Demonbreun et al. visualized muscle membrane repair in real time after laser-induced microdamage. Annexin proteins were observed to form a repair cap at the site of injury, supporting a shoulder-like structure containing EHD1, EHD2, dysferlin, and MG53.
Abstract
Disruption of the plasma membrane often accompanies cellular injury, and in muscle, plasma membrane resealing is essential for efficient recovery from injury. Muscle contraction, especially of lengthened muscle, disrupts the sarcolemma. To define the molecular machinery that directs repair, we applied laser wounding to live mammalian myofibers and assessed translocation of fluorescently tagged proteins using high-resolution microscopy. Within seconds of membrane disruption, annexins A1, A2, A5, and A6 formed a tight repair “cap.” Actin was recruited to the site of damage, and annexin A6 cap formation was both actin dependent and Ca2+ regulated. Repair proteins, including dysferlin, EHD1, EHD2, MG53, and BIN1, localized adjacent to the repair cap in a “shoulder” region enriched with phosphatidlyserine. Dye influx into muscle fibers lacking both dysferlin and the related protein myoferlin was substantially greater than control or individual null muscle fibers, underscoring the importance of shoulder-localized proteins. These data define the cap and shoulder as subdomains within the repair complex accumulating distinct and nonoverlapping components.
Introduction
The elongated nature of skeletal myofibers combined with the forces of muscle contraction renders the muscle plasma membrane prone to small lesions and tears (McNeil and Khakee, 1992). Disruption of this membrane simultaneously permits entry of extracellular Ca2+ and the leak of intracellular contents. Minimizing plasma membrane disruption and efficiently resealing membrane tears are each necessary for normal muscle function. Much of the membrane repair machinery is thought to be used by both muscle and nonmuscle cells, but mutations that disrupt these repair proteins often yield muscle defects and disease, underscoring the importance of membrane repair for muscle health (Bansal et al., 2003; Cai et al., 2009; Marg et al., 2012; Leung et al., 2013).
Plasma membrane resealing is thought to occur mainly by recruiting intracellular vesicles to the site of injury triggered by elevated Ca2+ concentration at or near the site of membrane disruption (McNeil and Khakee, 1992; McNeil et al., 2003). Vesicle recruitment to the lesion transports new membrane to the site of injury, providing materials to “patch” the membrane. Membrane repair is a critical mechanism for cell survival, and well-characterized proteins such as MG53 and dysferlin have been implicated in the resealing of human and mouse muscle plasma membrane. MG53 and dysferlin accumulate at the site of membrane damage in muscle fibers (Bansal et al., 2003; Cai et al., 2009; Marg et al., 2012). MG53, also known as TRIM72, localizes specifically to areas rich in phosphatidylserine, and dysferlin has been shown to bind negatively charged phospholipids in a Ca2+-dependent manner (Davis et al., 2002; Bansal et al., 2003; Cai et al., 2009; Therrien et al., 2009). Loss-of-function mutations in the gene encoding dysferlin lead to muscular dystrophy, whereas mice lacking MG53 display myopathy (Bashir et al., 1998; Liu et al., 1998; Cai et al., 2009). Muscle fibers lacking MG53 or dysferlin have increased levels of fluorescent dye influx after injury consistent with delayed resealing of the plasma membrane (Bansal et al., 2003; Cai et al., 2009). Notably, exogenous MG53 exposure rescues laser-induced membrane disruption, implicating MG53 as a molecular “bandage” (Weisleder et al., 2012).
More recently, the annexins, particularly annexin A6, have been implicated in muscle plasma membrane resealing (Roostalu and Strähle, 2012; Swaggart et al., 2014). In the C2C12 muscle cell line, MG53 colocalizes with annexin A5 during repair, whereas dysferlin colocalizes with annexins A1 and A2 (Lennon et al., 2003; Cai et al., 2009). The Epsin15 homology domain proteins have also been studied in plasma membrane resealing in cell culture models (Marg et al., 2012), and the Eps15 homology domain (EHD) proteins are known to interact with dysferlin and the highly related myoferlin protein (Doherty et al., 2008; Posey et al., 2011). The annexins are a broadly expressed family of proteins characterized by multiple annexin domains and the ability to bind phospholipids in the presence of Ca2+ (Gerke et al., 2005). Annexin A6 is a nonclassical annexin family member, containing eight annexin repeat domains, rather than four, and an atypical amino-terminal domain. In addition to binding phospholipids, annexin A6 binds to cholesterol-rich membranes as well as the cytoskeleton, positioning annexin A6 to modulate both membrane and cytoskeleton components (Cornely et al., 2011). Both annexin A1 and A2 bind dysferlin, and serum protein levels of A1 and A2 correlate with disease progression in dysferlinopathy patients, implicating A1 and A2 in muscle disease (Lennon et al., 2003; Cagliani et al., 2005). Recent work has shown annexins A1, A2, and A6 accumulate at the site of muscle membrane damage in zebrafish, suggesting a complex role for the annexin family in membrane repair (Roostalu and Strähle, 2012).
Here, we used a laser-wounding assay with refined parameters that generated reproducible injury in the plasma membrane of live muscle fibers. We combined this method with high-resolution imaging of the real-time translocation of fluorescently labeled proteins in mammalian muscle. Using this approach, we were able to delineate the dynamic formation of repair complex subdomains, including the annexin-rich cap and its interaction with “shoulder” proteins, including dysferlin, MG53, EHD1, EHD2, and BIN1. The formation of the annexin cap was actin dependent and Ca2+ regulated. We provide evidence that shoulder proteins are also critical for the repair process, because fibers lacking both myoferlin and dysferlin have significantly increased levels of fluorescent dye intake after laser-induced membrane disruption. Together, these data define two previously unidentified structures formed during plasma membrane repair, the repair cap and the shoulder, each with distinct protein components.
Results
Proteins implicated in muscle membrane repair enrich near transverse tubules in the uninjured state
Electroporation was used to introduce plasmids expressing fluorescently labeled proteins into muscle (DiFranco et al., 2009; Kerr et al., 2013). Plasmids expressing annexin A6, dysferlin, EHD1, EHD2, MG53, and BIN1 were introduced into myofibers to document their intracellular pattern of expression in the absence of muscle injury. The domain structures of these proteins, the position of the fluorescent tags and a schematic of the skeletal organization are shown in Fig. S1 (A and B). The intracellular position of the expressed proteins was monitored relative to F-actin, visualized with the LifeAct fluorescent protein, and coimaged with Di-8-ANEPPS, a lipophilic fast-responding membrane-potential dye that marks deep membrane invaginations of the transverse tubule (T-tubule; Lyon et al., 2009). Annexin A6 (A6) and Di-8-ANEPPS showed a similar pattern in myofibers (Fig. S1 C), reflective of the endogenous proteins (Lyon et al., 2009; Kerr et al., 2013; McDade et al., 2014; Posey et al., 2014). BIN1, dysferlin (DYSF), EHD1, and EHD2 were each individually expressed in muscle fibers, and each displayed a similar overlapping pattern with Di-8-ANEPPS in uninjured myofibers (Fig. S1 D). Many of these proteins also showed expression at the sarcolemma in addition to their T-tubule enrichment. Phospholipase C (PLC) is known to bind to phosphatidylinositol 4,5-bisphosphate (PIP2), and in the absence of PLC’s pleckstrin homology (PH) domain, this protein serves as an intracellular marker of PIP2 (Stauffer et al., 1998). Fluorescently labeled PLCΔPH displayed the same T-tubule pattern of localization, overlapping with Di-8-ANEPPS (Fig. S1 D). Therefore, in the uninjured muscle, the T-tubule appears to be a reservoir for many membrane repair proteins.
Distinct proteins in the membrane repair cap and shoulder
To directly evaluate the dynamic process of membrane repair in living myofibers, we used high-resolution microscopy of isolated, adherent muscle fibers that were subjected to laser-induced membrane ablation to create lesions in the sarcolemma of live myofibers. Previously established methods of laser-induced damage have focused on membrane damage of cultured cells and myotubes or targeted larger regions of damage, up to 25 µm2, in isolated myofibers (Bansal et al., 2003; Cai et al., 2009; Marg et al., 2012). In the current study, we directed the laser to the lateral edge of the myofiber. In the presence of FM4-64, laser ablation was applied to the sarcolemma, targeting one pixel within the sarcolemmal bilayer. At 405 nm through a 1.4-NA lens, this correlates to a diffraction limited spot approximated by a cylinder 0.25 × 0.75 µm. Given the laser energy required to induce sarcolemmal disruption, the region of injury extends significantly beyond the small targeted area. This protocol resulted in tightly controlled region of “microdamage,” after which most fibers remained affixed to the surface and did not undergo hypercontraction, which was defined as bending of the myofiber >15° (Table 1). Notably, muscle fibers remained alive >40 min after imaging. FM4-64 is a water-soluble dye that increases fluorescence upon exposure to negatively charged lipids within the membrane. With injury, the negatively charged phospholipid, phosphatidylserine (PS), is exposed at the site of injury, and FM4-64 fluorescence is triggered in the presence of PS (Zweifach, 2000; Cai et al., 2009; Yeung et al., 2009). Annexin A6 has a high affinity for PS (Lizarbe et al., 2013). After membrane injury, annexin A6 was recruited rapidly to the site of FM4-64 accumulation (Fig. 1). Annexin A6 aggregated into a tight structure that we termed the “repair cap.” The repair cap localized above a zone devoid of annexin A6, termed the “clearance zone.” In the merged image, the region of FM4-64 and annexin A6 enrichment was coincident with site of membrane disruption identified from the differential interference contrast image, where the cap was visualized on the exterior surface of the myofiber (Video 1). Adjacent to the repair cap is a region we refer to as the “shoulder.”
Table 1. Fiber bending at site of injury.
| Genotype | Number of fibers with a >15° bend | Total number of fibers | Percentage of fibers with a >15° bend |
|---|---|---|---|
| % | |||
| Wild type | 42 | 375 | 11 |
| Dysf | 5 | 50 | 10 |
| FER | 4 | 50 | 8 |
| MKO | 1 | 6 | 16 |
| Total | 52 | 481 | 11 |
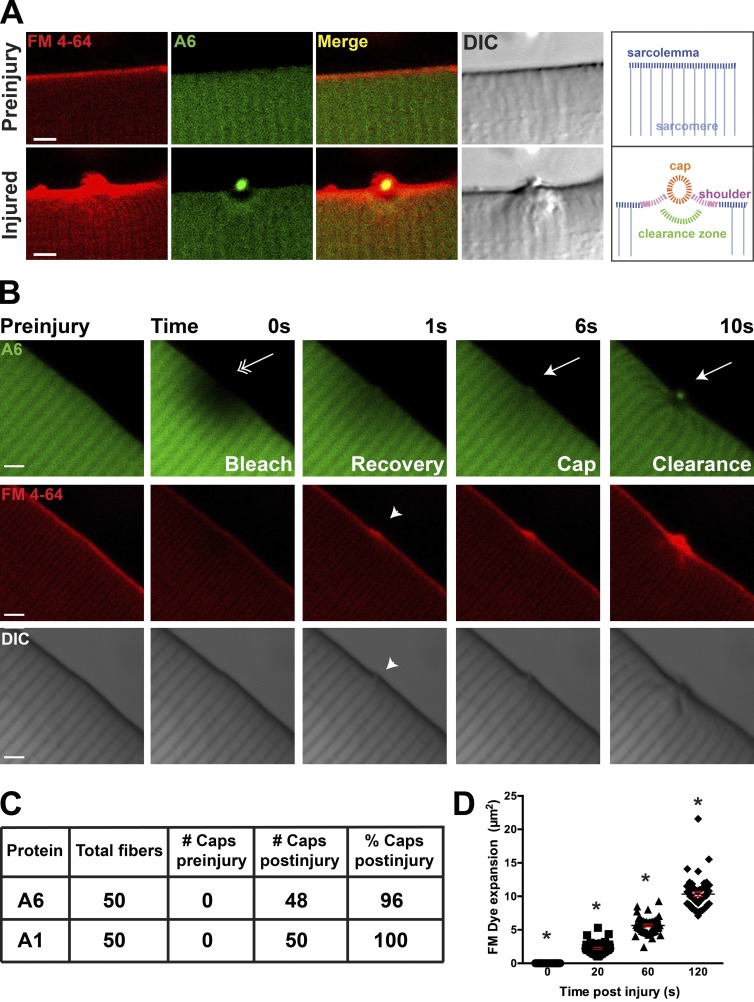
Figure 1.
Annexin A6 forms a repair cap at the site of myofiber sarcolemmal disruption. (A) High-resolution imaging of a single myofiber before laser ablation and then 80 s after sarcolemmal disruption. FM4-64 (red) increased fluorescence after binding lipids, particularly negatively charged lipids, like phosphatidylserine (Zweifach, 2000; Yeung et al., 2009). Upon membrane disruption, FM4-64 accumulated at the site of membrane disruption (bottom, left). Annexin A6 (A6)–GFP (green) was rapidly recruited to the site of FM4-64 aggregation, forming a repair cap after damage (yellow, merge). Within the myofiber, below the annexin A6 cap, was a region devoid of annexin A6 termed the clearance zone. A repair cap and shoulder visualized by differential interference contrast imaging. A schematic is shown indicated with labeling. (B) Laser-injury produced local photobleach that resolved by 1 s postinjury (double arrow). After fluorescence recovery, a repair cap and clearance zone formed (arrow). FM4-64 fluorescence accumulated at the lesion (arrowhead). Bars, 4 µm. (C) Myofibers expressing annexins A6 and A1 produced repair caps in 96% and 100% of damaged fibers, respectively. (D) The technique of laser damage is highly reproducible, as indicated by similar FM4-64 expansion zones after injury at each time point tested (*, P < 0.05; ∼10 myofibers per animal; n = 6 mice per condition). Error bars represent SEM.
To ensure the absence of annexin A6 in the clearance zone was not a result of photobleaching, we analyzed images for fluorescence recovery after photobleaching. Fig. 1 B illustrates the spatiotemporal effects of the laser within the myofiber after sarcolemmal ablation. Although the laser was directed at the edge of the sarcolemma, the photobleach area extended several microns in a radial diameter within the fiber spanning approximately six sarcomeres. At 1 s after laser damage, GFP fluorescence within the area was largely recovered. The annexin A6 repair cap and clearance zone were first seen at 6 and 10 s after injury, respectively, considerably after resolution of photobleaching. Fluorescently visible FM4-64 remained localized to a discreet region at the sarcolemma, suggesting that sarcolemmal disruption generated by this laser ablation did not extend far beyond the primary site of injury. Thus, the clearance zone is spatiotemporally distinct from and not the result of photobleaching. To estimate the degree of sarcomeric disruption produced from this injury protocol, α-actinin was labeled with GFP to visualize sarcomeres in the setting of laser injury and FM4-64. Fig. S2 indicates the limited extent of sarcomere disruption produced by the ablation protocol (arrow), consistent with the notion that laser ablation produces a limited degree of disruption in the myofiber itself (n = 12 myofibers from n = 3 mice per condition).
We further tested the consistency and reproducibility of this injury method. Myofibers were electroporated with annexin A1 or annexin A6 and subjected to laser-induced injury. Annexin A6 cap formation was present in 48/50 fibers (96%), and A1 cap formation was present in 50/50 (100%) of damaged myofibers (Fig. 1 C; ∼10 myofibers per animal, n = 6 mice per condition). The absence of annexin A6 caps in 2 of 50 fibers likely reflects misapplication of the laser with respect to the surface of the myofiber. FM4-64 fluorescence was analyzed for consistency in damaged myofibers at 0, 20, 60, and 120 s after injury. FM4-64 expanded within the myofibers at a similar rate (Fig. 1 D; *, P < 0.05, ∼10 myofibers per animal, n = 6 mice per condition). These data show that limiting laser power and exposure during ablation produced a highly consistent method of sarcolemma disruption resulting in the formation of a repair cap.
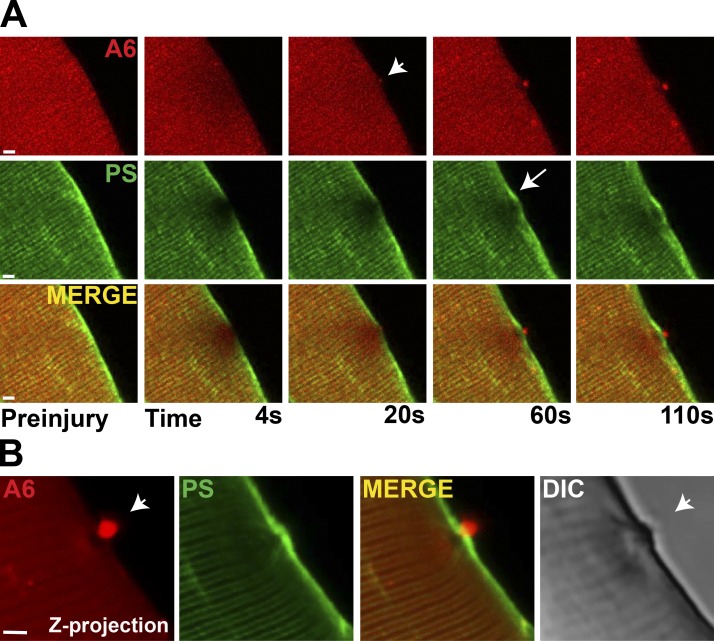
Annexin A5 itself is often used to mark PS localization (Cai et al., 2009). To examine PS localization independent of annexins, we used the Lact-C2-GFP plasmid. Lactcadherin is a glycoprotein found in milk that binds PS in a Ca2+-dependent manner through interaction with its C2 domain (Yeung et al., 2009). Lact-C2–GFP and annexin A6–mCherry were coelectroporated into muscle, and isolated muscle fibers were subjected to laser-induced damage. Annexin A6 formed a repair cap at the site of injury, whereas PS accumulated adjacent to the repair cap at the shoulder throughout the duration of imaging (110 s; Fig. 2 A). These distinct compartments of the cap and shoulder are apparent in the confocal z-projection of an injured myofiber (Fig. 2 B).
Figure 2.
Phosphatidylserine (PS) is adjacent to, but not overlapping with, the annexin A6 repair cap. (A) Myofibers were coelectroporated with plasmids expressing annexin A6 (A6) tagged with mCherry (red) and Lact-C2 representing PS and tagged with GFP (green) to visualize annexin A6 and PS translocation simultaneously. By 20 s postinjury, the annexin A6–rich cap was visualized (top, arrowhead). With membrane disruption, PS was concentrated in the plasma membrane juxtaposed, but not colocalized, with the annexin A6–rich cap (arrow). Both proteins continued to accumulate through 110 s of imaging. (B) A confocal z-projection illustrates the annexin A6 repair cap (white arrowhead) above an annexin-free clearance zone with PS localized at the shoulder. Bars, 4 µm.
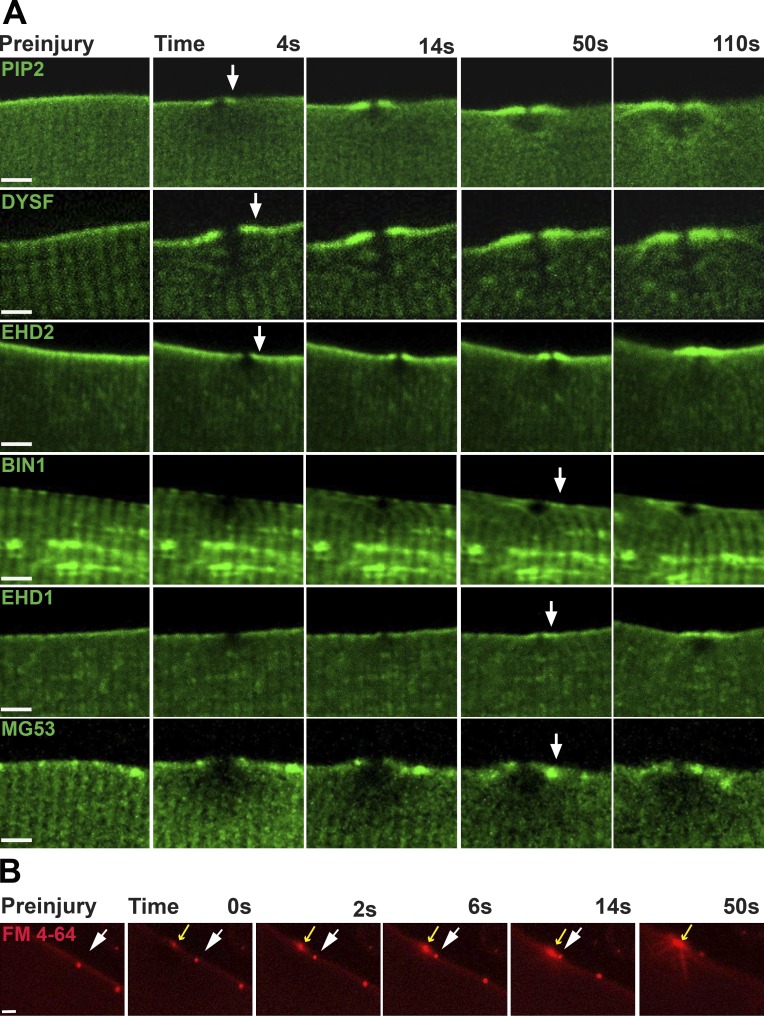
Plasmids encoding proteins implicated in membrane repair were similarly electroporated into skeletal muscle, and isolated myofibers were then subjected to the identical laser injury process. Dysferlin and EHD2 were among the earliest proteins recruited to the repair shoulder (Fig. 3). The PLCΔPH protein, reflecting PIP2 location, was similarly recruited to the shoulder between 4 and 14 s after membrane injury. EHD1, BIN1, and MG53 were seen enriched slightly later, after membrane injury, ∼50 s after membrane disruption. Increasing the size of the ablated region by increasing the laser power did not alter the translocation of the proteins to the site of damage. Along the plasma membrane and lateral to site of laser-induced injury, FM4-64 puncta would occasionally be seen, and these puncta were useful as points of reference to monitor the membrane movement relative to the injury site. Relative to the site of laser-induced disruption, FM4-64 puncta were observed to move laterally toward the site of membrane disruption (Fig. 3 B, white and yellow arrows; and Video 2). The lateral movement of membrane toward the damage site was also readily appreciated in differential interference contrast imaging of wounded muscle fibers (not depicted). The observation of lateral membrane recruitment does not exclude the possibility that internal vesicles are also recruited and fuse to the site of damage, although evidence for vesicle accumulation was not observed in Z-stack imaging. These data suggest lateral diffusion of membrane is at least one mechanism used to recruit membrane components to the site of membrane repair.
Figure 3.
T-Tubule–associated proteins are found at the repair shoulder. (A) PIP2, marked with PLCΔPH, accumulated at the shoulder region and was visualized as early as 4 s after sarcolemmal disruption. Dysferlin (DYSF) and EHD2 were also recruited to the shoulder region within 5–15 s after injury. BIN1, EHD1, and MG53 were recruited between 14 and 50 s after membrane disruption. (B) Aggregates of FM4-64 within the plasma membrane were observed to move laterally along the surface of injured myofibers toward the site of membrane disruption. Injury was conducted in the presence of FM4-64. Fibers containing small, static patches of FM4-64 were selected (white arrow) to provide a point of reference. The static patch of FM4-64 on the sarcolemma moved laterally toward the site of laser damage (yellow arrow) through the plasma membrane. Bars, 4 µm.
Multiple annexins contribute to the repair cap
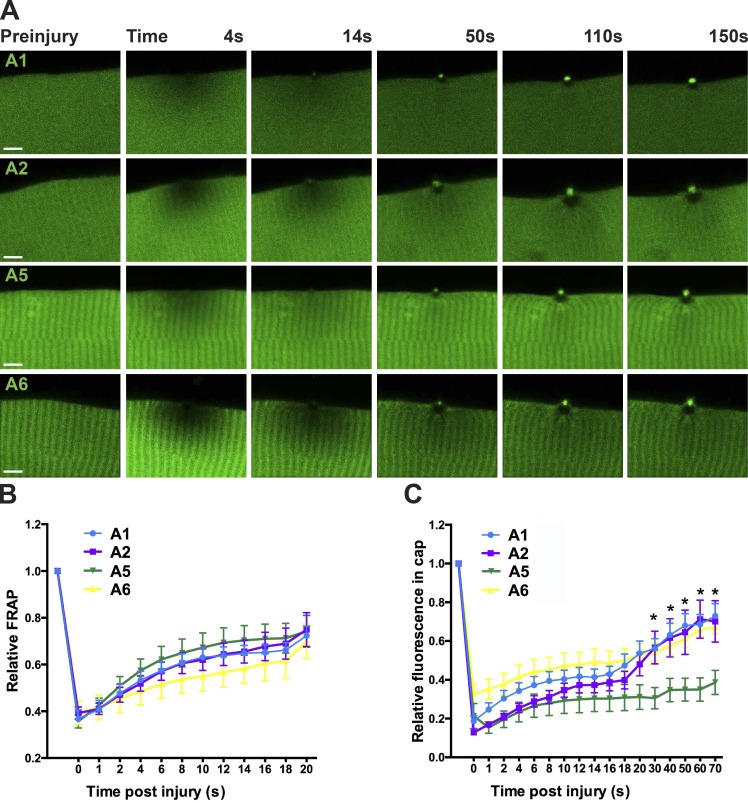
In uninjured muscle, fluorescently labeled annexins A2 and A5 each demonstrated a similar striated pattern to annexin A6 (Fig. 4 A, preinjury panel; n ≥ 14 myofibers from n ≥ 5 mice per condition). Within seconds of laser disruption, annexin A6 was rapidly recruited to the repair cap at the site of membrane damage, and the annexin-free zone was seen beneath the repair cap (Fig. 4, top row). Annexins A1, A2, and A5 also were recruited to repair cap, and in each case, an annexin-free zone formed under the repair cap. Annexin A6 caps were seen as early as 4 s after injury, persisting through the 150 s of imaging. Moreover, A6 caps persisted >40 min after laser injury (Fig. S3). FRAP analysis revealed that the motility of annexins A1, A2, A5, and A6 was comparable from 0 to 20 s after bleaching (Fig. 4 B; n ≥ 7 myofibers from n = 3 mice per condition). Annexin A1 and A2 fluorescence accumulated at the cap with similar kinetics as annexin A6 fluorescence and these caps persisted through the 150 s of imaging (Fig. 4 C; n ≥ 5 myofibers from n = 3 mice per condition). Annexin A5 appeared slightly later at repair caps compared with the other annexin family members (*, P < 0.05 at 30 s after damage; Fig. 4 C). The annexin-free clearance zone was distinctly devoid of annexin-GFP fluorescence and was clearly demarcated by the striated pattern of annexin-GFP within the myofiber.
Figure 4.
Multiple annexins contribute to repair cap formation. (A) Annexins A1, A2, A5, and A6 were rapidly recruited to the site of laser wounding and were seen between 4 and 14 s after membrane disruption. With each annexin repair cap, an annexin free repair zone was visible within the myofiber (n ≥ 14 myofibers from n ≥ 5 mice per condition). Bar, 4 µm. (B) Fluorescence recovery after photobleaching (FRAP) was performed with GFP-tagged annexins. The diffusion of the different annexins over the first 20 s after injury was comparable (n ≥ 7 myofibers from n = 3 mice per condition). (C) Kinetics for GFP-tagged annexins showed a rapid increase of annexin at the lesion reflecting recruitment to the repair cap (*, P < 0.05; n = 5 myofibers from n = 3 mice per condition). Error bars represent SEM.
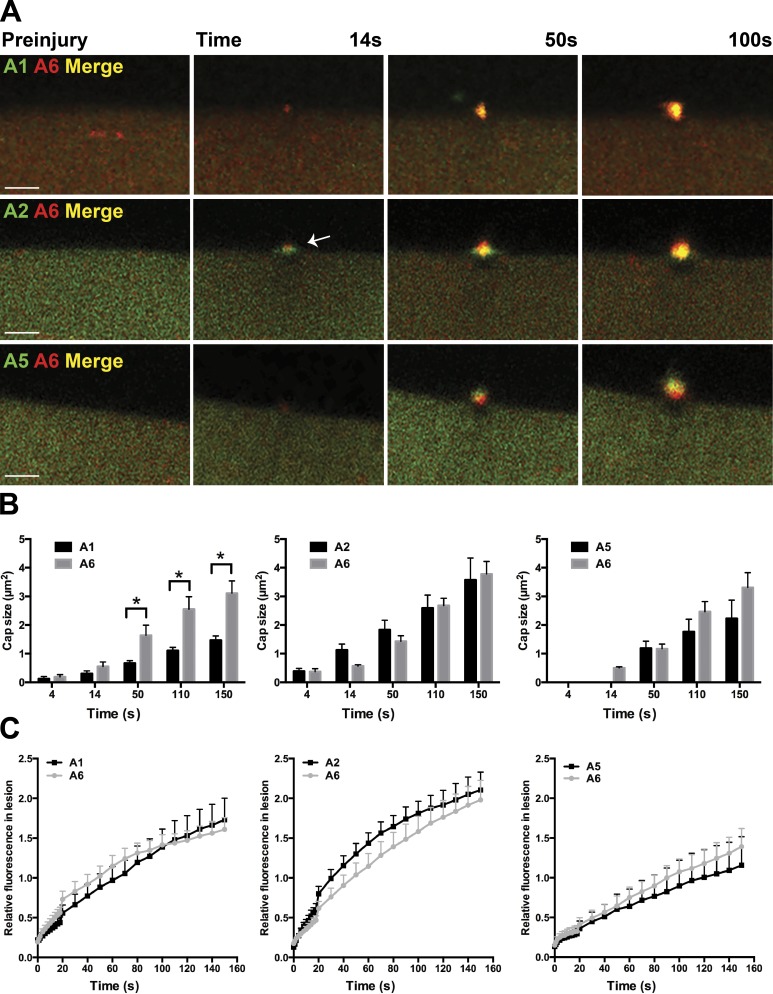
To better assess the sequential accumulation of annexin localization at the wound, we electroporated combinations of annexin subunits (A1 and A6, A2 and A6, or A5 and A6) and imaged membrane repair after laser-induced injury (Fig. 5 A; n ≥ 7 myofibers from n = 3 mice per condition). All annexin proteins accumulated at the repair cap and formed clearance zones at the lesion. Annexin A2 initially accumulated at the lesion forming a flat structure partially colocalizing with the annexin A6 cap (Fig. 5 A, arrow). By 100 s after injury, annexin A1 colocalized with annexin A6 in the repair cap. Annexin A1 caps were significantly smaller than annexin A6 repair caps at 50 s after injury (*, P < 0.05), whereas annexin A2 and A5 caps were similar in size to annexin A6 caps at all time periods analyzed (Fig. 5 B; n ≥ 7 myofibers from n = 3 mice per condition). The timing of each annexin subunit translocation was compared with annexin A6. Relative fluorescence at the lesion was similar between all pairwise comparisons, despite annexin A5 kinetics being delayed (Fig. 5 C; n ≥ 7 myofibers from n = 3 mice per condition). The assembly of annexins into a multimeric structure is consistent with the oligomerization properties of annexins (Hoque et al., 2014).
Figure 5.
Relative timing of annexin subunit translocation to the repair cap. Myofibers were coelectroporated with plasmids expressing annexin A6 and A1, A6 and A2, or A6 and A5 to compare the relative kinetics of annexin translocation to the site of sarcolemmal injury. (A) Representative images of electroporated myofibers illustrating each combination of annexin proteins localized to the repair cap. Annexin A2 localized to a broader structure at the base of the cap (white arrow) above the clearance zone just after damage and then formed a tighter repair cap that colocalized with annexin A6, and this was observed by 100s seconds after injury. Bars, 4 µm. (B) Annexin A1 cap size was reduced compared with annexin A6 cap size (*, P < 0.05 at 50 s), whereas there was no significant difference in annexin A2, A5, and A6 cap size (n ≥ 7 myofibers from n = 3 mice per condition). (C) All pairwise comparisons resulted in similar annexin kinetics compared with annexin A6 (n ≥ 7 myofibers from n = 3 mice per condition). Error bars represent SEM.
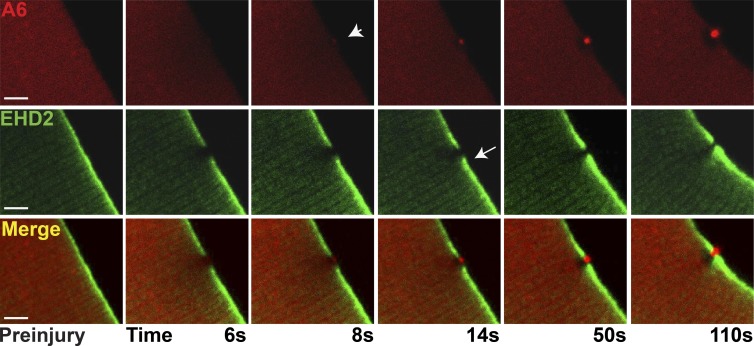
The sharp delineation of the repair cap and shoulder was visualized by coexpressing annexin A6 and EHD2, as both of these proteins were observed at very early time points after membrane disruption and serve as the earliest markers of the repair complex. Notably, the accumulation of these proteins at the repair complex preceded the accumulation of PS, suggesting that annexin A6 and EHD2 may recruit other components to the repair complex, including signaling components. Annexin A6, labeled with mCherry, was seen to form a repair cap, whereas EHD2, labeled with GFP, localized to the shoulder region. Notably, annexin A6 and EHD2 abutted but did not colocalize, consistent with these components being distinct substructures of the repair complex. Both proteins continued to accumulate in the cap and shoulder regions throughout the 110 s of imaging (Fig. 6 and Video 3; n = 12 myofibers from n = 4 mice). From this data, we conclude that there are at least two subdomains of the membrane repair complex and suggest these subdomains may perform different functions during repair.
Figure 6.
Distinct cap and shoulder repair proteins. Myofibers were coelectroporated with plasmids expressing annexin A6 tagged with mCherry (A6) and EHD2 tagged with GFP (EHD2) to visualize cap and shoulder protein simultaneously. By 8 s postinjury, the annexin A6–rich cap was visualized (arrowhead). The adjacent EHD2-containing shoulder protein was seen adjacent to, but not overlapping with, the A6-rich cap (arrow). The clear zone was devoid of both A6 and EHD2 proteins. Both proteins continued to accumulate through 110 s of imaging. This pattern of localization occurred in 12/12 myofibers collected from four animals. Bars, 4 µm.
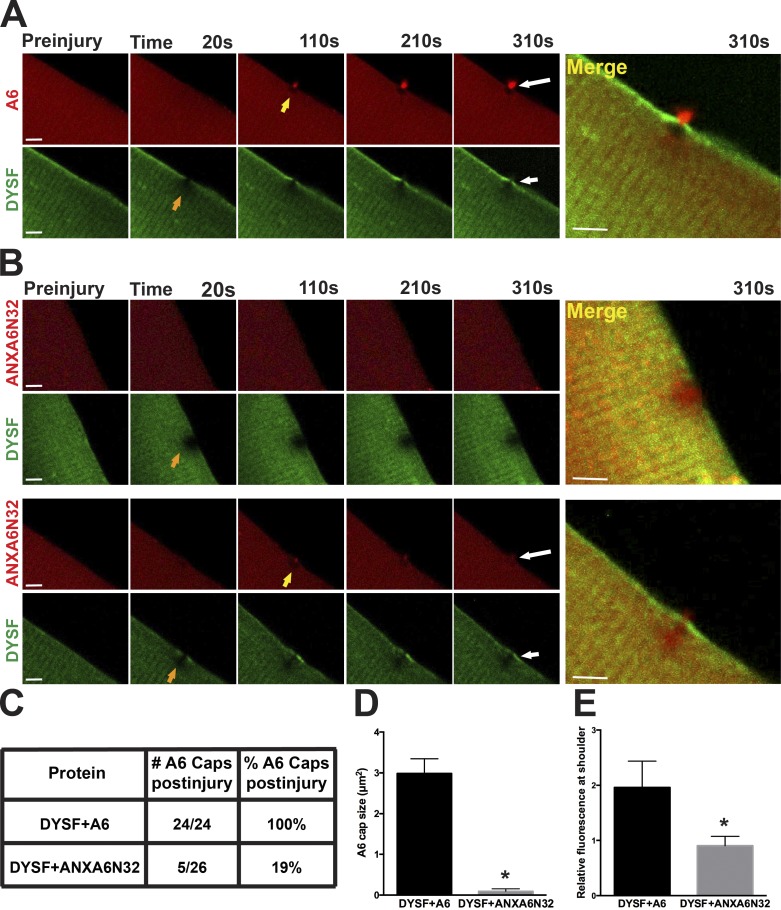
Dominant-negative annexin A6 expression disrupts dysferlin localization during membrane repair in vivo
Dysferlin was one of the first proteins implicated in membrane repair (Bansal et al., 2003), and it is known to bind annexins A1 and A2 (Lennon et al., 2003). In zebrafish, the loss of both annexin A6 and dysferlin resulted in a more severe myopathy-like phenotype (Roostalu and Strähle, 2012). Recently, a truncated form of annexin A6 that represents the amino-terminal 32 kD of annexin A6 (termed ANXA6N32) was discovered to exacerbate muscular dystrophy in mice by acting as a dominant-negative protein that inhibited the normal translocation of full-length annexin A6 (Swaggart et al., 2014). To test whether this dominant-negative ANXA6N32 also inhibited other repair components, wild-type myofibers were coelectroporated with mCherry-tagged annexin A6 or mCherry-tagged annexin ANXA6N32 along with Venus-tagged dysferlin. After laser damage, annexin A6 formed a repair cap at the site of damage and DYSF localized to the shoulder just beside the repair cap (Fig. 7 A). In contrast, damaged myofibers expressing truncated annexin A6, ANXA6N32, either did not form a repair cap or formed a much smaller repair cap (Fig. 7 B, top and bottom, respectively). The presence of ANXA6N32 resulted in a poorly demarcated annexin-free zone. When ANXA6N32 disrupted repair cap formation, it also inhibited the recruitment of dysferlin to the repair shoulder (Fig. 7 B, top). Annexin A6 formed a repair cap in 24/24 (100%) of damaged myofibers coelectroporated with dysferlin and annexin A6, whereas cap formation was seen in only 5/26 (19%) of damaged myofibers with coelectroporation of ANXA6N32 (Fig. 7 C; n = 7 mice per condition). Additionally, ANXA6N32 repair caps were smaller than annexin A6 repair caps (Fig. 7 D; *, P < 0.0001, n ≥ 13 myofibers from n = 4 mice per condition). Dysferlin accumulation at the shoulder was decreased in the presence of ANXA6N32 as compared with full-length A6 (Fig. 7 E; *, P < 0.05, n ≥ 7 myofibers from n = 3 mice per condition). The dominant-negative function of ANXA6N32 disrupts the formation of the annexin A6 repair cap, clearance zone, and recruitment of shoulder proteins in the repair complex.
Figure 7.
Truncated annexin A6 (ANXA6N32) disrupts dysferlin localization during membrane repair. Muscle fibers were coelectroporated with dysferlin (DYSF)-Venus and full-length annexin A6 (A6)–mCherry or truncated ANXA6N32-mCherry for live-cell imaging after laser damage. (A) Annexin A6 (red) formed a distinct repair cap (yellow arrow) at the site of membrane disruption, whereas dysferlin (green) localized to the shoulder (orange arrow), and a high-magnification image is shown to the right. (B) Fibers expressing ANXA6N32 either did not form a repair cap at all (example, top row) or formed a much smaller repair cap (example, bottom row). In both cases, the annexin-free zone was poorly demarcated compared with that seen with full-length annexin A6. ANXA6N32 disrupted the translocation of dysferlin to the shoulder, consistent with a dominant-negative effect. ANXA6N32 did not disrupt dysferlin’s formation of a clear zone under the annexin A6 cap. High-magnification images are shown on the right. Bars, 4 µm. n = 7 mice per condition. (C) The presence of ANXA6N32 was associated with decreased cap formation compared with full-length annexin A6 (n ≥ 24 myofibers from n = 7 mice per condition). (D) Coelectroporation of dysferlin and ANXA6N32 resulted in smaller annexin repair caps upon damage compared with full-length annexin A6 caps (*, P < 0.0001; n ≥ 13 myofibers from n = 4 mice per condition). (E) Coelectroporation of dysferlin and ANXA6N32 resulted in decreased DYSF-Venus fluorescence at the shoulder 150 s after damage (*, P < 0.05; n ≥ 7 myofibers from n = 3 mice per condition). Error bars represent SEM.
The annexin-free zone contains actin
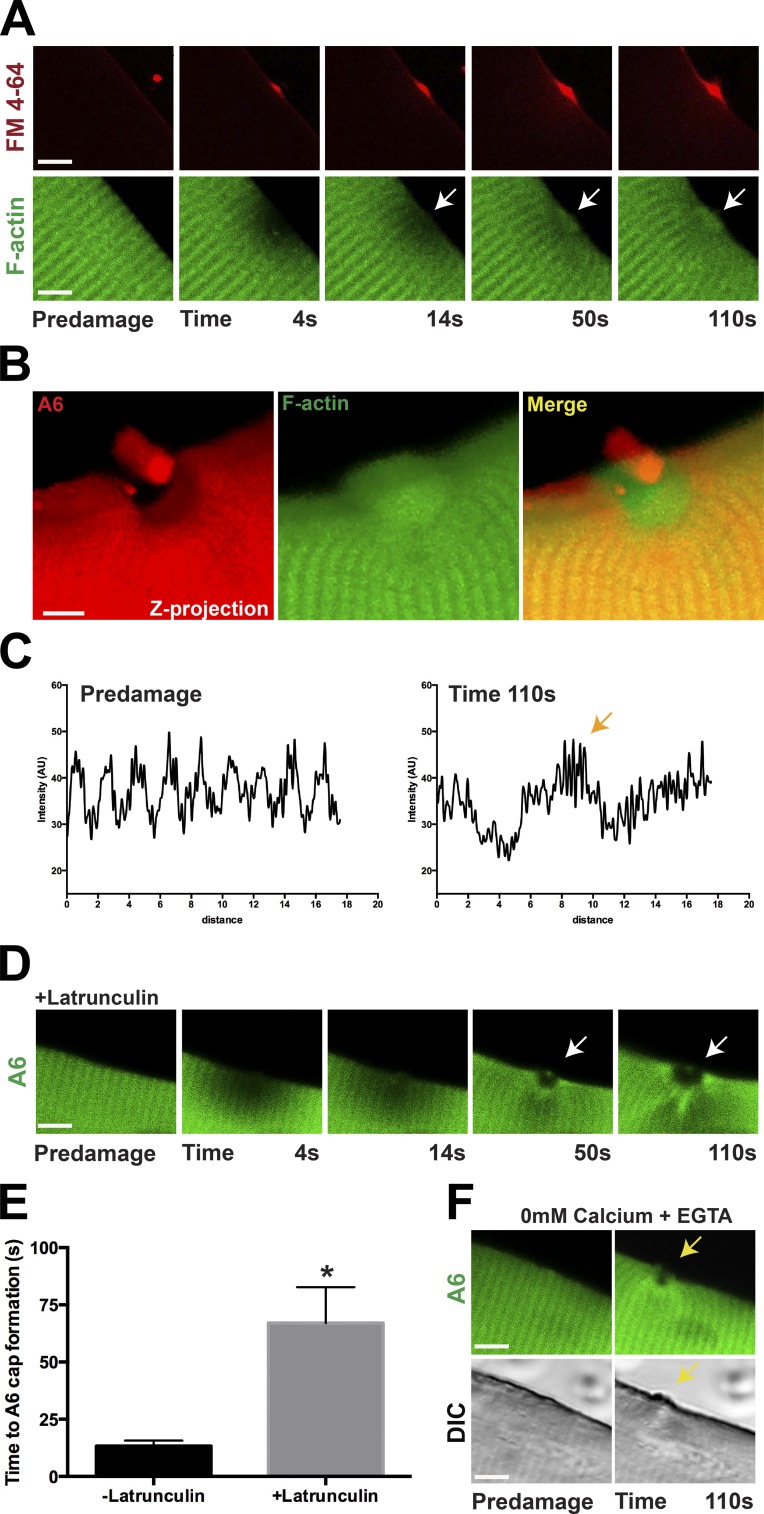
Reorganization of the cytoskeleton is known to occur during membrane resealing. After laser damage, F-actin is enriched at sites of membrane disruption in cultured muscle cells and oocytes (Mandato and Bement, 2001; Marg et al., 2012). To elucidate the role of F-actin in live myofibers during membrane repair, we expressed fluorescently tagged actin (LifeAct-mTurq2) into myofibers and performed laser-induced membrane damage. Upon injury, F-actin accumulated at the lesion immediately under the sarcolemma marked by FM4-64 fluorescence (Fig. 8 A; n = 15 myofibers from n = 4 mice). F-actin accumulated at the site of injury within 14 s of imaging (Fig. 8 A, arrow), similar to the timing of annexin A6 cap formation. F-actin accumulation was evident throughout the interval of imaging (110 s; Fig. 8 A, white arrow). F-actin accumulation was coincident with the region devoid of annexin A6 (Fig. 8 B and Video 4; n = 15 myofibers from n = 4 mice). Actin’s reorganization was monitored by evaluating a plot of its change in periodicity at the site of injury (Fig. 8 C, orange arrow). Annexin family members have been shown to bind actin (Hayes et al., 2004). To determine the necessity of F-actin for annexin A6 translocation to the repair cap, laser injury was similarly performed but in the presence of latrunculin. Latrunculin inhibits F-actin formation by sequestering monomeric G-actin, but it does not activate p53 signaling like cytochalasin D (Rasmussen et al., 2010). Latrunculin significantly delayed, but did not abolish, annexin A6 cap formation (Fig. 8, D and E; n = 7 myofibers from n = 3 mice per condition). In the absence of latrunculin, annexin A6 was normally seen at the repair cap within 13 s of injury. In the presence of latrunculin, annexin A6 caps were not seen until 67 s after injury (Fig. 8 E; *, P < 0.01).
Figure 8.
Efficient repair cap formation requires actin and Ca2+. (A) Myofibers were electroporated with actin (LifeAct-mTurq2) to visualize F-actin and FM4-64–marked laser-induced injury (red). Actin (green) slowly accumulated under the FM4-64–positive lesion (red) in response to laser damage (white arrow), beginning at ∼14 s but best visualized between 50 and 110 s (n = 15 myofibers from n = 4 mice). (B) Coelectroporation of actin (LifeAct-mTurq2) and annexin A6 demonstrated that F-actin accumulated in the annexin-free clearance zone (seen in 15/15 myofibers isolated from n = 4 mice). (C) F-actin reorganized at the site of repair as visualized by the change in periodicity in the plot profile (orange arrow). (D) Latrunculin, which disrupts actin filament formation, resulted in delayed annexin A6 cap formation (n ≥ 7 myofibers from n = 3 mice per condition). (E) The time for annexin A6 cap formation was 13 s compared with 67 s, with and without latrunculin (*, P < 0.01), indicating that rapid annexin A6 repair cap formation requires filamentous actin (n ≥ 7 myofibers from n = 3 mice per condition). (F) Annexin A6 caps failed to form in the absence of Ca2+. Incubation of fibers in 0 mM Ca2+ + EGTA inhibited annexin A6 cap formation (yellow arrow) up to 110 s of imaging (0 of 13 fibers formed caps). A few fibers retained the ability to form the annexin-free zone despite not forming a repair cap (n = 13 myofibers from n = 3 mice per condition). Bars, 4 µm. Error bars represent SEM.
ANXA6 cap formation requires extracellular Ca2+
Effective membrane resealing in nearly all cell types requires extracellular Ca2+ (McNeil et al., 2003). Like other cell types, myofibers have delayed resealing in Ca2+-free media as seen by increased FM dye uptake (Bansal et al., 2003). To evaluate the Ca2+ requirement for repair cap formation, EGTA was added to Ca2+-free media, and the injury assay was performed. The removal of Ca2+ abolished repair cap formation after injury (Fig. 8 F, yellow arrow). None of the 13 myofibers formed a repair cap, and 1 of 13 formed an annexin free zone in Ca2+-free solution. The representative image of the largest annexin-free zone is shown in (Fig. 8 F; n = 13 myofibers from n = 3 mice). This demonstrates that the formation of a normal repair cap and annexin-free zone is Ca2+ dependent.
Shoulder proteins are important for membrane repair
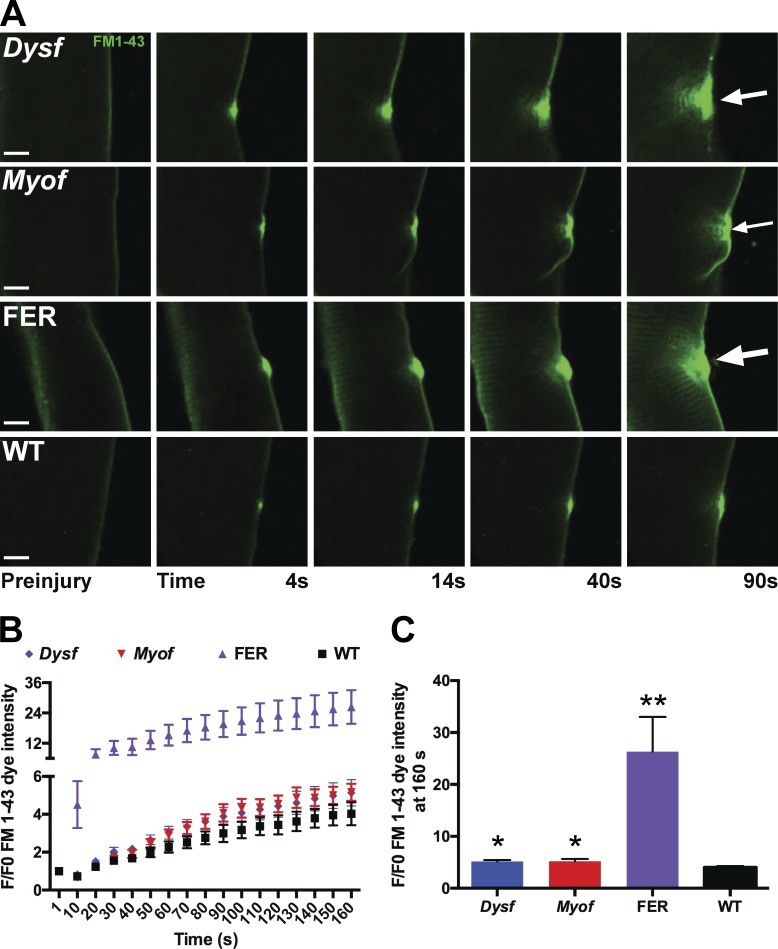
Dysferlin is a known protein required for membrane repair (Bansal et al., 2003), and we now showed that dysferlin is a key protein of the shoulder domain of the repair complex. To demonstrate the importance of shoulder proteins for resealing, we conducted laser injury on isolated fibers from dysferlin-null (Dysf), myoferlin-null (Myof), and fibers lacking both dysferlin and myoferlin (FER; Demonbreun et al., 2014). Of note, all these myofibers were isolated from mice in the same genetic background 129T2/SvEmsJ. We hypothesized if shoulder proteins were critical for normal membrane repair, then redundancy may be present between these related sequences. Dysferlin and myoferlin are ∼57% related at the amino acid level and have been implicated in receptor and vesicle trafficking, modulating muscle growth, regeneration, and repair (Davis et al., 2000; Doherty et al., 2005, 2008; Demonbreun et al., 2010; Posey et al., 2014). After laser-induced membrane damage, FM dye fluorescence was minimal in wild-type myofibers (Fig. 9; *, P < 0.05 at 160 s after injury). Myofibers lacking either dysferlin or myoferlin displayed the same degree of delayed resealing marked by increased FM-dye fluorescence, which was distinct from wild-type muscle. Myofibers lacking both dysferlin and myoferlin (FER fibers) demonstrated significantly elevated FM-dye fluorescence over time (Fig. 9; **, P < 0.01 at 160 s after injury, n = 6 myofibers from n = 2 mice per genotype). These data show that shoulder proteins are necessary for efficient membrane repair and that there is redundancy in these proteins.
Figure 9.
Dysferlin and myoferlin contribute to plasma membrane repair. Myofibers from dysferlin-null (Dysf), myoferlin-null (Myof), Dysf/Myof double-null (FER), and wild-type (WT) animals were isolated and subjected to laser-induced injury in the presence of FM1-43 fluorescence. Similar to FM4-64, FM1-43 is a lipophilic dye that increases fluorescence as it binds damaged membrane (Zweifach, 2000; Yeung et al., 2009). (A) Representative images are shown demonstrating FM1-43 uptake after injury. (B and C) Both Dysf and Myof fibers had increased FM1-43 uptake compared with wild type (*, P < 0.05). FER animals, lacking both Dysf and Myof, had significantly more FM1-43 dye uptake than either single mutant, consistent with a compensatory role of ferlin proteins (**, P < 0.01; n = 6 myofibers from n = 2 mice per genotype). Bars, 4 µm. Error bars represent SEM.
Discussion
Laser injury as a model for membrane repair in muscle
Membrane disruption is thought to occur in many forms of cellular injury, and muscle is thought to be unusually susceptible to plasma membrane disruption because of its elongated cell shape and susceptibility to contraction-induced injury (McNeil and Steinhardt, 1997). We now modified previous methods for sarcolemmal injury refining the spatiotemporal conditions for laser-induced injury. Previous studies described methods of laser-induced membrane-damage methods where the laser is focused on larger areas of the sarcolemma, ranging from as large as 25 µm2 (Bansal et al., 2003; Cai et al., 2009) to smaller regions of 2–6 µm2 (Marg et al., 2012; Defour et al., 2014). Although it is difficult to directly compare methods, given differences in microscopes and lasers, the methods used herein restricted the area of laser focus to a much smaller region of the sarcolemma, which resulted in living myofibers that could readily reseal. The degree of injury to the myofibers in this current system did not produce transection of fibers, nor did it produce significant deformation or hypercontraction of fibers. Using this point method of ablation, we were able to achieve a small and highly consistent area of membrane ablation. Combining this point method of microinjury with high-resolution microscopy permitted us to visualize subdomains of the repair structure and to image the timing of their formation. This imaging was replicated using microscopes from two manufacturers (Leica Biosystems and Nikon), indicating that this method can be readily adapted. How well these microdamage methods mirror in vivo muscle injury is not known, but it is reasonable to assume that more restricted injury is more physiological than larger injury methods, especially transection, from which myofibers do not recover.
Model for membrane repair complex
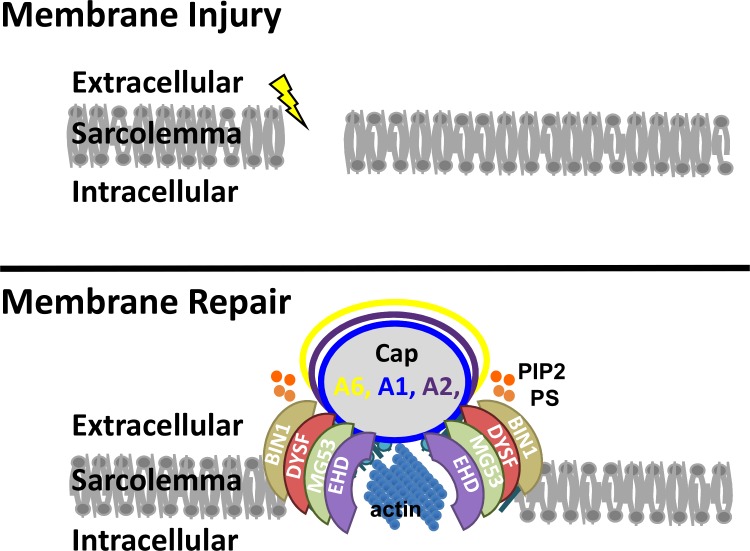
We propose a model of skeletal muscle membrane repair that is dependent on multiple membrane repair proteins, including dysferlin, annexins (A6, A1, A2, and A5), EHD proteins, and MG53, that fall within two subdomains, the repair cap and shoulder (Fig. 10). Disruption of the muscle plasma membrane is associated with recruitment of annexins to form a repair cap. This aggregation of annexins is consistent with studies in zebrafish (Roostalu and Strähle, 2012). The recruitment of multiple annexins (A6, A1, and A2) is consistent with higher-order oligomerization of an annexin-rich repair cap. Annexin A6 has a higher affinity for Ca2+ than other annexin proteins, which may contribute to the fast wound-healing response of the A6 protein (Enrich et al., 2011; Potez et al., 2011). These structures are supported by shoulder proteins within the plasma membrane and adjacent to the repair cap. These shoulder proteins, which include dysferlin, EHD1, EHD2, and MG53, are needed for efficient repair in the absence of dysferlin and the related protein myoferlin, there is significant delay in resealing. There is an interdependency of the subcomplexes within the repair complex, because dominant-negative annexin A6 was capable of disrupting recruitment of dysferlin to the shoulder. It should be appreciated that one limitation of this method is its reliance on overexpression of fluorescently tagged proteins. Either of these factors (overexpression or protein tagging) may interfere with normal translocation and function. However, we note that each of these tagged proteins assumed their normal intracellular location in uninjured myofibers and that electroporated myofibers demonstrated no gross structural abnormalities caused by electroporation or expression.
Figure 10.
Model of membrane repair. Upon plasma membrane injury, repair proteins localize to the site of damage and actin reorganizes, facilitating membrane repair. Annexins A1, A2, and A6 form a repair cap at the membrane lesion. Dysferlin (DYSF), MG53, BIN1, and EHD participate in forming a shoulder that abuts this repair cap. PIP2 and PS localize adjacent to the repair cap at the shoulder.
Several of these proteins, including dysferlin and annexin, are known to interact with negatively charged phospholipids, including PS (Gerke et al., 2005; Therrien et al., 2009). Using a sensor for PS binding, PS was enriched at the shoulder and temporally slightly after annexin A6, suggesting that phospholipid signaling is important for the coordination of shoulder proteins. We cannot exclude that phospholipid signaling is necessary within the annexin repair cap because the PH domain sensor may be excluded from this protein complex. With the influx of Ca2+ after membrane insult, PS is redistributed to the outer leaflet of the membrane (Williamson et al., 1992). This redistribution is thought to induce phagocytosis and inhibit the inflammatory response (Ramos et al., 2007). “Flipases” and “scramblases” are thought to promote such lipid redistribution, but this has not been definitively proven (Bevers and Williamson, 2010). Proteins from the anoctamin family have recently been evaluated for scramblase and reparative function, as loss-of-function mutations in anoctamin 5 result in muscular dystrophy in humans and membrane repair defects similar to the loss of dysferlin (Bolduc et al., 2010). Anoctamin 5 localizes to the site of membrane damage in HEK293 kidney and the CFBE lung cell line, implicating anoctamin 5 in the repair process (Tian et al., 2015). However, adeno-associated virus overexpression of anoctamin 5 in mice lacking dysferlin was not sufficient to rescue the membrane repair defects or the histopathology present within the dysferlin-null muscle, suggesting distinct roles for anoctamin 5 and dysferlin (Monjaret et al., 2013). Further studies elucidating the role of anoctamin 5 as a lipid regulator during muscle membrane repair are needed.
Several proteins implicated in membrane resealing are also implicated in muscle disease. Loss-of-function mutations in dysferlin lead to progressive muscular dystrophy (Bashir et al., 1998; Liu et al., 1998). Dysferlin localizes at the plasma membrane, where it was previously shown to be critical for the resealing process (Bansal et al., 2003). More recently, a role for dysferlin at the T-tubule has emerged (Klinge et al., 2010; Waddell et al., 2011; Kerr et al., 2013; Demonbreun et al., 2014). It is notable that proteins now implicated in the repair complex and its subdomains are normally found at or near the T-tubule. Despite the localization of these proteins in T-tubules, we did not observe direct recruitment from T-tubules to the plasma membrane during repair complex formation. Filamentous actin was enriched on cytoplasmic face in the annexin-clear zone. How such cytoskeletal organization supports vesicle-mediated resealing is not known. We did observe lateral recruitment of plasma membrane components to the repair complex similar to that reported by McDade et al. (McDade et al., 2014). Although it has been hypothesized that intracellular vesicles contribute membrane to the resealing process (Eddleman et al., 1997), we did not observe any evidence of intracellular vesicles participating in the resealing process.
Conservation of membrane repair
Maintaining cell integrity through membrane repair is a complex process required in the prevention of cellular death and disease. MG53, a TRIM family member, has been implicated in membrane repair, and loss of MG53 results in a mild myopathy in mouse models (Cai et al., 2009). MG53 targets the membrane during membrane repair localizing to the T-tubule in muscle and specifically binds to PS within the membrane. Unlike dysferlin and annexin, there is conflicting evidence on the requirement of Ca2+ in the recruitment of MG53 to the site of damage (Cai et al., 2009; Lek et al., 2013). MG53 oligomerizes and rapidly accumulates at the membrane, colocalizing with annexin A1, A5, and dysferlin (Cai et al., 2009; Waddell et al., 2011). Additionally, in muscle biopsies from human muscular dystrophy patients, including dysferlinopathy and Duchenne muscular dystrophy, MG53, dysferlin, and annexin A1 expression were up-regulated two to sixteen-fold compared with healthy control samples, suggesting a general up-regulation of repair proteins in muscle disease correlating with disease progression (Waddell et al., 2011). These results were replicated in muscle from the mdx mouse, a model of Duchenne muscular dystrophy. Membrane repair may be more essential to skeletal muscle as muscle undergoes contraction, which may induce sarcolemmal disruption and an increase in intracellular Ca2+ (McNeil and Khakee, 1992). However, membrane stress, chemical damage, and viral toxins create plasma membrane disruption in other cell types, resulting in the need for membrane repair in these cells. MG53 and annexins have been shown to regulate membrane repair in nonmuscle tissues, including heart, lung, kidney, MCF7, and HeLa cancer cells (Bouter et al., 2011; He et al., 2012; Jaiswal et al., 2014; Jia et al., 2014; Duann et al., 2015; Liu et al., 2015). This suggests the mechanism of membrane repair across tissues may be conserved. Understanding the process of membrane repair in the highly organized myofiber will provide a greater understanding of the repair process across numerous tissues and could potentially provide therapeutic benefit in several diseases.
Materials and methods
Animals
Mice were housed in a specific pathogen–free facility in accordance with the University of Chicago and Northwestern’s Institutional Animal Care and Use Committee regulations. Wild-type mice from the 129T2/SvEmsJ background were used for all studies unless otherwise noted. Dysf-null mice carrying the naturally occurring retrotransposon insertion in intron 4 found in the AJ strain housed at The Jackson Laboratory were bred into the 129T2/SvEmsJ background for more than six generations (Demonbreun et al., 2011). Myof-null mice were previously described and were bred into the 129Sv/EmsJ background for more than six generations (Doherty et al., 2005). Double-null mice (FER) were generated through crossing Myof-null and Dysf-null 129T2/SVEMsJ mice as previously described (Demonbreun et al., 2014). Male and female mice between the ages of 2 and 3 mo were used for studies.
Plasmids
Full-length EHD1 cDNA was previously described (Posey et al., 2014). Full-length EHD2-GFP was generated as described elsewhere (Doherty et al., 2008). Annexin A6–GFP and ANXA6N32 were generated as described previously (Swaggart et al., 2014). Annexin A6–mCherry and ANXA6N32-mCherry plasmids were generated by replacing the GFP tag with mCherry using BamHI and XhoI restriction sites. pEGFPC1-muscle amphiphysin II (BIN1 variant 8), GFP-C1-PLCΔ-PH, Lact-C2-GFP, pLifeAct-mTurquise2, pEGFP-N1 α-actinin1, and dysferlin-Venus were purchased from Addgene. Carboxy-terminal annexin A1-GFP, annexin A2-GFP, annexin A11-GFP, and MG53-GFP were obtained from OriGene.
Electroporation
Flexor digitorum brevis fibers were transfected by in vivo electroporation. Modifications to methods originally described in DiFranco et al. (2009) are as described below. In brief, the hindlimb footpad was injected with 10 µl hyaluronidase (8 U, H4272; Sigma-Aldrich). 2 h after injection, up to 20 µl of 2 µg/µl endotoxin-free plasmid was injected into the footpad. Electroporation was conducted by applying 20 pulses, 20 ms in duration each, at 1 Hz, at 100 V/cm. Animals were allowed to recover for a minimum of 7 d and not more than 10 d after electroporation to avoid examining injured muscle and to allow sufficient time for plasmid expression (Kerr et al., 2013). Flexor digitorum brevis muscle was removed and individual myofibers were isolated and imaged (Demonbreun and McNally, 2015).
Myofiber isolation and laser damage
Fibers were dissected and laser damaged as described previously (Swaggart et al., 2014; Demonbreun and McNally, 2015). In brief, fibers were dissociated in 0.2% BSA plus collagenase type II (catalog number 17101; Thermo Fisher Scientific) for up to 90 min with intermittent trituration at 37 degrees in 10% CO2. Fibers were then moved to Ringers solution and placed on MatTek confocal microscopy dishes (P35G-1.5-14-C; MatTek Corporation). After 1 h, fibers were adhered and the Ringers was replaced with 1 ml fresh Ringers solution. FM4-64 dye (T-13320; Molecular Probes) or FM1-43 (T-35356; Molecular Probes) was added to a final concentration of 2.5 µm before imaging.
Fibers were irradiated at room temperature using ana SP5 2 photon microscope (Leica Biosystems) equipped with a 63× 1.4 NA objective and LAS AF imaging software (Leica Biosystems) in the FRAP wizard protocol using a 405-nm laser using a Bleach Point for up to 5 s at 80% power. Images were acquired as follows: one image was acquired before damage (preinjury), one image upon laser damage (0 s), 10 images every 2 s after damage, and then one image every 10 s for 14 images. Alternatively, imaging and ablation was performed on the Nikon A1R laser scanning confocal equipped with GaSP detectors through a 60× Apo lambda 1.4-NA objective driven by Nikon Elements AR software. We ablated a single pixel set as 120 nm (0.0144 µm2) using the 405-nm laser at 100% power for up to 5 s. Images were acquired identically to that described for the Leica Biosystems microscope. Long-term imaging of annexin A6 was performed on the Nikon A1R as described previously in the presence of 2.5 µm FM4-64 with modifications as follows: one image was acquired preinjury and one image upon ablation (time 0 s), every 1 min for 6 min, and then every 5 min for 40 min (n = 3 myofibers from n = 3 mice). Single images were compiled into AVIs using Fiji displayed at seven frames per second.
z-stack projections were acquired from ∼35 consecutive acquisitions with 150-nm interval between each step, using the z-stack rendering built-in tool in Nis-elements AR (Nikon) or ImageJ. FRAP was performed using tagged annexin proteins on images identically acquired in a 5-µm2 region. FRAP measurements were calculated on individual myofibers using ImageJ. Averages were calculated using Prism GraphPad, and values were normalized to the prebleach intensity. Relative fluorescence within the lesion was calculated from images acquired as described above normalizing each frame to the prebleach intensity. Comparative measurements of translocation timing were compared pairwise against annexin A6 and individual analyses on images acquired as described above. All measurements were acquired from myofibers isolated from at least n = 3 mice and n ≥ 7 myofibers per condition, and error bars represent the standard error of the mean. Statistical analysis used Prism GraphPad and a two-way analysis of variance.
Quality control for myofibers selected for laser ablation was based several characteristics. Globally, only myofibers adherent to the MatTek dish from end to end were used and this was to prevent movement during and after laser ablation. Imaged fibers were required to have intact sarcomeres visible in brightfield, and the sarcolemma itself was required to be devoid of tears or ruptures induced during the isolation protocol. Fibers were also selected based on having an intermediate level of fluorescent protein expression that extended throughout the myofiber and have minimal to no visible protein aggregation. Fibers with autofluorescence were not used. The region of the myofiber selected for damage was required to be fully linear without visible deformation, including twists or bends in the membrane. Laser ablation was applied to areas without nuclei. On average, myofibers were 400 µm in length and 30 µm wide. However, a range of myofiber dimensions was present in each isolation.
For quantitative analysis of FM dye, fluorescence was measured at the site of injury in individual frames using ImageJ and adjusted to baseline fluorescence at time 0 calculated at the membrane before damage (F/F0). This method allows comparisons of all strains and reduces variability introduced by differences in dye uptake or binding at time 0. Fibers that severely bent during the damage process were excluded from the analysis, as the bending of membrane at the site of damage would falsely increase the dye measurement at the zone of interest. Statistics were performed using Prism GraphPad using a one-way or two-way analysis of variance. Quantitative analysis of dysferlin-Venus fluorescence at the shoulder was calculated at 150 s after damage. Fluorescent intensity was measured using ImageJ. Fluorescent intensity on both sides of the shoulder was measured and averaged. The mean shoulder fluorescence was then divided by the mean fluorescence of two points of sarcolemma. Statistical analysis used Prism GraphPad and an unpaired t test.
To determine the effect of actin inhibition, fibers were incubated with 5 µM latrunculin in Ringers solution loaded simultaneously with 2.5 µm FM4-64 before imaging. To determine the effect of Ca2+, fibers were incubated in Ca2+-free Ringers solution with 10 mM EGTA loaded simultaneously with 2.5 µm FM4-64 before imaging. Fibers were then subjected to the laser injury and imaging protocols. Movies were generated in ImageJ. Statistical analysis used Prism GraphPad and an unpaired t test.
Online supplemental material
Fig. S1 depicts domains of proteins as well as their intracellular localization after electroporation of plasmids in muscle. Fig. S2 demonstrates minimal local sarcomere disruption after laser-induced microinjury. Fig. S3 depicts captured images taken 40 min after laser injury when annexin A6 repair caps remained visible. Video 1 demonstrates an annexin A6 repair cap at the site of damage. Video 2 reveals lateral movement of membrane toward the site of sarcolemmal injury (red cluster moving in membrane). Video 3 shows that the repair cap remained distinct from the shoulder during repair, with annexin A6 at the repair cap (red) and EHD2 at the shoulder region (green). Video 4 shows F-actin (green) localized to the annexin A6 (red)–free zone. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201512022/DC1.
Supplementary Material
Acknowledgments
We acknowledge the outstanding support of Dr. Vytas Bindokas at the Integrated Microscopy Facility at the University of Chicago and Dr. Constadina Arvanitis at the Center for Advanced Microscopy at Northwestern University.
This work was supported by National Institutes of Health grants NS047726 and AR052646.
The authors declare no competing financial interests.
Footnotes
Abbreviations used in this paper:
- EHD
- Eps15 homology domain
- PIP2
- phosphatidylinositol 4,5-bisphosphate
- PS
- phosphatidylserine
- T-tubule
- transverse tubule
References
- Bansal D., Miyake K., Vogel S.S., Groh S., Chen C.C., Williamson R., McNeil P.L., and Campbell K.P.. 2003. Defective membrane repair in dysferlin-deficient muscular dystrophy. Nature. 423:168–172. 10.1038/nature01573 [DOI] [PubMed] [Google Scholar]
- Bashir R., Britton S., Strachan T., Keers S., Vafiadaki E., Lako M., Richard I., Marchand S., Bourg N., Argov Z., et al. 1998. A gene related to Caenorhabditis elegans spermatogenesis factor fer-1 is mutated in limb-girdle muscular dystrophy type 2B. Nat. Genet. 20:37–42. 10.1038/1689 [DOI] [PubMed] [Google Scholar]
- Bevers E.M., and Williamson P.L.. 2010. Phospholipid scramblase: an update. FEBS Lett. 584:2724–2730. 10.1016/j.febslet.2010.03.020 [DOI] [PubMed] [Google Scholar]
- Bolduc V., Marlow G., Boycott K.M., Saleki K., Inoue H., Kroon J., Itakura M., Robitaille Y., Parent L., Baas F., et al. 2010. Recessive mutations in the putative calcium-activated chloride channel Anoctamin 5 cause proximal LGMD2L and distal MMD3 muscular dystrophies. Am. J. Hum. Genet. 86:213–221. 10.1016/j.ajhg.2009.12.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouter A., Gounou C., Bérat R., Tan S., Gallois B., Granier T., d’Estaintot B.L., Pöschl E., Brachvogel B., and Brisson A.R.. 2011. Annexin-A5 assembled into two-dimensional arrays promotes cell membrane repair. Nat. Commun. 2:270 10.1038/ncomms1270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cagliani R., Magri F., Toscano A., Merlini L., Fortunato F., Lamperti C., Rodolico C., Prelle A., Sironi M., Aguennouz M., et al. 2005. Mutation finding in patients with dysferlin deficiency and role of the dysferlin interacting proteins annexin A1 and A2 in muscular dystrophies. Hum. Mutat. 26:283 10.1002/humu.9364 [DOI] [PubMed] [Google Scholar]
- Cai C., Masumiya H., Weisleder N., Matsuda N., Nishi M., Hwang M., Ko J.K., Lin P., Thornton A., Zhao X., et al. 2009. MG53 nucleates assembly of cell membrane repair machinery. Nat. Cell Biol. 11:56–64. 10.1038/ncb1812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornely R., Rentero C., Enrich C., Grewal T., and Gaus K.. 2011. Annexin A6 is an organizer of membrane microdomains to regulate receptor localization and signalling. IUBMB Life. 63:1009–1017. 10.1002/iub.540 [DOI] [PubMed] [Google Scholar]
- Davis D.B., Delmonte A.J., Ly C.T., and McNally E.M.. 2000. Myoferlin, a candidate gene and potential modifier of muscular dystrophy. Hum. Mol. Genet. 9:217–226. 10.1093/hmg/9.2.217 [DOI] [PubMed] [Google Scholar]
- Davis D.B., Doherty K.R., Delmonte A.J., and McNally E.M.. 2002. Calcium-sensitive phospholipid binding properties of normal and mutant ferlin C2 domains. J. Biol. Chem. 277:22883–22888. 10.1074/jbc.M201858200 [DOI] [PubMed] [Google Scholar]
- Defour A., Van der Meulen J.H., Bhat R., Bigot A., Bashir R., Nagaraju K., and Jaiswal J.K.. 2014. Dysferlin regulates cell membrane repair by facilitating injury-triggered acid sphingomyelinase secretion. Cell Death Dis. 5:e1306 10.1038/cddis.2014.272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demonbreun A.R., and McNally E.M.. 2015. DNA electroporation, isolation and imaging of myofibers. J. Vis. Exp. 106:e53551 10.3791/53551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demonbreun A.R., Posey A.D., Heretis K., Swaggart K.A., Earley J.U., Pytel P., and McNally E.M.. 2010. Myoferlin is required for insulin-like growth factor response and muscle growth. FASEB J. 24:1284–1295. 10.1096/fj.09-136309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demonbreun A.R., Fahrenbach J.P., Deveaux K., Earley J.U., Pytel P., and McNally E.M.. 2011. Impaired muscle growth and response to insulin-like growth factor 1 in dysferlin-mediated muscular dystrophy. Hum. Mol. Genet. 20:779–789. 10.1093/hmg/ddq522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demonbreun A.R., Rossi A.E., Alvarez M.G., Swanson K.E., Deveaux H.K., Earley J.U., Hadhazy M., Vohra R., Walter G.A., Pytel P., and McNally E.M.. 2014. Dysferlin and myoferlin regulate transverse tubule formation and glycerol sensitivity. Am. J. Pathol. 184:248–259. 10.1016/j.ajpath.2013.09.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiFranco M., Quinonez M., Capote J., and Vergara J.. 2009. DNA transfection of mammalian skeletal muscles using in vivo electroporation. J. Vis. Exp. 32:1520 10.3791/1520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doherty K.R., Cave A., Davis D.B., Delmonte A.J., Posey A., Earley J.U., Hadhazy M., and McNally E.M.. 2005. Normal myoblast fusion requires myoferlin. Development. 132:5565–5575. 10.1242/dev.02155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doherty K.R., Demonbreun A.R., Wallace G.Q., Cave A., Posey A.D., Heretis K., Pytel P., and McNally E.M.. 2008. The endocytic recycling protein EHD2 interacts with myoferlin to regulate myoblast fusion. J. Biol. Chem. 283:20252–20260. 10.1074/jbc.M802306200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duann P., Li H., Lin P., Tan T., Wang Z., Chen K., Zhou X., Gumpper K., Zhu H., Ludwig T., et al. 2015. MG53-mediated cell membrane repair protects against acute kidney injury. Sci. Transl. Med. 7:279ra36 10.1126/scitranslmed.3010755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddleman C.S., Ballinger M.L., Smyers M.E., Godell C.M., Fishman H.M., and Bittner G.D.. 1997. Repair of plasmalemmal lesions by vesicles. Proc. Natl. Acad. Sci. USA. 94:4745–4750. 10.1073/pnas.94.9.4745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enrich C., Rentero C., de Muga S.V., Reverter M., Mulay V., Wood P., Koese M., and Grewal T.. 2011. Annexin A6-Linking Ca(2+) signaling with cholesterol transport. Biochim. Biophys. Acta. 1813:935–947. 10.1016/j.bbamcr.2010.09.015 [DOI] [PubMed] [Google Scholar]
- Gerke V., Creutz C.E., and Moss S.E.. 2005. Annexins: linking Ca2+ signalling to membrane dynamics. Nat. Rev. Mol. Cell Biol. 6:449–461. 10.1038/nrm1661 [DOI] [PubMed] [Google Scholar]
- Hayes M.J., Rescher U., Gerke V., and Moss S.E.. 2004. Annexin-actin interactions. Traffic. 5:571–576. 10.1111/j.1600-0854.2004.00210.x [DOI] [PubMed] [Google Scholar]
- He B., Tang R.H., Weisleder N., Xiao B., Yuan Z., Cai C., Zhu H., Lin P., Qiao C., Li J., et al. 2012. Enhancing muscle membrane repair by gene delivery of MG53 ameliorates muscular dystrophy and heart failure in δ-Sarcoglycan-deficient hamsters. Mol. Ther. 20:727–735. 10.1038/mt.2012.5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoque M., Rentero C., Cairns R., Tebar F., Enrich C., and Grewal T.. 2014. Annexins - scaffolds modulating PKC localization and signaling. Cell. Signal. 26:1213–1225. 10.1016/j.cellsig.2014.02.012 [DOI] [PubMed] [Google Scholar]
- Jaiswal J.K., Lauritzen S.P., Scheffer L., Sakaguchi M., Bunkenborg J., Simon S.M., Kallunki T., Jäättelä M., and Nylandsted J.. 2014. S100A11 is required for efficient plasma membrane repair and survival of invasive cancer cells. Nat. Commun. 5:3795 10.1038/ncomms4795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia Y., Chen K., Lin P., Lieber G., Nishi M., Yan R., Wang Z., Yao Y., Li Y., Whitson B.A., et al. 2014. Treatment of acute lung injury by targeting MG53-mediated cell membrane repair. Nat. Commun. 5:4387 10.1038/ncomms5387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerr J.P., Ziman A.P., Mueller A.L., Muriel J.M., Kleinhans-Welte E., Gumerson J.D., Vogel S.S., Ward C.W., Roche J.A., and Bloch R.J.. 2013. Dysferlin stabilizes stress-induced Ca2+ signaling in the transverse tubule membrane. Proc. Natl. Acad. Sci. USA. 110:20831–20836. 10.1073/pnas.1307960110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klinge L., Harris J., Sewry C., Charlton R., Anderson L., Laval S., Chiu Y.H., Hornsey M., Straub V., Barresi R., et al. 2010. Dysferlin associates with the developing T-tubule system in rodent and human skeletal muscle. Muscle Nerve. 41:166–173. 10.1002/mus.21166 [DOI] [PubMed] [Google Scholar]
- Lek A., Evesson F.J., Lemckert F.A., Redpath G.M., Lueders A.K., Turnbull L., Whitchurch C.B., North K.N., and Cooper S.T.. 2013. Calpains, cleaved mini-dysferlinC72, and L-type channels underpin calcium-dependent muscle membrane repair. J. Neurosci. 33:5085–5094. 10.1523/JNEUROSCI.3560-12.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lennon N.J., Kho A., Bacskai B.J., Perlmutter S.L., Hyman B.T., and Brown R.H. Jr. 2003. Dysferlin interacts with annexins A1 and A2 and mediates sarcolemmal wound-healing. J. Biol. Chem. 278:50466–50473. 10.1074/jbc.M307247200 [DOI] [PubMed] [Google Scholar]
- Leung C., Yu C., Lin M.I., Tognon C., and Bernatchez P.. 2013. Expression of myoferlin in human and murine carcinoma tumors: role in membrane repair, cell proliferation, and tumorigenesis. Am. J. Pathol. 182:1900–1909. 10.1016/j.ajpath.2013.01.041 [DOI] [PubMed] [Google Scholar]
- Liu J., Aoki M., Illa I., Wu C., Fardeau M., Angelini C., Serrano C., Urtizberea J.A., Hentati F., Hamida M.B., et al. 1998. Dysferlin, a novel skeletal muscle gene, is mutated in Miyoshi myopathy and limb girdle muscular dystrophy. Nat. Genet. 20:31–36. 10.1038/1682 [DOI] [PubMed] [Google Scholar]
- Liu J., Zhu H., Zheng Y., Xu Z., Li L., Tan T., Park K.H., Hou J., Zhang C., Li D., et al. 2015. Cardioprotection of recombinant human MG53 protein in a porcine model of ischemia and reperfusion injury. J. Mol. Cell. Cardiol. 80:10–19. 10.1016/j.yjmcc.2014.12.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lizarbe M.A., Barrasa J.I., Olmo N., Gavilanes F., and Turnay J.. 2013. Annexin-phospholipid interactions. Functional implications. Int. J. Mol. Sci. 14:2652–2683. 10.3390/ijms14022652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyon A.R., MacLeod K.T., Zhang Y., Garcia E., Kanda G.K., Lab M.J., Korchev Y.E., Harding S.E., and Gorelik J.. 2009. Loss of T-tubules and other changes to surface topography in ventricular myocytes from failing human and rat heart. Proc. Natl. Acad. Sci. USA. 106:6854–6859. 10.1073/pnas.0809777106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandato C.A., and Bement W.M.. 2001. Contraction and polymerization cooperate to assemble and close actomyosin rings around Xenopus oocyte wounds. J. Cell Biol. 154:785–797. 10.1083/jcb.200103105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marg A., Schoewel V., Timmel T., Schulze A., Shah C., Daumke O., and Spuler S.. 2012. Sarcolemmal repair is a slow process and includes EHD2. Traffic. 13:1286–1294. 10.1111/j.1600-0854.2012.01386.x [DOI] [PubMed] [Google Scholar]
- McDade J.R., Archambeau A., and Michele D.E.. 2014. Rapid actin-cytoskeleton-dependent recruitment of plasma membrane-derived dysferlin at wounds is critical for muscle membrane repair. FASEB J. 28:3660–3670. 10.1096/fj.14-250191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNeil P.L., and Khakee R.. 1992. Disruptions of muscle fiber plasma membranes. Role in exercise-induced damage. Am. J. Pathol. 140:1097–1109. [PMC free article] [PubMed] [Google Scholar]
- McNeil P.L., and Steinhardt R.A.. 1997. Loss, restoration, and maintenance of plasma membrane integrity. J. Cell Biol. 137:1–4. 10.1083/jcb.137.1.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNeil P.L., Miyake K., and Vogel S.S.. 2003. The endomembrane requirement for cell surface repair. Proc. Natl. Acad. Sci. USA. 100:4592–4597. 10.1073/pnas.0736739100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monjaret F., Suel-Petat L., Bourg-Alibert N., Vihola A., Marchand S., Roudaut C., Gicquel E., Udd B., Richard I., and Charton K.. 2013. The phenotype of dysferlin-deficient mice is not rescued by adeno-associated virus-mediated transfer of anoctamin 5. Hum. Gene Ther. Clin. Dev. 24:65–76. 10.1089/humc.2012.217 [DOI] [PubMed] [Google Scholar]
- Posey A.D. Jr., Pytel P., Gardikiotes K., Demonbreun A.R., Rainey M., George M., Band H., and McNally E.M.. 2011. Endocytic recycling proteins EHD1 and EHD2 interact with fer-1-like-5 (Fer1L5) and mediate myoblast fusion. J. Biol. Chem. 286:7379–7388. 10.1074/jbc.M110.157222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posey A.D. Jr., Swanson K.E., Alvarez M.G., Krishnan S., Earley J.U., Band H., Pytel P., McNally E.M., and Demonbreun A.R.. 2014. EHD1 mediates vesicle trafficking required for normal muscle growth and transverse tubule development. Dev. Biol. 387:179–190. 10.1016/j.ydbio.2014.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potez S., Luginbühl M., Monastyrskaya K., Hostettler A., Draeger A., and Babiychuk E.B.. 2011. Tailored protection against plasmalemmal injury by annexins with different Ca2+ sensitivities. J. Biol. Chem. 286:17982–17991. 10.1074/jbc.M110.187625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos G.C., Fernandes D., Charão C.T., Souza D.G., Teixeira M.M., and Assreuy J.. 2007. Apoptotic mimicry: phosphatidylserine liposomes reduce inflammation through activation of peroxisome proliferator-activated receptors (PPARs) in vivo. Br. J. Pharmacol. 151:844–850. 10.1038/sj.bjp.0707302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen I., Pedersen L.H., Byg L., Suzuki K., Sumimoto H., and Vilhardt F.. 2010. Effects of F/G-actin ratio and actin turn-over rate on NADPH oxidase activity in microglia. BMC Immunol. 11:44 10.1186/1471-2172-11-44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roostalu U., and Strähle U.. 2012. In vivo imaging of molecular interactions at damaged sarcolemma. Dev. Cell. 22:515–529. 10.1016/j.devcel.2011.12.008 [DOI] [PubMed] [Google Scholar]
- Stauffer T.P., Ahn S., and Meyer T.. 1998. Receptor-induced transient reduction in plasma membrane PtdIns(4,5)P2 concentration monitored in living cells. Curr. Biol. 8:343–346. 10.1016/S0960-9822(98)70135-6 [DOI] [PubMed] [Google Scholar]
- Swaggart K.A., Demonbreun A.R., Vo A.H., Swanson K.E., Kim E.Y., Fahrenbach J.P., Holley-Cuthrell J., Eskin A., Chen Z., Squire K., et al. 2014. Annexin A6 modifies muscular dystrophy by mediating sarcolemmal repair. Proc. Natl. Acad. Sci. USA. 111:6004–6009. 10.1073/pnas.1324242111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Therrien C., Di Fulvio S., Pickles S., and Sinnreich M.. 2009. Characterization of lipid binding specificities of dysferlin C2 domains reveals novel interactions with phosphoinositides. Biochemistry. 48:2377–2384. 10.1021/bi802242r [DOI] [PubMed] [Google Scholar]
- Tian Y.W.J., Cebotaru L., Wang H., and Guggino W.B.. 2015. Anoctamin5 is related to plasma membrane repair. JSM Regenerative Medicine & Bioengineering. 3:6. [Google Scholar]
- Waddell L.B., Lemckert F.A., Zheng X.F., Tran J., Evesson F.J., Hawkes J.M., Lek A., Street N.E., Lin P., Clarke N.F., et al. 2011. Dysferlin, annexin A1, and mitsugumin 53 are upregulated in muscular dystrophy and localize to longitudinal tubules of the T-system with stretch. J. Neuropathol. Exp. Neurol. 70:302–313. 10.1097/NEN.0b013e31821350b0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisleder N., Takizawa N., Lin P., Wang X., Cao C., Zhang Y., Tan T., Ferrante C., Zhu H., Chen P.J., et al. 2012. Recombinant MG53 protein modulates therapeutic cell membrane repair in treatment of muscular dystrophy. Sci. Transl. Med. 4:139ra85 10.1126/scitranslmed.3003921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson P., Kulick A., Zachowski A., Schlegel R.A., and Devaux P.F.. 1992. Ca2+ induces transbilayer redistribution of all major phospholipids in human erythrocytes. Biochemistry. 31:6355–6360. 10.1021/bi00142a027 [DOI] [PubMed] [Google Scholar]
- Yeung T., Heit B., Dubuisson J.F., Fairn G.D., Chiu B., Inman R., Kapus A., Swanson M., and Grinstein S.. 2009. Contribution of phosphatidylserine to membrane surface charge and protein targeting during phagosome maturation. J. Cell Biol. 185:917–928. 10.1083/jcb.200903020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zweifach A. 2000. FM1-43 reports plasma membrane phospholipid scrambling in T-lymphocytes. Biochem. J. 349:255–260. 10.1042/bj3490255 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.