Encarnação et al. show that Rab3a, together with its newly identified effector NMHC IIA, mediates the positioning of peripheral lysosomes in nonsecretory cells, thereby promoting lysosome exocytosis and plasma membrane repair.
Abstract
Lysosome exocytosis plays a major role in resealing plasma membrane (PM) disruptions. This process involves two sequential steps. First, lysosomes are recruited to the periphery of the cell and then fuse with the damaged PM. However, the trafficking molecular machinery involved in lysosome exocytosis and PM repair (PMR) is poorly understood. We performed a systematic screen of the human Rab family to identify Rabs required for lysosome exocytosis and PMR. Rab3a, which partially localizes to peripheral lysosomes, was one of the most robust hits. Silencing of Rab3a or its effector, synaptotagmin-like protein 4a (Slp4-a), leads to the collapse of lysosomes to the perinuclear region and inhibition of PMR. Importantly, we have also identified a new Rab3 effector, nonmuscle myosin heavy chain IIA, as part of the complex formed by Rab3a and Slp4-a that is responsible for lysosome positioning at the cell periphery and lysosome exocytosis.
Introduction
Lysosomes are heterogenous organelles that are able to fuse with the plasma membrane (PM; Rodríguez et al., 1997). Although lysosome exocytosis was thought to be limited to secretory cells containing specialized lysosome-related organelles (LROs; Marks and Seabra, 2001; Blott and Griffiths, 2002), it was also known that conventional lysosomes from nonspecialized cells can also undergo secretion (Rodríguez et al., 1997). The best-documented example of this process occurs during PM repair (PMR; Andrews, 2002). PM damage can result from numerous threats, including infection with Mycobaterium tuberculosis (Mtb; Chen et al., 2008; Divangahi et al., 2009), biochemical agents such as pore-forming toxins, or mechanical stress triggering Ca2+-mediated signaling that recruits lysosomes to the vicinity of the damaged PM (Rodríguez et al., 1997; Jaiswal et al., 2002).
Lysosome translocation to the PM requires two sequential steps. First, lysosomes are recruited to the close proximity of the cell surface in a Ca2+-independent manner (Jaiswal et al., 2002). The pool of predocked lysosomes then fuses with the PM in response to Ca2+ elevation sensed by synaptotagmin VII (Syt VII), a Ca2+ sensor localized to lysosomes (Andrews, 2000; Martinez et al., 2000; Jaiswal et al., 2002; Tucker et al., 2004; Luzio et al., 2007; Cheng et al., 2010; Lloyd-Evans and Platt, 2011; Morgan et al., 2011; Pittman, 2011). The molecular trafficking machinery involved in these two steps is only partially known. Recently, Bonifacino and collaborators identified a multisubunit complex named BORC as a key component of the molecular machinery that regulates lysosome translocation to the periphery of the cell (Pu et al., 2015). BORC acts at an early stage, recruiting the small GTPase Arl8b to the lysosomal membrane and enabling the coupling to the SKIP–kinesin-1 complex that drives microtubule-guided movement of lysosomes toward the cell periphery (Bagshaw et al., 2006; Hofmann and Munro, 2006; Dumont et al., 2010; Rosa-Ferreira and Munro, 2011). Soluble N-ethylmaleimide sensitive factor attachment protein receptors, which mediate vesicle fusion, have also been identified as required for lysosome exocytosis. Indeed, the increase in Ca2+ levels causes a conformational change in Syt VII, enabling its interaction with VAMP-7 in lysosomes and the SNAP-23 and syntaxin-4 in the PM (Rao et al., 2004). However, so far, neither Rab small GTPases nor their effectors have been functionally implicated in PMR by lysosomes. Therefore, we decided to perform a screen to identify Rab proteins involved in the trafficking machinery that regulates lysosome exocytosis and PMR.
We identified Rab3a, a secretory Rab involved in regulated secretion, as one of the most robust hits. Silencing of Rab3a leads to a shift in lysosome positioning, culminating in the collapse of lysosomes to the perinuclear region and subsequent inhibition of PMR. This effect is mediated by its effector synaptotagmin-like protein 4a (Slp4-a). We also identified an actin molecular motor, the nonmuscle myosin heavy chain IIA (NMHC IIA), as part of the complex (that includes Rab3a and Slp4-a) responsible for the positioning of lysosomes at the periphery of the cell and lysosome exocytosis.
Results and discussion
Rab3a silencing changes the intracellular localization of lysosomes and blocks PRM
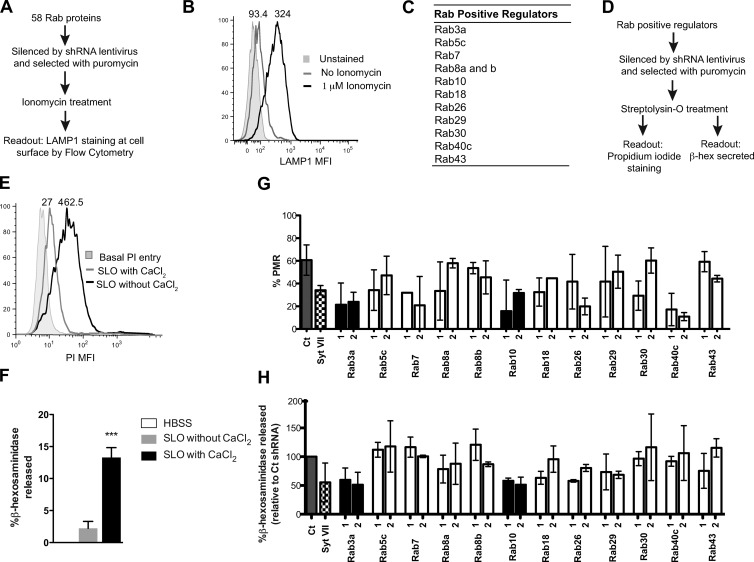
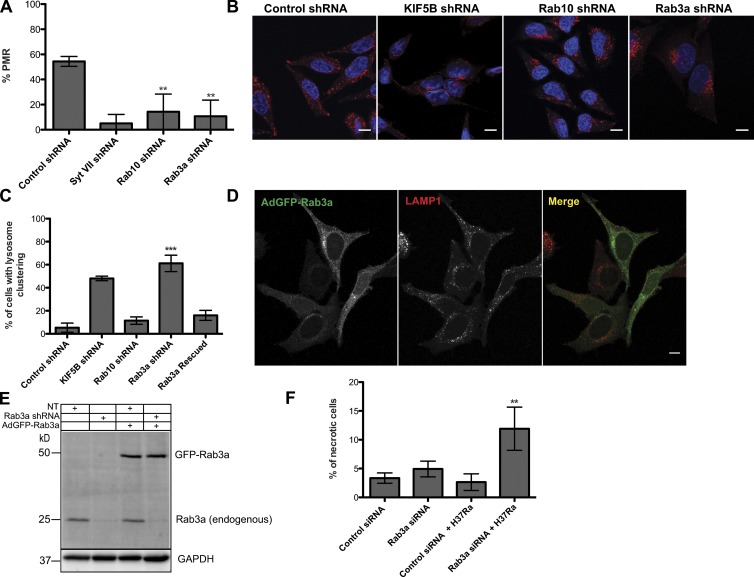
To identify RabGTPases involved in lysosome exocytosis and PMR we silenced 58 Rabs using a lentiviral shRNA library from the BROAD RNAi consortium. The primary screen was performed with a Ca2+ ionophore, ionomycin, that allows the formation of a transient channel on the PM, enabling Ca2+ influx and lysosome translocation to the PM. To assess lysosome exocytosis a flow cytometry-based assay to detect the lysosomal membrane marker lysosome-associated membrane protein 1 (LAMP1) was optimized using an antibody that recognizes a luminal domain of LAMP1 and a suspension cell line, THP-1 that exhibited robustness and good sensitivity to the readout used (a schematic of the screen performed is shown in Fig. 1 A). Ionomycin treatment of THP-1 cells in the presence of 4 mM Ca2+ led to lysosome exocytosis (Fig. 1 B). LAMP1 median fluorescence intensity (MFI) showed a significant shift in treated compared with untreated cells (324 vs. 93.4, respectively). Dead cells (propidium iodide [PI] positive) were excluded from the analysis. To minimize off-target effects we used a panel of unique shRNAs targeting the 58 Rabs along the length of the transcript, with four to five different shRNAs for each Rab (Table S1). Rabs that negatively impacted lysosome translocation to the PM were identified by plotting the % of LAMP1-positive cells at the cell surface in response to ionomycin in Rab-silenced cells (Fig. S1 A). Cells infected with viruses encoding a nontargeting shRNA or an shRNA against Syt VII were used as negative and positive controls, respectively. Rab proteins for which two or more shRNAs led to a decrease of 50% in LAMP1-positive cells when compared with the negative controls were considered positive regulators of Ca2+-dependent lysosome exocytosis. These positive hits were selected for a secondary screen (Fig. 1 C). Because our ultimate goal was identification of Rab proteins involved in Ca2+-dependent lysosome translocation upon PM injury, we performed a secondary screen with streptolysin-O (SLO) that form pores on the PM (Koffer and Gomperts, 1989; Idone et al., 2008). Rab proteins identified in the primary screen were silenced in the same manner as described above. As readouts, we measured cell permeability to PI and release of the lysosomal enzyme β-hexosaminidase (β-hex; Fig. 1 D). Cells that were not incubated with Ca2+ were PI positive (Fig. 1 E), and β-hex release was almost undetectable (Fig.1 F), indicating that membrane pores were not sealed. Conversely, cells treated with SLO in the presence of Ca2+ were able to repair the PM, as shown by the decrease of PI signal (from 462.5 to 27.5; Fig. 1 E) and the presence of β-hex in the supernatant (from 0.6% to 12.2%; Fig. 1 F). These results validated the use of both assays to identify Rabs required for lysosomal exocytosis and PMR. In this screen, the negative and positive controls were the same as described above. When both shRNAs, used for a given Rab, exhibited a significant reduction simultaneously in PMR and lysosome exocytosis, this Rab was considered a robust hit. Using these criteria Rab3a and Rab10 were identified as novel regulators of PMR by lysosomes and used for further studies (Fig. 1, G and H). Given that the library screening was performed with the THP1 cells, we extended this finding to HeLa cells as an adherent nonsecretory cell type (Fig. 2, A–D). We confirmed by RT-PCR that both shRNAs efficiently silenced Rab3a and Rab10 in HeLa cells (Fig. S1 B). Because both shRNAs for each Rab gave similar results, further experiments used a unique shRNA per Rab protein. Fig. 2 A shows that PMR was inhibited in the absence of either Rab3a or Rab10. The mechanism by which Rab3a and Rab10 regulate PMR was examined by analyzing the intracellular distribution of lysosomes in Rab3a- and Rab10-silenced cells. In control cells, lysosomes were distributed in two different regions: most localize to the perinuclear region, and a subset is present near the PM of the cell (Fig. 2 B, first image). A similar distribution was observed in Rab10-silenced cells (Fig. 2 B, third image). In contrast, in Rab3a-silenced cells, lysosomes were clustered only in the perinuclear region (Fig. 2 B, fourth image). Similar results were obtained in cells silenced for kinesin-1 heavy chain (KIF5B) used as a positive control, as its silencing leads to lysosome collapse in the perinuclear region (Fig. 2 B, second image; Tanaka et al., 1998; Pu et al., 2015). We quantified the percentage of cells with lysosome clustering and observed a statistically significant difference between Rab3a-silenced and control cells (Fig. 2 C). Because Rab10 silencing was not involved in lysosome distribution, we decided to focus our present work only on Rab3a. To exclude off-target effects, HeLa cells were transduced with Rab3a shRNA and subsequently infected with adenovirus carrying a mouse Rab3a resistant to the shRNA used to silence Rab3a. Overexpression of mouse GFP-Rab3a led to a significant increase in the percentage of cells showing rescue of lysosome positioning (quantified in Fig. 2 C and visualized in Fig. 2 D). A Western blot (Fig. 2 E) showed that Rab3a shRNA inhibited expression of endogenous Rab3a but had no effect on the expression levels of mouse GFP-Rab3a. Finally, independently of the levels of expression of mouse Rab3a, rescue of lysosome localization is observed in all cells (Fig. 2 D).
Figure 1.
Rab3a and Rab10 are required for Ca2+-dependent lysosomal exocytosis. (A) Scheme showing the different steps of the screening procedure (see Materials and methods). (B) A representative histogram of three independent experiments, with numbers indicating the LAMP1 MFI of the selected cell population. (C) Table with Rab-positive hits obtained from three independent experiments. (D) Scheme of the different steps and readouts used in the secondary screen. (E) Flow cytometry analysis of nontransduced THP-1 cells treated with SLO (see Materials and methods). Numbers in histograms indicate MFI of PI-positive cells. (F) Graph showing percent β-hex release upon SLO treatment with and without Ca2+. Error bars represent SD from five independent experiments (n = 5). (G) Graph showing the percentage PMR in Rab-silenced cells treated with SLO. Error bars represent SD from two to five independent experiments. (H) Graph showing percentage of β-hex release in the supernatant from Rab-silenced cells treated with SLO. Results were normalized to the negative control (Ct). Error bars represent SD from three independent experiments (n = 3). In G and H, only the best two shRNAs were used. Rab3a and Rab10 shRNA are marked as black bars in the graphs.
Figure 2.
Rab3a silencing induces lysosome clustering in the perinuclear region. (A) Percentage of PMR in HeLa cells silenced for Syt VII, Rab10 or Rab3a and control shRNA and challenged with SLO. (B) Representative confocal images of HeLa cells silenced for KIF5B, Rab10, or Rab3a stained for lysosomes, with LAMP1 antibodies (in red) and nuclei, with DAPI (in blue). Control shRNA and KIF5B were used as negative and positive control, respectively. Bars, 10 µm. (C) Quantification of the number of cells with lysosome clustering. This plot also includes the rescue of lysosome clustering in Rab3a-silenced cells infected with adenoviruses expressing the murine Rab3a. In A and C, error bars represent SD from three to four independent experiments. **, P < 0.01; ***, P < 0.001, comparing differences between control and Rab3- or Rab10-silenced cells. (D) Representative confocal images of Rab3a-silenced HeLa cells, infected by adenovirus expressing the murine Rab3a tagged with GFP and then immunostained for LAMP1. Bar, 10 μm. (E) Western blot showing endogenous and ectopical murine Rab3a levels in different experimental conditions. NT, nontransduced HeLa cells. GAPDH was used as loading control. (F) Percentage of necrotic cells in control and Rab3a-silenced primary human macrophages infected with H37Ra Mtb. The plot represents the mean ± SD of two independent experiments. **, P < 0.01, comparing differences between control and Rab3-silenced cells.
It is known that infection of macrophages with Mtb induces PM microdisruptions. Infection with avirulent Mtb (H37Ra) induces lysosome translocation to the PM allowing PMR, whereas infection with virulent H37Rv Mtb blocks these processes. As a result of this blockade, infected macrophages undergo necrosis rather than apoptosis (Chen et al., 2008; Divangahi et al., 2009). We assessed whether Rab3a silencing inhibited PMR in macrophages infected with H37Ra Mtb, leading to an increase in necrosis. To this end, the percentage of necrotic cells was assessed in Rab3a-silenced macrophages infected with H37Ra Mtb. We observed a 4.9-fold increase in the percentage of necrotic cells (Fig. 2 F) upon efficient silencing of Rab3a (Fig. S1 C). These results strongly suggest that Rab3a is required for the correct positioning of lysosomes at the cell periphery in different experimental settings, which is critical for PMR.
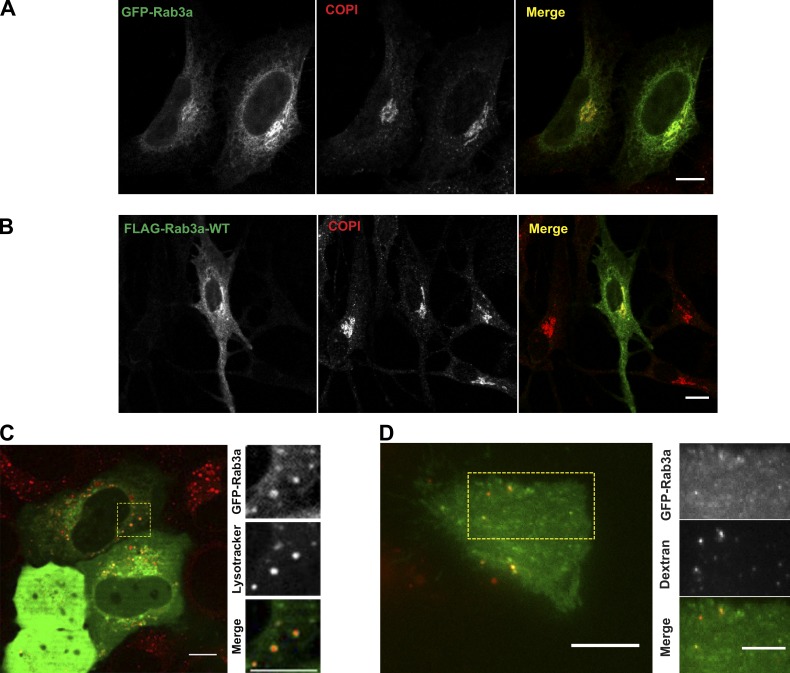
Rab GTPases localize to specific subcellular compartments, contributing to organelle identity and directing organelle trafficking. It is known that Rab3a localizes to synaptosomes and secretory granules, playing a critical role in regulated secretion (Coppola et al., 1999; Leenders et al., 2001; Tsuboi and Fukuda, 2006a; Coleman et al., 2007; Fukuda, 2008; Stevens et al., 2011). Because HeLa are nonsecretory cells and do not possess specialized secretory organelles, such as LROs, we started by comparing the localization of Rab3a in HeLa and Melan-ink4a melanocytes, which possess LROs, namely melanosomes. Because all tested antibodies failed to detect the endogenous Rab3a, we took advantage of tagged versions of Rab3a ectopically expressed. In addition, the overexpression of wild-type or constitutively active mutant (Rab3a-Q81L) was not modifying lysosomal positioning (Fig. S1 D). The intracellular localization of GFP-Rab3a was compared with that of markers for other organelles. In both cell lines (HeLa and melanocytes), Rab3a was mainly found in the perinuclear region, colocalizing with COPI, a Golgi marker (Fig. 3, A and B). To further exclude artifacts due the GFP tagged onto the N terminus of Rab3a, we ectopically expressed Rab3a conjugated with FLAG in HeLa cells that were then subjected to immunofluorescence with COPI antibodies. The tag did not change the intracellular localization of this Rab (Fig. S1 E).
Figure 3.
GFP-Rab3a associates with lysosomes near the PM. (A and B) Representative confocal images of HeLa cells (A) and Melan-ink4a melanocytes (B) overexpressing GFP-Rab3a or FLAG-Rab3a, respectively, and imunostained for the Golgi marker COPI. (C) Live-cell imaging of HeLa cells expressing GFP-Rab3a and stained with Lysotracker (red). The region outlined with a square was zoomed in and the channels were split. The zoomed-in region shows the colocalization of GFP-Rab3a and Lysotracker-loaded vesicles (visualized in yellow in the merge). (D) Representative image of TIRF microscopy of HeLa cells loaded with dextran 647 to stain lysosomes and transfected with GFP-Rab3a. The region outlined with a rectangle was zoomed in and the channels were split. The zoomed in region shows Rab3-positive lysosomes underneath the PM. Bars, 10 µm. WT, wild type.
The localization of Rab3a was not anticipated, because this Rab was required for lysosome exocytosis however, no colocalization with LAMP1 antibodies, in fixed cells, was observed. Therefore, we used confocal live cell imaging- and total internal reflection fluorescence (TIRF) microscopy to assess if Rab3a was present on lysosomes that were labeled with Lysotracker or dextran conjugated with a fluorophore (Fig. 3 C). A subset of acidic vesicles in the cytoplasm was positive for Rab3a (r = 0.79 ± 0.09, n = 52). Additionally, TIRF microscopy showed the existence of Rab3a-positive lysosomes underneath the PM (Fig. 3 D).
Rab3a induces lysosome clustering through the recruitment of the effector Slp4-a
When bound to GTP, Rab3a recruits protein effectors, such as Rab3-interacting protein (Rim), rabphilin 3A, Slp4-a, rabphilin 3A-like without C2 domains (Noc2), and myosin Va (MyoVa).
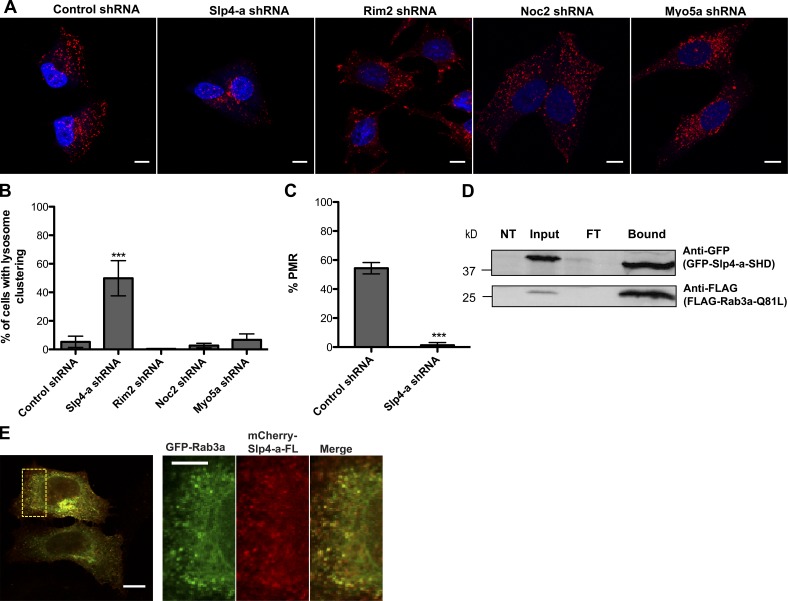
Because the role of Rab3a in lysosome exocytosis and PMR is likely to be mediated by an effector, we investigated if any of the known Rab3a effectors were required for lysosome exocytosis. HeLa cells were stably transduced with lentiviruses expressing shRNAs against Slp4-a, Rim2, Noc2, or MyoVa or control shRNA. The silencing was confirmed by RT-PCR (Fig. S1 F), and lysosome distribution was analyzed by immunostaining with anti-LAMP1 antibody. Among the effectors expressed in HeLa cells, Slp4-a was the only one whose silencing results in lysosome clustering at the perinuclear region (49.9 ± 12.4% against 5.2 ± 1.9% in control cells; Fig. 4, A and B).
Figure 4.
Silencing of the Rab3a effector Slp4-a induces lysosome clustering. (A) Representative images of HeLa cells silenced for Rab3a effectors Slp4-a, Rim2, Noc2, or MyoVa or transduced with control shRNA and then immunostained for the lysosomal marker LAMP1 (red) and DAPI (blue). (B) Quantification of the number of cells showing lysosome clustering (in percentage). More than 300 cells were analyzed per condition. (C) Effect of Slp4-a silencing in PMR in HeLa cells treated with SLO. In B and C, plots represent the mean ± SD of three independent experiments. ***, P < 0.001, comparing differences between control and Rab3-silenced cells. (D) Lysates from HeLa cells cotransfected with constitutively-active FLAG-Rab3a Q81L and GFP-Slp4-a-SHD and immunoprecipitated with GFP-Trap-A beads. Immunoprecipitation products were analyzed by immunoblotting using the indicated antibodies. NT, non-transfected; FT, flow-through. (E) Representative image of confocal microscopy of HeLa cells cotransfected with GFP-Rab3a and mCherry-Slp4-a-FL. The region outlined with a square was zoomed in and channels were split. The zoomed-in region shows colocalization of Rab3a and Slp4-a–positive vesicles (visualized in yellow in the merge). Bars: 10 µm; (zoomed region) 5 µm.
In contrast, Rim2 silencing induced lysosome dispersion with a visible accumulation in the cell tips (Fig. 4 A). On the other hand, neither Noc2 nor MyoVa silencing induced any change in lysosome distribution (Fig. 4, A and B). Importantly, PMR was dramatically impaired upon Slp4-a silencing (Fig. 4 C). To confirm that Rab3a was interacting with Slp4-a, HeLa cells were cotransfected with the Slp4-a Rab binding domain (GFP-Slp4-a-SHD) and FLAG-Rab3a-Q81L. We observed that Rab3a-Q81L coimmunoprecipitated with GFP-Slp4-a-SHD, suggesting that they interact (Fig. 4 D). Finally, we cotransfected HeLa cells with GFP-Rab3a and mCherry-Slp4-a (Fig. 4 E) and observed that they exhibited a striking colocalization (r = 0.86 ± 0.05, n = 100). Thus, silencing of Slp4-a was sufficient to phenocopy the effects of Rab3a silencing regarding lysosome clustering near the perinuclear region and the inhibition of PMR.
Lysosomes are positioned to the periphery of the cell by Rab3a, Slp4-a, and NMHC IIA
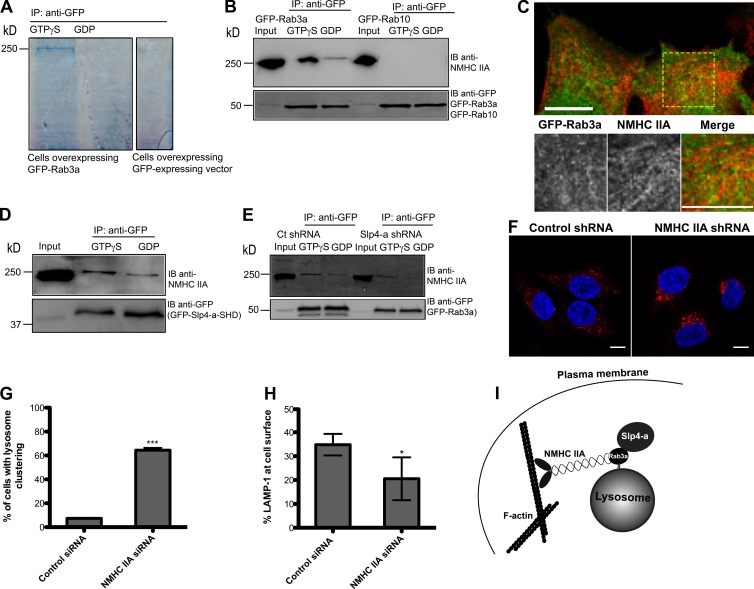
To gain further insights into the mechanism by which Rab3a regulated lysosome exocytosis, we performed Rab3a immunoprecipitations in order to identify novel effectors. For this, we used lysates of HeLa cells, expressing GFP-Rab3a, to immunoprecipitate Rab3a with GFP-Trap-A beads in the presence of a nonhydrolysable analog of GTP (GTPγS) or GDP as a control. We detected a band between 150 and 250 kD in one-dimensional SDS-PAGE that was present in the lane corresponding to GTPγS-loaded extract but absent in the lysates of GFP-overexpressing HeLa cells and GDP-loaded lane (Fig. 5 A). This band was then subjected to mass spectrometry and identified as NMHC IIA, an actin-based motor. Coimmunoprecipitation GFP-Rab3a with GFP-Trap-A beads in HeLa cells lysates using an anti-GFP antibody followed by the detection of NMHC IIA with a specific antibody confirmed the interaction between Rab3a and NMHC IIA. As expected, NMHC IIA was detected when the lysates were incubated with GTPγS, but not in the presence of GDP. This interaction was not observed for Rab10, the other positive hit obtained in our screen (Fig. 5 B).
Figure 5.
NMHCIIA is required for lysosome positioning. (A) Lysates from HeLa cells transfected with GFP-Rab3a were immunoprecipitated with GFP-Trap-A beads in the presence of nonhydrolysable GTP (GTPγS) or GDP. Bands visualized with Coomassie brilliant blue staining were excised and identified by mass spectrometry. (B) Validation of the NMHC IIA and GFP-Rab3a interaction by immunoprecipitation of HeLa cell lysates with GFP-Trap-A beads in the presence of GTPγS or GDP. Immunoprecipitation products were analyzed by immunoblotting using the indicated antibodies. (C) HeLa cells transfected with GFP-Rab3 and immunostained for the endogenous NMHC IIA. The region outlined with a square was zoomed in, and channels were split. (D) Lysates from HeLa cells transfected with GFP-Slp4-a-SHD were immunoprecipitated with GFP-Trap-A beads in the presence of nonhydrolysable GTP (GTPγS) or GDP. Immunoprecipitation products were analyzed by immunoblotting using the indicated antibodies. (E) Lysates from control shRNA and Slp4-a silenced HeLa cells transfected with GFP-Rab3a were immunoprecipitated with GFP-Trap-A beads in the presence of nonhydrolysable GTP (GTPγS) or GDP. Immunoprecipitation products were analyzed by immunoblotting using the indicated antibodies. (F) HeLa cells transfected with control siRNA or siRNA targeting NMHC IIA were fixed and stained with LAMP1 antibodies and DAPI to visualize lysosomes and the nucleus, respectively. Representative images for both are shown. Bars, 10 µm. (G) Quantification of the number of cells showing lysosome clustering (in percentage) in NMHC IIA–silenced and control cells. (H) Effect of NMHC IIA silencing in lysosomes exocytosis in cells challenged with ionomycin. Plots represent the mean ± SD of three independent experiments. *, P < 0.1; ***, P < 0.001, comparing differences between control and NMHC IIA–silenced cells. (I) Our working model postulates that lysosomes positive for Rab3a can recruit NMHCIIA when it is bound to GTP. This myosin motor is responsible for the positioning of lysosomes to the periphery of the cell. Slp4-a, another Rab3 effector, although not necessary for the recruitment of NMHC IIA, seems to be involved in peripheral lysosome positioning.
NMHC IIA is a primarily F-actin–interacting protein (Andzelm et al., 2007). However, a small amount of NMHC IIA was found to be colocalized with GFP-Rab3a as well, indicating that some NMHC IIA was localized in a position where it can participate in the positioning of Rab3-positive vesicles (Fig. 5 C). Biochemical analyses also showed coisolation of NMHC IIA with Slp4-a-SHD in the presence of GTPγS (Fig. 5 D). However, Slp4-a was not required for the interaction between NMHC IIA and Rab3a, suggesting a direct interaction between the Rab3a and NMHC IIA (Fig. 5 E).
As expected, we found that NMHC IIA depletion induced clustering of lysosomes in the perinuclear region (Fig. 5 F). We quantified this phenotype and found that in the case of NMHC IIA silencing, 64.4 ± 1.9% of the cells presented lysosome clustering in contrast with 7.3 ± 0.4% in the case of siRNA control (Fig. 5 G). Finally, successful depletion of NMHC IIA (89.7 ± 4.5%; Fig. S1 G) inhibited lysosome exocytosis in cells challenged with ionomycin, suggesting that NMHC IIA is also necessary for the positioning of lysosomes involved in PMR at the cell periphery, probably by tethering Rab3a-positive lysosomes to the cortical actin (Fig. 5 H).
NMHC IIA has two distinct but related functions. One is its ability to translocate actin filaments and resides in its globular motor domain and lever arm. The second function of NMHC IIA is structural and resides in the ability of the rod portion of the molecule to form filaments, which allows several myosin heads to maintain tension for long periods of time, similar to smooth muscle myosin (Conti and Adelstein, 2008; Bond et al., 2011). Both functions require binding to actin. In lysosome exocytosis in nonsecretory cells, we propose that NMHC IIA has a structural role, linking Rab3a peripheral lysosomes to the cortical actin cytoskeleton and keeping this pool of vesicles close to the PM (Fig. 5 I). Once the cells are damaged and intracellular Ca2+ increases, the actin cytoskeleton network undergoes a major reorganization, allowing the docking/tethering/priming and finally fusion of the Rab3a/Slp4-a–positive lysosomes with the PM. The role of Slp4-a in the positioning of lysosomes at the cell periphery is still an open question, although we cannot exclude an interaction between NMHC IIA and Slp4-a (via its linker domain), because the silencing of the latter was sufficient to induce lysosome clustering. Our proposed working model has some differences with those described for Rab27a in regulated secretion of LROs. In both cases, Mlph and MyRIP are linkers between Rab27 and motor proteins. However, in contrast with the Mlph and MyRIP, Slp4-a does not have a myosin-binding domain (Tsuboi and Fukuda, 2006b).
In conclusion, we have identified traffic molecular machinery involved in the positioning of peripheral lysosomes and complement existing knowledge concerning the machinery responsible for lysosome transport from the perinuclear region to the periphery that was recently identified (Pu et al., 2015).
Materials and methods
Cell culture and Mtb infection
HEK293T and HeLa cells were cultured in DMEM (containing 4.5 g/l D-glucose and 110 mg/l sodium pyruvate) with 10% FBS. THP-1 cells were maintained in RPMI supplemented with 10% FBS (Invitrogen), 2 mM L-glutamine, 10 mM Hepes, essential and nonessential amino acids, and 55 µM β-mercaptoethanol. Melan-ink4a melanocytes were grown in RPMI supplemented with 10% FBS, 200 pM cholera toxin, and 200 nM PMA. The experiments were performed between cell passages 3 and 10. All cells were kept at 37°C in a 5% CO2 atmosphere, and all cell lines were obtained from ATCC.
Human peripheral blood mononuclear cells were isolated from healthy donors as described (Duan et al., 2002). Macrophages were differentiated from PBMCs for 7 to 10 d in IMDM (Gibco) supplemeneted with 10% human serum AB (Gemini Bioproduct). For infection with Mycobacterium tuberculosis, the nonvirulent strain H37Ra was obtained from ATCC and cultured as previously described (Chen et al., 2006). Infections were performed at a multiplicity of infection of 10:1.
shRNA sequences, lentiviral production, adenoviral production, and cell transduction
Table S1 is a complete list of shRNA sequences used (Garg et al., 2011). All lentiviruses were produced in accordance with BL-2+ protocols, available at the Broad Institute RNAi Consortium website (http://www.broadinstitute.org/rnai/public/resources/protocols).
For infection, 104 THP-1 cells were mixed with 50 µl of lentivirus containing supernatant in the presence of 6 µg/ml polybrene (hexadimethrine bromide; Sigma-Aldrich), seeded into a well of a 96-well plate and centrifuged at 2,250 rpm for 30 min at 37°C. 24 h after infection, the medium was changed to normal culture medium with puromycin (final concentration, 2.5 µg/ml) for selection. All experiments were performed 7–8 d after infection.
To infect HeLa cells, 200 µl lentivirus supernatant was added to 2 × 105 cells (seeded the day before into a six-well plate) in the presence of polybrene. Cell selection was performed 24 h after infection as described in the previous paragraph. Cells were used for experiments 4 to 5 d after selection. The Adenovirus were produced accordingly to the Invitrogen protocol.
Ionomycin treatment and flow cytometry analysis
Transduced THP-1 or HeLa cells were centrifuged for 1 min at 2,000 rpm at 4°C. Then the cells were resuspended in 4 mM CaCl2 in the presence or not (control) of ionomycin (final concentration, 1 and 4 µM for THP-1 and HeLa cells, respectively; Sigma-Aldrich) or absence (control) and immediately incubated at 37°C for 10 min (for HeLa cells) or 2 min (for THP-1 cells). After incubation, the cells were shifted to 4°C, washed with flow cytometer buffer (1% FBS and 2 mM EDTA in PBS), and stained with mouse anti-LAMP1 antibody conjugated with FITC (BD) at 0.4 µg/ml for 30 min. Before analysis on a Canto II flow cytometer, cells were also stained with 50 µg/ml PI (Sigma-Aldrich). The software for acquisition was BD Cell Quest, and data from at least 5,000 cells were analyzed with FlowJo. All PI-positive cells were excluded for the quantification.
PM repair and β-hex assays
SLO was purified from BL21 Escherichia coli expressing 6× histidine-tagged SLO (cloned into pTrcHisA; Invitrogen; plasmid provided by N. Andrews, University of Maryland, College Park, MD). Transduced THP-1 or HeLa cells were washed with Ca2+- and Mg2+-free ice-cold HBSS. SLO was then added for 5 min at 4°C. The toxin-containing medium was removed and replaced by Ca2+- and Mg2+-free HBSS or 4 mM CaCl2-containing HBSS for 10 min at 37°C to allow PMR. Then, cells were washed twice with flow cytometry buffer, stained with 50 µg/ml PI for 30 s, and analyzed on a Canto II flow cytometer. The percentage of PMR was calculated according to the following formula: 1 − (% PI-positive cells with CaCl2/% PI-positive cells without Ca2+) × 100.
In parallel, β-hex release assay was performed as described elsewhere (Rodríguez et al., 1997; Kima et al., 2000). In brief, 106 THP-1 cells were seeded in a six-well plate and incubated overnight at 37°C. The next day, cells were collected and washed with ice-cold HBSS, 10 mM EGTA, and 200 µg/ml MgCl2. To allow pore-forming toxin SLO incorporation and PM pore formation, cells were resuspended in HBSS with 4 mM CaCl2 and 250 ng/ml SLO and incubated 5 min at 4°C followed by 10 min at 37°C. Cells were shifted to ice, and cell supernatant containing the released β-hex was collected. In parallel, cells were lysed with 1% IGEPAL/dH2O and diluted 1:5 in dH2O. β-Hex activity was determined by incubating cell supernatant or lysate in a 96-well plate with 6 mM of the substrate 4-methyl-umbelliferyl-N-acetyl-β-d-glucosaminide (4-MU-β-D-GlcNAc; Glycosynth) resuspended in HBSS with 40 mM sodium citrate and 88 mM Na2PO4, pH 4.5, for 15 min at 37°C. Fluorescence was measured in a plate-spectrofluorimeter at excitation 365 nm/emission 450 nm. Protein content from cell supernatant and lysate was determined using the BCA protein assay kit (Thermo Fisher Scientific), as described by the manufacturer. HBSS-SLO-CaCl2 and 1% IGEPAL 1%/dH2O were used as controls.
β-Hex activity is expressed as fluorescence (365 nm/450 nm) per milligram of cell protein. The total β-hex activity is the sum of the β-hex activity in the supernatant and 5 × (β-hex activity in the cell lysate). β-Hex released is expressed as the percentage of β-hex activity in the supernatant relative to the total β-hex activity.
Transfections and constructs
Cell transfections were performed using Turbofect (Thermo Fisher Scientific) or Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions, unless otherwise noted. Usually, the experiments were performed 14–16 h after transfection.
To silence NMHC IIA in HeLa cells, a siRNA smart pool and a nontargeting sequence siRNA (GE Healthcare) were used. In this case, cells were transfected with Dharmafect 1 (GE Healthcare) according to the manufacturer’s instructions. These experiments were performed 72 h after transfection with siRNAs.
Human Rab3a coding sequence was produced by RT-PCR amplification (forward primer, 5′-AGTGAATTCATGGCATCCGCCACAGACTCGCGC-3′; reverse primer, 5′-CATGGATCCTCAGCAGGCGCAGTCCTGGTGCGG-3′) using total RNA isolated from the SH-SY5Y (ATCC) cell line as a template, digested with EcoRI–BamHI and cloned into pEGFP-C2 (Takara Bio, Inc.) with the same restriction enzymes. FLAG-Rab3a and FLAG-Rab3a-Q81L were obtained from M. Fukuda (Tohoku University, Sendai, Japan). GFP-Slp4-a-SHD and mCherry-Slp4-a-FL were a gift of T. Carter (Medical Research Council, London, England, UK).
To rescue the lysosome clustering observed in Rab3a-silenced cells, adenovirus expressing the mouse Rab3a adenovirus expressing GFP-mouse Rab3a was produced according to Invitrogen’s protocol. Rab3a coding sequence was produced by RT-PCR amplification (forward primer, 5′-GCATGAATTCATGATGGCTTCCGCCACAGACTCTCGCTAT-3′; reverse primer, 5′-ACTCGTCGACCAGCAAGGTCCATTCGCTTTATTG-3′) using total RNA isolated from RAW264.7 (ATCC) cell line as a template, digested with EcoRI–SalI and cloned into an adapted Gateway mammalian expression vector pENTR-GFP previously described (Seixas et al., 2012).
The mammalian expressing cassette containing GFP-Rab3a was then easily and efficiently transferred into pAd vector (Invitrogen) by LR recombination following the manufacturer’s instructions. In the rescue experiments, HeLa cells were transduced with Rab3a shRNA as described above and subsequently infected with adenovirus carrying the mouse Rab3a resistant to the shRNA used for 24 h at a multiplicity of infection of 1:1. Then the cells were fixed and immunostained for LAMP1 antibodies.
Confocal microscopy and analysis
Cells (4–7 × 104) were seeded on glass coverslips. After each assay, cells were fixed in 4% PFA, quenched with 0.05 M NH4Cl, permeabilized in PBS with 0.05% saponin or ice-cold methanol, depending on the antibody, and blocked with 2.5% fish-gelatin (Sigma-Aldrich) according to standard procedures. Primary antibodies used were: mouse anti-LAMP1 (1:100; Hybridoma Bank), mouse anti-COPI (1:100, clone M3A5), and rabbit anti anti-FLAG (M2 antibody) conjugated with Alexa Fluor 555 (1:50, Cell Signaling Technology). Secondary antibodies conjugated to Cy3, Cy5 (1:800; Jackson ImmunoResearch Laboratories, Inc.), or Alexa Fluor 546 (1:1,000; Invitrogen) were used. DAPI was used to visualize nuclei (1:400; Fluka). Coverslips were mounted with mowiol/DABCO and examined using a LSM 510 or LSM 710 laser scanning confocal microscope (ZEISS) with a Plan-Apochromat 63× 1.4 NA oil-immersion objective; the 405-, 488-, 543-, and 633-nm laser lines; and spectral detection adjusted for the emission of DAPI, Cy3, and Cy5 fluorochromes. Digital images were analyzed by using LSM Image software or ImageJ.
For live-cell confocal imaging and TIRF, HeLa cells were seeded at 2 × 104 cells per well on glass-bottom eight-well (Labteks) and transfected overnight with the indicated plasmids using Fugene 6 transfection reagent (Promega). A long pulse and chase (5 and 2 h, respectively) of dextran conjugated with fluorophores (Thermo Fisher Scientific) was performed to mark lysosomes. Live-cell imaging was performed at 37°C using an Andor Revolution spinning disk confocal microscope (Andor Technology) equipped with a EMCCD camera with a Plan Apo VC PFS 60× objective (1.4 NA oil-immersion; Nikon). The system was controlled by iQ software (Andor Technology), and images were analyzed using ImageJ software. TIRF was performed at 37°C using a spinning-disk confocal microscope equipped with a TIRF module (Roper Technologies) with a Plan-Apochromat 100× 1.49 NA oil-immersion objective and a Photometric 512 EMCCD camera. A penetration depth of 120 nm was used. Images were obtained with Methamorph software and processed using ImageJ.
Lysosome clustering was scored by analyzing confocal images from different fields chosen in an unbiased manner or scored manually in a blinded fashion by four different co-authors. More than 300 cells were analyzed in each condition. Of note, no major differences were observed between the two different approaches.
For the statistical analysis of colocalization, vesicles with positive signal for both green and red channel were individually isolated. Cross-correlation analysis between both signals of the different regions of interest was conducted using ImageJ Jacop software for Pearson’s correlation coefficient. Vesicles accumulated around the nucleus and Golgi region were not individually distinguished and thus not considered for analysis.
Immunoblotting, immunoprecipitation, and mass spectrometry
GFP-Rab3a–transfected cells and Rab3a-silenced cells were lysed in ice-cold lysis buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 0.5 mM EDTA, and 1% IGEPAL) in the presence of protease inhibitors for 30 min on ice, followed by centrifugation at 20,000 g for 30 min at 4°C. Total protein concentration was determined using the DC protein assay kit (Bio-Rad), and samples were loaded on 10–12.5% SDS polyacrylamide gels and transferred onto activated PVDF membranes in transfer buffer (25 mM Tris, 192 mM glycine, and 20% methanol) for 2 h at 300 mA and 4°C. Membranes were blocked with blocking buffer (5% nonfat dry milk and 0.1% Tween-20 in PBS) and the antibodies incubated in the same buffer. Affinity-purified goat polyclonal anti-GFP (a gift from D. Drechsel, Max Planck Institute of Molecular Cell Biology and Genetics, Dresden, Germany) was used at 1 µg/ml, rabbit anti-NMHC IIA (Sigma-Aldrich) at 1 µg/ml, mouse anti-FLAG (Cell Signaling Technology) at 1:1,000 dilution, rabbit anti-Rab3a (sc-308; Santa Cruz Biotechnology, Inc.) at 0.2 µg/ml, goat anti-Rab3 (SICGEN) at 3 µg/ml, and goat anti-GAPDH (SICGEN) at 2 µg/ml. Alkaline phosphatase–conjugated secondary antibodies (Santa Cruz Biotechnology, Inc.) were used at 0.2 µg/ml. Blots were developed with ECF (GE Healthcare). Imager Chemidoc XRS (Bio-Rad Laboratories) was used to detect fluorescence. For immunoprecipitation, cell lysates (600 µg of total protein) were first incubated with GTPγS (Sigma-Aldrich) or GDP (Sigma-Aldrich) for 10 min at RT and 20–30 µl of GFP_TrapA beads (ChromoTek) were then added and mixed for 2 h at 4°C with rotation. Beads were recovered by centrifugation, washed three times with lysis buffer, and finally resuspended in 6× loading buffer. After boiling at 95°C for 10 min, the immunoprecipitates were resolved by 10–12.5% SDS-PAGE followed by immunoblotting with anti-GFP antibodies or visualized with Colloidal Coomassie BB (Neuhoff et al., 1988).
For mass spectrometry, the bands of interest excised from a 1D PAGE gel were treated as follows: destained, reduced, alkylated, and digested with trypsin (6.7 ng/µl; Promega) overnight at 37°C. The tryptic peptides were desalted and concentrated using POROS R2 (Applied Biosystems) and eluted directly onto the MALDI plate using 0.6 µl of 5 mg/ml CHCA (α-cyano-4-hydroxycinnamic acid; Sigma-Aldrich) in 50% (vol/vol) acetonitrile and 5% (vol/vol) formic acid. The data were acquired in positive reflector mass spectrometry (MS) and tandem MS (MS/MS) modes using the 4800plus MALDI-TOF/TOF (AB Sciex) mass spectrometer and 4000 Series Explorer Software v.3.5.3 (Applied Biosystems). External calibration was performed using Pepmix1 (Laser BioLabs). The 50 most intense precursor ions from the MS spectra were selected for MS/MS analysis.
The raw MS and MS/MS data were analyzed using Protein Pilot Software v. 4.5 (AB Sciex) with the Mascot search engine (MOWSE algorithm). The search parameters were as follows: monoisotopic peptide mass values were considered, maximum precursor mass tolerance (MS) of 50 ppm and a maximum fragment mass tolerance (MS/MS) of 0.3 D. The searches were performed against a protein database SwissProt (547,357 sequences; 194,874,700 residues) with taxonomic restriction to Homo sapiens (20,200 sequences). A maximum of two missed cleavages was allowed. Carboxyamidomethylation of cysteines and oxidation of methionines and N-Pyro Glu of the N-terminal Q were set as variable modifications. Protein identification was only accepted when significant protein homology scores were obtained (P < 0.05, protein scores > 56) and at least one peptide was fragmented with a significant individual ion score (P < 0.05).
Quantitative RT-PCR
Total RNA was extracted with RNeasy and QIAshredder kits (QIAGEN) and reverse transcribed with iScript cDNA synthesis kit (Bio-Rad Laboratories). Quantitative PCR was performed with SYBR green master mix (Bio-Rad Laboratories) and QuantiTect primers (QIAGEN). HPRT was used as housekeeping gene to normalize the expression.
Statistical analysis
Numerical data are presented as mean ± SD. One-way analysis of variance significance analysis followed by Tukey test was used for comparison relative to control with GraphPad Prism software (version 5).
Online supplemental material
Fig. S1 shows data supporting Figs. 1, 2, 3, 4, and 5. The data include a plot of percentage of LAMP1 at cell surface in Rab-silenced cells, knockdown efficiency of shRNAs or siRNAs used in Figs. 2, 4, and 5, and additional confocal images to support data presented in Fig. 3. Table S1 is a summary of shRNA sequences used in the primary and secondary sreenings. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201511093/DC1.
Supplementary Material
Acknowledgments
We would like to thank T. Pereira and N. Pimpão for technical assistance with microscopy. We also thank C. Seixas and N. Domingues for helping with some protocols.
This study was supported by the Portuguese Fundação para a Ciência e a Tecnologia (HMSP-ICT/0024/2010) and cofunded by the European Union (Fundo Europeu de Desenvolvimento Regional) through Programa Operacional Factores de Competitividade and Quadro de Referência Estratégico, iNOVA4Health (UID/Multi/04462/2013), National Institutes of Health grant R21 AI 112766-01A1, and Fundação para a Ciência e a Tecnologia fellowships (SFRH/BPD/87085/2012, SFRH/BD/90258/2012, and SFRH/BPD/78491/2011).
The authors declare no competing financial interests.
Footnotes
Abbreviations used in this paper:
- β-hex
- β-hexosaminidase
- LAMP1
- lysosomal-associated membrane protein 1
- LRO
- lysosome-related organelle
- MFI
- median fluorescence intensity
- MS
- mass spectrometry
- Mtb
- Mycobaterium tuberculosis
- MyoVa
- myosin Va
- NMHC IIA
- nonmuscle myosin heavy chain IIA
- Noc2
- rabphilin 3A-like without C2 domains
- PI
- propidium iodide
- PM
- plasma membrane
- PMR
- plasma membrane repair
- Rim
- Rab3-interacting protein
- SLO
- streptolysin-O
- Slp4-a
- synaptotagmin-like protein 4a
- Syt VII
- synaptotagmin VII
- TIRF
- total internal reflection fluorescence
References
- Andrews N.W. 2000. Regulated secretion of conventional lysosomes. Trends Cell Biol. 10:316–321. 10.1016/S0962-8924(00)01794-3 [DOI] [PubMed] [Google Scholar]
- Andrews N.W. 2002. Lysosomes and the plasma membrane: trypanosomes reveal a secret relationship. J. Cell Biol. 158:389–394. 10.1083/jcb.200205110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andzelm M.M., Chen X., Krzewski K., Orange J.S., and Strominger J.L.. 2007. Myosin IIA is required for cytolytic granule exocytosis in human NK cells. J. Exp. Med. 204:2285–2291. 10.1084/jem.20071143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagshaw R.D., Callahan J.W., and Mahuran D.J.. 2006. The Arf-family protein, Arl8b, is involved in the spatial distribution of lysosomes. Biochem. Biophys. Res. Commun. 344:1186–1191. 10.1016/j.bbrc.2006.03.221 [DOI] [PubMed] [Google Scholar]
- Blott E.J., and Griffiths G.M.. 2002. Secretory lysosomes. Nat. Rev. Mol. Cell Biol. 3:122–131. 10.1038/nrm732 [DOI] [PubMed] [Google Scholar]
- Bond L.M., Peden A.A., Kendrick-Jones J., Sellers J.R., and Buss F.. 2011. Myosin VI and its binding partner optineurin are involved in secretory vesicle fusion at the plasma membrane. Mol. Biol. Cell. 22:54–65. 10.1091/mbc.E10-06-0553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M., Gan H., and Remold H.G.. 2006. A mechanism of virulence: virulent Mycobacterium tuberculosis strain H37Rv, but not attenuated H37Ra, causes significant mitochondrial inner membrane disruption in macrophages leading to necrosis. J. Immunol. 176:3707–3716. 10.4049/jimmunol.176.6.3707 [DOI] [PubMed] [Google Scholar]
- Chen M., Divangahi M., Gan H., Shin D.S.J., Hong S., Lee D.M., Serhan C.N., Behar S.M., and Remold H.G.. 2008. Lipid mediators in innate immunity against tuberculosis: opposing roles of PGE2 and LXA4 in the induction of macrophage death. J. Exp. Med. 205:2791–2801. 10.1084/jem.20080767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng X., Shen D., Samie M., and Xu H.. 2010. Mucolipins: intracellular TRPML1-3 channels. FEBS Lett. 584:2013–2021. 10.1016/j.febslet.2009.12.056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman W.L., Bill C.A., and Bykhovskaia M.. 2007. Rab3a deletion reduces vesicle docking and transmitter release at the mouse diaphragm synapse. Neuroscience. 148:1–6. 10.1016/j.neuroscience.2007.06.011 [DOI] [PubMed] [Google Scholar]
- Conti M.A., and Adelstein R.S.. 2008. Nonmuscle myosin II moves in new directions. J. Cell Sci. 121:11–18. 10.1242/jcs.007112 [DOI] [PubMed] [Google Scholar]
- Coppola T., Perret-Menoud V., Lüthi S., Farnsworth C.C., Glomset J.A., and Regazzi R.. 1999. Disruption of Rab3-calmodulin interaction, but not other effector interactions, prevents Rab3 inhibition of exocytosis. EMBO J. 18:5885–5891. 10.1093/emboj/18.21.5885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Divangahi M., Chen M., Gan H., Desjardins D., Hickman T.T., Lee D.M., Fortune S., Behar S.M., and Remold H.G.. 2009. Mycobacterium tuberculosis evades macrophage defenses by inhibiting plasma membrane repair. Nat. Immunol. 10:899–906. 10.1038/ni.1758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan L., Gan H., Golan D.E., and Remold H.G.. 2002. Critical role of mitochondrial damage in determining outcome of macrophage infection with Mycobacterium tuberculosis. J. Immunol. 169:5181–5187. 10.4049/jimmunol.169.9.5181 [DOI] [PubMed] [Google Scholar]
- Dumont J., Oegema K., and Desai A.. 2010. A kinetochore-independent mechanism drives anaphase chromosome separation during acentrosomal meiosis. Nat. Cell Biol. 12:894–901. 10.1038/ncb2093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda M. 2008. Regulation of secretory vesicle traffic by Rab small GTPases. Cell. Mol. Life Sci. 65:2801–2813. 10.1007/s00018-008-8351-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garg S., Sharma M., Ung C., Tuli A., Barral D.C., Hava D.L., Veerapen N., Besra G.S., Hacohen N., and Brenner M.B.. 2011. Lysosomal trafficking, antigen presentation, and microbial killing are controlled by the Arf-like GTPase Arl8b. Immunity. 35:182–193. 10.1016/j.immuni.2011.06.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann I., and Munro S.. 2006. An N-terminally acetylated Arf-like GTPase is localised to lysosomes and affects their motility. J. Cell Sci. 119:1494–1503. 10.1242/jcs.02958 [DOI] [PubMed] [Google Scholar]
- Idone V., Tam C., Goss J.W., Toomre D., Pypaert M., and Andrews N.W.. 2008. Repair of injured plasma membrane by rapid Ca2+-dependent endocytosis. J. Cell Biol. 180:905–914. 10.1083/jcb.200708010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaiswal J.K., Andrews N.W., and Simon S.M.. 2002. Membrane proximal lysosomes are the major vesicles responsible for calcium-dependent exocytosis in nonsecretory cells. J. Cell Biol. 159:625–635. 10.1083/jcb.200208154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kima P.E., Burleigh B., and Andrews N.W.. 2000. Surface-targeted lysosomal membrane glycoprotein-1 (Lamp-1) enhances lysosome exocytosis and cell invasion by Trypanosoma cruzi. Cell. Microbiol. 2:477–486. 10.1046/j.1462-5822.2000.00071.x [DOI] [PubMed] [Google Scholar]
- Koffer A., and Gomperts B.D.. 1989. Soluble proteins as modulators of the exocytotic reaction of permeabilised rat mast cells. J. Cell. Sci. 94:585–591. [DOI] [PubMed] [Google Scholar]
- Leenders A.G., Lopes da Silva F.H., Ghijsen W.E., and Verhage M.. 2001. Rab3a is involved in transport of synaptic vesicles to the active zone in mouse brain nerve terminals. Mol. Biol. Cell. 12:3095–3102. 10.1091/mbc.12.10.3095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd-Evans E., and Platt F.M.. 2011. Lysosomal Ca(2+) homeostasis: role in pathogenesis of lysosomal storage diseases. Cell Calcium. 50:200–205. 10.1016/j.ceca.2011.03.010 [DOI] [PubMed] [Google Scholar]
- Luzio J.P., Bright N.A., and Pryor P.R.. 2007. The role of calcium and other ions in sorting and delivery in the late endocytic pathway. Biochem. Soc. Trans. 35:1088–1091. 10.1042/BST0351088 [DOI] [PubMed] [Google Scholar]
- Marks M.S., and Seabra M.C.. 2001. The melanosome: membrane dynamics in black and white. Nat. Rev. Mol. Cell Biol. 2:738–748. 10.1038/35096009 [DOI] [PubMed] [Google Scholar]
- Martinez I., Chakrabarti S., Hellevik T., Morehead J., Fowler K., and Andrews N.W.. 2000. Synaptotagmin VII regulates Ca(2+)-dependent exocytosis of lysosomes in fibroblasts. J. Cell Biol. 148:1141–1149. 10.1083/jcb.148.6.1141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan A.J., Platt F.M., Lloyd-Evans E., and Galione A.. 2011. Molecular mechanisms of endolysosomal Ca2+ signalling in health and disease. Biochem. J. 439:349–374. 10.1042/BJ20110949 [DOI] [PubMed] [Google Scholar]
- Neuhoff V., Arold N., Taube D., and Ehrhardt W.. 1988. Improved staining of proteins in polyacrylamide gels including isoelectric focusing gels with clear background at nanogram sensitivity using Coomassie Brilliant Blue G-250 and R-250. Electrophoresis. 9:255–262. 10.1002/elps.1150090603 [DOI] [PubMed] [Google Scholar]
- Pittman J.K. 2011. Vacuolar Ca(2+) uptake. Cell Calcium. 50:139–146. 10.1016/j.ceca.2011.01.004 [DOI] [PubMed] [Google Scholar]
- Pu J., Schindler C., Jia R., Jarnik M., Backlund P., and Bonifacino J.S.. 2015. BORC, a multisubunit complex that regulates lysosome positioning. Dev. Cell. 33:176–188. 10.1016/j.devcel.2015.02.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao S.K., Huynh C., Proux-Gillardeaux V., Galli T., and Andrews N.W.. 2004. Identification of SNAREs involved in synaptotagmin VII-regulated lysosomal exocytosis. J. Biol. Chem. 279:20471–20479. 10.1074/jbc.M400798200 [DOI] [PubMed] [Google Scholar]
- Rodríguez A., Webster P., Ortego J., and Andrews N.W.. 1997. Lysosomes behave as Ca2+-regulated exocytic vesicles in fibroblasts and epithelial cells. J. Cell Biol. 137:93–104. 10.1083/jcb.137.1.93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosa-Ferreira C., and Munro S.. 2011. Arl8 and SKIP act together to link lysosomes to kinesin-1. Dev. Cell. 21:1171–1178. 10.1016/j.devcel.2011.10.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seixas E., Ramalho J.S., Mota L.J., Barral D.C., and Seabra M.C.. 2012. Bacteria and protozoa differentially modulate the expression of Rab proteins. PLoS One. 7:e39858 10.1371/journal.pone.0039858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens D.R., Schirra C., Becherer U., and Rettig J.. 2011. Vesicle pools: lessons from adrenal chromaffin cells. Front. Synaptic Neurosci. 3:2 10.3389/fnsyn.2011.00002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka Y., Kanai Y., Okada Y., Nonaka S., Takeda S., Harada A., and Hirokawa N.. 1998. Targeted disruption of mouse conventional kinesin heavy chain, kif5B, results in abnormal perinuclear clustering of mitochondria. Cell. 93:1147–1158. 10.1016/S0092-8674(00)81459-2 [DOI] [PubMed] [Google Scholar]
- Tsuboi T., and Fukuda M.. 2006a Rab3A and Rab27A cooperatively regulate the docking step of dense-core vesicle exocytosis in PC12 cells. J. Cell Sci. 119:2196–2203. 10.1242/jcs.02962 [DOI] [PubMed] [Google Scholar]
- Tsuboi T., and Fukuda M.. 2006b The Slp4-a linker domain controls exocytosis through interaction with Munc18-1.syntaxin-1a complex. Mol. Biol. Cell. 17:2101–2112. 10.1091/mbc.E05-11-1047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker W.C., Weber T., and Chapman E.R.. 2004. Reconstitution of Ca2+-regulated membrane fusion by synaptotagmin and SNAREs. Science. 304:435–438. 10.1126/science.1097196 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.