Differentiation induces loss of centrosomal microtubule organizing activity in many cell types, though the underlying mechanisms are poorly understood. Using the epidermis, Muroyama et al. show that cell cycle exit causes loss of a Nedd1–γ-tubulin complex, which is required for anchoring microtubules at the centrosome. This defines a novel function for γ-tubulin complexes in microtubule anchoring at the centrosome.
Abstract
Differentiation induces the formation of noncentrosomal microtubule arrays in diverse tissues. The formation of these arrays requires loss of microtubule-organizing activity (MTOC) at the centrosome, but the mechanisms regulating this transition remain largely unexplored. Here, we use the robust loss of centrosomal MTOC activity in the epidermis to identify two pools of γ-tubulin that are biochemically and functionally distinct and differentially regulated. Nucleation-competent CDK5RAP2–γ-tubulin complexes were maintained at centrosomes upon initial epidermal differentiation. In contrast, Nedd1–γ-tubulin complexes did not promote nucleation but were required for anchoring of microtubules, a previously uncharacterized activity for this complex. Cell cycle exit specifically triggered loss of Nedd1–γ-tubulin complexes, providing a mechanistic link connecting MTOC activity and differentiation. Collectively, our studies demonstrate that distinct γ-tubulin complexes regulate different microtubule behaviors at the centrosome and show that differential regulation of these complexes drives loss of centrosomal MTOC activity.
Introduction
In most proliferative cells, the centrosome acts as the primary microtubule-organizing center (MTOC). Although it has been long appreciated that differentiation induces formation of noncentrosomal microtubule (MT) arrays in many tissues and cell types, including epithelium, neurons, and muscle, the mechanisms controlling inactivation of the centrosome during this process remain poorly characterized (Müsch, 2004; Bartolini and Gundersen, 2006; Srsen et al., 2009; Brodu et al., 2010; Nguyen et al., 2011; Feldman and Priess, 2012). In the proliferative basal cells of the mammalian epidermis, MTs are organized by the centrosome (Lechler and Fuchs, 2007). When these cells differentiate, MTs are no longer associated with the centrosome and instead are recruited to the cell cortex. Neither the molecular mechanism underlying loss of MTOC activity at the centrosome nor the specific signaling pathway that regulates this transition is known.
Centrosomal MTOC activity requires both MT nucleation and minus-end anchoring (Dammermann et al., 2003). Although previous work has identified several mechanisms that regulate MT nucleation, the molecular mechanisms underlying anchoring are just beginning to be elucidated. In some cell types, centrosomal subdistal appendages appear to be the preferred site for MT anchoring (Chrétien et al., 1997; Mogensen et al., 2000; Delgehyr et al., 2005; Guo et al., 2006; Ibi et al., 2011). In other cell types, however, loss of subdistal appendages does not affect centrosomal MTOC activity, and MTs appear to be more broadly anchored in the pericentriolar material (PCM) by unknown means (Ishikawa et al., 2005).
γ-Tubulin is a prominent component of the PCM and exists in two major complexes: the γ-tubulin small complex (γ-TuSC) and γ-tubulin ring complex (γ-TuRC). γ-TuRCs are the major MT nucleators at the centrosome, and they have also been proposed to play roles in minus-end capping (Moritz et al., 1995; Zheng et al., 1995; Wiese and Zheng, 2000; Anders and Sawin, 2011), but they have not been implicated in anchoring MTs at the centrosome. In addition to the core γ-TuRC components (GCP2-6), other γ-TuRC accessory factors such as Nedd1 and CDK5RAP2 have been more recently identified (Haren et al., 2006; Lüders et al., 2006; Fong et al., 2008; Choi et al., 2010). These proteins have been suggested to play roles in γ-tubulin recruitment to the centrosome, but these effects may be species and/or cell type dependent. For example, Nedd1 was originally shown to be necessary for γ-tubulin localization to centrosomes in human cancer cell lines but was not required for centrosomal γ-tubulin recruitment in Xenopus laevis or Drosophila melanogaster (Liu and Wiese, 2008; Zeng et al., 2009; Manning et al., 2010a; Reschen et al., 2012).
The presence of these accessory factors suggests that there may be biochemical heterogeneity of γ-TuRCs. However, whether different γ-TuRCs have distinct functions (e.g., nucleation versus minus-end anchoring) has not been addressed. CDK5RAP2 has been demonstrated to promote γ-TuRC’s MT nucleation activity in vitro (Choi et al., 2010). Although direct analysis of the effects of Nedd1 on γ-TuRC nucleation activity has not been reported, several studies have suggested that Nedd1 is required for centrosomal microtubule nucleation in interphase and in mitosis (Haren et al., 2006; Lüders et al., 2006; Gomez-Ferreria et al., 2012; Pinyol et al., 2013; Walia et al., 2014).
In this study, we report the isolation and identification of distinct γ-TuRCs from keratinocytes and show that these complexes are lost from centrosomes with different kinetics over the course of epidermal differentiation. CDK5RAP2–γ-TuRCs, which we demonstrate are potent MT nucleators in vivo, are maintained at centrosomes over the initial steps of differentiation. In contrast, Nedd1–γ-TuRCs do not nucleate MTs either in vitro or in vivo but are required for MT anchoring and are rapidly delocalized from centrosomes after cell cycle exit. Together, this work reveals that γ-TuRCs with separable functions exist in cells and elucidates a mechanism whereby MTOC activity at the centrosome is lost during tissue differentiation in mammals.
Results
Centrosomes intrinsically lose MTOC activity upon epidermal differentiation
Epidermal differentiation is associated with the reorganization of MTs from centrosomal to cortical arrays (Lechler and Fuchs, 2007; Sumigray et al., 2011, 2012). To ask whether these changes reflect intrinsic centrosomal changes or are caused by competition from a new cellular MTOC, we developed a protocol to purify centrosomes from proliferative and differentiated keratinocytes. A robust keratin network normally hinders purification of a pure centrosome pool from wild-type (WT) keratinocytes; we successfully circumvented this problem by isolating centrosomes from a keratinocyte line lacking keratin filaments, which was derived from mice in which the type II keratin locus was deleted (Seltmann et al., 2013; Fig. 1, A and B). As discussed in detail below, keratin-null cells exhibit similar centrosomal changes upon differentiation as WT cells (Fig. S1, A and B). To our knowledge, this is the first study to successfully isolate centrosomes from a defined cell population from different stages of differentiation.
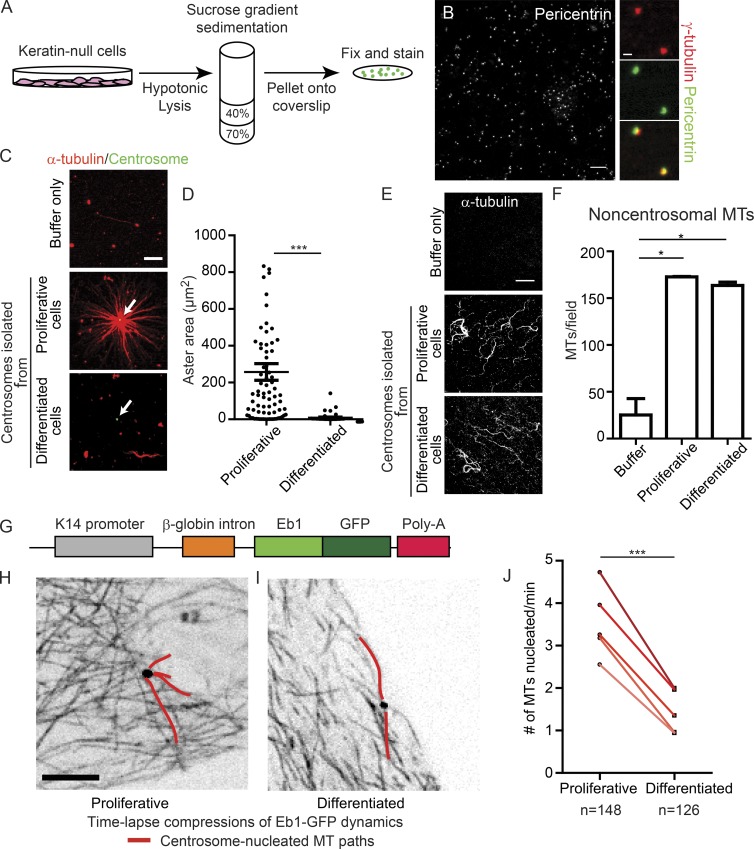
Figure 1.
Differentiated centrosomes lose MT-anchoring activity and have decreased nucleation activity. (A) Schematic of centrosome isolation from cultured keratin-null cells. (B) Field of isolated centrosomes stained for pericentrin. Bar, 10 µm. Images on the right show colocalization of γ-tubulin and pericentrin in isolated centrosomal puncta. Bar, 1 µm. (C) Representative images of MT assembly assays in the presence of buffer only, centrosomes from proliferative cells, and centrosomes from differentiated cells. Bar, 10 µm. White arrows indicate centrosomes. (D) Quantification of aster area surrounding purified centrosomes from proliferative and differentiated cells (n ≥ 54 centrosomes from three independent experiments). (E) Representative images of fields of free (noncentrosomal) MTs nucleated in reactions as in C. Bar, 10 µm. (F) Quantification of free (noncentrosomal) MTs in reactions as in C (n ≥ 9 fields from two independent experiments). (G) Diagram of construct used to generate Eb1-GFP expressing mice. (H and I) Compressions of Eb1-GFP movies in proliferative (H) or differentiation (I) keratinocytes derived from the Eb1-GFP transgenic mouse. Bar, 10 µm. (J) Quantification of centrosomal nucleation rate in proliferative and differentiated primary keratinocytes. Five separate replicates of paired samples derived from five independent mice are shown (n ≥ 126 centrosomes). A paired design was used because proliferative and differentiated centrosomes can be measured within the same dish. n.s., not significant; *, P < 0.05; **, P < 0.01; ***, P < 0.001. Data are presented as mean ± SEM.
To assess whether differentiation induces centrosomes to lose intrinsic MTOC function, we performed in vitro MT nucleation assays with purified centrosomes (Mitchison and Kirschner, 1984). Although reactions with tubulin alone contained few microtubules (Fig. 1, C–F), robust MT asters were observed in reactions containing centrosomes isolated from proliferative keratinocytes. In contrast, centrosomes purified from differentiated keratinocytes did not form MT asters, demonstrating that centrosomes intrinsically lose the ability to act as MTOCs upon epidermal differentiation. Importantly, although these differentiated centrosomes did not act as MTOCs, they did promote the formation of free (noncentrosomal) MTs (Fig. 1, E and F), suggesting that they still promote MT nucleation but are defective in MT anchoring.
Although our data suggested that nucleation still occurs from differentiated centrosomes, this assay is only semiquantitative and does not allow direct visualization of microtubule nucleation. To directly visualize and measure MT nucleation at the centrosome in cells, we generated transgenic mice that express the MT plus-tip tracking protein Eb1 tagged to GFP under the keratin 14-promoter (Fig. 1 G; Vasioukhin et al., 1999). These mice are viable, have no detectable skin defects, and allowed us to visualize MT growth dynamics in both intact tissue and isolated primary keratinocytes. Furthermore, by crossing these mice to a line expressing centrin-GFP (Lechler and Fuchs, 2005), we were able to quantify microtubule growth from centrosomes.
To quantify MT nucleation, we isolated primary keratinocytes (Eb1-GFP; centrin-GFP) from neonatal mouse backskin. Primary keratinocytes form two distinct layers in culture—a proliferative basal layer and a differentiated suprabasal layer that exhibit identical centrosomal changes as intact tissue (Fig. S1, C and D)—allowing us to image microtubule dynamics in proliferative and differentiated cells in the same experiment. Because a bona fide marker of a newly nucleated MT has not been identified, we scored Eb1-GFP comets that initiated at the centrin-GFP–labeled centrosome as newly nucleated MTs, as previously reported (Piehl et al., 2004). In five independent experiments, we observed an approximately twofold decrease in MT nucleation from centrosomes in differentiated cells (Fig. 1, H–J; and Videos 1 and 2). This reduction in MT nucleation cannot account for the total loss of MTOC activity we observed in the in vitro MT nucleation assays, suggesting that differentiation triggers a dramatic loss of MT anchoring as well as a less dramatic reduction in centrosomal MT nucleation.
Centrosomal proteins are delocalized upon epidermal differentiation
Because we observed centrosome-intrinsic loss of MTOC activity, we next assessed the localization of several key centrosomal proteins during epidermal differentiation, which occurs in a stereotypical manner from inner basal cells to the outer cornified envelope. Quantification of centrin-GFP puncta in the epidermis demonstrated that centrioles were maintained throughout the entire differentiation process (unpublished data). By assaying localization of key PCM components, we found that the PCM scaffolding protein pericentrin showed a modest decrease in centrosomal levels (∼25%) in the transition from proliferative basal cell to the immediate suprabasal cell layer (Fig. 2 B). As differentiation progressed, there was continued loss of pericentrin from the centrosome (Fig. 2, B and E). In contrast to pericentrin, a large fraction of centrosomal γ-tubulin was delocalized in the first suprabasal layer of cells that had initiated differentiation (Fig. 2, A and E). Quantification of the immunofluorescence signal revealed that over 70% of γ-tubulin was lost from centrosomes as cells exited the basal layer (Fig. 2 E). The remaining centrosomal γ-tubulin, like pericentrin, continued to decrease as differentiation proceeded. Therefore, (1) delocalization from the centrosome appears to be a general feature of centrosomal proteins during differentiation, and (2) the kinetics of loss differ significantly in a protein-specific manner. Here, we focus on the centrosomal changes as proliferative progenitors initiate differentiation.
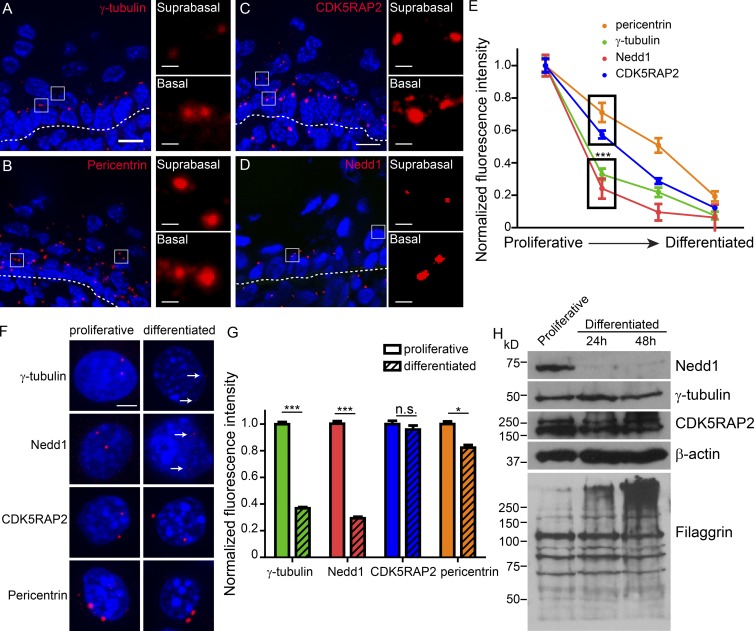
Figure 2.
Keratinocyte differentiation induces loss of centrosomal γ-tubulin and Nedd1. (A–D) Cryosections of neonatal mouse backskin were stained for centrosomal proteins, as indicated. Dashed lines represent the basement membrane separating underlying dermis from the epidermis. Insets of boxed regions show centrosomes at higher magnification. Bars: (main) 10 µm; (insets) 1 µm. (E) Quantification of fluorescence intensity of centrosomal proteins during epidermal differentiation. Proliferative indicates basal cells, the second time point is the first suprabasal cell layer, the third time point is upper spinous cells, and the fourth is granular cells. (F) Cultured mouse keratinocytes were grown under proliferative conditions or were induced to differentiate and then were stained for centrosomal proteins, as indicated. Arrows indicate low levels of residual centrosomal γ-tubulin and Nedd1 in differentiated keratinocytes. Bar, 5 µm. (G) Quantification of fluorescence intensity of centrosomal proteins in proliferative and differentiated cultured keratinocytes (n ≥ 280 centrosomes from three independent experiments). (H) Western blots of whole-cell lysates prepared from proliferative keratinocytes and keratinocytes at indicated time points after differentiation induction, blotted with antibodies as indicated. n.s., not significant; *, P < 0.05; ***, P < 0.001. Data are presented as mean ± SEM.
Differentiation induced similar changes at the centrosomes of cultured keratinocytes. Three days following differentiation induction, centrosomal pericentrin levels decreased only 20%, whereas γ-tubulin was much more dramatically lost (>60%; Fig. 2, F and G). Similar changes were also observed in keratin-null cells, centrosomes isolated from keratin null-cells, and isolated primary keratinocytes (Fig. S1, A–D). Therefore, both in intact epidermis and cultured keratinocytes, differentiation triggers a dramatic decrease in centrosomal γ-tubulin levels coincident with changes in centrosomal MTOC activity.
Distinct effects of differentiation on the localization of γ-tubulin interacting proteins
To identify potential upstream factors controlling γ-tubulin localization to centrosomes in keratinocytes, we assayed the localization of two proteins, Nedd1 and CDK5RAP2, which play roles in γ-tubulin localization to centrosomes in other cell lines (Haren et al., 2006; Lüders et al., 2006; Fong et al., 2008; Manning et al., 2010b). CDK5RAP2 localization to the centrosome was gradually decreased over epidermal differentiation (Fig. 2, C and E). In stark contrast, Nedd1 was abruptly lost from centrosomes upon differentiation both in intact epidermis and in cultured keratinocytes, similar to γ-tubulin (Fig. 2, D–G). These data suggested that a major pool of γ-tubulin requires Nedd1 for its centrosomal localization in keratinocytes and that this Nedd1–γ-tubulin pool is delocalized upon differentiation onset.
Assaying total protein levels in proliferative and differentiated keratinocytes revealed that the decrease in centrosomal γ-tubulin was caused by a relocalization to the cytoplasm, because total levels remained constant over the differentiation process (Fig. 2 H). Similarly, CDK5RAP2 protein levels remained unchanged (Fig. 2 H). Strikingly, total Nedd1 levels were dramatically reduced upon differentiation (Fig. 2 H). These data suggested that degradation of Nedd1 upon differentiation onset may be responsible for delocalizing a large pool of γ-tubulin.
To test whether Nedd1 was required to maintain γ-tubulin at centrosomes in keratinocytes, we used lentiviral-mediated knockdowns (KDs) to assess the relative contributions of CDK5RAP2 and Nedd1 to γ-tubulin centrosomal localization. At 96 h postinfection, centrosomal Nedd1 levels were reduced to ∼10% of WT levels (Fig. S2, A–C). Stable Nedd1 KD lines could not be maintained because of severe mitotic defects, as previously reported (Haren et al., 2006; Lüders et al., 2006; Fig. S2, E and F). Consistent with Nedd1 acting as the major recruiter of centrosomal γ-tubulin in keratinocytes, Nedd1 depletion severely reduced γ-tubulin localization to centrosomes without affecting CDK5RAP2 levels (Fig. 3, A and B; and Fig. S2 D). CDK5RAP2 depletion induced a small but statistically significant decrease in centrosomal γ-tubulin and Nedd1 levels (Fig. 3 B and Fig. S2, G and H). These data are consistent with a large fraction of γ-tubulin requiring Nedd1 for its centrosomal localization. Therefore, differentiation onset induces a profound loss of centrosomal Nedd1–γ-tubulin complexes coincident with loss of MT anchoring to centrosomes.
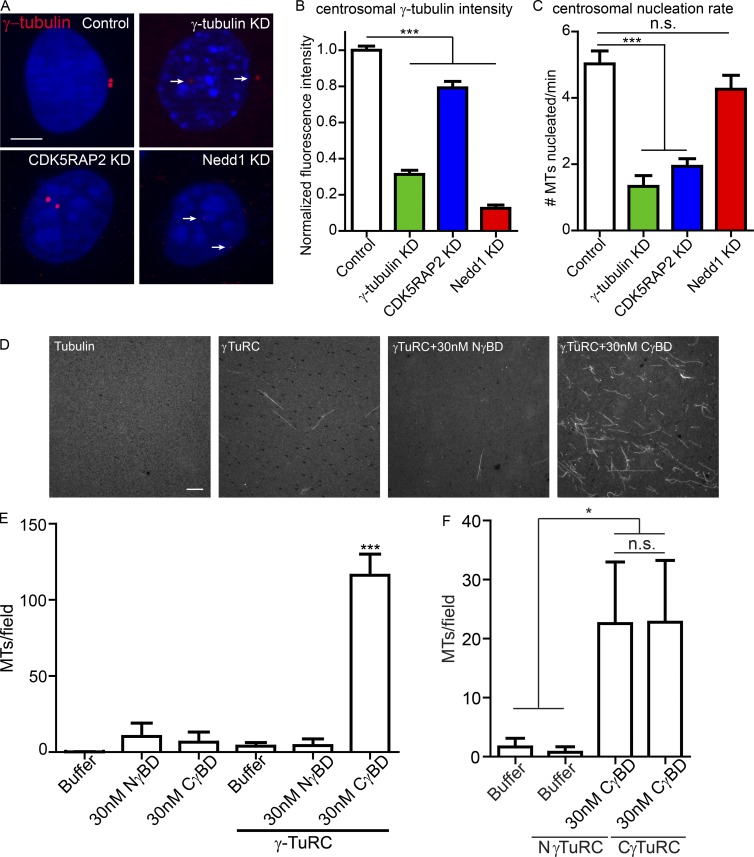
Figure 3.
Nedd1/γ-TuRC and CDK5RAP2/γ-TuRC have different properties in vivo and in vitro. (A) Images of γ-tubulin in control proliferative keratinocytes and those with lentiviral-mediated knockdown of γ-tubulin, CDK5RAP2, or Nedd1. Arrows indicate centrosomes. Bar, 5 µm. (B) Quantification of centrosomal γ-tubulin levels in control, γ-tubulin KD, CDK5RAP2 KD, and Nedd1 KD cells (n ≥ 70 centrosomes from at least two independently derived cell lines for each condition). (C) Centrosomal MT nucleation rate in control, γ-tubulin KD, CDK5RAP2 KD, and Nedd1 KD cells (n ≥ 18 cells from at least two independent experiments). (D) Representative images of fields from MT assembly assays with tubulin alone, γ-TuRC alone, or purified γ-TuRCs incubated with the γ-tubulin binding domain of either Nedd1 or CDK5RAP2. Bar, 10 µm. (E) Quantification of MTs per field in the MT assembly assays, as indicated (n = 40 total random fields for each condition from two independent experiments). Data are presented as mean ± SEM. (F) γ-TuRCs purified by affinity for Nedd1 (NγTuRCs) or CDK5RAP2 (CγTuRCs) were used in MT assembly assays with addition of the γ-tubulin binding domain of CDK5RAP2, as indicated. Quantification of MTs per field in the MT assembly assays, as indicated (n = 15 random fields for each condition from three independent experiments). n.s., not significant; *, P < 0.05; ***, P < 0.001. Data are presented as mean ± SD.
CDK5RAP2-bound and Nedd1-bound γ-TuRCs have different activities
Given the well-established function of γ-tubulin in MT nucleation and the dependence of centrosomal γ-tubulin levels on Nedd1, our results suggested that Nedd1 may be a major determinant of centrosomal MT nucleation activity. To test this, we began by assessing how much microtubule nucleation from the centrosome is γ-tubulin dependent. By using the Eb1-GFP/centrin-GFP assay described above, we found that lentiviral-mediated knockdown of γ-tubulin strongly decreased microtubule nucleation from the centrosome, clearly demonstrating that γ-tubulin is required for a least some of the microtubule nucleation at the centrosome (Fig. 3 C). However, because of our inability to completely deplete γ-tubulin from cells (Fig. 3 B), we are unable to conclude whether all microtubule nucleation from the centrosome requires γ-tubulin. Next, to test whether CDK5RAP2- and/or Nedd1-bound γ-tubulin is required for centrosomal microtubule nucleation, we measured Eb1-GFP growth from centrosomes in CDK5RAP2 KD and Nedd1 KD keratinocytes. Surprisingly, even though Nedd1 depletion was sufficient to cause significant γ-tubulin loss at centrosomes, the MT nucleation rate was not significantly decreased (Fig. 3, B and C). Conversely, CDK5RAP2 loss had a significant effect on microtubule nucleation from the centrosome despite having a smaller effect on γ-tubulin levels. These data indicate that the small pool of CDK5RAP2-bound γ-TuRCs at the centrosome is responsible for most MT nucleation.
Nedd1 and CDK5RAP2 form independent γ-TuRCs
These data demonstrate that CDK5RAP2 and Nedd1 differentially regulate γ-tubulin localization and centrosome nucleation activity. We next sought to further clarify the nature of these complexes. A GST fusion to the γ-tubulin binding domain of Nedd1 (GST-NγBD; Fig. S3 A) was able to pull down γ-tubulin and endogenous Nedd1, consistent with previous studies indicating that Nedd1 can oligomerize (Fig. S3 B; Manning et al., 2010b). No CDK5RAP2 was detected in GST-NγBD pull-downs. Additionally, a GST fusion to the γ-tubulin binding domain of CDK5RAP2 (GST-CγBD) could precipitate γ-tubulin, but not Nedd1 (Fig. S3 B).
Although both Nedd1 and CDK5RAP2 can directly bind to γ-TuRC, we found that in keratinocyte extracts a large fraction of Nedd1 cofractionated with γ-tubulin, whereas CDK5RAP2 did not (Fig. S3 C). We then isolated complexes that bound to the γ-tubulin binding domain of either Nedd1 or CDK5RAP2 by sucrose density gradient centrifugation. In both cases, the majority of associated γ-tubulin existed in a large complex, consistent with γ-TuRC, and we observed little to no interaction with γ-TuSC (Fig. S3 D). This was true both when keratinocyte or HEK293 extracts were used as the starting material and is consistent with an earlier study for CDK5RAP2 (Choi et al., 2010). Although they migrated similarly, it remained possible that these were compositionally distinct complexes.
To more sensitively assess the protein composition of Nedd1–γ-TuRCs, we then performed mass-spectrometry on NγBD-purified γ-TuRCs and identified peptides corresponding to all the core γ-TuRC components, as well as peptides corresponding to the recently identified MOZART1/2 and FME7 (Fig. S3 E). We did not identify any peptides in our sample corresponding to CDK5RAP2. Previous mass spectrometry analysis of CDK5RAP2-associated γ-TuRCs also identified all core γ-TuRC components, but, interestingly, not Nedd1 (Choi et al., 2010). In contrast, direct affinity isolation of γ-tubulin from cells identified both Nedd1 and CDK5RAP2 (Teixidó-Travesa et al., 2010). Therefore, our data are consistent with the existence of two distinct γ-TuRCs, one bound to CDK5RAP2 and one to Nedd1. However, it is important to note that this is in solution, and higher-order structures may form at the centrosome.
Nedd1 and CDK5RAP2 differentially affect γ-TuRC-mediated MT nucleation in vitro
We next tested whether CDK5RAP2 or Nedd1 could stimulate the MT nucleation activity of purified γ-TuRC. The affinity-purified γ-TuRCs were subjected to sucrose density gradient centrifugation, which removes most of the bound GST-tagged interacting protein (Choi et al., 2010). Because of loss of the Nedd1 and CDK5RAP2 recombinant fragments upon purification, we then supplemented the isolated γ-TuRCs with either GST-CγBD or GST-NγBD. Neither GST-CγBD nor GST-NγBD alone stimulated MT nucleation (Fig. 3, D and E). Addition of GST-CγBD resulted in an approximately eightfold increase in γ-TuRC nucleation activity, consistent with previous data (Fig. 3, D and E; Choi et al., 2010). This was true for both the γ-TuRCs purified by their affinity for either Nedd1 or CDK5RAP2 (Fig. 3, E and F), demonstrating that the complexes isolated by either binding partner are capable of being stimulated by CDK5RAP2. Interestingly, and in agreement with our live-imaging data from cultured keratinocytes, γ-TuRCs preincubated with GST-NγBD did not induce MT nucleation above levels seen with γ-TuRC alone (Fig. 3 E). Collectively, these data demonstrate that Nedd1–γ-TuRCs and CDK5RAP2–γ-TuRCs are functionally distinct and that Nedd1–γ-TuRCs are not potent MT nucleators in vitro.
CDK5RAP2–γ-TuRC and Nedd1–γ-TuRC have distinct activities in vivo
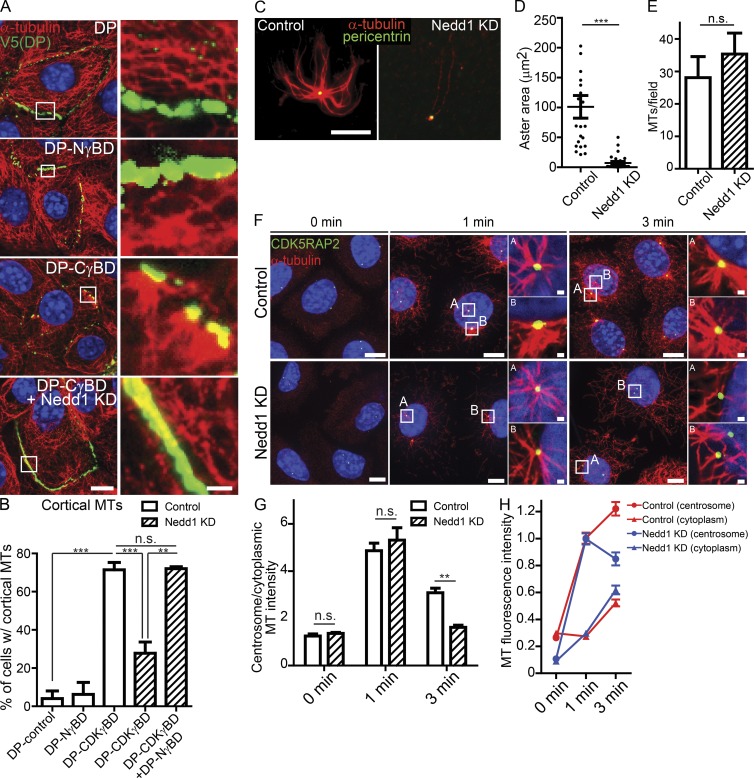
Analysis of protein function at the centrosome is confounded by the fact that nucleation, anchoring, and MT-dependent transport all occur at this site, making it difficult to specifically isolate the function of distinct complexes. We therefore developed a strategy to determine whether CDK5RAP2 and Nedd1 were sufficient for γ-TuRC recruitment and activity at a distinct cellular site. We fused the desmosome-targeting domain of desmoplakin (desmosomes are cell–cell adhesion structures at the cell cortex) to the Nedd1 or CDK5RAP2 γ-tubulin binding domain (DP-NγBD or DP-CγBD, respectively; Fig. 4 A). Transfection of desmoplakin-null keratinocytes (to eliminate desmoplakin-dependent effects on MTs; Lechler and Fuchs, 2007; Sumigray et al., 2011) with either DP-CγBD or DP-NγBD, but not with the desmosome-targeting domain of desmoplakin (DP) alone, resulted in γ-tubulin recruitment to desmosomes (Fig. 4 B). Thus, CDK5RAP2 and Nedd1 are each sufficient to recruit γ-tubulin in vivo. Fluorescence intensity measurements demonstrated that similar amounts of γ-tubulin were recruited to the cortex with either construct (Fig. 4 C). Consistent with our pull-down and mass-spectrometry data, DP-NγBD was able to recruit full-length Nedd1, but not CDK5RAP2, to the cortex (Fig. S4, A and B). Surprisingly, we found that DP-CγBD was able to recruit full-length Nedd1 to desmosomes (Fig. S4 A), suggesting that even though γ-TuRCs are isolated as distinct soluble complexes, higher-order structures containing both must form in cells. This is consistent with the loss of 30% of centrosomal Nedd1 upon CDK5RAP2 depletion (Fig. S2 H). To assess whether CDK5RAP2 is capable of recruiting γ-tubulin independent of Nedd1, we depleted endogenous Nedd1 from DP-CγBD-expressing cells; this did not significantly alter the levels of γ-tubulin at the cell cortex (Fig. 4, B and C). Thus, we have established an experimental system in which we can examine CDK5RAP2-bound γ-TuRCs, Nedd1-bound γ-TuRCs, or the combination of both and compare effects on MT nucleation and anchoring activity.
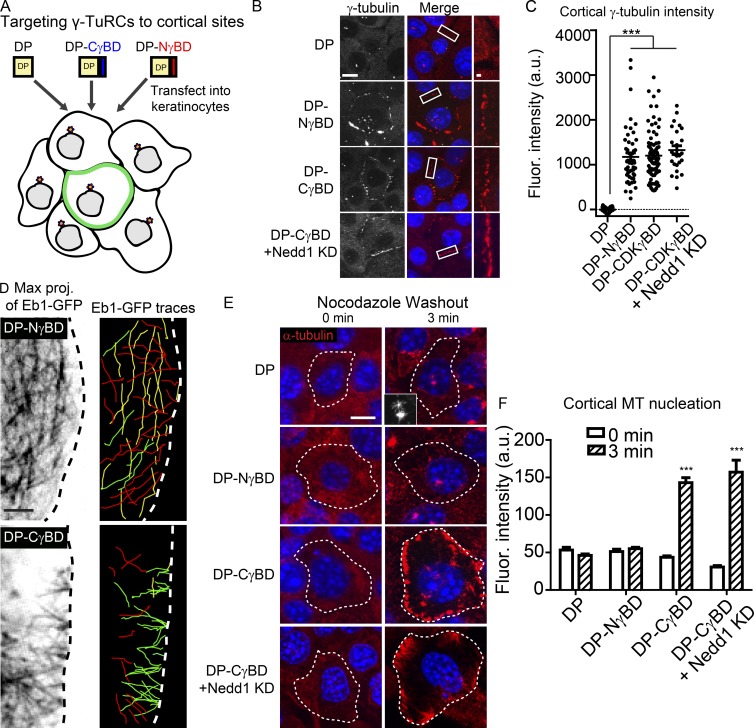
Figure 4.
CDK5RAP2, but not Nedd1, is sufficient to stimulate γ-TuRC–mediated MT nucleation in vivo. (A) Diagram of the experimental method. (B) γ-Tubulin localization in cells expressing DP alone, DP-NγBD, DP-CγBD, and DP-CγBD + Nedd1KD. Insets show zoomed in cortical regions. Bars: (main) 10 µm; (insets) 1 µm. (C) Quantification of cortical γ-tubulin levels in cells expressing DP, DP-NγBD, DP-CγBD (all n ≥ 50 cells from three independent experiments), and DP-CγBD + Nedd1KD (n = 25 cells from three independent experiments). (D) Compressions of movies (60 s) of GFP-Eb1 showing MT paths in cells expressing DP-NγBD or DP-CγBD (n ≥ 16 cells from three independent experiments). Images on the right are color coded by vectors of growth. Those growing toward the plasma membrane are red, those growing parallel to it are yellow, and those that are growing into the cytoplasm are green. (E) Representative images of transfected cells before and after nocodazole washout showing sites of MT nucleation. Inset shows centrosomal nucleation in control cells. Dotted line indicates the outline of the transfected cell. Bar, 10 µm. (F) Quantification of cortical α-tubulin intensity in control or DP-NγBD, DP-CγBD, or DP-CγBD + Nedd1KD cells 3 min after nocodazole washout (n = 23–92 cells from at least three independent experiments). ***, P < 0.001. Data are presented as mean ± SEM.
We first assayed how cortical CDK5RAP2 and Nedd1 complexes influence MT nucleation by imaging Eb1-GFP in cells transfected with either fusion construct under steady-state conditions. Consistent with CDK5RAP2 stimulating γ-TuRC–mediated nucleation, Eb1 comets originated at focal sites at the cell cortex and moved into the cytoplasm in DP-CγBD–expressing cells (Fig. 4 D, green lines in right panels). In contrast, few Eb1-GFP comets were observed emanating from the cortex in DP-NγBD–expressing cells (Fig. 4 D and Videos 3 and 4). To quantify MT nucleation in a larger number of cells, we assayed sites of new MT assembly after nocodazole washout (Fig. 4, E and F). Under these conditions, CDK5RAP2 robustly stimulated MT nucleation at cell–cell junctions independent of Nedd1. In mitotic cells, DP-CγBD cortical puncta even became functional “spindle poles,” with the DNA being pulled toward both the centrosomes and the cell cortex (Fig. S4 D). Cortically targeted DP-NγBD, in contrast, did not promote cortical nucleation after nocodazole washout, despite the presence of γ-tubulin at the cell cortex (Fig. 4, E and F). This was not because of requirements for other domains within Nedd1 as a full-length Nedd1 fusion that was targeted to desmosomes (DP-Nedd1FL) behaved identically to DP-NγBD (Fig. S4 C) and also did not promote MT nucleation there (Fig. S4 E). These data highlight that Nedd1–γ-TuRCs are not sufficient to promote MT nucleation in interphase cells and that γ-tubulin is not always a reliable marker of sites of MT nucleation in vivo.
Nedd1 is necessary for efficient microtubule minus-end anchoring
Because Nedd1–γ-TuRCs did not promote MT nucleation in vivo or in vitro, we wanted to determine whether they had other functions. Under steady-state conditions, we observed that MTs were anchored to the cell cortex in WT keratinocytes transfected with DP-CγBD, but not in DP-, DP-NγBD–, or DP-Nedd1FL–transfected cells (Fig. 5, A and B; and Fig. S4 F). However, when Nedd1 was depleted from DP-CγBD–transfected cells, MTs were no longer anchored at the cortical sites (Fig. 5, A and B), despite the fact that we saw comparable levels of nucleation after nocodazole washout (Fig. 4 E). Expression of DP-NγBD in these cells (DP-CγBD and Nedd1 KD) was able to rescue MT anchoring at the cortex, strongly suggesting that Nedd1 directly facilitates tethering of MTs through its γ-tubulin–interacting domain (Fig. 5 B). Notably, ninein and other centrosomal proteins such as pericentrin were not detected at cortical sites under these conditions (unpublished data). Together, these data support a model in which Nedd1–γ-TuRCs are required for MT anchoring.
Figure 5.
Nedd1 is required for MT anchoring. (A) Images of MT organization in control, DP-NγBD–, DP-CγBD–, or DP-CγBD + Nedd1KD–expressing cells under steady-state (nonperturbing) conditions. Insets show higher magnifications of cortical regions. Bars: (main) 10 µm; (insets) 2 µm. Note that cell junctions are acting as discrete MTOCs in DP-CγBD–expressing cells. (B) Quantification of the cortical MT-anchoring activity of cells expressing the DP fusion constructs. (C) Representative images of MT assembly assays using centrosomes purified from control and Nedd1 knockdown cells. Bar, 10 µm. (D) Quantification of MT aster area around control and Nedd1 knockdown centrosomes (n ≥ 23 centrosomes from two independent experiments). (E) Quantification of free (noncentrosomal) MTs nucleated by control and Nedd1 knockdown centrosomes (n = 3 independent experiments). (F) Representative images of control and Nedd1 KD cells at indicated time points after nocodazole washout. Insets show zoomed in centrosomes. Bars: (main) 10 µm; (insets) 1 µm. (G) Quantification of the ratio of α-tubulin intensity at the centrosome to intensity in the cytoplasm in control and Nedd1KD keratinocytes after nocodazole washout (n ≥ 146 centrosomes from three independent experiments). (H) Quantification of MT nucleation at the centrosome and cytoplasm after nocodazole washout in control and Nedd1 KD keratinocytes (n ≥ 146 centrosomes from three independent experiments). n.s., not significant; **, P < 0.01; ***, P < 0.001. Data are presented as mean ± SEM.
To directly test whether Nedd1 is required for microtubule anchoring to centrosomes, we isolated centrosomes from Nedd1 KD cells and performed in vitro MT assembly assays. Nedd1 KD centrosomes could not robustly anchor microtubules in vitro (Fig. 5, C and D), although they were still able to promote MT nucleation (Figs. 5 E and 3 C). In agreement with this data from isolated centrosomes, depletion of Nedd1 resulted in MT-anchoring defects at centrosomes in cultured keratinocytes. At steady state, the high density of MTs in keratinocytes makes analysis of MTOC activity difficult. However, we found that upon nocodazole washout, Nedd1-depleted cells have robust centrosomal nucleation, similar to control cells and consistent with our live-imaging data (Fig. 5, F–H). By 3 min after nocodazole washout, however, there was a decrease in centrosomal MTOC activity in Nedd1-depleted cells compared with controls (Fig. 5, F–H). The Nedd1 knockdown cells showed both a decrease in centrosomal MT intensity at 3 min after washout (as compared with 1 min after washout), as well as an increase in cytoplasmic MTs (Fig. 5 H).
Previous anchoring models were based on subdistal appendage proteins, like ninein, interacting with MT minus ends. Importantly, the Nedd1 KD centrosomes retained normal levels of ninein and CDK5RAP2 (Fig. S2, I–K), demonstrating that ninein levels do not correlate with anchoring activity. Thus, our data reveal a second way in which centrosomes can anchor MT ends that is dependent on Nedd1–γ-TuRCs.
γ-Tubulin localization correlates with cell cycle status in vivo
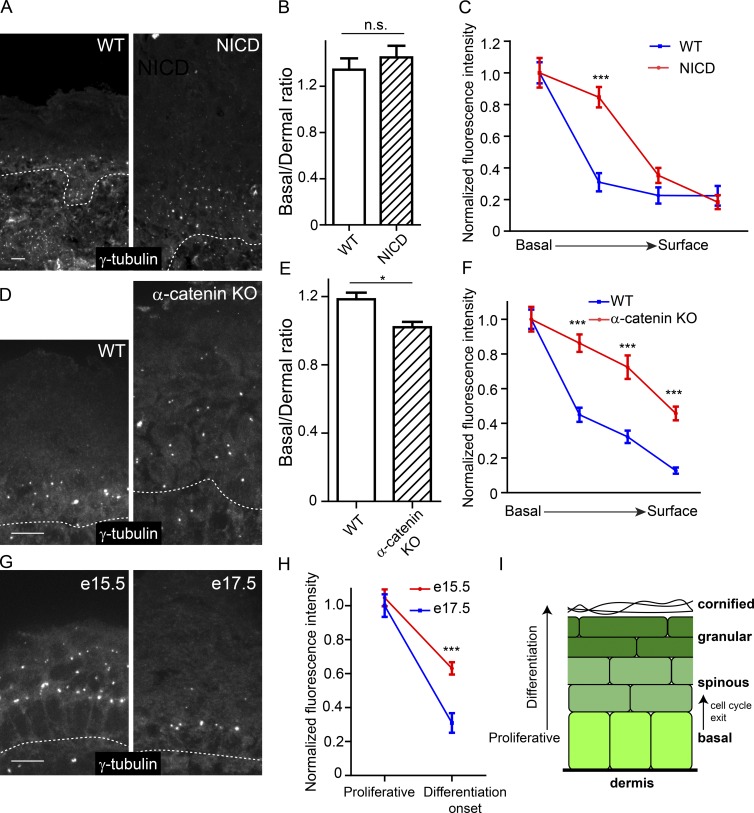
Having identified γ-TuRCs with distinct functions and distinct localization patterns after differentiation onset, we became interested in understanding which signaling pathways control their localization. Because of its critical role in inducing spinous cell fate upon epidermal differentiation (Rangarajan et al., 2001; Blanpain et al., 2006; Moriyama et al., 2008), we first investigated whether Notch signaling was sufficient to delocalize γ-tubulin from centrosomes. Expression of active Notch (NICD; Murtaugh et al., 2003) in epidermal progenitors induces markers of spinous cells (Blanpain et al., 2006) but did not change the ratio of centrosomal γ-tubulin intensity in basal keratinocytes versus dermal cells (Fig. 6, A–C). These data demonstrate that Notch signaling is not sufficient to induce γ-tubulin delocalization. Unexpectedly, we found that there was an expansion of γ-tubulin–enriched centrosomes to suprabasal cell layers in these NICD-expressing mice. In the NICD epidermis, proliferation is expanded into the suprabasal cell layers that are normally quiescent (Blanpain et al., 2006; Moriyama et al., 2008). This suggested that γ-tubulin was maintained at the centrosome in cells that were still actively cycling. In agreement with this, we observed maintenance of γ-tubulin at suprabasal centrosomes in the hyperproliferative α-catenin–null epidermis and also in intermediate cells, a transient population of proliferative suprabasal cells in embryonic day 14.5 (E14.5) to E15.5 embryos (Vasioukhin et al., 2001a; Lechler and Fuchs, 2005; Fig. 6, D–H).
Figure 6.
Centrosomal γ-tubulin levels correlate with cell-cycle status and not differentiation state. (A) Immunofluorescence localization of γ-tubulin in WT epidermis and epidermis expressing active Notch (NICD). (B) Quantification of the ratio of centrosomal γ-tubulin in basal cells versus underlying dermal cells (n = 2 control and NICD mice). (C) Quantification of γ-tubulin centrosomal localization from basal cells to terminally differentiated superficial cells. (D) Immunofluorescence localization of γ-tubulin in WT epidermis and α-catenin–null epidermis. (E) Quantification of the ratio of centrosomal γ-tubulin in basal cells verses underlying dermal cells (n = 2 control mice and n = 2 KO mice). (F) Quantification of γ-tubulin centrosomal localization from basal cells to superficial cells (n = 2 control mice and n = 2 KO mice). (G) Immunofluorescence localization of γ-tubulin in WT E15.5 and E17.5 epidermis. (H) Quantification of γ-tubulin centrosomal localization from basal cells and immediate suprabasal cells (n = 2 mice for each stage). Note that the data for the E17.5 mice are the WT data from the NICD mice in B. (I) Schematic of the epidermis showing the transition to a postmitotic state. n.s., not significant; *, P < 0.05; ***, P < 0.001. Data are presented as mean ± SEM. Bars, 10 µm. Dashed lines indicate the basement membrane.
Cell cycle exit is sufficient to induce γ-tubulin delocalization
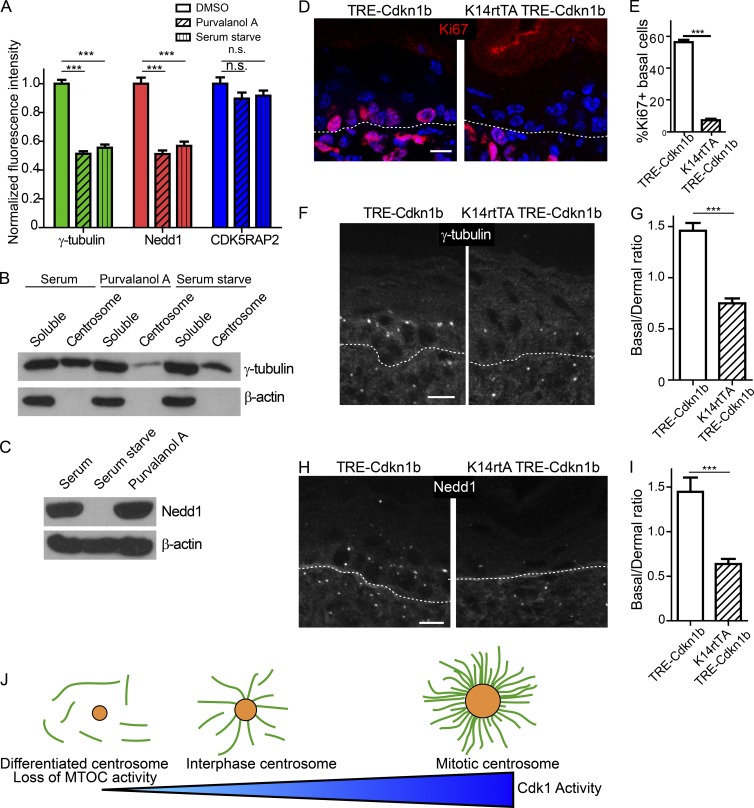
The mutant analysis suggested that it is cell cycle status and not differentiation per se that affects centrosomal composition and function (Fig. 6 I). To test whether altering cell cycle status was sufficient to induce changes in Nedd1 and γ-tubulin localization, we either serum starved keratinocytes or treated them with the CDK inhibitor Purvalanol A. Both treatments were sufficient to induce delocalization of Nedd1–γ-TuRCs from centrosomes over a period of hours (Fig. 7 A). No changes in pericentrin or CDK5RAP2 levels were observed after these treatments, demonstrating specificity (Fig. 7 A and not depicted). Furthermore, biochemical analyses revealed that Purvalanol A treatment or serum starvation was sufficient to induce a decrease in the amount of γ-tubulin at centrosomes and a corresponding increase in the cytoplasmic pool (Fig. 7 B). Although serum starvation was sufficient to induce Nedd1 degradation, short-term CDK inhibition did not (Fig. 7 C). This revealed two separable regulatory points; one resulted in loss of Nedd1 association with the centrosome, whereas the other resulted in Nedd1 degradation.
Figure 7.
Exit from the cell cycle induces centrosome composition changes. (A) Quantification of centrosomal levels of indicated proteins after serum starvation or Purvalanol A treatment (n ≥ 96 centrosomes from two independent experiments). (B) Western blots of γ-tubulin levels in cytoplasmic and centrosomal fractions from control, serum-starved, or Purvalanol A–treated cells. (C) Western blot of total levels of Nedd1 in control, serum-starved, or Purvalanol A–treated cells. (D) Backskin cryosections from postnatal day 1 K14rtTA TRE-Cdkn1b mice and littermate controls, stained for the proliferation marker Ki67. Dashed line marks the basement membrane. Note the loss of Ki67+ cells is specific to the epidermis. (E) Quantification of the percentage of Ki67+ basal cells in control and K14rtTA TRE-Cdkn1b mice (n = 3 mice of each genotype from two independent litters). (F) Immunofluorescence of γ-tubulin in backskins from control and K14rtTA TRE-Cdkn1b mice. (G) Quantification of the ratio of the fluorescence intensity of γ-tubulin on basal cell centrosomes to dermal centrosomes (n = 3 mice of each genotype from two independent litters). (H) Immunofluorescence of Nedd1 in backskins from control and K14rtTA TRE-Cdkn1b mice. (I) Quantification of the ratio of the fluorescence intensity of Nedd1 on basal cell centrosomes to dermal centrosomes (n = 3 mice each genotype from two independent litters). (J) Diagram of the effects of different levels of Cdk1 activity on centrosome morphology and function. Bars, 10 µm. n.s., not significant; ***, P < 0.001. Data are presented as mean ± SEM.
To test whether cell cycle exit is sufficient to induce these same centrosomal changes in vivo, we overexpressed the CDK inhibitor, Cdkn1b (p27), in basal cells of the epidermis (Nguyen et al., 2006; Pruitt et al., 2013). Doxycycline-induced overexpression of Cdkn1b dramatically decreased the percentage of actively cycling Ki67+ and BrdU+ cells in the epidermis, demonstrating that Cdkn1b overexpression is sufficient to induce quiescence (Fig. 7, D and E; and Fig. S5, A and B). Basal cells were still K5/14 positive and did not express the spinous marker K1 (Fig. S5 C). Cdkn1b expression decreased the amount of centrosomal Nedd1–γ-TuRC in basal cells of mutant mice relative to WT littermates whereas pericentrin levels were unchanged (Fig. 7, F–I; and Fig. S5 D). Collectively, these data indicate that it is not a differentiation signaling pathway directly, but rather cell cycle exit that induces changes in centrosome architecture, the first step of which is loss of Nedd1–γ-TuRC. As differentiation pathways induce cell cycle exit, this provides an elegant way to turn off centrosomal activity in post-mitotic cells.
Discussion
We have shown that loss of Nedd1 upon differentiation correlates with loss of MT anchoring activity, that Nedd1 is required for anchoring at an artificial cortical MTOC, and that depletion of Nedd1–γ-tubulin complexes at the centrosome results in loss of MT anchoring activity. Although other models for Nedd1 function exist, an intriguing possibility is that Nedd1–γ-TuRCs act directly to anchor MTs at the centrosome. Although γ-TuRCs have established roles in both nucleation and minus-end capping of MTs (Moritz et al., 1995; Zheng et al., 1995; Wiese and Zheng, 2000; Anders and Sawin, 2011), they were not previously implicated in MT anchoring. Whether anchoring and minus-end capping are linked or distinct activities will require further investigation. It is of note that our studies clearly demonstrate that γ-tubulin levels are not a good surrogate for MT nucleation activity. Molecularly, this is due, at least in part, to the presence of distinct γ-TuRCs with different activities: nucleation or anchoring. Our mass spectrometry data demonstrate that the difference between nucleating and anchoring γ-TuRCs are CDK5RAP2 and Nedd1, respectively, suggesting that these binding partners induce different conformations of γ-TuRC compatible with distinct MT activities. Although it is also possible that there are differences in stoichiometries of individual subunits in CDK5RAP2–γ-TuRC and Nedd1–γ-TuRC, these complexes migrate similarly on sucrose gradients and complexes purified based on affinity for Nedd1 can be activated by CDK5RAP2. Future work will be required to determine if and how γ-TuRC structure is altered upon binding either Nedd1 or CDK5RAP2. Our data rule out a simple model in which Nedd1–γ-TuRCs anchor MTs that are nucleated by this same complex. However, whether Nedd1 replaces CDK5RAP2 in γ-TuRCs with newly nucleated microtubules or whether they form a larger-order complex will require further investigation.
Our findings were particularly surprising given that Nedd1 has been implicated in γ-tubulin–dependent microtubule nucleation in plants, mitotic cells, and cultured cancer cell lines (Haren et al., 2006; Lüders et al., 2006; Pinyol et al., 2013; Walia et al., 2014; Scrofani et al., 2015). In keratinocytes, however, Nedd1 is not required for steady-state centrosomal MT nucleation or nucleation induced by nocodazole washout. In contrast to CDK5RAP2, it is also not sufficient to activate γ-TuRC nucleation by two distinct assays. These data suggest a context dependence to the activity of Nedd1–γ-TuRCs, i.e., they can be converted into different functional states by either posttranslational modifications or by the association of accessory factors. In support of this, Nedd1 undergoes complex mitotic phosphorylations that have been shown to influence its activity (Zhang et al., 2009; Gomez-Ferreria et al., 2012; Sdelci et al., 2012; Pinyol et al., 2013).
Molecular requirements for centrosomal MT anchoring remain poorly understood. Subdistal appendage proteins, like ninein, are required for centrosomal MTOC activity in some cell types (Delgehyr et al., 2005). However, our data suggest that these proteins are not sufficient for robust anchoring activity without Nedd1 in keratinocytes. It is likely that various cell types will use distinct anchoring complexes to different extents. However, the coincident loss of both Nedd1–γ-TuRCs and ninein from centrosomes upon differentiation likely allows for a very robust loss of anchoring activity in keratinocytes.
We have also found that differentiation regulates these functionally distinct γ-TuRC complexes in divergent ways. Nedd1–γ-TuRCs are lost from the centrosome immediately upon cell cycle exit, causing loss of MT anchoring, and thus loss of MTOC activity, at the centrosome. Although CDK up-regulation in mitosis has a well-defined role in centrosome maturation and increased nucleation, our work demonstrates an important interphase role for CDK activity in centrosome maintenance. In this way, CDK functions as a rheostat for centrosome activity rather than an on–off switch (Fig. 7 J). The role of CDK in regulating PCM in differentiating cells is consistent with studies in other organisms (Yang and Feldman, 2015). Nedd1 is regulated at two distinct levels: the association of Nedd1 with the centrosome and the degradation of Nedd1. These two mechanisms, combined with the dominant-negative effects of Nedd1 overexpression, have prevented simple reexpression experiments to rescue Nedd1 levels and centrosome function. Understanding the molecular underpinnings of these two steps should give greater insight into the mechanisms of MTOC shutoff. Although γ-tubulin loss appears common during differentiation (Brodu et al., 2010; Feldman and Priess, 2012), our data highlight that examining specific γ-tubulin complexes may be more informative than examining bulk γ-tubulin. Finally, our data suggest that distinct γ-tubulin complexes may be independently used by different tissues to generate cell type–specific noncentrosomal MT arrays. Examining these distinct γ-tubulin complexes may be informative in understanding the mechanisms regulating noncentrosomal MT arrays in other tissues.
Materials and methods
Mice and tissues
All mice were maintained in accordance with Duke Institutional Animal Care and Use Committee–approved protocols. For in vivo centrosomal quantifications, K14-centrin-GFP mouse epidermis (Lechler and Fuchs, 2005) was isolated from postnatal day 0 mice, embedded in optimal cutting temperature compound, and sectioned and processed for immunofluorescence. To assess γ-tubulin localization in vivo in mutant conditions, the following mouse lines were used: α-catenin (Vasioukhin et al., 2001a; gift from E. Fuchs, Rockefeller University, New York, NY), NICD (Murtaugh et al., 2003), K14-rtTA (Nguyen et al., 2006), TRE-Cdkn1b (Pruitt et al., 2013; gift from S. Pruitt, Roswell Park Cancer Center, Buffalo, NY), and K14Cre (Vasioukhin et al., 1999). For TRE-Cdkn1b experiments, pregnant dams were fed 200 mg/kg doxycycline chow (Bioserv) at E17.5 or E18.5; the pups were sacrificed 48 h later, and backskins were embedded and processed for immunofluorescence. For BrdU experiments, mice were injected with 10 mg/kg BrdU (Sigma-Aldrich) and sacrificed 1 h after injection, and backskins were embedded and processed for immunofluorescence. For α-catenin and NICD experiments, E17.5 embryos were used. K14-Eb1-GFP transgenic mice were generated by the Duke Transgenic Core by pronuclear injection.
Constructs
Nedd1 (TRCN0000248316), γ-tubulin (TRCN0000089907), and CDK5RAP2 (TRCN0000363878) MISSION shRNA constructs were purchased from Sigma-Aldrich. The “DP” construct used as the control construct for all DP fusion experiments was made by cloning the first 1,056 amino acids of the human desmoplakin gene with NheI and EcoRI sites on the 3′ end into pEF6/V5-His TOPO. This construct served as the foundation for cloning all DP fusion constructs. The DP–Nedd1 full-length fusion construct was generated by amplifying all 660 amino acids of the human Nedd1 gene from pcDNA3-GFP-Nedd1 (gift from L. Pelletier, University of Toronto, Toronto, Canada) with an NheI site on the 5′ end and NotI site on the 3′ end and cloning into the DP control construct digested with the same. The DP–NγBD fusion construct included amino acids 597–660 of human Nedd1 and was cloned into the DP control construct using NheI and EcoRI sites. GST-NγBD was generated by cloning the identical Nedd1 fragment into pGEX-4T1 using the BamHI site. DP-CγBD was cloned by PCR amplification of amino acids 51–100 of human CDK5RAP2 (derived from Addgene plasmid #41152) with an NheI site on the 5′ end and EcoRI site on the 3′ end; this CγBD fragment was then inserted into the DP control construct digested with the same restriction enzymes. GST-CγBD was generated by cloning the same fragment of CDK5RAP2 into pGEX-4T1 using BamHI and EcoRI sites. All constructs were verified by sequencing.
Cell culture
All keratinocytes were maintained in E low Ca media at 37°C. Differentiation was induced by addition of 1.2 mM Ca2+ to the culture media for 48–72 h. To derive all primary keratinocytes, postnatal day 0 mouse backskin was placed in 1.2 U/ml dispase overnight at 4°C. Isolated epidermal sheets were placed in 1:1 versene/0.25% trypsin-EDTA for 15 min at 37°C, and cells were strained (70 µm pore) before being plated. Desmoplakin-null and keratin-null keratinocytes were derived from DP-null (Vasioukhin et al., 2001b) and keratin-null (Seltmann et al., 2013) epidermis, respectively. For imaging microtubule dynamics in proliferative and differentiated cells, primary keratinocytes were isolated from K14-centrin-GFP; K14-Eb1-GFP mice and maintained in E low Ca media supplemented with 0.5 mM Ca2+.
Drug treatments
For quiescence experiments, 15 µM Purvalanol A (Abcam) was added to a confluent plate of keratinocytes for 5 h and the cells were processed for Western blot analysis or immunofluorescence. For serum-starvation experiments, confluent keratinocytes were washed three times with PBS, placed into DMEM ± FBS for 8 h, and processed for Western blot analysis or immunofluorescence.
Centrosome isolation
Centrosome isolation was performed similarly to previously described protocols (Mitchison and Kirschner, 1984). Keratin-null keratinocytes (Seltmann et al., 2013) were grown to confluence in either proliferative or differentiated cell media. Cells were treated with 10 µM nocodazole (Sigma-Aldrich) and 5 µg/ml cytochalasin B (Cayman Chemical) for 1.5 h before lysis. Keratinocytes were washed sequentially with 1× PBS, 0.1× PBS + 8% sucrose, 8% sucrose, and LB (1 mM Tris, pH 8.0, and 8 mM β-mercaptoethanol), and then lysed in LB + 0.5% NP-40 (hypotonic lysis). Lysate was pooled into conical tubes and made to 1 mM EDTA and 10 mM Pipes, pH 7.2, and then pelleted at 3,000 g for 10 min. The soluble fraction was loaded onto a 70%/40% discontinuous sucrose gradient in HB100 (50 mM Hepes, pH 8.0, 100 mM NaCl, 1 mM MgCl2, 1 mM EGTA, 1 mM βME, 0.1 mM GTP, 1× protease inhibitor [Roche], and 1 mM DTT) and spun in a SW41 rotor for 30 min at 25,000 g. The 40%/70% interface was collected and centrosomes were desalted into 1× BRB80 buffer (80 mM Pipes, pH 6.8, 1 mM MgCl2, and 1 mM EGTA) using Zeba columns (Thermo Fisher Scientific) or by dialysis and aliquots were snap frozen. Centrosomes were assayed by pelleting onto poly-l-lysine–coated coverslips in an ultracentrifuge in an SW41 rotor at 30,000 g for 30 min using a custom-made polydimethylsiloxane coverslip platform. After pelleting, coverslips were removed and fixed in MeOH at −20°C and processed for staining. For isolating centrosome and soluble fractions for Western blot analysis, keratin-null cells were used. Centrosome isolation was performed as previously described up to the 3,000 g pelleting step. At this point, the soluble fraction was spun in a rotor (TLA100.1) for 45 min at 100,000 g to pellet centrosomes. Centrosome pellets were resuspended in HB100 in the same volume as the soluble fraction to make ratios directly comparable.
γ-TuRC isolation
To isolate CDK5RAP2–γ-TuRCs and Nedd1–γ-TuRCs, cell lysates from HEK293 cells were made in γ-TuRC lysis buffer (50 mM Hepes, pH 7.2, 150 mM NaCl, 1 mM EGTA, 1 mM MgCl2, 1 mM DTT, 0.5% NP-40, 0.1 mM GTP, and 1× protease inhibitor). Lysates were cleared in an ultracentrifuge at 100,000 g for 30 min. GST-NγBD and GST-CγBD were bound to glutathione-agarose beads (Thermo Fisher Scientific) and incubated in cell lysate overnight at 4°C. Beads were pelleted and washed two times with HB100 and two times with HB250. Complexes were eluted in elution buffer (γ-TuRC lysis buffer and 5 mM glutathione). Eluent was put on a 5–40% continuous sucrose gradient and spun in a TLS-55 rotor for 3 h and 15 min at 200,000 g. Then, 150-µl fractions were collected from the top of the gradient and samples were taken for Western blotting. Peak γ-tubulin fractions were pooled and desalted into 1× BRB80 using Zeba columns. Purified complexes were immediately aliquoted and snap frozen. The approximate γ-TuRC concentration was determined by Western blot using a standard with known γ-tubulin concentration.
Pull-down assay
For pull-down assays, GST, GST-CγBD, or GST-NγBD were bound to glutathione-agarose beads for 1 h with rotating at 4°C. Beads were washed three times with PBS and incubated with precleared keratinocyte cell lysate for 1 h at 4°C with rotating. Beads were washed two times with HB150 (50 mM Hepes, pH 7.2, 150 mM NaCl, 1 mM EGTA, 1 mM MgCl2, 1 mM DTT, 0.01% NP-40, and 0.1 mM GTP) and two times with HB250 (50 mM Hepes, pH 7.2, 250 mM NaCl, 1 mM EGTA, 1 mM MgCl2, 1 mM DTT, 0.01% NP-40, and 0.1 mM GTP). Pelleted beads were boiled in sample buffer (63 mM Tris, pH 6.8, 0.7% SDS, 10% glycerol, and 0.01% bromophenol blue solution) to use for Western blots.
DP construct nocodazole washout and EB1 nucleation from the cortex
For all experiments with the DP fusion constructs, DP−/− keratinocytes were transfected in the morning and 1.2 mM Ca2+ was added 8 h later to allow cell–cell junctions to form overnight. To monitor microtubule dynamics in DP fusion construct-transfected cells, keratinocytes were cotransfected with either DP-CγBD or DP-NγBD along with Eb1-GFP. For nocodazole washout experiments, 10 µM nocodazole was added to the culture media of transfected cells that had formed cell–cell junctions overnight for 1 h at 37°C. Cells were washed three times with PBS and placed into fresh media, and microtubule regrowth was monitored by fixing coverslips in −20°C methanol at 0, 1, and 3 min after nocodazole washout.
Antibodies
The following primary antibodies were used in this study: rat anti–α-tubulin (YL1/2), mouse anti-Nedd1 (39-J; both from Santa Cruz Biotechnology, Inc.), mouse anti–β-tubulin (clone TUB2.1), mouse anti–γ-tubulin (clone GTU-88), mouse anti-β-actin (clone AC-15; all from Sigma-Aldrich), mouse anti-V5 (clone 2F11F7; Invitrogen), rabbit anti-pericentrin (ab4448), rabbit anti-Ki67 (ab15580), rat anti-BrdU (ab6326), mouse anti-Nedd1 (ab57336; all from Abcam), rabbit anti-CDK5RAP2 (06–1398; EMD Millipore), chicken anti-K5/14 (generated in laboratory), rabbit anti-K1 (gift from C. Jamora, InStem, Bangalore, India), rabbit anti–GST-HRP (A190-122P-9; Bethyl Laboratories, Inc.), and rabbit antifilaggrin (PRB-417P; Covance).
Image acquisition and quantification
All fixed images were acquired on an Axio Imager microscope (ZEISS) with Apotome attachment with the following objective lenses: 20× Plan-Apo 0.8 NA lens, 40× Plan-Neofluar 1.3 NA oil lens, and 63× Plan-Apo 1.4 NA oil lens. Images on the Axio Imager microscope were acquired using AxioVision software. All images within one experiment were taken with identical exposure times. Movies of Eb1-GFP in primary keratinocytes and in DP fusion transfected cells were taken on a microscope (DMI6000; Leica Biosystems) at 37°C and 5% CO2 using a 63× Plan-Apo 1.4–0.6 NA oil objective. Images were acquired using ZEN software. Movies of Eb1-GFP in the knockdown cells were acquired on an XD revolution spinning disc confocal microscope (Andor) at 37°C and 5% CO2 using a 60× Plan-Apo 1.2 NA water objective. Images on the spinning disc were acquired using MetaMorph software.
Centrosomes were quantified in vivo by identifying centrin-GFP puncta and finding the mean intensity in a 15 × 15-pixel box around the centrosome. For all centrosomal quantifications, a background value was calculated by averaging 10 random 15 × 15 boxes within the vicinity of the centrosome. The average background value was subtracted from each centrosome value to calculate the mean centrosomal intensity value. Values were normalized by setting the mean centrosomal intensity in the basal cells or proliferative cell population to 1. Cortical MT regrowth after nocodazole washout was quantified by manually drawing a line around the cortical region of a given transfected cell and finding the mean α-tubulin intensity across the line. Nocodazole washout data in Nedd1KD cells was quantified by finding the ratio of α-tubulin intensity around the centrosome (20 × 20 pixel circle) to a randomly chosen area (also 20 × 20-pixel circle) in the cytoplasm. Normalized MT intensity was quantified by setting the mean α-tubulin intensity at the centrosome at 1min to a value of 1.0 and plotting the α-tubulin intensities at the centrosome relative to the 1-min time point. All image processing and quantification was done using Fiji software.
Statistical analysis
For statistical tests, GraphPad Prism and JMP Pro 11 were used. All t tests were unpaired two-tailed Student’s t tests, except for analysis of EB1-GFP in primary keratinocytes, where a paired t test was used. For experiments with multiple variables, analysis of variances were used. If interactions were statistically significant, post hoc comparisons were performed using Tukey’s post hoc test.
Mass spectrometry analysis
Nedd1-bound γ-TuRCs were desalted into 50 mM ammonium bicarbonate using Zeba columns (Thermo Fisher Scientific) and supplemented with Rapigest SF surfactant (Waters) to 0.1%. The sample was reduced with 10 mM dithiolthreitol for 20 min at 80°C and alkylated with 20 mM iodoacetamide for 45 min at room temperature before digesting with sequencing-grade modified trypsin (Promega) at 37°C overnight. After hydrolysis of Rapigest in 0.1% TFA for 2 h, peptides were lyophilized to dryness and then resuspended in 12 µl 1% TFA/2% acetonitrile. Liquid chromatography (LC) tandem mass spectrometry (MS/MS) was performed on 5 µl of the sample using a nanoAcquity UPLC system (Waters) coupled to a Thermo QExactive Plus high-resolution accurate mass tandem mass spectrometer (Thermo Fisher Scientific) via a nanoelectrospray ionization source. In brief, the sample was first trapped on a Symmetry C18 300 mm × 180 mm trapping column (5 µl/min at 99.9/0.1 vol/vol water/acetonitrile), after which the analytical separation was performed using a 1.7 µm Acquity BEH130 C18 75 mm × 250 mm column (Waters) using a 60-min gradient of 5 to 40% acetonitrile with 0.1% formic acid at a flow rate of 400 nl/min with a column temperature of 55°C. Data collection was performed on a QExactive Plus mass spectrometer in a data-dependent acquisition mode with an r = 70,000 (at m/z 200) full MS scan from m/z 375–1,600 with a target 1e6 ions AGC value. This was followed by 10 MS/MS scans at r-17,500 (at m/z 200) at a target 5e4 ions AGC value. Coverage depth was increased with a 20-s dynamic exclusion.
Proteome Discoverer was used to process the raw data files. The LC-MS/MS data files were then submitted to independent Mascot searches (Matrix Science) against a SwissProt database (Human taxonomy, 20,347 forward entries) consisting of both forward and reverse entries for each protein. The following tolerances were used: 5 ppm for precursor ions and 0.02 D for product ions with trypsin specificity with up to two missed cleavages. Carbamidomethylation (+57.0214 D on C) was set as a fixed modification. Oxidation (+15.9949 D on M) and deamidation (+0.98 D on NQ) were set as dynamic modifications. Scaffold (v4.4, Proteome Software) was used to visualize and search the data. To realize a protein false discovery rate of 1%, scoring thresholds were modified using the PeptideProphet algorithm.
MT nucleation from centrosomes
MT nucleation from centrosomes was performed as previously described (Mitchison and Kirschner, 1984). Isolated centrosomes were incubated with 1× BRB80 (80 mM Pipes, pH 6.8, 1 mM EGTA, and 1 mM MgCl2) and either 97% or 99% tubulin (Cytoskeleton, Inc.; final concentration, 1 mg/ml) for 5 min on ice. Identical results were obtained when using unlabeled tubulin or a 1:6 mix of rhodamine-conjugated tubulin (Cytoskeleton, Inc.)/unlabeled tubulin. GTP was added to 1 mM, and the reaction was incubated for an additional 5 min on ice. Reactions were allowed to proceed for 5 min at 37°C, and reactions were fixed by addition of 10× reaction volumes of prewarmed glutaraldehyde fixation buffer (1% glutaraldehyde in 1× BRB80). Reactions were pelleted onto coverslips for aster and microtubule detection.
MT nucleation with soluble complexes
In vitro MT nucleation experiments were performed as previously described (Choi et al., 2010). In brief, 99% tubulin (Cytoskeleton, Inc.) was clarified at 200,000 g in a TLA200.1 rotor for 30 min. γ-TuRCs were isolated by affinity purification over GST-NγBD or GST-CγBD followed by sucrose gradient centrifugation, which causes loss of the GST fusion proteins. Then, 30 nM γ-TuRCs (estimated based on γ-tubulin concentration) was preincubated on ice with either 1× BRB80 alone or with 30 nM GST-CγBD or GST-NγBD for 5 min. Tubulin (final concentration of 1 mg/ml) was added to the reaction, and tubes were incubated for 5 min on ice. GTP was added to 1 mM and the reactions were incubated at 37°C for 3 min. Reactions were fixed with addition of 1% glutaraldehyde fixation buffer. A fraction of the reaction was pelleted through 1× BRB80 onto coverslips for quantification. MTs were quantified by randomly choosing fields under the microscope and averaging all the fields to generate a mean number of MT per field per experiment.
Online supplemental material
Fig. S1 shows the quantification of γ-tubulin and pericentrin centrosomal levels in keratin-null cells, primary cells, and isolated centrosomes. Fig. S2 shows additional characterization of Nedd1 KD and CDK5RAP2 KD cells. Fig. S3 shows results from GST-CγBD and GST-NγBD pull-downs and further characterization of γ-TuRC isolations. Fig. S3 also contains mass-spectrometry data from γ-TuRCs isolated using GST-NγBD. Fig. S4 contains further characterization of DP fusion–transfected cells. Fig. S5 contains further characterization of K14rtTA TRE-Cdkn1b epidermis. Videos 1 and 2 show MT dynamics in proliferative and differentiated keratinocytes, respectively. Videos 3 and 4 show Eb1-GFP dynamics in DP-NγBD- and DP-CγBD-expressing cells, respectively. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201601099/DC1.
Supplementary Material
Acknowledgments
We thank Scott Pruitt, Elaine Fuchs, and Ben Stanger for mice; Thomas Magin for keratin-null cells; Laurence Pelletier for Nedd1 plasmids; Julie Underwood for mouse care; and Scott Soderling, and Michel Bagnat and members of the Lechler laboratory for comments on the manuscript. We also thank the Duke Transgenic and Proteomics facilities, which are part of the Duke Cancer Center.
This work was supported by a National Science Foundation predoctoral fellowship to A. Muroyama and by National Institutes of Health grant R01GM111336 to T. Lechler.
The authors declare no competing financial interests.
Footnotes
Abbreviations used in this paper:
- DP
- desmoplakin
- E
- embryonic day
- γ-TuRC
- γ-tubulin ring complex
- γ-TuSC
- γ-tubulin small complex
- KD
- knockdown
- LC
- liquid chromatography
- MS/MS
- tandem mass spectrometry
- MT
- microtubule
- MTOC
- microtubule-organizing center
- PCM
- pericentriolar material
- WT
- wild type
References
- Anders A., and Sawin K.E.. 2011. Microtubule stabilization in vivo by nucleation-incompetent γ-tubulin complex. J. Cell Sci. 124:1207–1213. 10.1242/jcs.083741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartolini F., and Gundersen G.G.. 2006. Generation of noncentrosomal microtubule arrays. J. Cell Sci. 119:4155–4163. 10.1242/jcs.03227 [DOI] [PubMed] [Google Scholar]
- Blanpain C., Lowry W.E., Pasolli H.A., and Fuchs E.. 2006. Canonical notch signaling functions as a commitment switch in the epidermal lineage. Genes Dev. 20:3022–3035. 10.1101/gad.1477606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodu V., Baffet A.D., Le Droguen P.M., Casanova J., and Guichet A.. 2010. A developmentally regulated two-step process generates a noncentrosomal microtubule network in Drosophila tracheal cells. Dev. Cell. 18:790–801. 10.1016/j.devcel.2010.03.015 [DOI] [PubMed] [Google Scholar]
- Choi Y.K., Liu P., Sze S.K., Dai C., and Qi R.Z.. 2010. CDK5RAP2 stimulates microtubule nucleation by the gamma-tubulin ring complex. J. Cell Biol. 191:1089–1095. 10.1083/jcb.201007030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chrétien D., Buendia B., Fuller S.D., and Karsenti E.. 1997. Reconstruction of the centrosome cycle from cryoelectron micrographs. J. Struct. Biol. 120:117–133. 10.1006/jsbi.1997.3928 [DOI] [PubMed] [Google Scholar]
- Dammermann A., Desai A., and Oegema K.. 2003. The minus end in sight. Curr. Biol. 13:R614–R624. 10.1016/S0960-9822(03)00530-X [DOI] [PubMed] [Google Scholar]
- Delgehyr N., Sillibourne J., and Bornens M.. 2005. Microtubule nucleation and anchoring at the centrosome are independent processes linked by ninein function. J. Cell Sci. 118:1565–1575. 10.1242/jcs.02302 [DOI] [PubMed] [Google Scholar]
- Feldman J.L., and Priess J.R.. 2012. A role for the centrosome and PAR-3 in the hand-off of MTOC function during epithelial polarization. Curr. Biol. 22:575–582. 10.1016/j.cub.2012.02.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong K.W., Choi Y.K., Rattner J.B., and Qi R.Z.. 2008. CDK5RAP2 is a pericentriolar protein that functions in centrosomal attachment of the gamma-tubulin ring complex. Mol. Biol. Cell. 19:115–125. 10.1091/mbc.E07-04-0371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Ferreria M.A., Bashkurov M., Helbig A.O., Larsen B., Pawson T., Gingras A.C., and Pelletier L.. 2012. Novel NEDD1 phosphorylation sites regulate γ-tubulin binding and mitotic spindle assembly. J. Cell Sci. 125:3745–3751. 10.1242/jcs.105130 [DOI] [PubMed] [Google Scholar]
- Guo J., Yang Z., Song W., Chen Q., Wang F., Zhang Q., and Zhu X.. 2006. Nudel contributes to microtubule anchoring at the mother centriole and is involved in both dynein-dependent and -independent centrosomal protein assembly. Mol. Biol. Cell. 17:680–689. 10.1091/mbc.E05-04-0360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haren L., Remy M.H., Bazin I., Callebaut I., Wright M., and Merdes A.. 2006. NEDD1-dependent recruitment of the gamma-tubulin ring complex to the centrosome is necessary for centriole duplication and spindle assembly. J. Cell Biol. 172:505–515. 10.1083/jcb.200510028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibi M., Zou P., Inoko A., Shiromizu T., Matsuyama M., Hayashi Y., Enomoto M., Mori D., Hirotsune S., Kiyono T., et al. 2011. Trichoplein controls microtubule anchoring at the centrosome by binding to Odf2 and ninein. J. Cell Sci. 124:857–864. 10.1242/jcs.075705 [DOI] [PubMed] [Google Scholar]
- Ishikawa H., Kubo A., Tsukita S., and Tsukita S.. 2005. Odf2-deficient mother centrioles lack distal/subdistal appendages and the ability to generate primary cilia. Nat. Cell Biol. 7:517–524. 10.1038/ncb1251 [DOI] [PubMed] [Google Scholar]
- Lechler T., and Fuchs E.. 2005. Asymmetric cell divisions promote stratification and differentiation of mammalian skin. Nature. 437:275–280. 10.1038/nature03922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechler T., and Fuchs E.. 2007. Desmoplakin: an unexpected regulator of microtubule organization in the epidermis. J. Cell Biol. 176:147–154. 10.1083/jcb.200609109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., and Wiese C.. 2008. Xenopus NEDD1 is required for microtubule organization in Xenopus egg extracts. J. Cell Sci. 121:578–589. 10.1242/jcs.018937 [DOI] [PubMed] [Google Scholar]
- Lüders J., Patel U.K., and Stearns T.. 2006. GCP-WD is a gamma-tubulin targeting factor required for centrosomal and chromatin-mediated microtubule nucleation. Nat. Cell Biol. 8:137–147. 10.1038/ncb1349 [DOI] [PubMed] [Google Scholar]
- Manning J.A., Lewis M., Koblar S.A., and Kumar S.. 2010a An essential function for the centrosomal protein NEDD1 in zebrafish development. Cell Death Differ. 17:1302–1314. 10.1038/cdd.2010.12 [DOI] [PubMed] [Google Scholar]
- Manning J.A., Shalini S., Risk J.M., Day C.L., and Kumar S.. 2010b A direct interaction with NEDD1 regulates gamma-tubulin recruitment to the centrosome. PLoS One. 5:e9618 10.1371/journal.pone.0009618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchison T., and Kirschner M.. 1984. Microtubule assembly nucleated by isolated centrosomes. Nature. 312:232–237. 10.1038/312232a0 [DOI] [PubMed] [Google Scholar]
- Mogensen M.M., Malik A., Piel M., Bouckson-Castaing V., and Bornens M.. 2000. Microtubule minus-end anchorage at centrosomal and non-centrosomal sites: the role of ninein. J. Cell Sci. 113:3013–3023. [DOI] [PubMed] [Google Scholar]
- Moritz M., Braunfeld M.B., Sedat J.W., Alberts B., and Agard D.A.. 1995. Microtubule nucleation by gamma-tubulin-containing rings in the centrosome. Nature. 378:638–640. 10.1038/378638a0 [DOI] [PubMed] [Google Scholar]
- Moriyama M., Durham A.D., Moriyama H., Hasegawa K., Nishikawa S., Radtke F., and Osawa M.. 2008. Multiple roles of Notch signaling in the regulation of epidermal development. Dev. Cell. 14:594–604. 10.1016/j.devcel.2008.01.017 [DOI] [PubMed] [Google Scholar]
- Murtaugh L.C., Stanger B.Z., Kwan K.M., and Melton D.A.. 2003. Notch signaling controls multiple steps of pancreatic differentiation. Proc. Natl. Acad. Sci. USA. 100:14920–14925. 10.1073/pnas.2436557100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müsch A. 2004. Microtubule organization and function in epithelial cells. Traffic. 5:1–9. 10.1111/j.1600-0854.2003.00149.x [DOI] [PubMed] [Google Scholar]
- Nguyen H., Rendl M., and Fuchs E.. 2006. Tcf3 governs stem cell features and represses cell fate determination in skin. Cell. 127:171–183. 10.1016/j.cell.2006.07.036 [DOI] [PubMed] [Google Scholar]
- Nguyen M.M., Stone M.C., and Rolls M.M.. 2011. Microtubules are organized independently of the centrosome in Drosophila neurons. Neural Dev. 6:38 10.1186/1749-8104-6-38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piehl M., Tulu U.S., Wadsworth P., and Cassimeris L.. 2004. Centrosome maturation: measurement of microtubule nucleation throughout the cell cycle by using GFP-tagged EB1. Proc. Natl. Acad. Sci. USA. 101:1584–1588. 10.1073/pnas.0308205100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinyol R., Scrofani J., and Vernos I.. 2013. The role of NEDD1 phosphorylation by Aurora A in chromosomal microtubule nucleation and spindle function. Curr. Biol. 23:143–149. 10.1016/j.cub.2012.11.046 [DOI] [PubMed] [Google Scholar]
- Pruitt S.C., Freeland A., Rusiniak M.E., Kunnev D., and Cady G.K.. 2013. Cdkn1b overexpression in adult mice alters the balance between genome and tissue ageing. Nat. Commun. 4:2626 10.1038/ncomms3626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rangarajan A., Talora C., Okuyama R., Nicolas M., Mammucari C., Oh H., Aster J.C., Krishna S., Metzger D., Chambon P., et al. 2001. Notch signaling is a direct determinant of keratinocyte growth arrest and entry into differentiation. EMBO J. 20:3427–3436. 10.1093/emboj/20.13.3427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reschen R.F., Colombie N., Wheatley L., Dobbelaere J., St Johnston D., Ohkura H., and Raff J.W.. 2012. Dgp71WD is required for the assembly of the acentrosomal Meiosis I spindle, and is not a general targeting factor for the γ-TuRC. Biol. Open. 1:422–429. 10.1242/bio.2012596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scrofani J., Sardon T., Meunier S., and Vernos I.. 2015. Microtubule nucleation in mitosis by a RanGTP-dependent protein complex. Curr. Biol. 25:131–140. 10.1016/j.cub.2014.11.025 [DOI] [PubMed] [Google Scholar]
- Sdelci S., Schütz M., Pinyol R., Bertran M.T., Regué L., Caelles C., Vernos I., and Roig J.. 2012. Nek9 phosphorylation of NEDD1/GCP-WD contributes to Plk1 control of γ-tubulin recruitment to the mitotic centrosome. Curr. Biol. 22:1516–1523. 10.1016/j.cub.2012.06.027 [DOI] [PubMed] [Google Scholar]
- Seltmann K., Roth W., Kröger C., Loschke F., Lederer M., Hüttelmaier S., and Magin T.M.. 2013. Keratins mediate localization of hemidesmosomes and repress cell motility. J. Invest. Dermatol. 133:181–190. 10.1038/jid.2012.256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srsen V., Fant X., Heald R., Rabouille C., and Merdes A.. 2009. Centrosome proteins form an insoluble perinuclear matrix during muscle cell differentiation. BMC Cell Biol. 10:28 10.1186/1471-2121-10-28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumigray K.D., Chen H., and Lechler T.. 2011. Lis1 is essential for cortical microtubule organization and desmosome stability in the epidermis. J. Cell Biol. 194:631–642. 10.1083/jcb.201104009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumigray K.D., Foote H.P., and Lechler T.. 2012. Noncentrosomal microtubules and type II myosins potentiate epidermal cell adhesion and barrier formation. J. Cell Biol. 199:513–525. 10.1083/jcb.201206143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixidó-Travesa N., Villén J., Lacasa C., Bertran M.T., Archinti M., Gygi S.P., Caelles C., Roig J., and Lüders J.. 2010. The gammaTuRC revisited: a comparative analysis of interphase and mitotic human gammaTuRC redefines the set of core components and identifies the novel subunit GCP8. Mol. Biol. Cell. 21:3963–3972. 10.1091/mbc.E10-05-0408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasioukhin V., Degenstein L., Wise B., and Fuchs E.. 1999. The magical touch: genome targeting in epidermal stem cells induced by tamoxifen application to mouse skin. Proc. Natl. Acad. Sci. USA. 96:8551–8556. 10.1073/pnas.96.15.8551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasioukhin V., Bauer C., Degenstein L., Wise B., and Fuchs E.. 2001a Hyperproliferation and defects in epithelial polarity upon conditional ablation of alpha-catenin in skin. Cell. 104:605–617. 10.1016/S0092-8674(01)00246-X [DOI] [PubMed] [Google Scholar]
- Vasioukhin V., Bowers E., Bauer C., Degenstein L., and Fuchs E.. 2001b Desmoplakin is essential in epidermal sheet formation. Nat. Cell Biol. 3:1076–1085. 10.1038/ncb1201-1076 [DOI] [PubMed] [Google Scholar]
- Walia A., Nakamura M., Moss D., Kirik V., Hashimoto T., and Ehrhardt D.W.. 2014. GCP-WD mediates γ-TuRC recruitment and the geometry of microtubule nucleation in interphase arrays of Arabidopsis. Curr. Biol. 24:2548–2555. 10.1016/j.cub.2014.09.013 [DOI] [PubMed] [Google Scholar]
- Wiese C., and Zheng Y.. 2000. A new function for the gamma-tubulin ring complex as a microtubule minus-end cap. Nat. Cell Biol. 2:358–364. 10.1038/35014051 [DOI] [PubMed] [Google Scholar]
- Yang R., and Feldman J.L.. 2015. SPD-2/CEP192 and CDK Are limiting for microtubule-organizing center function at the centrosome. Curr. Biol. 25:1924–1931. 10.1016/j.cub.2015.06.001 [DOI] [PubMed] [Google Scholar]
- Zeng C.J., Lee Y.R., and Liu B.. 2009. The WD40 repeat protein NEDD1 functions in microtubule organization during cell division in Arabidopsis thaliana. Plant Cell. 21:1129–1140. 10.1105/tpc.109.065953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Chen Q., Feng J., Hou J., Yang F., Liu J., Jiang Q., and Zhang C.. 2009. Sequential phosphorylation of Nedd1 by Cdk1 and Plk1 is required for targeting of the gammaTuRC to the centrosome. J. Cell Sci. 122:2240–2251. 10.1242/jcs.042747 [DOI] [PubMed] [Google Scholar]
- Zheng Y., Wong M.L., Alberts B., and Mitchison T.. 1995. Nucleation of microtubule assembly by a gamma-tubulin-containing ring complex. Nature. 378:578–583. 10.1038/378578a0 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.