriguez IR, Alam S, Lee JW. Cytotoxicity of oxidized low-density lipoprotein in cultured RPE cells is dependent on the formation of 7-ketocholesterol. Invest Ophthalmol Vis Sci. 2004;45:2830–7. doi: 10.1167/iovs.04-0075. [DOI] [PubMed] [Google Scholar]
- 98.Chang MK, Binder CJ, Torzewski M, Witztum JL. C-reactive protein binds to both oxidized LDL and apoptotic cells through recognition of a common ligand: Phosphorylcholine of oxidized phospholipids. Proc Natl Acad Sci U S A. 2002;99:13043–8. doi: 10.1073/pnas.192399699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Rodriguez IR, Larrayoz IM. Cholesterol oxidation in the retina: implications of 7KCh formation in chronic inflammation and age-related macular degeneration. J Lipid Res. 2010;51:2847–62. doi: 10.1194/jlr.R004820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Toomey CB, Kelly U, Saban DR, Bowes Rickman C. Regulation of age-related macular degeneration-like pathology by complement factor H. Proc Natl Acad Sci U S A. 2015;112:E3040–9. doi: 10.1073/pnas.1424391112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kew RR, Ghebrehiwet B, Janoff A. Cigarette smoke can activate the alternative pathway of complement in vitro by modifying the third component of complement. J Clin Invest. 1985;75:1000–7. doi: 10.1172/JCI111760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Gass JD. Drusen and disciform macular detachment and degeneration. Arch Ophthal. 1973;90:206–17. doi: 10.1001/archopht.1973.01000050208006. [DOI] [PubMed] [Google Scholar]
- 103.Cirulli ET, Goldstein DB. Uncovering the roles of rare variants in common disease through whole-genome sequencing. Nat Rev Genet. 2010;11:415–25. doi: 10.1038/nrg2779. [DOI] [PubMed] [Google Scholar]
- 104.Reich DE, Lander ES. On the allelic spectrum of human disease. Trends Genet. 2001;17:502–10. doi: 10.1016/S0168-9525(01)02410-6. [DOI] [PubMed] [Google Scholar]
- 105.Schork NJ, Murray SS, Frazer KA, Topol EJ. Common vs. rare allele hypotheses for complex diseases. Curr Opin Genet Dev. 2009;19:212–9. doi: 10.1016/j.gde.2009.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Ding JD, et al. Expression of human complement factor H prevents age-related macular degeneration-like retina damage and kidney abnormalities in aged Cfh knockout mice. Am J Pathol. 2015;185:29–42. doi: 10.1016/j.ajpath.2014.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007. 447, 661-678. doi:10.1038/nature05911. [DOI] [PMC free article] [PubMed]
- 108.Zeggini E, et al. Meta-analysis of genome-wide association data and large-scale replication identifies additional susceptibility loci for type 2 diabetes. Nat Genet. 2008;40:638–45. doi: 10.1038/ng.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Cohen JC, et al. Multiple rare alleles contribute to low plasma levels of HDL cholesterol. Science (New York, NY) 2004;305:869–72. doi: 10.1126/science.1099870. [DOI] [PubMed] [Google Scholar]
- 110.Ji W, et al. Rare independent mutations in renal salt handling genes contribute to blood pressure variation. Nat Genet. 2008;40:592–9. doi: 10.1038/ng.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.McCarroll SA, et al. Deletion polymorphism upstream of IRGM associated with altered IRGM expression and Crohn’s disease. Nat Genet. 2008;40:1107–12. doi: 10.1038/ng.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Schulte EC, et al. Targeted resequencing and systematic in vivo functional testing identifies rare variants in MEIS1 as significant contributors to restless legs syndrome. Am J Hum Genet. 2014;95:85–95. doi: 10.1016/j.ajhg.2014.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Emison ES, et al. A common sex-dependent mutation in a RET enhancer underlies Hirschsprung disease risk. Nature. 2005;434:857–63. doi: 10.1038/nature03467. [DOI] [PubMed] [Google Scholar]
- 114.Helgadottir A, et al. A common variant on chromosome 9p21 affects the risk of myocardial infarction. Science (New York, NY) 2007;316:1491–3. doi: 10.1126/science.1142842. [DOI] [PubMed] [Google Scholar]
- 115.McPherson R, et al. A common allele on chromosome 9 associated with coronary heart disease. Science (New York, NY) 2007;316:1488–91. doi: 10.1126/science.1142447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Samani NJ, et al. Genomewide association analysis of coronary artery disease. N Engl J Med. 2007;357:443–53. doi: 10.1056/NEJMoa072366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Jordan DM, et al. Identification of cis-suppression of human disease mutations by comparative genomics. Nature. 2015;524:225–9. doi: 10.1038/nature14497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Hobbs RP, Bernstein PS. Nutrient supplementation for age-related macular degeneration, cataract, and dry eye. J Ophthalmic Vis Res. 2014;9:487–93. doi: 10.4103/2008-322X.150829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Botto M, et al. Complement in human diseases: lessons from complement deficiencies. Mol Immunol. 2009;46:2774–83. doi: 10.1016/j.molimm.2009.04.029. [DOI] [PubMed] [Google Scholar]
- 120.Fisher SA, et al. Meta-analysis of genome scans of age-related macular degeneration. Hum Mol Genet. 2005;14:2257–64. doi: 10.1093/hmg/ddi230. [DOI] [PubMed] [Google Scholar]
- 121.Nilsson UR, Nilsson B. Simplified assays of hemolytic activity of the classical and alternative complement pathways. J Immunol Methods. 1984;72:49–59. doi: 10.1016/0022-1759(84)90432-0. [DOI] [PubMed] [Google Scholar]
- 122.Tortajada A, et al. C3 glomerulopathy-associated CFHR1 mutation alters FHR oligomerization and complement regulation. J Clin Invest. 2013;123:2434–46. doi: 10.1172/JCI68280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Yoshida Y, et al. A novel quantitative hemolytic assay coupled with restriction fragment length polymorphisms analysis enabled early diagnosis of atypical hemolytic uremic syndrome and identified unique predisposing mutations in Japan. PLoS One. 2015;10:e0124655. doi: 10.1371/journal.pone.0124655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Neale BM, et al. Genome-wide association study of advanced age-related macular degeneration identifies a role of the hepatic lipase gene (LIPC). Proc Natl Acad Sci USA. 2010;107:7395-7400. doi:10.1073/pnas.0912019107. [DOI] [PMC free article] [PubMed]
- 125.Chen W, et al. Genetic variants near TIMP3 and high-density lipoprotein-associated loci influence susceptibility to age-related macular degeneration. Proc Natl Acad Sci USA. 2010;107:7401-6. doi:10.1073/pnas.0912702107. [DOI] [PMC free article] [PubMed]
- 126.Kopplin LJ, et al. Genome-wide association identifies SKIV2L and MYRIP as protective factors for age-related macular degeneration. Genes and immunity. 2010;11:609–21. doi:10.1038/gene.2010.39. [DOI] [PMC free article] [PubMed]
- 127.Arakawa S, et al. Genome-wide association study identifies two susceptibility loci for exudative agerelated macular degeneration in the Japanese population. Nat Genet. 2011;43:1001–4. doi:10.1038/ng.938. [DOI] [PubMed]
- 128.Yu Y, et al. Common variants near FRK/COL10A1 and VEGFA are associated with advanced agerelated macular degeneration. Hum Mol Genet. 2011;20:3699-709. doi:10.1093/hmg/ddr270. [DOI] [PMC free article] [PubMed]
- 129.Fritsche LG, et al. Seven new loci associated with age-related macular degeneration. Nat Genet. 2013;45:433-439. 439e431–432, doi:10.1038/ng.2578. [DOI] [PMC free article] [PubMed]
- 130.Montes T, Tortajada A, Morgan BP, Rodriguez de Cordoba S, Harris, CL. Functional basis of protection against age-related macular degeneration conferred by a common polymorphism in complement factor B. Proc Natl Acad Sci USA. 2009;106:4366–71. doi:10.1073/pnas.0812584106. [DOI] [PMC free article] [PubMed]
- 131.Herbert AP, et al. Structure shows that a glycosaminoglycan and protein recognition site in factor H is perturbed by age-related macular degeneration-linked single nucleotide polymorphism. The Journal of biological chemistry. 2007;282:18960–8. doi:10.1074/jbc.M609636200. [DOI] [PubMed]
- 132.Eberhardt HU, et al. Human factor H-related protein 2 (CFHR2) regulates complement activation. PLoS One. 2013;8:e78617. doi:10.1371/journal.pone.0078617. [DOI] [PMC free article] [PubMed]
- 133.Fritsche LG, et al. An imbalance of human complement regulatory proteins CFHR1, CFHR3 and factor H influences risk for age-related macular degeneration (AMD). Hum Mol Genet. 2010;19:4694–704. doi:10.1093/hmg/ddq399. [DOI] [PubMed]
- 134.Hughes AE, et al. A common CFH haplotype, with deletion of CFHR1 and CFHR3, is associated with lower risk of age-related macular degeneration. Nat Genet. 2006;38:1173–7. doi:10.1038/ng1890. [DOI] [PubMed]
- 135.Spencer KL, et al. Deletion of CFHR3 and CFHR1 genes in age-related macular degeneration. Hum Mol Genet. 2008;17:971–7. doi:10.1093/hmg/ddm369. [DOI] [PubMed]


