Abstract
Over the past decade, numerous advances have been made in the role and regulation of inflammasomes during pathogenic and sterile insults. An inflammasome complex comprises a sensor, an adaptor, and a zymogen procaspase-1. The functional output of inflammasome activation includes secretion of cytokines, IL-1β and IL-18, and induction of an inflammatory form of cell death called pyroptosis. Recent studies have highlighted the intersection of this inflammatory response with fundamental cellular processes. Novel modulators and functions of inflammasome activation conventionally associated with the maintenance of homeostatic biological functions have been uncovered. In this review, we discuss the biological processes involved in the activation and regulation of the inflammasome.
Introduction
Inflammasomes are multimeric complexes formed in response to a variety of physiological and pathogenic stimuli. Inflammasome activation is an essential component of the innate immune response and is critical for the clearance of pathogens or damaged cells. However, overt inflammasome activation is also a major driver of autoimmune and metabolic disorders, underlying the importance of understanding this process in physiological and pathological contexts.
The inflammasome sensors are grouped according to their structural features into nucleotide-binding domain–like receptors (NLRs), absent in melanoma 2–like receptors (ALRs), and the recently identified pyrin. These receptors have the ability to assemble inflammasomes and activate the cysteine protease caspase-1. In addition to the sensor (NLR, ALR, or pyrin) and enzymatic component (caspase-1), most inflammasomes also use an adaptor molecule known as ASC (apoptosis-associated speck-like protein containing a caspase activation and recruitment domain). Upon detecting specific stimuli, the activated sensor nucleates ASC to form a discrete foci, or “speck,” within the activated cell. Nucleated ASC sequentially recruits caspase-1, which undergoes proximity induced autocatalytic cleavage to produce the active subunits p10 and p20. These active caspase-1 subunits can then proteolytically process cytokines, IL-1β and IL-18, and induce a specific form of inflammatory cell death termed pyroptosis. Activated caspase-1 thus provides the host cell with dual defense mechanisms through the release of mature cytokines and removal of the infected or damaged cell. Inflammasome assembly is thus a coordinated signaling event essential for mounting an appropriate immune response after pathogenic or sterile insults. Although most studies have used in vitro culture systems and reconstitution assays to analyze inflammasome activation, recent advances have allowed visualization of these processes in vivo after an infectious insult at the level of a single cell (Sagoo et al., 2016). Recent studies have also highlighted the existence of an NLRP3 inflammasome pathway, mediated by caspase-11 (Kayagaki et al., 2011).
Inflammasome activation is tightly regulated to provide defense against pathogenic insults and avoid damage to the host. Multiple molecular and cellular signals are therefore involved in maintaining the balance between inflammatory response and resolution. In this review, we provide an overview of the cellular and molecular mechanisms involved in the regulation of inflammasome activation.
NLR family
NLR family members all share a central nucleotide-binding domain (NBD), and most members have a C-terminal leucine-rich repeat (LRR) domain and a variable N-terminal domain. The NLR family can be subdivided into NLRP or NLRC based on whether the N terminus contains a pyrin or caspase activation and recruitment domain (CARD), respectively. Certain members of the family, including NLRP1, NLRP3, and NLRC4, have been well established as NLRs capable of forming inflammasomes, whereas other members, like NLRP6 and NLRP12, are still considered putative inflammasome sensors. It remains to be seen if other members of the NLR family are capable of forming or regulating inflammasome assembly in response to some unknown stimuli.
NLRP1
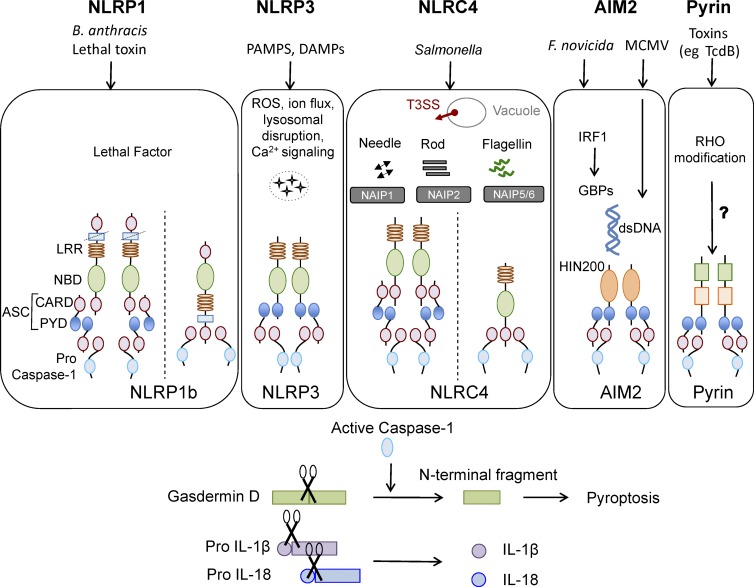
NLRP1 was the first cytosolic receptor identified for its ability to form a caspase-1 activating platform (Martinon et al., 2002). Human NLRP1 contains the canonical NBD and LRR domains, a pyrin domain (PYD), as well as a function-to-find and a C-terminal CARD domain. The mouse genome encodes three paralogs, NLRP1(a–c), which all lack the PYD. NLRP1b is activated by anthrax lethal toxin produced by Bacillus anthrax. Lethal toxin comprises protective antigen and lethal factor; protective antigen generates pores in the host cell membrane, whereas lethal factor enters the cell and cleaves NLRP1b at an N-terminal site, leading to the induction of inflammasome activation (Fig. 1; Chavarría-Smith and Vance, 2013). This cleavage is essential and sufficient to induce NLRP1b activation (Chavarría-Smith and Vance, 2013). Proteolytic cleavage within the NLRP1b function-to-find domain is also required for assembly of a functional inflammasome (Frew et al., 2012). Based on these observations, it has been hypothesized that NLRP1b could act as a sensor of protease activity caused by various bacterial toxins, although this has not been rigorously tested or shown. NLRP1b activation also promotes parasite clearance and provides protection against mortality in response to Toxoplasma gondii infection in mice (Gorfu et al., 2014). Toxoplasma-induced NLRP1 activation is observed in susceptible rat strains (Cirelli et al., 2014) and is a major determinant of parasite control in infected macrophages (Cavailles et al., 2014), although activation of NLRP1b through this parasite does not result in detectable levels of an NLRP1b cleavage product (Ewald et al., 2014). This suggests that either NLRP1b processing is not a prerequisite for inflammasome activation as posited or a thorough analysis for a potentially unstable cleavage product is required to reconcile this discrepancy. The link between immune response to Toxoplasma infection and NLRP1 inflammasome is clinically relevant even in humans, as congenital toxoplasmosis is associated with single-nucleotide polymorphisms in the NLRP1 gene and loss of NLRP1 in human monocytic cell lines promotes Toxoplasma infectivity (Witola et al., 2011).
Figure 1.
Canonical inflammasomes NLRP1, NLRP3, NLRC4, AIM2, and pyrin. Ligands and upstream mediators involved in inflammasome activation. The NLRP1b inflammasome responds to lethal factor produced by B. anthracis and assembles an inflammasome by recruiting caspase-1 through its CARD domain or through ASC as an adaptor. The NLRP3 inflammasome responds to intracellular damage induced by pathogenic or sterile insults. The NLRC4 inflammasome assembles in response to recognition of bacterial flagellin or components of the type III secretion system via NAIPs and can form complexes with or without recruiting ASC. AIM2 inflammasome senses double-stranded DNA through its HIN200 domain. Pyrin responds to Rho modification induced by bacterial toxins. Inflammasome activation leads to caspase-1 activation that in turn cleaves its downstream effectors: the newly identified pyroptosis executioner gasdermin D and pro-form of cytokines IL-1β and IL-18. DAMP, danger associated molecular pattern; GBP, guanylate binding protein; PAMP, pathogen associated molecular pattern; T3SS, type III secretion systems.
Evidence for the ability of NLRP1a to induce inflammasome activation comes from a genetic model, wherein mice harboring a mutation in the NLRP1a gene (Q593P) develop a caspase-1– and IL-1β–mediated systemic inflammatory disease (Masters et al., 2012). These mice also exhibit a change in myelopoiesis, such that the number and function of hematopoietic progenitor cells are markedly altered in a caspase-1–dependent but IL-1R signaling–independent manner. Thus, a significant loss of hematopoietic progenitors is observed because of aberrant inflammasome activation and cell-intrinsic pyroptosis. However, the specific triggers or mechanisms regulating the NLRP1a inflammasome are as yet unknown. Genetic studies in humans have identified NLRP1 mutations linked to autoinflammatory diseases including vitiligo (Jin et al., 2007), Addison’s disease (Zurawek et al., 2010), rheumatoid arthritis, systemic sclerosis, and Crohn’s disease (Finger et al., 2012); however, further studies that define the molecular mechanisms and structural conformations involved in NLRP1 activation will be needed to deduce the link between NLRP1 and these autoinflammatory diseases.
NLRP3
NLRP3 was first shown to be associated with hereditary autoinflammatory syndromes called cryopyrin-associated periodic syndromes, which are characterized by skin rashes and episodes of fever (Hoffman et al., 2001). In fact, over 90 disease-associated mutations have since been observed in humans in and around the NBDs that render NLRP3 constitutively active (Masters et al., 2009). NLRP3 is an inflammasome-forming NLR that responds to a wide range of infectious and endogenous ligands and is implicated in the pathogenesis of several autoinflammatory diseases, including arthritis, gout, diabetes, obesity, and Alzheimer’s disease (Guo et al., 2015).
The triggers that have been shown to induce NLRP3 activation include pathogen-derived ligands such as microbial cell wall components, nucleic acids, and pore-forming toxins; environmental crystalline pollutants like silica, asbestos, and alum; and endogenous danger signals like ATP, serum amyloid A, and uric acid crystals (Man and Kanneganti, 2015). In fact, the diversity of its activators is one of the most distinctive features of the NLRP3 inflammasome and makes the possibility of a direct interaction with each activator unlikely. It is therefore assumed that the NLRP3 inflammasome either senses a common secondary activator downstream of these stimuli or responds to cellular stress associated with infection or physiological damage. Production of reactive oxygen species (ROS), potassium efflux, changes in cell volume, calcium signaling, and lysosomal disruption have all been proposed as critical upstream signals required for NLRP3 activation (Cruz et al., 2007; Pétrilli et al., 2007; Cassel et al., 2008; Dostert et al., 2008; Halle et al., 2008; Hornung et al., 2008; Schorn et al., 2011; Zhou et al., 2011; Compan et al., 2012; Lee et al., 2012; Murakami et al., 2012; Muñoz-Planillo et al., 2013). Even though potassium efflux has been reported by multiple groups to be the unifying mechanism regulating NLRP3 activation, potassium efflux is also observed in response to NLRP1b activation (Pétrilli et al., 2007), raising doubt over the specificity of potassium efflux for NLRP3 inflammasome activation. Nevertheless, all of these features are associated with cellular stress, suggesting NLRP3 could be a global sensor of cellular damage (Fig. 1).
Although the precise trigger is unknown, recent studies have identified multiple cellular mechanisms that regulate activation of the NLRP3 inflammasome. Under resting conditions, ASC is found in the mitochondria, cytosol, and nucleus, whereas NLRP3 associates with the ER (Zhou et al., 2011). Nucleation of the inflammasome requires a change in the subcellular localization of these molecules to facilitate their interaction. This change in spatial arrangement is brought about by dynein-mediated transport of mitochondria toward the ER, bringing ASC and NLRP3 in close proximity (Misawa et al., 2013). Further, mitochondrial dysfunction induces microtubule alterations that enable the juxtaposition of NLRP3 and ASC, suggesting that diverse cellular functions involved in mitochondrial homeostasis and cytoskeletal modulations coordinate and converge on NLRP3 activation (Misawa et al., 2013). Although colocalization of inflammasome components is required for all inflammasomes to be activated, inhibitors of microtubule alterations specifically block the NLRP3 inflammasome, suggesting that distinct cytoskeletal signatures are involved in activation of certain inflammasomes (Misawa et al., 2013).
In addition to cytoskeletal rearrangements, recent studies have identified NEK7, a protein involved in cell-cycle progression, as a novel regulator of the NLRP3 inflammasome (He et al., 2016; Schmid-Burgk et al., 2016; Shi et al., 2016). NEK7 is a serine/threonine kinase that is known to promote mitotic spindle formation and cytokinesis and is required for progression through mitosis (O’Regan and Fry, 2009). NEK7 interacts with the LRR domain of NLRP3, independent of its kinase activity, and is required for NLRP3 inflammasome activation (He et al., 2016). This interaction additionally restricts NLRP3 inflammasome activation to cells in interphase (Shi et al., 2016). It was hypothesized that this cell cycle–mediated restriction of NLRP3 activation is caused by the limited cellular concentration of NEK7, such that once a cell enters mitosis, NEK7 is localized to mitotic spindles and NLRP3 activation is attenuated (Shi et al., 2016). The significance of this intersection between NLRP3 activation and cell cycle progression is currently unknown, though it could be posited that this is a safeguard cells use to dampen the inflammasome while undergoing cell division, during which the cell may be enriched in endogenous ligands that could be misrepresented as cellular stress.
NLRC4
NLRC4 activates procaspase-1 through its CARD domain to induce cell death (Poyet et al., 2001) in response to Salmonella infection (Mariathasan et al., 2004). Bacterial flagellin (Franchi et al., 2006; Miao et al., 2006; Molofsky et al., 2006; Ren et al., 2006) and multiple components of the bacterial type III secretion systems (Miao et al., 2010b; Zhao et al., 2011; Yang et al., 2013) activate the NLRC4 inflammasome, enabling it to provide host defense against a diverse range of pathogens. However, NLRC4 does not interact directly with its activators; instead, a family of proteins, termed NAIPs (NLR family, apoptosis inhibitory proteins), act as sensors that recognize the ligand and induce activation of the NLRC4 inflammasome. Mouse NAIP1 and NAIP2 recognize bacterial needle and inner rod proteins, respectively, whereas NAIP5 and NAIP6 bind to flagellin (Fig. 1; Kofoed and Vance, 2011; Zhao et al., 2011; Rayamajhi et al., 2013; Yang et al., 2013). The human genome encodes only one known NAIP (Endrizzi et al., 2000), which can sense both flagellin and needle proteins, suggesting a similarity in the mode of recognition of the two structurally related ligands (Yang et al., 2013; Kortmann et al., 2015).
NLRC4 contains a winged-helix domain, which together with the NBD stabilizes NLRC4 in a closed conformation under basal conditions, whereas the LRR domain provides steric hindrance to its oligomerization (Tenthorey et al., 2014). These two inhibitory mechanisms thus maintain NLRC4 in an inactive state in the absence of a ligand. Support for this theory comes from structural modeling based on cryo-EM analysis of NAIP5-NLRC4 multimers and a crystal structure of dormant NLRC4 (Diebolder et al., 2015). This study suggested that rotation of the LRR domain would be required to allow NLRC4 to fit into the active oligomeric form detected by cryo-EM (Diebolder et al., 2015). A recently characterized cryo-EM structure of the NLRC4 inflammasome in response to bacterial type III secretion systems rod protein PrgJ revealed that nucleation is initiated from a single, activated NAIP2 molecule that subsequently induces NLRC4 polymerization. The inflammasome assembly follows a domino-like reaction, wherein the activated NAIP2 molecule provides a platform for self-oligomerization of NLRC4 to form a wheel- or disc-like structure (Hu et al., 2015; Zhang et al., 2015).
In addition to removal of infected cells through pyroptosis and perpetuation of inflammation through IL-1β and IL-18 release, NLRC4 activation affects other aspects of cell biology that are critical in host defense. In macrophages infected with Salmonella, NLRC4 induces an actin polymerization response that prevents further bacterial uptake and increases intracellular ROS production to enhance intracellular killing and lower bacterial dissemination (Man et al., 2014a). NLRC4 activation in epithelial cells is specifically required to control pathogen load during Salmonella infection. In accordance with this, epithelium-specific deletion of inflammasome components (Naip 1–6, NLRC4, and caspase-1/caspase-11) results in higher bacterial load and extraintestinal dissemination (Sellin et al., 2014). A surprising feature of NLRC4 activation is observed in a mouse model using intraperitoneal delivery of flagellin, which induces rapid host mortality (within 30 min). This striking inflammasome-dependent response is independent of IL-1β or IL-18 and instead relies on the production of eicosanoids, including prostaglandins and leukotrienes. This inflammasome-dependent eicosanoid production leads to rapid vascular fluid loss and mortality (von Moltke et al., 2012). How the NLRC4 inflammasome activates biosynthesis of eicosanoids and whether this function of eicosanoid production can be extended to all inflammasomes are currently unknown. Overall, these studies highlight the importance of NLRC4 inflammasome in cellular responses beyond the conventional roles of cytokine release and pyroptosis.
Human genetic studies have identified gain-of-function mutations in NLRC4 that are associated with autoinflammation and enterocolitis (Canna et al., 2014; Kitamura et al., 2014; Romberg et al., 2014). Because NLRC4 has been ascribed such diverse functions in animal models of infection, it remains to be seen whether human patients recapitulate any specific features of NLRC4 activation.
NLRP6
NLRP6 is uncharacteristic by virtue of the unusual ligand it recognizes and its observed downstream effects. One of the earliest studies identified NLRP6 as a negative regulator of the mitogen-activated protein kinase and NF-κB pathway in macrophages infected with Listeria monocytogenes, Escherichia coli, and Salmonella (Anand et al., 2012). Thereafter, the role of NLRP6 in activation of caspase-1 was hypothesized as being required to maintain optimal mucosal response to chemical damage and pathogens (Elinav et al., 2011; Wlodarska et al., 2014). In the gut, NLRP6 antagonizes microbial dysbiosis through mucus production (Elinav et al., 2011). In the absence of NLRP6, the intestinal barrier integrity is compromised, rendering the animal susceptible to pathogenic and chemical insults (Chen et al., 2011; Elinav et al., 2011; Huber et al., 2012; Hu et al., 2013; Nowarski et al., 2015). The identity of the ligand that activates NLRP6 remained elusive until a recent study identified taurine, a microbial metabolite, as its activator (Levy et al., 2015). The authors demonstrated that NLRP6–ASC–caspase-1 axis, through IL-18 production, promotes induction of antimicrobial peptides that resist microbial dysbiosis. The normal microbiota in turn produces a specific metabolite profile enriched in taurine that engages NLRP6 in the gut to provide mucosal immunity (Levy et al., 2015). NLRP6 therefore has a central role in gut homeostasis and associated disorders (Henao-Mejia et al., 2012). It should be noted that altered or dysbiotic microbiota is also observed in mice lacking inflammasome sensors NLRP3 (Henao-Mejia et al., 2012) and AIM2 (Man et al., 2015b), suggesting that the inflammasome plays a nonredundant role in maintaining gut bacterial symbiosis. The role of NLRP6 at mucosal sites is relevant even in the context of viral infection. During systemic infection with encephalomyocarditis and norovirus, NLRP6 affected viral loads specifically in the gastrointestinal tract, and oral infection with the same viruses required NLRP6-mediated defense to confer survival (Wang et al., 2015). However, direct proof for the importance of NLRP6 at nonmucosal sites is yet to be demonstrated and further investigation is needed to uncover the molecular mechanisms governing this putative inflammasome.
NLRP12
NLRP12 was initially characterized as an inhibitor of noncanonical NF-κB signaling (Williams et al., 2005; Lich et al., 2007). It was also involved in caspase-1 activation in response to Yersinia pestis and Plasmodium infection (Vladimer et al., 2012; Ataide et al., 2014). The role of NLRP12 in NF-κB–mediated signaling makes it an important regulator of immune response after Salmonella infection and during colorectal cancer (Zaki et al., 2011, 2014). Further, it modulates NF-κB signaling intrinsically in T cells during their activation, thereby affecting their colitogenic and encephalitogenic potential (Lukens et al., 2015). However, the ligands that activate NLRP12 and its regulatory mechanisms remain poorly understood.
ALR family
The ALR family is named for its most well-characterized member, AIM2 (absent in melanoma 2). The members of the ALR family are characterized by an N-terminal PYD and a C-terminal hematopoietic interferon-inducible nuclear protein with 200–amino acid repeat (HIN200) domain. The expression of ALRs is restricted to and conserved among mammalian species (Cridland et al., 2012). AIM2 has a distinct cytosolic localization by virtue of lacking a nuclear localization signal that is present in all other members. Further, the PYD of AIM2 is functionally distinct from other ALRs in its ability to interact with the PYD of ASC (Hornung et al., 2009). It is because of this specific interaction that AIM2 is the only member of this family known to be capable of forming an inflammasome. Other mouse ALRs have been shown to interact with ASC, making them potential regulators of inflammasome responses (Brunette et al., 2012), and another human ALR, γ-IFN–inducible protein 16, is considered a putative inflammasome (Kerur et al., 2011). However, most major studies have concerned the identification and regulation of the AIM2 inflammasome.
AIM2
AIM2 was initially identified as a γ-IFN–inducible protein in a tumor suppressor screen. A later study redefined it as a nucleic acid sensor that assembles into an inflammasome, specifically in response to double-stranded DNA (dsDNA; Fig. 1; Bürckstümmer et al., 2009; Fernandes-Alnemri et al., 2009; Hornung et al., 2009; Roberts et al., 2009). AIM2 is critical to immune responses after infection with various viral and bacterial infectious agents, such as vaccinia virus, mouse cytomegalovirus (Rathinam et al., 2010), Francisella tularensis (Fernandes-Alnemri et al., 2010; Jones et al., 2010; Rathinam et al., 2010), and Listeria (Kim et al., 2010; Rathinam et al., 2010).
The HIN200 domain of AIM2 is involved in ligand binding and mediates dsDNA recognition, independent of sequence identity, but requires the dsDNA to be of a certain length (∼80 bp; Jin et al., 2012). Structural and biophysical studies suggest existence of sequence-neutral electrostatic interactions between the HIN200 domain and DNA bases (Jin et al., 2012; Li et al., 2014). Structural analysis also ascribes an autoinhibitory role to the HIN200 domain, because in the absence of a ligand, it interacts with the PYD of AIM2. DNA binding by the HIN200 domain relieves this autoinhibition, allowing the PYD of AIM2 to undergo homotypic interaction with the adaptor ASC (Jin et al., 2013b). Reconstituted AIM2–ASC–caspase-1 inflammasome, viewed by EM, exhibits a star-shaped structure with multiple filaments radiating from a central hub. In this complex, AIM2 and ASC form the center, whereas caspase-1 makes up the filaments (Lu et al., 2014).
Although AIM2 binds to both viral and bacterial DNA, there is a distinct difference in their inflammasome engagement. Type I IFN signaling is required for AIM2 activation in response to bacterial stimuli, such as Francisella, but not viral stimuli, like mouse cytomegalovirus (Man et al., 2015a; Meunier et al., 2015). Type I IFN signaling induces expression of IFN regulatory genes that mediate bacterial cytosolic escape and bacterial lysis, releasing the ligand for AIM2 activation (Man et al., 2015a; Meunier et al., 2015). Despite the ubiquitous nature of this ligand, not all bacteria and viruses activate the AIM2 inflammasome. Recent studies indicate that certain bacteria, such as Mycobacterium and Legionella, encode virulence factors that can reduce dsDNA release and thereby evade detection by AIM2 (Peng et al., 2011; Ge et al., 2012; Shah et al., 2013). The AIM2 inflammasome is also inhibited by host-encoded decoy molecules and dominant-negative regulators (Dombrowski et al., 2011; Morizane et al., 2012; Khare et al., 2014). Another member of the HIN200 family, p202, lacks the N-terminal PYD and acts as a negative regulator of the AIM2 inflammasome, perhaps by sequestering dsDNA and/or AIM2 through heterodimerization (Roberts et al., 2009; Yin et al., 2013). Recognition of nucleic acids also grants AIM2 the ability to respond to host DNA released in response to cellular damage. This recognition and the ensuing inflammasome activation underlie various autoinflammatory diseases, such as psoriasis, systemic lupus erythematosus, and abdominal aortic aneurysm (Dombrowski et al., 2011; Choubey, 2012; Dihlmann et al., 2014), raising the possibility of AIM2 as a therapeutic target for these autoimmune disorders.
Pyrin
Pyrin is associated with an autoinflammatory disorder called familial Mediterranean fever and was only recently identified as an inflammasome-forming protein. Pyrin was first implicated in inflammasome activation from a mouse model expressing familial Mediterranean fever mutation–containing pyrin; these mice exhibited an ASC and IL-1–mediated autoinflammatory disorder (Chae et al., 2011). A more recent study provided definitive proof by showing that the pyrin inflammasome assembles in response to Rho-modifying toxins produced by various bacterial species, including Clostridium difficile (TcdB), Vibrio parahemolyticus (VopS), Histophilus somni (IbpA), Clostridium botulinum (C3), and Burkholderia cenocepacia (Fig. 1; Xu et al., 2014). These toxins induce covalent modifications that include glycosylation, adenylylation, and ADP ribosylation within the Switch I region of Rho family members, suggesting a universal signature detected by the pyrin inflammasome. In another study, pertussis toxin, through its ADP-ribosyltransferase activity, was shown to engage the pyrin inflammasome (Dumas et al., 2014). Although a direct interaction between Rho and pyrin has not been detected, Rho modification seems to be essential for pyrin inflammasome activation, suggesting that pyrin responds to the functional activity of Rho. However, the consequence of Rho modification and the intermediary steps involved in pyrin activation are currently unknown. As Rho modifications are closely associated with multiple cellular functions, including division and migration, pyrin activation is likely kept in check during these processes. A recent study identified 14-3-3 proteins as regulators of pyrin activity until a pyrin-activating stimuli releases pyrin from this inhibition (Masters et al., 2016). Pyrin phosphorylation targets 14-3-3 for binding under basal conditions and dephosphorylation attenuates 14-3-3 binding. Further, a human mutation in this phosphosite recapitulates pyrin activation, even in the absence of an exogenous stimuli (Masters et al., 2016). The molecular regulators controlling pyrin phosphorylation and dephosphorylation are still being elucidated. 14-3-3 proteins are critical scaffolds involved in cell cycle progression and cell death, so it remains to be seen whether 14-3-3 interaction forms a nexus between these cellular processes and pyrin activation.
ASC oligomerization and the “ASC speck”
The interaction between major inflammasome components occurs through homotypic PYD–PYD or CARD–CARD interactions. The PYD and CARD domains possess nucleating ability that allows them to induce oligomerization, forming the structural basis for assembly of inflammasomes (Cai et al., 2014; Lu et al., 2014; Sborgi et al., 2015). Based on these domains, inflammasome sensors assemble into complexes with or without the adaptor ASC. CARD-containing NLRP1 and NLRC4 directly recruit caspase-1 (Nour et al., 2009; Jin et al., 2013a; Ponomareva et al., 2013), whereas PYD-containing NLRP3, AIM2, and pyrin recruit ASC for inflammasome assembly. Even though NLRP1 and NLRC4 can induce pyroptosis in an ASC-independent manner, efficient cytokine processing depends on ASC-mediated supramolecular complex assembly (Mariathasan et al., 2004; Broz et al., 2010b; Guey et al., 2014; Van Opdenbosch et al., 2014). Because of the homotypic interactions underlying inflammasome assembly, pyrin-only proteins and CARD-only proteins can act as dominant-negative regulators to block transduction of inflammasome signaling (Matusiak et al., 2015).
ASC-dependent NLRP3 and AIM2 activation have been recapitulated in vitro and proceeds through two subsequent nucleation events. First, the sensor nucleates ASC to form oligomeric filaments. Second, ASC nucleates caspase-1, such that the sensor, the adaptor, and the enzyme are present at increasing concentrations within the larger complex to effectuate signal amplification (Lu et al., 2014). ASC specks generated in response to inflammasome activation can be visualized as micrometer-sized foci and can be recapitulated in vitro. Under physiological pH, ASC assembles into filaments through electrostatic and hydrophobic interactions among the PYD domains, as observed by nuclear magnetic resonance spectroscopy and cryo-EM (Moriya et al., 2005; Sborgi et al., 2015). The CARD domain in these filaments is dynamic to facilitate interaction with the CARD domain of caspase-1. Mutational studies pinpointed multiple interacting surfaces between ASC–CARD and caspase-1–CARD, and inflammasome activity is dependent on these interfaces (Proell et al., 2013). Although large strides have been made in determining the structural features of inflammasomes, the architecture of the inflammasome in intact cells remains unresolved.
Activation of inflammasome is an all-or-none phenomenon (Liu et al., 2014), and a pathogen can simultaneously engage multiple inflammasome sensors within a single cell (Broz et al., 2010a; Man et al., 2014b; Karki et al., 2015). It is notable that irrespective of activation of multiple inflammasomes, they all converge on a single supramolecular structure and ASC speck per cell (Broz et al., 2010a; Man et al., 2014b; Karki et al., 2015). It was recently demonstrated that in response to Salmonella infection, NLRC4 recruits NLRP3 and ASC to promote inflammasome activation, highlighting crosstalk between the inflammasome sensors (Qu et al., 2016). Salmonella activates NLRP3 and NLRC4 through distinct stimuli (Broz et al., 2010a), so it remains to be seen how and at what stage these signals are integrated. It is possible that the two sensors, once in close proximity, somehow interact and merge to form one complex, though this remains to be confirmed through direct visualization of such events.
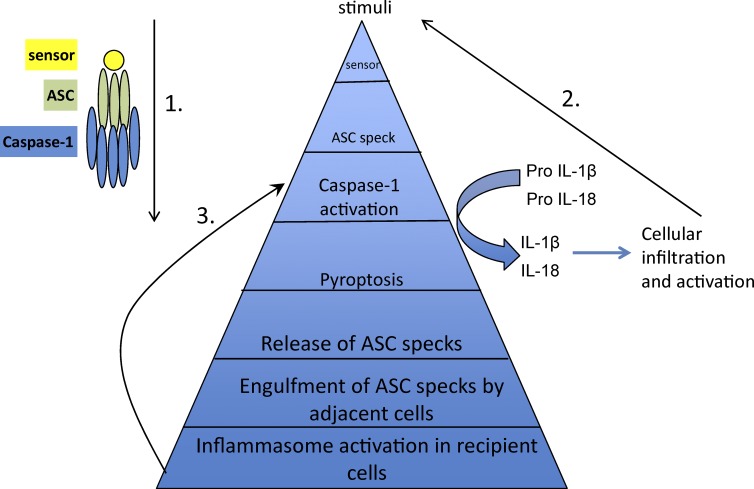
In addition to driving pyroptosis and cytokine maturation, inflammasome activation leads to release of ASC specks that are phagocytosed by adjacent cells. This drives ASC nucleation, demonstrating the ability of specks to act as inflammasome-perpetuating signals in recipient cells (Baroja-Mazo et al., 2014; Franklin et al., 2014). Given the ability of PYD and CARD domains to assemble into filamentous structures, the initial nucleation event might allow an ASC speck to act as a scaffold for addition of soluble ASC monomers, via a prionic, self-propagating mechanism (Cheng et al., 2010; Cai et al., 2014; Franklin et al., 2014). Thus, the ASC speck can potentiate inflammation by amplifying the inflammasome response through multiple mechanisms (Fig. 2).
Figure 2.
ASC-mediated amplification of inflammasome activation. ASC mediates amplification of inflammasome signals through at least three distinct mechanisms. (1) Signal transduction during inflammasome assembly, such that the sensor, ASC, and caspase-1 are present at increasing concentrations and each sensor activates a much greater number of the enzyme molecules. (2) Caspase-1–mediated maturation and release of bioactive cytokines affects activation and further infiltration of immune cells, eventually amplifying the overall inflammatory response. (3) The released ASC speck can be taken up by neighboring cells and promotes ASC assembly in the recipient cells, consequently providing another mode of inflammasome amplification.
Caspase-11 and the NLRP3 inflammasome
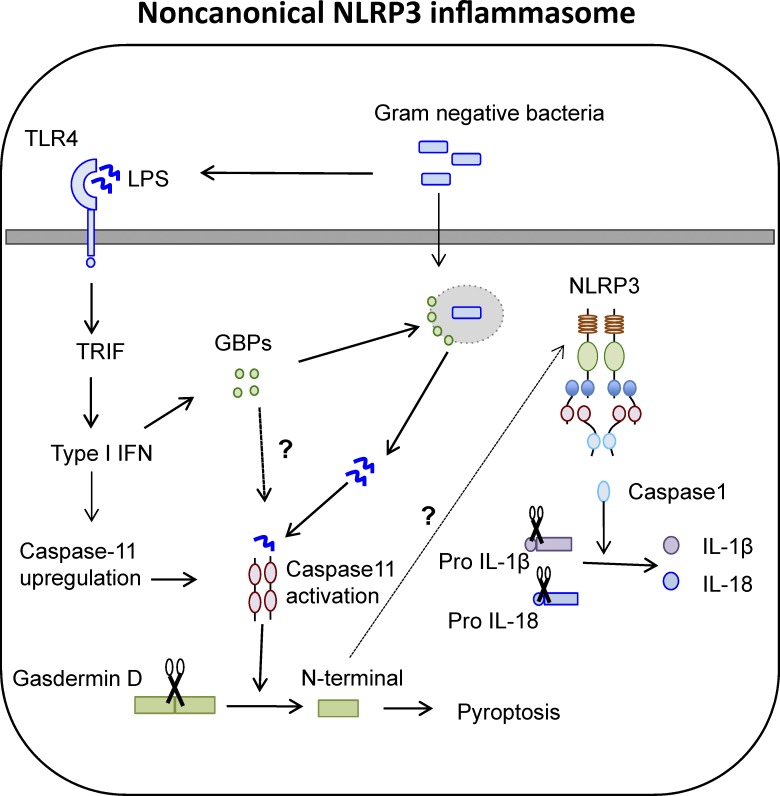
Caspase-11 is an inflammatory caspase identified as a binding partner and mediator of caspase-1 activation (Wang et al., 1998). The functional significance of caspase-11 was masked for a long time because of its high genetic linkage to caspase-1 (Kayagaki et al., 2011). It was eventually identified as an inducer of pyroptosis independent of caspase-1 and was further required for NLRP3 inflammasome activation specifically in response to gram-negative bacteria through its CARD domain (Shi et al., 2014). Caspase-11 acts as both the sensor and the inducer of lipopolysaccharide (LPS)-induced pyroptotic responses and further triggers assembly of the NLRP3 inflammasome. This distinguishes the two functions of inflammasome activation, such that pyroptosis proceeds through activation of caspase-11, whereas cytokine processing occurs through NLRP3-mediated caspase-1 activation (Fig. 3).
Figure 3.
Caspase-11–regulated NLRP3 inflammasome. The NLRP3 inflammasome can be assembled in response to caspase-11–mediated recognition of cytosolic LPS. The up-regulation of caspase-11 and induction of guanylate binding proteins (GBPs), involved in vacuolar lysis for LPS release, is mediated through a TLR4–TRIF–type I IFN signaling module. Caspase-11 directly cleaves gasdermin D to induce pyroptosis and activate NLRP3 through an as-of-yet-unknown mechanism.
How caspase-11 promotes the noncanonical NLRP3 activation is not entirely known. Although caspase-11 can interact with caspase-1 (Wang et al., 1998), loss of caspase-11 does not affect canonical NLRP3–ASC complex assembly, disproving its role as an essential adaptor. Another possibility is that caspase-11 employs a downstream molecule to instigate NLRP3 inflammasome assembly. For example, caspase-11 activation causes a drop in intracellular potassium levels, suggesting that potassium efflux could bridge caspase-11 and NLRP3 activation (Rühl and Broz, 2015; Schmid-Burgk et al., 2015). Pannexin-1 is also a caspase-11 target, and pannexin-1 cleavage induces activation of the NLRP3 inflammasome through a drop in intracellular potassium levels (Yang et al., 2015). However, the specificity of potassium efflux to NLRP3 activation has not been established (Pétrilli et al., 2007), so it remains to be seen whether intracellular potassium is the exclusive link between caspase-11 and NLRP3 activation.
Pyroptosis and GSDMD
Inflammasome activation leads to a distinct form of programmed cell death termed pyroptosis, characterized by cellular lysis, release of intracellular components, and an inflammatory response. To date, caspase-induced pyroptosis has been demonstrated in macrophages (Fink et al., 2008), dendritic cells (Edgeworth et al., 2002), enterocytes (Sellin et al., 2014), and hematopoietic progenitors (Masters et al., 2012), whereas neutrophils and monocytes do not undergo pyroptosis after inflammasome activation (Miao et al., 2010a; Chen et al., 2014; Gaidt et al., 2016). Pyroptosis also appears to be evolutionarily conserved, as loss of macrophages caused by pyroptosis has been visualized in zebrafish larvae after Listeria infection, marked by ASC speck and nuclear condensation (Vincent et al., 2016). Pyroptosis is therefore a critical feature of inflammasome activation in a wide variety of cells.
Pyroptosis is a distinct form of cell death but shares certain features with both apoptosis and necrosis. Similar to necrosis, pyroptosis involves the formation of 1- to 2-nm pores on the cell membrane (Fink and Cookson, 2006). It further proceeds through cytoplasmic swelling, osmotic lysis, and release of intracellular contents. Similar to apoptosis are the features of nuclear condensation and DNA damage (Monack et al., 1996; Fink and Cookson, 2006; Lamkanfi et al., 2008). Furthermore, processing of executioner caspase-3 and -7 is observed in both apoptosis and pyroptosis (Lamkanfi et al., 2008; Akhter et al., 2009), though it is important to note that, during pyroptosis, membrane permeabilization and DNA fragmentation can occur even in the absence of executioner caspases. These studies highlight both the similarities and distinctions in these cell death pathways.
Until recently the mediators driving cell death after caspase-1 or caspase-11 activation were completely unknown. Although caspase-7 was identified as a caspase-1 target (Lamkanfi et al., 2008; Akhter et al., 2009), pyroptosis can occur independently of executioner caspases, ruling out its role in pyroptosis. A missing link between caspase-1 activation and pyroptosis was later identified as GSDMD by three independent groups (He et al., 2015; Kayagaki et al., 2015; Shi et al., 2015). GSDMD was found to be a direct target of inflammatory caspase-1, caspase-4, and caspase-11 and is essential for pyroptosis and IL-1β release after inflammasome activation (Kayagaki et al., 2015; Shi et al., 2015). Caspase-1, -4, -5, and -11 recognize and cleave GSDMD at the same site (Kayagaki et al., 2015; Shi et al., 2015). GSDMD cleavage is required for its activity, and the N terminus is sufficient to induce pyroptosis, suggestive of an autoinhibitory role for the GSDMD C terminus (Shi et al., 2015). These studies highlight the importance of GSDMD as a critical inducer of pyroptosis and provide one of the missing links between activation of inflammatory caspases and cell death. These studies also shed light on the process of IL-1β secretion; although maturation is caspase-1 dependent, release requires pyroptosis (He et al., 2015; Kayagaki et al., 2015; Shi et al., 2015). This idea of dying cells as the major source of secreted IL-1β was put forth and demonstrated in another study that characterized caspase-1 activation as an all-or-none signal (Liu et al., 2014). However, how GSDMD executes cell death is still not known. Nevertheless, identification of GSDMD will now allow investigators to assess the physiological relevance of pyroptosis independent of other aspects of inflammasome activation.
Caspase-8 and inflammasomes
Although caspase-8 has long been considered to be in the domain of apoptosis, recent studies have highlighted its contribution to inflammasome activation, IL-1β production, and cell death in response to a range of ligands. Fungal, mycobacterial, or β-glycan recognition through dectin-1 induces caspase-8–dependent IL-1β processing (Gringhuis et al., 2012) via a complex composed of caspase-8, MALT1, and ASC (Gringhuis et al., 2012). Although this complex employs the inflammasome adaptor ASC and leads to processing of IL-1β, a typical inflammasome readout, it does not proceed through activation of inflammatory caspase-1 or caspase-11. For this reason, it is not widely accepted as an inflammasome platform (Man and Kanneganti, 2016). In response to NLRC4 activation by Salmonella, caspase-8 is recruited to the inflammasome via ASC and contributes to IL-1β processing (Man et al., 2013). Caspase-8 was also shown to have a role in both priming and activation of the NLRP3 inflammasome (Gurung et al., 2014). This role for caspase-8 in mediating inflammasome activation is supported by another study in which caspase-8 inhibition reduced NLRP3 and AIM2-induced caspase-1 activation (Wu et al., 2014). Caspase-8 additionally acts in concert with NLRP3 inflammasome to drive IL-1β–mediated osteomyelitis in PSTPIP2cmo mice (Lukens et al., 2014; Gurung et al., 2016). ASC-mediated caspase-8 recruitment and activation additionally leads to apoptosis in response to AIM2 and NLRP3 activation. This study proposed that although pyroptosis is a rapid form of cell death, cells lacking caspase-1 undergo caspase-8–mediated apoptosis induced with delayed kinetics (Sagulenko et al., 2013). This ASC/caspase-8–mediated apoptosis in response to AIM2 stimulation is also observed in response to Francisella infection (Pierini et al., 2012). Even though caspase-8 is known to be a determinant of cell survival or cell death through apoptosis, these studies have ascribed a pivotal role for caspase-8 in inflammasome activation.
Autophagy and inflammasomes
Autophagy involves the degradation of damaged organelles and recycling of cellular metabolites. Inflammasomes, on the other hand, respond to stimuli that mark cellular damage through aberrant physiological functions or in response to infectious agents. The intersection of these two cellular responses is therefore inevitable and critical to the maintenance of cellular physiology. Cells activate autophagy in response to a pathogenic insult to contain the infection and promote clearance of the pathogen (Shi et al., 2012). Autophagy also clears damaged organelles generated as a byproduct of homeostatic or activatory conditions (Jabir et al., 2015) and thereby attenuates the inflammasome response. Mechanisms so far identified to mediate inflammasome inhibition via autophagy include the dampening of ROS production (Zhou et al., 2011; Lupfer et al., 2013), removal of damaged mitochondria (Jabir et al., 2015), degradation of ASC aggregates (Shi et al., 2012), and sequestration of pro–IL-1β (Harris et al., 2011). Conversely, blocking autophagy promotes inflammasome activation through accumulation of ROS-generating mitochondria (Zhou et al., 2011; Jabir et al., 2015). Further, cells lacking components of autophagosome assembly, such as ATG16L, LC3B, and beclin-1 (Saitoh et al., 2008; Nakahira et al., 2011), show increased inflammasome activation. In addition, IL-1β promotes autophagy, highlighting a negative feedback mechanism for inflammasome activation (Peral de Castro et al., 2012).
Conversely, autophagy can be regulated by inflammasome components. NLRP3 and NLRC4 can interact with beclin 1, an important regulator of autophagy. Association with NLRC4 inhibits autophagy under both physiological conditions and bacterial infection. NLRC4 additionally interacts with the class C vacuolar protein-sorting complex and inhibits autophagosome maturation (Jounai et al., 2011). Caspase-1 activation can also inhibit autophagy in response to inflammasome activation after bacterial stimuli. Toll/Interleukin receptor domain-containing adapter-inducing interferon β (TRIF) is a caspase-1 substrate, such that caspase-1 activation reduces TRIF-dependent type I IFN–mediated autophagy (Jabir et al., 2014). The functional consequence of this modulation is not entirely known and seems to be a mechanism to amplify the inflammatory response by blocking autophagy-mediated bacterial clearance and removal of endogenous inflammasome activators. In addition to autophagy, inflammasome activation can affect other physiological processes associated with cellular metabolism. Glycolytic enzymes are direct caspase-1 substrates, and reduced glycolytic flux is observed in response to inflammasome activation (Shao et al., 2007). Inflammasome activation additionally induces lipid synthesis and membrane biogenesis through activation of sterol regulatory element binding proteins (Gurcel et al., 2006). This serves as a means to promote cell membrane repair and consequent cell survival.
These studies highlight that inflammasome activation controls multiple processes involved in cellular biology. In addition to inducing cell death, caspase-1 activation affects signals regulating cell metabolism and survival. It is tempting to hypothesize that in response to a cellular stress, multiple signaling modules are affected, including pyroptosis/apoptosis and cellular metabolism, such that based on the degree of damage, the overall fate of the affected cell could be repair or death.
Conclusion and future directions
Inflammasome activation is intricately intertwined with basic cellular functions. In addition to removal of damaged cells, inflammasomes are also involved in cell repair, metabolism, and proliferation. Various molecules believed to be involved in the maintenance of cellular homeostasis have been demonstrated to act as critical regulators of inflammasome function and vice versa. Newly uncovered functions for the inflammasome in cell metabolism and proliferation require further investigation. In addition, several questions still remain: What are the endogenous triggers of inflammasome activation, and how are they generated during physiological processes? What is the link between cell division and inflammasome activation? What regulates inflammasome assembly in response to multiple-sensor activation to permit one ASC speck per cell? What are the cytoskeletal changes involved in inflammasome activation, and how are they brought about? What are the molecules involved in GSDMD-mediated cell rupture? What regulates the crosstalk between different cell death mechanisms? Investigating these and other research avenues highlighted in this review will expand our current understanding of the cell biology of inflammasomes and fuel the interest of the research community for years to come.
Acknowledgments
We would like to thank Drs. Si Ming Man and Prajwal Gurung for their helpful suggestions in writing and editing this manuscript. We apologize to our colleagues whose work we could not cite because of space limitations.
Work from the laboratory is supported by grants from the National Institutes of Health (AR056296, CA163507, AI101935, and AI124346) and the American Lebanese Syrian Associated Charities (to T.-D. Kanneganti).
The authors declare no competing financial interests.
Footnotes
Abbreviations used in this paper:
- ALR
- absent in melanoma 2–like receptor
- CARD
- caspase activation and recruitment domain
- dsDNA
- double-stranded DNA
- GSDMD
- gasdermin D
- HIN200
- hematopoietic interferon-inducible nuclear proteins with 200–amino acid repeat
- LPS
- lipopolysaccharide
- LRR
- leucine-rich repeat
- NBD
- nucleotide-binding domain
- NLR
- nucleotide-binding domain–like receptor
- PYD
- pyrin domain
- ROS
- reactive oxygen species
- TRIF
- Toll/Interleukin receptor domain-containing adapter-inducing interferon β
References
- Akhter A., Gavrilin M.A., Frantz L., Washington S., Ditty C., Limoli D., Day C., Sarkar A., Newland C., Butchar J., et al. 2009. Caspase-7 activation by the Nlrc4/Ipaf inflammasome restricts Legionella pneumophila infection. PLoS Pathog. 5:e1000361 10.1371/journal.ppat.1000361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anand P.K., Malireddi R.K., Lukens J.R., Vogel P., Bertin J., Lamkanfi M., and Kanneganti T.D.. 2012. NLRP6 negatively regulates innate immunity and host defence against bacterial pathogens. Nature. 488:389–393. 10.1038/nature11250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ataide M.A., Andrade W.A., Zamboni D.S., Wang D., Souza M.C., Franklin B.S., Elian S., Martins F.S., Pereira D., Reed G., et al. 2014. Malaria-induced NLRP12/NLRP3-dependent caspase-1 activation mediates inflammation and hypersensitivity to bacterial superinfection. PLoS Pathog. 10:e1003885 (published erratum appears in PLoS Pathog. 2014. 10:e1004258) 10.1371/journal.ppat.1003885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baroja-Mazo A., Martín-Sánchez F., Gomez A.I., Martínez C.M., Amores-Iniesta J., Compan V., Barberà-Cremades M., Yagüe J., Ruiz-Ortiz E., Antón J., et al. 2014. The NLRP3 inflammasome is released as a particulate danger signal that amplifies the inflammatory response. Nat. Immunol. 15:738–748. 10.1038/ni.2919 [DOI] [PubMed] [Google Scholar]
- Broz P., Newton K., Lamkanfi M., Mariathasan S., Dixit V.M., and Monack D.M.. 2010a Redundant roles for inflammasome receptors NLRP3 and NLRC4 in host defense against Salmonella. J. Exp. Med. 207:1745–1755. 10.1084/jem.20100257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broz P., von Moltke J., Jones J.W., Vance R.E., and Monack D.M.. 2010b Differential requirement for Caspase-1 autoproteolysis in pathogen-induced cell death and cytokine processing. Cell Host Microbe. 8:471–483. 10.1016/j.chom.2010.11.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunette R.L., Young J.M., Whitley D.G., Brodsky I.E., Malik H.S., and Stetson D.B.. 2012. Extensive evolutionary and functional diversity among mammalian AIM2-like receptors. J. Exp. Med. 209:1969–1983. 10.1084/jem.20121960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bürckstümmer T., Baumann C., Blüml S., Dixit E., Dürnberger G., Jahn H., Planyavsky M., Bilban M., Colinge J., Bennett K.L., and Superti-Furga G.. 2009. An orthogonal proteomic-genomic screen identifies AIM2 as a cytoplasmic DNA sensor for the inflammasome. Nat. Immunol. 10:266–272. 10.1038/ni.1702 [DOI] [PubMed] [Google Scholar]
- Cai X., Chen J., Xu H., Liu S., Jiang Q.X., Halfmann R., and Chen Z.J.. 2014. Prion-like polymerization underlies signal transduction in antiviral immune defense and inflammasome activation. Cell. 156:1207–1222. 10.1016/j.cell.2014.01.063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canna S.W., de Jesus A.A., Gouni S., Brooks S.R., Marrero B., Liu Y., DiMattia M.A., Zaal K.J., Sanchez G.A., Kim H., et al. 2014. An activating NLRC4 inflammasome mutation causes autoinflammation with recurrent macrophage activation syndrome. Nat. Genet. 46:1140–1146. 10.1038/ng.3089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassel S.L., Eisenbarth S.C., Iyer S.S., Sadler J.J., Colegio O.R., Tephly L.A., Carter A.B., Rothman P.B., Flavell R.A., and Sutterwala F.S.. 2008. The Nalp3 inflammasome is essential for the development of silicosis. Proc. Natl. Acad. Sci. USA. 105:9035–9040. 10.1073/pnas.0803933105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavailles P., Flori P., Papapietro O., Bisanz C., Lagrange D., Pilloux L., Massera C., Cristinelli S., Jublot D., Bastien O., et al. 2014. A highly conserved Toxo1 haplotype directs resistance to toxoplasmosis and its associated caspase-1 dependent killing of parasite and host macrophage. PLoS Pathog. 10:e1004005 10.1371/journal.ppat.1004005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chae J.J., Cho Y.H., Lee G.S., Cheng J., Liu P.P., Feigenbaum L., Katz S.I., and Kastner D.L.. 2011. Gain-of-function Pyrin mutations induce NLRP3 protein-independent interleukin-1β activation and severe autoinflammation in mice. Immunity. 34:755–768. 10.1016/j.immuni.2011.02.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavarría-Smith J., and Vance R.E.. 2013. Direct proteolytic cleavage of NLRP1B is necessary and sufficient for inflammasome activation by anthrax lethal factor. PLoS Pathog. 9:e1003452 10.1371/journal.ppat.1003452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G.Y., Liu M., Wang F., Bertin J., and Núñez G.. 2011. A functional role for Nlrp6 in intestinal inflammation and tumorigenesis. J. Immunol. 186:7187–7194. 10.4049/jimmunol.1100412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K.W., Groß C.J., Sotomayor F.V., Stacey K.J., Tschopp J., Sweet M.J., and Schroder K.. 2014. The neutrophil NLRC4 inflammasome selectively promotes IL-1β maturation without pyroptosis during acute Salmonella challenge. Cell Reports. 8:570–582. 10.1016/j.celrep.2014.06.028 [DOI] [PubMed] [Google Scholar]
- Cheng J., Waite A.L., Tkaczyk E.R., Ke K., Richards N., Hunt A.J., and Gumucio D.L.. 2010. Kinetic properties of ASC protein aggregation in epithelial cells. J. Cell. Physiol. 222:738–747. 10.1002/jcp.22005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choubey D. 2012. Interferon-inducible Ifi200-family genes as modifiers of lupus susceptibility. Immunol. Lett. 147:10–17. 10.1016/j.imlet.2012.07.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirelli K.M., Gorfu G., Hassan M.A., Printz M., Crown D., Leppla S.H., Grigg M.E., Saeij J.P., and Moayeri M.. 2014. Inflammasome sensor NLRP1 controls rat macrophage susceptibility to Toxoplasma gondii. PLoS Pathog. 10:e1003927 10.1371/journal.ppat.1003927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Compan V., Baroja-Mazo A., López-Castejón G., Gomez A.I., Martínez C.M., Angosto D., Montero M.T., Herranz A.S., Bazán E., Reimers D., et al. 2012. Cell volume regulation modulates NLRP3 inflammasome activation. Immunity. 37:487–500. 10.1016/j.immuni.2012.06.013 [DOI] [PubMed] [Google Scholar]
- Cridland J.A., Curley E.Z., Wykes M.N., Schroder K., Sweet M.J., Roberts T.L., Ragan M.A., Kassahn K.S., and Stacey K.J.. 2012. The mammalian PYHIN gene family: phylogeny, evolution and expression. BMC Evol. Biol. 12:140 10.1186/1471-2148-12-140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz C.M., Rinna A., Forman H.J., Ventura A.L., Persechini P.M., and Ojcius D.M.. 2007. ATP activates a reactive oxygen species-dependent oxidative stress response and secretion of proinflammatory cytokines in macrophages. J. Biol. Chem. 282:2871–2879. 10.1074/jbc.M608083200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diebolder C.A., Halff E.F., Koster A.J., Huizinga E.G., and Koning R.I.. 2015. Cryoelectron tomography of the NAIP5/NLRC4 inflammasome: implications for NLR Activation. Structure. 23:2349–2357. 10.1016/j.str.2015.10.001 [DOI] [PubMed] [Google Scholar]
- Dihlmann S., Erhart P., Mehrabi A., Nickkholgh A., Lasitschka F., Böckler D., and Hakimi M.. 2014. Increased expression and activation of absent in melanoma 2 inflammasome components in lymphocytic infiltrates of abdominal aortic aneurysms. Mol. Med. 20:230–237. 10.2119/molmed.2013.00162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dombrowski Y., Peric M., Koglin S., Kammerbauer C., Göss C., Anz D., Simanski M., Gläser R., Harder J., Hornung V., et al. 2011. Cytosolic DNA triggers inflammasome activation in keratinocytes in psoriatic lesions. Sci. Transl. Med. 3:82ra38 10.1126/scitranslmed.3002001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dostert C., Pétrilli V., Van Bruggen R., Steele C., Mossman B.T., and Tschopp J.. 2008. Innate immune activation through Nalp3 inflammasome sensing of asbestos and silica. Science. 320:674–677. 10.1126/science.1156995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumas A., Amiable N., de Rivero Vaccari J.P., Chae J.J., Keane R.W., Lacroix S., and Vallières L.. 2014. The inflammasome pyrin contributes to pertussis toxin-induced IL-1β synthesis, neutrophil intravascular crawling and autoimmune encephalomyelitis. PLoS Pathog. 10:e1004150 10.1371/journal.ppat.1004150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgeworth J.D., Spencer J., Phalipon A., Griffin G.E., and Sansonetti P.J.. 2002. Cytotoxicity and interleukin-1beta processing following Shigella flexneri infection of human monocyte-derived dendritic cells. Eur. J. Immunol. 32:1464–1471. [DOI] [PubMed] [Google Scholar]
- Elinav E., Strowig T., Kau A.L., Henao-Mejia J., Thaiss C.A., Booth C.J., Peaper D.R., Bertin J., Eisenbarth S.C., Gordon J.I., and Flavell R.A.. 2011. NLRP6 inflammasome regulates colonic microbial ecology and risk for colitis. Cell. 145:745–757. 10.1016/j.cell.2011.04.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endrizzi M.G., Hadinoto V., Growney J.D., Miller W., and Dietrich W.F.. 2000. Genomic sequence analysis of the mouse Naip gene array. Genome Res. 10:1095–1102. 10.1101/gr.10.8.1095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewald S.E., Chavarria-Smith J., and Boothroyd J.C.. 2014. NLRP1 is an inflammasome sensor for Toxoplasma gondii. Infect. Immun. 82:460–468. 10.1128/IAI.01170-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes-Alnemri T., Yu J.W., Datta P., Wu J., and Alnemri E.S.. 2009. AIM2 activates the inflammasome and cell death in response to cytoplasmic DNA. Nature. 458:509–513. 10.1038/nature07710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes-Alnemri T., Yu J.W., Juliana C., Solorzano L., Kang S., Wu J., Datta P., McCormick M., Huang L., McDermott E., et al. 2010. The AIM2 inflammasome is critical for innate immunity to Francisella tularensis. Nat. Immunol. 11:385–393. 10.1038/ni.1859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finger J.N., Lich J.D., Dare L.C., Cook M.N., Brown K.K., Duraiswami C., Bertin J., and Gough P.J.. 2012. Autolytic proteolysis within the function to find domain (FIIND) is required for NLRP1 inflammasome activity. J. Biol. Chem. 287:25030–25037. 10.1074/jbc.M112.378323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fink S.L., and Cookson B.T.. 2006. Caspase-1-dependent pore formation during pyroptosis leads to osmotic lysis of infected host macrophages. Cell. Microbiol. 8:1812–1825. 10.1111/j.1462-5822.2006.00751.x [DOI] [PubMed] [Google Scholar]
- Fink S.L., Bergsbaken T., and Cookson B.T.. 2008. Anthrax lethal toxin and Salmonella elicit the common cell death pathway of caspase-1-dependent pyroptosis via distinct mechanisms. Proc. Natl. Acad. Sci. USA. 105:4312–4317. 10.1073/pnas.0707370105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franchi L., Amer A., Body-Malapel M., Kanneganti T.D., Ozören N., Jagirdar R., Inohara N., Vandenabeele P., Bertin J., Coyle A., et al. 2006. Cytosolic flagellin requires Ipaf for activation of caspase-1 and interleukin 1beta in salmonella-infected macrophages. Nat. Immunol. 7:576–582. 10.1038/ni1346 [DOI] [PubMed] [Google Scholar]
- Franklin B.S., Bossaller L., De Nardo D., Ratter J.M., Stutz A., Engels G., Brenker C., Nordhoff M., Mirandola S.R., Al-Amoudi A., et al. 2014. The adaptor ASC has extracellular and ‘prionoid’ activities that propagate inflammation. Nat. Immunol. 15:727–737. 10.1038/ni.2913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frew B.C., Joag V.R., and Mogridge J.. 2012. Proteolytic processing of Nlrp1b is required for inflammasome activity. PLoS Pathog. 8:e1002659 10.1371/journal.ppat.1002659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaidt M.M., Ebert T.S., Chauhan D., Schmidt T., Schmid-Burgk J.L., Rapino F., Robertson A.A., Cooper M.A., Graf T., and Hornung V.. 2016. Human monocytes engage an alternative inflammasome pathway. Immunity. 44:833–846. 10.1016/j.immuni.2016.01.012 [DOI] [PubMed] [Google Scholar]
- Ge J., Gong Y.N., Xu Y., and Shao F.. 2012. Preventing bacterial DNA release and absent in melanoma 2 inflammasome activation by a Legionella effector functioning in membrane trafficking. Proc. Natl. Acad. Sci. USA. 109:6193–6198. 10.1073/pnas.1117490109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorfu G., Cirelli K.M., Melo M.B., Mayer-Barber K., Crown D., Koller B.H., Masters S., Sher A., Leppla S.H., Moayeri M., et al. 2014. Dual role for inflammasome sensors NLRP1 and NLRP3 in murine resistance to Toxoplasma gondii. MBio. 5: e01117–e01113. 10.1128/mBio.01117-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gringhuis S.I., Kaptein T.M., Wevers B.A., Theelen B., van der Vlist M., Boekhout T., and Geijtenbeek T.B.. 2012. Dectin-1 is an extracellular pathogen sensor for the induction and processing of IL-1β via a noncanonical caspase-8 inflammasome. Nat. Immunol. 13:246–254. 10.1038/ni.2222 [DOI] [PubMed] [Google Scholar]
- Guey B., Bodnar M., Manié S.N., Tardivel A., and Petrilli V.. 2014. Caspase-1 autoproteolysis is differentially required for NLRP1b and NLRP3 inflammasome function. Proc. Natl. Acad. Sci. USA. 111:17254–17259. 10.1073/pnas.1415756111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H., Callaway J.B., and Ting J.P.. 2015. Inflammasomes: mechanism of action, role in disease, and therapeutics. Nat. Med. 21:677–687. 10.1038/nm.3893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurcel L., Abrami L., Girardin S., Tschopp J., and van der Goot F.G.. 2006. Caspase-1 activation of lipid metabolic pathways in response to bacterial pore-forming toxins promotes cell survival. Cell. 126:1135–1145. 10.1016/j.cell.2006.07.033 [DOI] [PubMed] [Google Scholar]
- Gurung P., Anand P.K., Malireddi R.K., Vande Walle L., Van Opdenbosch N., Dillon C.P., Weinlich R., Green D.R., Lamkanfi M., and Kanneganti T.D.. 2014. FADD and caspase-8 mediate priming and activation of the canonical and noncanonical Nlrp3 inflammasomes. J. Immunol. 192:1835–1846. 10.4049/jimmunol.1302839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurung P., Burton A., and Kanneganti T.D.. 2016. NLRP3 inflammasome plays a redundant role with caspase 8 to promote IL-1β-mediated osteomyelitis. Proc. Natl. Acad. Sci. USA. 113:4452–4457. 10.1073/pnas.1601636113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halle A., Hornung V., Petzold G.C., Stewart C.R., Monks B.G., Reinheckel T., Fitzgerald K.A., Latz E., Moore K.J., and Golenbock D.T.. 2008. The NALP3 inflammasome is involved in the innate immune response to amyloid-beta. Nat. Immunol. 9:857–865. 10.1038/ni.1636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris J., Hartman M., Roche C., Zeng S.G., O’Shea A., Sharp F.A., Lambe E.M., Creagh E.M., Golenbock D.T., Tschopp J., et al. 2011. Autophagy controls IL-1beta secretion by targeting pro-IL-1beta for degradation. J. Biol. Chem. 286:9587–9597. 10.1074/jbc.M110.202911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He W.T., Wan H., Hu L., Chen P., Wang X., Huang Z., Yang Z.H., Zhong C.Q., and Han J.. 2015. Gasdermin D is an executor of pyroptosis and required for interleukin-1β secretion. Cell Res. 25:1285–1298. 10.1038/cr.2015.139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y., Zeng M.Y., Yang D., Motro B., and Núñez G.. 2016. NEK7 is an essential mediator of NLRP3 activation downstream of potassium efflux. Nature. 530:354–357. 10.1038/nature16959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henao-Mejia J., Elinav E., Jin C., Hao L., Mehal W.Z., Strowig T., Thaiss C.A., Kau A.L., Eisenbarth S.C., Jurczak M.J., et al. 2012. Inflammasome-mediated dysbiosis regulates progression of NAFLD and obesity. Nature. 482:179–185. 10.1038/nature10809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman H.M., Mueller J.L., Broide D.H., Wanderer A.A., and Kolodner R.D.. 2001. Mutation of a new gene encoding a putative pyrin-like protein causes familial cold autoinflammatory syndrome and Muckle-Wells syndrome. Nat. Genet. 29:301–305. 10.1038/ng756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornung V., Bauernfeind F., Halle A., Samstad E.O., Kono H., Rock K.L., Fitzgerald K.A., and Latz E.. 2008. Silica crystals and aluminum salts activate the NALP3 inflammasome through phagosomal destabilization. Nat. Immunol. 9:847–856. 10.1038/ni.1631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornung V., Ablasser A., Charrel-Dennis M., Bauernfeind F., Horvath G., Caffrey D.R., Latz E., and Fitzgerald K.A.. 2009. AIM2 recognizes cytosolic dsDNA and forms a caspase-1-activating inflammasome with ASC. Nature. 458:514–518. 10.1038/nature07725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu B., Elinav E., Huber S., Strowig T., Hao L., Hafemann A., Jin C., Wunderlich C., Wunderlich T., Eisenbarth S.C., and Flavell R.A.. 2013. Microbiota-induced activation of epithelial IL-6 signaling links inflammasome-driven inflammation with transmissible cancer. Proc. Natl. Acad. Sci. USA. 110:9862–9867. 10.1073/pnas.1307575110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z., Zhou Q., Zhang C., Fan S., Cheng W., Zhao Y., Shao F., Wang H.W., Sui S.F., and Chai J.. 2015. Structural and biochemical basis for induced self-propagation of NLRC4. Science. 350:399–404. 10.1126/science.aac5489 [DOI] [PubMed] [Google Scholar]
- Huber S., Gagliani N., Zenewicz L.A., Huber F.J., Bosurgi L., Hu B., Hedl M., Zhang W., O’Connor W. Jr., Murphy A.J., et al. 2012. IL-22BP is regulated by the inflammasome and modulates tumorigenesis in the intestine. Nature. 491:259–263. 10.1038/nature11535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jabir M.S., Ritchie N.D., Li D., Bayes H.K., Tourlomousis P., Puleston D., Lupton A., Hopkins L., Simon A.K., Bryant C., and Evans T.J.. 2014. Caspase-1 cleavage of the TLR adaptor TRIF inhibits autophagy and β-interferon production during Pseudomonas aeruginosa infection. Cell Host Microbe. 15:214–227. 10.1016/j.chom.2014.01.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jabir M.S., Hopkins L., Ritchie N.D., Ullah I., Bayes H.K., Li D., Tourlomousis P., Lupton A., Puleston D., Simon A.K., et al. 2015. Mitochondrial damage contributes to Pseudomonas aeruginosa activation of the inflammasome and is downregulated by autophagy. Autophagy. 11:166–182. 10.4161/15548627.2014.981915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin T., Perry A., Jiang J., Smith P., Curry J.A., Unterholzner L., Jiang Z., Horvath G., Rathinam V.A., Johnstone R.W., et al. 2012. Structures of the HIN domain:DNA complexes reveal ligand binding and activation mechanisms of the AIM2 inflammasome and IFI16 receptor. Immunity. 36:561–571. 10.1016/j.immuni.2012.02.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin T., Curry J., Smith P., Jiang J., and Xiao T.S.. 2013a Structure of the NLRP1 caspase recruitment domain suggests potential mechanisms for its association with procaspase-1. Proteins. 81:1266–1270. 10.1002/prot.24287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin T., Perry A., Smith P., Jiang J., and Xiao T.S.. 2013b Structure of the absent in melanoma 2 (AIM2) pyrin domain provides insights into the mechanisms of AIM2 autoinhibition and inflammasome assembly. J. Biol. Chem. 288:13225–13235. 10.1074/jbc.M113.468033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Y., Mailloux C.M., Gowan K., Riccardi S.L., LaBerge G., Bennett D.C., Fain P.R., and Spritz R.A.. 2007. NALP1 in vitiligo-associated multiple autoimmune disease. N. Engl. J. Med. 356:1216–1225. 10.1056/NEJMoa061592 [DOI] [PubMed] [Google Scholar]
- Jones J.W., Kayagaki N., Broz P., Henry T., Newton K., O’Rourke K., Chan S., Dong J., Qu Y., Roose-Girma M., et al. 2010. Absent in melanoma 2 is required for innate immune recognition of Francisella tularensis. Proc. Natl. Acad. Sci. USA. 107:9771–9776. 10.1073/pnas.1003738107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jounai N., Kobiyama K., Shiina M., Ogata K., Ishii K.J., and Takeshita F.. 2011. NLRP4 negatively regulates autophagic processes through an association with beclin1. J. Immunol. 186:1646–1655. 10.4049/jimmunol.1001654 [DOI] [PubMed] [Google Scholar]
- Karki R., Man S.M., Malireddi R.K., Gurung P., Vogel P., Lamkanfi M., and Kanneganti T.D.. 2015. Concerted activation of the AIM2 and NLRP3 inflammasomes orchestrates host protection against Aspergillus infection. Cell Host Microbe. 17:357–368. 10.1016/j.chom.2015.01.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayagaki N., Warming S., Lamkanfi M., Vande Walle L., Louie S., Dong J., Newton K., Qu Y., Liu J., Heldens S., et al. 2011. Non-canonical inflammasome activation targets caspase-11. Nature. 479:117–121. 10.1038/nature10558 [DOI] [PubMed] [Google Scholar]
- Kayagaki N., Stowe I.B., Lee B.L., O’Rourke K., Anderson K., Warming S., Cuellar T., Haley B., Roose-Girma M., Phung Q.T., et al. 2015. Caspase-11 cleaves gasdermin D for non-canonical inflammasome signalling. Nature. 526:666–671. 10.1038/nature15541 [DOI] [PubMed] [Google Scholar]
- Kerur N., Veettil M.V., Sharma-Walia N., Bottero V., Sadagopan S., Otageri P., and Chandran B.. 2011. IFI16 acts as a nuclear pathogen sensor to induce the inflammasome in response to Kaposi Sarcoma-associated herpesvirus infection. Cell Host Microbe. 9:363–375. 10.1016/j.chom.2011.04.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khare S., Ratsimandresy R.A., de Almeida L., Cuda C.M., Rellick S.L., Misharin A.V., Wallin M.C., Gangopadhyay A., Forte E., Gottwein E., et al. 2014. The PYRIN domain-only protein POP3 inhibits ALR inflammasomes and regulates responses to infection with DNA viruses. Nat. Immunol. 15:343–353. 10.1038/ni.2829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S., Bauernfeind F., Ablasser A., Hartmann G., Fitzgerald K.A., Latz E., and Hornung V.. 2010. Listeria monocytogenes is sensed by the NLRP3 and AIM2 inflammasome. Eur. J. Immunol. 40:1545–1551. 10.1002/eji.201040425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitamura A., Sasaki Y., Abe T., Kano H., and Yasutomo K.. 2014. An inherited mutation in NLRC4 causes autoinflammation in human and mice. J. Exp. Med. 211:2385–2396. 10.1084/jem.20141091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kofoed E.M., and Vance R.E.. 2011. Innate immune recognition of bacterial ligands by NAIPs determines inflammasome specificity. Nature. 477:592–595. 10.1038/nature10394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kortmann J., Brubaker S.W., and Monack D.M.. 2015. Cutting edge: inflammasome activation in primary human macrophages is dependent on flagellin. J. Immunol. 195:815–819. 10.4049/jimmunol.1403100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamkanfi M., Kanneganti T.D., Van Damme P., Vanden Berghe T., Vanoverberghe I., Vandekerckhove J., Vandenabeele P., Gevaert K., and Núñez G.. 2008. Targeted peptidecentric proteomics reveals caspase-7 as a substrate of the caspase-1 inflammasomes. Mol. Cell. Proteomics. 7:2350–2363. 10.1074/mcp.M800132-MCP200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee G.S., Subramanian N., Kim A.I., Aksentijevich I., Goldbach-Mansky R., Sacks D.B., Germain R.N., Kastner D.L., and Chae J.J.. 2012. The calcium-sensing receptor regulates the NLRP3 inflammasome through Ca2+ and cAMP. Nature. 492:123–127. 10.1038/nature11588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy M., Thaiss C.A., Zeevi D., Dohnalová L., Zilberman-Schapira G., Mahdi J.A., David E., Savidor A., Korem T., Herzig Y., et al. 2015. Microbiota-modulated metabolites shape the intestinal microenvironment by regulating NLRP6 inflammasome signaling. Cell. 163:1428–1443. 10.1016/j.cell.2015.10.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Wang J., Wang J., Cao L.S., Wang Z.X., and Wu J.W.. 2014. Structural mechanism of DNA recognition by the p202 HINa domain: insights into the inhibition of Aim2-mediated inflammatory signalling. Acta Crystallogr. F Struct. Biol. Commun. 70:21–29. 10.1107/S2053230X1303135X [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lich J.D., Williams K.L., Moore C.B., Arthur J.C., Davis B.K., Taxman D.J., and Ting J.P.. 2007. Monarch-1 suppresses non-canonical NF-kappaB activation and p52-dependent chemokine expression in monocytes. J. Immunol. 178:1256–1260. 10.4049/jimmunol.178.3.1256 [DOI] [PubMed] [Google Scholar]
- Liu T., Yamaguchi Y., Shirasaki Y., Shikada K., Yamagishi M., Hoshino K., Kaisho T., Takemoto K., Suzuki T., Kuranaga E., et al. 2014. Single-cell imaging of caspase-1 dynamics reveals an all-or-none inflammasome signaling response. Cell Reports. 8:974–982. 10.1016/j.celrep.2014.07.012 [DOI] [PubMed] [Google Scholar]
- Lu A., Magupalli V.G., Ruan J., Yin Q., Atianand M.K., Vos M.R., Schröder G.F., Fitzgerald K.A., Wu H., and Egelman E.H.. 2014. Unified polymerization mechanism for the assembly of ASC-dependent inflammasomes. Cell. 156:1193–1206. 10.1016/j.cell.2014.02.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukens J.R., Gurung P., Vogel P., Johnson G.R., Carter R.A., McGoldrick D.J., Bandi S.R., Calabrese C.R., Vande Walle L., Lamkanfi M., and Kanneganti T.D.. 2014. Dietary modulation of the microbiome affects autoinflammatory disease. Nature. 516:246–249. 10.1038/nature13788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukens J.R., Gurung P., Shaw P.J., Barr M.J., Zaki M.H., Brown S.A., Vogel P., Chi H., and Kanneganti T.D.. 2015. The NLRP12 Sensor negatively regulates autoinflammatory disease by modulating interleukin-4 production in T cells. Immunity. 42:654–664. 10.1016/j.immuni.2015.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupfer C., Thomas P.G., Anand P.K., Vogel P., Milasta S., Martinez J., Huang G., Green M., Kundu M., Chi H., et al. 2013. Receptor interacting protein kinase 2-mediated mitophagy regulates inflammasome activation during virus infection. Nat. Immunol. 14:480–488. 10.1038/ni.2563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Man S.M., and Kanneganti T.D.. 2015. Regulation of inflammasome activation. Immunol. Rev. 265:6–21. 10.1111/imr.12296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Man S.M., and Kanneganti T.-D.. 2016. Converging roles of caspases in inflammasome activation, cell death and innate immunity. Nat. Rev. Immunol. 16:7–21. 10.1038/nri.2015.7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Man S.M., Tourlomousis P., Hopkins L., Monie T.P., Fitzgerald K.A., and Bryant C.E.. 2013. Salmonella infection induces recruitment of Caspase-8 to the inflammasome to modulate IL-1β production. J. Immunol. 191:5239–5246. 10.4049/jimmunol.1301581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Man S.M., Ekpenyong A., Tourlomousis P., Achouri S., Cammarota E., Hughes K., Rizzo A., Ng G., Wright J.A., Cicuta P., et al. 2014a Actin polymerization as a key innate immune effector mechanism to control Salmonella infection. Proc. Natl. Acad. Sci. USA. 111:17588–17593. 10.1073/pnas.1419925111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Man S.M., Hopkins L.J., Nugent E., Cox S., Glück I.M., Tourlomousis P., Wright J.A., Cicuta P., Monie T.P., and Bryant C.E.. 2014b Inflammasome activation causes dual recruitment of NLRC4 and NLRP3 to the same macromolecular complex. Proc. Natl. Acad. Sci. USA. 111:7403–7408. 10.1073/pnas.1402911111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Man S.M., Karki R., Malireddi R.K., Neale G., Vogel P., Yamamoto M., Lamkanfi M., and Kanneganti T.D.. 2015a The transcription factor IRF1 and guanylate-binding proteins target activation of the AIM2 inflammasome by Francisella infection. Nat. Immunol. 16:467–475. 10.1038/ni.3118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Man S.M., Zhu Q., Zhu L., Liu Z., Karki R., Malik A., Sharma D., Li L., Malireddi R.K., Gurung P., et al. 2015b Critical role for the DNA sensor AIM2 in stem cell proliferation and cancer. Cell. 162:45–58. 10.1016/j.cell.2015.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariathasan S., Newton K., Monack D.M., Vucic D., French D.M., Lee W.P., Roose-Girma M., Erickson S., and Dixit V.M.. 2004. Differential activation of the inflammasome by caspase-1 adaptors ASC and Ipaf. Nature. 430:213–218. 10.1038/nature02664 [DOI] [PubMed] [Google Scholar]
- Martinon F., Burns K., and Tschopp J.. 2002. The inflammasome: a molecular platform triggering activation of inflammatory caspases and processing of proIL-beta. Mol. Cell. 10:417–426. 10.1016/S1097-2765(02)00599-3 [DOI] [PubMed] [Google Scholar]
- Masters S.L., Simon A., Aksentijevich I., and Kastner D.L.. 2009. Horror autoinflammaticus: the molecular pathophysiology of autoinflammatory disease. Annu. Rev. Immunol. 27:621–668. 10.1146/annurev.immunol.25.022106.141627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masters S.L., Gerlic M., Metcalf D., Preston S., Pellegrini M., O’Donnell J.A., McArthur K., Baldwin T.M., Chevrier S., Nowell C.J., et al. 2012. NLRP1 inflammasome activation induces pyroptosis of hematopoietic progenitor cells. Immunity. 37:1009–1023. 10.1016/j.immuni.2012.08.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masters S.L., Lagou V., Jéru I., Baker P.J., Van Eyck L., Parry D.A., Lawless D., De Nardo D., Garcia-Perez J.E., Dagley L.F., et al. 2016. Familial autoinflammation with neutrophilic dermatosis reveals a regulatory mechanism of pyrin activation. Sci. Transl. Med. 8:332ra45 10.1126/scitranslmed.aaf1471 [DOI] [PubMed] [Google Scholar]
- Matusiak M., Van Opdenbosch N., and Lamkanfi M.. 2015. CARD- and pyrin-only proteins regulating inflammasome activation and immunity. Immunol. Rev. 265:217–230. 10.1111/imr.12282 [DOI] [PubMed] [Google Scholar]
- Meunier E., Wallet P., Dreier R.F., Costanzo S., Anton L., Rühl S., Dussurgey S., Dick M.S., Kistner A., Rigard M., et al. 2015. Guanylate-binding proteins promote activation of the AIM2 inflammasome during infection with Francisella novicida. Nat. Immunol. 16:476–484. 10.1038/ni.3119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao E.A., Alpuche-Aranda C.M., Dors M., Clark A.E., Bader M.W., Miller S.I., and Aderem A.. 2006. Cytoplasmic flagellin activates caspase-1 and secretion of interleukin 1beta via Ipaf. Nat. Immunol. 7:569–575. 10.1038/ni1344 [DOI] [PubMed] [Google Scholar]
- Miao E.A., Leaf I.A., Treuting P.M., Mao D.P., Dors M., Sarkar A., Warren S.E., Wewers M.D., and Aderem A.. 2010a Caspase-1-induced pyroptosis is an innate immune effector mechanism against intracellular bacteria. Nat. Immunol. 11:1136–1142. 10.1038/ni.1960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao E.A., Mao D.P., Yudkovsky N., Bonneau R., Lorang C.G., Warren S.E., Leaf I.A., and Aderem A.. 2010b Innate immune detection of the type III secretion apparatus through the NLRC4 inflammasome. Proc. Natl. Acad. Sci. USA. 107:3076–3080. 10.1073/pnas.0913087107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misawa T., Takahama M., Kozaki T., Lee H., Zou J., Saitoh T., and Akira S.. 2013. Microtubule-driven spatial arrangement of mitochondria promotes activation of the NLRP3 inflammasome. Nat. Immunol. 14:454–460. 10.1038/ni.2550 [DOI] [PubMed] [Google Scholar]
- Molofsky A.B., Byrne B.G., Whitfield N.N., Madigan C.A., Fuse E.T., Tateda K., and Swanson M.S.. 2006. Cytosolic recognition of flagellin by mouse macrophages restricts Legionella pneumophila infection. J. Exp. Med. 203:1093–1104. 10.1084/jem.20051659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monack D.M., Raupach B., Hromockyj A.E., and Falkow S.. 1996. Salmonella typhimurium invasion induces apoptosis in infected macrophages. Proc. Natl. Acad. Sci. USA. 93:9833–9838. 10.1073/pnas.93.18.9833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriya M., Taniguchi S., Wu P., Liepinsh E., Otting G., and Sagara J.. 2005. Role of charged and hydrophobic residues in the oligomerization of the PYRIN domain of ASC. Biochemistry. 44:575–583. 10.1021/bi048374i [DOI] [PubMed] [Google Scholar]
- Morizane S., Yamasaki K., Mühleisen B., Kotol P.F., Murakami M., Aoyama Y., Iwatsuki K., Hata T., and Gallo R.L.. 2012. Cathelicidin antimicrobial peptide LL-37 in psoriasis enables keratinocyte reactivity against TLR9 ligands. J. Invest. Dermatol. 132:135–143. 10.1038/jid.2011.259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muñoz-Planillo R., Kuffa P., Martínez-Colón G., Smith B.L., Rajendiran T.M., and Núñez G.. 2013. K+ efflux is the common trigger of NLRP3 inflammasome activation by bacterial toxins and particulate matter. Immunity. 38:1142–1153. 10.1016/j.immuni.2013.05.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami T., Ockinger J., Yu J., Byles V., McColl A., Hofer A.M., and Horng T.. 2012. Critical role for calcium mobilization in activation of the NLRP3 inflammasome. Proc. Natl. Acad. Sci. USA. 109:11282–11287. 10.1073/pnas.1117765109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakahira K., Haspel J.A., Rathinam V.A.K., Lee S.-J., Dolinay T., Lam H.C., Englert J.A., Rabinovitch M., Cernadas M., Kim H.P., et al. 2011. Autophagy proteins regulate innate immune responses by inhibiting the release of mitochondrial DNA mediated by the NALP3 inflammasome. Nat. Immunol. 12:222–230. 10.1038/ni.1980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nour A.M., Yeung Y.G., Santambrogio L., Boyden E.D., Stanley E.R., and Brojatsch J.. 2009. Anthrax lethal toxin triggers the formation of a membrane-associated inflammasome complex in murine macrophages. Infect. Immun. 77:1262–1271. 10.1128/IAI.01032-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowarski R., Jackson R., Gagliani N., de Zoete M.R., Palm N.W., Bailis W., Low J.S., Harman C.C., Graham M., Elinav E., and Flavell R.A.. 2015. Epithelial IL-18 equilibrium controls barrier function in colitis. Cell. 163:1444–1456. 10.1016/j.cell.2015.10.072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Regan L., and Fry A.M.. 2009. The Nek6 and Nek7 protein kinases are required for robust mitotic spindle formation and cytokinesis. Mol. Cell. Biol. 29:3975–3990. 10.1128/MCB.01867-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng K., Broz P., Jones J., Joubert L.M., and Monack D.. 2011. Elevated AIM2-mediated pyroptosis triggered by hypercytotoxic Francisella mutant strains is attributed to increased intracellular bacteriolysis. Cell. Microbiol. 13:1586–1600. 10.1111/j.1462-5822.2011.01643.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peral de Castro C., Jones S.A., Ní Cheallaigh C., Hearnden C.A., Williams L., Winter J., Lavelle E.C., Mills K.H.G., and Harris J.. 2012. Autophagy regulates IL-23 secretion and innate T cell responses through effects on IL-1 secretion. J. Immunol. 189:4144–4153. 10.4049/jimmunol.1201946 [DOI] [PubMed] [Google Scholar]
- Pétrilli V., Papin S., Dostert C., Mayor A., Martinon F., and Tschopp J.. 2007. Activation of the NALP3 inflammasome is triggered by low intracellular potassium concentration. Cell Death Differ. 14:1583–1589. 10.1038/sj.cdd.4402195 [DOI] [PubMed] [Google Scholar]
- Pierini R., Juruj C., Perret M., Jones C.L., Mangeot P., Weiss D.S., and Henry T.. 2012. AIM2/ASC triggers caspase-8-dependent apoptosis in Francisella-infected caspase-1-deficient macrophages. Cell Death Differ. 19:1709–1721. 10.1038/cdd.2012.51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponomareva L., Liu H., Duan X., Dickerson E., Shen H., Panchanathan R., and Choubey D.. 2013. AIM2, an IFN-inducible cytosolic DNA sensor, in the development of benign prostate hyperplasia and prostate cancer. Mol. Cancer Res. 11:1193–1202. 10.1158/1541-7786.MCR-13-0145 [DOI] [PubMed] [Google Scholar]
- Poyet J.L., Srinivasula S.M., Tnani M., Razmara M., Fernandes-Alnemri T., and Alnemri E.S.. 2001. Identification of Ipaf, a human caspase-1-activating protein related to Apaf-1. J. Biol. Chem. 276:28309–28313. 10.1074/jbc.C100250200 [DOI] [PubMed] [Google Scholar]
- Proell M., Gerlic M., Mace P.D., Reed J.C., and Riedl S.J.. 2013. The CARD plays a critical role in ASC foci formation and inflammasome signalling. Biochem. J. 449:613–621. 10.1042/BJ20121198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu Y., Misaghi S., Newton K., Maltzman A., Izrael-Tomasevic A., Arnott D., and Dixit V.M.. 2016. NLRP3 recruitment by NLRC4 during Salmonella infection. J. Exp. Med. 213:877–885. 10.1084/jem.20132234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rathinam V.A., Jiang Z., Waggoner S.N., Sharma S., Cole L.E., Waggoner L., Vanaja S.K., Monks B.G., Ganesan S., Latz E., et al. 2010. The AIM2 inflammasome is essential for host defense against cytosolic bacteria and DNA viruses. Nat. Immunol. 11:395–402. 10.1038/ni.1864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayamajhi M., Zak D.E., Chavarria-Smith J., Vance R.E., and Miao E.A.. 2013. Cutting edge: Mouse NAIP1 detects the type III secretion system needle protein. J. Immunol. 191:3986–3989. 10.4049/jimmunol.1301549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren T., Zamboni D.S., Roy C.R., Dietrich W.F., and Vance R.E.. 2006. Flagellin-deficient Legionella mutants evade caspase-1- and Naip5-mediated macrophage immunity. PLoS Pathog. 2:e18 10.1371/journal.ppat.0020018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts T.L., Idris A., Dunn J.A., Kelly G.M., Burnton C.M., Hodgson S., Hardy L.L., Garceau V., Sweet M.J., Ross I.L., et al. 2009. HIN-200 proteins regulate caspase activation in response to foreign cytoplasmic DNA. Science. 323:1057–1060. 10.1126/science.1169841 [DOI] [PubMed] [Google Scholar]
- Romberg N., Al Moussawi K., Nelson-Williams C., Stiegler A.L., Loring E., Choi M., Overton J., Meffre E., Khokha M.K., Huttner A.J., et al. 2014. Mutation of NLRC4 causes a syndrome of enterocolitis and autoinflammation. Nat. Genet. 46:1135–1139. 10.1038/ng.3066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rühl S., and Broz P.. 2015. Caspase-11 activates a canonical NLRP3 inflammasome by promoting K+ efflux. Eur. J. Immunol. 45:2927–2936. 10.1002/eji.201545772 [DOI] [PubMed] [Google Scholar]
- Sagoo P., Garcia Z., Breart B., Lemaître F., Michonneau D., Albert M.L., Levy Y., and Bousso P.. 2016. In vivo imaging of inflammasome activation reveals a subcapsular macrophage burst response that mobilizes innate and adaptive immunity. Nat. Med. 22:64–71. 10.1038/nm.4016 [DOI] [PubMed] [Google Scholar]
- Sagulenko V., Thygesen S.J., Sester D.P., Idris A., Cridland J.A., Vajjhala P.R., Roberts T.L., Schroder K., Vince J.E., Hill J.M., et al. 2013. AIM2 and NLRP3 inflammasomes activate both apoptotic and pyroptotic death pathways via ASC. Cell Death Differ. 20:1149–1160. 10.1038/cdd.2013.37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitoh T., Fujita N., Jang M.H., Uematsu S., Yang B.G., Satoh T., Omori H., Noda T., Yamamoto N., Komatsu M., et al. 2008. Loss of the autophagy protein Atg16L1 enhances endotoxin-induced IL-1β production. Nature. 456:264–268. 10.1038/nature07383 [DOI] [PubMed] [Google Scholar]
- Sborgi L., Ravotti F., Dandey V.P., Dick M.S., Mazur A., Reckel S., Chami M., Scherer S., Huber M., Böckmann A., et al. 2015. Structure and assembly of the mouse ASC inflammasome by combined NMR spectroscopy and cryo-electron microscopy. Proc. Natl. Acad. Sci. USA. 112:13237–13242. 10.1073/pnas.1507579112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid-Burgk J.L., Gaidt M.M., Schmidt T., Ebert T.S., Bartok E., and Hornung V.. 2015. Caspase-4 mediates non-canonical activation of the NLRP3 inflammasome in human myeloid cells. Eur. J. Immunol. 45:2911–2917. 10.1002/eji.201545523 [DOI] [PubMed] [Google Scholar]
- Schmid-Burgk J.L., Chauhan D., Schmidt T., Ebert T.S., Reinhardt J., Endl E., and Hornung V.. 2016. A genome-wide CRISPR (clustered regularly interspaced short palindromic repeats) screen identifies NEK7 as an essential component of NLRP3 inflammasome activation. J. Biol. Chem. 291:103–109. 10.1074/jbc.C115.700492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schorn C., Frey B., Lauber K., Janko C., Strysio M., Keppeler H., Gaipl U.S., Voll R.E., Springer E., Munoz L.E., et al. 2011. Sodium overload and water influx activate the NALP3 inflammasome. J. Biol. Chem. 286:35–41. 10.1074/jbc.M110.139048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sellin M.E., Müller A.A., Felmy B., Dolowschiak T., Diard M., Tardivel A., Maslowski K.M., and Hardt W.D.. 2014. Epithelium-intrinsic NAIP/NLRC4 inflammasome drives infected enterocyte expulsion to restrict Salmonella replication in the intestinal mucosa. Cell Host Microbe. 16:237–248. 10.1016/j.chom.2014.07.001 [DOI] [PubMed] [Google Scholar]
- Shah S., Bohsali A., Ahlbrand S.E., Srinivasan L., Rathinam V.A., Vogel S.N., Fitzgerald K.A., Sutterwala F.S., and Briken V.. 2013. Cutting edge: Mycobacterium tuberculosis but not nonvirulent mycobacteria inhibits IFN-β and AIM2 inflammasome-dependent IL-1β production via its ESX-1 secretion system. J. Immunol. 191:3514–3518. 10.4049/jimmunol.1301331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao W., Yeretssian G., Doiron K., Hussain S.N., and Saleh M.. 2007. The caspase-1 digestome identifies the glycolysis pathway as a target during infection and septic shock. J. Biol. Chem. 282:36321–36329. 10.1074/jbc.M708182200 [DOI] [PubMed] [Google Scholar]
- Shi C.-S., Shenderov K., Huang N.-N., Kabat J., Abu-Asab M., Fitzgerald K.A., Sher A., and Kehrl J.H.. 2012. Activation of autophagy by inflammatory signals limits IL-1β production by targeting ubiquitinated inflammasomes for destruction. Nat. Immunol. 13:255–263. 10.1038/ni.2215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H., Wang Y., Li X., Zhan X., Tang M., Fina M., Su L., Pratt D., Bu C.H., Hildebrand S., et al. 2016. NLRP3 activation and mitosis are mutually exclusive events coordinated by NEK7, a new inflammasome component. Nat. Immunol. 17:250–258. 10.1038/ni.3333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J., Zhao Y., Wang Y., Gao W., Ding J., Li P., Hu L., and Shao F.. 2014. Inflammatory caspases are innate immune receptors for intracellular LPS. Nature. 514:187–192. 10.1038/nature13683 [DOI] [PubMed] [Google Scholar]
- Shi J., Zhao Y., Wang K., Shi X., Wang Y., Huang H., Zhuang Y., Cai T., Wang F., and Shao F.. 2015. Cleavage of GSDMD by inflammatory caspases determines pyroptotic cell death. Nature. 526:660–665. 10.1038/nature15514 [DOI] [PubMed] [Google Scholar]
- Tenthorey J.L., Kofoed E.M., Daugherty M.D., Malik H.S., and Vance R.E.. 2014. Molecular basis for specific recognition of bacterial ligands by NAIP/NLRC4 inflammasomes. Mol. Cell. 54:17–29. 10.1016/j.molcel.2014.02.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Opdenbosch N., Gurung P., Vande Walle L., Fossoul A., Kanneganti T.D., and Lamkanfi M.. 2014. Activation of the NLRP1b inflammasome independently of ASC-mediated caspase-1 autoproteolysis and speck formation. Nat. Commun. 5:3209 10.1038/ncomms4209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent W.J., Freisinger C.M., Lam P.Y., Huttenlocher A., and Sauer J.D.. 2016. Macrophages mediate flagellin induced inflammasome activation and host defense in zebrafish. Cell. Microbiol. 18:591–604. 10.1111/cmi.12536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vladimer G.I., Weng D., Paquette S.W., Vanaja S.K., Rathinam V.A., Aune M.H., Conlon J.E., Burbage J.J., Proulx M.K., Liu Q., et al. 2012. The NLRP12 inflammasome recognizes Yersinia pestis. Immunity. 37:96–107. 10.1016/j.immuni.2012.07.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Moltke J., Trinidad N.J., Moayeri M., Kintzer A.F., Wang S.B., van Rooijen N., Brown C.R., Krantz B.A., Leppla S.H., Gronert K., and Vance R.E.. 2012. Rapid induction of inflammatory lipid mediators by the inflammasome in vivo. Nature. 490:107–111. 10.1038/nature11351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P., Zhu S., Yang L., Cui S., Pan W., Jackson R., Zheng Y., Rongvaux A., Sun Q., Yang G., et al. 2015. Nlrp6 regulates intestinal antiviral innate immunity. Science. 350:826–830. 10.1126/science.aab3145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Miura M., Jung Y.K., Zhu H., Li E., and Yuan J.. 1998. Murine caspase-11, an ICE-interacting protease, is essential for the activation of ICE. Cell. 92:501–509. 10.1016/S0092-8674(00)80943-5 [DOI] [PubMed] [Google Scholar]
- Williams K.L., Lich J.D., Duncan J.A., Reed W., Rallabhandi P., Moore C., Kurtz S., Coffield V.M., Accavitti-Loper M.A., Su L., et al. 2005. The CATERPILLER protein monarch-1 is an antagonist of toll-like receptor-, tumor necrosis factor α-, and Mycobacterium tuberculosis-induced pro-inflammatory signals. J. Biol. Chem. 280:39914–39924. 10.1074/jbc.M502820200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witola W.H., Mui E., Hargrave A., Liu S., Hypolite M., Montpetit A., Cavailles P., Bisanz C., Cesbron-Delauw M.F., Fournié G.J., and McLeod R.. 2011. NALP1 influences susceptibility to human congenital toxoplasmosis, proinflammatory cytokine response, and fate of Toxoplasma gondii-infected monocytic cells. Infect. Immun. 79:756–766. 10.1128/IAI.00898-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wlodarska M., Thaiss C.A., Nowarski R., Henao-Mejia J., Zhang J.P., Brown E.M., Frankel G., Levy M., Katz M.N., Philbrick W.M., et al. 2014. NLRP6 inflammasome orchestrates the colonic host-microbial interface by regulating goblet cell mucus secretion. Cell. 156:1045–1059. 10.1016/j.cell.2014.01.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y.H., Kuo W.C., Wu Y.J., Yang K.T., Chen S.T., Jiang S.T., Gordy C., He Y.W., and Lai M.Z.. 2014. Participation of c-FLIP in NLRP3 and AIM2 inflammasome activation. Cell Death Differ. 21:451–461. 10.1038/cdd.2013.165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H., Yang J., Gao W., Li L., Li P., Zhang L., Gong Y.-N., Peng X., Xi J.J., Chen S., et al. 2014. Innate immune sensing of bacterial modifications of Rho GTPases by the Pyrin inflammasome. Nature. 513:237–241. 10.1038/nature13449 [DOI] [PubMed] [Google Scholar]
- Yang D., He Y., Muñoz-Planillo R., Liu Q., and Núñez G.. 2015. Caspase-11 requires the pannexin-1 channel and the purinergic P2X7 pore to mediate pyroptosis and endotoxic shock. Immunity. 43:923–932. 10.1016/j.immuni.2015.10.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Zhao Y., Shi J., and Shao F.. 2013. Human NAIP and mouse NAIP1 recognize bacterial type III secretion needle protein for inflammasome activation. Proc. Natl. Acad. Sci. USA. 110:14408–14413. 10.1073/pnas.1306376110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Q., Sester D.P., Tian Y., Hsiao Y.S., Lu A., Cridland J.A., Sagulenko V., Thygesen S.J., Choubey D., Hornung V., et al. 2013. Molecular mechanism for p202-mediated specific inhibition of AIM2 inflammasome activation. Cell Reports. 4:327–339. 10.1016/j.celrep.2013.06.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaki M.H., Vogel P., Malireddi R.K., Body-Malapel M., Anand P.K., Bertin J., Green D.R., Lamkanfi M., and Kanneganti T.D.. 2011. The NOD-like receptor NLRP12 attenuates colon inflammation and tumorigenesis. Cancer Cell. 20:649–660. 10.1016/j.ccr.2011.10.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaki M.H., Man S.M., Vogel P., Lamkanfi M., and Kanneganti T.D.. 2014. Salmonella exploits NLRP12-dependent innate immune signaling to suppress host defenses during infection. Proc. Natl. Acad. Sci. USA. 111:385–390. 10.1073/pnas.1317643111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Chen S., Ruan J., Wu J., Tong A.B., Yin Q., Li Y., David L., Lu A., Wang W.L., et al. 2015. Cryo-EM structure of the activated NAIP2-NLRC4 inflammasome reveals nucleated polymerization. Science. 350:404–409. 10.1126/science.aac5789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Yang J., Shi J., Gong Y.N., Lu Q., Xu H., Liu L., and Shao F.. 2011. The NLRC4 inflammasome receptors for bacterial flagellin and type III secretion apparatus. Nature. 477:596–600. 10.1038/nature10510 [DOI] [PubMed] [Google Scholar]
- Zhou R., Yazdi A.S., Menu P., and Tschopp J.. 2011. A role for mitochondria in NLRP3 inflammasome activation. Nature. 469:221–225. 10.1038/nature09663 [DOI] [PubMed] [Google Scholar]
- Zurawek M., Fichna M., Januszkiewicz-Lewandowska D., Gryczyńska M., Fichna P., and Nowak J.. 2010. A coding variant in NLRP1 is associated with autoimmune Addison’s disease. Hum. Immunol. 71:530–534. 10.1016/j.humimm.2010.02.004 [DOI] [PubMed] [Google Scholar]