Abstract
Background
Epidemiological studies have evaluated the associations of CD16 158F>V and CD32 131H>R gene polymorphisms with the risk of idiopathic thrombocytopenic purpura (ITP).
Material/Methods
Published studies on CD16 158F>V and CD32 131H>R polymorphisms with susceptibility to ITP were systematically reviewed until April 1, 2014. The Cochrane Library Database, Medline, CINAHL, EMBASE, Web of Science, and Chinese Biomedical Database (CBM) were used to search for relevant studies and then a meta-analysis was conducted by using Stata 12.0 software in order to produce consistent statistical results.
Results
In total, 10 clinical case-control studies with 741 ITP patients and 1092 healthy controls were enrolled for quantitative data analysis. Results of this meta-analysis suggest that CD16 158F>V polymorphism had strong correlations with the susceptibility to ITP under 5 genetic models (all P<0.05). However, no similar associations were found between CD32 131H>R polymorphism and the susceptibility to ITP (all P>0.05). Subgroup analysis by ethnicity revealed that CD16 158F>V polymorphism was associated with the increased risk of ITP among both Caucasian and non-Caucasian populations. Nevertheless, no statistically significant correlations between CD32 131H>R polymorphism and the risk of ITP were observed among Caucasians and non-Caucasians (all P>0.05).
Conclusions
Our findings indicate that CD16 158F>V polymorphism may contribute to the increased risk of ITP, whereas CD32 131H>R polymorphism may not be an important risk factor for ITP.
MeSH Keywords: Junctional Adhesion Molecule A; Meta-Analysis; Polymorphism, Genetic
Background
Idiopathic thrombocytopenic purpura (ITP) is the most common type of thrombocytopenic purpura and is often diagnosed as a type of autoimmune disorder [1]. Due to the destruction of immune-mediated platelets, ITP is also regarded as a type of hemorrhagic disease, resulting in low platelet count in the peripheral blood [2]. ITP is often expressed as extensive hemorrhage of skin, mucous membranes, and internal organs, persistent low platelet count, short platelet survival time, and presence of anti-platelet auto-antibodies [3]. The overall adult ITP incidence is approximately 4.4 female cases per 100 000 population per year and 3.4 male cases per 100 000 population per year in the UK, having especially prominent differences by sex in persons under 65 years old [4]. ITP is also considered to be a multifactorial disease caused by interactions between genetic and non-genetic factors [5,6]. A wide range of environmental risk factors for ITP have been identified, including viral infections, autoimmune diseases (particularly antiphospholipid syndromes), and drug inductions [7–9]. Apart from environmental risk factors, genetic factors also have been revealed to play pivotal roles in the susceptibility to ITP by numerous genes that might be associated with the development and progression of ITP [10].
Human Fcγ receptors (FcγRs) are glycoproteins that bind the Fc portion of immunoglobulin G (IgG) and are involved in preserving antibodies [11]. Human FcγRs are expressed on effector cells that have functions of autoantibody-sensitized platelets and phagocytes, as well as antibody-dependent cell-mediated cytotoxicity [12]. FcγRs play pivotal roles in the pathogenesis of autoimmune disease and they act as essential substances between the humoral and cell-mediated immune responses in order to generate inflammation [13]. Importantly, FcγRs modulated antibody production using B cells and participated in phagocytosis, antibody-dependent cellular cytotoxicity, and mediator release and are considered to be correlated with the increased risk of systemic lupus erythematosus, lupus nephritis, and ITP [14,15]. Therefore, the blocking of the FcγRs might cause reduced platelet destruction due to autoimmune diseases, including ITP. Based on structural homology, there are 3 major classes of receptors in the human FcγRs family: Fcγ RI, Fcγ RIIa (CD32); Fcγ RIIb, Fcγ RIIIa (CD16); and Fcγ RIIIb, which differ in their antibody affinities due to their different molecular structures. Low-affinity activating receptors, such as Fcγ RII and Fcγ RIII, only bind immune-complexed IgG [16,17]. Several studies have revealed that gene polymorphisms of FcγRs (CD32 and CD16) and amino acid differences may alter the receptor affinity to bind immunoglobulins [18,19]. Therefore, it was hypothesized that CD32 and CD16 might also be associated with the increased risk of ITP [20]. However, other studies showed contradictory results concerning the potential association between CD32 or CD16 and the susceptibility to ITP [21,22]. For the sake of obtaining consistent results, we performed the present meta-analysis of all available studies to determine the association between gene polymorphisms in the CD32 and CD16 genes and the susceptibility to ITP.
Material and Methods
Search strategy
Studies concerning the association between CD32 and CD16 gene polymorphisms and the susceptibility to ITP were retrieved from: Cochrane Library Database, Medline, EMBASE, CINAHL, Web of Science, PubMed, and Chinese Biomedical Database (CBM). A diverse combination of MeSH terms and keywords was used for selecting relevant studies: (“genetic polymorphism” or “SNP” or “variation” or “single nucleotide polymorphism” or “polymorphism” or “mutation” or “variant”) and (“Fc gamma receptor IIA” or “FCGR3A protein, human” or “FCGR2B protein, human” or “Fc gamma receptor IIA” or “FcgammaRIIA” or “FcgammaRIIIA” or “FcgammaRIIB” or “FCGR3A” or “FCGR2B” or “FcgammaRIIB protein”) and (“Purpura, Thrombocytopenic, Idiopathic” or “immune thrombocytopenic purpura” or “Werlhof’s Disease” or “Werlhofs Disease” or “Autoimmune Thrombocytopenic Purpura” or “Idiopathic Thrombocytopenic Purpura” or “Immune Thrombocytopenic Purpura” or “Autoimmune Thrombocytopenia”). In addition to electronic searching, other relevant studies were manually identified using references in enrolled papers obtained from the electronic search and abstracts presented at meetings of relevant scientific societies.
Inclusion criteria
To determine the trial eligibility for the meta-analysis, 4 criteria were considered: (1) Trials should be either clinically published or nested case-control studies focusing on the association between CD32 and CD16 SNPs and the risk of ITP; (2) All included subjects must be diagnosed with ITP regarded as the case group, and other comparable healthy people at the same period were chosen as the control group; and (3) Sufficient information on CD32 and CD16 polymorphisms should be supplied by eligible studies.
Data extraction and quality score assessment
Information was systematically pooled from selected publications by 2 investigators based on the inclusion criteria described above. The following data were collected for all studies: first author, countries, ethnicity, geographical locations, languages, study design, case numbers, age, sample size, sources of the subjects, genotype detection methods, and genotype polymorphism distributions.
The qualities of selected trials were assessed by 2 independent investigators using the Newcastle-Ottawa Scale (NOS) criteria [23]. The NOS criteria use a star rating system for quality assessments: (1) subject selections: 0~4; (2) subject comparability: 0~2; and (3) clinical outcomes: 0~3. NOS scores range from 0 to 9; studies with scores of more than 7 were considered as high-quality studies.
Statistical analysis
Version 12.0 of the STATA software (Stata Corporation, College Station, TX, USA) was used to process data to achieve integrity and rigorousness of statistical analysis. Associations between gene polymorphisms and the risk of ITP were assessed by odds ratios (OR) and 95% confidence interval (95%CI). The Z test was used to evaluate the statistical significance of pooled ORs. Heterogeneity across studies was assessed using Cochran’s Q-statistic and I2 tests [24]. A P-value <0.05 or I2 >50% indicates heterogeneity across all studies and either a random-effects model or a fixed-effects model was applied to the studies. Subgroup analysis was performed by ethnicity and disease base. Apart from that, sensitivity analysis was used to further investigate heterogeneity, and potential publication bias was assessed with the use of funnel plots together with Egger’s test [25].
Results
Characteristics of included studies
Fifty-six articles were initially selected based on the search strategy described above, and 24 articles were excluded after reviewing their titles and abstracts. After that, another 20 articles were excluded based on systematic reviews of their contents, and another 2 articles were also excluded due to incomplete data. As a result of this, a total of 10 clinical case-control studies with 741 ITP patients and 1092 healthy controls were selected for quantitative data analysis [10,20–22,26–31]. All eligible studies were published during the period from 1998 to 2013. Seven of them were carried out among Caucasians, 2 of them were carried out among Asians, and only 1 of them was carried out among Africans. The classical polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP), Envision, Allele-specific PCR (AS-PCR), direct sequencing, and multiplex ligation-dependent probe amplification (MLPA) assay methods were utilized in our meta-analysis. No studies deviated from Hardy-Weinberg equilibrium (HWE) (all P>0.05). The scores of all selected trials were more than 5 using NOS. The baseline characteristics and quality are displayed in Table 1.
Table 1.
Baseline characteristics and methodological quality of all included studies.
| First author | Year | Ethnicity | Disease | Number | Gender (M/F) | Age (years) | Genotype method | Gene | SNP | NOS score | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Case | Control | Case | Control | Case | Control | ||||||||
| Papagianni A [30] | 2013 | Caucasians | Children | 53 | 45 | 26/27 | – | 5.9±3.9 | – | PCR-RFLP | CD16 | 158 F>V | 6 |
| Nourse JP [29] | 2012 | Caucasians | Total | 100 | 100 | – | – | 51 (18~85) | 39 (21~66) | AS-PCR | CD16 | 158 F>V | 6 |
| Eyada TK [20] | 2012 | Africans | Children | 92 | 90 | 44/48 | 40/50 | 8.3±4.5 | 6.2±3.5 | PCR-RFLP | CD16 | 131 H>R | 8 |
| AS-PCR | CD16 | 158 F>V | |||||||||||
| Amorim DM [10] | 2012 | Caucasians | Children | 39 | 78 | 18/21 | 36/42 | 7.3±3.2 | 5.4±4.0 | PCR-RFLP | CD16 | 158 F>V | 7 |
| 131 H>R | |||||||||||||
| Breunis WB [21] | 2008 | Caucasians | Adult | 44 | 100 | 7/37 | – | – | – | MLPA assay | CD32 | 131 H>R | 6 |
| Children | 72 | 100 | 35/37 | – | – | – | MLPA assay | CD32 | 131 H>R | 6 | |||
| Wang JH [31] | 2007 | Asians | Total | 74 | 111 | 22/52 | 45/66 | 34.5±13.3 | 39.0±16.3 | AS-PCR | CD16 | 158 F>V | 7 |
| Carcao MD [26] | 2003 | Caucasians | Children | 98 | 130 | 46/52 | – | 0.5±16.9 | – | PCR-RFLP | CD32 | 158 F>V | 8 |
| 131 H>R | |||||||||||||
| Fujimoto TT [27] | 2001 | Asians | Total | 104 | 59 | 28/76 | 30/29 | 26 | 54.2 | PCR-RFLP | CD32 | 158 F>V | 8 |
| 131 H>R | |||||||||||||
| Foster CB [22] | 2001 | Caucasians | Children/chronic | 36 | 218 | – | – | – | – | PCR-RFLP | CD32 | 158 F>V | 6 |
| 131 H>R | |||||||||||||
| Williams Y [32] | 1998 | Caucasians | Total | 29 | 61 | 30/60 | 10/19 | 7~75 | 20~55 | AS-PCR | CD32 | 131 H>R | 6 |
M – male; F – female; SNP – single nucleotide polymorphism; NOS – Newcastle-Ottawa Scale; PCR-RFLP – polymerase chain reaction-restriction fragment length polymorphism; AS-PCR – allele-specific PCR, MLPA multiplex ligation-dependent probe amplification.
Quantitative data synthesis
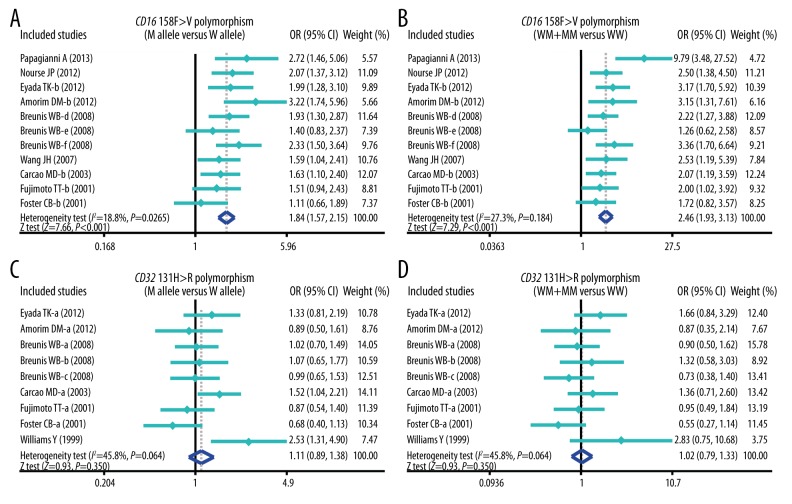
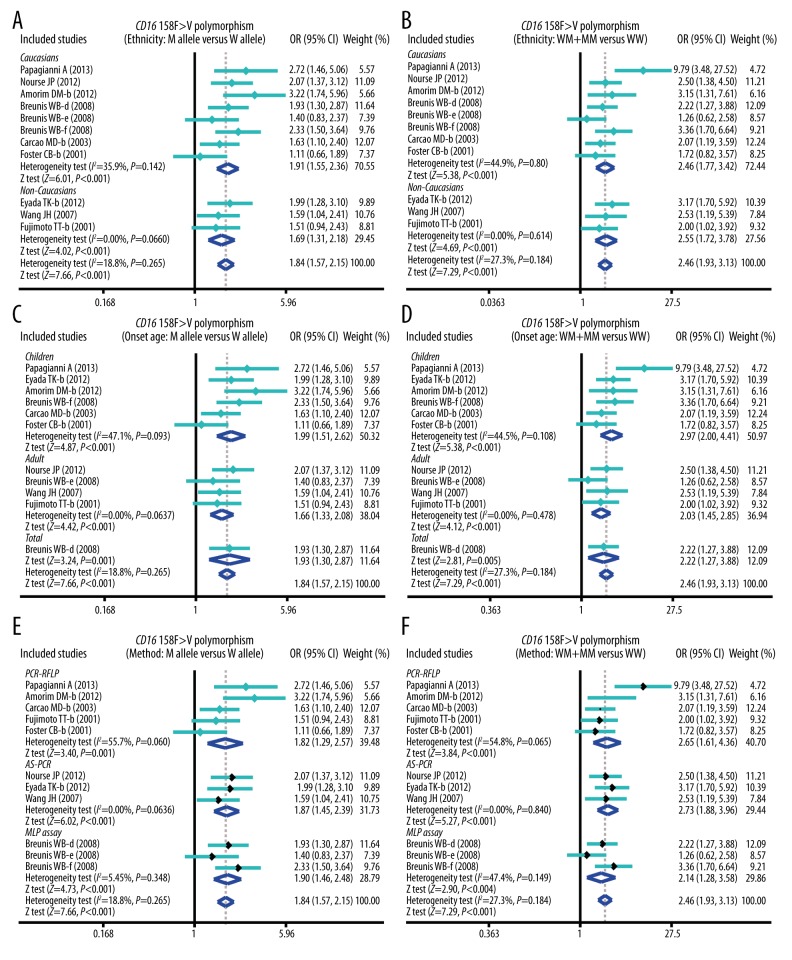
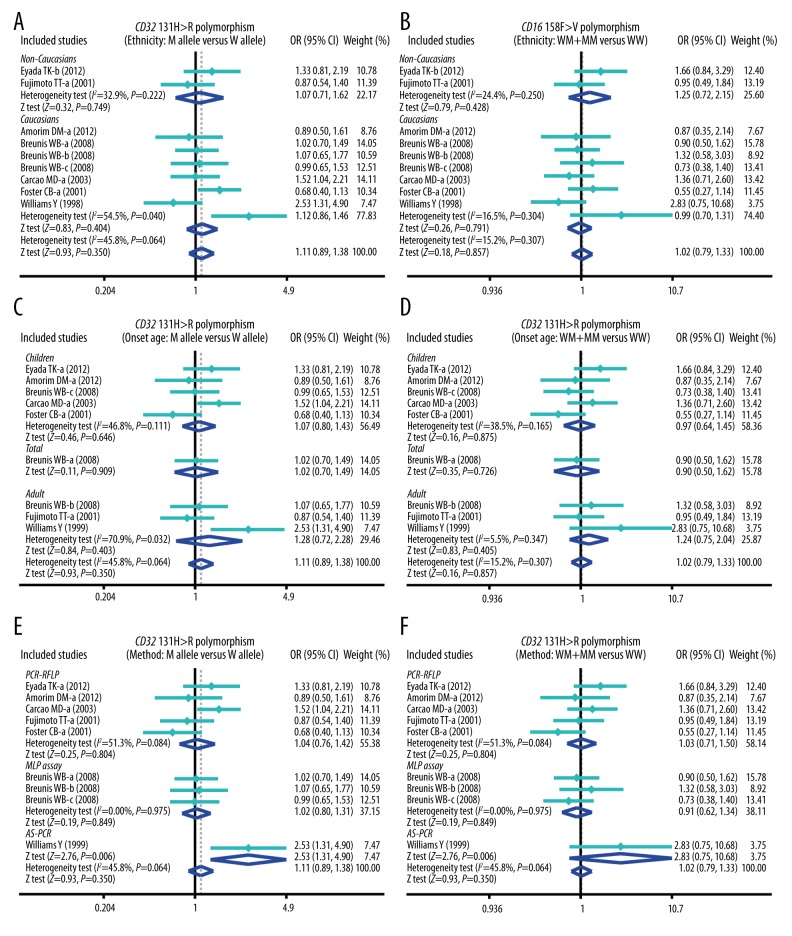
The results of this meta-analysis of relationships of CD32 131H>R and CD16 158F>V genetic polymorphisms with susceptibility to ITP are shown in Table 2. The results showed that CD16 158F>V polymorphism was significantly associated with the increased risk of ITP (P < 0.05) (Figure 1), whereas CD32 131H>R polymorphism was not significantly associated with the susceptibility to ITP (all P>0.05) (Figure 1). Furthermore, subgroup analysis was performed to investigate the independent effects of ethnicity, onset age, and genotype methods on the risk of ITP. Ethnicity-stratified analysis revealed positive correlations between 158F>V SNP and the increased risk of ITP among both Caucasian and non-Caucasian groups (P<0.05), whereas no such associations were found between 131H>R polymorphism and the risk of ITP (all P>0.05) (Figure 2). With regard to onset age, CD16 158F>V polymorphism was correlated with increased risk of ITP in most of the pediatric and adult groups (P<0.05), whereas CD32 131H>R polymorphism was not associated with the risk of ITP in pediatric and adult groups (all P>0.05) (Figure 3). Stratified analysis by genotype methods revealed that CD16 158F>V polymorphism was associated with the increased risk of ITP in the PCR-RFLP, AS-PCR, and MLPA assay groups under 4 genetic models (all P<0.05) (Figure 2). However, we found no significant associations between CD32 polymorphisms in the PCR-RFLP and AS-PCR groups (all P>0.05), except for the MLPA assay group under 4 genetic models (Allele model: OR=2.53, 95%CI=1.31~4.90, P=0.006; Recessive model: OR=4.24, 95%CI=1.60~11.28, P=0.004; Homozygous model: OR=6.36, 95%CI=1.46~27.67, P=0.014; and Heterozygous model: OR=3.71, 95%CI=1.33~10.36, P=0.012; respectively) (Table 2).
Table 2.
Meta-analysis of the relationships of CD16 158F>V and CD32 131H>R polymorphisms with the immune thrombocytopenic purpura.
| M allele vs. W (allele model) | WM + MM vs. WW (dominant model) | MM vs. WW + WM (recessive model) | MM vs. WW (homozygous model) | MM vs. WM (heterozygous model) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| OR | 95%CI | P | OR | 95%CI | P | OR | 95%CI | P | OR | 95%CI | P | OR | 95%CI | P | |
| 158F>V | 1.84 | 1.57–2.15 | <0.001 | 2.46 | 1.93–3.13 | <0.001 | 2.12 | 1.56–2.88 | <0.001 | 3.31 | 2.38–4.59 | <0.001 | 1.53 | 1.12–2.09 | 0.008 |
| Ethnicity | |||||||||||||||
| Caucasians | 1.91 | 1.55–2.36 | <0.001 | 2.46 | 1.77–3.42 | <0.001 | 2.36 | 1.56–3.57 | <0.001 | 3.44 | 2.23–5.30 | <0.001 | 1.70 | 1.11–2.60 | 0.015 |
| Non-Caucasians | 1.69 | 1.31–2.18 | <0.001 | 2.55 | 1.72–3.78 | <0.001 | 1.60 | 0.94–2.74 | 0.084 | 2.90 | 1.55–5.42 | 0.001 | 1.18 | 0.68–2.05 | 0.558 |
| Onset age | |||||||||||||||
| Children | 1.99 | 1.51–2.62 | <0.001 | 2.97 | 2.00–4.41 | <0.001 | 1.94 | 1.03–3.64 | 0.039 | 3.27 | 1.75–6.12 | <0.001 | 1.27 | 0.68–2.37 | 0.457 |
| Adult | 1.66 | 1.33–2.08 | <0.001 | 2.03 | 1.45–2.85 | <0.001 | 2.02 | 1.29–3.18 | 0.002 | 3.10 | 1.85–5.20 | <0.001 | 1.61 | 1.00–2.58 | 0.049 |
| Genotype method | |||||||||||||||
| PCR-RFLP | 1.82 | 1.29–2.57 | 0.001 | 2.65 | 1.61–4.36 | <0.001 | 1.63 | 0.71–3.75 | 0.250 | 2.59 | 1.19–5.63 | 0.016 | 1.14 | 0.50–2.58 | 0.762 |
| AS-PCR | 1.87 | 1.46–2.39 | <0.001 | 2.73 | 1.88–3.96 | <0.001 | 1.97 | 1.20–3.22 | 0.007 | 3.58 | 2.02–6.34 | <0.001 | 1.42 | 0.85–2.38 | 0.176 |
| MLPA assay | 1.90 | 1.46–2.48 | <0.001 | 2.14 | 1.28–3.58 | 0.004 | 2.61 | 1.56–4.39 | <0.001 | 3.70 | 2.11–6.50 | <0.001 | 1.98 | 1.15–3.43 | 0.014 |
| 13 H>R | 1.11 | 0.89–1.38 | 0.350 | 1.02 | 0.79–1.33 | 0.857 | 1.25 | 0.87–1.79 | 0.232 | 1.15 | 0.78–1.69 | 0.489 | 1.25 | 0.80–1.93 | 0.323 |
| Ethnicity | |||||||||||||||
| Non-Caucasians | 1.07 | 0.71–1.62 | 0.749 | 1.25 | 0.72–2.15 | 0.428 | 0.85 | 0.47–1.56 | 0.605 | 0.93 | 0.47–1.83 | 0.828 | 0.28 | 0.04–1.89 | 0.192 |
| Caucasians | 1.12 | 0.86–1.46 | 0.404 | 0.96 | 0.70–1.31 | 0.791 | 1.39 | 0.93–2.09 | 0.112 | 1.24 | 0.77–2.01 | 0.378 | 1.49 | 1.04–2.14 | 0.031 |
| Onset age | |||||||||||||||
| Children | 1.07 | 0.80–1.43 | 0.646 | 0.97 | 0.64–1.45 | 0.875 | 1.27 | 0.85–1.87 | 0.239 | 1.11 | 0.71–1.74 | 0.644 | 1.27 | 0.69–2.32 | 0.447 |
| Adult | 1.28 | 0.72–2.28 | 0.403 | 1.24 | 0.75–2.04 | 0.405 | 1.28 | 0.39–4.26 | 0.687 | 1.47 | 0.40–5.38 | 0.562 | 1.17 | 0.37–3.69 | 0.786 |
| Genotype method | |||||||||||||||
| PCR-RFLP | 1.04 | 0.76–1.42 | 0.804 | 1.03 | 0.71–1.50 | 0.880 | 1.04 | 0.63–1.74 | 0.871 | 0.99 | 0.58–1.70 | 0.971 | 0.90 | 0.42–1.92 | 0.776 |
| MLPA assay | 1.02 | 0.80–1.31 | 0.849 | 0.91 | 0.62–1.34 | 0.632 | 1.20 | 0.78–1.84 | 0.398 | 1.08 | 0.66–1.78 | 0.756 | 1.29 | 0.82–2.03 | 0.272 |
| AS-PCR | 2.53 | 1.31–4.90 | 0.006 | 2.83 | 0.75–10.68 | 0.126 | 4.24 | 1.60–11.28 | 0.004 | 6.36 | 1.46–27.67 | 0.014 | 3.71 | 1.33–10.36 | 0.012 |
W – wild allele; M – mutant allele; WW – wild homozygote; WM – heterozygote; MM – mutant homozygote; OR – odds ratio; 95%CI – 95% confidence interval; AS-PCR – allele-specific PCR; PCR-RFLP – polymerase chain reaction-restriction fragment length polymorphism; MLPA – multiplex ligation-dependent probe amplification.
Figure 1.
Forest plots for the correlation between CD16 158F>V and CD32 131H>R polymorphisms and the risk of idiopathic thrombocytopenic purpura under allele and dominant models.
Figure 2.
Subgroup analysis by ethnicity, onset age, and genotype methods of the correlation between CD16 158F>V polymorphism and the risk of idiopathic thrombocytopenic purpura under allele and dominant models.
Figure 3.
Subgroup analysis by ethnicity, onset age, and genotype methods of the correlation between CD32 131H>R polymorphism and the risk of idiopathic thrombocytopenic purpura under allele and dominant models.
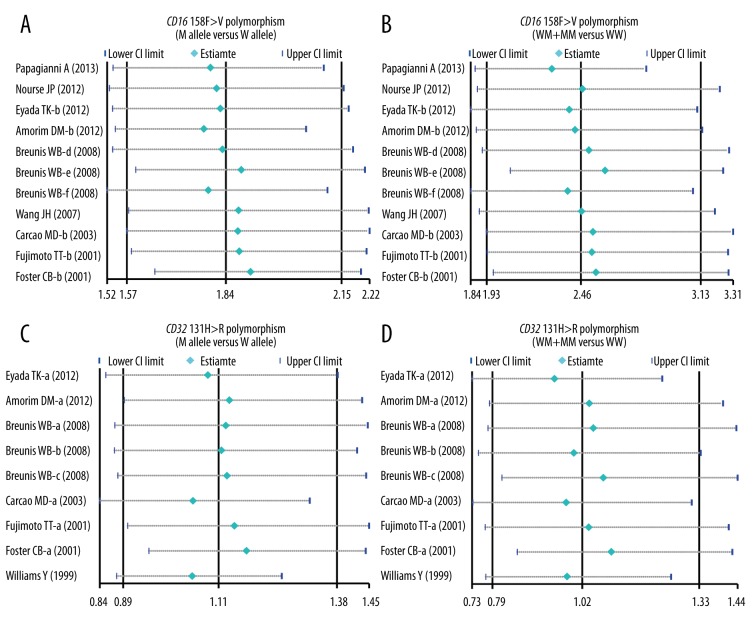
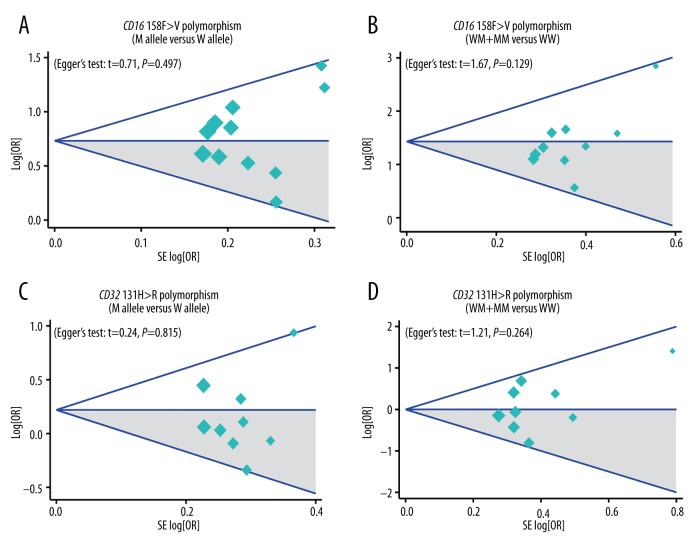
Sensitivity analysis and publication bias evaluation
Table 3 of the meta-regression analysis illustrates that publication year, ethnicity, onset age, and genotype method were not potential sources of heterogeneity for determining associations between gene polymorphisms and the risk of ITP. Figure 4 reveals the results from sensitivity analysis, which suggested that the overall ORs were not substantially influenced by any individual study. Finally, Figure 5 shows that there was no significant evidence of asymmetry patterns in the funnel plots, and Egger’s test also showed no publication bias (all P>0.05).
Table 3.
Univariate and multivariate meta-regression analyses of potential source of heterogeneity.
| Heterogeneity factors | 158 F>V | 131 H>R | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Coefficient | SE | Z | P | 95%CI | Coefficient | SE | Z | P | 95%CI | |||
| LL | UL | LL | UL | |||||||||
| Publication year | ||||||||||||
| Univariate | 0.056 | 0.029 | 1.94 | 0.052 | −0.001 | 0.112 | 0.052 | 0.015 | 0.36 | 0.722 | −0.024 | 0.034 |
| Multivariate | 0.071 | 0.051 | 1.37 | 0.169 | −0.030 | 0.172 | 0.012 | 0.020 | 0.59 | 0.552 | −0.028 | 0.052 |
| Ethnicity | ||||||||||||
| Univariate | 0.247 | 0.242 | 1.02 | 0.307 | −0.227 | 0.721 | −0.054 | 0.124 | −0.44 | 0.661 | −0.298 | 0.189 |
| Multivariate | 0.026 | 0.289 | 0.09 | 0.928 | −0.536 | 0.588 | 0.113 | 0.223 | 0.51 | 0.613 | −0.324 | 0.550 |
| Onset age | ||||||||||||
| Univariate | 0.252 | 0.201 | 1.25 | 0.210 | −0.141 | 0.645 | 0.015 | 0.011 | 1.33 | 0.184 | −0.007 | 0.038 |
| Multivariate | 0.135 | 0.328 | 0.41 | 0.681 | −0.507 | 0.777 | 0.016 | 0.011 | 1.36 | 0.174 | −0.007 | 0.038 |
| Genotyping method | ||||||||||||
| Univariate | 0.180 | 0.142 | 1.27 | 0.206 | −0.099 | 0.460 | 0.038 | 0.040 | 0.93 | 0.350 | −0.041 | 0.116 |
| Multivariate | −0.172 | 0.310 | −0.56 | 0.578 | −0.780 | 0.435 | 0.059 | 0.059 | 1.00 | 0.315 | −0.056 | 0.174 |
SE – standard error; 95%CI – 95% confidence interval; UL – upper limit; LL – lower limit.
Figure 4.
Sensitivity analysis of the correlation between CD16 158F>V and CD32 131H>R polymorphisms and the risk of idiopathic thrombocytopenic purpura under allele and dominant models.
Figure 5.
Funnel plots for publication biases against the correlation of CD16 158F>V and CD32 131H>R polymorphisms with the risk of idiopathic thrombocytopenic purpura under allele and dominant models.
Discussion
Our meta-analysis explored potential associations between CD32 131H>R and CD16 158F>V SNPs and the susceptibility to ITP. Our results suggest that CD16 158F>V polymorphism is closely associated with the increased risk of ITP, whereas CD32 131H>R polymorphism is not associated with the susceptibility to ITP, suggesting that the 158F>V polymorphism can be considered as a potential predictor of ITP development. A large body of evidence suggests that ITP development might be attributable to a key gene change that leads to the dysfunction of the immunologic system and even to thrombocytopenia [32]. There is a wide range of well-known pathologies that could explain thrombocytopenia, such as B and T cell responses, cytokine equilibrium alterations, anti-platelet antibody productions, phagocytic cell activations, surface molecule transformations, and cell immunity dysfunctions with variation of the Th1/Th2 ratio [33,34]. The underlying mechanism of thrombocytopenia remains unclear, although platelet destruction caused by phagocytic cells is one of the key mechanisms. It is well established that low-affinity receptors of CD32 and CD16 interact with multimeric or complexed IgG, such as IgG1 and IgG3, because CD32 is the only Fcγ able to bind IgG2 [27]. Variant Fcγ alleles are also risk factors for systemic autoimmune diseases, and the increased activation of CD32 and CD16 can trigger the processes of phagocytosis and of antibody-dependent cellular cytotoxicity and the release of inflammatory mediators [35]. In general, SNPs of Fcγ receptors accompanied with the loss of inhibitory Fcγ expression and altered functions may result in an unbalanced immunity and subsequently cause auto-inflammation, which might be associated with the susceptibility to ITP [21]. Specifically for CD16, the presence of valine (V) instead of phenylalanine (F) in codon 158 may alter the affinity of IgG1 and IgG3 receptors [10]. Consistent with our findings, Eyada et al. also concluded that CD16 gene polymorphisms may contribute to susceptibility to ITP, which is consistent with results obtained from the present meta-analysis, and this further suggests the crucial role of CD16 158F>V polymorphisms in ITP development [20]. On the other hand, CD32 gene polymorphisms caused by the substitution of a single nucleotide (A–G), which codes for the amino acids histidine (H) instead of arginine (R) at position 131 (131H>R), may alter the ability of its receptor to bind to IgG2 [30]. However, our study results do not suggest any associations between CD32 131H>R polymorphisms and the risk of ITP. Subgroup analysis by ethnicity suggests that CD16 158F>V polymorphisms may drive the susceptibility to ITP among both Caucasians and non-Caucasians. Nevertheless, it is essential to discuss some statistical limitations of our meta-analysis in order to improve the power of our study. Firstly, the possible confounding effects of sex, disease, and ethnicity was not controlled in selected trials, which might cause confounding effects or affect medications, and these restrict the power of our meta-analysis. Secondly, the power of our meta-analysis is limited due to the small number of studies and their small sample sizes, which in turn could affect our conclusion regarding the association between gene polymorphism (CD16 and CD 32) and the risk of ITP. Thirdly, it was not possible to determine the effects of genetic interactions and interactions between genes and the environment on the association due to the limitations of the original data; for example, our study only includes articles in English or Chinese, which might have caused selection biases. However, this is the first meta-analysis concerning the association between CD32 and CD16 SNPs with ITP. More importantly, with the use of a statistical approach to combine the results from various studies, and then quantifying and analyzing those inconsistent results in our meta-analysis, it is possible to achieve more reliable results.
Conclusions
Our results suggest that CD16 158F>V may contribute to the increased risk of ITP. However, this conclusion should be drawn with caution due to the small sample size. In addition, our study does not suggest any associations between CD32 131H>R polymorphisms and the risk of ITP. As a result of this, it is critical to implement well-designed case-control studies that include subjects with the same ethnic background and tissue-specific biochemical and biological characterization to verify the results obtained from the present meta-analysis.
Acknowledgments
We acknowledge and appreciate our colleagues for their valuable efforts and comments on this paper.
Footnotes
Source of support: Departmental sources
Conflict of interest
The authors have declared that no competing interests.
References
- 1.Hunt CW. Immune thrombocytopenia purpura. Medsurg Nurs. 2010;19:237–39. [PubMed] [Google Scholar]
- 2.Del Vecchio GC, Giordano P, Tesse R, et al. Clinical significance of serum cytokine levels and thrombopoietic markers in childhood idiopathic thrombocytopenic purpura. Blood Transfus. 2012;10:194–99. doi: 10.2450/2011.0055-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stasi R, Evangelista ML, Stipa E, et al. Idiopathic thrombocytopenic purpura: current concepts in pathophysiology and management. Thromb Haemost. 2008;99:4–13. doi: 10.1160/TH07-08-0513. [DOI] [PubMed] [Google Scholar]
- 4.Deane S, Teuber SS, Gershwin ME. The geoepidemiology of immune thrombocytopenic purpura. Autoimmun Rev. 2010;9:A342–49. doi: 10.1016/j.autrev.2009.11.020. [DOI] [PubMed] [Google Scholar]
- 5.Cines DB, Bussel JB, Liebman HA, et al. The ITP syndrome: pathogenic and clinical diversity. Blood. 2009;113:6511–21. doi: 10.1182/blood-2009-01-129155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gernsheimer T. Chronic idiopathic thrombocytopenic purpura: mechanisms of pathogenesis. Oncologist. 2009;14:12–21. doi: 10.1634/theoncologist.2008-0132. [DOI] [PubMed] [Google Scholar]
- 7.Neunert C, Lim W, Crowther M, et al. The American Society of Hematology 2011 evidence-based practice guideline for immune thrombocytopenia. Blood. 2011;117:4190–207. doi: 10.1182/blood-2010-08-302984. [DOI] [PubMed] [Google Scholar]
- 8.Liebman HA. Viral-associated immune thrombocytopenic purpura. Hematology Am Soc Hematol Educ Program. 2008:212–18. doi: 10.1182/asheducation-2008.1.212. [DOI] [PubMed] [Google Scholar]
- 9.Johnsen J. Pathogenesis in immune thrombocytopenia: new insights. Hematology Am Soc Hematol Educ Program. 2012;2012:306–12. doi: 10.1182/asheducation-2012.1.306. [DOI] [PubMed] [Google Scholar]
- 10.Amorim DM, Silveira Vda S, Scrideli CA, et al. Fcgamma receptor gene polymorphisms in childhood immune thrombocytopenic purpura. J Pediatr Hematol Oncol. 2012;34:349–52. doi: 10.1097/MPH.0b013e3182580908. [DOI] [PubMed] [Google Scholar]
- 11.Daeron M. Fc receptor biology. Annu Rev Immunol. 1997;15:203–34. doi: 10.1146/annurev.immunol.15.1.203. [DOI] [PubMed] [Google Scholar]
- 12.Nimmerjahn F, Ravetch JV. Fcgamma receptors: old friends and new family members. Immunity. 2006;24:19–28. doi: 10.1016/j.immuni.2005.11.010. [DOI] [PubMed] [Google Scholar]
- 13.Li X, Ptacek TS, Brown EE, et al. Fcgamma receptors: structure, function and role as genetic risk factors in SLE. Genes Immun. 2009;10:380–89. doi: 10.1038/gene.2009.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Espeli M, Niederer HA, Traherne JA, et al. Genetic variation, Fcgamma receptors, KIRs and infection: the evolution of autoimmunity. Curr Opin Immunol. 2010;22:715–22. doi: 10.1016/j.coi.2010.10.003. [DOI] [PubMed] [Google Scholar]
- 15.Lee YH, Ji JD, Song GG. Fcgamma receptor IIB and IIIB polymorphisms and susceptibility to systemic lupus erythematosus and lupus nephritis: A meta-analysis. Lupus. 2009;18:727–34. doi: 10.1177/0961203309104020. [DOI] [PubMed] [Google Scholar]
- 16.Nimmerjahn F, Ravetch JV. Fcgamma receptors as regulators of immune responses. Nat Rev Immunol. 2008;8:34–47. doi: 10.1038/nri2206. [DOI] [PubMed] [Google Scholar]
- 17.Hoemberg M, Stahl D, Schlenke P, et al. The isotype of autoantibodies influences the phagocytosis of antibody-coated platelets in autoimmune thrombocytopenic purpura. Scand J Immunol. 2011;74:489–95. doi: 10.1111/j.1365-3083.2011.02600.x. [DOI] [PubMed] [Google Scholar]
- 18.Chai L, Song YQ, Leung WK. Genetic polymorphism studies in periodontitis and Fcgamma receptors. J Periodontal Res. 2012;47:273–85. doi: 10.1111/j.1600-0765.2011.01437.x. [DOI] [PubMed] [Google Scholar]
- 19.Bruhns P, Iannascoli B, England P, et al. Specificity and affinity of human Fcgamma receptors and their polymorphic variants for human IgG subclasses. Blood. 2009;113:3716–25. doi: 10.1182/blood-2008-09-179754. [DOI] [PubMed] [Google Scholar]
- 20.Eyada TK, Farawela HM, Khorshied MM, et al. FcgammaRIIa and FcgammaRIIIa genetic polymorphisms in a group of pediatric immune thrombocytopenic purpura in Egypt. Blood Coagul Fibrinolysis. 2012;23:64–68. doi: 10.1097/MBC.0b013e32834ddf2f. [DOI] [PubMed] [Google Scholar]
- 21.Breunis WB, van Mirre E, Bruin M, et al. Copy number variation of the activating FCGR2C gene predisposes to idiopathic thrombocytopenic purpura. Blood. 2008;111:1029–38. doi: 10.1182/blood-2007-03-079913. [DOI] [PubMed] [Google Scholar]
- 22.Foster CB, Zhu S, Erichsen HC, et al. Polymorphisms in inflammatory cytokines and Fcgamma receptors in childhood chronic immune thrombocytopenic purpura: a pilot study. Br J Haematol. 2001;113:596–99. doi: 10.1046/j.1365-2141.2001.02807.x. [DOI] [PubMed] [Google Scholar]
- 23.Stang A. Critical evaluation of the Newcastle-Ottawa scale for the assessment of the quality of nonrandomized studies in meta-analyses. Eur J Epidemiol. 2010;25:603–5. doi: 10.1007/s10654-010-9491-z. [DOI] [PubMed] [Google Scholar]
- 24.Zintzaras E, Ioannidis JP. HEGESMA: Genome search meta-analysis and heterogeneity testing. Bioinformatics. 2005;21:3672–73. doi: 10.1093/bioinformatics/bti536. [DOI] [PubMed] [Google Scholar]
- 25.Peters JL, Sutton AJ, Jones DR, et al. Comparison of two methods to detect publication bias in meta-analysis. JAMA. 2006;295:676–80. doi: 10.1001/jama.295.6.676. [DOI] [PubMed] [Google Scholar]
- 26.Wang JH, Xie R, Wang L, et al. [FcgammaRIIIA-158V/F polymorphisms in idiopathic thrombocytopenic purpura patients]. Chin J Internal Med. 2007:937–38. [in Chinese] [Google Scholar]
- 27.Carcao MD, Blanchette VS, Wakefield CD, et al. Fcgamma receptor IIa and IIIa polymorphisms in childhood immune thrombocytopenic purpura. Br J Haematol. 2003;120:135–41. doi: 10.1046/j.1365-2141.2003.04033.x. [DOI] [PubMed] [Google Scholar]
- 28.Fujimoto TT, Inoue M, Shimomura T, et al. Involvement of Fc gamma receptor polymorphism in the therapeutic response of idiopathic thrombocytopenic purpura. Br J Haematol. 2001;115:125–30. doi: 10.1046/j.1365-2141.2001.03109.x. [DOI] [PubMed] [Google Scholar]
- 29.Nourse JP, Lea R, Crooks P, et al. The KIR2DS2/DL2 genotype is associated with adult persistent/chronic and relapsed immune thrombocytopenia independently of FCGR3a-158 polymorphisms. Blood Coagul Fibrinolysis. 2012;23:45–50. doi: 10.1097/MBC.0b013e32834d7ce3. [DOI] [PubMed] [Google Scholar]
- 30.Papagianni A, Economou M, Tragiannidis A, et al. FcgammaRIIa and FcgammaRIIIa polymorphisms in childhood primary immune thrombocytopenia: implications for disease pathogenesis and outcome. Blood Coagul Fibrinolysis. 2013;24:35–39. doi: 10.1097/MBC.0b013e328359bc3b. [DOI] [PubMed] [Google Scholar]
- 31.Williams Y, Lynch S, McCann S, et al. Correlation of platelet Fc gammaRIIA polymorphism in refractory idiopathic (immune) thrombocytopenic purpura. Br J Haematol. 1998;101:779–82. doi: 10.1046/j.1365-2141.1998.00802.x. [DOI] [PubMed] [Google Scholar]
- 32.Podolanczuk A, Lazarus AH, Crow AR, et al. Of mice and men: an open-label pilot study for treatment of immune thrombocytopenic purpura by an inhibitor of Syk. Blood. 2009;113:3154–60. doi: 10.1182/blood-2008-07-166439. [DOI] [PubMed] [Google Scholar]
- 33.Zhou B, Zhao H, Yang RC, et al. Multi-dysfunctional pathophysiology in ITP. Crit Rev Oncol Hematol. 2005;54:107–16. doi: 10.1016/j.critrevonc.2004.12.004. [DOI] [PubMed] [Google Scholar]
- 34.Emmerich F, Bal G, Barakat A, et al. High-level serum B-cell activating factor and promoter polymorphisms in patients with idiopathic thrombocytopenic purpura. Br J Haematol. 2007;136:309–14. doi: 10.1111/j.1365-2141.2006.06431.x. [DOI] [PubMed] [Google Scholar]
- 35.Karassa FB, Trikalinos TA, Ioannidis JP. The role of FcgammaRIIA and IIIA polymorphisms in autoimmune diseases. Biomed Pharmacother. 2004;58:286–91. doi: 10.1016/j.biopha.2004.04.004. [DOI] [PubMed] [Google Scholar]