Abstract
Insulin/insulin-like growth factor (IGF)-1 signaling (IIS) pathway regulates aging in many organisms, ranging from simple invertebrates to mammals, including humans. Many seminal discoveries regarding the roles of IIS in aging and longevity have been made by using the roundworm Caenorhabditis elegans and the fruit fly Drosophila melanogaster. In this review, we describe the mechanisms by which various IIS components regulate aging in C. elegans and D. melanogaster. We also cover systemic and tissue-specific effects of the IIS components on the regulation of lifespan. We further discuss IIS-mediated physiological processes other than aging and their effects on human disease models focusing on C. elegans studies. As both C. elegans and D. melanogaster have been essential for key findings regarding the effects of IIS on organismal aging in general, these invertebrate models will continue to serve as workhorses to help our understanding of mammalian aging. [BMB Reports 2016; 49(2): 81-92]
Keywords: Aging, C. elegans, D. melanogaster, Insulin/IGF-1 signaling, Longevity
INTRODUCTION
The roundworm Caenorhabditis elegans and the fruit fly Drosophila melanogaster have been used as two most popular invertebrate models for studying aging and longevity (1, 2). In particular, their short lifespan together with their low cost and easy handling has established these invertebrates as excellent systems for research on molecular mechanisms regulating animal aging. Many important discoveries regarding evolutionarily conserved aging-regulatory pathways have been made using C. elegans and D. melanogaster. One of such pathways is the insulin/insulin-like growth factor (IGF)-1 signaling (IIS) pathway, which was first shown to regulate longevity in C. elegans, and subsequently confirmed by using D. melanogaster. Importantly, the findings using these two invertebrate model organisms stimulated research on the role of IIS in mammalian aging, and led to discoveries showing that IIS also regulates aging in mammals, including mice and humans (3, 4). In this review, we will describe which components of IIS regulate lifespan, and how IIS modulates aging processes in these two model organisms. We will also review endocrine signaling and the importance of insulin-like peptides (ILPs) for systemic longevity regulation. Overall, our review will provide useful information regarding the conserved roles of IIS pathway in the aging of model organisms, which will eventually pave the way for understanding the mystery of human aging.
THE ROLE OF INSULIN/IGF-1 SIGNALING IN C. elegans AGING
Insulin/IGF-1 signaling pathway components that regulate the lifespan of C. elegans
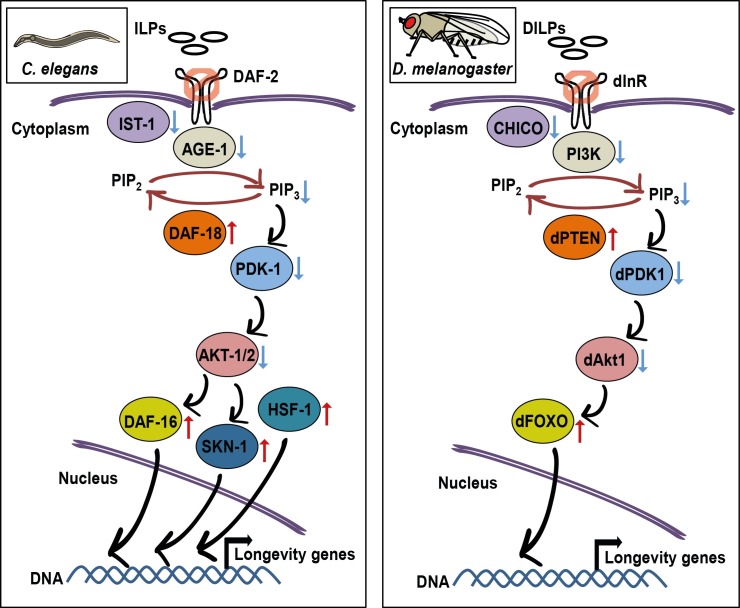
The insulin/IGF-1 signaling (IIS) pathway contains many evolutionarily conserved components that regulate aging (Fig. 1). The gerontogenes daf-2 and age-1 encode the sole insulin/IGF-1 receptor and phosphatidylinositol-3-OH kinase (PI3K) (5, 6), respectively. DAF-2 and AGE-1 are two key upstream components of IIS that regulate various physiological aspects, including aging and adult lifespan. Two of the most important discoveries in the field of aging research were perhaps the findings demonstrating that inhibition of daf-2 or age-1 dramatically extended lifespan in C. elegans (7-9). These discoveries stimulated many subsequent studies on the role of IIS in lifespan regulation, not only in C. elegans but also in D. melanogaster and mammals.
Fig. 1. Conserved longevity-regulatory components of insulin/IGF-1 signaling pathway in C. elegans and D. melanogaster. Insulin-like peptides (ILPs in Caenorhabditis elegans and DILPs in Drosophila melanogaster) bind to insulin/IGF-1 receptor (DAF-2 in C. elegans and dInR in D. melanogaster) and lead to its phosphorylation. Inhibition of insulin/IGF-1 receptor results in decreased binding to the insulin receptor substrate (IST-1 in C. elegans and CHICO in D. melanogaster), which in turn decreases the activity of phosphoinositide-3 kinase (AGE-1 in C. elegans and PI3K in D. melanogaster) that converts PIP2 to PIP3; conversely, the PTEN phosphatase (DAF-18 in C. elegans and dPTEN in D. melanogaster) functions to antagonize the activity of the phosphoinositide-3 kinase by converting PIP3 to PIP2. Decreased PIP3 levels lead to decreased activities of phosphoinositide-dependent kinase 1 (PDK-1 in C. elegans and dPDK1 in D. melanogaster) and the serine/threonine-specific protein kinase B (AKT-1/-2 in C. elegans and dAkt1 in D. melanogaster), and the activation of downstream transcription factor FOXO (DAF-16 in C. elegans and dFOXO in D. melanogaster). Reduced insulin/IGF-1 signaling in C. elegans also increases the activities of heat shock transcription factor-1 (HSF-1) and SKN-1 (NRF2). These transcription factors regulate the expression of target genes, which contribute to longevity.
IIS transduces signals through a combination of wellorganized sequential events, depending on environmental conditions. Under favorable conditions, IIS is activated and this confers normal development and adult lifespan. Specifically, agonist insulin-like peptides (ILPs) bind to their receptor, DAF-2, which in turn recruits an insulin receptor substrate (IRS)/IST-1 (10). This leads to the activation of the AGE-1/PI3K, which increases the level of phosphatidylinositol (3,4,5)-trisphosphate (PIP3) (5, 11); this event is antagonistically balanced by DAF-18/PTEN phosphatase that promotes the conversion of PIP3 to phosphatidylinositol (4,5)-bisphosphate (PIP2) (12-19). The signals provided by PIP3 activate the downstream kinase cascade, composed of 3-phosphoinositide-dependent protein kinase 1 (PDK-1) (2), protein kinase B (AKT-1/-2) (21), and serum- and glucocorticoid-inducible kinase-1 (SGK-1) (22; but see also 23, 24). This in turn phosphorylates and inactivates DAF-16/FOXO transcription factor, by promoting its nucleusto-cytosol translocation (22, 25-30). Conversely, in unfavorable conditions, IIS is down-regulated and leads to the activation of DAF-16/FOXO via enhancing its translocation from the cytoplasm to the nucleus, where it switches on the expression of genes that promote longevity. Thus, C. elegans IIS pathway acts as a system in which many components transduce signals to modulate the aging processes, depending on extracellular conditions.
Three most important downstream lifespan-regulatory transcription factors of IIS that have been identified so far are DAF-16/FOXO, heat shock transcription factor 1 (HSF-1) and SKN-1/nuclear factor erythroid 2 (NRF2). DAF-16/FOXO regulates aging processes downstream of the canonical IIS cascade as described above. In addition, Jun-N-terminal kinase (JNK/JNK-1) (31), AMP-activated protein kinase (AMPK/AAK-2) (32-34), and Ste20-like protein kinase (MST1/CST-1) (35) activate DAF-16/FOXO via phosphorylation. Other non-kinase proteins have been shown to regulate C. elegans DAF-16/FOXO. A serine/threonine-protein phosphatase 4-regulatory subunit SMK-1 (36), and an RNA helicase HEL-1 (37), extend longevity by acting together with DAF-16/FOXO. DAF-16/FOXO is acetylated by an acetyl-transferase CBP-1/CREB binding protein (CBP), whose inhibition leads to constitutive nuclear localization of DAF-16/FOXO (38). Host cell factor 1 (HCF-1) and enhancer of akt-1 null 7 (EAK-7) are other regulatory factors that inhibit DAF-16/FOXO activity without altering its subcellular localization (39-41). DAF-16/FOXO also interacts with two highly homologous 14-3-3 protein family members, FTT-1/PAR-5 and FTT-2 (42, 43). The 14-3-3 proteins modulate the interaction between DAF-16/FOXO and other co-factors, such as SIR-2.1/sirtuin 1, an NAD-dependent deacetylase (42, 44). These diverse interactions and posttranslational modifications may help differentially regulate the activity of DAF-16/FOXO upon various environmental changes.
Downstream targets of DAF-16/FOXO were identified by using various approaches such as chromatin immunoprecipitation, bioinformatics, microarray and mRNA sequencing (31, 45-51). The DAF-16/FOXO target genes collectively contribute to longevity by enhancing cellular maintenance in animals with reduced IIS. Since many regulatory modes and targets of FOXO transcription factors are conserved among species, the longevity-regulatory modes of C. elegans DAF-16/FOXO are likely to be recapitulated in IIS-medicated longevity in mammals.
SKN-1, an oxidative stress-responsive NRF transcription factor, also contributes to the longevity conferred by reduced IIS (52, 53). Similar to DAF-16/FOXO, SKN-1 is sequestered in the cytoplasm by phosphorylation via the canonical IIS protein kinases, including AKT-1/-2 (52). SKN-1 mediates the expression of genes involved in detoxification and stress responses (52, 54-63). Overexpression of constitutively nuclear SKN-1 extends lifespan in a DAF-16/FOXO-independent manner (52). SKN-1 also promotes protein homeostasis through regulating proteasome production, which contributes to a longer lifespan (55, 57, 64). In addition, SKN-1 promotes longevity of animals with reduced IIS through remodeling of extracellular matrix (65).
HSF-1 is another important transcription factor acting downstream of IIS, and is essential for the longevity of animals with reduced IIS (66-70). The function of HSF-1 in promoting longevity and reducing proteotoxicity is closely associated with the conserved IIS pathway. Genetic inhibition of hsf-1 accelerates tissue aging, thereby shortening the lifespan (71). Knockdown of hsf-1 also suppresses the longevity phenotype of daf-2 and age-1 mutants; conversely, the overexpression of hsf-1 is sufficient to extend lifespan (51, 66, 67, 69, 72, 73). HSF-1 binds to specific regions of DNA containing heat shock elements (HSEs) (74-76). The binding of HSF-1 to HSEs triggers the induction of genes encoding molecular chaperones, such as HSP-70 and HSP-16, whose overexpression extends lifespan (77, 78). Thus, HSF-1 appears to lead to longevity by upregulating the chaperone network that enhances the proper folding of various proteins (66, 67). DDL-1 (the C. elegans homolog of human coiled-coil domain-containing protein 53: CCDC53), DDL-2 (the C. elegans homolog of human Wiskott-Aldrich syndrome protein and SCAR homolog: WASH2), and HSB-1 (heat-shock factor binding protein-1), form a complex with HSF-1 and regulate lifespan by inhibiting the activity of HSF-1 (69). Overall, HSF-1 and SKN-1 appear to promote longevity mainly through the induction of target genes that increase resistance to various stresses.
Systemic regulation of insulin/IGF-1 signaling-mediated longevity in C. elegans
As the IIS pathway consists of many potential endocrine components, it is likely that IIS regulates lifespan in a systemic manner. The C. elegans genome encodes 40 ILPs, which appear to act as extracellular endocrine signals in C. elegans (79, 80). Functional studies on several ILPs, including ins-6 (81, 82), ins-7 (47, 83, 84) and daf-28 (80, 85, 86), have been conducted. However, the majority of the 40 ILPs, which potentially regulate longevity and development, are yet to be characterized in detail. This is perhaps because many possible combinations of the interactions between ILPs and DAF-2/insulin/IGF-1 receptor make it difficult to dissect the specific functions of each ILP. A recent study indicates that ILPs can function in a combinatorial manner to coordinate various physiological processes (87). This finding is different from the previous notion that ILPs generally confer a functional redundancy due to their structural similarities (79, 80, 88-91). Therefore, some individuals or a group of ILPs may have a profound effect on longevity.
Most of the ILPs are expressed in neurons, although some ILPs are expressed in non-neuronal tissues such as hypodermis and intestine (79-83, 88-90, 92-94). Overexpression of ins-7 in the intestine decreases the activity of DAF-16/FOXO in non-intestinal tissues and shortens lifespan (83), suggesting an endocrine tissue-nonautonomous role of INS-7 in longevity. The expression of daf-2 in neurons is largely responsible for the longevity of daf-2 mutants (95, 96), pointing to the endocrine regulation of longevity by neuronal IIS. Together, it appears that IIS can systemically regulate lifespan from various tissues, via endocrine signaling.
The role of insulin/IGF-1 signaling in C. elegans physiology and age-related disease models
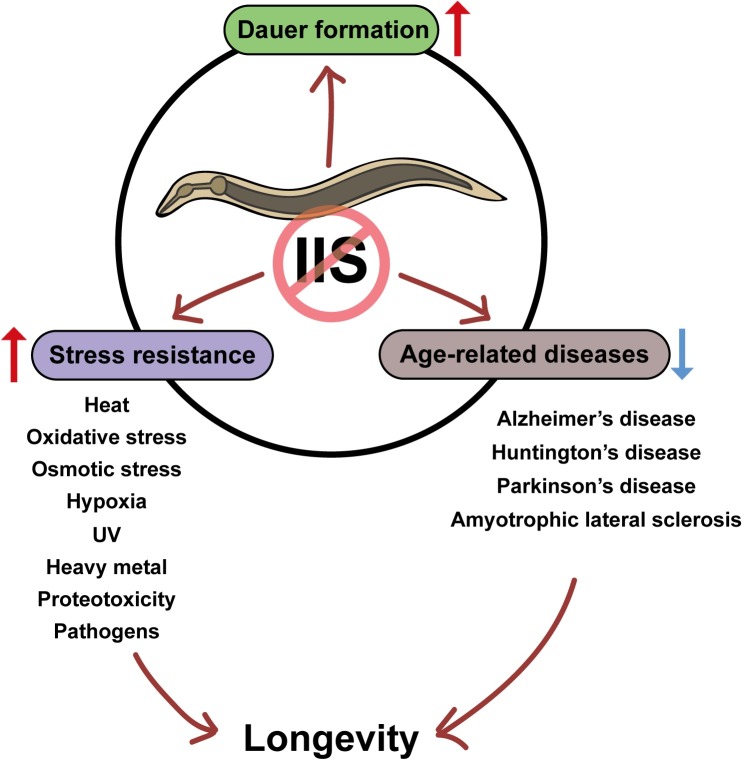
C. elegans that is exposed to unfavorable environmental conditions such as reduced food availability, extreme temperatures and a high population during development, enters an alternative diapause stage called dauer (97, 98). IIS is one of extensively studied signaling pathways that govern this dauer developmental decision (Fig. 2). Genetic inhibition of daf-2 or age-1, which extends adult lifespan, can cause constitutive dauer formation even under favorable conditions (97, 98). This dauer formation requires key downstream effectors in IIS, including DAF-18/PTEN and DAF-16/FOXO (97, 98). Reduced IIS activates a transcriptional program through DAF-16/FOXO, which leads to dauer formation. These findings raise the possibility that IIS may regulate dauer decision and lifespan using same effectors. However, the regulation of longevity and dauer formation by IIS can be uncoupled. Neuronal DAF-16/FOXO plays a more important role in the dauer decision than in lifespan regulation, whereas intestinal DAF-16/FOXO has a more profound effect on the lifespan extension than on the dauer decision (99). In addition, IIS pathway regulates the lifespan exclusively during adulthood, while it regulates the dauer formation during early larval development (100). Thus, spatiotemporal regulation of IIS differentially influences two separate aspects of animal physiology, development and adult lifespan.
Fig. 2. The role of insulin/IGF-1 signaling in C. elegans physiology and age-related disease models. Insulin/IGF-1 signaling (IIS) regulates dauer formation, stress resistance, and the models of age-related diseases in C. elegans. Reduced IIS promotes dauer formation and enhances resistance to various external and internal stresses, and pathogens. Inhibition of IIS also ameliorates defects associated with various human disease models. These protective effects of reduced IIS contribute to organismal longevity.
IIS also regulates resistance to a variety of stresses. C. elegans with reduced IIS displays enhanced resistance to environmental stresses such as oxidative stress (Fig. 2) (52, 101- 103), heat stress (104-106), hypoxic stress (107, 108), osmotic stress (109, 110), ultraviolet (UV) stress (36, 111), and heavy metal toxicity (112). Moreover, reduced IIS promotes better maintenance of internal homeostasis against cytosolic proteotoxicity (66, 113) and endoplasmic reticulum (ER) stress (114). The key downstream transcription factors of IIS that contribute to longevity, including DAF-16/FOXO (25, 26, 36, 66, 103, 106-116), HSF-1 (66, 67) and SKN-1 (52, 53), regulate these stress resistance phenotypes as well. Thus, proper regulation of IIS is crucial for the protection of C. elegans from both external and internal stresses.
Bacteria serve as a major food source for C. elegans, and are likely to be abundant in the natural habitats of C. elegans, such as rotten fruits. Therefore, it seems likely that C. elegans constantly comes in contact with various bacterial species, which may include pathogenic bacteria. To combat infection by pathogens, C. elegans is equipped with an innate immune system, and IIS is one of the most prominent innate immune signaling pathways (117). C. elegans with reduced IIS displays enhanced pathogen resistance, which is mediated by DAF-16/FOXO, HSF-1, and SKN-1 (84, 118-122). Reduced IIS leads to the induction of several antimicrobial genes (47), and reduction in bacterial packing in the intestine (123). Interestingly, Pseudomonas aeruginosa, a popular model bacterial pathogen in C. elegans, activates IIS to counteract the host immunity (124). Therefore, IIS may be located at the front of constant battles between the host C. elegans and its bacterial pathogens
Because of its powerful genetics, C. elegans has also been widely used for modeling various human diseases, especially neurodegenerative diseases. The disease models of C. elegans were established by generating transgenic animals expressing various human disease-associated proteins; these include β-amyloid peptides (Aβ) for Alzheimer’s disease (125-127), polyglutamine (polyQ) proteins for Huntington’s disease (128-132), α-synuclein for Parkinson’s disease (133-137), and a mutant superoxide dismutase 1 (SOD1) for amyotrophic lateral sclerosis (ALS) (138-141). The Alzheimer’s disease model C. elegans, which expresses Aβ1-42 in body wall muscles, is paralyzed and displays the accumulation of protein aggregates (68, 125, 142). Reduced IIS relieves these phenotypes via activating DAF-16/FOXO and HSF-1 (68), and inducing autophagic degradation of the protein aggregates (142). Reduced IIS also suppresses the short lifespan of Aβ1-42-expressing animals (68). The C. elegans model for Huntington’s disease has been widely used for studying proteotoxicity caused by aggregation of polyQ proteins (113, 128-131, 143-150). The polyQ-expressing worms display progressive neurodegeneration, neuronal dysfunction, retarded development, and defective motility (113, 128-131, 143-150). The daf-2 and age-1 mutations ameliorate a gradual age-dependent increase in toxicity resulting from polyQ aggregation through HSF-1 and DAF-16/FOXO (66, 113, 132, 146, 149). Parkinson’s disease patients suffer from degeneration of dopaminergic neurons, which display accumulated protein inclusions that contain α-synuclein (151). Similarly, the C. elegans models for Parkinson’s disease, which express wild-type or mutant human α-synuclein proteins in neurons, display the loss of dopaminergic neurons (133, 134, 137, 152). Reduced IIS by daf-2 mutations dramatically suppresses this neurodegeneration phenotype (152). ALS, which is characterized by progressive motor neuron degeneration (153), has also been studied using a C. elegans model (138-141). Familial ALS is associated with mutations in the gene encoding SOD1 (154, 155). Neuronal expression of a mutant human SOD1 causes locomotion defects (140) and paralysis (141) in C. elegans. daf-2 mutations protect the ALS model worms from the paralysis (141). Collectively, the results using C. elegans models indicate that IIS plays a crucial role in the pathophysiology of a majority of neurodegenerative diseases (Fig. 2). These findings imply that IIS modulates protein homeostasis to regulate normal neuronal functions, which may be essential for a long and healthy life.
INSULIN/IGF-1 SIGNALING PATHWAY AND Drosophila melanogaster AGING
Insulin/IGF-1 signaling components implicated in the longevity of D. melanogaster
The IIS pathway of Drosophila melanogaster consists of many components (Fig. 1), including the insulin/IGF receptor (dInR), the insulin receptor substrate (CHICO), the phosphatidylinositol 3-kinase (PI3K) Dp110/p60, 3-phosphoinositide-dependent protein kinase 1 (dPDK1) and the protein kinase B (PKB), also known as dAkt1, and the transcription factor Drosophila FOXO (dFOXO) (156-171). The activation mechanism of the IIS pathway in Drosophila has substantial similarities to that in C. elegans. Basically, the activation of dInR leads to up-regulation of a cascade of intracellular phosphorylation events, subsequently leading to the phosphorylation of dFOXO protein (160-162). dInR conveys signals from Drosophila insulin-like peptides (DILPs), directly to PI3K or to CHICO, the insulin receptor substrate (156, 172). PI3K, which converts PIP2 to PIP3, has a catalytic subunit, Dp110, and a regulatory subunit, Dp60 (158, 159). The action of PI3K is antagonized by the activity of dPTEN (173-175), which catalyzes PIP3 to PIP2. PIP3 acts as an intracellular second messenger that activates a cascade of protein kinases, including dPDK1 and PKB/dAkt, which subsequently lead to the phosphorylation and the nuclear exclusion of dFOXO (162, 170). Conversely, reduced IIS through dInR or CHICO mutations, or overexpression of dPTEN, causes the translocation of dFOXO from the cytoplasm to the nucleus, where it up-regulates genes involved in longevity and stress resistance (160, 162, 176, 177).
In Drosophila, the IIS pathway regulates various physiological processes, including lifespan, stress responses, growth and development. Genetic inhibition of negative regulators of dFOXO, including several DILPs (178, 179), dInR (180), the IRS/CHICO (181, 182), or 14-3-3 epsilon (183), extends the lifespan of Drosophila. Conversely, overexpression of antagonistic IIS regulators, such as dPTEN or dFOXO, also extends the lifespan and/or delay heart aging (177, 184-186; but see also 187). Overall, these findings using Drosophila have remarkable similarities with those of C. elegans, highlighting the evolutionarily conserved nature of lifespan regulation by IIS components.
Endocrine regulation of lifespan by Drosophila insulin/IGF-1 signaling
The Drosophila melanogaster genome encodes eight DILPs (172, 188-190). The dilp genes display distinct temporal expression patterns. For example, dilp2 is expressed from embryo to adult stages, whereas dilp4 is expressed only during development prior to adulthood (172, 179, 191-195). In addition, the expression sites of the eight dilp genes are diverse (196). Notably, the major site of DILP production is the median neurosecretory cells (mNSCs) in the brain, also called insulin-producing cells (IPCs), where dilp1, dilp2, dilp3 and dilp5 are expressed (172, 179, 191-194).
Cell non-autonomous regulation of lifespan by Drosophila IIS was proposed based on findings using tissue-specific overexpression of IIS components. Up-regulation of dFOXO in the adult head fat body is sufficient to promote longevity and oxidative stress resistance (177). Muscle-specific overexpression of either dPTEN, dFOXO, or 4E-BP (a dFOXO target), also significantly increases lifespan (184). The ablation of IPCs lengthens lifespan (179, 197), which corroborates the endocrine regulation of lifespan by IIS; this is also reminiscent of lifespan extension by the ablation of sensory neurons in C. elegans (reviewed in 198). Among the DILPs expressed in the IPCs, DILP2 has been extensively explored for its implication in lifespan regulation, since it has the highest homology with human insulin (172, 177-179, 199-201). The dilp2 null-mutant flies live long (178), and the expression of dilp2 in the IPCs is reduced by the activation of dFOXO (177, 199). Therefore, Drosophila IIS appears to regulate the expression of DILPs and longevity via a feedback mechanism. Furthermore, reduced dilp2 expression and inhibited IIS in the fat body are associated with lifespan extension conferred by transgenic expression of a dominant-negative p53 (200). DILP6, which is predominantly produced in the fat body, is another endocrine lifespan regulator (200-204). Surprisingly, overexpression of dilp6 in the abdominal fat body leads to the repression of dilp2 in the brain, suggesting synergistic effects on lifespan regulation by a potential dInR antagonist DILP6 and an agonist DILP2 (204). Collectively, lifespan regulation by IIS is controlled systemically by the action of DILPs that transmit signals, at least between the brain and fat body.
CONCLUSIONS
In this review, we have described findings regarding mechanisms by which IIS influences lifespan in two representative invertebrate models, C. elegans and D. melanogaster. The roles of many IIS components in aging are remarkably well conserved between C. elegans and D. melanogaster, and the intervention of the IIS leads to an extended lifespan in both animals. This suggests that the role of IIS in aging is likely to be conserved across phyla beyond these two species. Indeed, many findings using these invertebrate models have led to the discoveries demonstrating that changes in IIS can extend lifespan in mammals. For example, heterozygous IGF1 receptor- knockout mice (Igf1r+/−) live longer than wild-type (205), and lower circulating IGF1 level correlates with mouse longevity (206). In addition, genetic variants of IIS components, including IGF1 receptor and FOXO3A, are associated with human longevity (4, 207). Thus, the evidence for the evolutionarily conserved nature of IIS-mediated longevity is extremely strong, ranging from invertebrates to humans.
Both C. elegans and D. melanogaster have been invaluable for the identification of IIS components and their roles in aging at the organism level. Still, much remains to be discovered regarding the regulatory mechanisms of aging and longevity at the molecular level. As we can make new discoveries regarding organismal aging using these invertebrate models much faster than using vertebrates, C. elegans and D. melanogaster will continue to serve as indispensable tools for broadening our knowledge in aging. The progresses made by using these invertebrate models will eventually lead to the promotion of long and healthy human lives, and the prevention of age-associated diseases.
Acknowledgments
We thank the Lee laboratory members for critical comments on the manuscript. This work was supported by Basic Research Laboratory Grants NRF-2012R1A4A1028200 and NRF-2013R1A1A2014754 funded by the Korean Government (MSIP) through the National Research Foundation of Korea (NRF) to S-J.V.L.
References
- 1.Lee Y, An S, Artan M, et al. Genes and Pathways That Influence Longevity in Caenorhabditis elegans; in Aging Mechanisms, Mori N and Mook-Jung I (eds) Springer; Japan: (2015). pp. 123–169. [Google Scholar]
- 2.Giannakou ME, Partridge L. Role of insulin-like signalling in Drosophila lifespan. Trends Biochem Sci. (2007);32:180–188. doi: 10.1016/j.tibs.2007.02.007. [DOI] [PubMed] [Google Scholar]
- 3.Fontana L, Partridge L, Longo VD. Extending healthy life span--from yeast to humans. Science. (2010);328:321–326. doi: 10.1126/science.1172539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kenyon CJ. The genetics of ageing. Nature. (2010);464:504–512. doi: 10.1038/nature08980. [DOI] [PubMed] [Google Scholar]
- 5.Morris JZ, Tissenbaum HA, Ruvkun G. A phosphatidylinositol-3-OH kinase family member regulating longevity and diapause in Caenorhabditis elegans. Nature. (1996);382:536–539. doi: 10.1038/382536a0. [DOI] [PubMed] [Google Scholar]
- 6.Kimura KD, Tissenbaum HA, Liu Y, Ruvkun G. daf-2, an insulin receptor-like gene that regulates longevity and diapause in Caenorhabditis elegans. Science. (1997);277:942–946. doi: 10.1126/science.277.5328.942. [DOI] [PubMed] [Google Scholar]
- 7.Friedman DB, Johnson TE. A mutation in the age-1 gene in Caenorhabditis elegans lengthens life and reduces hermaphrodite fertility. Genetics. 1988;118:75–86. doi: 10.1093/genetics/118.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kenyon C, Chang J, Gensch E, Rudner A, Tabtiang R. A C. elegans mutant that lives twice as long as wild type. Nature. (1993);366:461–464. doi: 10.1038/366461a0. [DOI] [PubMed] [Google Scholar]
- 9.Klass MR. A method for the isolation of longevity mutants in the nematode Caenorhabditis elegans and initial results. Mech Ageing Dev. (1983);22:279–5/9. doi: 10.1016/0047-6374(83)90082-9. [DOI] [PubMed] [Google Scholar]
- 10.Wolkow CA, Munoz MJ, Riddle DL, Ruvkun G. Insulin receptor substrate and p55 orthologous adaptor proteins function in the Caenorhabditis elegans daf-2/insulin-like signaling pathway. J Biol Chem. (2002);277:49591–49597. doi: 10.1074/jbc.M207866200. [DOI] [PubMed] [Google Scholar]
- 11.Zhou K, Pandol S, Bokoch G, Traynor-Kaplan AE. Disruption of Dictyostelium PI3K genes reduces [32P]phosphatidylinositol 3,4 bisphosphate and [32P]phosphatidylinositol trisphosphate levels, alters F-actin distribution and impairs pinocytosis. J Cell Sci. (1998);111(Pt 2):283–294. doi: 10.1242/jcs.111.2.283. [DOI] [PubMed] [Google Scholar]
- 12.Ogg S, Ruvkun G. The C. elegans PTEN homolog, DAF-18, acts in the insulin receptor-like metabolic signaling pathway. Mol Cell. (1998);2:887–893. doi: 10.1016/S1097-2765(00)80303-2. [DOI] [PubMed] [Google Scholar]
- 13.Gil EB, Malone Link E, Liu LX, Johnson CD, Lees JA. Regulation of the insulin-like developmental pathway of Caenorhabditis elegans by a homolog of the PTEN tumor suppressor gene. Proc Natl Acad Sci U S A. (1999);96:2925–2930. doi: 10.1073/pnas.96.6.2925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mihaylova VT, Borland CZ, Manjarrez L, Stern MJ, Sun H. The PTEN tumor suppressor homolog in Caenorhabditis elegans regulates longevity and dauer formation in an insulin receptor-like signaling pathway. Proc Natl Acad Sci U S A. (1999);96:7427–7432. doi: 10.1073/pnas.96.13.7427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rouault JP, Kuwabara PE, Sinilnikova OM, Duret L, Thierry-Mieg D, Billaud M. Regulation of dauer larva development in Caenorhabditis elegans by daf-18, a homologue of the tumour suppressor PTEN. Curr Biol. (1999);9:329–332. doi: 10.1016/S0960-9822(99)80143-2. [DOI] [PubMed] [Google Scholar]
- 16.Dorman JB, Albinder B, Shroyer T, Kenyon C. The age-1 and daf-2 genes function in a common pathway to control the lifespan of Caenorhabditis elegans. Genetics. (1995);141:1399–1406. doi: 10.1093/genetics/141.4.1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Larsen PL, Albert PS, Riddle DL. Genes that regulate both development and longevity in Caenorhabditis elegans. Genetics. (1995);139:1567–1583. doi: 10.1093/genetics/139.4.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gottlieb S, Ruvkun G. daf-2, daf-16 and daf-23: genetically interacting genes controlling Dauer formation in Caenorhabditis elegans. Genetics. (1994);137:107–120. doi: 10.1093/genetics/137.1.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Solari F, Bourbon-Piffaut A, Masse I, Payrastre B, Chan AM, Billaud M. The human tumour suppressor PTEN regulates longevity and dauer formation in Caenorhabditis elegans. Oncogene. (2005);24:20–27. doi: 10.1038/sj.onc.1207978. [DOI] [PubMed] [Google Scholar]
- 20.Paradis S, Ailion M, Toker A, Thomas JH, Ruvkun G. A PDK1 homolog is necessary and sufficient to transduce AGE-1 PI3 kinase signals that regulate diapause in Caenorhabditis elegans. Genes Dev. (1999);13:1438–1452. doi: 10.1101/gad.13.11.1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Paradis S, Ruvkun G. Caenorhabditis elegans Akt/PKB transduces insulin receptor-like signals from AGE-1 PI3 kinase to the DAF-16 transcription factor. Genes Dev. (1998);12:2488–2498. doi: 10.1101/gad.12.16.2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hertweck M, Gobel C, Baumeister R. C. elegans SGK-1 is the critical component in the Akt/PKB kinase complex to control stress response and life span. Dev Cell. (2004);6:577–588. doi: 10.1016/S1534-5807(04)00095-4. [DOI] [PubMed] [Google Scholar]
- 23.Chen AT, Guo C, Dumas KJ, Ashrafi K, Hu PJ. Effects of Caenorhabditis elegans sgk-1 mutations on lifespan, stress resistance, and DAF-16/FoxO regulation. Aging Cell. (2013);12:932–940. doi: 10.1111/acel.12120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xiao R, Zhang B, Dong Y, et al. A genetic program promotes C. elegans longevity at cold temperatures via a thermosensitive TRP channel. Cell. (2013);152:806–817. doi: 10.1016/j.cell.2013.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Henderson ST, Johnson TE. daf-16 integrates developmental and environmental inputs to mediate aging in the nematode Caenorhabditis elegans. Curr Biol. (2001);11:1975–1980. doi: 10.1016/S0960-9822(01)00594-2. [DOI] [PubMed] [Google Scholar]
- 26.Lee RY, Hench J, Ruvkun G. Regulation of C. elegans DAF-16 and its human ortholog FKHRL1 by the daf-2 insulin-like signaling pathway. Curr Biol. (2001);11:1950–1957. doi: 10.1016/S0960-9822(01)00595-4. [DOI] [PubMed] [Google Scholar]
- 27.Lin K, Hsin H, Libina N, Kenyon C. Regulation of the Caenorhabditis elegans longevity protein DAF-16 by insulin/IGF-1 and germline signaling. Nat Genet. (2001);28:139–145. doi: 10.1038/88850. [DOI] [PubMed] [Google Scholar]
- 28.Cahill CM, Tzivion G, Nasrin N, et al. Phosphatidylinositol 3-kinase signaling inhibits DAF-16 DNA binding and function via 14-3-3-dependent and 14-3-3-independent pathways. J Biol Chem. (2001);276:13402–13410. doi: 10.1074/jbc.M010042200. [DOI] [PubMed] [Google Scholar]
- 29.Ogg S, Paradis S, Gottlieb S, et al. The Fork head transcription factor DAF-16 transduces insulin-like metabolic and longevity signals in C. elegans. Nature. (1997);389:994–999. doi: 10.1038/40194. [DOI] [PubMed] [Google Scholar]
- 30.Lin K, Dorman JB, Rodan A, Kenyon C. daf-16: An HNF-3/forkhead family member that can function to double the life-span of Caenorhabditis elegans. Science. (1997);278:1319–1322. doi: 10.1126/science.278.5341.1319. [DOI] [PubMed] [Google Scholar]
- 31.Oh SW, Mukhopadhyay A, Svrzikapa N, Jiang F, Davis RJ, Tissenbaum HA. JNK regulates lifespan in Caenorhabditis elegans by modulating nuclear translocation of forkhead transcription factor/DAF-16. Proc Natl Acad Sci U S A. (2005);102:4494–4499. doi: 10.1073/pnas.0500749102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Curtis R, O G, DiStefano PS. Aging networks in Caenorhabditis elegans: AMP-activated protein kinase (aak-2) links multiple aging and metabolism pathways. Aging Cell. (2006);5:119–126. doi: 10.1111/j.1474-9726.2006.00205.x. [DOI] [PubMed] [Google Scholar]
- 33.Greer EL, Dowlatshahi D, Banko MR, et al. An AMPK-FOXO pathway mediates longevity induced by a novel method of dietary restriction in C. elegans. elegans. Curr Biol. (2007);17:1646–1656. doi: 10.1016/j.cub.2007.08.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Apfeld J, O G, McDonagh T, DiStefano PS, Curtis R. The AMP-activated protein kinase AAK-2 links energy levels and insulin-like signals to lifespan in C. elegans. Genes Dev. (2004);18:3004–3009. doi: 10.1101/gad.1255404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lehtinen MK, Yuan Z, Boag PR, et al. A conserved MST-FOXO signaling pathway mediates oxidative-stress responses and extends life span. Cell. (2006);125:987–1001. doi: 10.1016/j.cell.2006.03.046. [DOI] [PubMed] [Google Scholar]
- 36.Wolff S, Ma H, Burch D, Maciel GA, Hunter T, Dillin A. SMK-1, an essential regulator of DAF-16-mediated longevity. Cell. (2006);124:1039–1053. doi: 10.1016/j.cell.2005.12.042. [DOI] [PubMed] [Google Scholar]
- 37.Seo M, Seo K, Hwang W, et al. RNA helicase HEL-1 promotes longevity by specifically activating DAF-16/FOXO transcription factor signaling in Caenorhabditis elegans. Proc Natl Acad Sci U S A. (2015);112:E4246–E4255. doi: 10.1073/pnas.1505451112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chiang WC, Tishkoff DX, Yang B, et al. C. elegans SIRT6/7 homolog SIR-2.4 promotes DAF-16 relocalization and function during stress. PLoS Genet. (2012);8:e1002948. doi: 10.1371/journal.pgen.1002948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hu PJ, Xu J, Ruvkun G. Two membrane-associated tyrosine phosphatase homologs potentiate C. elegans AKT-1/PKB signaling. PLoS Genet. (2006);2:e99. doi: 10.1371/journal.pgen.0020099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li J, Ebata A, Dong Y, Rizki G, Iwata T, Lee SS. Caenorhabditis elegans HCF-1 functions in longevity maintenance as a DAF-16 regulator. PLoS Biol. (2008);6:e233. doi: 10.1371/journal.pbio.0060233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Alam H, Williams TW, Dumas KJ, et al. EAK-7 controls development and life span by regulating nuclear DAF-16/FoxO activity. Cell Metab. (2010);12:30–41. doi: 10.1016/j.cmet.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Berdichevsky A, Viswanathan M, Horvitz HR, Guarente L. C. elegans SIR-2.1 interacts with 14-3-3 proteins to activate DAF-16 and extend life span. Cell. (2006);125:1165–1177. doi: 10.1016/j.cell.2006.04.036. [DOI] [PubMed] [Google Scholar]
- 43.Li J, Tewari M, Vidal M, Lee SS. The 14-3-3 protein FTT-2 regulates DAF-16 in Caenorhabditis elegans. Dev Biol. (2007);301:82–91. doi: 10.1016/j.ydbio.2006.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang Y, Oh SW, Deplancke B, Luo J, Walhout AJ, Tissenbaum HA. C. elegans 14-3-3 proteins regulate life span and interact with SIR-2.1 and DAF-16/FOXO. Mech Ageing Dev. (2006);127:741–747. doi: 10.1016/j.mad.2006.05.005. [DOI] [PubMed] [Google Scholar]
- 45.Lee SS, Kennedy S, Tolonen AC, Ruvkun G. DAF-16 target genes that control C. elegans life-span and metabolism. Science. (2003);300:644–647. doi: 10.1126/science.1083614. [DOI] [PubMed] [Google Scholar]
- 46.Ookuma S, Fukuda M, Nishida Identification of a DAF-16 transcriptional target gene, scl-1, that regulates longevity and stress resistance in Caenorhabditis elegans. Curr Biol. (2003);13:427–431. doi: 10.1016/S0960-9822(03)00108-8. [DOI] [PubMed] [Google Scholar]
- 47.Murphy CT, McCarroll SA, Bargmann CI, et al. Genes that act downstream of DAF-16 to influence the lifespan of Caenorhabditis elegans. Nature. (2003);424:277–283. doi: 10.1038/nature01789. [DOI] [PubMed] [Google Scholar]
- 48.McElwee J, Bubb K, Thomas JH. Transcriptional outputs of the Caenorhabditis elegans forkhead protein DAF-16. Aging Cell. (2003);2:111–121. doi: 10.1046/j.1474-9728.2003.00043.x. [DOI] [PubMed] [Google Scholar]
- 49.Golden TR, Melov S. Microarray analysis of gene expression with age in individual nematodes. Aging Cell. (2004);3:111–124. doi: 10.1111/j.1474-9728.2004.00095.x. [DOI] [PubMed] [Google Scholar]
- 50.Halaschek-Wiener J, Khattra JS, McKay S, et al. Analysis of long-lived <italic>C. Genome Res. (2005);15:603–615. doi: 10.1101/gr.3274805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lee SJ, Murphy CT, Kenyon C. Glucose shortens the life span of C. elegans by downregulating DAF-16/FOXO activity and aquaporin gene expression. Cell Metab. (2009);10:379–391. doi: 10.1016/j.cmet.2009.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tullet JM, Hertweck M, An JH, et al. Direct inhibition of the longevity-promoting factor SKN-1 by insulin-like signaling in C. elegans. Cell. (2008);132:1025–1038. doi: 10.1016/j.cell.2008.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.An JH, Blackwell TK. SKN-1 links C. elegans mesendodermal specification to a conserved oxidative stress response. Genes Dev. (2003);17:1882–1893. doi: 10.1101/gad.1107803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.An JH, Vranas K, Lucke M, et al. Regulation of the Caenorhabditis elegans oxidative stress defense protein SKN-1 by glycogen synthase kinase-3. Proc Natl Acad Sci U S A. (2005);102:16275–16280. doi: 10.1073/pnas.0508105102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kahn NW, Rea SL, Moyle S, Kell A, Johnson TE. Proteasomal dysfunction activates the transcription factor SKN-1 and produces a selective oxidativestress response in Caenorhabditis elegans. Biochem J. (2008);409:205–213. doi: 10.1042/BJ20070521. [DOI] [PubMed] [Google Scholar]
- 56.Oliveira RP, Porter Abate J, Dilks K, et al. Condition-adapted stress and longevity gene regulation by Caenorhabditis elegans SKN-1/Nrf. Aging Cell. (2009);8:524–541. doi: 10.1111/j.1474-9726.2009.00501.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang J, Robida-Stubbs S, Tullet JM, Rual JF, Vidal M, Blackwell TK. RNAi screening implicates a SKN-1-dependent transcriptional response in stress resistance and longevity deriving from translation inhibition. PLoS Genet. (2010);6:e1001048. doi: 10.1371/journal.pgen.1001048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Staab TA, Griffen TC, Corcoran C, Evgrafov O, Knowles JA, Sieburth D. The conserved SKN-1/Nrf2 stress response pathway regulates synaptic function in Caenorhabditis elegans. PLoS Genet. 2013;9:e1003354. doi: 10.1371/journal.pgen.1003354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Glover-Cutter KM, Lin S, Blackwell TK. Integration of the unfolded protein and oxidative stress responses through SKN-1/Nrf. PLoS Genet. (2013);9:e1003701. doi: 10.1371/journal.pgen.1003701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Choe KP, Przybysz AJ, Strange K. The WD40 repeat protein WDR-23 functions with the CUL4/DDB1 ubiquitin ligase to regulate nuclear abundance and activity of SKN-1 in Caenorhabditis elegans. Mol Cell Biol. e1003701;29:2704–2715. doi: 10.1128/MCB.01811-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Park SK, Tedesco PM, Johnson TE. Oxidative stress and longevity in Caenorhabditis elegans as mediated by SKN-1. Aging Cell. (2009);8:258–269. doi: 10.1111/j.1474-9726.2009.00473.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pang S, Lynn DA, Lo JY, Paek J, Curran SP. SKN-1 and Nrf2 couples proline catabolism with lipid metabolism during nutrient deprivation. Nat Commun. (2014);5:5048. doi: 10.1038/ncomms6048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kell A, Ventura N, Kahn N, Johnson TE. Activation of SKN-1 by novel kinases in Caenorhabditis elegans. Free Radic Biol Med. (2007);43:1560–1566. doi: 10.1016/j.freeradbiomed.2007.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Li X, Matilainen O, Jin C, Glover-Cutter KM, Holmberg CI, Blackwell TK. Specific SKN-1/Nrf stress responses to perturbations in translation elongation and proteasome activity. PLoS Genet. (2011);7:e1002119. doi: 10.1371/journal.pgen.1002119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ewald CY, Landis JN, Porter Abate J, Murphy CT, Blackwell TK. Dauer-independent insulin/IGF-1-signalling implicates collagen remodelling in longevity. Nature. (2015);519:97–101. doi: 10.1038/nature14021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hsu AL, Murphy CT, Kenyon C. Regulation of aging and age-related disease by DAF-16 and heat-shock factor. Science. (2003);300:1142–1145. doi: 10.1126/science.1083701. [DOI] [PubMed] [Google Scholar]
- 67.Morley JF, Morimoto RI. Regulation of longevity in Caenorhabditis elegans by heat shock factor and molecular chaperones. Mol Biol Cell. (2004);15:657–664. doi: 10.1091/mbc.E03-07-0532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cohen E, Bieschke J, Perciavalle RM, Kelly JW, Dillin A. Opposing activities protect against age-onset proteotoxicity. Science. (2006);313:1604–1610. doi: 10.1126/science.1124646. [DOI] [PubMed] [Google Scholar]
- 69.Chiang WC, Ching TT, Lee HC, Mousigian C, Hsu AL. HSF-1 regulators DDL-1/2 link insulin-like signaling to heat-shock responses and modulation of longevity. Cell. (2012);148:322–334. doi: 10.1016/j.cell.2011.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Seo K, Choi E, Lee D, Jeong DE, Jang SK, Lee SJ. Heat shock factor 1 mediates the longevity conferred by inhibition of TOR and insulin/IGF-1 signaling pathways in C. elegans. Aging Cell. (2013);12:1073–1081. doi: 10.1111/acel.12140. [DOI] [PubMed] [Google Scholar]
- 71.Garigan D, Hsu AL, Fraser AG, Kamath RS, Ahringer J, Kenyon C. Genetic analysis of tissue aging in Caenorhabditis elegans: a role for heat-shock factor and bacterial proliferation. Genetics. (2002);161:1101–1112. doi: 10.1093/genetics/161.3.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Douglas PM, Baird NA, Simic MS, et al. Heterotypic signals from neural HSF-1 separate thermotolerance from longevity. Cell Rep. (2015);12:1196–1204. doi: 10.1016/j.celrep.2015.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Baird NA, Douglas PM, Simic MS, et al. HSF-1-mediated cytoskeletal integrity determines thermotolerance and life span. Science. (2015);346:360–363. doi: 10.1126/science.1253168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Amin J, Ananthan J, Voellmy R. Key features of heat shock regulatory elements. Mol Cell Biol. (1988);8:3761–3769. doi: 10.1128/MCB.8.9.3761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kay RJ, Boissy RJ, Russnak RH, Candido EP. Efficient transcription of a Caenorhabditis elegans heat shock gene pair in mouse fibroblasts is dependent on multiple promoter elements which can function bidirectionally. Mol Cell Biol. (1986);6:3134–3143. doi: 10.1128/MCB.6.9.3134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Russnak RH, Candido EP. Locus encoding a family of small heat shock genes in Caenorhabditis elegans: two genes duplicated to form a 3.8-kilobase inverted repeat. Mol Cell Biol. (1985);5:1268–1278. doi: 10.1128/MCB.5.6.1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yokoyama K, Fukumoto K, Murakami T, et al. Extended longevity of Caenorhabditis elegans by knocking in extra copies of hsp70F, a homolog of mot-2 (mortalin)/mthsp70/Grp75. FEBS Lett. (2002);516:53–57. doi: 10.1016/s0014-5793(02)02470-5. [DOI] [PubMed] [Google Scholar]
- 78.Walker GA, Lithgow GJ. Lifespan extension in C. elegans by a molecular chaperone dependent upon insulin-like signals. Aging Cell. (2003);2:131–139. doi: 10.1046/j.1474-9728.2003.00045.x. [DOI] [PubMed] [Google Scholar]
- 79.Pierce SB, Costa M, Wisotzkey R, et al. Regulation of DAF-2 receptor signaling by human insulin and ins-1, a member of the unusually large and diverse C. elegans insulin gene family. Genes Dev. 2001;15:672–686. doi: 10.1101/gad.867301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Li W, Kennedy SG, Ruvkun G. daf-28 encodes a C. elegans insulin superfamily member that is regulated by environmental cues and acts in the DAF-2 signaling pathway. Genes Dev. (2003);17:844–858. doi: 10.1101/gad.1066503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Chen Z, Hendricks M, Cornils A, Maier W, Alcedo J, Zhang Y. Two insulin-like peptides antagonistically regulate aversive olfactory learning in C. elegans. Neuron. (2013);77:572–585. doi: 10.1016/j.neuron.2012.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Cornils A, Gloeck M, Chen Z, Zhang Y, Alcedo J. Specific insulin-like peptides encode sensory information to regulate distinct developmental processes. Development. (2011);138:1183–1193. doi: 10.1242/dev.060905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Murphy CT, Lee SJ, Kenyon C. Tissue entrainment by feedback regulation of insulin gene expression in the endoderm of Caenorhabditis elegans. Proc Natl Acad Sci U S A. (2007);104:19046–19050. doi: 10.1073/pnas.0709613104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kawli T, Tan MW. Neuroendocrine signals modulate the innate immunity of Caenorhabditis elegans through insulin signaling. Nat Immunol. (2008);9:1415–1424. doi: 10.1038/ni.1672. [DOI] [PubMed] [Google Scholar]
- 85.Malone EA, Inoue T, Thomas JH. Genetic analysis of the roles of daf-28 and age-1 in regulating Caenorhabditis elegans dauer formation. Genetics. (1996);143:1193–1205. doi: 10.1093/genetics/143.3.1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Malone EA, Thomas JH. A screen for nonconditional dauer-constitutive mutations in Caenorhabditis elegans. Genetics. (1994);136:879–886. doi: 10.1093/genetics/136.3.879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Fernandes de Abreu DA, Caballero A, Fardel P, et al. An insulin-to-insulin regulatory network orchestrates phenotypic specificity in development and physiology. PLoS Genet. (2014);10:e1004225. doi: 10.1371/journal.pgen.1004225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ritter AD, Shen Y, Fuxman Bass J, et al. Complex expression dynamics and robustness in C. elegans insulin networks. Genome Res. (2013);23:954–965. doi: 10.1101/gr.150466.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hung WL, Wang Y, Chitturi J, Zhen M. A Caenorhabditis elegans developmental decision requires insulin signaling-mediated neuron-intestine communication. Development. (2014);141:1767–1779. doi: 10.1242/dev.103846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Chen Y, Baugh LR. Ins-4 and daf-28 function redundantly to regulate C. elegans L1 arrest. Dev Biol. (2014);394:314–326. doi: 10.1016/j.ydbio.2014.08.002. [DOI] [PubMed] [Google Scholar]
- 91.Duret L, Guex N, Peitsch MC, Bairoch A. New insulin-like proteins with atypical disulfide bond pattern characterized in Caenorhabditis elegans by comparative sequence analysis and homology modeling. Genome Res. (1998);8:348–353. doi: 10.1101/gr.8.4.348. [DOI] [PubMed] [Google Scholar]
- 92.Michaelson D, Korta DZ, Capua Y, Hubbard EJ. Insulin signaling promotes germline proliferation in C. elegans. Development. (2010);137:671–680. doi: 10.1242/dev.042523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Leinwand SG, Chalasani SH. Neuropeptide signaling remodels chemosensory circuit composition in Caenorhabditis elegans. Nat Neurosci. (2013);16:1461–1467. doi: 10.1038/nn.3511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ohta A, Ujisawa T, Sonoda S, Kuhara A. Light and pheromone-sensing neurons regulates cold habituation through insulin signalling in Caenorhabditis elegans. Nat Commun. (2014);5:4412. doi: 10.1038/ncomms5412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wolkow CA, Kimura KD, Lee MS, Ruvkun G. Regulation of C. elegans life-span by insulinlike signaling in the nervous system. Science. (2000);290:147–150. doi: 10.1126/science.290.5489.147. [DOI] [PubMed] [Google Scholar]
- 96.Iser WB, Gami MS, Wolkow CA. Insulin signaling in Caenorhabditis elegans regulates both endocrine-like and cell-autonomous outputs. Dev Biol. (2007);303:434–447. doi: 10.1016/j.ydbio.2006.04.467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Hu PJ. Dauer. WormBook: the online review of C. elegans biology. (2007). pp. 1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Riddle DL, Albert PS. Genetic and Environmental Regulation of Dauer Larva Development; in C. elegans II, Riddle DL, Blumenthal T, Meyer BJ et al (eds) Cold Spring Harbor Laboratory Press; Cold Spring Harbor (NY): (1997). [PubMed] [Google Scholar]
- 99.Libina N, Berman JR, Kenyon C. Tissue-specific activities of C. elegans DAF-16 in the regulation of lifespan. Cell. (2003);115:489–502. doi: 10.1016/S0092-8674(03)00889-4. [DOI] [PubMed] [Google Scholar]
- 100.Dillin A, Crawford DK, Kenyon C. Timing requirements for insulin/IGF-1 signaling in C. elegans. Science. (2002);298:830–834. doi: 10.1126/science.1074240. [DOI] [PubMed] [Google Scholar]
- 101.Larsen PL. Aging and resistance to oxidative damage in Caenorhabditis elegans. Proc Natl Acad Sci U S A. (1993);90:8905–8909. doi: 10.1073/pnas.90.19.8905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Vanfleteren JR. Oxidative stress and ageing in Caenorhabditis elegans. Biochem J. (1993);292(Pt 2):605–608. doi: 10.1042/bj2920605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Honda Y, Honda S. The daf-2 gene network for longevity regulates oxidative stress resistance and Mn-superoxide dismutase gene expression in Caenorhabditis elegans. FASEB J. (1999);13:1385–1393. [PubMed] [Google Scholar]
- 104.Gems D, Sutton AJ, Sundermeyer ML, et al. Two pleiotropic classes of daf-2 mutation affect larval arrest, adult behavior, reproduction and longevity in Caenorhabditis elegans. Genetics. (1998);150:129–155. doi: 10.1093/genetics/150.1.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Lithgow GJ, White TM, Melov S, Johnson TE. Thermotolerance and extended life-span conferred by single-gene mutations and induced by thermal stress. Proc Natl Acad Sci U S A. (1995);92:7540–7544. doi: 10.1073/pnas.92.16.7540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.McColl G, Rogers AN, Alavez S, et al. Insulin-like signaling determines survival during stress via posttranscriptional mechanisms in C. elegans. Cell Metab. (2010);12:260–272. doi: 10.1016/j.cmet.2010.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Scott BA, Avidan MS, Crowder CM. Regulation of hypoxic death in C. elegans by the insulin/IGF receptor homolog DAF-2. Science. (2002);296:2388–2391. doi: 10.1126/science.1072302. [DOI] [PubMed] [Google Scholar]
- 108.Mabon ME, Scott BA, Crowder CM. Divergent mechanisms controlling hypoxic sensitivity and lifespan by the DAF-2/insulin/IGF-receptor pathway. PLoS One. (2009);4:e7937. doi: 10.1371/journal.pone.0007937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Lamitina ST, Strange K. Transcriptional targets of DAF-16 insulin signaling pathway protect C. elegans from extreme hypertonic stress. Am J Physiol Cell Physiol. (2005);288:C467–C474. doi: 10.1152/ajpcell.00451.2004. [DOI] [PubMed] [Google Scholar]
- 110.Burkewitz K, Choe K, Strange K. Hypertonic stress induces rapid and widespread protein damage in C. elegans. Am J Physiol Cell Physiol. (2011);301:C566–C576. doi: 10.1152/ajpcell.00030.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Murakami S, Johnson TE. A genetic pathway conferring life extension and resistance to UV stress in Caenorhabditis elegans. Genetics. (1996);143:1207–1218. doi: 10.1093/genetics/143.3.1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Barsyte D, Lovejoy DA, Lithgow GJ. Longevity and heavy metal resistance in daf-2 and age-1 long-lived mutants of Caenorhabditis elegans. FASEB J. (2001);15:627–634. doi: 10.1096/fj.99-0966com. [DOI] [PubMed] [Google Scholar]
- 113.Morley JF, Brignull HR, Weyers JJ, Morimoto RI. The threshold for polyglutamine-expansion protein aggregation and cellular toxicity is dynamic and influenced by aging in Caenorhabditis elegans. Proc Natl Acad Sci U S A. (2002);99:10417–10422. doi: 10.1073/pnas.152161099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Henis-Korenblit S, Zhang PC, Hansen M, et al. Insulin/IGF-1 signaling mutants reprogram ER stress response regulators to promote longevity. Proc Natl Acad Sci U S A. (2010);107:9730–9735. doi: 10.1073/pnas.1002575107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Essers MA, de Vries-Smits LM, Barker N, Polderman PE, Burgering BM, Korswagen HC. Functional interaction between beta-catenin and FOXO in oxidative stress signaling. Science. (2005);308:181–1184. doi: 10.1126/science.1109083. [DOI] [PubMed] [Google Scholar]
- 116.Mueller MM, Castells-Roca L, Babu V, et al. DAF-16/FOXO and EGL-27/GATA promote developmental growth in response to persistent somatic DNA damage. Nat Cell Biol. (2014);16:1168–1179. doi: 10.1038/ncb3071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Ermolaeva MA, Schumacher B. Insights from the worm: the C. elegans model for innate immunity. Semin Immunol. (2014);26:303–309. doi: 10.1016/j.smim.2014.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Garsin DA, Villanueva JM, Begun J, et al. Long-lived C. elegans daf-2 mutants are resistant to bacterial pathogens. Science. (2003);300:1921. doi: 10.1126/science.1080147. [DOI] [PubMed] [Google Scholar]
- 119.Kerry S, TeKippe M, Gaddis NC, Aballay A. GATA transcription factor required for immunity to bacterial and fungal pathogens. PLoS One. (2006);1:e77. doi: 10.1371/journal.pone.0000077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Singh V, Aballay A. Heat-shock transcription factor (HSF)-1 pathway required for Caenorhabditis elegans immunity. Proc Natl Acad Sci U S A. (2006);103:13092–13097. doi: 10.1073/pnas.0604050103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Papp D, Csermely P, Soti C. A role for SKN-1/Nrf in pathogen resistance and immunosenescence in Caenorhabditis elegans. PLoS Pathog. (2012);8:e1002673. doi: 10.1371/journal.ppat.1002673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Evans EA, Chen WC, Tan MW. The DAF-2 insulin-like signaling pathway independently regulates aging and immunity in C. elegans. Aging Cell. (2008);7:879–893. doi: 10.1111/j.1474-9726.2008.00435.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Portal-Celhay C, Bradley ER, Blaser MJ. Control of intestinal bacterial proliferation in regulation of lifespan in Caenorhabditis elegans. BMC Micobiol. (2012);12:49. doi: 10.1186/1471-2180-12-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Evans EA, Kawli T, Tan M-W. Pseudomonas aeruginosa suppresses host immunity by activating the DAF-2 insulin-like signaling pathway in Caenorhabditis elegans. PLoS Pathog. (2008);4:e1000175. doi: 10.1371/journal.ppat.1000175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Link CD. Expression of human β-amyloid peptide in transgenic Caenorhabditis elegans. Proc Natl Acad Sci U S A. (1995);92:9368–9372. doi: 10.1073/pnas.92.20.9368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Fay DS, Fluet A, Johnson CJ, Link CD. In Vivo Aggregation of β‐Amyloid Peptide Variants. J Neurochem. (1998);71:1616–1625. doi: 10.1046/j.1471-4159.1998.71041616.x. [DOI] [PubMed] [Google Scholar]
- 127.Link CD, Taft A, Kapulkin V, et al. Gene expression analysis in a transgenic Caenorhabditis elegans Alzheimer’s disease model. Neurobiol Aging. (2003);24:397–413. doi: 10.1016/S0197-4580(02)00224-5. [DOI] [PubMed] [Google Scholar]
- 128.Faber PW, Alter JR, MacDonald ME, Hart AC. Polyglutamine-mediated dysfunction and apoptotic death of a Caenorhabditis elegans sensory neuron. Proc Natl Acad Sci U S A. (1999);96:179–184. doi: 10.1073/pnas.96.1.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Satyal SH, Schmidt E, Kitagawa K, et al. Polyglutamine aggregates alter protein folding homeostasis in Caenorhabditis elegans. Proc Natl Acad Sci U S A. (2000);97:5750–5755. doi: 10.1073/pnas.100107297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Parker JA, Connolly JB, Wellington C, Hayden M, Dausset J, Neri C. Expanded polyglutamines in Caenorhabditis elegans cause axonal abnormalities and severe dysfunction of PLM mechanosensory neurons without cell death. Proc Natl Acad Sci U S A. (2001);98:13318–13323. doi: 10.1073/pnas.231476398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Brignull HR, Moore FE, Tang SJ, Morimoto RI. Polyglutamine proteins at the pathogenic threshold display neuron-specific aggregation in a pan-neuronal Caenorhabditis elegans model. J Neurosci. (2006);26:7597–7606. doi: 10.1523/JNEUROSCI.0990-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Mohri-Shiomi A, Garsin DA. Insulin signaling and the heat shock response modulate protein homeostasis in the Caenorhabditis elegans intestine during infection. J Biol Chem. (2008);283:194–201. doi: 10.1074/jbc.M707956200. [DOI] [PubMed] [Google Scholar]
- 133.Lakso M, Vartiainen S, Moilanen AM, et al. Dopaminergic neuronal loss and motor deficits in Caenorhabditis elegans overexpressing human alpha-synuclein. J Neurochem. (2003);86:165–172. doi: 10.1046/j.1471-4159.2003.01809.x. [DOI] [PubMed] [Google Scholar]
- 134.Cao S, Gelwix CC, Caldwell KA, Caldwell GA. Torsin-mediated protection from cellular stress in the dopaminergic neurons of Caenorhabditis elegans. J Neurosci. (2005);25:3801–3812. doi: 10.1523/JNEUROSCI.5157-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Kuwahara T, Koyama A, Gengyo-Ando K, et al. Familial Parkinson mutant α-synuclein causes dopamine neuron dysfunction in transgenic Caenorhabditis elegans. J Biol Chem. (2006);281:334–340. doi: 10.1074/jbc.M504860200. [DOI] [PubMed] [Google Scholar]
- 136.Kuwahara T, Koyama A, Koyama S, et al. A systematic RNAi screen reveals involvement of endocytic pathway in neuronal dysfunction in ^-synuclein transgenic C. elegans. Hum Mol Genet. (2008);17:2997–3009. doi: 10.1093/hmg/ddn198. [DOI] [PubMed] [Google Scholar]
- 137.Hamamichi S, Rivas RN, Knight AL, Cao S, Caldwell KA, Caldwell GA. Hypothesis-based RNAi screening identifies neuroprotective genes in a Parkinson's disease model. Proc Natl Acad Sci U S A. (2008);105:728–733. doi: 10.1073/pnas.0711018105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Oeda T, Shimohama S, Kitagawa N, et al. Oxidative stress causes abnormal accumulation of familial amyotrophic lateral sclerosis-related mutant SOD1 in transgenic Caenorhabditis elegans. Hum Mol Genet. (2001);10:203–2023. doi: 10.1093/hmg/10.19.2013. [DOI] [PubMed] [Google Scholar]
- 139.Gidalevitz T, Krupinski T, Garcia S, Morimoto RI. Destabilizing protein polymorphisms in the genetic background direct phenotypic expression of mutant SOD1 toxicity. PLoS Genet. (2009);5:e1000399. doi: 10.1371/journal.pgen.1000399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Wang J, Farr GW, Hall DH, et al. An ALS-linked mutant SOD1 produces a locomotor defect associated with aggregation and synaptic dysfunction when expressed in neurons of Caenorhabditis elegans. PLoS Genet. (2009);5:e1000350. doi: 10.1371/journal.pgen.1000350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Li J, Huang KX, Le WD. Establishing a novel C. elegans model to investigate the role of autophagy in amyotrophic lateral sclerosis. Acta Pharmacol Sin. (2013);34:644–650. doi: 10.1038/aps.2012.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Florez-McClure ML, Hohsfield LA, Fonte G, Bealor MT, Link CD. Decreased insulin-receptor signaling promotes the autophagic degradation of β-amyloid peptide in C. elegans. Autophagy. (2007);3:569–580. doi: 10.4161/auto.4776. [DOI] [PubMed] [Google Scholar]
- 143.David DC, Ollikainen N, Trinidad JC, Cary MP, Burlingame AL, Kenyon C. Widespread protein aggregation as an inherent part of aging in C. elegans. PLoS Biol. (2010);8:e1000450. doi: 10.1371/journal.pbio.1000450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Wang H, Lim PJ, Yin C, Rieckher M, Vogel BE, Monteiro MJ. Suppression of polyglutamine-induced toxicity in cell and animal models of Huntington's disease by ubiquilin. Hum Mol Genet. (2006);15:1025–1041. doi: 10.1093/hmg/ddl017. [DOI] [PubMed] [Google Scholar]
- 145.Wang H, Lim PJ, Karbowski M, Monteiro MJ. Effects of overexpression of huntingtin proteins on mitochondrial integrity. Hum Mol Genet. (2009);18:737–752. doi: 10.1093/hmg/ddn404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Steinkraus KA, Smith ED, Davis C, et al. Dietary restriction suppresses proteotoxicity and enhances longevity by an hsf-1-dependent mechanism in Caenorhabditis elegans. Aging Cell. (2008);7:394–404. doi: 10.1111/j.1474-9726.2008.00385.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Faber PW, Voisine C, King DC, Bates EA, Hart AC. Glutamine/proline-rich PQE-1 proteins protect Caenorhabditis elegans neurons from huntingtin polyglutamine neurotoxicity. Proc Natl Acad Sci U S A. (2002);99:17131–17136. doi: 10.1073/pnas.262544899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Parker JA, Metzler M, Georgiou J, et al. Huntingtininteracting protein 1 influences worm and mouse presynaptic function and protects Caenorhabditis elegans neurons against mutant polyglutamine toxicity. J Neurosci. (2007);27:11056–11064. doi: 10.1523/JNEUROSCI.1941-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Parker JA, Arango M, Abderrahmane S, et al. Resveratrol rescues mutant polyglutamine cytotoxicity in nematode and mammalian neurons. Nat Genet. (2005);37:349–350. doi: 10.1038/ng1534. [DOI] [PubMed] [Google Scholar]
- 150.Bates EA, Victor M, Jones AK, Shi Y, Hart AC. Differential contributions of Caenorhabditis elegans histone deacetylases to huntingtin polyglutamine toxicity. J Neurosci. (2006);26:2830–2838. doi: 10.1523/JNEUROSCI.3344-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Schapira AH, Jenner P. Etiology and pathogenesis of Parkinson's disease. Mov Disord. (2011);26:1049–1055. doi: 10.1002/mds.23732. [DOI] [PubMed] [Google Scholar]
- 152.Knight AL, Yan X, Hamamichi S, et al. The glycolytic enzyme, GPI, is a functionally conserved modifier of dopaminergic neurodegeneration in Parkinson's models. Cell Metab. (2014);20:145–157. doi: 10.1016/j.cmet.2014.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Pasinelli P, Brown RH. Molecular biology of amyotrophic lateral sclerosis: insights from genetics. Nat Rev Neurosci. (2006);7:710–723. doi: 10.1038/nrn1971. [DOI] [PubMed] [Google Scholar]
- 154.Rosen DR, Siddique T, Patterson D, et al. Mutations in Cu/Zn superoxide dismutase gene are associated with familial amyotrophic lateral sclerosis. Nature. (1993);362 doi: 10.1038/362059a0. [DOI] [PubMed] [Google Scholar]
- 155.Cudkowicz M, McKenna‐Yasek D, Sapp P, et al. Epidemiology of mutations in superoxide dismutase in amyotrophic lateal sclerosis. Ann Neurol. (1997);41:210–221. doi: 10.1002/ana.410410212. [DOI] [PubMed] [Google Scholar]
- 156.Bohni R, Riesgo-Escovar J, Oldham S, et al. Autonomous control of cell and organ size by CHICO, a Drosophila homolog of vertebrate IRS1-4. Cell. (1999);97:865–875. doi: 10.1016/S0092-8674(00)80799-0. [DOI] [PubMed] [Google Scholar]
- 157.Verdu J, Buratovich MA, Wilder EL, Birnbaum MJ. Cell-autonomous regulation of cell and organ growth in Drosophila by Akt/PKB. Nat Cell Biol. (1999);1:500–506. doi: 10.1038/70293. [DOI] [PubMed] [Google Scholar]
- 158.Weinkove D, Neufeld TP, Twardzik T, Waterfield MD, Leevers SJ. Regulation of imaginal disc cell size, cell number and organ size by Drosophila class I(A) phosphoinositide 3-kinase and its adaptor. Curr Biol. (1999);9:1019–1029. doi: 10.1016/S0960-9822(99)80450-3. [DOI] [PubMed] [Google Scholar]
- 159.Leevers SJ, Weinkove D, MacDougall LK, Hafen E, Waterfield MD. The Drosophila phosphoinositide 3-kinase Dp110 promotes cell growth. EMBO J. (1996);15:6584–6594. [PMC free article] [PubMed] [Google Scholar]
- 160.Junger MA, Rintelen F, Stocker H, et al. The Drosophila forkhead transcription factor FOXO mediates the reduction in cell number associated with reduced insulin signaling. J Biol. (2003);2:20. doi: 10.1186/1475-4924-2-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Kramer JM, Davidge JT, Lockyer JM, Staveley BE. Expression of Drosophila FOXO regulates growth and can phenocopy starvation. BMC Dev Biol. (2003);3:5. doi: 10.1186/1471-213X-3-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Puig O, Marr MT, Ruhf ML, Tjian R. Control of cell number by Drosophila FOXO: downstream and feedback regulation of the insulin receptor pathway. Genes Dev. (2003);17:2006–2020. doi: 10.1101/gad.1098703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Staveley BE, Ruel L, Jin J, et al. Genetic analysis of protein kinase B (AKT) in Drosophila. Curr Biol. (1998);8:599–602. doi: 10.1016/S0960-9822(98)70231-3. [DOI] [PubMed] [Google Scholar]
- 164.Poltilove RM, Jacobs AR, Haft CR, Xu P, Taylor SI. Characterization of Drosophila insulin receptor substrate. J Biol Chem. (2000);275:23346–23354. doi: 10.1074/jbc.M003579200. [DOI] [PubMed] [Google Scholar]
- 165.Petruzzelli L, Herrera R, Arenas-Garcia R, Fernandez R, Birnbaum MJ, Rosen OM. Isolation of a Drosophila genomic sequence homologous to the kinase domain of the human insulin receptor and detection of the phosphorylated Drosophila receptor with an anti-peptide antibody. Proc Natl Acad Sci U S A. (1986);83:4710–4714. doi: 10.1073/pnas.83.13.4710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Fernandez-Almonacid R, Rosen OM. Structure and ligand specificity of the Drosophila melanogaster insulin receptor. Mol Cell Biol. (1987);7:2718–2727. doi: 10.1128/MCB.7.8.2718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Fernandez R, Tabarini D, Azpiazu N, Frasch M, Schlessinger J. The Drosophila insulin receptor homolog: a gene essential for embryonic development encodes two receptor isoforms with different signaling potential. EMBO J. (1995);14:3373–3384. doi: 10.1002/j.1460-2075.1995.tb07343.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Marin-Hincapie M, Garofalo RS. The carboxyl terminal extension of the Drosophila insulin receptor homologue binds IRS-1 and influences cell survival. J Biol Chem. (1999);274:24987–24994. doi: 10.1074/jbc.274.35.24987. [DOI] [PubMed] [Google Scholar]
- 169.Franke TF, Tartof KD, Tsichlis PN. The SH2-like Akt homology (AH) domain of c-akt is present in multiple copies in the genome of vertebrate and invertebrate eucaryotes. Cloning and characterization of the Drosophila melanogaster c-akt homolog Dakt1. Oncogene. (1994);9:141–148. [PubMed] [Google Scholar]
- 170.Cho KS, Lee JH, Kim S, et al. Drosophila phosphoinositide-dependent kinase-1 regulates apoptosis and growth via the phosphoinositide 3-kinase-dependent signaling pathway. Proc Natl Acad Sci U S A. (2001);98:6144–6149. doi: 10.1073/pnas.101596998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Linassier C, MacDougall LK, Domin J, Waterfield MD. Molecular cloning and biochemical characterization of a Drosophila phosphatidylinositol-specific phosphoinositide 3-kinase. Biochem J. (1997);321(Pt 3):849–856. doi: 10.1042/bj3210849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Brogiolo W, Stocker H, Ikeya T, Rintelen F, Fernandez R, Hafen E. An evolutionarily conserved function of the Drosophila insulin receptor and insulin-like peptides in growth control. Curr Biol. (2001);11:213–221. doi: 10.1016/S0960-9822(01)00068-9. [DOI] [PubMed] [Google Scholar]
- 173.Gao X, Neufeld TP, Pan D. Drosophila PTEN regulates cell growth and proliferation through PI3K-dependent and -independent pathways. Dev Biol. (2000);221:404–418. doi: 10.1006/dbio.2000.9680. [DOI] [PubMed] [Google Scholar]
- 174.Goberdhan DC, Paricio N, Goodman EC, Mlodzik M, Wilson C. Drosophila tumor suppressor PTEN controls cell size and number by antagonizing the Chico/PI3-kinase signaling pathway. Genes Dev. (1999);13:3244–3258. doi: 10.1101/gad.13.24.3244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Huang H, Potter CJ, Tao W, et al. PTEN affects cell size, cell proliferation and apoptosis during Drosophila eye development. Development. (1999);126:5365–5372. doi: 10.1242/dev.126.23.5365. [DOI] [PubMed] [Google Scholar]
- 176.Bai H, Kang P, Hernandez AM, Tatar M. Activin signaling targeted by insulin/dFOXO regulates aging and muscle proteostasis in Drosophila. PLoS Genet. (2013);9:e1003941. doi: 10.1371/journal.pgen.1003941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Hwangbo DS, Gershman B, Tu MP, Palmer M, Tatar M. Drosophila dFOXO controls lifespan and regulates insulin signalling in brain and fat body. Nature. (2004);429:562–566. doi: 10.1038/nature02549. [DOI] [PubMed] [Google Scholar]
- 178.Gronke S, Clarke DF, Broughton S, Andrews TD, Partridge L. Molecular evolution and functional characterization of Drosophila insulin-like peptides. PLoS Genet. 2010;6:e1000857. doi: 10.1371/journal.pgen.1000857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Broughton SJ, Piper MD, Ikeya T, et al. Longer lifespan, altered metabolism, and stress resistance in Drosophila from ablation of cells making insulin-like ligands. Proc Natl Acad Sci U S A. (2005);102:3105–3110. doi: 10.1073/pnas.0405775102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Tatar M, Kopelman A, Epstein D, Tu MP, Yin CM, Garofalo RS. A mutant Drosophila insulin receptor homolog that extends life-span and impairs neuroendocrine function. Science. (2001);292:107–110. doi: 10.1126/science.1057987. [DOI] [PubMed] [Google Scholar]
- 181.Clancy DJ, Gems D, Harshman LG, et al. Extension of life-span by loss of CHICO, a Drosophila insulin receptor substrate protein. Science. (2001);292:104–106. doi: 10.1126/science.1057991. [DOI] [PubMed] [Google Scholar]
- 182.Tu MP, Epstein D, Tatar M. The demography of slow aging in male and female Drosophila mutant for the insulin-receptor substrate homologue chico. Aging Cell. (2002);1:75–80. doi: 10.1046/j.1474-9728.2002.00010.x. [DOI] [PubMed] [Google Scholar]
- 183.Nielsen MD, Luo X, Biteau B, Syverson K, Jasper H. 14-3-3ϵ antagonizes FoxO to control growth, apoptosis and longevity in Drosophila. Aging Cell. (2008);7:688–699. doi: 10.1111/j.1474-9726.2008.00420.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Demontis F, Perrimon N. FOXO/4E-BP signaling in Drosophila muscles regulates organism-wide proteostasis during aging. Cell. (2008);143:813–825. doi: 10.1016/j.cell.2010.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Giannakou ME, Goss M, Junger MA, Hafen E, Leevers SJ, Partridge L. Long-lived Drosophila with overexpressed dFOXO in adult fat body. Science. (2004);305:361. doi: 10.1126/science.1098219. [DOI] [PubMed] [Google Scholar]
- 186.Wessells RJ, Fitzgerald E, Cypser JR, Tatar M, Bodmer R. Insulin regulation of heart function in aging fruit flies. Nat Genet. (2004);36:1275–1281. doi: 10.1038/ng1476. [DOI] [PubMed] [Google Scholar]
- 187.Ford D, Hoe N, Landis GN, et al. Alteration of Drosophila life span using conditional, tissue-specific expression of transgenes triggered by doxycyline or RU486/Mifepristone. Exp Gerontol. (2007);42:483–497. doi: 10.1016/j.exger.2007.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Broeck JV. Neuropeptides and their precursors in the fruitfly, Drosophila melanogaster. Peptides. (2001);22:241–254. doi: 10.1016/S0196-9781(00)00376-4. [DOI] [PubMed] [Google Scholar]
- 189.Colombani J, Andersen DS, Léopold P. Secreted peptide Dilp8 coordinates Drosophila tissue growth with developmental timing. Science. (2012);336:582–585. doi: 10.1126/science.1216689. [DOI] [PubMed] [Google Scholar]
- 190.Garelli A, Gontijo AM, Miguela V, Caparros E, Dominguez M. Imaginal discs secrete insulin-like peptide 8 to mediate plasticity of growth and maturation. Science. (2012);336:579–582. doi: 10.1126/science.1216735. [DOI] [PubMed] [Google Scholar]
- 191.Cao C, Brown MR. Localization of an insulin-like peptide in brains of two flies. Cell Tissue Res. (2001);304:317–321. doi: 10.1007/s004410100367. [DOI] [PubMed] [Google Scholar]
- 192.Ikeya T, Galic M, Belawat P, Nairz K, Hafen E. Nutrient-dependent expression of insulin-like peptides from neuroendocrine cells in the CNS contributes to growth regulation in Drosophila. Curr Biol. (2002);12:1293–1300. doi: 10.1016/S0960-9822(02)01043-6. [DOI] [PubMed] [Google Scholar]
- 193.Rulifson EJ, Kim SK, Nusse R. Ablation of insulin-producing neurons in flies: growth and diabetic phenotypes. Science. (2002);296:1118–1120. doi: 10.1126/science.1070058. [DOI] [PubMed] [Google Scholar]
- 194.Lee KS, Kwon OY, Lee JH, et al. Drosophila short neuropeptide F signalling regulates growth by ERK-mediated insulin signalling. Nat Cell Biol. (2008);10:468–475. doi: 10.1038/ncb1710. [DOI] [PubMed] [Google Scholar]
- 195.Grönke S, Partridge L. The functions of insulin-like peptides in insects; in IGFs: Local Repair and Survival Factors Throughout Life Span, Clemmons DR, Robinson I and Christen Y (eds) Springer; (2010). pp. 105–124. [Google Scholar]
- 196.Nassel DR, Liu Y, Luo J. Insulin/IGF signaling and its regulation in Drosophila. Gen Comp Endocrinol. (2015);221:255–266. doi: 10.1016/j.ygcen.2014.11.021. [DOI] [PubMed] [Google Scholar]
- 197.Haselton A, Sharmin E, Schrader J, Sah M, Poon P, Fridell Y-WC. Partial ablation of adult Drosophila insulin-producing neurons modulates glucose homeostasis and extends life span without insulin resistance. Cell Cycle. (2010);9:3135–3143. doi: 10.4161/cc.9.15.12458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Jeong DE, Artan M, Seo K, Lee SJ. Regulation of lifespan by chemosensory and thermosensory systems: findings in invertebrates and their implications in mammalian aging. Front Genet. (2012);3:218. doi: 10.3389/fgene.2012.00218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Wang MC, Bohmann D, Jasper H. JNK extends life span and limits growth by antagonizing cellular and organism-wide responses to insulin signaling. Cell. (2005);121:115–125. doi: 10.1016/j.cell.2005.02.030. [DOI] [PubMed] [Google Scholar]
- 200.Bauer JH, Chang C, Morris SNS, et al. Expression of dominant-negative Dmp53 in the adult fly brain inhibits insulin signaling. Proc Natl Acad Sci U S A. (2007);104:13355–13360. doi: 10.1073/pnas.0706121104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Broughton S, Alic N, Slack C, et al. Reduction of DILP2 in Drosophila triages a metabolic phenotype from lifespan revealing redundancy and compensation among DILPs. PLoS One. (2008);3:e3721. doi: 10.1371/journal.pone.0003721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Okamoto N, Yamanaka N, Yagi Y, et al. A fat body-derived IGF-like peptide regulates postfeeding growth in Drosophila. Dev Cell. (2009);17:885–891. doi: 10.1016/j.devcel.2009.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Slaidina M, Delanoue R, Gronke S, Partridge L, L#233;opold P. A Drosophila insulin-like peptide promotes growth during nonfeeding states. Dev Cell. (2009);17:874–884. doi: 10.1016/j.devcel.2009.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Bai H, Kang P, Tatar M. Drosophila insulin‐like peptide‐6 (dilp6) expression from fat body extends lifespan and represses secretion of Drosophila insulin‐like peptide‐2 from the brain. Aging cell. (2012);11:978–985. doi: 10.1111/acel.12000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Holzenberger M, Dupont J, Ducos B, et al. IGF-1 receptor regulates lifespan and resistance to oxidative stress in mice. Nature. (2003);421:182–187. doi: 10.1038/nature01298. [DOI] [PubMed] [Google Scholar]
- 206.Yuan R, Tsaih SW, Petkova SB, et al. Aging in inbred strains of mice: study design and interim report on median lifespans and circulating IGF1 levels. Aging Cell. (2009);8:277–287. doi: 10.1111/j.1474-9726.2009.00478.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Tazearslan C, Cho M, Suh Y. Discovery of functional gene variants associated with human longevity: opportunities and challenges. J Gerontol A Biol Sci Med Sci. (2012);67:376–383. doi: 10.1093/gerona/glr200. [DOI] [PMC free article] [PubMed] [Google Scholar]