Abstract
Spa-typing and microarray techniques were used to study epidemiological changes in methicillin-resistant Staphylococcus aureus (MRSA) in South-East Austria. The population structure of 327 MRSA isolated between 2002 and 2012 was investigated. MRSA was assigned to 58 different spa types and 14 different MLST CC (multilocus sequence type clonal complexes); in particular, between 2007 and 2012, an increasing diversity in MRSA clones could be observed. The most abundant clonal complex was CC5. On the respective SCCmec cassettes, the CC5 isolates differed clearly within this decade and CC5/SCCmecI, the South German MRSA, predominant in 2002, was replaced by CC5/SCCmecII, the Rhine-Hesse MRSA in 2012. Whereas in many European countries MLST CC22-MRSA (EMRSA 15, the Barnim epidemic MRSA) is predominant, this clone occurred in Austria nearly 10 years later than in neighbouring countries. CC45, the Berlin EMRSA, epidemic in Germany, was only sporadically found in South-East Austria. The Irish ST8-MRSA-II represented by spa-type t190 was frequently found in 2002 and 2007, but disappeared in 2012. Our results demonstrate clonal replacement of MRSA clones within the last years in Austria. Ongoing surveillance is warranted for detection of changes within the MRSA population.
Keywords: MRSA, Austria, mecC, PVL, microarray, epidemiology
Changes in the epidemiology of MRSA clones in Austria.
INTRODUCTION
Over the last six decades, methicillin-resistant Staphylococcus aureus (MRSA) spread over the whole world and has become a global public health threat. Primarily MRSA was restricted to hospitals (hospital-acquired MRSA), where few, multiresistant strains dominated (Grundmann et al. 2010; Kinnevey et al. 2014; Stryjewski and Corey 2014). For the last three decades, MRSA could also be found outside this setting (community-acquired MRSA, CA-MRSA) (Kock et al. 2014). Most of these CA-MRSA strains carry the Panton-Valentine leukocidin (PVL), a virulence factor associated with soft tissue infection or skin infections (Chambers and Deleo 2009; Albrecht et al. 2011; Otto 2013; Kock et al. 2014). Another development was the occurrence of so-called livestock-associated MRSA (LA-MRSA) linked with mainly pig farming and is therefore found besides the animals, in people with contact to these. Spread from human to human is rarely seen in LA-MRSA, and LA-MRSA seems to have a reduced repertoire of virulence factors. In Austria, the first LA-MRSA was detected in 2004, with increasing numbers up to now (Huijsdens et al. 2006; Witte et al. 2007; Zarfel et al. 2013). While these changes within the MRSA population were observed, during the last 10 years MRSA predominantly belonging to only a few multilocus sequence typing (MLST) clonal complexes (CC) (CC5, CC22, CC8, CC1 or CC398) emerged worldwide, but appeared at different times, replacing other epidemic MRSA clones (Chambers and Deleo 2009; Wyllie, Paul and Crook 2011). In a recently described example from Germany, CC22-MRSA (Barnim Epidemic strain, UK-EMRSA15) was first detected in 2001 and increased up to 58.6% of the investigated MRSA isolates in 2010, in contrast to CC5/ST228-MRSA I decreased from nearly 50% in 2000 to 2.3% in 2010 (Albrecht et al. 2011). Further examples of a replacement of MRSA clones were reported from many countries all over the world, such as Ireland, UK, Portugal or the USA (Enright et al. 2002; Amorim et al. 2007; Chambers and Deleo 2009; Grundmann et al. 2010).
There are a few publications on MRSA epidemiology from Austria, but they mainly focused on CA-MRSA or LA-MRSA (Ruppitsch et al. 2006; Krziwanek et al. 2007, 2008; Grisold et al. 2009).
This study is the first study for Austria, investigating the genetic background of all detected MRSA primary isolates from 2002 up to 2012 in the South-East Austria to determine epidemiological changes. A total of 327 MRSA isolates detected in 2002, 2007 and 2012 in the South-East Austria was spa typed, analysed with microarray techniques and assigned to epidemic strains.
MATERIAL AND METHODS
Bacterial isolates
The study was performed at the Institute of Hygiene, Microbiology and Environmental Medicine, Medical University Graz, Austria. Clinical samples were obtained from the University Hospital of Graz (approximately 1200 beds), from eight peripheral hospitals and from local practitioners in the district Styria, in the South-East Austria. MRSA isolates of every fifth year were included, resulting in 2629 Staphylococcus aureus single-patient isolates in the year 2002, 2823 in the year 2007 and 2772 in the year 2012. From those, MRSA isolates were found in 96 patients (3.65%) in 2002, 72 (2.55%) in 2007 and 159 (5.74%) in 2012. Over study period, only one MRSA isolate per patient was included. MRSA isolates were from hospitalised or from outpatients, distribution was as follows: hospitalised/outpatient was 45 (46.9%)/51 (53.3%) in 2002, 37 (51.4%)/35 (48.6%) in 2007 and 71 (44.6%)/88 (65.4%) in 2012. Distribution to gender was as follows: male/female was 62 (64.6%)/ 34 (35.4%) in 2002, 41 (56.9%)/31 (43.1%) in 2007 and 89 (56.0%)/70 (44.0%) in 2012. Bacterial identification and antibiotic susceptibility testing was performed by using the semi-automated VITEK II instrument (bioMérieux, Marcy l'Etoile, France). Since 2002 all MRSA primary isolates were routinely stored at –70°C. For this study, MRSA were retested, and resistance to cefoxitin was confirmed by Etest (AB Biodisk, Solna, Sweden).
Spa typing
Spa typing, DNA purification and PCR were performed as described previously (Grisold et al. 2009). The spa types and appropriate BURP clusters were assigned by using Ridom StaphType software (http://www.ridom.de/staphtype).
DNA microarray
For genetic characterization, diagnostic DNA microarray (Identibac, UK; StaphyType, Alere Technologies GmbH, Germany) was used, analysing the MRSA isolates for the presence of over 300 different genes. Protocols have been previously described in detail and are also provided by the kit's manufacturer (Monecke et al. 2007).
Statistical analyses
The statistical analyses were conducted using R® Version 3.2.1, a free software environment for statistical computing (www.r-project.org). Group-specific proportions were tested on their equality by a two-sided binomial test.
RESULTS
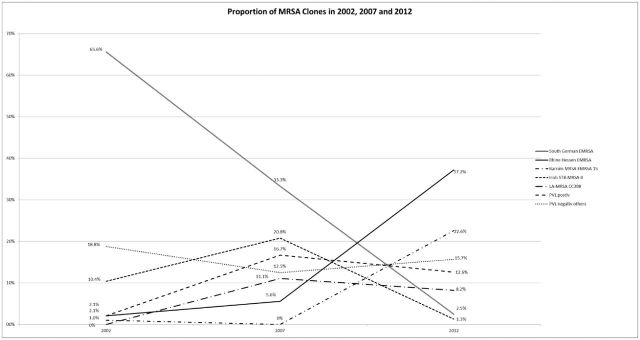
A total of 327 MRSA isolates from Austria detected between 2002 and 2012 were investigated, including 96 MRSA isolates from 2002, 72 from 2007 and 159 from 2012. A significant reduction in MRSA cases was observed between 2002 and 2007 (P = 0.023). In contrast, a significant increase in the prevalence of MRSA was observed between 2007 and 2010 (P < 0.01). MRSA isolates were spa typed and analysed with microarray chip. All together, the analysed MRSA isolates belonged to 58 different spa types and 14 different MLST clonal complexes (CCs). Concerning spa types and MLST CCs in 2002, 14 different spa types and six MLST CCs were detected. Eight of these spa types were found in 2002 only. In 2007, 20 spa types and eight MLST CCs were detected, seven of these spa types were found in 2007 only. In 2012, 40 spa types and 13 MLST CCs were detected. Notably 28 of these spa types and five MLST CCs could be detected in 2012 only. The proportion of the different MRSA clones and the temporal changes are shown in Fig. 1.
Figure 1.
Temporal changes in the MRSA population; PVL-negative MRSA (others) include all MRSA isolates that are not represented in one of the other groups.
MLST CC5-MRSA
In all three investigated years (2002, 2007 and 2012), MLST CC5 was the dominant clonal complex with 160 of 327 investigated MRSA isolates (48.92%), representing 65/96 isolates (67.71%) in 2002, 28/72 (38.89%) in 2007 and 67/159 (42.14%) in 2012. On the respective SCCmec cassettes, the CC5 isolates differed clearly within the three years. In 2002, the CC5/SCCmecI (South German EMRSA) was predominant with 63 isolates, represented by the spa types t001, t041, t12555 and t12898 and in 2007 with 24 isolates, represented by the spa types t001, t002, t041 and t1301. In 2012, CC5/SCCmecI was found in four isolates (t001, t010, t041). The decline of the South German EMRSA was significant in both observed time periods, with P < 0.01 for each period. The CC5/SCCmecII (Rhine-Hesse EMRSA) was found in two isolates (t002) in 2002 and four isolates in 2007 (t002, t003, t014). In 2012, CC5/SCCmecII was found as the dominant clone, with 58/159 (37.18%) isolates (t002, t003, t014, t626, t10303), representing a significant increase between 2007 and 2012 (P < 0.01). In 2012, also three CC5-MRSA with SCCmecIV were detected, two of them t002, which were also positive for the PVL toxin (the only PVL-positive CC5 isolates) and one t579 without PVL. In 2012, also two CC5-MRSA isolates harbouring SCCmecV were detected, both of which were assigned to spa-type t688.
MLST CC22-MRSA
The MLST CC22/SCCmecIV (EMRSA 15, the Barnim epidemic MRSA) was detected, with one exception, in 2012. In total, there were 36 isolates (22.64%) of CC22/SCCmecIV (t022, t032, t223, t515, t845, t11769) occurring in 2012 only, which means a significant increase (P < 0.01). In 2002, a single isolate could be assigned to CC22/SCCmecIV (t005). One CC22 isolate (t032) in 2012 harboured a SCCmecIV and additionally a SCCmecV cassette.
MLST CC8-MRSA
The MLST CC8 was the second most frequent clonal complex with 26/96 isolates (27.08%) in 2002 and 20/72 isolates (27.78%) in 2007. In 2012, 14/159 isolates (8.80%) were assigned to MLST CC8. SCCmec cassettes I-V could be found in isolates from this clonal complex.
The most frequent CC8 clone was CC8/SCCmecII with 10 isolates (Irish ST8-MRSA-II clone) in 2002 and 15 isolates in 2007, represented by spa-type t190 (no significant increase). In 2012, no isolate with t190 could be found. CC8/SCCmecII was represented by two t334 isolates. CC8/SCCmecIV (t008, t024, t036, t064, t622, t10437) was detected in a constant number throughout the investigated three years (ten isolates in 2002, four isolates in 2007 and nine isolates in 2012). Within the CC8/SCCmecIV isolates, the PVL toxin (US300 clone) was found in five isolates in 2012 (all t008), one isolate in 2007 (t622), but no PVL-positive CC8/SCCmecIV isolate was found in 2002. CC8 harbouring SCCmecI was represented by only three isolates in 2002 (t008, t051) and one in 2007 (t2023); CC8 with SCCmecIII was found in three isolates (t030) in 2002 and three isolates (t037) in 2012. The t037 isolates harboured additionally to the SCCmecIII and a SCCmecV cassette.
MLST CC1-MRSA
In total, MLST CC1 could be assigned to 11 MRSA isolates. Similar to the CC22-MRSA, MLST CC1 was detected, with one single exception, in 2012. In detail, there were 10 CC1 isolates with SCCmecIV (t127, t386, t693, t1784), including the single CC1/SCCmecIV isolate (t127) from 2007. One CC1 isolate in 2012 (t127) harboured a SCCmecV cassette. This isolate and one CC1/SCCmecIV (t1784) isolate were PVL toxin positive.
MLST CC398-MRSA
There was no CC398- MRSA in 2002. In 2007 and 2012, CC398 (LA-MRSA) was represented by 8/72 (11.11%) and 13/159 (8.17%) isolates, respectively. A total of 20 isolates (t011, t034, t1250, t1451) harboured SCCmecV; one isolate (t034) revealed SCCmecIV, and all isolates were PVL negative. The proportion of CC398-MRSA remained stable between 2007 and 2012 (P = 0.63).
PVL-positive isolates
Within the study period, the number of the PVL-positive isolates increased, with 2/96 (2.08%) PVL-positive isolates in 2002 and 12/72 (16.67%) in 2007 (significant increase P < 0.01). No significant change in the percentage of PVL-positive isolates was found between 2007 and 2012 with 20/159 (12.58%) isolates (P = 0.53) in 2012. The dominant clones were the US300 clone CC8/SCCmecIV, the European CA-MRSA clone CC80/SCCmecIV and the Pacific clone CC30/SCCmecIV. The European CA-MRSA clone CC80/SCCmecIV (t044, t692, t1339, t4152) was detected in all three years with one isolate in 2002 and six isolates each in 2007 and 2012. The Pacific clone could be assigned to eight isolates (all t019), four isolates each in 2007 and 2012. There were two PVL-positive clonal complexes, MLST CC152 and CC88 which occurred only sporadically. The MLST CC152 was assigned to three isolates (one in 2002 and two in 2007), that was the only MLST type, which in this study did not occur in 2012.
Sporadic PVL-negative isolates
Six MLST CCs were represented by less than 10 isolates within the study period. CC45 was assigned to six isolates; five CC45 isolates with SCCmecIV; one (t004) found in 2002, two (t015) in 2007, two (t026, t371) in 2012 and one CC45 isolate (t331) with SCCmecV (2012). Two CC30/SCCmecIV (t190, t253) isolates without PVL were found in 2007.
All other sporadic PVL-negative MLST CCs were represented by one or two isolates and were all detected in 2012: two CC361/SCCmecV (t315), one CC59/SCCmecIV (t216), one CC6/SCCmecV (t701) and one isolate CC130/SCCmecXI (t4335). This isolate was the only one with SCCmecXI, so therefore it did not harbour the mecA gene but the mecC gene.
DISCUSSION
Infections caused by MRSA have globally reached epidemic proportions and are well established both in the healthcare setting and in the community (Diederen and Kluytmans 2006; Chambers and Deleo 2009; Stryjewski and Corey 2014). In some countries such as the United States, MRSA is among the most common causes of nosocomial infections (Uhlemann et al. 2014). The prevalence rate of MRSA in bloodstream infections (BSIs) in Europe is monitored by the European Antimicrobial Resistance Surveillance Network (EARS-Net). Annual rates of MRSA from BSIs reach from 0.7% for Norway to 64.5% for Romania. For Austria, MRSA in BSI was reported with 9.2% in 2013 (European Centre for Disease Prevention and Control 2010, 2014).
With the occurrence of MRSA strains in different settings, hospitals as well as in outpatients or livestock associated, an increasing number of circulating MRSA clones was observed, the clones obviously in competition with each other (Huijsdens et al. 2006; Chambers and Deleo 2009; Otto 2013).
At the beginning of this century, the South German type (CC5) was very common in different parts of Europe, Germany or Croatia; also for Austria, the South German MRSA was the dominant clone in 2002 (Budimir et al. 2010). But within the next five years, the South German type disappeared and was replaced mainly by the Rhine-Hesse type; this clone was already common in other parts of Europe such as Great Britain or Ireland (UK-EMRSA-3) (Hookev, Richardson and Cookson 1998; Aucken et al. 2002) For Austria, as found in this study, the South German type was still the predominant clone until 2007 but disappeared later, with only four MRSA isolates in 2012. It could be noticed that the South German type was replaced by the Rhine-Hesse type, which was the predominant clone in 2012 with 38.18% of all investigated MRSA isolates (Enright et al. 2002; Chambers and Deleo 2009; Albrecht et al. 2011).
A similar delay could be observed for EMRSA 15. In many European countries, such as Germany, Ireland or Great Britain, EMRSA 15 is the predominant clone since the middle of the last decade (Amorim et al. 2007; Marchese et al. 2009; Grundmann et al. 2010; Albrecht et al. 2011). In Austria, the EMRSA 15 was found in one isolate only until 2012, but was the second most common clone in 2012, with 22.64%.
Beside CC5 and CC22, the CC8-MRSA, including the Irish ST8-MRSA-II and the USA300 clone, was continuously found throughout the study period. The Irish ST8-MRSA-II, represented by spa-type t190, was frequently found in 2002 (10.42%) and 2007 (20.83%); this spa type could not be found in any isolate in 2012. The high occurrence of spa190 in Austria was confirmed by a study from Ruppitsch et al. (2006) and Schmid et al. (2013), investigating MRSA from other parts of Austria, whereas there is no report of spa190 from neighbouring countries such as Germany (Ruppitsch et al. 2006; Cookson and HARMONY participants 2008; Albrecht et al. 2011; Schmid et al. 2013). In contrast CC45, the Berlin EMRSA clone which is epidemic in Germany was only found sporadically in Austria throughout the study period.
Although some other PVL-negative MRSA clones occurred sporadically throughout the study period, e.g. the USA600 (CC45) clone. The highest diversity of PVL-negative MRSA clones was found in 2012, including one CC130 isolate harbouring SCCmecXI with the new mecC variant, up to date only reported once for Austria. Isolates with SCCmecXI are described since only a few years, found both in humans and animals, such as cows (Smith and Cook 2005; Albrecht et al. 2011; García-Álvarez et al.2011; Loncaric et al. 2013; Kerschner et al. 2014).
CA-MRSA showed high clonal diversity in this study with the majority of the strains belonging to a few predominant clones. The most epidemic clonal type among CA-MRSA in Europe is the European CA-MRSA clone (CC80/SCCmecIV), found in at least 11 European countries, as described by Rolo et al. For Austria, this clone was described first in 2001 and was the dominant CA-MRSA clone throughout the observed time period in this study (Krziwanek et al. 2007; Rolo et al. 2012).
The USA300 clone (CC8) is epidemic in the USA, where it became a predominant strain. Additionally to the USA, the USA300 spread all over the world, with reports from Asia, Australia, South America and throughout Europe, but this clone did not become the predominant CA-MRSA in these countries. For Austria, the first report was published in 2003 and USA300 is found up to now constantly, but in annually low numbers (Krziwanek et al. 2007; Chambers and Deleo 2009; Grisold et al. 2009). Since 2004, LA-MRSA/CC398 is also constantly found, mainly concentrated in two regions in Austria (Upper Austria and South-East Austria) with high density of pig farming (Krziwanek et al. 2009; Grisold et al. 2010). Eight CC1/ SCCmecIV with spa-type t127 and PVL toxin negative were detected. This LA-MRSA clone is known for harbouring numerous virulence factors and was first described in Austria in 2008, in that report associated with horses. In other European countries, the MRSA t127 clone was also isolated from pigs and cattle (Cuny et al. 2008; Franco et al. 2011; Alba et al. 2015).
This study investigated for the first time the molecular epidemiology of MRSA isolates in Austria within a 10-year period, but there are some limitations of this study. Although all primary MRSA isolates of a huge region in the South-East Austria were investigated, data from other Austrian regions could not be included. It was also not aim of this study to focus in detail on the clinical background or investigate the outcome of hospital hygiene regimes.
To summarise the diversity within the MRSA, population in South-East Austria increased within the observed period from 2002 to 2012. The results showed that the occurrence of some MRSA clones follows the European trends, but with a delay of five to ten years. CC22-MRSA (the Barnim clone), mostly represented in neighbouring countries such as Austria, did not appear before 2012, which means nearly 10 years later than in Germany. Local specifics like the occurrence of the spa-type t190 could be observed. Following the dynamics of MRSA, this study documents that new MRSA clones with new features could emerge at any time highlighting the importance of closely monitoring the epidemiology and genetic background of MRSA.
FUNDING
This study was supported by (Pfizer reference number ).
ETHICAL STATEMENT
Isolates were obtained as part of routine diagnostic testing and were analysed anonymously. All data were collected in accordance with the European Parliament and Council decision for the epidemiological surveillance and control of communicable diseases in the European community (The European Parliament and the Council of the European Union. 1998; European Commission. 1999). Ethical approval and informed consent were thus not required. Patients were not physically involved and privacy of patients was provided by coding the tested specimens.
REFERENCES
- Alba P, Feltrin F, Cordaro G, et al. Livestock-associated methicillin resistant and methicillin susceptible Staphylococcus aureus sequence type (CC)1 in European farmed animals: high genetic relatedness of isolates from Italian cattle herds and humans. PLoS One. 2015;10:e0137143. doi: 10.1371/journal.pone.0137143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albrecht N, Jatzwauk L, Slickers P, et al. Clonal replacement of epidemic methicillin-resistant Staphylococcus aureus strains in a German university hospital over a period of eleven years. PLoS One. 2011;6:e28189. doi: 10.1371/journal.pone.0028189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amorim ML, Faria NA, Oliveira DC, et al. Changes in the clonal nature and antibiotic resistance profiles of methicillin-resistant Staphylococcus aureus isolates associated with spread of the EMRSA-15 clone in a tertiary care Portuguese hospital. J Clin Microbiol. 2007;45:2881–8. doi: 10.1128/JCM.00603-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aucken HM, Ganner M, Murchan S, et al. A new UK strain of epidemic methicillin-resistant Staphylococcus aureus (EMRSA-17) resistant to multiple antibiotics. J Antimicrob Chemoth. 2002;50:171–5. doi: 10.1093/jac/dkf117. [DOI] [PubMed] [Google Scholar]
- Budimir A, Deurenberg RH, Bosnjak Z, et al. A variant of the Southern German clone of methicillin-resistant Staphylococcus aureus is predominant in Croatia. Clin Microbiol Infect. 2010;16:1077–83. doi: 10.1111/j.1469-0691.2009.03042.x. [DOI] [PubMed] [Google Scholar]
- Chambers HF, Deleo FR. Waves of resistance: Staphylococcus aureus in the antibiotic era. Nat Rev Microbiol. 2009;7:629–41. doi: 10.1038/nrmicro2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cookson B, HARMONY participants. HARMONY–the International Union of Microbiology Societies' European Staphylococcal Typing Network. Euro Surveill. 2008;13:18860. [PubMed] [Google Scholar]
- Cuny C, Strommenger B, Witte W, et al. Clusters of infections in horses with MRSA ST1, ST254, and ST398 in a veterinary hospital. Microb Drug Resist. 2008;14:307–10. doi: 10.1089/mdr.2008.0845. [DOI] [PubMed] [Google Scholar]
- Diederen BM, Kluytmans JA. The emergence of infections with community-associated methicillin resistant Staphylococcus aureus. J Infect. 2006;52:157–68. doi: 10.1016/j.jinf.2005.09.001. [DOI] [PubMed] [Google Scholar]
- Enright MC, Robinson DA, Randle G, et al. The evolutionary history of methicillin-resistant Staphylococcus aureus (MRSA) P Natl Acad Sci USA. 2002;99:7687–92. doi: 10.1073/pnas.122108599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- European Centre for Disease Prevention and Control. Annual Report of the European Antimicrobial Resistance Surveillance Network (EARS-Net) 2010. Antimicrobial resistance surveillance in Europe 2009. http://ecdc.europa.eu/en/publications/Publications/1011_SUR_annual_EARS_Net_2009.pdf. [Google Scholar]
- European Centre for Disease Prevention and Control. Annual Report of the European Antimicrobial Resistance Surveillance Network (EARS-Net) 2014. Antimicrobial resistance surveillance in Europe 2013. http://ecdc.europa.eu/en/publications/Publications/antimicrobial-resistance-surveillance-europe-2013.pdf. [Google Scholar]
- European Commission. Official Journal of the European Union. 1999. Decision of 22 December 1999 on the communicable diseases to be progressively covered by the Community network under Decision No 2119/98/EC of the European Parliament and of the Council (notified under document number C(1999) 4015) (2000/96/EC) http://eur-lex.europa.eu/LexUriServ/LexUriServ.do?uri=CONSLEG:2000D0096:20120905:EN:PDF (28 May 2016, date last accessed) [Google Scholar]
- Franco A, Hasman H, Iurescia M, et al. Molecular characterization of spa type t127, sequence type 1 methicillin-resistant Staphylococcus aureus from pigs. J Antimicrob Chemoth. 2011;66:1231–5. doi: 10.1093/jac/dkr115. [DOI] [PubMed] [Google Scholar]
- García-Álvarez L, Holden MT, Lindsay H, et al. Meticillin-resistant Staphylococcus aureus with a novel mecA homologue in human and bovine populations in the UK and Denmark: a descriptive study. Lancet Infect Dis. 2011;11:595–603. doi: 10.1016/S1473-3099(11)70126-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grisold AJ, Zarfel G, Hoenigl M, et al. Occurrence and genotyping using automated repetitive-sequence-based PCR of methicillin-resistant Staphylococcus aureus ST398 in Southeast Austria. Diagn Microbiol Infect Dis. 2010;66:217–21. doi: 10.1016/j.diagmicrobio.2009.09.006. [DOI] [PubMed] [Google Scholar]
- Grisold AJ, Zarfel G, Stoeger A, et al. Emergence of community-associated methicillin-resistant Staphylococcus aureus (CA-MRSA) in Southeast Austria. J Infect. 2009;58:168–70. doi: 10.1016/j.jinf.2008.12.004. [DOI] [PubMed] [Google Scholar]
- Grundmann H, Aanensen DM, van den Wijngaard CC, et al. European Staphylococcal Reference Laboratory Working Group. Geographic distribution of Staphylococcus aureus causing invasive infections in Europe: a molecular-epidemiological analysis. PLoS Med. 2010;7:e1000215. doi: 10.1371/journal.pmed.1000215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hookev JV, Richardson JF, Cookson BD. Molecular typing of Staphylococcus aureus based on PCR restriction fragment length polymorphism and DNA sequence analysis of the coagulase gene. J Clin Microbiol. 1998;36:1083–9. doi: 10.1128/jcm.36.4.1083-1089.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huijsdens XW, van Dijke BJ, Spalburg E, et al. Community-acquired MRSA and pig-farming. Ann Clin Microbiol Antimicrob. 2006;5:26. doi: 10.1186/1476-0711-5-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerschner H, Harrison EM, Hartl R, et al. First report of mecC MRSA in human samples from Austria: molecular characteristics and clinical data. New Microbes New Infect. 2014;3:4–9. doi: 10.1016/j.nmni.2014.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinnevey PM, Shore AC, Brennan GI, et al. Extensive genetic diversity identified among sporadic methicillin-resistant Staphylococcus aureus isolates recovered in Irish hospitals between 2000 and 2012. Antimicrob Agents Ch. 2014;58:1907–17. doi: 10.1128/AAC.02653-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kock R, Becker K, Cookson B, et al. Systematic literature analysis and review of targeted preventive measures to limit healthcare-associated infections by meticillin-resistant Staphylococcus aureus. Euro Surveill. 2014;19:20860. doi: 10.2807/1560-7917.es2014.19.29.20860. [DOI] [PubMed] [Google Scholar]
- Krziwanek K, Luger C, Sammer B, et al. PVL-positive MRSA in Austria. Eur J Clin Microbiol. 2007;26:931–5. doi: 10.1007/s10096-007-0391-4. [DOI] [PubMed] [Google Scholar]
- Krziwanek K, Luger C, Sammer B, et al. MRSA in Austria–an overview. Clin Microbiol Infect. 2008;14:250–9. doi: 10.1111/j.1469-0691.2007.01896.x. [DOI] [PubMed] [Google Scholar]
- Krziwanek K, Metz-Gercek S, Mittermayer H. Methicillin-resistant Staphylococcus aureus ST398 from human patients, Upper Austria. Emerg Infect Dis. 2009;15:766–9. doi: 10.3201/eid1505.080326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loncaric I, Kübber-Heiss A, Posautz A, et al. Characterization of methicillin-resistant Staphylococcus spp. carrying the mecC gene, isolated from wildlife. J Antimicrob Chemoth. 2013;68:2222–5. doi: 10.1093/jac/dkt186. [DOI] [PubMed] [Google Scholar]
- Marchese A, Gualco L, Maioli E, et al. Molecular analysis and susceptibility patterns of meticillin-resistant Staphylococcus aureus (MRSA) strains circulating in the community in the Ligurian area, a northern region of Italy: emergence of USA300 and EMRSA-15 clones. Int J Antimicrob Ag. 2009;34:424–8. doi: 10.1016/j.ijantimicag.2009.06.016. [DOI] [PubMed] [Google Scholar]
- Monecke S, Kuhnert P, Hotzel H, et al. Microarray based study on virulence-associated genes and resistance determinants of Staphylococcus aureus isolates from cattle. Vet Microbiol. 2007;125:128–40. doi: 10.1016/j.vetmic.2007.05.016. [DOI] [PubMed] [Google Scholar]
- Otto M. Community-associated MRSA: what makes them special? Int J Med Microbiol. 2013;303:324–30. doi: 10.1016/j.ijmm.2013.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolo J, Miragaia M, Turlej-Rogacka A, et al. High genetic diversity among community-associated Staphylococcus aureus in Europe: results from a multicenter study. PLoS One. 2012;7:e34768. doi: 10.1371/journal.pone.0034768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruppitsch W, Indra A, Stoger A, et al. Classifying spa types in complexes improves interpretation of typing results for methicillin-resistant Staphylococcus aureus. J Clin Microbiol. 2006;44:2442–8. doi: 10.1128/JCM.00113-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid D, Simons E, Ruppitsch W, et al. Limited value of routine spa typing: a cross-sectional study of methicillin-resistant Staphylococcus aureus-positive patients in an Austrian hospital. Am J Infect Control. 2013;41:617–24. doi: 10.1016/j.ajic.2012.09.013. [DOI] [PubMed] [Google Scholar]
- Smith JM, Cook GM. A decade of community MRSA in New Zealand. Epidemiol Infect. 2005;133:899–904. doi: 10.1017/S0950268805004024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stryjewski ME, Corey GR. Methicillin-resistant Staphylococcus aureus: an evolving pathogen. Clin Infect Dis. 2014;58(Suppl 1):S10–9. doi: 10.1093/cid/cit613. [DOI] [PubMed] [Google Scholar]
- The European Parliament and the Council of the European Union. Decision number 2119/98/EC of the European Parliament and of the Council of 24 September 1998: setting up a network for the epidemiological surveillance and control of communicable diseases in the community. Official Journal of the European Union. 1998 http://eur-lex.europa.eu/LexUriServ/LexUriServ.do?uri=OJ:L:1998:268:0001:0006:EN:PDF (28 May 2016, date last accessed) [Google Scholar]
- Uhlemann AC, Otto M, Lowy FD, et al. Evolution of community- and healthcare-associated methicillin-resistant Staphylococcus aureus. Infect Genet Evol. 2014;21:563–74. doi: 10.1016/j.meegid.2013.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witte W, Strommenger B, Stanek C, et al. Methicillin-resistant Staphylococcus aureus ST398 in humans and animals, Central Europe. Emerg Infect Dis. 2007;13:255–8. doi: 10.3201/eid1302.060924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyllie D, Paul J, Crook D. Waves of trouble: MRSA strain dynamics and assessment of the impact of infection control. J Antimicrob Chemoth. 2011;66:2685–8. doi: 10.1093/jac/dkr392. [DOI] [PubMed] [Google Scholar]
- Zarfel G, Krziwanek K, Johler S, et al. Virulence and antimicrobial resistance genes in human MRSA ST398 isolates in Austria. Epidemiol Infect. 2013;141:888–92. doi: 10.1017/S0950268812001343. [DOI] [PMC free article] [PubMed] [Google Scholar]