Abstract
The CRISPR-Cas system has emerged as a fascinating and important genome editing tool. It is now widely used in biology, biotechnology, and biomedical research in both academic and industrial settings. To improve the specificity and efficiency of Cas nucleases and to extend the applications of these systems for other areas of research, an understanding of their precise working mechanisms is crucial. In this review, we summarize current studies on the molecular structures and dynamic functions of type I and type II Cas nucleases, with a focus on target DNA searching and cleavage processes as revealed by single-molecule observations. [BMB Reports 2016; 49(4): 201-207]
Keywords: CRISPR-Cas system, Crystal structure, Single molecule, Type I CRISPR, Type II CRISPR
INTRODUCTION
Since it was first successfully applied to eukaryotic genome editing in 2013 (1-5), the clustered regularly interspaced short palindromic repeats (CRISPR) – CRISPR associated proteins (Cas) system (referred to as the CRISPR-Cas system) has completely revolutionized genome engineering, greatly facilitating techniques such as gene knockout, gene knock-in, and targeted modification of genetic regulatory elements (6-10). Described as a “molecular marvel” for genome editing in various organisms, the CRISPR-Cas system was selected as Science’s ‘Breakthrough of the Year’ twice in 2013 and 2015 (11). In comparison with other programmable nucleases, such as ZFNs (zinc finger nucleases) and TALENs (transcription activator-like effector nucleases), CRISPR-Cas derived RNA-guided endonucleases (RGENs) have substantial advantages; they are both easy to use and inexpensive (10).
CRISPR-Cas is an adaptive immune system present in most archaea and 40% of bacteria, used for defending against attack by foreign viruses or plasmids (12). It functions by cleaving the invading DNA with a protein-guide RNA complex (13-15). This dynamic process is composed of three different steps: acquisition, expression, and interference (15). During the acquisition step, Cas proteins incorporate the invading DNAs into the repeat region of the CRISPR locus. At the expression stage, precursor transcripts (called pre-CRISPR RNAs; pre-crRNAs) are generated by transcription of the incorporated endogenous CRISPR array and then cleaved into individual crRNAs by other Cas proteins. During the final interference step, a Cas nuclease-crRNA complex searches for target DNA sequences complementary to the crRNA and then cleaves them. The CRISPR-Cas system completely discriminates between exogenous and self DNA by recognizing an adjacent DNA sequence called the protospacer adjacent motif (PAM), present in the invading DNA but not at the CRISPR locus (16).
CRISPR-Cas systems are classified into three major types (I, II, III) according to the participating proteins (15). For most CRISPR-Cas systems, Cas1 and Cas2 proteins are primarily responsible for the acquisition process (17). These proteins recognize invading foreign DNAs and incorporate them into the CRISPR locus (18, 19). After incorporation, pre-crRNAs are transcribed from the CRISPR locus and Cas6 (for type I and III) and RNaseIII (for type II) proteins are recruited to the transcripts to execute further trimming processes (20, 21). Lastly, processed crRNA binds to the target DNA with the help of the Cascade-Cas3 complex (type I), Cas9 endonuclease and trans-activating crRNA (tracrRNA) (type II), or the Csm-Cmr complex (type III), after which the target DNA is cleaved (22).
For CRISPR-based genome editing, researchers have only adopted Cas proteins from the last interference step of the CRISPR-Cas system. Thus, to increase genome editing efficiency and reduce off-target effects, it is important to understand the precise working mechanisms of Cas components that function during that step. Here, we review structural studies of components from the type I and II CRISPR-Cas systems that function in target DNA cleavage. We also review mechanisms of Cas protein and guide RNA complex recognition and cleavage of DNA targets, as revealed by dynamical information from single-molecule observations.
STRUCTURAL VIEWS OF CRISPR-CAS TARGET DNA CLEAVAGE COMPONENTS
Type I-E CRISPR-Cas system (Cascade-Cas3 complex)
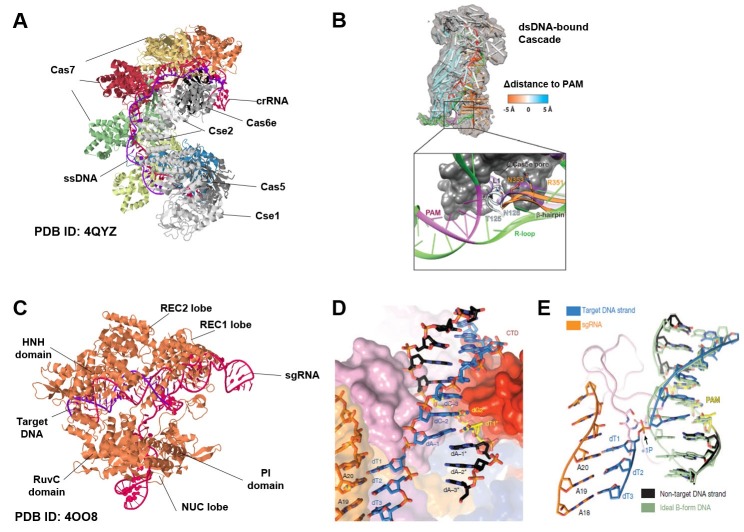
In the interference step of the type I-E CRISPR-Cas system, the multisubunit Cascade complex reportedly interrogates target DNA sequences for binding sites and recruits the Cas3 protein to the target site for cleavage via an interaction with the Cse1 subunit (15, 23). The Cascade complex is composed of five different Cas proteins, present in single or multiple copies: Cse1, Cse2 (two copies), Cas5, Cas6e, and Cas7 (six copies) (24, 25). Recent studies unraveled the structure of the Cascade complex, showing that Cas6 and Cas5 bind to the 3’ end and 5’ stem loop sequences of the guide crRNA, respectively, and in the middle of the complex, six Cas7 subunits form a filament that specifically binds the crRNA-target DNA heteroduplex (Fig. 1A) (24, 26-28), giving a seahorse-like shape to the overall structure. To interact with the target DNA sequence, Cascade uses the L1 loop (residues 125-131) of the Cse1 subunit (Fig. 1B) to interact with the PAM sequence (5’-CAT-3’ or 5’-AAT-3’) in the target DNA (29). In addition to this interaction, Cascade uses β-hairpin residues (343-366) and the L1 loop in the Cse1 subunit together to destabilize the target DNA (Fig. 1B) (29, 30). Once crRNA binds to the target DNA to make a DNA-RNA hybrid, the Watson-Crick base pairing results in the repulsion of the non-complement ssDNA (30, 31), inducing an R-loop structure. The Cas3 cleavage protein is then recruited to the target DNA through the interaction with the Cse1 subunit (23).
Fig. 1. Structures of the CRISPR components involved in PAM recognition and R-loop formation. (A) Overall structure of the Cascade complex, shown with crRNA hybridized with the target DNA to form a duplex. ssDNA is bound to Cascade, resulting in the seahorse-shaped architecture shown. Each Cascade subunit is shown in a different color. The figure is taken from (Sabin Mulepati et al. Science, 2014) (B) Molecular dynamic flexible fitting of the Cascade complex with the crRNA-DNA heteroduplex. In the simulation, the L1-helix is positioned proximal to the PAM and the β-hairpin is positioned between single-stranded regions of the DNA target. The figure is taken from (Paul B.G. van Erp et al. Nucleic Acids Res, 2015). (C) Overall structure of the Cas9 protein complexed with sgRNA. The figure is taken from (Hiroshi Nishimasu et al. Cell, 2014). (D) Close-up view of the Cas9 PAM binding region. (E) Superimposition of the unwound target DNA strand on an ideal B-form DNA duplex (green). The figure is taken from (Carolin Anders et al. Nature, 2014).
Type II CRISPR-Cas system (Cas9)
In the interference step of the type II CRISPR-Cas system, the Cas9 nuclease, with the help of two guide RNAs (crRNA and tracrRNA) or single-chain guide RNA (sgRNA), is the only component needed for cleavage (15, 16). The Cas9 protein from Streptococcus pyogenes (SpCas9) has a molecular weight of about 150 kDa and a structure that is mainly composed of two distinguishable domains: a REC lobe for DNA-RNA helix recognition and a NUC lobe for cleavage of target DNA (32) (Fig. 1C). The REC lobe is structurally divided into two subdomains, the REC1 and REC2 lobes; both are required for recognition of the backbone of a sgRNA (Fig. 1C). The NUC lobe is composed of an HNH domain and a RuvC domain, which are connected with a flexible linker so that the two domains have mobility for ligand binding (32). The HNH domain has a binding pocket similar to that of a normal endonuclease (33), so it likewise binds target DNA with a Mg2+ ion cofactor. The ability of the Cas9 protein to induce double-strand breaks (DSBs) at target DNA sequences, involves residues from two domain; D10 from the HNH domain and H840 from the RuvC domain (34). In addition to the cleavage domains, Cas9 has a PAM (5’-NGG-3’) recognition domain (residues 1099-1368) at the C-terminus of the NUC lobe (32) and it uses this domain for DNA recognition. A recent study determined that the Cas9-sgRNA complex binds to the PAM sequence of target DNA via two conserved arginine residues (R1333, R1335) in the C-terminal region of Cas9 (35). These residues play a major role in the recognition of the GG dinucleotide in the non-complementary strand of the target DNA through interaction with the major groove (Fig. 1D) (35). After PAM recognition, an interaction between the K1107 and S1109 loop (the phosphate lock loop) in Cas9, and the +1 phosphate upstream of the dinucleotide GG in the complementary strand induces target strand kinking (Fig. 1D, E), explaining R-loop formation in the RNA-DNA duplex and the mechanism for its propagation (30, 35-37). Recently, the crystal structure of a smaller ortholog, Cas9 from Staphylococcus aureus (SaCas9), was revealed; it shows a similar mechanism for target DNA opening (38).
DYNAMIC OBSERVATIONS OF DNA CLEAVAGE ACTIVITY OF CRISPR-CAS SYSTEMS
In addition to conformational knowledge gained from crystalized structures and biochemical assays, real-time dynamics of the interference step of the type II CRISPR-Cas system (e.g., target DNA searching, DNA binding, R-loop formation in the DNA-RNA complex, and the DNA cleavage process) have been observed at the single-molecule level. Tracking single protein dynamics enables highly sensitive real-time observations, which give precise spatial and temporal informations (39).
Target DNA searching mechanism of the Cas9-RNA complex (Type II CRISPR-Cas system)
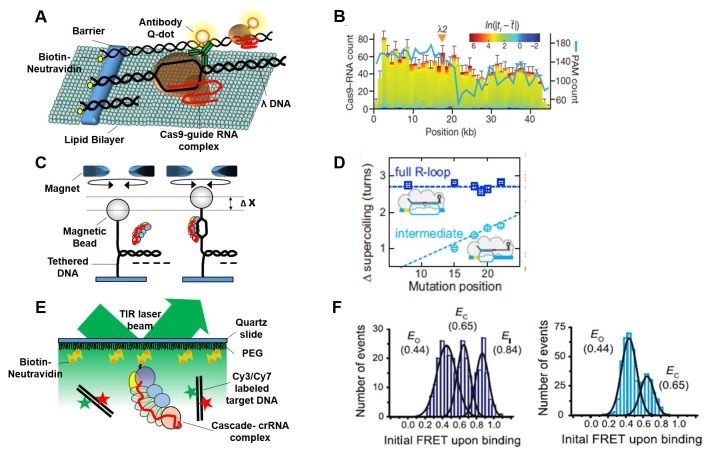
Eric Greene and colleagues have described how Cas9-RNA complexes of the type II CRISPR-Cas system search for their target DNA on long DNA sequences (36). In this study, the movement of single Cas9 proteins (with C-terminal Q-dot labeling) was tracked using a tethered long DNA curtain assay (Fig. 2A). Cas9-RNA complexes were observed to find target DNA sites not by one-dimensional hopping or sliding but by a three-dimensional (3D) diffusion binding mechanism. The authors found that there are two distinguishable binding modes; one involves longer binding times at the on-target site (Cas9-RNA remains tightly bound to target DNA, acting as a single-turnover enzyme) and the other is non-specific short binding (∼3.3 s or ∼58 s at 25 mM KCl)). In addition, non-specific bindings correlated very well with the distribution of PAM sequences (5’-NGG-3’) on the long λ DNA sequence used in the assay (Fig. 2B), indicating that Cas9-RNA complexes weakly interact with PAM sequences of the target DNA. Another recently published study elucidated the structural origin of these weak interactions (35), which are generated between conserved arginine residues in the C-terminus of Cas9 and the major groove of the non-target DNA strand at the ‘GG’ dinucleotide. From the above structural and dynamic information, the authors suggested that non-specific binding of the Cas9-RNA complex to off-target sites, which differ from each on-target site by several nucleotides, induces fast dissociation from DNA, thus facilitating the search for the on-target sequence. These observations elucidate how Cas9-RNA complexes find their on-target sites precisely among billions of possible nucleotides.
Fig. 2. Various single-molecule approaches for dynamic observation of the CRISPR-Cas system. (A) Tethered DNA curtain-based observation of the movement of a single Cas9 protein on target DNA. Cas9 movement was followed using 3X-FLAG tag-mediated Q-dot labeling. The crRNA-tracrRNA was designed for target sequence (λ2) binding on λ-DNA. (B) Distribution of Cas9–RNA binding events (n = 2,330) and PAM sites. Color-coding indicates the binding dwell time (ti) relative to the mean dwell time (t). Figure taken from (Samuel H. Sternberg et al. Nature, 2014). (C) Magnetic tweezers-based twisting assay. R-loop formation induced by the Cascade complex on negatively supercoiled DNA causes local DNA untwisting. Compensatory overtwisting of the DNA changes the supercoiling, resulting in a change in DNA length (Δx). Each mutant DNA template contains mutations at various locus from the PAM in wild-type DNA template (40). (D) Mean supercoiling changes associated with full (blue) and intermediate R-loop formation (light blue). Mutation position indicates the distance between PAM and mismatch position. Figure taken from (Marius Rutkauskas et al. Cell Reports, 2015). (E) Schematic of smFRET experiment for monitoring Cascade binding to labeled DNA substrates. Dual color (Cy3 and Cy7, shown in green and red star) labeled bona fide target construct contains a 15 bp flank, a PAM, and a protospacer (43). (F) FRET histogram from binding traces of Cascade to bona fide (Left) and PAM-mutated templates (Right) respectively. Figure taken from (Timothy R. Blosser et al. 2015, Mol Cell).
ATP-independent R-loop formation by Cas9 (Type II CRISPR-Cas system)
Cas9-RNA complexes can bind stably to on-target DNA sites to make R-loop structures and induce DNA cleavage. The detailed process of R-loop formation by the Cas9-RNA complex has been directly observed in real time at the single-molecule level in experiments involving magnetic tweezers (37). In this study, the authors used a 2.1 kbp DNA, containing a single copy of the on-target site, which was surface-tethered at one end and conjugated to a magnetic bead at the other to allow torque generation (Fig. 2C). The authors could track R-loop formation and dissociation by measuring the height of the bead under different torque conditions. In these experiments, they observed that i) R-loop formation is dependent on the Cas9-RNA complex concentration but rupture is independent and ii) PAM proximal sequences affect R-loop formation whereas PAM distal sequences affect R-loop stability but not formation, suggesting that the two regions discretely regulate Cas9 binding to target DNA.
Directional R-loop formation by the Cascade complex (Type I-E CRISPR-Cas system)
R-loop formation induced by a single Cascade complex of the type I-E CRISPR-Cas system has also been followed in real time in experiments using magnetic tweezers (Fig. 2C) (40). The authors first checked whether R-loops might be more stable at off-target versus on-target sites. They measured R-loop formation after changing the DNA template to give a mismatch between the target DNA sequence and the guiding crRNA within the Cascade complex. Whereas complete R-loop formation was observed in perfectly matching DNA templates, intermediate states (partial loops) were measured in mismatch-containing DNA templates (Fig. 2D). The difference between supercoiling values for the intermediate and full locked R-loop states is proportional to the distance between the mismatched position and the position of the PAM sequence, suggesting that the R-loop propagates from the PAM sequence to the PAM distal region. In further experiments, the authors showed that a mismatch-induced intermediate R-loop structure is easily ruptured under negative torque, suggesting that the unlocked state and small size of the intermediate R-loop make it highly unstable. In addition to studying R-loop formation, the authors also conducted cleavage experiments and showed that once the Cascade complex forms locked R-loop structures at target sites, Cas3 of Cascade can be recruited to induce the target DNA cleavage.
Different DNA binding modes of the Cascade complex (Type I-E CRISPR-Cas system)
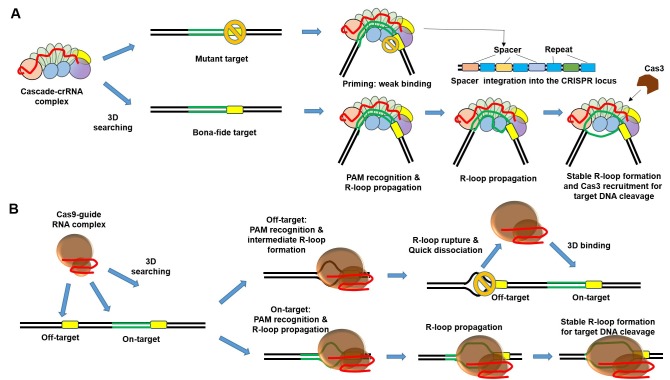
In the type I-E CRISPR-Cas system, the Cascade complex functions in two immune pathways; one is a priming step involving recognition and incorporation of the exogenous viral DNA to CRISPR locus (41, 42) and the other is the interference step that induces target DNA cleavage. A recently published study showed directly how Cascade can function in both pathways by using a single-molecule FRET (smFRET) assay (43). The authors first used biotinylated Cse1 subunits to immobilize Cascade complexes on a PEG-coated surface that was placed in a flow-cell chamber, after which dual dye (donor/acceptor) labeled target DNA templates were flowed into the chamber (Fig. 2E). Binding of a single Cascade complex to target DNA could be monitored in real time by measuring the fluorescence signals. For bona fide DNA targets that perfectly match the crRNA sequence and have an exact PAM sequence, three distinct FRET histograms were obtained (Fig. 2F, Left). They were interpreted to represent an initial high FRET state (FRET = 0.84, dwell time = 1.6 ± 0.4 s) assigned to transient bent DNA conformation and other two different states: long-lived (dwell time=about 1000s) middle FRET states thought to represent typical R-loop formation during the interference step (open formation: FRET = 0.44) and Cascade unbound state of the DNA (closed formation: FRET = 0.65). This pattern was interpreted to be the typical interference binding mode of the Cascade complex. On the other hand, for DNA targets that perfectly match the crRNA sequence but have a mutated PAM sequence, there were short-lived (dwell time = 24.8 ± 8.9 s) middle FRET (FRET = 0.44) states representing partial R-loop formation (Fig. 2F, Right); these were interpreted to represent atypical Cascade binding in the priming mode. Further experiments with mismatched DNA sequences in PAM proximal or PAM distal regions suggested that the priming mode of Cascade binding covers more exogenous DNA targets than the interference binding mode, and also suggested a mechanism by which the host E. coli immune system efficiently recognizes and cleaves exogenous viral DNAs (Fig. 3A).
Fig. 3. Schematic model of target searching and R-loop formation by Cascade or Cas9 on long stretches of DNA. (A) Model of target searching and discrimination by the Cascade complex. The Cascade–crRNA complex searches for the target sequence through random 3D collisions. When the complex recognizes a bona fide target, R-loop propagation starts from the PAM proximal region; the full R-loop structure is stabilized by DNA-RNA hybridization for target cleavage. In contrast, when Cascade complex binds weakly to a mutated target, Cascade processing the exogenous mutant DNA incorporation to the CRISPR spacer locus called priming. Target and PAM sequence is shown in green and yellow box respectively. DNA sequence mutation is shown in orange circle. (B) Model of target searching and R-loop formation by Cas9. The Cas9-guide RNA complex scans the target sequence through random 3D collisions. Non-specific binding to off-target sites induces the formation of an intermediate R-loop structure; failure to maintain the full R-loop structure generates a quick dissociation from the off-target sequence. This weak binding helps make searching for the on-target sequence efficient. On-target Cas9 binding promotes R-loop formation, similar to the Cascade complex, and stabilization of the R-loop structure allows cleavage of the DNA target.
CONCLUSIONS AND PERSPECTIVE
Taken together, crystal structures and dynamic observations suggest mechanisms for target DNA recognition and cleavage by type I (Fig. 3A) and type II CRISPR-Cas systems (Fig. 3B). Both systems induce R-loop formation during the DNA recognition process and have longer dwell times on on-target versus off-target DNA sites, even though the proteins involved are wholly different. To date, many outstanding in vitro studies about the CRISPR-Cas system have been reported, but the details of in vivo genome editing by CRISPR-Cas nucleases in human cells are still unclear. In the future, to obtain answers for questions such as ‘How do Cascade complexes or Cas9 nucleases search for their DNA target sites among the billions of nucleotides in the human genome?’ or ‘How fast do Cascade complexes or Cas9 nucleases find their DNA target sites in the chromatin structures of human cells?’, it will be necessary to directly observe Cascade complexes or Cas9 nucleases in vivo at the single cell level. Such an understanding of the working mechanisms of the CRISPR-Cas system in vitro and in vivo will be helpful for designing CRISPR-based tools for genome engineering and for expanding CRISPR applications further.
Acknowledgments
This work was supported by grants from the Plant Molecular Breeding Center of Next Generation BioGreen 21 Program (PJ01119201) and the Young Faculty supporting fund of Hanyang University to S.B.
References
- 1.Mali P, Yang L, Esvelt KM, et al. RNA-guided human genome engineering via Cas9. Science. (2013);339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jinek M, East A, Cheng A, Lin S, Ma E, Doudna J. RNA-programmed genome editing in human cells. eLife. (2013);2:e00471. doi: 10.7554/eLife.00471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hwang WY1, Fu Y, Reyon D, et al. Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat Biotechnol. (2013);31:227–229. doi: 10.1038/nbt.2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cong L, Ran FA, Cox D, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. (2013);339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cho SW, Kim S, Kim JM, Kim JS. Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nat Biotechnol. (2013);31:230–232. doi: 10.1038/nbt.2507. [DOI] [PubMed] [Google Scholar]
- 6.Barrangou R. RNA events. Cas9 targeting and the CRISPR revolution. Science. (2014);344:707–708. doi: 10.1126/science.1252964. [DOI] [PubMed] [Google Scholar]
- 7.Doudna JA, Charpentier E. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science. (2014);346:1258096. doi: 10.1126/science.1258096. [DOI] [PubMed] [Google Scholar]
- 8.Shalem O, Sanjana NE, Zhang F. High-throughput functional genomics using CRISPR-Cas9. Nat Rev Genet. (2015);16:299–311. doi: 10.1038/nrg3899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sander JD, Joung JK. CRISPR-Cas systems for editing, regulating and targeting genomes. Nat Biotechnol. (2014);32:347–355. doi: 10.1038/nbt.2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim H, Kim JS. A guide to genome engineering with programmable nucleases. Nat Rev Genet. (2014);15:321–334. doi: 10.1038/nrg3686. [DOI] [PubMed] [Google Scholar]
- 11.Travis J. Making the cut. Science. (2015);350:1456–1457. doi: 10.1126/science.350.6267.1456. [DOI] [PubMed] [Google Scholar]
- 12.Marraffini LA, Sontheimer EJ. CRISPR interference: RNA-directed adaptive immunity in bacteria and archaea. Nat Rev Genet. (2010);11:181–190. doi: 10.1038/nrg2749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wiedenheft B, Sternberg SH, Doudna JA. RNAguided genetic silencing systems in bacteria and archaea. Nature. (2012);482:331–338. doi: 10.1038/nature10886. [DOI] [PubMed] [Google Scholar]
- 14.Bhaya D, Davison M, Barrangou R. CRISPR-Cas systems in bacteria and archaea: versatile small RNAs for adaptive defense and regulation. Annu Rev Genet. (2011);45:273–297. doi: 10.1146/annurev-genet-110410-132430. [DOI] [PubMed] [Google Scholar]
- 15.van der Oost J, Westra ER, Jackson RN, Wiedenheft B. Unravelling the structural and mechanistic basis of CRISPR-Cas systems. Nat Rev Microbiol. (2014);12:479–492. doi: 10.1038/nrmicro3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. (2012);337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nuñez JK, Kranzusch PJ, Noeske J, Wright AV, Davies CW, Doudna JA. Cas1-Cas2 complex formation mediates spacer acquisition during CRISPR-Cas adaptive immunity. Nat Struct Mol Biol. (2014);21:528–534. doi: 10.1038/nsmb.2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mojica FJ, Diez-Villasenor C, Garcia-Martinez J, Almendros C. Short motif sequences determine the targets of the prokaryotic CRISPR defence system. Microbiology. (2009);155:733–740. doi: 10.1099/mic.0.023960-0. [DOI] [PubMed] [Google Scholar]
- 19.Deveau H, Barrangou R, Garneau JE, et al. Phage response to CRISPR-encoded resistance in Streptococcus thermophilus. J Bacteriol. (2008);190:1390–1400. doi: 10.1128/JB.01412-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brouns SJ, Jore MM, Lundgren M, et al. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science. (2008);321:960–964. doi: 10.1126/science.1159689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Carte J, Wang RY, Li H, Terns RM, Terns MP. Cas6 is an endoribonuclease that generates guide RNAs for invader defense in prokaryotes. Gene Dev. (2008);22:3489–3496. doi: 10.1101/gad.1742908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tsui TK, Li H. Structure Principles of CRISPR-Cas Surveillance and Effector Complexes. Annu Rev Biophys. (2015);44:229–255. doi: 10.1146/annurev-biophys-060414-033939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Westra ER, van Erp PB, Künne T, et al. CRISPR immunity relies on the consecutive binding and degradation of negatively supercoiled invader DNA by Cascade and Cas3. Mol Cell. (2012);46:595–605. doi: 10.1016/j.molcel.2012.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jackson RN, Golden SM, van Erp PB, et al. Structural biology. Crystal structure of the CRISPR RNA-guided surveillance complex from Escherichia coli. Science. (2014);345:1473–1479. doi: 10.1126/science.1256328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jore MM, Lundgren M, van Duijn E, et al. Structural basis for CRISPR RNA-guided DNA recognition by Cascade. Nat Struct Mol Biol. (2011);18:529–U141. doi: 10.1038/nsmb.2019. [DOI] [PubMed] [Google Scholar]
- 26.Mulepati S, Heroux A, Bailey S. Structural biology. Crystal structure of a CRISPR RNA-guided surveillance complex bound to a ssDNA target. Science. (2014);345:1479–1484. doi: 10.1126/science.1256996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhao H, Sheng G, Wang J, et al. Crystal structure of the RNA-guided immune surveillance Cascade complex in Escherichia coli. Nature. (2014);515:147–150. doi: 10.1038/nature13733. [DOI] [PubMed] [Google Scholar]
- 28.Wiedenheft B, Lander GC, Zhou K, et al. Structures of the RNA-guided surveillance complex from a bacterial immune system. Nature. (2011);477:486–489. doi: 10.1038/nature10402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van Erp PB, Jackson RN, Carter J, Golden SM, Bailey S, Wiedenheft B. Mechanism of CRISPR-RNA guided recognition of DNA targets in Escherichia coli. Nucleic Acids Res. (2015);43:8381–8391. doi: 10.1093/nar/gkv793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thomas M, White RL, Davis RW. Hybridization of RNA to double-stranded DNA: formation of R-loops. Proc Natl Acad Sci U S A. (1976);73:2294–2298. doi: 10.1073/pnas.73.7.2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wilson-Sali T, Hsieh TS. Preferential cleavage of plasmid-based R-loops and D-loops by Drosophila topoisomerase IIIbeta. Proc Natl Acad Sci U S A. 2002;99:7974–7979. doi: 10.1073/pnas.122007999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nishimasu H, Ran FA, Hsu PD, et al. Crystal structure of Cas9 in complex with guide RNA and target DNA. Cell. (2014);156:935–949. doi: 10.1016/j.cell.2014.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Biertumpfel C, Yang W, Suck D. Crystal structure of T4 endonuclease VII resolving a Holliday junction. Nature. (2007);449:616–620. doi: 10.1038/nature06152. [DOI] [PubMed] [Google Scholar]
- 34.Sternberg SH, LaFrance B, Kaplan M, Doudna JA. Conformational control of DNA target cleavage by CRISPR-Cas9. Nature. (2015);527:110–113. doi: 10.1038/nature15544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Anders C, Niewoehner O, Duerst A, Jinek M. Structural basis of PAM-dependent target DNA recognition by the Cas9 endonuclease. Nature. (2014);513:569–573. doi: 10.1038/nature13579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sternberg SH, Redding S, Jinek M, Greene EC, Doudna JA. DNA interrogation by the CRISPR RNA-guided endonuclease Cas9. Nature. (2014);507:62–67. doi: 10.1038/nature13011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Szczelkun MD, Tikhomirova MS, Sinkunas T, et al. Direct observation of R-loop formation by single RNAguided Cas9 and Cascade effector complexes. Proc Natl Acad Sci U S A. (2014);111:9798–9803. doi: 10.1073/pnas.1402597111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nishimasu H, Cong L, Yan WX, et al. Crystal Structure of Staphylococcus aureus Cas9. Cell. (2015);162:1113–1126. doi: 10.1016/j.cell.2015.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Joo C, Balci H, Ishitsuka Y, Buranachai C, Ha T. Advances in single-molecule fluorescence methods for molecular biology. Annu Rev Biochem. (2008);77:51–76. doi: 10.1146/annurev.biochem.77.070606.101543. [DOI] [PubMed] [Google Scholar]
- 40.Rutkauskas M, Sinkunas T, Songailiene I, Tikhomirova MS, Siksnys V, Seidel R. Directional R-Loop Formation by the CRISPR-Cas Surveillance Complex Cascade Provides Efficient Off-Target Site Rejection. Cell Rep. (2015) doi: 10.1016/j.celrep.2015.01.067. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 41.Datsenko KA, Pougach K, Tikhonov A, Wanner BL, Severinov K, Semenova E. Molecular memory of prior infections activates the CRISPR/Cas adaptive bacterial immunity system. Nat Commun. (2012);3:945. doi: 10.1038/ncomms1937. [DOI] [PubMed] [Google Scholar]
- 42.Fineran PC, Gerritzen MJ, Suárez-Diez M, et al. Degenerate target sites mediate rapid primed CRISPR adaptation. Proc Natl Acad Sci U S A. (2014);111:E1629–1638. doi: 10.1073/pnas.1400071111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Blosser TR, Loeff L, Westra ER, et al. Two distinct DNA binding modes guide dual roles of a CRISPR-Cas protein complex. Mol Cell. (2015);58:60–70. doi: 10.1016/j.molcel.2015.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]