SUMMARY
Transfer RNAs (tRNAs) are primarily viewed as static contributors to gene expression. By developing a high-throughput tRNA profiling method, we find that specific tRNAs are upregulated in human breast cancer cells as they gain metastatic activity. Through loss-of-function, gain-of-function, and clinical-association studies, we implicate tRNAGluUUC and tRNAArgCCG as promoters of breast cancer metastasis. Upregulation of these tRNAs enhances stability and ribosome occupancy of transcripts enriched for their cognate codons. Specifically, tRNAGluUUC promotes metastatic progression by directly enhancing EXOSC2 expression and enhancing GRIPAP1—constituting an “inducible” pathway driven by a tRNA. The cellular proteomic shift toward a prometastatic state mirrors global tRNA shifts, allowing for cell-state and cell-type transgene expression optimization through codon content quantification. TRNA modulation represents a mechanism by which cells achieve altered expression of specific transcripts and proteins. TRNAs are thus dynamic regulators of gene expression and the tRNA codon landscape can causally and specifically impact disease progression.
Graphical Abstract
INTRODUCTION
Transfer RNAs and the genetic code underlying protein synthesis are universal to all domains of life (Dever and Green, 2012). Despite this universality, genomes exhibit substantial variations in their preference for specific codons across their coding sequences. The source of this bias, though still debated, likely reflects selection for translational efficiency and accuracy (Drummond and Wilke, 2008; Plotkin and Kudla, 2011; Shah and Gilchrist, 2011). Importantly, even the genes within the same genome show high levels of variation in their codon preferences and synonymous codon usage bias. While rigorous proof remains lacking, there is substantial evidence linking these observed variations to different aspects of cellular biology. Given the link between protein synthesis rates, protein concentration, and optimized growth and function (Han et al., 2014; Li et al., 2014), it is conceivable that the components of translation machinery may affect protein expression levels in a concerted fashion. In Saccharomyces cerevisiae, the estimated translational speeds determined across all genes showed a significant correlation between codon usage bias and tRNA abundances, highlighting codon usage as an optimizing factor in overall cellular efficiency (Qian et al., 2012). At the same time, the role of tRNAs as direct modulator of translation efficiency in yeast has been challenged (Pop et al., 2014).
On the other hand, microarray-based analysis of tRNA abundances in various tissues has shown a significant correlation between tRNA content and codon usage bias of highly expressed tissue-specific proteins (Dittmar et al., 2006). Given that protein synthesis rate is correlated with tRNA abundance in transgene overexpression experiments (Zouridis and Hatzimanikatis, 2008), it is further hypothesized that tRNA content may effectively regulate the rate of translation for a subset of endogenous proteins (Gustafsson et al., 2004). In contrast, measurements of in vivo translational speeds in S. cerevisiae have shown that preferentially used codons may not necessarily be translated faster (Qian et al., 2012) and may instead stem from optimization for efficiency at a cellular level rather than for translational speed. The significant role of tRNAs in regulating gene expression is further supported by recent experiments revealing that variations in active tRNA content of bacteria play an adaptive role in response to environmental cues (Subramaniam et al., 2013). Importantly, a recent study based on microarray profiling of tRNA contents in various human cell lines and samples characterized distinct tRNA signatures to correlate with proliferation and differentiation, two distinct cellular programs (Gingold et al., 2014).
It has been hypothesized that modulations in tRNA content may impact the protein expression landscape of the cell (Begley et al., 2007; Chan et al., 2010; Dittmar et al., 2006; Pavon-Eternod et al., 2009). Moreover, rare codon bias has been shown to impact Kras-driven tumorigenesis (Pershing et al., 2015). More recently, codon optimality was also reported as a major determinant of mRNA stability (Presnyak et al., 2015). Additionally, mutation of a tissue-specific tRNA expressed in the mouse nervous system was implicated as the cause of neurodegeneration (Ishimura et al., 2014). However, the regulatory consequences of tRNA modulations and their potential direct roles in gene expression control and human disease remain poorly characterized. Here, we have performed an unbiased study of tRNA abundances in malignant and non-malignant human cell lines. We find that highly metastatic sublines derived from distinct parental cancer cell populations exhibit similar modulations in their tRNA content relative to their isogenic poorly metastatic parental lines. Through loss-of-function and gain-of-function experiments, we establish a causal role for two specific tRNA species as promoters of breast cancer metastasis. Increased expression of these specific tRNAs reshapes protein expression through the direct modulation of ribosome occupancy and/or transcript stability of specific transcripts enriched for codon complementary to these tRNAs. We reveal that increased expression of a specific tRNA enhances the expression of a direct target gene downstream of the tRNA in a codon-specific manner. The downstream target(s) of this tRNA constitute novel promoters of metastasis and, in combination with their upstream regulatory tRNA, form a tRNA-activated pathway that drives cancer progression.
RESULTS
Metastatic Progression and Modulations in the tRNA Expression Landscape
To create a snapshot of the tRNA landscape in different cellular contexts, we measured the cellular content of various tRNA species in five different human cell lines (Minn et al., 2005; Tavazoie et al., 2008): a non-tumorigenic epithelial cell line (MCF10a), poorly metastatic breast cancer lines MDA-231 and CN34, and their respective highly metastatic sub-lines MDA-LM2 and CN-LM1a (Figure 1A). To do this, we developed an approach based on hybridization and subsequent ligation of complementary DNA probes. Due to their strong secondary structures and extensive base modifications, tRNAs are not suitable substrates for reverse transcription. Thus, their quantification using common cDNA-based approaches results in unpredictable biases and spurious measurements (Dittmar et al., 2006). While enzymatic removal of certain tRNA modifications and the application of highly processive reverse transcriptase enzymes provide a promising avenue for tRNA-sequencing and quantification (Zheng et al., 2015), high-quality tRNA profiling is still a need, and tRNA content and its regulatory roles remain poorly studied. Here, by relying on the hybridization and quantification of tRNA-specific probes, we have bypassed the first-strand cDNA synthesis step (Nilsson et al., 2000). Briefly, for each tRNA species, a pair of probes were designed that upon hybridization to their cognate tRNAs would provide a nick at the site of the anticodon. The nick was then repaired in a ligation reaction, giving rise to a first-strand cDNA. Biotinylation of the tRNA population and streptavidin-mediated co-precipitation steps were included to achieve a higher signal-to-noise ratio (Figures S1A–S1E; see Experimental Procedures for details). Following the successful splinted ligation of probes on their cognate tRNAs, high-throughput sequencing or qPCR can be used for relative quantification of tRNA levels (Figure 1A).
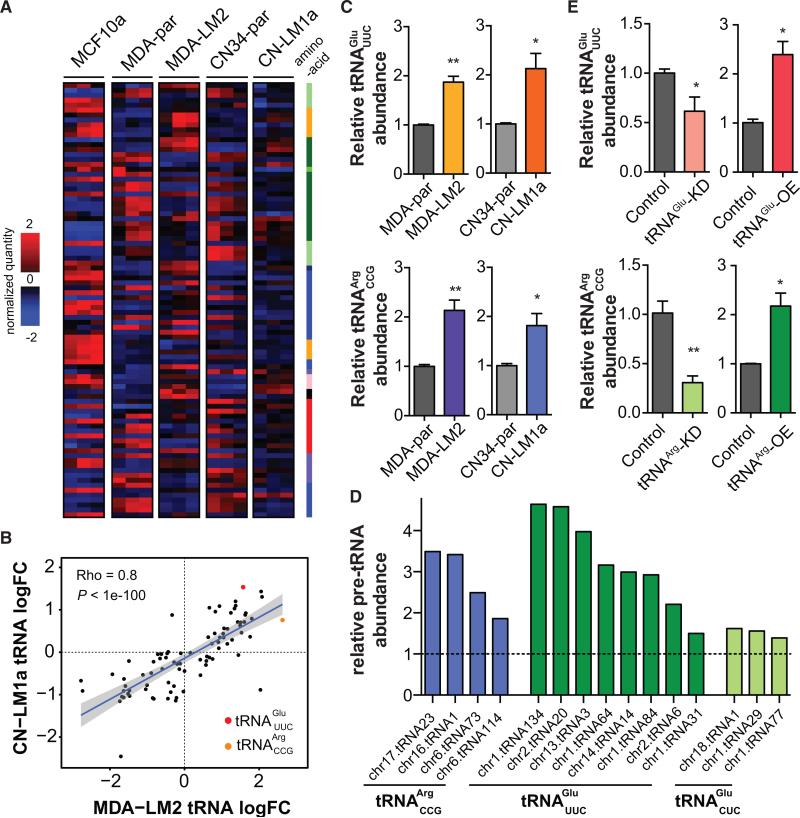
Figure 1. Transfer RNA Profiling of Metastatic and Non-metastatic Breast Cancer Lines.
(A) Whole-genome tRNA profiling was performed for MCF10a, MDA-par, MDA-LM2, CN34-par, and CN-LM1a cell lines. Hierarchical clustering was used to cluster the resulting profiles. The tRNAs are labeled based on their cognate amino acid: A, G: light green; C: green; D, E, N, Q: dark green; I, L, M, V: blue; F, W, Y: lilac; H: dark blue; K, R: orange; P: pink; S, T: red.
(B) Correlation plot for changes in tRNA levels between MDA-LM2 and CN-LM1a cells. Strong positive correlation suggests these two distinct metastatic derivatives employ similar approach in modulating tRNA levels to attain metastatic phenotypes. TRNAArgCCG and tRNAGluUUC are among the most highly upregulated in both MDA-LM2 and CN-LM1a.
(C) Quantitative PCR-based tRNA quantification validated the changes in the abundance of tRNAArgCCG and tRNAGluUUC in metastatic MDA-LM2 and CN-LM1a cells relative to their respective parental lines.
(D) Relative pre-tRNA abundances for tRNAArgCCG and tRNAGluUUC across multiple genetic loci as determined by quantitative RT-PCR. tRNAGluCUC, pre-tRNAs for which deregulated expression was not observed, were also included for comparison.
(E) TRNAGluUUC and tRNAArgCCG were successfully overexpressed and knocked down as revealed by quantitative PCR. Note that manipulation of the levels of these two tRNAs occurs within the physiological boundaries of the parental or metastatic backgrounds. One-tailed Student's t test was used to measure statistical significance between the two samples in each experiment. Error bars indicate SEM. *p < 0.05 and **p < 0.01.
TRNA profiling revealed that tRNA expression levels in breast cancer lines were different from those of non-tumorigenic cells (Figure 1A). And more importantly, while cells from the MDA-231 and CN34 populations showed distinct tRNA profiles, we observed substantial similarities between the two cell lines with respect to their differential tRNA content when comparing each parental cell line and its metastatic derivative sub-line (Figures 1A and 1B). In other words, in vivo selection of MDA-231 and CN34 parental cancer cell populations for higher metastatic capacity selected for similar modulations in tRNA abundances. Such concerted modulation of tRNA levels suggested a potentially direct role for tRNAs in promoting cancer progression.
Among the tRNAs deregulated in the metastatic cells, tRNAArgCCG and tRNAGluUUC were consistently upregulated in both MDA-LM2 and CN-LM1a highly metastatic cells relative to their parental lines (Figure 1B). We further validated this upregulation using both splinted-ligation followed by qPCR and Northern blot analysis (Figures 1C and S1F). Importantly, qRT-PCR assays for pre- tRNAArgCCG and pre- tRNAGluUUC species revealed a significant upregulation in levels of the pre-cursors of these tRNAs as well (Figure 1D). Consistent with the increased pre-tRNA and mature tRNA levels expressed in highly metastatic cells, increased genomic copy number of tRNAArgCCG and tRNAGluUUC loci were observed in highly metastatic cells (Figure S1G).
To measure the phenotypic and molecular consequences of modulated levels of these tRNAs, we first tested whether stable cell lines overexpressing or knocked down for these tRNAs could be generated. Endogenous levels of tRNAArgCCG and tRNAGluUUC in the highly metastatic MDA-LM2 background could be effectively reduced by expressing short hairpins targeting these tRNAs respectively (Figure 1E). Similarly, expression levels of these tRNAs in MDA-parental cells were enhanced upon stable integration of additional copies of these tRNAs driven by a U6 promoter (Figure 1E). It should be noted that these observed overexpression and knockdowns (~2-fold as measured by quantitative PCR-based tRNA quantification) in the levels of these tRNAs were within physiological boundaries of their levels of endogenous modulation between the parental and the in vivo-selected highly metastatic cells (Figures 1C and 1E).
tRNAGluUUC and tRNAArgCCG Promote Metastatic Progression
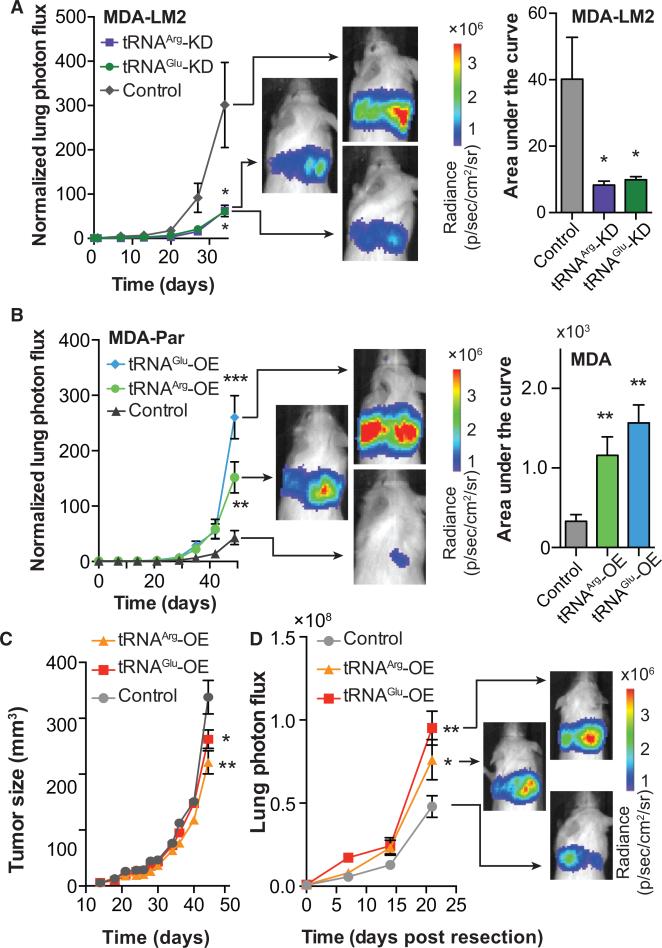
To test whether the increased expression levels of tRNAArgCCG and tRNAGluUUC play direct roles in conferring higher metastatic capacity, we employed short hairpins targeting tRNAArgCCG and tRNAGluUUC in metastatic MDA-LM2 cells. Reducing the levels of tRNAGluUUC and tRNAArgCCG by 40% and 70%, respectively, in metastatic MDA-LM2 cells (Figure 1E) to physiologically similar levels observed in poorly metastatic parental cells significantly reduced their lung colonization capacity (Figure 2A). Gross histology of extracted lungs also revealed significantly fewer metastatic nodules relative to MDA-LM2 control cells (Figure S2A). Consistent with these observations, overexpression of tRNAGluUUC and tRNAArgCCG in poorly metastatic MDA-parental cells significantly enhanced metastatic progression (Figures 2B and S2B). More strikingly, tRNA overexpressing cells exhibited enhanced orthotopic metastasis compared to control cells despite their significantly reduced primary tumor growth rates (Figures 2C and 2D). These results reveal that increased abundance of specific tRNAs can promote the metastatic phenotype of breast cancer cells.
Figure 2. tRNAGluUUC and tRNAArgCCG Promote Metastatic Breast Cancer.
(A) Bioluminescence imaging plot of lung colonization by MDA-LM2 cells expressing short hairpins targeting tRNAGluUUC, tRNAArgCCG, or a control hairpin (shControl); n = 5 in each cohort. Area-under-the-curve was also calculated for each mouse.
(B) Bioluminescence imaging plot of lung colonization by tRNAGluUUC or tRNAArgCCG overexpressing lines, as compared to control in MDA-parental cells; n = 5 in each cohort. Area-under-the-curve was also calculated for each mouse.
(C) Primary tumor growth measurement after orthotopic injection of Control, tRNAGluUUC, or tRNAArgCCG overexpressing cells into the mammary fat pads of mice; n = 5 in each cohort.
(D) Orthotopic metastasis bioluminescence imaging plot of mice after primary tumor resection; n = 5 in each cohort. For comparing lung colonization, primary tumor growth, and orthotopic metastasis assays, two-way ANOVA was used to measure statistical significance. One-tailed Mann-Whitney test was used to measure statistical significance between the areas under the curves. Error bars indicate SEM. *p < 0.05, **p < 0.01, and ***p < 0.001.
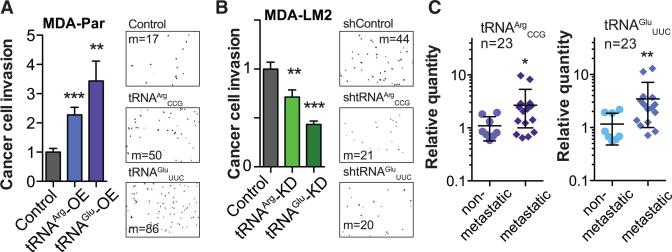
Given that cancer cell invasion is a key phenotypic attribute required for metastatic progression from the mammary gland in vivo, we asked whether modulations in the levels of tRNAArgCCG or tRNAGluUUC could affect the invasiveness of these cells. We performed in vitro cancer cell invasion assays for tRNAArgCCG or tRNAGluUUC overexpressing cells (Figure 3A) and observed significant increases in invasion capacity by cells overexpressing either of these tRNAs. Conversely, highly metastatic MDA-LM2 cells expressing shRNAs targeting either of these tRNAs exhibited significantly reduced invasive capacity (Figure 3B). The observed increase in in vitro invasion upon up-regulation of these tRNAs was not due to a general increase in proliferative capacity, as overexpression of these tRNAs actually slightly decreased in vitro proliferation rates (Figure S2C). These in vitro findings are consistent with the tumor growth and metastatic phenotypes observed in vivo and provide further support to the notion that individual tRNAs can have specific, pro-metastatic phenotypic consequences.
Figure 3. In Vitro Characterization of tRNAGluUUC and tRNAArgCCG and Their Clinical Associations with Breast Cancer Progression.
(A) Overexpression of tRNAGluUUC or tRNAArgCCG in MDA-parental cells significantly increased cancer cell invasion. Also included are representative fields from the invasion inserts along with the median number of cells observed in each cohort.
(B) Conversely, tRNAGluUUC or tRNAArgCCG knockdown in MDA-LM2 cells significantly decreased cancer cell invasion. Also included are representative fields from the invasion inserts along with the median number of cells observed in each cohort.
(C) The relative abundance of tRNAArgCCG and tRNAGluUUC in primary breast tumor samples from patients who either developed metastatic relapse (n = 15) or remained disease-free (n = 8), measured using quantitative PCR. One-tailed Mann-Whitney test was used to establish statistical significance between the two cohorts. Error bars indicate SEM. *p < 0.05, **p < 0.01, and ***p < 0.001.
To ensure the broad biological relevance of these findings and to rule out off-target effects due to shRNA expression, we performed the following experiments. First, we functionally tested the phenotypic effects of tRNA knockdown in an independent cell line—the metastatic CN-LM1a sub-line. Knockdown of each of these two tRNAs also reduced metastatic capacity of this breast cancer cell population as well (Figures S2D and S2E). We then overexpressed and knocked down each tRNA simultaneously and measured their metastatic capacity in vivo relative to control cells. Consistent with on-target effect of shRNAs, overexpressing each tRNA prevented the reduced metastatic phenotype observed upon depletion of each tRNA (Figure S3A). Lastly, to ensure that the observed phenotypes were specific to tRNAArgCCG and tRNAGluUUC and that varying the levels of another tRNA, which was not observed to be modulated in cancer cells upon selection for enhanced metastatic activity, would not manifest similar effects, we also varied the levels of tRNATyrGUA. Modulating tRNATyrGUA in both knockdown and overexpression experiments did not significantly affect the metastatic activity of MDA-LM2 cells (Figure S3B).
To further ascertain whether these associations are clinically relevant to human breast cancer progression, we quantified the levels of these tRNAs in a cohort of primary tumors that had not metastasized as well as in a set that had exhibited clinical metastasis (n = 23). We observed significant upregulations in tRNAArgCCG and tRNAGluUUC levels across the metastatic primary tumors relative to non-metastatic primary tumors (Figure 3C). These findings reveal that the expression levels of tRNAArgCCG and tRNAGluUUC in human breast cancers correlate with and predict metastatic propensity.
tRNAGluUUC and tRNAArgCCG Overexpression Impacts Transcript Stability and Translation
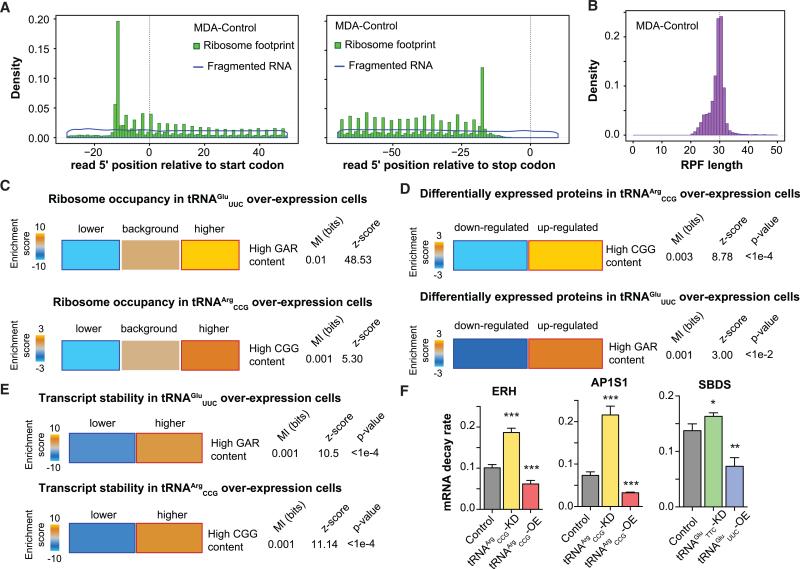
As previously mentioned, modulations in tRNA levels may impact the gene expression landscape of the cell. To assess the regulatory consequences of upregulating these tRNAs, we systematically measured their impact on post-transcriptional regulatory processes. First, we performed ribosome profiling in control as well as tRNAArgCCG and tRNAGluUUC overexpressing cells to provide a snapshot of changes in active translation (Ingolia et al., 2014). We observed a substantial positive correlation between relative ribosome occupancy of transcripts across biological replicates (Figure S4A). Importantly, the periodicity and length generally noted for ribosome protected fragments by other investigators were observed in this dataset as well (Figures 4A, 4B, S4B, and S4C). For each gene, we calculated a corrected ribosome-occupancy score to compare active translation in tRNA overexpressing cells relative to control cells (MDA-parental background; see Experimental Procedures). We subsequently asked whether the frequency of codons cognate to each overexpressed tRNA was informative of the observed changes in active translation. We observed that, in tRNAGluUUC overexpressing cells, genes with high GAR content in their coding sequence (GAA and GAG codons, since tRNAGluYUC can Wobble-base pair at the third nucleotide) were significantly enriched among those with higher relative ribosome occupancy (Figures 4C and S4D). Similarly, higher CGG content was associated with higher active translation in tRNAArgCCG overexpressing cells (Figure 4C).
Figure 4. Post-Transcriptional Consequences of tRNAGluUUC and tRNAArgCCG Upregulation.
(A) A hallmark of ribosome profiling libraries is a 3-nt periodicity. As an example, we have included the coverage of the 5′-end of reads along the coding sequence with respect to the start (left) or stop codon (right). In comparison, the total RNA library (fragmented RNA) did not exhibit this periodicity.
(B) Given the footprint of ribosomes on mRNAs, ribosome protected fragments (RPF) of ~30-nt are expected. Here, as an example, we have shown the RPF length distribution for our control samples.
(C) Genes with higher GAR and CGG contents exhibited significant enrichment among transcripts with increased ribosomal occupancy in tRNAGluUUC and tRNAArgCCG overexpressing cells, respectively.
(D) Genes with a high abundance of GAG and GAA (GAR) codons were significantly enriched among proteins significantly upregulated (corrected for their transcript changes) in tRNAGluUUC overexpression cells. Similarly, genes with higher CGG content exhibited a significant enrichment among the proteins upregulated (after correction for transcript changes) upon overexpression of tRNAArgCCG. The statistical significance of these enrichments was assessed using mutual-information calculations and associated Z score (based on randomized input vectors). Also included is the χ2 p value for the associated contingency table. The heatmap was generated using the –log of the hypergeometric p value for enrichment and log of p value for depletion (collectively termed the enrichment score). The red and dark-blue borders indicate the statistical significance of the calculated hypergeometric p values (for details, see Goodarzi et al., 2009).
(E) Whole-genome transcript stability measurements reveal significant enrichment for genes with higher GAR content among those strongly stablized in tRNAGluUUC overexpressing line. Similarly, stability of transcripts with higher CGG content is also significantly enhanced in the context of tRNAArgCCG overexpression.
(F) ERH, AP1S1, and SBDS were chosen to validate by qRT-PCR the impact overexpressing or knocking down corresponding tRNAs has on mRNA stability as a function of decay rate.
To directly assess the impact of tRNA modulations on proteomic output, we measured the expression levels of roughly 4,000 proteins using mass-spectrometry-based quantification in tRNAArgCCG and tRNAGluUUC, as well as control cells (MDA-parental background). In tRNA overexpressing cells, we identified hundreds of proteins that were significantly altered in their expression levels (Figures S4E and S4F). To correct for changes in transcript levels, we performed transcriptomic measurements in tRNAArgCCG and tRNAGluUUC overexpressing as well as control cells. We then corrected the fold-changes in protein expression levels with those of their corresponding transcripts. As shown in Figure 4D, similar to ribosome profiling results, proteins with high GAR content in their genes (GAA and GAG codons) were significantly enriched among those upregulated in the tRNAGluUUC overexpressing cells. Moreover, proteins with high CGG content showed a significant enrichment among the upregulated genes in the tRNAArgCCG overexpressing line (Figure 4D). These observations are consistent with the hypothesis that protein expression levels can be modulated as a function of their codon usage and cellular tRNA content.
In addition to a direct impact on active translation, differential ribosome occupancy may also affect other aspects of RNA life cycle, specifically RNA stability (Huch and Nissan, 2014). To test whether tRNA abundance can likewise impact transcript stability, we performed α-amanitin-mediated whole-genome mRNA stability measurements in tRNA overexpressing and control cell lines. Consistent with a positive association between translation and mRNA stability (Coller and Parker, 2005; Huch and Nissan, 2014; Muhlrad et al., 1995), we observed a general stabilization of transcripts with higher GAR and CGG content in tRNAGluUUC and tRNAArgCCG overexpressing cells, respectively (Figure 4E). We also validated these observations by measuring transcript decay rates using qRT-PCR for a set of mRNAs showing differential stability in tRNA overexpressing cells (Figure 4F). Taken together, modulations in tRNA levels can have broad regulatory consequences across the proteome of cancer cells, which can partly be explained by variations in codon usage of the target proteins.
Codon-Specific Modulation of tRNAGluUUC Downstream Targets and Clinical Associations
Given the regulatory consequences of tRNA modulations in cancer cells and their impact on metastatic capacity, we hypothesized that a set of core target transcripts may be the major drivers of the metastatic phenotype downstream of tRNAGluUUC as a global regulator. To identify these targets, we systematically combined the various whole-genome datasets we had compiled as part of this study. We specifically focused on the ribosome profiling data in tRNAGluUUC overexpressing cell line to identify potential targets that exhibited a higher rate of active translation when tRNAGluUUC was more abundant. We identified EXOSC2 and GRIPAP1 as such targets. Consistent with their higher ribosome occupancy in tRNAGluUUC overexpressing cell, these genes also displayed higher protein levels in highly metastatic cells relative to their poorly metastatic parental cells (both MDA and CN34 cell line backgrounds, data not shown). Quantitative western blots further validate the increased expression of these genes in tRNAGluUUC overexpressing background (Figure 5A). It should be emphasized that neither of these genes exhibited a significant upregulation at the transcript level (log fold-change of < 0.1 for both genes in both MDA- and CN34- backgrounds), highlighting that the observed upregulation for these genes was primarily post-transcriptionally mediated.
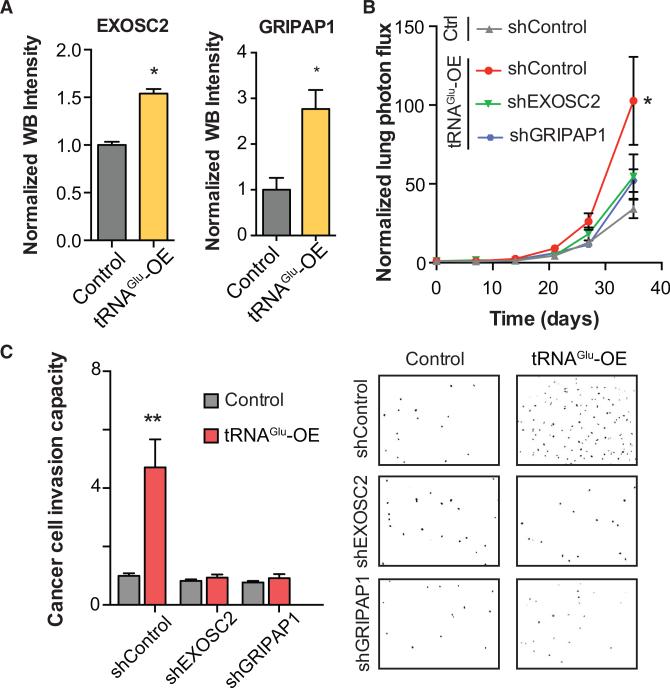
Figure 5. Variations in tRNAGluUUC Levels Post-Transcriptionally Modulate Expression of Breast Cancer Metastasis Promoters EXOSC2 and GRIPAP1.
(A) Endogenous EXSOC2 and GRIPAP1 protein levels as measured by quantitative western blotting (see Experimental Procedures) in tRNAGluUUC overexpressing or control lines.
(B) Bioluminescence imaging plot of lung colonization by EXOSC2 and GRIPAP1 knocked-down cells in MDA-parental overexpressing tRNAGluUUC relative to control cells expressing a control hairpin, shControl; n = 5 in each cohort. Two-way ANOVA was used to measure statistical significance.
(C) Knockdown of EXOSC2 or GRIPAP1 abrogated the enhanced invasion capacity of tRNAGluUUC overexpressing line. Also included are representative fields from the invasion inserts along with the median number of cells observed in each cohort. Error bars indicate SEM. *p < 0.05 and **p < 0.01 by one-tailed Student's t test.
To test whether these genes impact metastatic progression downstream of tRNAGluUUC, we conducted epistasis experiments in xenograft mouse models. EXOSC2 and GRIPAP1 were stably knocked down in the context of tRNAGluUUC overexpression, as well as in the control line, and these cells were injected into the tail-veins of NSG mice. Reduced EXOSC2 and GRIPAP1 expression levels substantially abrogated the enhanced metastatic colonization outcome caused by tRNAGluUUC overexpression (Figure 5B and data also shown as separated plots in Figure S5). Consistent with this, the in vitro invasiveness of tRNAGluUUC overexpressing line was also dramatically decreased upon silencing of EXOSC2 and GRIPAP1 (Figure 5C). It should be noted that reduced lung colonization and invasiveness were not observed in the control MDA-parental background upon EXOSC2 or GRIPAP1 depletion. This suggests that EXOSC2 and GRIPAP1 are downstream targets of tRNAGluUUC overexpression and that in the context of elevated tRNAGluUUC, cancer cells can exploit these proteins to elicit a pro-metastatic phenotype. Furthermore, silencing EXOSC2 and GRIPAP1 did not completely abolish the metastatic phenotype of tRNAGluUUC overexpression, supporting the likely possibility that additional genes may operate downstream of this tRNA in driving metastasis.
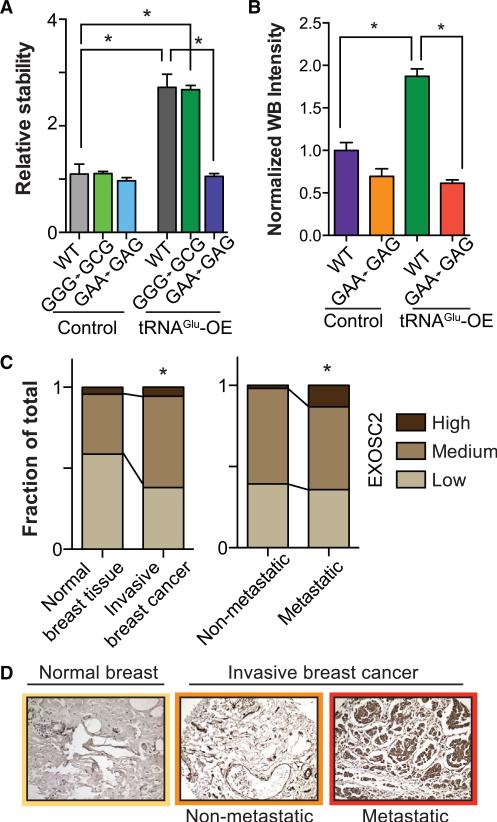
To elucidate the importance of codon specificity on the effect of tRNAGluUUC on its targets, we performed a codon mutagenesis experiment in which every preferred cognate GAA codon in the EXOSC2 coding sequence was switched to the synonymous GAG codon. As a negative control, we instead mutated every instance of a non-deregulated codon, Gly-GGG to Gly-GGC (see Figure S6A and Methods for details). Transcript stability as-says for exogenously transfected wild-type, Gly-mutated, and Glu-mutated versions of EXOSC2 revealed significant loss of the stabilizing effect of tRNAGluUUC overexpression in the GAA-to-GAG mutant (Figure 6A). Reduction in protein expression of the GAA-to-GAG (Glu) codon mutated transcript, and not of the GGG-to-GGC (Gly) codon mutated transcript (data not shown), further validates the codon-specific mechanism through which tRNAGluUUC confers its effect (Figures 6B, S6B, and S6C). These mutagenesis studies establish the direct interaction of a specific tRNA with its downstream target of regulation.
Figure 6. Codon-Specific Modulation of EXOSC2 and Its Clinical Association.
(A) Relative transcript stability measured by qRT-PCR (see Experimental Procedures) of exogenous wild-type, GGG-to-GCG (Gly) codon mutated, and GAA-to-GAG (Glu) codon mutated transcript in control and tRNAGluUUC overexpressing backgrounds. While overexpression of tRNAGluUUC significantly stabilized wild-type and GGG-to-GCG (negative control) transcripts, such an effect was absent when the specific cognate Glu codons were mutated.
(B) Quantitative western blot demonstrated similar loss of translational enhancement brought about by overexpressing tRNAGluUUC when its cognate codons were mutated. Error bars indicate SEM. *p < 0.05 by one-tailed Student's t test.
(C) Stacked bars representing the fraction of tissue samples from TMA with respectively low, medium, and high intensity of EXOSC2 in normal breast tissues and invasive breast cancer tissues (n = 46 and 160, respectively). Also shown are fractions of tissues of different EXOSC2 intensity in breast cancer from patients without metastasis and those with detected metastasis in distant organs (n = 107 and 53, respectively). Hypergeometric p values were calculated to assess the significance of the increase in the frequency of samples with higher intensities; *p < 0.05.
(D) Shown are representative tissue-microarray immunohistochemical images of stained tissues of median score from normal breast, non-metastatic invasive breast cancer, and metastatic breast cancer tissues. Higher EXOSC2 intensity positively correlated with disease progression stage.
Because EXOSC2 and GRIPAP1 have not been previously implicated in breast cancer metastasis, we sought clinical as sociation evidence for our experimental findings. Although we could not establish specificity for multiple commercially available GRIPAP1 antibodies, we identified an EXOSC2 antibody, which exhibited specificity in fixed tissue immunohistochemistry (IHC). Immunohistochemical staining of breast cancer progression tissue microarrays (TMA) for EXOSC2 revealed a positive association between its expression and clinical breast cancer progression. EXOSC2 protein expression was significantly higher in invasive breast cancer relative to normal breast tissues (Figures 6C, 6D, and S6D). More importantly, EXOSC2 protein levels were also significantly higher in primary tumors of patients with distant metastases compared to earlier stage tumors (Figures 6C, 6D, and S6D). These clinical association results not only support our in vitro and in vivo findings regarding the role of tRNA modulation in promoting metastasis but also support this tRNA-based pathway discovery approach as a means for identifying post-transcriptionally regulated targets that might have been otherwise missed by traditional transcriptomic profiling methods.
Association among tRNA Preference, Ribosomal Occupancy, and Protein Expression
Given their crucial roles in translation, deregulations in tRNA abundance could strongly impact the protein expression landscape of the cell. While tRNAArgCCG and tRNAGluUUC were significantly upregulated in highly metastatic cells, the modifications in the tRNA profiles of highly metastatic cells relative to poorly metastatic cells were not limited to these two tRNAs. To test the consequences of the broader tRNA modulations, we first asked whether the observed changes in tRNA content, across all measured tRNA species, were informative of expression levels of proteins based on their coding sequences alone. As mentioned before, differential tRNA expression levels can impact translational outcome by affecting the abundance of ribosome-bound transcripts. We thus sought to quantify the likely impact of variations in tRNA content on active translation and protein expression given the frequency of each codon for every gene. For this, we defined tRNA preference scores as the sum of changes in the tRNA content in each background (log-ratio in MDA-231 and CN34 backgrounds) across all codons of a given gene (MDA- and CN- preference scores; see Experimental Procedures). In this scoring scheme, genes whose codons are favored, based on the upregulation of their cognate tRNAs, are assigned positive scores, and those whose codons on average are associated with downregulated tRNAs are assigned negative scores.
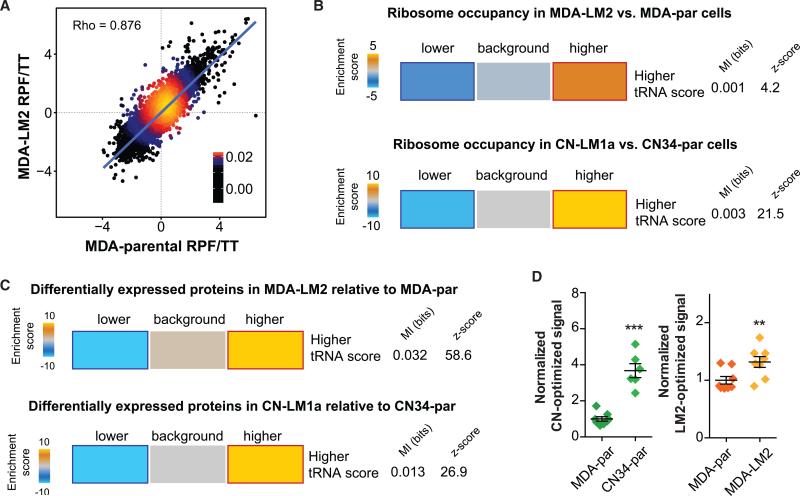
We performed whole-genome ribosome profiling on poorly metastatic parental lines, MDA-par and CN34-par, as well as their highly metastatic derivatives, MDA-LM2 and CN34-LM1a. The ribosome-protected fragments (RPF) were normalized to total RNA (TT) for every cell line to account for differences in gene expression. As expected, the RPF/TT ratio between parental and derivative lines, as well as between biological duplicates, demonstrated a strong positive correlation (Figures 7A and S7A). TRNA preference scores of every gene were then overlapped with its respective corrected ribosome footprints. Interestingly, transcripts with higher tRNA preference scores were strongly enriched among those bound more by ribosomes (Figure 7B). We subsequently measured differential protein levels between the metastatic and parental lines using stable-isotope labeling by amino acids in cell culture (SILAC, Ong et al., 2003; also see Figure S7B). To correct for protein expression changes due to variations in transcript abundances, we normalized the change in protein expression for each gene to its transcript level in each background. In accordance with the increased ribosomal occupancy, we observed a significant enrichment of genes with high tRNA preference scores among those translationally upregulated in the highly metastatic sub-lines in both MDA-231 and CN34 backgrounds (Figures 7C). Importantly, consistent with the observed correlation between the changes in tRNA abundance in the MDA and CN34 backgrounds, we also observed a highly significant correlation between tRNA preference scores calculated across all coding sequences (Figure S7C).
Figure 7. tRNA Preference Scores Were Informative of Differential Ribosome Occupancy and Protein Expression.
(A) As an example, we have shown the linear regression of MDA-parental and MDA-LM2 ribosome protected fragment to total RNA ratio (RPF/TT). RPF reads were normalized to TT reads to correct for variation in transcript expression in each cell line.
(B) Genes with positive tRNA preference score (based on derivative versus parental tRNA profiling results) were significantly enriched among transcripts with higher corrected ribosome occupancy values in both MDA-LM2 and CN-LM1a relative to MDA-par and CN34-par, respectively (see Experimental Procedures). In other words, coding sequences with more favorable codon content, based on changes in tRNA abundance between parental cells and their highly metastatic derivatives, exhibited a more active translation.
(C) Genes with positive tRNA preference score were significantly enriched among the proteins upregulated in MDA-LM2 cells and CN-LM1a compared to MDA-par and CN34-par, respectively. The significance of these enrichments was determined by calculating mutual-information values and their associated Z scores (based on randomized input values). Also included is the χ2 p value for the associated contingency table. The enrichment score, based on which the heatmap was generated, is defined as the –log of hypergeometric p value for enrichment (gold) and log of p value for depletion (blue). The red and dark-blue borders indicate the statistical significance of the calculated hypergeometric p values (Goodarzi et al., 2009).
(D) Normalized relative luciferase activity for CN-optimized and LM2-optimized luciferase constructs.
Our findings reveal that differential tRNA expression is informative of changes in translational landscapes. However, to show that there in fact may be a causal link between global tRNA content and protein expression, we took advantage of two synthetic constructs based on the coding sequence of Renilla luciferase. We designed a comparative luciferase coding sequence by scanning the gene and inserting the codon variant whose cognate tRNA had the highest relative expression in CN34 versus MDA-parental cells (CN-optimized luciferase). We should emphasize that the chosen codons were not the ones with highest tRNA levels but rather the ones with highest CN34 to MDA-231 ratios. We similarly constructed an LM2-optimized luciferase coding sequence comparing MDA-LM2 tRNA levels to those of the parental MDA-231. We then measured luciferase activity of the CN-optimized construct in both MDA- and CN34-parental cells. Interestingly, we observed a substantially higher luciferase signal in CN34-parental cells (Figure 7D). We next focused on cancer cell populations originating from the same patient (MDA and MDA-LM2 cells). Expression of the LM2-optimized construct similarly showed a higher signal in MDA-LM2 line relative to MDA-parental cells (Figure 7D). The magnitude of the change also supports the level of variation we had initially observed with the tRNA profile in CN34 being more distinct from the MDA lines and MDA-231 and MDA-LM2 showing more similar tRNA profiles. These findings reveal that, even within the same cancer type, the proteomic output of cells obtained from two distinct patients is constrained by the cells’ tRNA contents. Moreover, even within a given patient, cancer subpopulations’ expression output can be significantly impacted by the tRNA codon landscape within cell sub-populations.
DISCUSSION
Collectively, our findings demonstrate that changes in tRNA abundance can modulate protein expression in the cell and that cancer cells can evolve to fine-tune the expression of multiple promoters of cancer progression through modulations in tRNA levels. Consistent with previous reports (Pavon-Eternod et al., 2009), we observed that the tRNA profiles of breast cancer lines can markedly differ from non-cancerous epithelial cells. More importantly, the tRNA profile is further modified en route to higher metastatic capacity. TRNAArgCCG and tRNAGluUUC were significantly upregulated in breast cancer lines relative to epithelial cells, but this increase was further augmented in metastatic lines relative to their parental population. Moreover, we find that the levels of these tRNAs are higher in metastatic human tumors relative to non-metastatic ones. We have shown that the increased expression of these tRNAs contributes to the metastatic phenotype. We propose that modulations in specific tRNA levels enhance the translational efficiency of genes that are promoters of metastasis in a codon-dependent manner. A detailed analysis of differential protein expression and its relationship to codon preference revealed a modest albeit highly significant correlation between these parameters.
Variations in the abundance of tRNA isoacceptors impact the rate of translation in vivo (Fredrick and Ibba, 2010). Transient pauses at codons with rare cognate tRNAs can also affect protein folding and translocation (Zhang and Ignatova, 2011). As a result, even synonymous substitutions can alter the rate of translation and impact protein dynamics (Kirchner and Ignatova, 2015). In humans, tissue-specific tRNA expression mirrors the codon usage of tissue-specific proteins, indicating an association between the active tRNA pool and translational demand (Dittmar et al., 2006). Recently, it was also hypothesized that the translational machinery can be locked into a “proliferation” versus a “differentiation” program (Gingold et al., 2014). Our results, however, indicate that changes in tRNA expression states in the context of pathophysiology are more dynamic and their modulations can have specific phenotypic consequences such as enhanced invasiveness. As previously mentioned, the role of global tRNA content in translation efficiency and the role of codon usage in concerted regulation of protein expression have been debated (dos Reis et al., 2004). Based on our results, our model sides with those reporting a significant selection on codon usage adaptation with respect to tRNA content (Comeron, 2004; Gingold et al., 2014). It was based on this tRNA program that we successfully identified and functionally validated novel promoters of metastatic colonization.
In many organisms, such as bacteria, yeast, and even human, tRNA gene copy number is correlated with codon usage of highly expressed genes (Novoa and Ribas de Pouplana, 2012; Novoa et al., 2012). However, in yeast, modulating the levels of a rare tRNAArgCCU did not impact elongation rate or translational efficiency, while tRNAThrUGU knockdown, achieved by deleting three of the four copies of the heavily used tRNAThrUGU, had a modest effect on efficiency (Pop et al., 2014). Thus, the observed correlation between codon bias and efficiency may have arisen from the selection on highly expressed genes to utilize translation machinery efficiently (Pop et al., 2014). In our data, the association between tRNA abundance and codon usage were more pronounced among highly expressed genes. For example, limiting our analysis to roughly 300 genes with one SD above average expression (based on whole-transcriptomic measurement), a significant correlation between codon CGG content and protein levels in tRNAArgCCG overexpressing cells (Rho = 0.12, p = 0.03) was observed—a correlation that is otherwise less pronounced across all the detected proteins (Rho = 0.06). Another point to consider is that higher levels of these specific tRNAs were specifically selected for in the context of pathological disease progression. It is possible that modulating other tRNAs, as was the case for tRNATyr, will not result in significant changes in protein expression levels or may not be tolerated by the cells under study. Furthermore, it is plausible that the link between tRNA levels and translational efficiency is more pronounced in mammalian cells selected in vivo than it is in yeast cells grown in culture. More importantly, in our data, codon usage explains a fraction of protein expression changes, implying that (1) tRNA levels may affect protein expression in ways other than direct translation (Lee et al., 2006) and stability, and/or (2) positive and negative feedback loops may amplify and propagate the regulatory consequences of modulations in tRNA levels.
Taken together, our study puts forth the notion that cancer cells, in addition to many known regulatory mechanisms, exhibit tRNA landscape modulations that modify the expression of promoters of cancer progression. Our work reveals that specific tRNAs can form “inducible” pathways with their direct target transcripts, which are enriched for their cognate codons. Such target transcripts can become stabilized in the context of their favored tRNA content or can be more effectively translated—ultimately yielding greater protein output. It should be noted that, while we have focused on breast cancer metastasis, this approach based on tRNA profiling is general in concept and can be employed to study other diseases and can be extended to other models and species, as well as to developmental processes where similar tRNA modulations may govern developmental gene expression programs and phenotypic outcomes.
EXPERIMENTAL PROCEDURES
Transfer RNA Profiling
For each family of mature tRNAs (with introns removed and containing terminal CCA) with a similar consensus sequence and a common anticodon, a pair of probes is designed so that, upon annealing to the complementary tRNA, the resulting nick in the DNA-RNA hybrid is located at the site of the anticodon. We designed a total of 67 probe-pairs to cover the majority of cytosolic tRNAs. The downstream probes were 5′-phosphorylated to enable enzymatic ligation. Small-RNAs extracted from cells were subjected to deacylation and biotinylation, followed by hybridization with the probe library. The DNA/RNA hybrids were then subjected to overnight ligation with T4 DNA ligase. MyOne-C1 Streptavidin Dynabeads (Invitrogen) were then used to purify biotinylated DNA/RNA hybrids, and the ligated probes were then eluted after incubation with RNase H and RNase A followed by incubation with an elution buffer (50 mM Tris [pH 8], 10 mM EDTA, 1% SDS; incubate at 65°C for 30 min with intermittent vortexing). Eluted probes were subsequently cleaned up and minimally PCR amplified (12–15 cycles) for high-throughput sequencing.
Low-Throughput tRNA Quantification
The preparation of samples for low-throughput tRNA quantification was identical to the high-throughput tRNA profiling protocol described above, with the following exception: (1) instead of using a library of probes, the RNA samples were hybridized to a single probe-pair (matching the tRNA of interest); (2) the amplification step is replaced with quantitative PCR using SYBR Green (Life Technologies) per manufacturer's instructions.
Whole-Genome Ribosomal Occupancy Profiling
The procedure was performed with Truseq Ribo Profile for mammalian cells (Illumina) per manufacturer's instructions. An input of 50 × 106 cells were harvested for each replicate (biological duplicates for every cell line profiled), and libraries were sequenced using Illumina Nextseq 500 at the Rockefeller Genomics Center.
Codon-Specific Mutational Assays
Wild-type and mutated versions of EXOSC2 coding sequences were synthesized by IDT. Every Glu-GAA codon was mutated to Glu-GAG. As a negative control, another construct in which every Gly-GGG codon was mutated to Gly-GGC was used (Figure S6A). Gly-GGG and Gly-GGC codons were chosen because there was no significant difference in their respective tRNA levels based on our high-throughput tRNA profiling analysis. Each synthetic coding sequence also contained a 3′-FLAG-tag. The constructs were then cloned into the psiCHECK2 backbone (replacing synthetic Renilla Luciferase gene) together with an upstream Tetracycline-response element.
Animal Studies
All mouse studies were conducted according to a protocol approved by the Institutional Animal Care and Use Committee (IACUC) at the Rockefeller University.
Supplementary Material
Highlights.
Specific tRNAs are upregulated in highly metastatic breast cancer cells
TRNAs promote stability and translation of transcripts enriched for their codons
TRNAGluUUC drives metastasis by directly upregulating EXOSC2 and enhancing GRIPAP1
In Brief
A new tRNA profiling method reveals that specific tRNAs are upregulated in metastatic breast cancer cells and drive metastasis by enhancing stability and translation of transcripts enriched for their cognate codons.
ACKNOWLEDGMENTS
We thank C. Alarcon for technical help in performing Northern blot experiments. We also thank P. Freddolino, C. Alarcon, H. Najafabadi, and A. Nguyen for comments on previous versions of this manuscript. We are grateful to C. Zhao, C. Lai, and N. Nnatubeugo of the Rockefeller Genomics Resource Center for assistance with next-generation RNA sequencing and microarray profiling. H.G. was previously supported by a Ruth L. Kirschstein National Research Service Award (T32CA009673) and is currently supported by an NIH Pathway to Independence Award (K99CA194077). S.F.T. is a Department of Defense Era of Hope Scholar and a Department of Defense Breast Cancer Collaborative Scholars and Innovators Award recipient.
Footnotes
AUTHOR CONTRIBUTIONS
H.G. and S.F.T. conceived the project. S.F.T. supervised all research. S.F.T., H.G., and H.C.B.N. wrote the manuscript. H.G., H.C.B.N., S.Z., B.D.D., and H.M. designed, performed, and analyzed the experiments.
ACCESSION NUMBERS
The data for high-throughput sequencing and microarray profiling experiments are deposited at GEO under the accession number GSE77401.
SUPPLEMENTAL INFORMATION
Supplemental Information includes Supplemental Experimental Procedures and seven figures and can be found with this article online at http://dx.doi.org/10.1016/j.cell.2016.05.046.
REFERENCES
- Begley U, Dyavaiah M, Patil A, Rooney JP, DiRenzo D, Young CM, Conklin DS, Zitomer RS, Begley TJ. Trm9-catalyzed tRNA modifications link translation to the DNA damage response. Mol. Cell. 2007;28:860–870. doi: 10.1016/j.molcel.2007.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan CT, Dyavaiah M, DeMott MS, Taghizadeh K, Dedon PC, Begley TJ. A quantitative systems approach reveals dynamic control of tRNA modifications during cellular stress. PLoS Genet. 2010;6:e1001247. doi: 10.1371/journal.pgen.1001247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coller J, Parker R. General translational repression by activators of mRNA decapping. Cell. 2005;122:875–886. doi: 10.1016/j.cell.2005.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comeron JM. Selective and mutational patterns associated with gene expression in humans: influences on synonymous composition and intron presence. Genetics. 2004;167:1293–1304. doi: 10.1534/genetics.104.026351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dever TE, Green R. The elongation, termination, and recycling phases of translation in eukaryotes. Cold Spring Harb. Perspect. Biol. 2012;4:a013706. doi: 10.1101/cshperspect.a013706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dittmar KA, Goodenbour JM, Pan T. Tissue-specific differences in human transfer RNA expression. PLoS Genet. 2006;2:e221. doi: 10.1371/journal.pgen.0020221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- dos Reis M, Savva R, Wernisch L. Solving the riddle of codon usage preferences: a test for translational selection. Nucleic Acids Res. 2004;32:5036–5044. doi: 10.1093/nar/gkh834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond DA, Wilke CO. Mistranslation-induced protein mis-folding as a dominant constraint on coding-sequence evolution. Cell. 2008;134:341–352. doi: 10.1016/j.cell.2008.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fredrick K, Ibba M. How the sequence of a gene can tune its translation. Cell. 2010;141:227–229. doi: 10.1016/j.cell.2010.03.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gingold H, Tehler D, Christoffersen NR, Nielsen MM, Asmar F, Kooistra SM, Christophersen NS, Christensen LL, Borre M, Sørensen KD, et al. A dual program for translation regulation in cellular proliferation and differentiation. Cell. 2014;158:1281–1292. doi: 10.1016/j.cell.2014.08.011. [DOI] [PubMed] [Google Scholar]
- Goodarzi H, Elemento O, Tavazoie S. Revealing global regulatory perturbations across human cancers. Mol. Cell. 2009;36:900–911. doi: 10.1016/j.molcel.2009.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustafsson C, Govindarajan S, Minshull J. Codon bias and heterologous protein expression. Trends Biotechnol. 2004;22:346–353. doi: 10.1016/j.tibtech.2004.04.006. [DOI] [PubMed] [Google Scholar]
- Han K, Jaimovich A, Dey G, Ruggero D, Meyuhas O, Sonenberg N, Meyer T. Parallel measurement of dynamic changes in translation rates in single cells. Nat. Methods. 2014;11:86–93. doi: 10.1038/nmeth.2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huch S, Nissan T. Interrelations between translation and general mRNA degradation in yeast. Wiley Interdiscip. Rev. RNA. 2014;5:747–763. doi: 10.1002/wrna.1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia NT, Brar GA, Stern-Ginossar N, Harris MS, Talhouarne GJ, Jackson SE, Wills MR, Weissman JS. Ribosome profiling reveals pervasive translation outside of annotated protein-coding genes. Cell Rep. 2014;8:1365–1379. doi: 10.1016/j.celrep.2014.07.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishimura R, Nagy G, Dotu I, Zhou H, Yang XL, Schimmel P, Senju S, Nishimura Y, Chuang JH, Ackerman SL. RNA function. Ribo-some stalling induced by mutation of a CNS-specific tRNA causes neurode-generation. Science. 2014;345:455–459. doi: 10.1126/science.1249749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchner S, Ignatova Z. Emerging roles of tRNA in adaptive translation, signalling dynamics and disease. Nat. Rev. Genet. 2015;16:98–112. doi: 10.1038/nrg3861. [DOI] [PubMed] [Google Scholar]
- Lee JW, Beebe K, Nangle LA, Jang J, Longo-Guess CM, Cook SA, Davisson MT, Sundberg JP, Schimmel P, Ackerman SL. Editing-defective tRNA synthetase causes protein misfolding and neurode-generation. Nature. 2006;443:50–55. doi: 10.1038/nature05096. [DOI] [PubMed] [Google Scholar]
- Li GW, Burkhardt D, Gross C, Weissman JS. Quantifying absolute protein synthesis rates reveals principles underlying allocation of cellular resources. Cell. 2014;157:624–635. doi: 10.1016/j.cell.2014.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minn AJ, Gupta GP, Siegel PM, Bos PD, Shu W, Giri DD, Viale A, Olshen AB, Gerald WL, Massagué J. Genes that mediate breast cancer metastasis to lung. Nature. 2005;436:518–524. doi: 10.1038/nature03799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhlrad D, Decker CJ, Parker R. Turnover mechanisms of the stable yeast PGK1 mRNA. Mol. Cell. Biol. 1995;15:2145–2156. doi: 10.1128/mcb.15.4.2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson M, Barbany G, Antson DO, Gertow K, Landegren U. Enhanced detection and distinction of RNA by enzymatic probe ligation. Nat. Biotechnol. 2000;18:791–793. doi: 10.1038/77367. [DOI] [PubMed] [Google Scholar]
- Novoa EM, Ribas de Pouplana L. Speeding with control: codon usage, tRNAs, and ribosomes. Trends Genet. 2012;28:574–581. doi: 10.1016/j.tig.2012.07.006. [DOI] [PubMed] [Google Scholar]
- Novoa EM, Pavon-Eternod M, Pan T, Ribas de Pouplana L. A role for tRNA modifications in genome structure and codon usage. Cell. 2012;149:202–213. doi: 10.1016/j.cell.2012.01.050. [DOI] [PubMed] [Google Scholar]
- Ong SE, Kratchmarova I, Mann M. Properties of 13C-substituted arginine in stable isotope labeling by amino acids in cell culture (SILAC). J. Proteome Res. 2003;2:173–181. doi: 10.1021/pr0255708. [DOI] [PubMed] [Google Scholar]
- Pavon-Eternod M, Gomes S, Geslain R, Dai Q, Rosner MR, Pan T. tRNA over-expression in breast cancer and functional consequences. Nucleic Acids Res. 2009;37:7268–7280. doi: 10.1093/nar/gkp787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pershing NL, Lampson BL, Belsky JA, Kaltenbrun E, MacAlpine DM, Counter CM. Rare codons capacitate Kras-driven de novo tumor-igenesis. J. Clin. Invest. 2015;125:222–233. doi: 10.1172/JCI77627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plotkin JB, Kudla G. Synonymous but not the same: the causes and consequences of codon bias. Nat. Rev. Genet. 2011;12:32–42. doi: 10.1038/nrg2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pop C, Rouskin S, Ingolia NT, Han L, Phizicky EM, Weissman JS, Koller D. Causal signals between codon bias, mRNA structure, and the efficiency of translation and elongation. Mol. Syst. Biol. 2014;10:770. doi: 10.15252/msb.20145524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Presnyak V, Alhusaini N, Chen YH, Martin S, Morris N, Kline N, Olson S, Weinberg D, Baker KE, Graveley BR, Coller J. Codon optimality is a major determinant of mRNA stability. Cell. 2015;160:1111–1124. doi: 10.1016/j.cell.2015.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian W, Yang JR, Pearson NM, Maclean C, Zhang J. Balanced codon usage optimizes eukaryotic translational efficiency. PLoS Genet. 2012;8:e1002603. doi: 10.1371/journal.pgen.1002603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah P, Gilchrist MA. Explaining complex codon usage patterns with selection for translational efficiency, mutation bias, and genetic drift. Proc. Natl. Acad. Sci. USA. 2011;108:10231–10236. doi: 10.1073/pnas.1016719108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramaniam AR, Pan T, Cluzel P. Environmental perturbations lift the degeneracy of the genetic code to regulate protein levels in bacteria. Proc. Natl. Acad. Sci. USA. 2013;110:2419–2424. doi: 10.1073/pnas.1211077110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavazoie SF, Alarcón C, Oskarsson T, Padua D, Wang Q, Bos PD, Gerald WL, Massagué J. Endogenous human microRNAs that suppress breast cancer metastasis. Nature. 2008;451:147–152. doi: 10.1038/nature06487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G, Ignatova Z. Folding at the birth of the nascent chain: coordinating translation with co-translational folding. Curr. Opin. Struct. Biol. 2011;21:25–31. doi: 10.1016/j.sbi.2010.10.008. [DOI] [PubMed] [Google Scholar]
- Zheng G, Qin Y, Clark WC, Dai Q, Yi C, He C, Lambowitz AM, Pan T. Efficient and quantitative high-throughput tRNA sequencing. Nat. Methods. 2015;12:835–837. doi: 10.1038/nmeth.3478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zouridis H, Hatzimanikatis V. Effects of codon distributions and tRNA competition on protein translation. Biophys. J. 2008;95:1018–1033. doi: 10.1529/biophysj.107.126128. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.