Abstract
Focal cortical dysplasia type II (FCDII) is a focal malformation of the developing cerebral cortex and the major cause of intractable epilepsy. However, since the molecular genetic etiology of FCD has remained enigmatic, the effective therapeutic target for this condition has remained poorly understood. Our recent study on FCD utilizing various deep sequencing platforms identified somatic mutations in MTOR (existing as low as 1% allelic frequency) only in the affected brain tissues. We observed that these mutations induced hyperactivation of the mTOR kinase. In addition, focal cortical expression of mutant MTOR using in utero electroporation in mice, recapitulated the neuropathological features of FCDII, such as migration defect, cytomegalic neuron and spontaneous seizures. Furthermore, seizures and dysmorphic neurons were rescued by the administration of mTOR inhibitor, rapamycin. This study provides the first evidence that brain somatic activating mutations in MTOR cause FCD, and suggests the potential drug target for intractable epilepsy in FCD patients. [BMB Reports 2016; 49(2): 71-72]
Keywords: Focal cortical dysplasia, Mechanistic target of rapamycin, Mouse model, Somatic mutation
Focal cortical dysplasia (FCD) is a subgroup of malformation of cortical development (MCD), and is characterized by abnormal cortical lamination and dysmorphic neurons. FCD is a common cause of intractable epilepsy that is difficult to treat with available antiepileptic drugs; it has been diagnosed in 20-25% of patients with symptomatic focal epilepsy. Although epilepsy surgery is a treatment option for controlling drug-resistant seizures, many patients continue to have convulsions despite surgical intervention. Thus, it is necessary to discover druggable targets for FCD by understanding its molecular genetic causes.
According to the consensus classification proposed by the International League Against Epilepsy (ILAE), FCD can be classified into 3 types, based on histopathologic examination, clinical manifestations and neuroimaging. Among the FCD classifications, focal cortical dysplasia type II (FCD II) shows the relatively clear histologic phenotype characterized by cytologic abnormalities in the affected cortical area, such as dysmorphic neurons or balloon cells. Over the last decade, there has been accumulating evidence that FCDII is associated with the aberrant activation of the mTOR pathway, and it belongs to a spectrum of disorders described as “TORopathies”. These investigations showed that the phosphorylation of the mTOR downstream effectors such as ribosomal S6, S6 kinase (S6K), and 4E-BP1 protein, was increased in the brain tissues resected from FCDII patients. mTOR is a key protein kinase in the mTOR pathway, which controls cells growth in response to growth factors and nutrients. In addition, the PI3K-AKT-mTOR pathway plays a critical role in regulating cortical development. Although the hyperactivation of mTOR pathway is known to be implicated in the pathogenesis of FCDII, the exact molecular genetic cause of mTOR hyperactivation is unknown. Recently, increasing evidence supports the importance of the brain somatic mutations in neurodevelopmental disorders, including hemimegalencephaly (HME), showing histologic findings similar to that seen in FCD. In addition, the occurrence of FCD is sporadic; it shows a funnel-shaped appearance on neuroimaging, implying that dispersed abnormal neurons are derived from progenitors at the ventricular zone. Based on these findings, it should be reasonable to hypothesize that FCDII is caused by brain somatic mutations in MTOR itself, or in other key genes regulating the mTOR pathways.
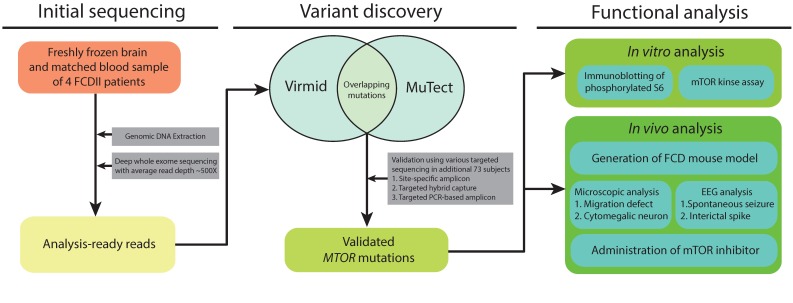
To elucidate our hypothesis, we first performed the deep whole exome sequencing with paired blood-brain DNA samples on four FCDII patients, and jointly analyzed the paired blood-brain sequencing data to detect any brain specific somatic mutations. For bioinformatic analysis, we utilized the recently developed algorithms, MuTect and Virmid, which are particularly designed for identifying somatic mutations with low allelic frequencies (Fig. 1). After validating the identified mutations with site-specific deep amplicon sequencing for minimizing the sequencing artifact or erroneous calls, we found brain somatic mutation of MTOR in two out of the four patients. Next, we performed deep targeted sequencing of the MTOR gene in a large FCDII cohort comprising an additional 73 patients, using two different sequencing platforms, namely hybrid capture and PCR based amplicon sequencing. In total, 15.6% of the participants (12 of 77) were positive for 9 different brain somatic mutations in MTOR.
Fig. 1. Schematic presentation of the experimental design. Deep whole exome sequencing (WES) was performed in four FCDII patients, and the raw sequencing data were analyzed to discover an alternative allele. Overlapping mutations in both Virmid and MuTect algorithms were selected as candidate single nucleotide variants (SNVs). Subsequently, we performed deep sequencing of the MTOR gene with various sequencing platforms in a large FCDII cohort comprising an additional 73 patients. Finally, the validated MTOR mutations were subjected to in vitro and in vivo functional analysis.
Subsequently, we tested the biological and pathological functions of the identified mutations at the in vitro and in vivo levels (Fig. 1). We performed in vitro analysis including immunoblotting of S6 phosphorylation, a well-known biomarker of mTOR activation, as well as mTOR kinase assays. Our results indicated that the activity of mTOR kinase was abnormally increased by the identified mutations. Next, to examine the pathogenic role of the identified mutations at the in vivo level, one of the identified MTOR mutations recurrently found in patients was introduced into the developing mouse cortex using in utero electroporation; we successfully generated the mouse model recapitulating all neuropathological features of FCDII patients, such as migration defect, cytomegalic neuron and spontaneous seizures. Notably, the spontaneous convulsive seizures and dysmorphic neurons in mice carrying the MTOR mutant were almost completely rescued by mTOR inhibitor. These findings suggest that brain somatic activating mutation of MTOR induced a hyperactivation of the mTOR pathway, resulting in FCD pathology, leading to intractable epilepsy.
The present study not only elucidated the brain somatic mutations in MTOR as the molecular genetic cause of FCD, but also led to the discovery of the genetically validated target and a clinically available drug. Moreover, our findings suggest that even such a low-level somatic mutation in a focal area of brain is sufficient to cause a neurodevelopmental disorder by disrupting the brain connectivity. However, the present study focused on somatic missense mutations in protein coding regions covering only ∼3% of the entire human genome. Future studies utilizing whole genome sequencing with high depth might reveal pathogenic mutations in non-coding regions. Nevertheless, our findings will provide the foundation for future scientific and clinical research of FCD patients.
Acknowledgments
This work was supported by a grant of the Korean Health Technology R&D Project, Ministry of Health & Welfare, Republic of Korea (A121070, HI13C0208 to J.H.L.), the Brain Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Science, ICT & Future Planning (2013M3C7A1056564 to J.H.L.), the KAIST Future Systems Healthcare Project from the Ministry of Science, ICT and Future Planning (to J.H.L.).