Abstract
BACKGROUND:
Embryonic aneuploidy may result in miscarriage, implantation failure, or birth defects. Thus, it is clinically necessary to avoid the selection of aneuploid embryos during in vitro fertilization treatment.
AIM:
The aim of this study was to identify the morphokinetic differences by analyzing the development of euploid and aneuploid embryos using a time-lapse technology. We also checked the accuracy of a previously described model for selection of euploid embryos based on morphokinetics in our study population.
MATERIALS AND METHODS:
It is a retrospective study of 29 cycles undergoing preimplantation genetic screening from October 2013 to April 2015 at our center. Of 253 embryos, 167 suitable for biopsy embryos were analyzed for their chromosomal status using array-comparative genome hybridization (CGH). The morphokinetic behavior of these embryos was further analyzed in embryoscope using time-lapse technology.
RESULTS:
Among the analyzed embryos, 41 had normal and 126 had abnormal chromosome content. No significant difference in morphokinetics was found between euploid and aneuploid embryos. The percentage of embryos with blastulation was similar in the euploid (65.85%, 27/41) and aneuploid (60.31%, 76/126) embryos (P = 0.76). Although hard to define, majority of the chromosomal defects might be due to meiotic errors. On applying embryo selection model from Basile et al., embryos falling within optimal ranges for time to division to 5 cells (t5), time period of the third cell cycle (CC3), and time from 2 cell division to 5 cell division (t5-t2) exhibited greater proportion of normal embryos than those falling outside the optimal ranges (28.6%, 25.9%, and 26.7% vs. 17.5%, 20.8%, and 14.3%).
CONCLUSION:
Keeping a track of time interval between two stages can help us recognize aneuploid embryos at an earlier stage and prevent their selection of transfer. However, it cannot be used as a substitute for array CGH to select euploid embryos for transfer.
KEY WORDS: Aneuploidy, embryos, euploid, in vitro fertilization, morphokinetics
INTRODUCTION
Embryonic aneuploidy is one of the major factors affecting embryo implantation. Aneuploidy may result in miscarriage or implantation failure[1] and is considered as the main cause of most of the birth defects.[2,3] Premature division and nondisjunction of chromatids during meiosis along with several contributory factors such as paternal, mitotic, or meiotic errors leads to aneuploidy of human embryos.[1,4] Chromosomes containing fragments/micronuclei may contribute to aneuploidy that may have negative consequences on the normal development process.[4] Hence, it is clinically necessary to avoid the selection of aneuploid embryos during the in vitro fertilization (IVF) procedure.[1]
Embryo selection for transfer is the most critical step during an IVF procedure.[5] Earlier conventional procedures were used for embryo grading and selection based on the static observations and critical assessment of morphologic parameters. However, these subjective observations had large flaws such as inter- and intra-observer variation and the dynamic process of the embryo development were also not defined largely.[5,6,7] Further, these observations require repeated removal of the embryos from incubator environment, which may result in undesired temperature and change of pH in the embryo culture.[5]
Kinetic behavior of a normal and an abnormal embryo is different. A qualitative morphology observation of an individual embryo along with monitoring of the dynamics in the development of embryo is referred to as morphokinetic study.[1] The introduction of time-lapse technology in the clinical IVF laboratory has enabled more detailed observations on the development of embryo.[5]
A relationship exists between time-lapse parameters and embryo viability.[8] The precise timing of specific events is an indicator of developmental potential of embryo.[5] The continuous monitoring of morphokinetic parameters such as pronuclear formation, syngamy, cleavage events, synchronicity of cell division, cell cycle intervals, and initiation of blastulation can yield valuable information that can aid in selecting the best embryos for transfer.[5] Morphokinetic markers combined with preimplantation genetic screening (PGS) may help in better embryo screening and selection.[9]
There are contradicting reports on morphokinetic behavior of embryos. Basile et al. had shown that the division times for embryo to 5 cells (t5), the time of third cell cycle (t5-t3) from 3 cell division to 5 cell division (CC3) and time from 2 cell division to 5 cell division (t5-t2) are significantly different in euploid versus aneuploid embryos.[10] Chawla et al. in their retrospective cohort study observed a significant difference in the mean duration of pronuclei fading (PNf), time to division to 2 cells (t2), t5, the time of second cell cycle (t3-t2) from 2 to 3 cells (CC2), and CC3 between normal and abnormal embryos. They also suggested that CC3 could be favourable factor for detecting significantly higher number of normal embryos (optimum time of 10 h more than CC3).[11] Basile et al.[9] reported a greater proportion of chromosomally normal embryos within their optimal ranges proposed for t5-t2 and CC3. Chavez et al. observed that all normal embryos displayed strict clustered cell cycle parameters up to the 4-cell stage while only 30% of aneuploid embryos exhibited values within normal time ranges. They reported a significant difference in three dynamic cell cycle parameters, viz., duration of the first cytokinesis from the first cleavage, time from two cells to the appearance of third cell (which in turn is our CC2), and time from three to four cells appearance between the normal and abnormal embryos (P < 0.05).[4] In contrast, Yang et al. reported no significant differences between aneuploid and euploid embryos in 10 morphokinetic parameters analyzed by combined time-lapse microscopy and array comparative genome hybridization (CGH).[12] Similarly, in a study by Campbell et al., no significant difference was observed between euploid and aneuploid embryos during early stages of development.[1] There is a paucity of such data available from India.
The purpose of the present study was to analyze the morphokinetic behavior and differences between euploid and aneuploid embryos by means of the time-lapse imaging. At the same time, we checked the accuracy of a previously proposed selection model of euploid embryos by Basile et al. based on time-lapse observations in the same database.[9]
MATERIALS AND METHODS
Study population
Patients with advance maternal age, recurrent implantation failure, recurrent miscarriage, and undergoing PGS were included in the study. The cases where <5 mature oocytes were retrieved were excluded from the study.
Study design and procedure
A total of 167 embryos were biopsied and analyzed for their chromosomal status using array-CGH. This is a retrospective analysis of the morphokinetic behavior of these 167 biopsied embryos in the Embryoscope™ (Unisense FertiliTech, Denmark) using the time-lapse technology. In addition, we applied the similar model as Basile et al. to check its accuracy for identifying chromosomally abnormal embryos.[9]
Oocyte retrieval
All patients underwent ovarian stimulation from day 2 of cycle on the flexible antagonist protocol. Oocyte retrieval was performed 35 h after human chorionic gonadotropin (hCG) administration under general anesthesia using transvaginal ultrasound guidance with single lumen aspiration needle (Cook, Australia). Oocyte-cumulus complexes were scanned and separated in G-MOPS media (Vitrolife G5 Series). Later, surplus cumulus was trimmed from the surrounding of oocytes with only 4–5 layers remaining and then was cultured in G-IVF plus media (Vitrolife G5 Series) that was covered with mineral oil (Cryobiosystems, France).
Embryo culture and biopsy
Following intracytoplasmic sperm injection (ICSI), the oocytes were kept in embryoscope in G1 media (Vitrolife G5 Series) till day 3 of development. On day 3, the embryos with at least six blastomeres and <30% fragmentation were selected for biopsy.
Embryos were kept in Ca2+/Mg2+ free media (G-PGD, Vitrolife) and then zona was perforated using laser (OCTAX). Single blastomere biopsy was performed using blastomere biopsy pipette (Humagen, USA) and the blastomere was loaded in polymerase chain reaction tubes, fixed at −20°C for at least an hour and couriered for comprehensive chromosome screening through array CGH. Further, the embryos were cultured in G2 media till day 5 of the development. Euploid embryos were transferred/frozen on day 5. Following culture, the embryos were either transferred or vitrified. Aneuploid embryos were not cultured further. Of 167 biopsied embryos, only 1 embryo was degenerated on day 5.
Array comparative genome hybridization
Single cell array CGH analysis was performed using Sureplex™ kit for whole genome amplification and Platform 24sure™ (BlueGnome Ltd., Cambridge, UK) for 24 chromosome aneuploidy screening. BlueFuse Multi V3.1 (BlueGnome Ltd., Cambridge, UK) software was used to analyze the data.[13]
Time-lapse analysis
Embryos were annotated in embryoscope from time of ICSI to PN appearance, PNf, two cell division (t2) to eight cell division (t8), and start of blastulation (tSB) till hatching of blastocyst (iHB).
Hierarchical model
Embryos were grouped using the hierarchical model [Figure 1] previously proposed by Basile et al.[9]
Figure 1.
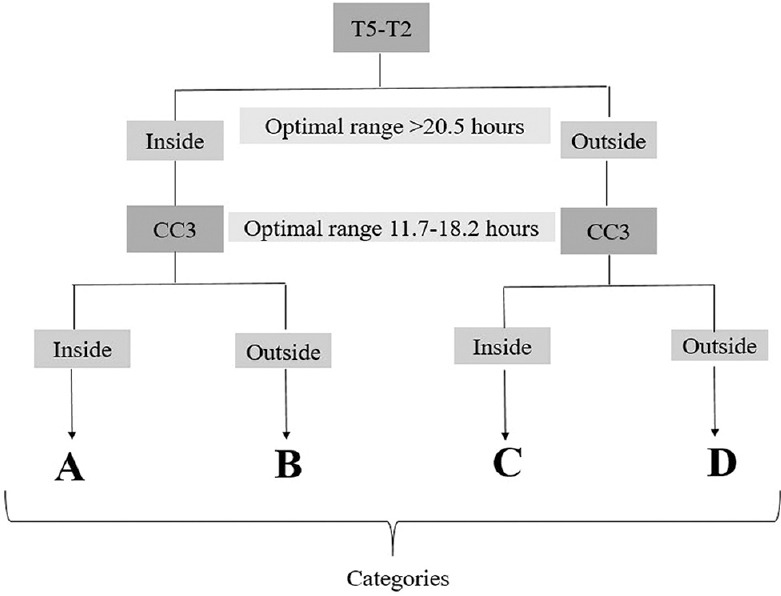
Hierarchical classification of embryos based on interval t5-t2, and the duration of the third cell cycle (CC3)
Statistical analysis
Descriptive statistics were used for analysis. Chi-square test was used to compare the two groups. Student's t-test was used to calculate P values between two groups of unequal variance. The optimal range for each time variable was defined as the combined range spanned by the two quartiles or by the one quartile with the highest proportion of normal embryo rates. A binary variable was defined by the value inside if the value of the time variable was inside the optimal range, and outside if the value of the time variable was outside the optimal range.
The odds ratio (OR) of the effect of binary variables generated for embryonic chromosomal normality was expressed in terms of the 95% confidence interval (CI) and significance. The effect of optimal ranges and binary variables on chromosomal normality was quantified by logistic regression analysis using the Cox and Snell R2 and the Nagelkerke R2.
To test the predictive value of all variables included with respect to chromosomal normality, receiver operating characteristic curves were used that provided area under the curve values between 0.5 and 1.
RESULTS
Subjects
During October 2013 to April 2015, 26 patients with 29 ICSI cycles (380 mature oocytes, 277 fertilized, 253 embryos formed, 167 biopsied) undergoing PGS at our center were included in the analysis. The cases where <5 mature oocytes (n = 6) were retrieved were excluded from the analysis. The patients with advance maternal age (≥35 years, n = 9), recurrent implantation failure (n = 10), and recurrent miscarriage (n = 7) undergoing PGS were included in the study. The average age of the patients was 32.94 ± 3.19 years (mean ± standard deviation [SD]) having an average of 3.00 ± 1.72 (mean ± SD) previously failed cycles. The average number of oocytes obtained was 15.55 ± 9.35 (mean ± SD). Of total 29 cycles for PGS, 7 cycles had all abnormal embryos resulting in cancellation of embryo transfer. Thus, no embryo transfer was done for aneuploid embryos. High number of aneuploid embryos (n = 126) was obtained. Of total 26 embryo transfers (including fresh and frozen), 11 cycles (42.31%) were β-hCG positive and later 5 cycles (19.23%) had miscarriage finally resulting into nine livebirths (three twins and three singletons).
Outcomes
Among the analyzed embryos, 41 (24.55%) had normal and 126 (75.45%) had abnormal chromosome content. The mean time for each cell division and for the intervals between consecutive divisions is shown in Table 1. No significant difference was observed between euploid and aneuploid embryos for the analyzed variables.
Table 1.
Morphokinetic parameters in euploid and aneuploid embryos
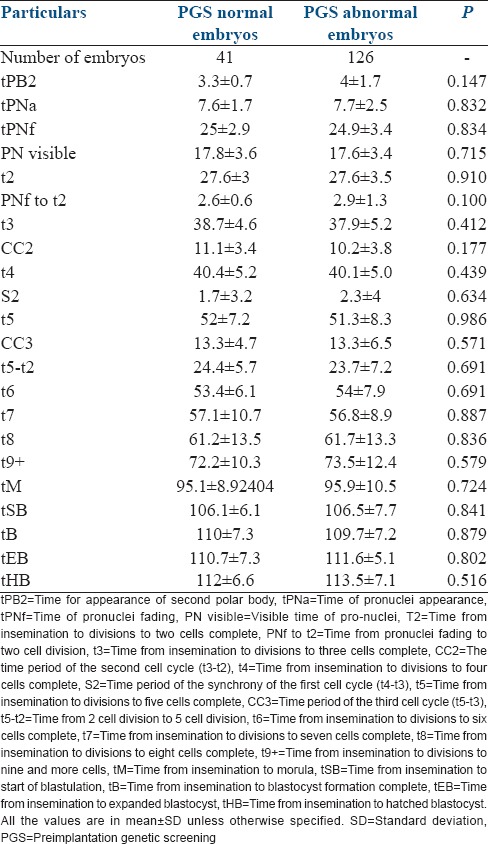
Similarly, there was no significant difference in (CC2), and (CC3) between euploid and aneuploid embryos. The percentage of embryos with blastulation in the euploid embryos was 65.85% (27/41) and 60.31% (76/126) in the aneuploid embryos (P = 0.76).
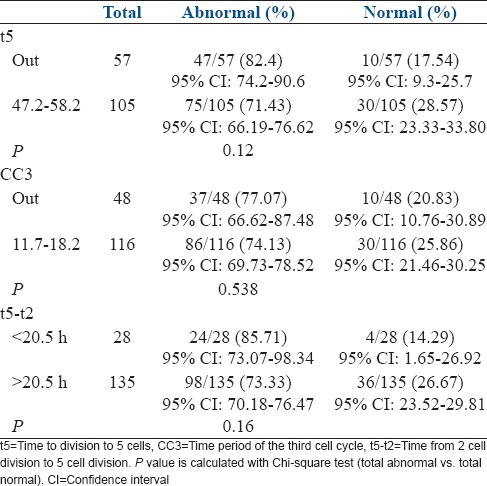
Based on the application of Basile's model, the percentage of normal and abnormal embryos, in and out of defined optimal ranges for t5, CC2, CC3, and t5-t2, is given in Table 2. Embryos falling within optimal ranges for t5, CC3, and t5-t2 exhibited a greater proportion of normal embryos than those falling outside the optimal ranges (28.6%, 25.9%, and 26.7% vs. 17.5%, 20.8%, and 14.3%, P values - 0.12, 0.538, and 0.16, respectively). A higher number of abnormal embryos fell outside the optimal range as compared with the normal embryos (t5-82.4% vs. 17.54%, CC3-77.07% vs. 20.83%, and t5-t2-85.71% vs. 14.29%).
Table 2.
Percentage of abnormal and normal embryos in and out of defined optimal ranges
The PGS model based on logistic regression identified t5-t2 (OR = 9.0) and CC3 (OR = 0.08, 95% CI, 0.007–0.895) as the most relevant variables related to normal chromosomal content. PGS model is described in Figure 2. High number of normal embryos (96.7%) were under category A, followed by category B - 7 (70%), category C - 3 (30%), and category D - 1 (3.3%).
Figure 2.
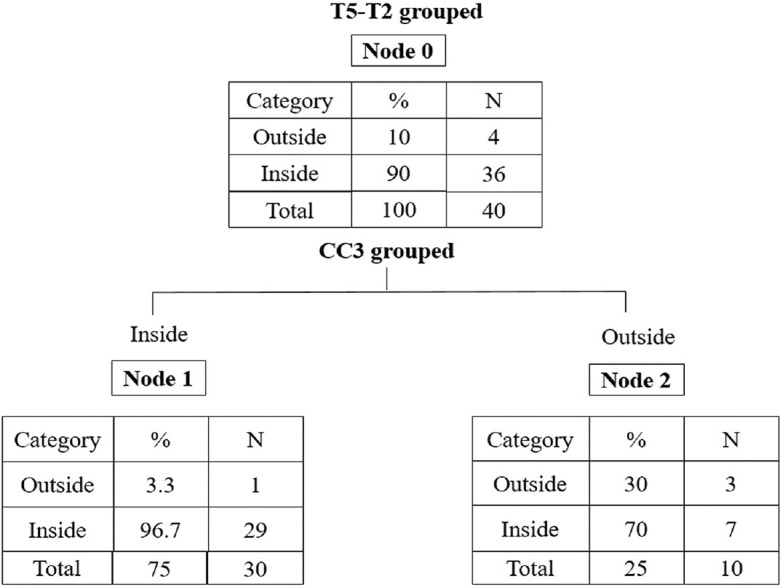
Preimplantation genetic screening model
DISCUSSION
Time-lapse imaging has provided new insights into the early embryo development and its selection. It is a novel technology that permits the recording and retrospective analysis of morphokinetic variables. It allows the observation of zygotes and embryos at each stage starting from fertilization and syngamy to cleavage, compaction, blastulation, and hatching.[14] The present study analyzed the morphokinetic behavior of chromosomally normal and abnormal embryos to determine whether it can be used to noninvasively select chromosomally normal embryos. A high number of abnormal embryos were obtained (126/167, 75.45%). It is hard to calculate the real mosaicism rate in preimplantation embryos but, currently, it has been estimated to range from 10% to 15%, therefore most of the chromosome errors found in this work might be meiotic errors. The array CGH technique used in the present work to perform PGS allows a comprehensive chromosome screening by detecting not only full chromosome gains and losses but also partial or segmental chromosome gains and losses. However, array CGH technique does not allow to distinguish between meiotic or mitotic chromosomal errors. Nevertheless, the current knowledge about chromosomal defects in preimplantation embryos explains that meiotic errors imply that all the cells of the embryo carry the same chromosomal error while mitotic errors imply that some cells will have different chromosomal content (mosaic embryos).[15,16,17,18]
Most pre- and post-implantation losses are associated predominantly with aneuploidies in the embryos.[19,20,21] Aneuploid embryo formation is the main reason for an unsuccessful IVF.[5] Depending upon the age of female, more than half of the embryos are likely to be aneuploid.[1] Various studies have reported a high rate of aneuploidy in in vitro-generated embryos.[4,20] Fragouli et al. reported aneuploidy rate of >60% in in vitro-produced embryos that resulted in implantation failures and spontaneous abortions.[20] Similarly, Chavez et al. reported aneuploidy in 75% (n = 75) of the embryos analyzed. They determined that the high frequency of aneuploidy was not associated with a subset of chromosomes, but all 23 pairs of chromosomes were affected.[4] In our study, majority of the embryos (126/167) were aneuploid as analyzed by array CGH.
Hence, it is crucial to find a safe method of selecting euploid blastocysts with high in vivo developmental competence to improve the overall efficiency.[22] In IVF technology, a morphokinetic era has started as chromosomal abnormality can be a deciding factor for implantation of embryo.[2] Morphokinetic analysis of in vitro embryo development has become most attractive advancement in human embryology.[22] Time-lapse imaging is a new and potentially powerful tool used to assess the embryo quality.[10] Continuous observation of morphokinetic parameters with the help of time-lapse system has been proposed as a noninvasive and reliable approach to predict the probability of euploidy.[23] Various studies have reported that the time-lapse monitoring in embryoscope does not compromise with the development of embryo, embryo quality, blastocyst development or viability and allows evaluation of morphokinetic data with high consistency[24,25,26,27] and with spatial/temporal analysis of embryo development.[24] In the present study, a total of 167 embryos kept in embryoscope were analyzed through combination of time-lapse technology and PGS.
Aneuploid and euploid embryos have different morphokinetic behavior as demonstrated through time-lapse technique under standardized IVF culture,[1] making an early prediction of aneuploid embryos possible. To classify the risk of aneuploidy effectively, (tSB) and the time from insemination to the formation of a “full blastocyst” (tB) are used in a predictive algorithm,[19] but the retrospective data from time-lapse patients were compared with the non-PGS patients. In our study, tSB was found to be earlier in normal embryos than abnormal, but the difference was not statistically significant. Similarly, in a study by Campbell et al., tSB was observed earlier in euploid than the aneuploid embryos. They demonstrated that aneuploid embryos show delayed periblastulation phase as compared with the euploid embryos.[1] Other parameters such as PNf, t2, t5, CC2, and CC3 have been studied to predict morphokinetic behavior of embryos in previous studies. Basile et al. found a higher proportion of normal embryos within the optimal ranges defined for t5 (47.2–58.2 h), CC3 (11.7–18.2 h), and t5-t2 (>20.5 h).[10] We also observed a higher proportion of normal embryos within the optimal ranges (t5: 17.54%, CC3: 20.83%, and t5-t2: 14.29%), but our results could not achieve a statistical significance. In our study, we did not observe any significant difference in any of the parameters between the normal and abnormal embryos. The studies conducted so far have presented an equivocal observation in the difference between the normal and abnormal embryos.[1,4,9,12]
Our study has certain limitations. Embryo biopsy was performed on day 3 which may have affected the results due to the probability of mosaicism in blastomeres. Chavez et al. had observed complex mosaicism in the majority of the embryos.[4] Fragouli et al. analyzed 52 blastomeres through array CGH and observed 42.3% euploid, 30% aneuploid, and 32.4% mosaic blastocysts. Most mosaic blastocysts contained no normal cells. However, it is estimated that only 6% of potentially viable embryos are at a risk of misdiagnosis due to mosaicism.[20] At the same time, we have included variables t5-t2 and CC3 in our algorithm and according to Basile et al., this algorithm could be applied regardless of the day of biopsy.[9] Also, our result of abnormality detection by day 3 of biopsy was in line with the result of a study by Garcia-Velasco et al. where abnormality was diagnosed with biopsy on day 5.[28]
CONCLUSION
We did not observe any significant difference in the parameters between the normal and abnormal embryos. However, this could be due to a smaller sample size and other limitations as majority of our patients had a history of repeated implantation failures or advanced maternal age, that could result in variable timings of embryo development. Hence, larger data are required to test whether the difference in time of cleavage is effective in predicting aneuploidy.
However, keeping a track of time interval between two stages can help us in recognizing aneuploid embryos at an earlier stage and prevent their selection of transfer. Nevertheless, it cannot be used as a substitute for array CGH to select euploid embryos for transfer, since so far only models based on risk or probabilities have been proposed.
It is required to carry out more studies with a bigger sample size so that a differential morphokinetic pattern can be established. Maybe in that moment, the time-lapse technology can replace the current invasive techniques used for genetic analysis.
Financial support and sponsorship
Nil.
Conflicts of interest
There are no conflicts of interest.
Acknowledgment
The authors would like to acknowledge the Knowledge Isotopes Pvt. Ltd. (www.knowledgeisotopes.com) for writing support.
REFERENCES
- 1.Campbell A, Fishel S, Bowman N, Duffy S, Sedler M, Hickman CF. Modelling a risk classification of aneuploidy in human embryos using non-invasive morphokinetics. Reprod Biomed Online. 2013;26:477–85. doi: 10.1016/j.rbmo.2013.02.006. [DOI] [PubMed] [Google Scholar]
- 2.Hassold T, Hunt P. To err (meiotically) is human: The genesis of human aneuploidy. Nat Rev Genet. 2001;2:280–91. doi: 10.1038/35066065. [DOI] [PubMed] [Google Scholar]
- 3.Wang WH, Sun QY. Meiotic spindle, spindle checkpoint and embryonic aneuploidy. Front Biosci. 2006;11:620–36. doi: 10.2741/1822. [DOI] [PubMed] [Google Scholar]
- 4.Chavez SL, Loewke KE, Han J, Moussavi F, Colls P, Munne S, et al. Dynamic blastomere behaviour reflects human embryo ploidy by the four-cell stage. Nat Commun. 2012;3:1251. doi: 10.1038/ncomms2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Desai N, Ploskonka S, Goodman LR, Austin C, Goldberg J, Falcone T. Analysis of embryo morphokinetics, multinucleation and cleavage anomalies using continuous time-lapse monitoring in blastocyst transfer cycles. Reprod Biol Endocrinol. 2014;12:54. doi: 10.1186/1477-7827-12-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baxter Bendus AE, Mayer JF, Shipley SK, Catherino WH. Interobserver and intraobserver variation in day 3 embryo grading. Fertil Steril. 2006;86:1608–15. doi: 10.1016/j.fertnstert.2006.05.037. [DOI] [PubMed] [Google Scholar]
- 7.Scott L. The biological basis of non-invasive strategies for selection of human oocytes and embryos. Hum Reprod Update. 2003;9:237–49. doi: 10.1093/humupd/dmg023. [DOI] [PubMed] [Google Scholar]
- 8.Kirkegaard K, Ahlström A, Ingerslev HJ, Hardarson T. Choosing the best embryo by time lapse versus standard morphology. Fertil Steril. 2015;103:323–32. doi: 10.1016/j.fertnstert.2014.11.003. [DOI] [PubMed] [Google Scholar]
- 9.Basile N, Nogales Mdel C, Bronet F, Florensa M, Riqueiros M, Rodrigo L, et al. Increasing the probability of selecting chromosomally normal embryos by time-lapse morphokinetics analysis. Fertil Steril. 2014;101:699–704. doi: 10.1016/j.fertnstert.2013.12.005. [DOI] [PubMed] [Google Scholar]
- 10.Basile N, Morbeck D, García-Velasco J, Bronet F, Meseguer M. Type of culture media does not affect embryo kinetics: A time-lapse analysis of sibling oocytes. Hum Reprod. 2013;28:634–41. doi: 10.1093/humrep/des462. [DOI] [PubMed] [Google Scholar]
- 11.Chawla M, Fakih M, Shunnar A, Bayram A, Hellani A, Perumal V, et al. Morphokinetic analysis of cleavage stage embryos and its relationship to aneuploidy in a retrospective time-lapse imaging study. J Assist Reprod Genet. 2015;32:69–75. doi: 10.1007/s10815-014-0372-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang Z, Zhang J, Salem SA, Liu X, Kuang Y, Salem RD, et al. Selection of competent blastocysts for transfer by combining time-lapse monitoring and array CGH testing for patients undergoing preimplantation genetic screening: A prospective study with sibling oocytes. BMC Med Genomics. 2014;7:38. doi: 10.1186/1755-8794-7-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mir P, Rodrigo L, Mercader A, Buendía P, Mateu E, Milán-Sánchez M, et al. False positive rate of an arrayCGH platform for single-cell preimplantation genetic screening and subsequent clinical application on day-3. J Assist Reprod Genet. 2013;30:143–9. doi: 10.1007/s10815-012-9918-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Meseguer M, Herrero J, Tejera A, Hilligsøe KM, Ramsing NB, Remohí J. The use of morphokinetics as a predictor of embryo implantation. Hum Reprod. 2011;26:2658–71. doi: 10.1093/humrep/der256. [DOI] [PubMed] [Google Scholar]
- 15.Mir P, Mateu E, Mercader A, Herrer R, Rodrigo L, Vera M, et al. Confirmation rates of array-CGH in day-3 embryo and blastocyst biopsies for preimplantation genetic screening. J Assist Reprod Genet. 2016;33:59–66. doi: 10.1007/s10815-015-0605-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gardner DK, Meseguer M, Rubio C, Treff NR. Diagnosis of human preimplantation embryo viability. Hum Reprod Update. 2015;21:727–47. doi: 10.1093/humupd/dmu064. [DOI] [PubMed] [Google Scholar]
- 17.Mamas T, Gordon A, Brown A, Harper J, Sengupta S. Detection of aneuploidy by array comparative genomic hybridization using cell lines to mimic a mosaic trophectoderm biopsy. Fertil Steril. 2012;97:943–7. doi: 10.1016/j.fertnstert.2011.12.048. [DOI] [PubMed] [Google Scholar]
- 18.Novik V, Moulton EB, Sisson ME, Shrestha SL, Tran KD, Stern HJ, et al. The accuracy of chromosomal microarray testing for identification of embryonic mosaicism in human blastocysts. Mol Cytogenet. 2014;7:18. doi: 10.1186/1755-8166-7-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Campbell A, Fishel S, Bowman N, Duffy S, Sedler M, Thornton S. Retrospective analysis of outcomes after IVF using an aneuploidy risk model derived from time-lapse imaging without PGS. Reprod Biomed Online. 2013;27:140–6. doi: 10.1016/j.rbmo.2013.04.013. [DOI] [PubMed] [Google Scholar]
- 20.Fragouli E, Alfarawati S, Daphnis DD, Goodall NN, Mania A, Griffiths T, et al. Cytogenetic analysis of human blastocysts with the use of FISH, CGH and aCGH: Scientific data and technical evaluation. Hum Reprod. 2011;26:480–90. doi: 10.1093/humrep/deq344. [DOI] [PubMed] [Google Scholar]
- 21.Fragouli E, Wells D. Aneuploidy screening for embryo selection. Semin Reprod Med. 2012;30:289–301. doi: 10.1055/s-0032-1313908. [DOI] [PubMed] [Google Scholar]
- 22.Rienzi L, Capalbo A, Stoppa M, Romano S, Maggiulli R, Albricci L, et al. No evidence of association between blastocyst aneuploidy and morphokinetic assessment in a selected population of poor-prognosis patients: A longitudinal cohort study. Reprod Biomed Online. 2015;30:57–66. doi: 10.1016/j.rbmo.2014.09.012. [DOI] [PubMed] [Google Scholar]
- 23.Montag M, Toth B, Strowitzki T. New approaches to embryo selection. Reprod Biomed Online. 2013;27:539–46. doi: 10.1016/j.rbmo.2013.05.013. [DOI] [PubMed] [Google Scholar]
- 24.Cruz M, Gadea B, Garrido N, Pedersen KS, Martínez M, Pérez-Cano I, et al. Embryo quality, blastocyst and ongoing pregnancy rates in oocyte donation patients whose embryos were monitored by time-lapse imaging. J Assist Reprod Genet. 2011;28:569–73. doi: 10.1007/s10815-011-9549-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kirkegaard K, Hindkjaer JJ, Grøndahl ML, Kesmodel US, Ingerslev HJ. A randomized clinical trial comparing embryo culture in a conventional incubator with a time-lapse incubator. J Assist Reprod Genet. 2012;29:565–72. doi: 10.1007/s10815-012-9750-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nakahara T, Iwase A, Goto M, Harata T, Suzuki M, Ienaga M, et al. Evaluation of the safety of time-lapse observations for human embryos. J Assist Reprod Genet. 2010;27:93–6. doi: 10.1007/s10815-010-9385-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sundvall L, Ingerslev HJ, Breth Knudsen U, Kirkegaard K. Inter- and intra-observer variability of time-lapse annotations. Hum Reprod. 2013;28:3215–21. doi: 10.1093/humrep/det366. [DOI] [PubMed] [Google Scholar]
- 28.Garcia-Velasco JA, Bermejo A, Ruiz F, Martinez-Salazar J, Requena A, Pellicer A. Cycle scheduling with oral contraceptive pills in the GnRH antagonist protocol vs the long protocol: A randomized, controlled trial. Fertil Steril. 2011;96:590–3. doi: 10.1016/j.fertnstert.2011.06.022. [DOI] [PubMed] [Google Scholar]


