Abstract
A proportion of MYC translocation positive diffuse large B‐cell lymphomas (DLBCL) harbour a BCL2 and/or BCL6 translocation, known as double‐hit DLBCL, and are clinically aggressive. It is unknown whether there are other genetic abnormalities that cooperate with MYC translocation and form double‐hit DLBCL, and whether there is a difference in clinical outcome between the double‐hit DLBCL and those with an isolated MYC translocation. We investigated TP53 gene mutations along with BCL2 and BCL6 translocations in a total of 234 cases of DLBCL, including 81 with MYC translocation. TP53 mutations were investigated by PCR and sequencing, while BCL2 and BCL6 translocation was studied by interphase fluorescence in situ hybridization. The majority of MYC translocation positive DLBCLs (60/81 = 74%) had at least one additional genetic hit. In MYC translocation positive DLBCL treated by R‐CHOP (n = 67), TP53 mutation and BCL2, but not BCL6 translocation had an adverse effect on patient overall survival. In comparison with DLBCL with an isolated MYC translocation, cases with MYC/TP53 double‐hits had the worst overall survival, followed by those with MYC/BCL2 double‐hits. In MYC translocation negative DLBCL treated by R‐CHOP (n = 101), TP53 mutation, BCL2 and BCL6 translocation had no impact on patient survival. The prognosis of MYC translocation positive DLBCL critically depends on the second hit, with TP53 mutations and BCL2 translocation contributing to an adverse prognosis. It is pivotal to investigate both TP53 mutations and BCL2 translocations in MYC translocation positive DLBCL, and to distinguish double‐hit DLBCLs from those with an isolated MYC translocation.
Keywords: DLBCL, chromosome translocation, TP53 mutation, double‐hit, overall survival
Introduction
Diffuse large B‐cell lymphoma (DLBCL) represents 30–45% of adult cases of non‐Hodgkin lymphoma (https://www.hmrn.org/statistics) 1, accounting for more than 80% of aggressive lymphomas. The addition of rituximab has significantly improved the treatment outcome of patients with DLBCL. Nonetheless, a significant proportion of DLBCLs show primary treatment failure (∼10%), partial response (∼15%) or relapse after initial response (20–30%) to R‐CHOP (rituximab, cyclophosphamide, doxorubicin, vincristine and prednisone), the current first line treatment for this malignancy 2. A number of biomarkers have been investigated with the aim of predicting treatment outcome at diagnosis and identifying those that may benefit from novel therapeutic strategies, but only a few have proven to be clinically useful due to lack of reproducibility (immunohistochemistry‐based markers) and/or difficulty of routine clinical application (molecular subtypes by gene expression profiling) on formalin‐fixed paraffin‐embedded diagnostic tissue biopsies 3, 4, 5. Among the many biomarkers investigated, MYC chromosome translocation is widely accepted and used in routine clinical practice.
The MYC translocation occurs in 5–15% of DLBCL, and is usually associated with a complex pattern of genomic alterations 6, 7, 8, 9, 10, 11, 12. A proportion (21–83%) of DLBCLs with MYC translocation also harbour a BCL2 and/or BCL6 translocation, known as ‘double‐hit’ or ‘triple‐hit’ lymphoma. Patients with ‘double‐hit’ DLBCL commonly show aggressive clinical features and respond poorly to currently available treatments, with a median survival less than 1.5 years 6. However, it remains controversial whether DLBCL with MYC single translocation has a different prognosis from that with MYC/BCL2 double translocation. For example, the recent studies by Cuccuini et al 13, Aukema et al 6 and Valera et al 14 showed a similar poor overall survival between DLBCL with MYC single translocation and those with MYC double translocations. In contrast, the studies by Johnson et al, Green et al and Landsburg et al demonstrated no adverse impact of MYC single translocation in DLBCL, while cases with MYC/BCL2 double translocations had a very poor outcome 15, 16, 17. Although the reasons underlying the discrepancies between these studies are unclear, potential factors that may account for the discrepancies could include the small numbers of cases investigated, variations in clinicopathological parameters (age, stage, international prognostic index [IPI]) and a variable presence of additional genetic changes such as TP53 mutation that modifies the prognostic value of MYC translocation.
MYC drives cell proliferation but also sensitizes cells to apoptotic stimuli, which provides a safeguard to prevent any potential MYC induced malignant transformation. The MYC mediated proapoptotic activity is largely through the activation of the p19(ARF)‐MDM2‐TP53 pathway and repression of the apoptosis inhibitor BCL2 18, 19. There is extensive literature showing that MYC requires cooperating events to abrogate its proapoptotic activities to exert its full oncogenic potential, and both expression of BCL2 and loss of TP53 function cooperate with MYC translocation in lymphomagenesis 18, 19, 20, 21. In DLBCL, TP53 mutations are found in ∼20% of cases and are significantly and independently associated with poor overall survival of both activated B‐cell like (ABC) and germinal centre B‐cell like (GCB) DLBCL treated with R‐CHOP 22, 23. TP53 mutations have been reported in cases of DLBCL with MYC translocation 24, 25, but their frequency in MYC translocation positive DLBCL and their combined clinical impact are unclear. In this study, we have investigated TP53 mutations and BCL2 translocations in a large cohort of DLBCL, and analysed their association and clinical impact in the presence and absence of MYC translocation.
Materials and Methods
Patients and tissue materials
A total of 234 cases of de novo DLBCL were investigated in this study. 168 cases were retrieved from the Haematological Malignancy Diagnostic Service (HMDS) at St James's University Hospital, Leeds (n = 145) and Addenbrooke's hospital, Cambridge (n = 23), based on the availability of lymphoma tissue specimens. 153 of these cases have been classified by cell of origin (COO) using the Illumina WG‐DASL assay used in previous studies 7, 26. The remaining 66 cases were positive for MYC translocation by interphase fluorescence in situ hybridisation (FISH) and identified from five participating centres where the assessment of MYC translocation status is a part of the routine diagnostic workup of DLBCL 27. The diagnosis in each case was established by two expert haematopathologists, and those defined as B‐cell lymphoma, unclassifiable, with features intermediate between DLBCL and Burkitt lymphoma, in the 2008 WHO classification were included in this study 28. Burkitt lymphoma, DLBCL potentially transformed from a low grade lymphoma, cases with HIV or primary CNS lymphoma were excluded from this study. All the laboratory investigations described below were based on the initial diagnostic lymphoma tissue specimens. Ethical guidelines were followed for the use of archival tissues for research with the approval of the ethics committees of the involved institutions.
Microdissection and DNA preparation
Haematoxylin and eosin slides were reviewed for all cases, and crude microdissection was performed where indicated to enrich tumour cells, ensuring that the tissue area containing >60% of tumour cells was used for DNA preparation. DNA was extracted using the QIAamp DNA Micro Kit (QIAGEN, Crawley, UK). The quality of the DNA samples was assessed by polymerase chain reaction (PCR) of variably sized genomic fragments 29, and those with successful amplifications of >300 bp were used for mutational screening.
PCR and Sanger sequencing
Mutations in TP53 coding exons 5–10, commonly targeted by somatic mutation in human cancer, were investigated by PCR and Sanger sequencing using primers and conditions detailed in supplementary material Table S1. In each case, sequence change was confirmed by at least two independent PCR and sequencing experiments. The somatic mutation was ascertained by excluding germline changes through SNP database search and analysis of DNA prepared from microdissected normal cells, where possible.
Interphase FISH
Chromosome translocations involving the MYC, BCL2 and BCL6 loci were investigated using dual‐colour break‐apart probes (Vysis/Abbott Laboratories, UK) 30. For each probe, the mean plus three standard deviations of false positive signals in 100 nuclei from 8 to 10 reactive tonsils was used as the cut‐off value for the diagnosis of chromosomal translocation. The cut‐off value for MYC, BCL2 and BCL6 break‐apart probes is very similar, all being <6%.
Statistical analyses
Rates of genetic alterations and molecular subtypes by COO classification were compared using difference‐of‐proportions tests. Survival analyses were performed using the Kaplan–Meier method with log‐rank tests and by using the Cox proportional hazards model.
Results
We investigated a total of 234 cases of primary DLBCL, including 168 cases selected based on their tissue availability, and 66 additional cases based on their positivity for MYC translocation. Among the 168 cases selected based on tissue availability, the frequencies of MYC (15/168 = 8.9%), BCL2 (30/168 = 17.8%) and BCL6 (49/167 = 29.3%) translocation and TP53 mutation (26/166 = 15.7%) were in line with those reported in the literature 6, 22, 23, 31, 32. To understand the potential oncogenic cooperation of these genetic changes, we analysed associations across the entire patient group.
High frequencies of TP53 mutation and BCL2 translocation in MYC translocation positive DLBCL
The frequency of TP53 mutation was significantly higher in cases with MYC translocation than in those without the translocation (27/81 = 33.3% versus 23/151 = 15.2%, p = 0.001, Figure 1A,B). However, there was no significant difference in the frequency of TP53 mutation between DLBCL with an isolated MYC translocation and those with double translocations (MYC plus BCL2 or BCL6) (14/35 = 40% versus 13/46 = 28.3%, p = 0.38). Five cases (6% within the MYC translocation positive series) were found to have concurrent translocations of MYC, BCL2 and BCL6, and none of these had a TP53 mutation (Figure 1A). In addition, no differences in the spectrum of TP53 mutations were observed between DLBCL with MYC translocation and those without (supplementary material Figure S1).
Figure 1.
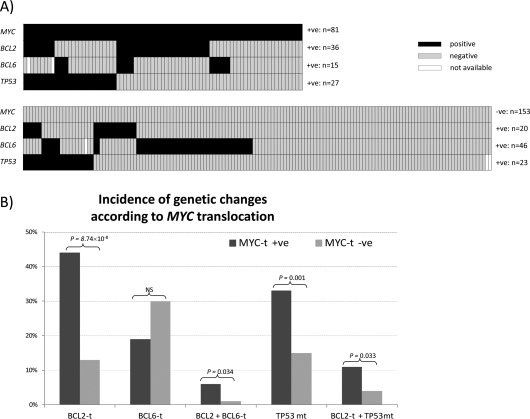
Correlation of TP53 mutation, MYC, BCL2 and BCL6 translocation in primary DLBCL. (A) Distribution of TP53 mutation, BCL2 and BCL6 translocation in DLBCLs with and without MYC translocation. The majority of MYC translocation positive DLBCLs harbour at least one additional genetic abnormality, frequently TP53 mutation or BCL2 translocation, and occasionally BCL2 translocation plus TP53 mutation or BCL6 translocation. Black cell: positive for the genetic abnormality indicated; Grey cell: negative for the genetic abnormality indicated; White cell: data not available. (B) Incidence of TP53 mutation, BCL2 and BCL6 translocation in DLBCLs with and without MYC translocation. The frequency of TP53 mutation and BCL2 translocation is significantly higher in cases with MYC translocation. t: translocation; +ve: positive; ‐ve: negative; NS: no significance.
The frequency of BCL2 translocation was also significantly higher in DLBCL with MYC translocation than in those without (36/81 = 44.4% versus 20/153 = 13.1%, p = 8.74 × 10−8, Figure 1B). In contrast, the frequency of BCL6 translocation was lower in DLBCL with MYC translocation than in those without (15/79 = 18.9% versus 46/152 = 30.0%, p = 0.065, Figure 1B).
In cases with MYC translocation, there was no evidence of correlation among TP53 mutation, BCL2 and BCL6 translocation (supplementary material Table S2). In cases without MYC translocation, there was a positive correlation between BCL2 translocation and TP53 mutation (p = 0.048), and a negative correlation between BCL2 and BCL6 translocation (p = 0.034) (supplementary material Table S2).
Among the 234 cases of DLBCL investigated, 153 had data on COO‐classification by Illumina WG‐DASL array from a previous study and 140 of these cases were MYC translocation negative 4. We, thus, correlated COO subtypes with genetic abnormalities in cases without MYC translocation. As expected 33, BCL2 translocation was significantly associated with GCB‐DLBCL (p = 0.004), although no significant association was seen between BCL6 translocation and COO subtype (p = 0.079, supplementary material Table S2). There was no correlation between TP53 mutation and COO molecular subtype.
Distinct impact of TP53 mutations, BCL2 and BCL6 translocations on prognosis of MYC translocation positive DLBCL
Of the 81 MYC translocation positive DLBCLs investigated, 60 (74.1%) had at least one additional genetic abnormality, namely TP53 mutation, BCL2 or BCL6 translocation. Of these 60 cases, 18 had three abnormalities, including nine cases with MYC/BCL2 double translocation and TP53 mutation, four cases with MYC/BCL6 double translocation and TP53 mutation, and a further five cases with triple translocations (Figure 1A). The remaining 42 cases with a single additional genetic abnormality included 22 with BCL2 translocation, 14 with TP53 mutation, and six with BCL6 translocation (Figure 1A).
To investigate whether the prognosis of these MYC translocation positive DLBCLs was affected by the number and nature of additional genetic abnormalities, we performed a series of survival analyses. These were carried out exclusively on cases treated with R‐CHOP or equivalent regimens with a curative intent, and included 67 cases with MYC translocation and 101 cases without MYC translocation.
We first focused on the MYC translocation positive DLBCL, subdivided according to the number (1 or 2) of additional genetic abnormalities regardless of their nature, and examined whether the number of additional genetic abnormalities impacted on patient survival. To our surprise, cases with one, but not those with two additional genetic abnormalities, showed a worse survival than those with an isolated MYC translocation. Such difference might be due to an insufficient number of the cases with two additional genetic abnormalities for comparison. Alternatively, the findings suggest that the nature rather than the number of the additional genetic abnormalities may be more important in influencing the prognosis of MYC translocation positive DLBCL.
We next examined the survival of patients with MYC translocation positive DLBCL purely according to the status of TP53 mutation, BCL2 and BCL6 translocation. The MYC translocation positive DLBCL with TP53 mutation had a significantly worse overall survival than those without TP53 mutation by multivariate Cox regression analysis adjusted for age (p = 0.036, supplementary material Figure S2C). Similarly, MYC translocation positive DLBCL with BCL2 translocation showed a worse survival, albeit not statistically significant, than those lacking the translocation (supplementary material Figure S2A). In contrast, MYC translocation positive DLBCL with BCL6 translocation had a significantly better overall survival than those without BCL6 translocation by multivariate Cox regression analysis adjusted for age (p = 0.035, supplementary material Figure S2B). Despite that the reference subgroup in each of the above analyses comprised of mixed cases that included not only those with isolated MYC translocation but also cases with other second hit, the analyses, nonetheless, suggested that TP53 mutation and possibly BCL2, but not BCL6 translocation may have an adverse effect on patients survival. In view of this and the small number of cases with only MYC and BCL6 translocation, our subsequent analyses focused on TP53 mutation and BCL2 translocation.
We divided MYC translocation positive DLBCL into the following subgroups according to TP53 mutation and BCL2 translocation status: MYC/BCL2 double translocation with TP53 mutation, MYC single translocation with TP53 mutation, MYC/BCL2 double translocation, and isolated MYC translocation. In comparison with the subgroup with isolated MYC translocation, all other three subgroups had a significantly worse overall survival by logrank test (Figure 2A). Interestingly, the patients with MYC/BCL2/TP53 triple hit and the cases with MYC/TP53 double‐hit appeared to be separated from those with the MYC/BCL2 double‐hit (Figure 2A). In view of these findings, we combined all cases with TP53 mutation irrespective of the BCL2 translocation status, and these cases had the worst overall survival (Figure 2B, supplementary material Table S3). Further univariate and multivariate Cox regression analyses adjusted for age confirmed the significant worse overall survival of patients with MYC/TP53 (n = 22, p = 0.0095 and p = 0.0059 respectively) and cases with MYC/BCL2 (n = 25, p = 0.03 and p = 0.019, respectively) double‐hit in comparison with those with isolated MYC translocation (n = 20) Figure 2B, Table 1).
Figure 2.
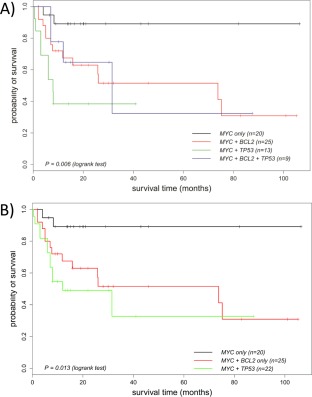
Impact of TP53 mutation and BCL2 translocation on overall survival of patients with MYC translocation positive DLBCL. (A) MYC translocation positive DLBCL are divided into four subgroups: MYC/BCL2 translocation/TP53 mutation, MYC translocation/TP53 mutation, MYC/BCL2 translocation, and MYC translocation only. The cases with the MYC/BCL2 /TP53 triple‐hit show the worst overall survival, followed by cases with the MYC/TP53 and those with the MYC/BCL2 double‐hit. The significant difference in overall survival between cases with the MYC/BCL2 /TP53 triple‐hit and those with isolated MYC translocation is also shown by Cox proportional hazards regression adjusted for age. (B) The cases with TP53 mutation are combined together irrespective of their BCL2 translocation status. The cases with TP53 mutation show a worse overall survival than those with an isolated MYC translocation, being statistically significant by Cox regression model adjusted for age (p = 0.0059, Table 1). The cases with BCL2 translocation also show a significantly worse overall survival than those with isolated MYC translocation by Cox regression model adjusted for age (p = 0.019, Table 1).
Table 1.
Impact of TP53 mutation and BCL2 translocation on overall survival of patients with MYC translocation positive DLBCL by Cox proportional hazards regression.
| Univariate analysis a | Multivariate analysis adjusted for age | |||||
|---|---|---|---|---|---|---|
| HR | 95% CI | P value | HR | 95% CI | P value | |
| MYC + BCL2 translocation(n=25) | 5.11 | 1.15‐22.7 | 0.03 | 6.17 | 1.34‐28.3 | 0.019 |
| MYC translocation + TP53 mutation (irrespective of BCL2 translocation status)(n=22) | 7.26 | 1.62‐32.5 | 0.0095 | 8.74 | 1.87–40.8 | 0.0059 |
In comparison with DLBCL with isolated MYC translocation; HR: Hazard ratio; CI: confidence interval.
Prognostic value of TP53 mutation, BCL2 and BCL6 translocation in MYC translocation negative DLBCL
All MYC translocation negative DLBCLs were retrieved from HMDS, St James's University Hospital, Leeds and Addenbrooke's hospital, Cambridge, and 101 of these cases treated with R‐CHOP had clinical follow up data. As expected, COO‐classification clearly separated these cases into distinct prognostic groups with the ABC‐DLBCL showing a worse overall survival by logrank test (p = 0.011, supplementary material Figure S3D). To our surprise, neither isolated TP53 mutation nor single BCL2 translocation had a significant impact on overall survival (supplementary material Figure S3A,C). BCL6 translocation appeared to be associated with a worse overall survival; however, this was not statistically significant by Cox regression analysis after age adjustment (supplementary material Figure S3B).
Discussion
This is the first study combining chromosome translocations and TP53 mutation analysis in a large cohort of DLBCL. We have demonstrated that MYC translocation positive DLBCL had a significantly higher frequency of TP53 mutation and BCL2 translocation 7, and that DLBCL with MYC translocation and TP53 mutation had the worst overall survival, followed by cases with MYC/BCL2 double‐hits. These findings have significant implications for using these biomarkers in the prognostic evaluation of patients with DLBCL.
By investigating retrospectively a large cohort of MYC translocation positive DLBCL, we showed that the prognostic value of MYC translocation was critically influenced by the presence of the second hit, essentially its oncogenic cooperating events. The patients with MYC/TP53 abnormalities irrespective of BCL2 translocation status had a significantly worse overall survival than those with an isolated MYC translocation. The cases with MYC/BCL2 double translocations but wild type TP53 showed an intermediate overall survival, appearing better than those with MYC/TP53 abnormalities, but worse than those with an isolated MYC translocation (Figure 2B). These findings suggest that TP53 mutation may have a more adverse impact on prognosis than BCL2 translocation in MYC translocation positive DLBCL. The importance of the second hit in prognosis of MYC translocation positive DLBCL is further emphasized by the observation that cases with an isolated MYC translocation appeared to have an overall survival similar to those without MYC translocation (supplementary material Figure S4). No adverse impact of MYC single translocation in DLBCL was also reported in some previous studies 15, 16, 17, but not supported by others 6, 13, 14. However, these previous studies did not investigate TP53 mutation and it is unknown whether the discrepancy was caused by a variable presence of TP53 mutation in cases with single MYC translocation among these studies. In view of the retrospective nature of the current study, it remains necessary to confirm these findings in a prospective study containing a large cohort of MYC translocation positive DLBCL.
Both TP53 mutation and BCL2 translocation are known to cooperate with MYC translocation in lymphomagenesis by impeding the proapoptotic activities of MYC. In Eµ‐Myc transgenic animals, lymphoma development requires the acquisition of additional genetic alterations, which commonly comprise of disruption of the p19ARF‐Mdm2‐p53 pathway or overexpression of Bcl2 34, 35. In addition, Eµ‐Myc mice with lymphoma showing a loss of p53 function have a significantly worse survival than those with lymphoma overexpressing Bcl2 36. These findings together with our observations in this study indicate that there is a strong selection of genetic events that cooperates with MYC translocation during lymphoma development, and these cooperating events are a major determining factor in modulating the prognostic impact of MYC translocation in DLBCL.
The prognostic value of BCL6 translocation in MYC translocation positive DLBCL is unclear. The majority of DLBCL with MYC and BCL6 translocation also had BCL2 translocation or TP53 mutation, and the number of cases with only MYC and BCL6 translocation is small for assessing the prognostic impact of BCL6 translocation in MYC translocation positive DLBCL. There was no association between MYC and BCL6 translocation in DLBCL. In addition, there is no direct evidence supporting critical oncogenic cooperation between MYC and BCL6 translocation in lymphoma development. Findings from the present and a previous study 37 suggest that BCL6 translocation appears to be associated with a better prognosis in MYC translocation positive DLBCL, although this differs from that observed by Pillai et al 38. There are several potential reasons that might account for the discrepancy among these studies, and these include small numbers of cases with MYC/BCL6 translocation, difference/potential bias in case selection, variations in clinicopathological features (age, stage, IPI) and treatment, and potential differences in the genetic abnormalities associated with MYC translocation among these studies. Therefore, it is important to investigate the prognostic value of BCL6 translocation in MYC translocation positive DLBCL in a prospective study.
The term ‘double‐hit’ lymphoma was originally used to describe aggressive DLBCL with MYC and BCL2 translocation, then subsequently extended to include those with MYC and BCL6 translocation. In light of the findings in this study and the discussion above, the term ‘double‐hit’ lymphoma should be extended to include those with MYC translocation and TP53 mutation. Our data clearly highlight the prognostic significance of TP53 mutation, even more so than BCL2 translocation, in MYC translocation positive DLBCL. It is therefore pivotal to confirm these findings in a prospective study and investigate TP53 mutation status in routine clinical practice for cases of MYC translocation positive DLBCL, in addition to the current standard investigation for BCL2 translocation.
MYC and BCL2 translocations are commonly investigated by interphase FISH, while TP53 mutation can be readily screened for by PCR and Sanger sequencing, or a next generation sequencing‐based approach. Alternatively, DLBCL with MYC/BCL2 or MYC/TP53 double‐hit might be screened by combined immunohistochemistry for MYC, BCL2 and TP53 15, 16, 23, 31, 32. The challenges for such an immunohistochemistry‐based approach are reproducibility, accurate assessment of the extent and percentage of staining positivity, and the identification of the best cut‐off value for dichotomy between positive and negative staining results. In addition, combined MYC and BCL2 immunohistochemistry also identifies ∼20% of DLBCL that show concurrent MYC and BCL2 protein expression, but no evidence of their involvement in translocation. This group of DLBCL appears to show an overall survival better than the cases with MYC/BCL2 translocation, but worse than those without these translocations 15, 16, 32, and thus, should be regarded as a separate prognostic group. In contrast, combined MYC and TP53 immunohistochemistry would under‐detect DLBCL with MYC translocation and TP53 mutation, as a high proportion (∼17%) of TP53 mutations such as frameshift and nonsense mutations result in a truncated protein product that is unlikely to be detectable by immunohistochemistry 23.
Our findings also highlight the importance of separating MYC translocation positive DLBCL from negative cases in biomarker validation, particularly the genetic events associated with MYC translocation, to avoid the confounding effect of MYC translocation. TP53 mutations have been shown to be significantly associated with poor overall survival in both ABC and GCB‐DLBCL treated with R‐CHOP 22, 23. In line with this, we also found a significant association of TP53 mutation with poor overall survival in DLBCL when MYC translocation positive cases were included (based on unselected cases from HMDS, Leeds /Addenbrooke's Hospital). However, this significant association disappeared after excluding the MYC translocation positive cases, indicating a confounding effect of the translocation. Nonetheless, the number of MYC translocation negative cases investigated in this study is relatively small and the prognostic value of TP53 mutation in MYC translocation negative DLBCL remains to be elucidated.
In summary, we have shown that MYC translocation positive DLBCL has a significantly higher frequency of TP53 mutation and BCL2 translocation, and that the cases with MYC translocation and TP53 mutation had the worst overall survival, followed by cases with MYC/BCL2 double‐hits. It is critical to investigate both TP53 mutation and BCL2 rearrangement in MYC translocation positive DLBCL, and to distinguish double‐hit DLBCLs from those with an isolated MYC translocation in routine clinical practice.
Contract/grant details
The research in MQD lab was supported by grants from Leukaemia & Lymphoma Research, U.K. Kay. Kendall Leukaemia Fund (KKLF), The Pathological Society of UK & Ireland and the Addenbrooke's Charitable Trust (ACT). NG was supported by a KKLF and an ACT fellowship. Research in JG's lab was funded by Tenovus Tayside; JB was supported by a research grant from the Paul Abrahams Fund.
Author contributions
AC, SB, NZ, HL, SK, MW & YH collected and analyzed laboratory data; SC performed statistical analyses; NFG collected clinical data; LW assisted with sample preparation; JG, JB, MN, PF, BW, JWG, PW, HED, GAF, ER, PWMJ, AJW and AJ provided cases, diagnosis and clinical follow up data. MQD supervised and coordinated the study and wrote the manuscript. All authors have commented on the manuscript and approved its final version for submission. The authors declare no conflict of interest.
Supporting information
Figure S1. Nature and distribution of TP53 mutations in primary DLBCL with and without MYC translocation. All mutations are reported in the COSMIC somatic mutation database, with the exception of c.672+1G>T, c.783‐1G>A and R333C. There is no apparent difference in the nature and distribution of TP53 mutation found in primary DLBCL with and without MYC translocation. trans+ve: translocation positive; trans‐ve: translocation negative; Mutations seen in the same case are indicated by the same colour scheme with the exception of those in black.
Figure S2. Impact of TP53 mutation, BCL2 and BCL6 translocation on overall survival of patients with MYC translocation positive DLBCL. trans+ve: translocation positive; trans‐ve: translocation negative.
Figure S3. Impact of TP53 mutation, BCL2 and BCL6 translocation, and COO molecular subtype on the overall survival of patients with MYC translocation negative DLBCL. These cases are from the Haematological Malignancy Diagnostic Service (HMDS) at St James's University Hospital, Leeds and Addenbrooke's hospital, Cambridge, retrieved based on the availability of lymphoma tissue specimens. All cases included in the survival analysis were treated with R‐CHOP or a rituximab‐containing equivalent regimen. trans+ve: translocation positive; trans‐ve: translocation negative; COO: cell of origin
Figure S4. Comparison of overall survival between DLBCL with isolated MYC translocation (absence of TP53 mutation, BCL2 and BCL6 translocation) and those without MYC translocation. These cases are selected based on the availability of lymphoma tissue specimens from the Haematological Malignancy Diagnostic Service (HMDS) at St James's University Hospital, Leeds, and Addenbrooke's hospital, Cambridge, and all cases included in this figure were treated with R‐CHOP or equivalent regimens. Dotted lines indicate 95% confidence intervals.
Supporting Information Table S1
Supporting Information Table S2
Supporting Information Table S3
Acknowledgements
The authors would like to thank Dr. Ian McFarlane, the Microarray CoreLab, David Withers, IMS Metabolic Research Laboratories, National Institute of Health Research, Cambridge Comprehensive Biomedical Research Centre for their help in DNA sequencing. We also thank Dr. Ramesh Bulusu, Dr. Kanchan Regge, Dr. Muttuswamy Sivakumaran, Dr. Sateesh Nagumantry, Dr. Andrew Hodson, Dr. Mamtha Karanth, Dr. Sandra Young‐Min, Dr. Jane Keidan, and Dr. Martin Lewis for providing clinical data.
No conflicts of interest were declared.
References
- 1. Young KH, Medeiros LJ, Chan WC. Diffuse large B‐cell lymphoma In Knowles' Neoplastic Hematopathology, Orazi A, Weiss LM, Foucar K, Knowles DM. (eds). Wolters Kluwer, Lippincott Williams & Wilkins: New York, London, Hong Kong, Sydney, Tokyo, 2014; 502–565. [Google Scholar]
- 2. Thieblemont C, Gisselbrecht C. Second‐line treatment paradigms for diffuse large B‐cell lymphomas. Curr Oncol Rep 2009; 11: 386–393. [DOI] [PubMed] [Google Scholar]
- 3. Coutinho R, Clear A, Owen A, et al Poor concordance among nine immunohistochemistry classifiers of cell‐of‐origin for Diffuse Large B‐cell Lymphoma: implications for therapeutic strategies. Clin Cancer Res 2013; 19: 6686–6695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Barrans SL, Crouch S, Care MA, et al Whole genome expression profiling based on paraffin embedded tissue can be used to classify diffuse large B‐cell lymphoma and predict clinical outcome. Br J Haematol 2012; 159: 441–453. [DOI] [PubMed] [Google Scholar]
- 5. Scott DW, Wright GW, Williams PM, et al Determining cell‐of‐origin subtypes of diffuse large B‐cell lymphoma using gene expression in formalin‐fixed paraffin‐embedded tissue. Blood 2014; 123: 1214–1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Aukema SM, Siebert R, Schuuring E, et al Double‐hit B‐cell lymphomas. Blood 2011; 117: 2319–2331. [DOI] [PubMed] [Google Scholar]
- 7. Barrans S, Crouch S, Smith A, et al Rearrangement of MYC is associated with poor prognosis in patients with diffuse large B‐cell lymphoma treated in the era of rituximab. J Clin Oncol 2010; 28: 3360–3365. [DOI] [PubMed] [Google Scholar]
- 8. Obermann EC, Csato M, Dirnhofer S, et al Aberrations of the MYC gene in unselected cases of diffuse large B‐cell lymphoma are rare and unpredictable by morphological or immunohistochemical assessment. J Clin Pathol 2009; 62: 754–756. [DOI] [PubMed] [Google Scholar]
- 9. Copie‐Bergman C, Gaulard P, Leroy K, et al Immuno‐fluorescence in situ hybridization index predicts survival in patients with diffuse large B‐cell lymphoma treated with R‐CHOP: a GELA study. J Clin Oncol 2009; 27: 5573–5579. [DOI] [PubMed] [Google Scholar]
- 10. Savage KJ, Johnson NA, Ben Neriah S, et al MYC gene rearrangements are associated with a poor prognosis in diffuse large B‐cell lymphoma patients treated with R‐CHOP chemotherapy. Blood 2009; 114: 3533–3537. [DOI] [PubMed] [Google Scholar]
- 11. Yoon SO, Jeon YK, Paik JH, et al MYC translocation and an increased copy number predict poor prognosis in adult diffuse large B‐cell lymphoma (DLBCL), especially in germinal centre‐like B cell (GCB) type. Histopathology 2008; 53: 205–217. [DOI] [PubMed] [Google Scholar]
- 12. Klapper W, Stoecklein H, Zeynalova S, et al Structural aberrations affecting the MYC locus indicate a poor prognosis independent of clinical risk factors in diffuse large B‐cell lymphomas treated within randomized trials of the German High‐Grade Non‐Hodgkin's Lymphoma Study Group (DSHNHL). Leukemia 2008; 22: 2226–2229. [DOI] [PubMed] [Google Scholar]
- 13. Cuccuini W, Briere J, Mounier N, et al MYC+ diffuse large B‐cell lymphoma is not salvaged by classical R‐ICE or R‐DHAP followed by BEAM plus autologous stem cell transplantation. Blood 2012; 119: 4619–4624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Valera A, Lopez‐Guillermo A, Cardesa‐Salzmann T, et al MYC protein expression and genetic alterations have prognostic impact in patients with diffuse large B‐cell lymphoma treated with immunochemotherapy. Haematologica 2013; 98: 1554–1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Johnson NA, Slack GW, Savage KJ, et al Concurrent Expression of MYC and BCL2 in Diffuse Large B‐Cell Lymphoma Treated With Rituximab Plus Cyclophosphamide, Doxorubicin, Vincristine, and Prednisone. J Clin Oncol 2012; 30: 3452–3459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Green TM, Young KH, Visco C, et al Immunohistochemical double‐hit score is a strong predictor of outcome in patients with diffuse large B‐cell lymphoma treated with rituximab plus cyclophosphamide, doxorubicin, vincristine, and prednisone. J Clin Oncol 2012; 30: 3460–3467. [DOI] [PubMed] [Google Scholar]
- 17. Landsburg DJ, Nasta SD, Svoboda J, et al ‘Double‐Hit’ cytogenetic status may not be predicted by baseline clinicopathological characteristics and is highly associated with overall survival in B cell lymphoma patients. Br J Haematol 2014; 166: 369–374. [DOI] [PubMed] [Google Scholar]
- 18. Klapproth K, Wirth T. Advances in the understanding of MYC‐induced lymphomagenesis. Br J Haematol 2010; 149: 484–497. [DOI] [PubMed] [Google Scholar]
- 19. Luscher B, Vervoorts J. Regulation of gene transcription by the oncoprotein MYC. Gene 2012; 494: 145–160. [DOI] [PubMed] [Google Scholar]
- 20. Meyer N, Kim SS, Penn LZ. The Oscar‐worthy role of Myc in apoptosis. Semin Cancer Biol 2006; 16: 275–287. [DOI] [PubMed] [Google Scholar]
- 21. Meyer N, Penn LZ. Reflecting on 25 years with MYC. Nat Rev Cancer 2008; 8: 976–990. [DOI] [PubMed] [Google Scholar]
- 22. Young KH, Leroy K, Moller MB, et al Structural profiles of TP53 gene mutations predict clinical outcome in diffuse large B‐cell lymphoma: an international collaborative study. Blood 2008; 112: 3088–3098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Xu‐Monette ZY, Wu L, Visco C, et al Mutational profile and prognostic significance of TP53 in diffuse large B‐cell lymphoma patients treated with R‐CHOP: report from an International DLBCL Rituximab‐CHOP Consortium Program Study. Blood 2012; 120: 3986–3996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Akasaka T, Akasaka H, Ueda C, et al Molecular and clinical features of non‐Burkitt's, diffuse large‐cell lymphoma of B‐cell type associated with the c‐MYC/immunoglobulin heavy‐chain fusion gene. J Clin Oncol 2000; 18: 510–518. [DOI] [PubMed] [Google Scholar]
- 25. Gebauer N, Bernard V, Gebauer W, et al TP53 mutations are frequent events in double‐hit B‐cell lymphomas with MYC and BCL2 but not MYC and BCL6 translocations. Leuk Lymphoma 2014, in press, doi: 10.3109/10428194.2014.907896. [DOI] [PubMed] [Google Scholar]
- 26. Care MA, Barrans S, Worrillow L, et al A microarray platform‐independent classification tool for cell of origin class allows comparative analysis of gene expression in diffuse large B‐cell lymphoma. PLoS One 2013; 8: e55895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Foot NJ, Dunn RG, Geoghegan H, et al Fluorescence in situ hybridisation analysis of formalin‐fixed paraffin‐embedded tissue sections in the diagnostic work‐up of non‐Burkitt high grade B‐cell non‐Hodgkin's lymphoma: a single centre's experience. J Clin Pathol 2011; 64: 802–808. [DOI] [PubMed] [Google Scholar]
- 28. Stein H, Warnke RA, Chan WC, et al Diffuse large B‐cell lymphoma, not otherwise specified In WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues, Swerdlow SH, Campo E, Harris NL, et al. (eds). International Agency for Research on Cancer: Lyon, 2008; 233–237. [Google Scholar]
- 29. Liu H, Bench AJ, Bacon CM, et al A practical strategy for the routine use of BIOMED‐2 PCR assays for detection of B‐ and T‐cell clonality in diagnostic haematopathology. Br J Haematol 2007; 138: 31–43. [DOI] [PubMed] [Google Scholar]
- 30. Nakamura S, Ye H, Bacon CM, et al Translocations involving the immunoglobulin heavy chain gene locus predict better survival in gastric diffuse large B‐cell lymphoma. Clin Cancer Res 2008; 14: 3002–3010. [DOI] [PubMed] [Google Scholar]
- 31. Horn H, Ziepert M, Becher C, et al MYC status in concert with BCL2 and BCL6 expression predicts outcome in diffuse large B‐cell lymphoma. Blood 2013; 121: 2253–2263. [DOI] [PubMed] [Google Scholar]
- 32. Hu S, Xu‐Monette ZY, Tzankov A, et al MYC/BCL2 protein coexpression contributes to the inferior survival of activated B‐cell subtype of diffuse large B‐cell lymphoma and demonstrates high‐risk gene expression signatures: a report from The International DLBCL Rituximab‐CHOP Consortium Program. Blood 2013; 121: 4021–4031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lenz G, Staudt LM. Aggressive lymphomas. N Engl J Med 2010; 362: 1417–1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Eischen CM, Weber JD, Roussel MF, et al Disruption of the ARF‐Mdm2‐p53 tumor suppressor pathway in Myc‐induced lymphomagenesis. Genes Dev 1999; 13: 2658–2669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Strasser A, Harris AW, Bath ML, et al Novel primitive lymphoid tumours induced in transgenic mice by cooperation between myc and bcl‐2. Nature 1990; 348: 331–333. [DOI] [PubMed] [Google Scholar]
- 36. Schuster C, Berger A, Hoelzl MA, et al The cooperating mutation or "second hit" determines the immunologic visibility toward MYC‐induced murine lymphomas. Blood 2011; 118: 4635–4645. [DOI] [PubMed] [Google Scholar]
- 37. Tzankov A, Xu‐Monette ZY, Gerhard M, et al Rearrangements of MYC gene facilitate risk stratification in diffuse large B‐cell lymphoma patients treated with rituximab‐CHOP. Mod Pathol 2014; 27: 958–971. [DOI] [PubMed] [Google Scholar]
- 38. Pillai RK, Sathanoori M, Van Oss SB, et al Double‐hit B‐cell lymphomas with BCL6 and MYC translocations are aggressive, frequently extranodal lymphomas distinct from BCL2 double‐hit B‐cell lymphomas. Am J Surg Pathol 2013; 37: 323–332. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Nature and distribution of TP53 mutations in primary DLBCL with and without MYC translocation. All mutations are reported in the COSMIC somatic mutation database, with the exception of c.672+1G>T, c.783‐1G>A and R333C. There is no apparent difference in the nature and distribution of TP53 mutation found in primary DLBCL with and without MYC translocation. trans+ve: translocation positive; trans‐ve: translocation negative; Mutations seen in the same case are indicated by the same colour scheme with the exception of those in black.
Figure S2. Impact of TP53 mutation, BCL2 and BCL6 translocation on overall survival of patients with MYC translocation positive DLBCL. trans+ve: translocation positive; trans‐ve: translocation negative.
Figure S3. Impact of TP53 mutation, BCL2 and BCL6 translocation, and COO molecular subtype on the overall survival of patients with MYC translocation negative DLBCL. These cases are from the Haematological Malignancy Diagnostic Service (HMDS) at St James's University Hospital, Leeds and Addenbrooke's hospital, Cambridge, retrieved based on the availability of lymphoma tissue specimens. All cases included in the survival analysis were treated with R‐CHOP or a rituximab‐containing equivalent regimen. trans+ve: translocation positive; trans‐ve: translocation negative; COO: cell of origin
Figure S4. Comparison of overall survival between DLBCL with isolated MYC translocation (absence of TP53 mutation, BCL2 and BCL6 translocation) and those without MYC translocation. These cases are selected based on the availability of lymphoma tissue specimens from the Haematological Malignancy Diagnostic Service (HMDS) at St James's University Hospital, Leeds, and Addenbrooke's hospital, Cambridge, and all cases included in this figure were treated with R‐CHOP or equivalent regimens. Dotted lines indicate 95% confidence intervals.
Supporting Information Table S1
Supporting Information Table S2
Supporting Information Table S3


