Abstract
Alzheimer's disease (AD) is the most common form of dementia and exhibits a considerable level of heritability. The bridging integrator 1 (BIN1) gene has recently been identified in several large genome-wide association studies (GWAS) as the second most important risk locus for AD following apolipoprotein E (APOE). However, how and when the established genetic risk locus BIN1 rs744373 confers risk to late-onset AD has yet to be determined. Here using an imaging genetic strategy in large-sample Chinese subjects, we show that healthy homozygous carriers of the rs744373 risk allele exhibit worse high-load working memory (WM) performance, larger hippocampal volume and lower functional connectivity between the bilateral hippocampus and the right dorsolateral prefrontal cortex (DLPFC), mirroring clinical evidence of disturbed memory and connectivity in patients. Our findings demonstrate that rs744373 itself or a variation in linkage disequilibrium may provide a neurogenetic mechanism for BIN1 while further validating the possibility of combining genetic and neuroimaging strategies to monitor individuals at risk for AD.
INTRODUCTION
Alzheimer's disease (AD) is the leading cause of dementia and is characterized by a slowly progressive decline in cognitive functions that typically begins with a deterioration in memory (Holtzman et al, 2011). To date, no effective treatment is available to delay the onset or slow the progression of AD. Given that the disease process begins years before the onset of observable cognitive problems, there is an increasing push to identify the earliest disease stages and the at-risk individuals who will benefit most from disease-modifying interventions and prevention (Jiang et al, 2012). Unfortunately, there is currently no valid method to identify asymptomatic adults at risk for developing late-onset AD (LOAD). A combination of genetic, neuropsychological, and neuroimaging strategies may prove useful in this regard (Wishart et al, 2006).
The majority of AD is LOAD, which is genetically complex with heritability estimates as high as 80% (Gatz et al, 2006). Apolipoprotein E (APOE) has been clearly demonstrated to be the major genetic risk factor for LOAD, exhibiting semidominant inheritance. In recent years, large genome-wide association studies (GWAS) have identified several other genes/loci that, along with APOE ɛ4, contribute to a high proportion of genetic risk for LOAD. Among them is the bridging integrator 1 (BIN1) gene, located on chromosome 2q14.3, which has been identified as the most significant risk locus for LOAD after APOE (Bertram et al, 2007). Moreover, our previous studies also found that genetic variants in BIN1 were significantly associated with LOAD in the Han Chinese population (Tan et al, 2013a, 2014). BIN1 rs744373 is one of the most significant and best replicated single-nucleotide polymorphisms (SNPs) associated with AD in Caucasian populations, and it is located >25 kB upstream from the BIN1 coding region (Lambert et al, 2013; Seshadri et al, 2010). AlzGene meta-analysis revealed that the G allele confers a 1.17 greater odds of developing LOAD than does the A allele (http://www.alzgene.org/meta.asp?geneID=708). Recently, the results of pooled analysis and meta-analysis indicated that the rs744373 polymorphism contributed to AD with similar genetic risk in East Asian and Caucasian populations (Liu et al, 2013). The SNP rs744373 is in almost complete linkage disequilibrium (LD) (D'=0.98, r2=0.94) with functional rs59335482, which is associated with an increase in BIN1 transcriptional activity (Chapuis et al, 2013). In the brains of patients with AD, BIN1 protein expression levels increase in several regions, especially the hippocampus, and are correlated with tau pathology (Holler et al, 2014). Although BIN1 has been implicated in the pathophysiology of AD (Lambert et al, 2013), little is known about the neural mechanisms of the gene and how its protein product contributes to the manifestation of disease.
Although deficits in episodic memory characterize AD, there is increasing evidence that working memory (WM), which involves the short-term online storage and manipulation of information, is also impaired during the earliest stages of disease (Huntley and Howard, 2010). Deficits in WM have also been demonstrated using specialized testing in APOE ɛ4 carriers (Rosen et al, 2002). Interestingly, a high-density GWAS followed by replication studies, functional magnetic resonance imaging (MRI), and expression data suggest a role for several loci located on chromosome 2q in human short-term memory (Papassotiropoulos et al, 2011). GWAS SNPs were selected to tag underlying variation. Hence, there should be other as yet unidentified functional variants in LD with them located in chromosome 2q. BIN1 is also located in chromosome 2q and have an important role in the genetics and pathogenesis of AD (Tan et al, 2013b), hence, might associate with human short-term memory.
Evaluating the extent of AD pathology using neuroimaging biomarkers in patients with mild cognitive impairment (MCI) could provide clues regarding the biological mechanisms underlying progression to AD and assist with early identification of patients with greatest risk to progress to an AD diagnosis, which will be important for clinical trials and treatment development (Risacher and Saykin, 2013). Using structural MRI methods, numerous studies have demonstrated that MCI patients show significant atrophy in the medial temporal lobe (MTL), particularly in the entorhinal cortex and hippocampus, and focal cortical atrophy, particularly in the parietal, temporal, and frontal lobes (Chetelat et al, 2002; Hamalainen et al, 2007; Whitwell et al, 2008). In addition to detect differences in MCI patients, increased annual hippocampal atrophy rates and reduced baseline hippocampal volume accurately predicted MCI to probable AD conversion (Devanand et al, 2008; Jack et al, 1999). Moreover, studies using resting-state functional MRI and connectivity analyses, which are designed to evaluate intrinsic brain networks, have shown significant alterations in default mode network (DMN) connectivity and the connectivity of MTL regions in patients with MCI. Furthermore, more impaired MCI patients show impaired connectivity between the DMN and the hippocampus (episodic memory network) at rest (Buckner et al, 2005; Greicius et al, 2004). In summary, use of neuroimaging biomarkers, including notable brain atrophy and alterations in brain function, in clinical settings to detect and predict outcomes in MCI patients may soon be warranted as key tools for personalization of diagnostics and therapeutics (Risacher and Saykin, 2013). Therefore, brain structural and functional measures based on multi-modal neuroimaging techniques may be promising intermediate phenotypes to study the specific mechanisms of BIN1 genetic variant on risk of AD.
To increase our understanding of the neural mechanisms by which the common AD risk variant rs744373 in BIN1 may contribute to disease, we combined human genetics with memory assessments, structural MRI and resting-state functional MRI (fMRI) to trace the effects of this risk variant on WM, brain structure, and functional connectivity in a young healthy population.
MATERIALS AND METHODS
Subjects
We recruited 360 young healthy Chinese university students (186 males and 174 females; mean age=19.41±1.09 years, range=17–24 years; school education=12.33±0.80 years, range=10–16 years) (Table 1). The study was approved by the Ethics Committee of School of Life Science and Technology at University of Electronic Science and Technology of China, and all participants gave written informed consent. All of the participants were carefully screened to exclude individuals with a history of neurological or psychological diseases in the subjects or their third-degree relatives, psychiatric treatment, drug or alcohol abuse, traumatic brain injury, or visible brain lesions on conventional MRI. All subjects were examined using the Chinese Revised Wechsler Adult Intelligence Scale (WAIS-RC).
Table 1. Sociodemographic Features and General Brain Morphology of the Three Genotype Groups.
| AA | AG | GG | P | |
|---|---|---|---|---|
| Subjects | 155 | 172 | 33 | |
| Sex (male/female) | 79/76 | 84/88 | 23/10 | 0.087 |
| Handedness (right/left) | 144/11 | 158/14 | 30/3 | 0.839 |
| Age (years) | 19.41±1.06 | 19.39±1.14 | 19.47±0.99 | 0.934 |
| School education (years) | 12.34±0.71 | 12.33±0.86 | 12.30±0.88 | 0.968 |
| Full-scale intelligence quotient | 109.68±11.52 | 109.27±11.31 | 111.45±9.96 | 0.594 |
| Working memory performance (2-back condition) | 89.61±5.77 | 89.04±6.27 | 88.15±7.75 | 0.418 |
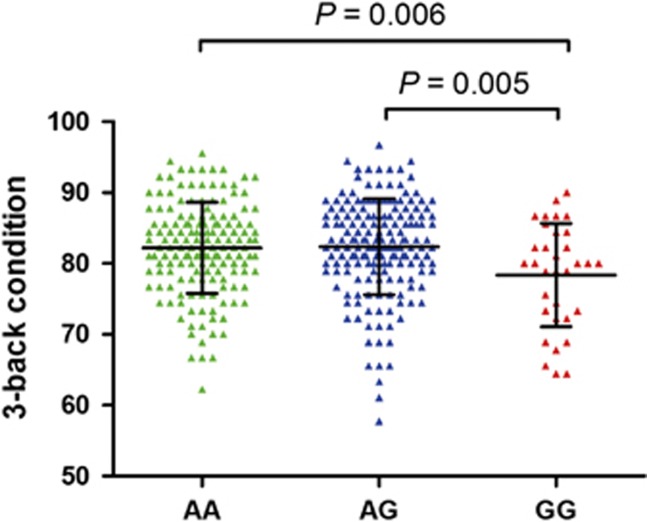
| Working memory performance (3-back condition) | 82.19±6.44 | 82.34±6.78 | 78.35±7.30 | 0.006 |
| Gray-matter volume (GMV) | 715.40±61.39 | 711.03±60.72 | 726.85±63.13 | 0.383 |
| White-matter volume (WMV) | 501.36±56.42 | 502.94±59.70 | 523.72±57.17 | 0.125 |
| CSF volume (CSFV) | 207.12±26.47 | 208.70±27.94 | 209.00±22.78 | 0.850 |
| Total intracranial volume (TIV) | 1423.88±129.50 | 1422.67±133.34 | 1459.56±127.95 | 0.317 |
| Normalized GMVa | 0.5029±0.0153 | 0.5004±0.0159 | 0.4982±0.0150 | 0.185 |
| Normalized WMVa | 0.3517±0.0156 | 0.3530±0.0168 | 0.3584±0.0160 | 0.098 |
| Normalized CSFVa | 0.1454±0.0123 | 0.1466±0.0123 | 0.1434±0.0115 | 0.337 |
| APOE status (APOEɛ4−/+) | 133/22 | 139/33 | 30/3 | 0.538 |
Values denote mean±standard deviation or number of subjects; P-values refer to one-way ANOVAs (parametric data) and Fisher–Freeman–Halton exact tests (categorical data).
The normalization of tissue volumes was calculated by dividing the GM, WM, and CSF volume by the TIV.
WM Performance
Individual WM capacity was evaluated using the n-back task, which has been widely used in previous studies on WM (Owen et al, 2005). This task was performed on a computer in a quiet room outside the MRI scanner before performing magnetic resonance scanning for each subject, and the data were evaluated using E-Prime, Version 2.0 (http://www.pstnet.com/eprime.cfm) as described in our previous study (Liu et al, 2014).
DNA Extraction and Genotyping
Ethylene Diamine Tetraacetic Acid (EDTA) anti-coagulated venous blood samples were collected from all individuals. Genomic DNA was extracted from whole blood using the EZgene Blood gDNA Miniprep Kit (Biomiga, San Diego, CA) according to the manufacturer's recommendations. Genotype data for BIN1 (rs744373) and APOE (rs429358 and rs7412) were obtained using the standard Illumina genotyping protocol (Illumina).
MRI Data Acquisition
MRI scans were performed on a MR750 3.0-Tesla magnetic resonance scanner (GE Healthcare). Resting-state functional imaging data were acquired using a gradient-echo echo-planar-imaging (GRE-EPI) sequence with the following parameters: repetition time (TR)=2000 ms, echo time (TE)=30 ms, field of view (FOV)=240 × 240 mm2, matrix=64 × 64, flip angle=90°, voxel size=3.75 × 3.75 × 4.0 mm3, 39 slices, and 255 volumes. High-resolution 3D T1-weighted brain volume (BRAVO) MRI sequence was subsequently performed with the following parameters: TR=8.16 ms, TE=3.18 ms, flip angle=7°, FOV=256 × 256 mm2, voxel size=1 × 1 × 1 mm3, and 188 slices. Before the scanning, all subjects were informed that they should move as little as possible, keep their eyes closed, think of nothing in particular and avoid falling asleep. Then, subjects were asked whether they fell asleep during and after the scanning to confirm that the included subjects did not fall asleep.
Structural MRI Preprocessing
A Diffeomorphic Anatomical Registration Through the Exponentiated Lie Algebra (DARTEL)-based T1 voxel-based morphometry (VBM) approach was used for preprocessing and subsequent analysis of whole-brain T1-weighted volumetric images (Ashburner, 2007). Using default settings, individual T1-weighted volumetric images were analyzed using the VBM8 toolbox (http://dbm.neuro.uni-hen.de/vbm) with Statistical Parametric Mapping 8 (SPM8, http://www.fil.ion.ucl.ac.uk/spm) executed in MATLAB 2013a (The MathWorks, Natick, MA). All images for each participant were carefully checked by an experienced radiologist to ensure the absence of scanner artifacts, motion problems, and gross anatomical abnormalities.
The structural magnetic resonance images were segmented into gray matter (GM), white matter, and cerebrospinal fluid (CSF) using the standard unified segmentation model in SPM8. Following segmentation, GM population templates were generated from the entire image data set using the DARTEL technique (Ashburner, 2007). After an initial affine registration of the GM DARTEL template to the tissue probability map in Montreal Neurological Institute (MNI) space (http://www.mni.mcgill.ca/), non-linear warping of the GM images was performed to the DARTEL GM template in MNI space with a resolution of 1.5 mm3 (as recommended for the DARTEL procedure). Gray-matter segments were modulated by the affine and non-linear components to preserve actual gray-matter values locally (modulated GM volumes). The homogeneity of the gray-matter images was checked according to the covariance structure of each image with all other images, as implemented in the check data quality function. Segmented tissue volumes (ie, GM, white matter, and CSF) were estimated in mm3 by counting the voxels representing GM, white matter, and CSF in standard space. Total intracranial volume (TIV) was determined as the sum of the GM, white matter, and CSF volumes. Finally, to compensate for residual between-subjects anatomical differences, the gray-matter images were smoothed with a Gaussian kernel of 8 mm full-width-at-half-maximal. After spatial preprocessing, the normalized, modulated, and smoothed gray-matter maps were used for statistical analysis.
Resting-State fMRI Preprocessing
All the raw fMRI data were inspected by two experienced radiologists who were blinded to the genotype information. Data preprocessing was completed using DPARSFA (Data Processing Assistant for Resting State fMRI Advanced Edition, http://www.restfmri.net/forum/DPARSF). The first 10 volumes of each scan were discarded to allow magnetization equilibrium. Subsequent preprocessing included slice timing, head motion correction, spatial normalizing to the EPI template, resampling to 3 × 3 × 3 mm3, smoothing with a 4-mm Gaussian kernel to decrease spatial noise, linear regressing to remove the effects of linear trends, temporal bandpass filtering, and regressing out nuisance signals (including head motion parameters and white matter, CSF, and global signals). Specifically, 27 subjects who exhibited a maximum displacement in any of the cardinal directions (x, y, z) of >2 mm or a maximum spin (x, y, z) of >2° were excluded from subsequent analyses. In the end, 333 subjects were included in the functional connectivity analysis (AA=144; AG=158; GG=31).
Statistical Analysis
We used Pearson's χ2 test to check for effects of gender, handedness, and APOE status differences; one-way analysis of variance (ANOVA) and analysis of covariance (ANCOVA) were used to check for differences in continuous sociodemographic variables, cognitive performance, and brain volumes. Hardy–Weinberg equilibrium between expected and observed genotype distributions were tested using the χ2 test. Correlations between WM performance, GM volume, and functional connectivity were computed using two-tailed Pearson correlations. These analysis were performed using the Statistical Package for the Social Sciences, version 18.0 (SPSS, Chicago, IL) for Windows. An alpha of P<0.05 was considered as significant.
Voxel-wise two-sample t-tests with five covariates (age, gender, education, TIV, and APOE status) were implemented in SPM8 to investigate the regional GM volume (GMV) differences. For each genotype group, Pearson's correlation analysis was further performed to test the association between the identified cluster volume and WM performance.
Based on the region that showed significant group differences in the whole brain structure analysis and a functional association with cognitive performance, we defined the bilateral hippocampus as regions-of-interest (ROIs) using the automated anatomical labeling (AAL) templates implemented in Wake Forest University PickAtlas. (Version 2.4) (Maldjian et al, 2003). Then, we computed Pearson correlation coefficients between the mean time series of the ROI and those of all the voxels throughout the whole brain and converted the correlation coefficient to z values using Fisher's r-to-z transformation to improve normality. To identify the clusters that showed significant functional connectivity with the ROI, we conducted a one-sample t-test on the z-functional connectivity maps of each individual to detect whether they were significantly different from zero (P<0.05, family-wise error (FWE) correction). Two-sample t-tests were used to compute the effects of the BIN1 genotype on the functional connectivity within the GM mask, with age, sex, education level, and APOE status as covariates. All statistical steps were completed using SPM8. For each genotype group, Pearson's correlation analysis was further performed to test the association between functional connectivity of the identified clusters and WM performance.
To control for multiple statistical testing in both the structural and functional connectivity analyses, we maintained a cluster-level false-positive detection rate at P<0.05 using a voxel-level threshold of P<0.005 with a cluster extent (k) empirically determined using Monte Carlo simulations (n=1000 iterations). This calculation was performed using the AlphaSim procedure implemented in the Resting-state fMRI Data Analysis Toolkit (REST) (http://restfmri.net/forum/index.php).
RESULTS
Genotyping and Demographics
Genotyping and demographic information are shown in Table 1. Genotyping for BIN1 rs744373 identified 155 individuals carrying an AA homozygous genotype, 172 individuals with an AG heterozygous genotype, and 33 individuals with a GG homozygous genotype. Genotype distributions for both BIN1 and APOE did not deviate from Hardy–Weinberg equilibrium (P>0.05). There were no significant differences in sex, handedness, age, education, full-scale intelligence quotient (IQ), and APOE ɛ4 status between the BIN1 rs744373 genotype groups (all P>0.05; Table 1). We found similar results among the 333 healthy participants included in the resting-state fMRI analysis (Supplementary Table S1).
Working Memory
We examined associations separately between rs744373 status and accuracy rates on the 2-back and 3-back WM tasks (Table 1). Group accuracies for the 3-back task showed significant differences among the three groups (P=0.006). However, group accuracies for the 2-back task did not differ among the three groups (P=0.418). We also found significant differences in the group accuracies for the 3-back task (P=0.002) and no significant differences in the group accuracies for 2-back task (P=0.373) among the 333 healthy participants included in the resting-state fMRI analysis (Supplementary Table S1). Post hoc analysis revealed that this association was the consequence of a reduced accuracy rate on the 3-back task in GG homozygotes compared with individuals who carried one or two A alleles (Figure 1). Hence, according to the BIN1 genotype, the subjects were divided into A allele carriers (AA+AG) and GG homozygotes in the subsequent structure MRI and resting-state fMRI analyses.
Figure 1.
Results of BIN1 rs744373 on the 3-back condition (mean±SD).
Structural MRI
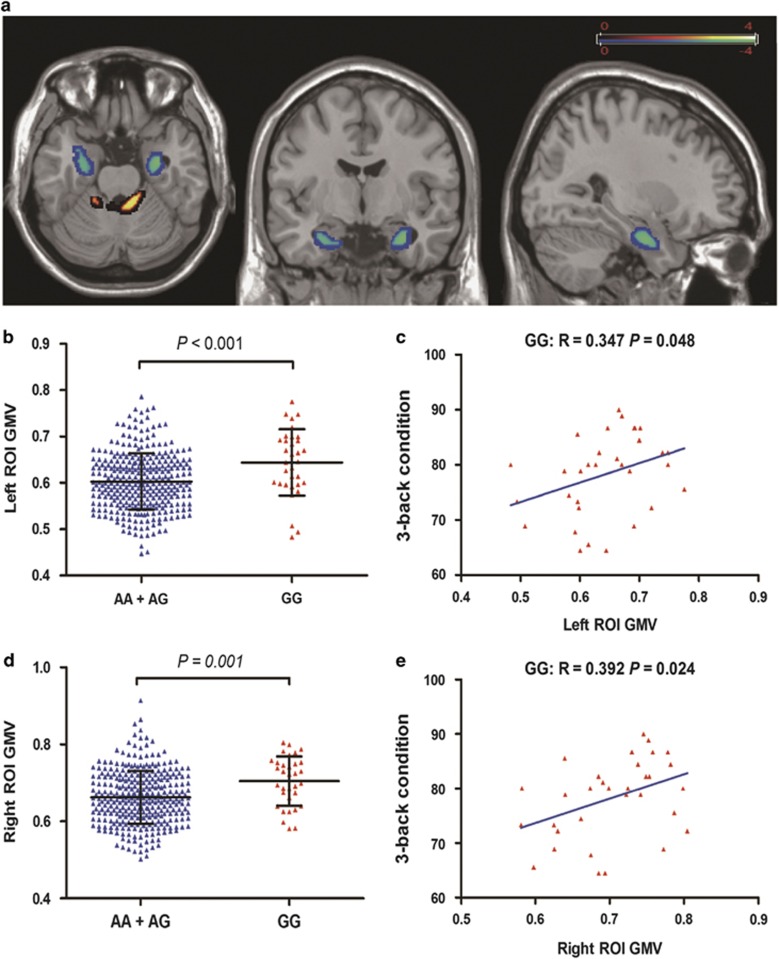
We did not find any significant results with respect to differences in GMV, white matter volume (WMV), CSF volume, their normalized volumes and TIV among the BIN1 genotypes (Table 1). The whole-brain VBM analyses demonstrated that GG homozygotes exhibited a significantly increased GMV compared with A allele carriers in the bilateral medial temporal cortex, including the hippocampus, amygdala, and parahippocampal gyrus (left: peak MNI coordinates x=−31.5, y=−6, z=−25.5, 300 voxels, peak T=4.28, P<0.005; right: peak MNI coordinates x=25.5, y=−12, z=−27, 312 voxels, peak T=3.21, P<0.005) (Figure 2a, b, and d), and significantly decreased GMV in the cerebellum (peak MNI coordinates x=−3, y=−40.5, z=−16.5, 897 voxels, peak T=−4.15, P<0.005) (Supplementary Figure S1). Comparing AA group with GG group, we got similar results (Supplementary Figure S2).
Figure 2.
Significant gray-matter volume (GMV) differences between A allele carriers and individuals who are homozygous for the risk allele (GG). (a) Whole-brain voxel-based morphometry showed that A allele carriers exhibit significantly (AlphaSim corrected P<0.05, using a voxel-level threshold of P<0.005 and a cluster threshold of k=300 voxels) increased GMV in the bilateral medial temporal cortex, including hippocampus, amygdala, and parahippocampal gyrus, and reduced GMV in the cerebellum compared with GG homozygotes. The average GMV of left (b) and right (c) affected medial temporal cortex was displayed for the two different genotype groups (mean±SD). Individual GMV within the left (c) and right (e) affected medial temporal cortex positively correlated with individual 3-back working memory performance.
To examine the influence of significant difference in sample size between two different groups on the results, we performed 10 000 iterations of simulation-based comparisons. For each iteration, we randomly took 33 out of the 327A allele carriers as one random subsample, and then compared the GMV of the bilateral medial temporal cortex of the random subsample with that of TG subjects. The same procedure was used in our previous study (Liu et al, 2014). A total of 9108/9094 out of the 10 000 comparisons made between groups of equal sample sizes indicated that the GMV of the left/right medial temporal cortex was significantly lower in the A allele carrier subsample than those in the GG group ( P<0.05), whereas none of the comparisons showed the inverse result (two-sample t-tests, All P>0.05).
Interestingly, the GM volumes of the bilateral affected regions were significantly correlated with the outcome of the 3-back condition in GG individuals (left: r=0.347, P=0.048, Figure 2c; right: r=0.392, P=0.024, Figure 2e), whereas this association disappeared in individuals carrying an A allele (AA+AG) (left: r=0.035, P=0.522; right: r=0.015, P=0.781). The correlations were positive, with better performance associated with greater GM volumes.
Resting-State fMRI
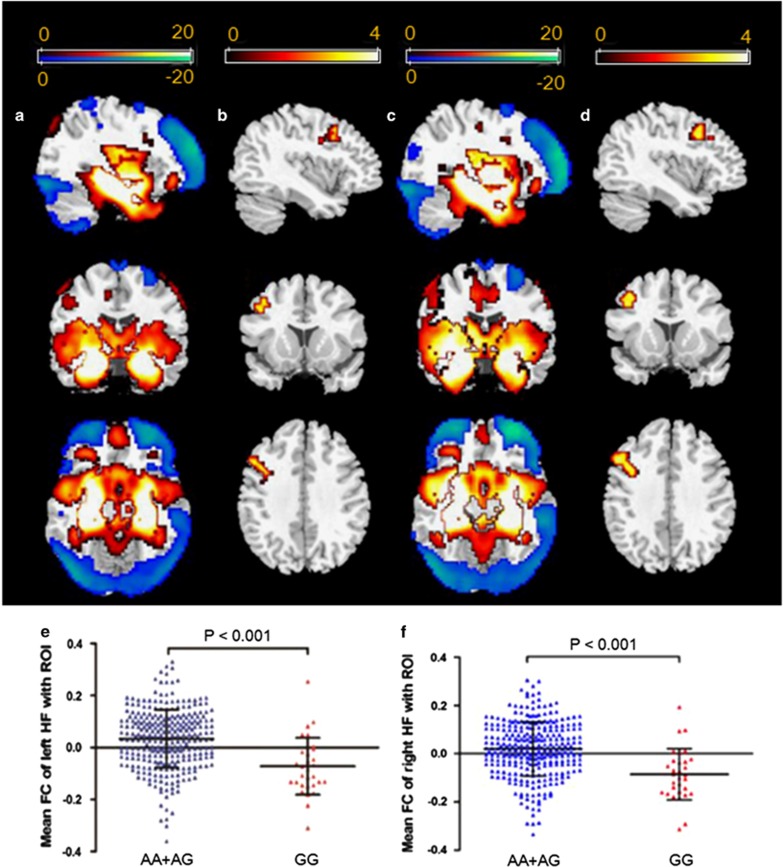
In the 333 healthy participants included in the resting-state fMRI analysis (AA=144; AG=158; GG=31), we investigated the influence of BIN1 genotype on the connectivity of the seed regions to the whole brain. Here, we extracted the bilateral hippocampus as the seed regions because of the significant difference observed in GMV between genotypes and the known relationship of the hippocampus with WM and AD. A one-sample t-test (FWE, P<0.05) was initially performed for the whole group (left hippocampus, Figure 3a; right hippocampus, Figure 3c), AA group, AG group, and GG group. The findings revealed that the whole group, AA group, and AG group exhibited similar hippocampal functional connectivity patterns (Supplementary Figure S3).
Figure 3.
Effect of BIN1 rs744373 on the resting-state bilateral hippocampal functional connectivity: (a) left hippocampus and (c) right hippocampus functional connectivity patterns obtained using one-sample t-tests for the whole group (family-wise error corrected to P<0.05); (b) left hippocampus and (d) right hippocampus functional connectivity analyses revealed decreased hippocampal connectivity with the right dorsolateral prefrontal cortex (DLPFC) in individuals who were homozygous for the risk allele (GG) compared with A allele carriers (AlphaSim corrected P<0.05, using a voxel-level threshold of P<0.005 and a cluster threshold of k=11 voxels); (e) independent region-of-interest (ROI) analyses by taking the structural left hippocampal formation (HF) as ROI further supported that the DLPFC-HF functional connectivity was different between the two different genotype groups (mean±SD); (f) independent ROI analyses by taking the structural right HF as ROI further supported that the DLPFC-HF functional connectivity was different between the two different genotype groups (mean±SD).
Comparisons of the functional connectivity between A allele carriers and GG homozygotes were performed using two-sample t-test analyses after correcting for sex, age, education, and APOE ɛ4 status. When the left hippocampus was treated as the seed region, there was a significant effect of genotype on the connectivity between the left hippocampus and the right dorsolateral prefrontal cortex (DLPFC) (peak MNI coordinates: x=54, y=21, z=48; 137 voxels; peak T=4.13; P<0.001) (Figure 3b). Similarly, when the right hippocampus was treated as the seed region, there was also a significant effect of genotype on the connectivity between the seed region and the right DLPFC (peak MNI coordinates: x=54, y=21, z=48; 184 voxels; peak T=4.67; P<0.001) (Figure 3d). For the functional connectivity between the bilateral hippocampal formation (HF) and the right DLPFC, GG homozygotes exhibited decreased functional connectivity (FC) compared with A allele carriers (left HF: P<0.001, Figure 3e; right HF: P<0.001, Figure 3f).
Furthermore, we analyzed the association between functional connectivity and brain volume as well as the association between functional connectivity and WM performance. Among these analyses, we only found that the functional connectivity between the right hippocampus and the affected right DLPFC was significantly correlated with the outcome of 2-back condition in AA+AG group (r=−0.117, p=0.043). To test the potential influence of global signal regression on our findings, we reanalyzed our data without global signal regression and confirmed that the main findings remained consistent with the results that included global signal regression (Supplementary Figure S4). To test the structure–function relationship, we repeated the functional connectivity analysis while controlling for the GMV of the seed region and continued to not find any significant changes.
DISCUSSION
By using a behavioral genetics approach combined with structural and resting-state functional MRI in a large sample of young healthy individuals, we found that healthy homozygous carriers of the rs744373 risk allele exhibit worse high-load WM performance, larger hippocampal volume and lower functional connectivity between the bilateral hippocampus and the right DLPFC. The current findings provide important implications regarding individual differences in the genetic contribution of BIN1 to memory, brain structure, and function, as well as the risk of developing AD.
Interestingly, this study is the first to show that the rs744373 AD risk variant in BIN1 significantly affects the n-back WM performance in a recessive genetic model of young healthy individuals. Although early and significant episodic memory impairment constitutes the clinical core diagnostic criterion of AD (Dubois et al, 2014), there is increasing evidence of early deficits in WM and executive function in patients with MCI and AD (Crawford et al, 2013; Gagnon and Belleville, 2011; Huntley and Howard, 2010; Kessels et al, 2011; Koppel et al, 2014). Moreover, fMRI studies have also suggested a possible subclinical impairment of WM capacity in healthy APOE ɛ4 carriers (Chen et al, 2013). Here, we investigated the effects of BIN1 rs744373 on the performance of n-back WM tasks in young healthy individuals. The results indicated that during the 2-back WM task, performance was not different between groups. However, as the WM load increased to a 3-back task, individuals who were homozygous for the G risk allele at rs744373 exhibited worse performance than did demographically matched A allele carriers. One possible explanation for this phenomenon is a mild impairment of WM capacity in the G risk allele homozygotes. During low-load WM (2-back condition), the GG homozygotes recruited additional processing resources to compensate for processing inefficiencies. As the WM load increased (3-back condition), the GG homozygotes, however, had already recruited most of their available resources (which are presumably finite), resulting in worse performance.
As the BIN rs744373 polymorphism affects performance during the 3-back WM task in a recessive genetic model, recessive model analysis was performed following the structural MRI and resting-state fMRI studies. The recessive contrast model of the whole-brain VBM analyses demonstrates that individuals who are homozygous for the G risk allele at rs744373 compared with A allele carriers show significantly increased GMV in the bilateral regions associated with WM and AD, namely the medial temporal cortex, including the hippocampus, amygdala, and parahippocampal gyrus. One previous GWAS has found nominal association of risk allele in BIN1 rs744373 with hippocampal volume (Bis et al, 2012). However, the association was not replicated in another GWAS (Stein et al, 2012), the risk allele of BIN1 rs744373 showed no significant associations with hippocampal volume controlling for other factors in all subjects and only in normal subjects (All P>0.05, data can be obtained from the website of ENIGMA: http://enigma.ini.usc.edu/enigma-vis/). One study has also investigated whether the BIN1 rs7561528 polymorphism, which is different from rs744373 and rs59335482, influences brain structure in AD and MCI patients and in elderly controls (Biffi et al, 2010). The study found a trend toward a significant effect on decreased hippocampal volume (corrected P=0.08) in the combined samples of AD, MCI patients, and elderly controls. Decreased hippocampal volume is an early feature of AD (Schott et al, 2003). Past VBM studies reported decreased GMV bilaterally in the MTL, including the hippocampus, in ɛ4 homozygotes compared with non-carriers in healthy elderly subjects (Lemaitre et al, 2005). Interestingly, in contrast to the APOE gene, homozygotes of the BIN1 risk allele at rs744373 were found to exhibit increased bilateral hippocampal GMV compared with A allele carriers. The increased bilateral hippocampal GMV might reflect a compensatory recruitment of neural resources to help maintain normal cognition in individuals with a genetic risk for AD (Bookheimer et al, 2000). This interpretation is supported by the observation of a positive correlation between affected hippocampal volume and WM performance in GG groups, reflecting better compensatory recruitment of neural resources due to the larger hippocampal volume; however, a similar correlation was not found in A allele carriers. It will be important to assess the compensatory hypothesis through continued longitudinal research that combines structural and functional neuroimaging approaches with genetics and neuropsychological testing.
Furthermore, we investigated the influence of the BIN1 genotype on the functional connectivity of the bilateral hippocampus to the whole brain. We found that the increased hippocampal volume in GG individuals was accompanied by a significant reduction in functional connectivity between the bilateral HF and the right DLPFC. Previous studies have showed the laterality effects for prefrontal effects in normal controls (Van Horn et al, 1996), and the right DLPFC showed greater recruitment during the WM task in siblings of schizophrenia compared with normal controls (MacDonald et al, 2009). The connectivity of only right DLPFC with HF was found to be disrupted in schizophrenia patients (Meyer-Lindenberg et al, 2005) and high-risk individuals (Liu et al, 2014; Rasetti et al, 2011). The effect of BIN1 genotype on brain structure and FC can be interpreted by the hypothesis that the reduction in HF-DLPFC functional connectivity leads to a compensatory increase in hippocampal volume to maintain performance. One previous study also proposed a similar mechanism that hypersensitivity to negative stimuli in the DLPFC might lead to hippocampal atrophy by loss of or inhibited neurogenesis or synaptogenesis of neurons (Suzuki et al, 2013). Our finding of altered functional connectivity has been supported by cognitive neuroscience evidence indicating that the HF and the DLPFC form part of a distributed functional network of regions involved in AD (Allen et al, 2007; Wang et al, 2006) and memory (Preston and Eichenbaum, 2013) and that interactions between these regions are particularly important for WM (Axmacher et al, 2008; Ranganath, 2006). Using seeds in the bilateral hippocampus, patients with AD demonstrated markedly reduced functional connectivity with the prefrontal cortex (Allen et al, 2007; Wang et al, 2006). Hippocampal-prefrontal connectivity has been shown to contribute to WM in several functional connectivity studies of fMRI data (Axmacher et al, 2008; Petersson et al, 2006; Rissman et al, 2008). More specifically, evidence suggests that these two regions serve complementary roles during WM processes (Hasselmo and Stern, 2006; Stern et al, 2001). Whereas the prefrontal cortex (PFC) appears to be predominantly important for WM maintenance of familiar items, such as words, the hippocampus may have a role in maintaining unfamiliar novel items. Reduced functional connectivity between the hippocampus and DLPFC as a neural mechanism of risk is further supported by clinical evidence, with reduced or even absent hippocampal-prefrontal connectivity reported during memory retrieval in amnestic MCI (Bai et al, 2009) and AD patients (Allen et al, 2007; Grady et al, 2001). It has been suggested that memory breakdown during AD is associated with a marked reduction in integrated activity within a distributed network that includes the hippocampus and DLPFC (Grady et al, 2001). Moreover, fMRI studies have also provided evidence for significantly reduced coupling between the hippocampus and DLPFC during episodic memory retrieval (Erk et al, 2011), as well as aberrant activation in the same regions, during WM performance in healthy young individuals carrying the CLU risk allele (C allele of 11136000) for AD (Lancaster et al, 2011). Although they are in a different cognitive domain, these results additionally emphasize the relevance of hippocampal–prefrontal interaction to the pathology of AD risk.
The molecular and cellular mechanisms responsible for the observed changes in WM performance, hippocampal volume, and hippocampus-DLPFC functional connectivity remain to be elucidated. However, our data indicate that BIN1 rs744373 may be functional in the human brain even in young healthy individuals and provide a mechanism for the genetic findings suggested by the GWAS that are congruent with current findings on the pathophysiology of AD. It is important to stress that the rs744373 polymorphism is most likely a marker SNP that is in LD with one or more functional variations located elsewhere within BIN1 that also affects the current findings. Chapuis et al, (2013) reported that the SNP rs744373 was in almost complete LD (D'=0.98, r2=0.94) with the functional insertion/deletion rs59335482 in Caucasian populations. The functional rs59335482 insertion risk allele was found to be associated with an increase in BIN1 transcriptional activity in vitro, BIN1 expression levels in the human brain and AD risk. In any genetic association study, the core finding is the identification of a gene, rather than a specific variation, associated with the phenotype of interest. The exact pathogenic mechanisms of BIN1 in the AD pathophysiological process remain to be determined. Emerging data suggest that BIN1 might modulate microtubule stability, Tau phosphorylation/aggregation, or neurofibrillary tangle formation (Chapuis et al, 2013). BIN1 has also been identified as a regulator of endocytosis and trafficking, immunity and inflammation of the brain, transient calcium potentials, and apoptosis (Lambert et al, 2013). Additionally, while we did not find any interaction effect between APOE and BIN1 on GMV and FC, the various pathophysiological effects between APOE and BIN1 may explain the wide range of presentations regarding compensatory neuronal resources in healthy individuals. BIN1 contributes to AD pathogenesis primarily through a modulation of tau pathology (Lambert et al, 2013). These various pathophysiological effects on AD lead to a reduced hippocampal GMV (Lemaitre et al, 2005) with higher levels of neural activity in the right DLPFC (Wishart et al, 2006) for APOE; however, larger hippocampal GMV with lower functional connectivity with the right DLPFC for BIN1 was observed in the current study, both of which help maintain normal cognition in individuals who carry genetic risk for AD.
Taken together, our findings add to the growing body of evidence supporting the presence of preclinical cognitive, structure, and functional neural changes in cognitively healthy individuals who, as a group, are at genetic risk for AD. BIN1 rs744373 itself (or an unknown variant that is in LD with this polymorphism) is functional in the human brain even in young healthy individuals and offers a mechanism for the genetic findings indicated by GWAS that are congruent with current findings on the pathophysiology of AD. The current finding further provide possible neural mechanisms underlying the association between the BIN1 polymorphism and risk for AD (Lambert et al, 2013) along with altered hippocampal volume and HF-DLPFC connectivity that contribute to the neurogenetic architecture implicated in AD susceptibility. Quantitative mapping of brain structural and functional connectivity differences in those at genetic risk for AD is crucial for evaluating treatment and prevention strategies. Moreover, targeting adults at greatest risk for cognitive deterioration can also improve the power of clinical trials with the development of disease-modifying drugs (Sperling et al, 2014). Because our current findings track an established genetic risk factor in young healthy individuals, they might be useful as a means to identify an intermediate phenotype or biomarker to determine those at risk, develop treatments designed to prevent the development of the disease, achieve early detection of disease susceptibility and monitor treatment studies, as well as potentially even identifying a novel potential therapeutic target.
FUNDING AND DISCLOSURE
The authors declare no conflict of interest.
Acknowledgments
This work was supported by the National Key Basic Research and Development Program (973) (Grant No. 2011CB707800), the Strategic Priority Research Program of the Chinese Academy of Sciences (Grant No. XDB02030300), the Natural Science Foundation of China (Grant Nos. 91232718 and 91132301), and CAS Youth Innovation Promotion Association and Excellent Young Scientist Program.
Footnotes
Supplementary Information accompanies the paper on the Neuropsychopharmacology website (http://www.nature.com/npp)
Supplementary Material
References
- Allen G, Barnard H, McColl R, Hester AL, Fields JA, Weiner MF et al (2007). Reduced hippocampal functional connectivity in Alzheimer disease. Arch Neurol 64: 1482–1487. [DOI] [PubMed] [Google Scholar]
- Ashburner J (2007). A fast diffeomorphic image registration algorithm. Neuroimage 38: 95–113. [DOI] [PubMed] [Google Scholar]
- Axmacher N, Schmitz DP, Wagner T, Elger CE, Fell J (2008). Interactions between medial temporal lobe, prefrontal cortex, and inferior temporal regions during visual working memory: a combined intracranial EEG and functional magnetic resonance imaging study. J Neurosci 28: 7304–7312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai F, Zhang Z, Watson DR, Yu H, Shi Y, Yuan Y et al (2009). Abnormal functional connectivity of hippocampus during episodic memory retrieval processing network in amnestic mild cognitive impairment. Biol Psychiatry 65: 951–958. [DOI] [PubMed] [Google Scholar]
- Bertram L, McQueen MB, Mullin K, Blacker D, Tanzi RE (2007). Systematic meta-analyses of Alzheimer disease genetic association studies: the AlzGene database. Nat Genet 39: 17–23. [DOI] [PubMed] [Google Scholar]
- Biffi A, Anderson CD, Desikan RS, Sabuncu M, Cortellini L, Schmansky N et al (2010). Genetic variation and neuroimaging measures in Alzheimer disease. Arch Neurol 67: 677–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bis JC, DeCarli C, Smith AV, van der Lijn F, Crivello F, Fornage M et al (2012). Common variants at 12q14 and 12q24 are associated with hippocampal volume. Nat Genet 44: 545–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bookheimer SY, Strojwas MH, Cohen MS, Saunders AM, Pericak-Vance MA, Mazziotta JC et al (2000). Patterns of brain activation in people at risk for Alzheimer's disease. N Engl J Med 343: 450–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckner RL, Snyder AZ, Shannon BJ, LaRossa G, Sachs R, Fotenos AF et al (2005). Molecular, structural, and functional characterization of Alzheimer's disease: evidence for a relationship between default activity, amyloid, and memory. J Neurosci 25: 7709–7717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapuis J, Hansmannel F, Gistelinck M, Mounier A, Van Cauwenberghe C, Kolen KV et al (2013). Increased expression of BIN1 mediates Alzheimer genetic risk by modulating tau pathology. Mol Psychiatry 18: 1225–1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CJ, Chen CC, Wu D, Chi NF, Chen PC, Liao YP et al (2013). Effects of the apolipoprotein E epsilon4 allele on functional MRI during n-back working memory tasks in healthy middle-aged adults. AJNR Am J Neuroradiol 34: 1197–1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chetelat G, Desgranges B, de la Sayette V, Viader F, Eustache F, Baron JC (2002). Mapping gray matter loss with voxel-based morphometry in mild cognitive impairment. Neuroreport 13: 1939–1943. [DOI] [PubMed] [Google Scholar]
- Crawford TJ, Higham S, Mayes J, Dale M, Shaunak S, Lekwuwa G (2013). The role of working memory and attentional disengagement on inhibitory control: effects of aging and Alzheimer's disease. Age 35: 1637–1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devanand DP, Liu XH, Tabert MH, Pradhaban G, Cuasay K, Bell K et al (2008). Combining early markers strongly predicts conversion from mild cognitive impairment to Alzheimer's disease. Biol Psychiatry 64: 871–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubois B, Feldman HH, Jacova C, Hampel H, Molinuevo JL, Blennow K et al (2014). Advancing research diagnostic criteria for Alzheimer's disease: the IWG-2 criteria. Lancet Neurol 13: 614–629. [DOI] [PubMed] [Google Scholar]
- Erk S, Meyer-Lindenberg A, Opitz von Boberfeld C, Esslinger C, Schnell K, Kirsch P et al (2011). Hippocampal function in healthy carriers of the CLU Alzheimer's disease risk variant. J Neurosci 31: 18180–18184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagnon LG, Belleville S (2011). Working memory in mild cognitive impairment and Alzheimer's disease: contribution of forgetting and predictive value of complex span tasks. Neuropsychology 25: 226–236. [DOI] [PubMed] [Google Scholar]
- Gatz M, Reynolds CA, Fratiglioni L, Johansson B, Mortimer JA, Berg S et al (2006). Role of genes and environments for explaining Alzheimer disease. Arch Gen Psychiatry 63: 168–174. [DOI] [PubMed] [Google Scholar]
- Grady CL, Furey ML, Pietrini P, Horwitz B, Rapoport SI (2001). Altered brain functional connectivity and impaired short-term memory in Alzheimer's disease. Brain 124: 739–756. [DOI] [PubMed] [Google Scholar]
- Greicius MD, Srivastava G, Reiss AL, Menon V (2004). Default-mode network activity distinguishes Alzheimer's disease from healthy aging: Evidence from functional MRI. Proc Natl Acad Sci USA 101: 4637–4642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamalainen A, Tervo S, Grau-Olivares M, Niskanen E, Pennanen C, Huuskonen J et al (2007). Voxel-based morphometry to detect brain atrophy in progressive mild cognitive impairment. Neuroimage 37: 1122–1131. [DOI] [PubMed] [Google Scholar]
- Hasselmo ME, Stern CE (2006). Mechanisms underlying working memory for novel information. Trends Cogn Sci 10: 487–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holler CJ, Davis PR, Beckett TL, Platt TL, Webb RL, Head E et al (2014). Bridging Integrator 1 (BIN1) protein expression increases in the Alzheimer's disease brain and correlates with neurofibrillary tangle pathology. J Alzheimers Dis. 42: 1221–1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holtzman DM, Morris JC, Goate AM (2011). Alzheimer's disease: the challenge of the second century. Sci Transl Med 3: 77sr71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huntley JD, Howard RJ (2010). Working memory in early Alzheimer's disease: a neuropsychological review. Int J Geriatr Psychiatry 25: 121–132. [DOI] [PubMed] [Google Scholar]
- Jack CR, Petersen RC, Xu YC, O'Brien PC, Smith GE, Ivnik RJ et al (1999). Prediction of AD with MRI-based hippocampal volume in mild cognitive impairment. Neurology 52: 1397–1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang T, Yu JT, Tan L (2012). Novel disease-modifying therapies for Alzheimer's disease. J Alzheimers Dis 31: 475–492. [DOI] [PubMed] [Google Scholar]
- Kessels RP, Molleman PW, Oosterman JM (2011). Assessment of working-memory deficits in patients with mild cognitive impairment and Alzheimer's dementia using Wechsler's Working Memory Index. Aging Clin Exp Res 23: 487–490. [DOI] [PubMed] [Google Scholar]
- Koppel J, Sunday S, Goldberg TE, Davies P, Christen E, Greenwald BS (2014). Psychosis in Alzheimer's disease is associated with frontal metabolic impairment and accelerated decline in working memory: findings from the Alzheimer's Disease Neuroimaging Initiative. Am J Geriatr Psychiatry 22: 698–707. [DOI] [PubMed] [Google Scholar]
- Lambert JC, Ibrahim-Verbaas CA, Harold D, Naj AC, Sims R, Bellenguez C et al (2013). Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer's disease. Nat Genet 45: 1452–1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancaster TM, Baird A, Wolf C, Jackson MC, Johnston SJ, Donev R et al (2011). Neural hyperactivation in carriers of the Alzheimer's risk variant on the clusterin gene. Eur Neuropsychopharmacol 21: 880–884. [DOI] [PubMed] [Google Scholar]
- Lemaitre H, Crivello F, Dufouil C, Grassiot B, Tzourio C, Alperovitch A et al (2005). No epsilon4 gene dose effect on hippocampal atrophy in a large MRI database of healthy elderly subjects. Neuroimage 24: 1205–1213. [DOI] [PubMed] [Google Scholar]
- Liu B, Zhang X, Hou B, Li J, Qiu C, Qin W et al (2014). The impact of MIR137 on dorsolateral prefrontal-hippocampal functional connectivity in healthy subjects. Neuropsychopharmacology 39: 2153–2160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G, Zhang S, Cai Z, Li Y, Cui L, Ma G et al (2013). BIN1 gene rs744373 polymorphism contributes to Alzheimer's disease in East Asian population. Neurosci Lett 544: 47–51. [DOI] [PubMed] [Google Scholar]
- MacDonald AW 3rd, Thermenos HW, Barch DM, Seidman LJ (2009). Imaging genetic liability to schizophrenia: systematic review of FMRI studies of patients' nonpsychotic relatives. Schizophr Bull 35: 1142–1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maldjian JA, Laurienti PJ, Kraft RA, Burdette JH (2003). An automated method for neuroanatomic and cytoarchitectonic atlas-based interrogation of fMRI data sets. Neuroimage 19: 1233–1239. [DOI] [PubMed] [Google Scholar]
- Meyer-Lindenberg AS, Olsen RK, Kohn PD, Brown T, Egan MF, Weinberger DR et al (2005). Regionally specific disturbance of dorsolateral prefrontal–hippocampal functional connectivity in schizophrenia. Arch Gen Psychiatry 62: 379–386. [DOI] [PubMed] [Google Scholar]
- Owen AM, McMillan KM, Laird AR, Bullmore E (2005). N-back working memory paradigm: a meta-analysis of normative functional neuroimaging studies. Hum Brain Mapp 25: 46–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papassotiropoulos A, Henke K, Stefanova E, Aerni A, Muller A, Demougin P et al (2011). A genome-wide survey of human short-term memory. Mol Psychiatry 16: 184–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersson KM, Gisselgard J, Gretzer M, Ingvar M (2006). Interaction between a verbal working memory network and the medial temporal lobe. Neuroimage 33: 1207–1217. [DOI] [PubMed] [Google Scholar]
- Preston AR, Eichenbaum H (2013). Interplay of hippocampus and prefrontal cortex in memory. Curr Biol 23: R764–R773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranganath C (2006). Working memory for visual objects: complementary roles of inferior temporal, medial temporal, and prefrontal cortex. Neuroscience 139: 277–289. [DOI] [PubMed] [Google Scholar]
- Rasetti R, Sambataro F, Chen Q, Callicott JH, Mattay VS, Weinberger DR (2011). Altered cortical network dynamics: a potential intermediate phenotype for schizophrenia and association with ZNF804A. Arch Gen Psychiatry 68: 1207–1217. [DOI] [PubMed] [Google Scholar]
- Risacher SL, Saykin AJ (2013). Neuroimaging and other biomarkers for Alzheimer's disease: the changing landscape of early detection. Annu Rev Clin Psychol 9: 621–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rissman J, Gazzaley A, D'Esposito M (2008). Dynamic adjustments in prefrontal, hippocampal, and inferior temporal interactions with increasing visual working memory load. Cereb Cortex 18: 1618–1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosen VM, Bergeson JL, Putnam K, Harwell A, Sunderland T (2002). Working memory and apolipoprotein E: what's the connection? Neuropsychologia 40: 2226–2233. [DOI] [PubMed] [Google Scholar]
- Schott JM, Fox NC, Frost C, Scahill RI, Janssen JC, Chan D et al (2003). Assessing the onset of structural change in familial Alzheimer's disease. Ann Neurol 53: 181–188. [DOI] [PubMed] [Google Scholar]
- Seshadri S, Fitzpatrick AL, Ikram MA, DeStefano AL, Gudnason V, Boada M et al (2010). Genome-wide analysis of genetic loci associated with Alzheimer disease. JAMA 303: 1832–1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperling RA, Rentz DM, Johnson KA, Karlawish J, Donohue M, Salmon DP et al (2014). The A4 study: stopping AD before symptoms begin? Sci Transl Med 6: 228fs213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein JL, Medland SE, Vasquez AA, Hibar DP, Senstad RE, Winkler AM et al (2012). Identification of common variants associated with human hippocampal and intracranial volumes. Nat Genet 44: 552–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern CE, Sherman SJ, Kirchhoff BA, Hasselmo ME (2001). Medial temporal and prefrontal contributions to working memory tasks with novel and familiar stimuli. Hippocampus 11: 337–346. [DOI] [PubMed] [Google Scholar]
- Suzuki H, Botteron KN, Luby JL, Belden AC, Gaffrey MS, Babb CM et al (2013). Structural-functional correlations between hippocampal volume and cortico-limbic emotional responses in depressed children. Cogn Affect Behav Neurosci 13: 135–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan L, Yu JT, Zhang W, Wu ZC, Zhang Q, Liu QY et al (2013. a). Association of GWAS-linked loci with late-onset Alzheimer's disease in a northern Han Chinese population. Alzheimers Dement 9: 546–553. [DOI] [PubMed] [Google Scholar]
- Tan MS, Yu JT, Jiang T, Zhu XC, Guan HS, Tan L (2014). Genetic variation in BIN1 gene and Alzheimer's disease risk in Han Chinese individuals. Neurobiol Aging 35: 1781 e1781–1781 e1788. [DOI] [PubMed] [Google Scholar]
- Tan MS, Yu JT, Tan L (2013. b). Bridging integrator 1 (BIN1): form, function, and Alzheimer's disease. Trends Mol Med 19: 594–603. [DOI] [PubMed] [Google Scholar]
- Van Horn JD, Berman KF, Weinberger DR (1996). Functional lateralization of the prefrontal cortex during traditional frontal lobe tasks. Biol Psychiatry 39: 389–399. [DOI] [PubMed] [Google Scholar]
- Wang L, Zang Y, He Y, Liang M, Zhang X, Tian L et al (2006). Changes in hippocampal connectivity in the early stages of Alzheimer's disease: evidence from resting state fMRI. Neuroimage 31: 496–504. [DOI] [PubMed] [Google Scholar]
- Whitwell JL, Shiung MM, Przybelski SA, Weigand SD, Knopman DS, Boeve BF et al (2008). MRI patterns of atrophy associated with progression to AD in amnestic mild cognitive impairment. Neurology 70: 512–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wishart HA, Saykin AJ, Rabin LA, Santulli RB, Flashman LA, Guerin SJ et al (2006). Increased brain activation during working memory in cognitively intact adults with the APOE epsilon4 allele. Am J Psychiatry 163: 1603–1610. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.