Abstract
Background
We screened RARβ methylation in primary glioblastoma multiforme (GBM) and the results were evaluated based on the clinical data and treatment type.
Objective
The objective of this study was to find new areas for the usage of MS-HRM applications in the determination of methylation levels in primary GBM samples and it shows the association of RARβ methylation with the clinical outcome.
Methods
In our study, tumor samples were collected during surgical resection by the Department of Neurosurgery. The clinical and radiologic data was carefully reviewed, compared, and evaluated with the histological results. The methylation status of RARβ was determined by using MS-HRM.
Results
RARβ gene methylation was detected in 24 out of 40 cases (60%), with different quantitative methylation levels. The mean survival time was 19 months form ethylated cases and 15 months for the non-methylated cases. The survival time of the patients who received treatment was 25 months and the survival time of the patients who received radiotherapy alone or where no treatment protocol applied was 15–20 months. Therefore, a significant difference in survival rates has been observed (P<0.05). This study indicates a potential prognostic value for GBM treatment planning.
Conclusion
Our study is the first study to investigate RARβ methylation in primary GBMs. We conclude that the RARβ gene could be a new prognostic and predictive candidate marker to designate the treatment protocol for primary GBMs.
Keywords: RARβ, primary glioblastoma multiforme, methylation, MS-HRM
Introduction
Glioblastoma (WHO grade IV), or glioblastoma multiforme (GBM), is the most frequent and malignant form of brain tumor, accounting for approximately 51% of all primary gliomas (CBTRUS, 2010). The median survival time is 12 months. Depending on clinical and biological differences, GBMs can be divided into two distinct subtypes: primary (or de novo) glioblastomas that develop without the presence of any precursor neoplastic lesion and manifest after a short clinical history; and secondary glioblastomas that develop from lower grade glial tumors13.
Methylation is one of the most known and studied epigenetic mechanisms. It is the oldest and most widely used epigenetic mechanism used to identify expressions of the cancer specific genes. Many genes in glioblastomas have epigenetic characteristics that include hypermethylation of the MGMT, RASSF1A, p16/INK4A and p15/INK4. These hypermethylated genes have been reported as a marker for prognosis, treatment protocol selection and survival for GBM patients1,7.
Abnormal DNA methylation of brain tumors was first discovered 20 years ago and previous methylation studies for candidate genes in brain tissue have concentrated on:
Cell cycle genes that include: RB, CDKN2A, CDKN2B, p73, and PTEN
Genes which are important in cell-cell interactions that include: EMP3, E-Cadherin and RASSF1A
DNA repair and drug resistance genes that include: MGMT, SOX10, apoptosis genes, DAPkinase, angiogenesis genes THSB1
However, the impact of methylation is still under debate1,4,14.
RARβ receptors bind with retinoic acid, which is a biologically active form of vitamin A. RARβ is important for the embryonic period during morphogenesis, cell growth and differentiation. The expression of RARβ shows diversity and is expressed at different times during the embryonic development of a mouse's nervous system8. The methylation of the RARβ gene has been reported in intracranial ependymomas during the childhood period and in choroid plexus tumors14. However, the role of the hypermethylated RARβ gene has to this point not been associated with pathogenesis of GBMs. The RARβ gene methylation within GBMs was only reported by Piperi and colleagues and detected 58.8% RARβ methylation in 23 cases of grade II-IV tumors. A positive correlation between RARβ methylation and tumor grade in the GBMs was demonstrated and they also showed an association between the RARβ methylation and the aggressive phenotype in the primary GBM11. However, there is not sufficient data regarding the role of RARβ in GBM.
In our study, we aimed to determine the relationship between the methylation of the tumor suppressor gene RARβ with tumor growth, treatment response and survival time in GBM. The additional purpose of this study was to determine the effectiveness of the Methylation-Specific High Resolution Melting (MS-HRM) method for evaluation of the methylation, which is a newly developed application. According to our knowledge, there has been only one publication in the literature regarding RARβ methylation in GBM cases. Therefore, our study group will form a basis for global literature for determining the RARβ methylation frequency in GBM patients.
Materials and method
We studied the frequency of RARβ methylation in a series of 40 primary GBM patients in the Turkish population according to patient age, gender, GBM type and survival time. Tumor samples were collected during surgical resection by the Department of Neurosurgery. Informed consent was obtained from each of the patients and the study was approved by the local ethics committee. Both the clinical and radiologic data was carefully reviewed, compared, and correlated with the histopathological results. Clinical data included gender, date of birth, Karnofsky performance score (KPS) and type of treatment (surgical resection — gross total, subtotal, partial, as per radiologic report; radiotherapy and/or chemotherapy). The samples included frozen tissues, which were collected upon surgical removal. Our study group included female and male patients, where the average age of the women was 51.71 ± 3.98 and the average age of the men was 58.07 ± 2.71.
The DNA isolation and bisulfate modification methods were performed as described previously6. A Light-Cycler ® 480 real time PCR (Roche) device was used during implementation of the processes at this stage. PCR amplification and high resolution melting analysis was conducted using the “LightCycler ® 480 High Resolution Melting Master” kit, which is compatible with the device, and was applied according to the manufacturer's protocol. In our study, we examined the methylation pattern of the RARβ gene by the below primer sequences:
5′-TCGAGAACGCGAGCGATTCG-3′ (sense)
methylated RARβ primer
5′-GACCAATCCAACCGAAACGA-3′ (antisense)
5′-TTGAGAATGTGAGTGATTTGA-3′ (sense)
unmethylated RARβ primer
5′-AACCAATCCAACCAAAACAA-3′ (antisense)
A mixture was prepared for the RARβ promoter methylation analysis using the DNA samples with completed bisulfite modification, as described in the user guide for the LightCycler Probe Master Mix.
Determination of RARβ methylation status
The bisulfite modification was applied according to the manufacturers' protocol. After conversion, the DNA was eluted in buffer (Qiagen) to a final concentration of 30 ng/µl. universal methylated (25%, 50%, 75% and 100% methylated control) and unmethylated DNA (Chemicon International, Temecula, CA, USA) were used as methylated and unmethylated controls. We used MS-HRM to detect the methylation levels of our patients. The MS-HRM analysis was performed according to the LightCycler®480 High Resolution Melting Master kits' protocol. The methylation levels for the RARβ gene were compared with the databases “Tm Calling”, “Gene Scanning” and “Difference plot”. Similar findings were obtained from all three databases for each of the samples. The Tm Calling database was used during the presentation of the data. In this analysis method, normalized melting profiles were compared directly along with the methylation level of the DNA sample and the methylated / unmethylatedrates were evaluated in comparison to established standard melting profiles. In this algorithm, the melting temperature curve of PCR products of the selected cases was compared with the melting temperature curve of the control PCR products with a known rate of unmethylated to methylated controls. The results from the methylated and unmethylated control DNA peaks were compared with the DNA samples of the patients and consequently the methylation pattern of each sample was determined.
Statistical analyses
The statistical analyses were performed by chi-square test (x2), two-tailed Fisher's exact test and T-test. The overall survival (OS) time was calculated as the time in months between surgery and day of death. The logrank test was used for univariate analysis to estimate differences in survival time for RARβ methylation status according to the Kaplan-Meier method. Calculations were performed using SPSS 15.0 software (SPSS, Chicago, IL, USA), with a statistical significance of p<0.05.
Results
Methylation Specific HRM was performed on 40 primary GBM patients and RARβ methylation status was successfully determined in all of the cases. The average age of the female patients was 51.71 ± 3.98 and the average age of the male patients was 58.07 ± 2.71. RARβ methylation was detected in 24 (60%) out of 40 cases with glioblastoma multiforme in different quantitative ratios (Figure 1).
Fig. 1.
Methylated and unmethylated peaks of RARβ gene for the patient groups
(The peaks obtained in different Tm degrees for the unmethylated and methylated cases are shown).
Both T-test and chi-square test were conducted to evaluate RARβ methylation according to the age and gender of the patients, but the results did not show a significant relationship (p=0.329, p=0.099). In addition, all the samples in our study group were grade IV type tumor samples and therefore this parameter was not included in the statistical evaluation.
The melting curves of positive/methylated control and negative/unmethylated control samples and their 75%, 50% and 25%, interpolated samples, according to their fluorescent peak, are shown in Figure 2.
Fig. 2.
The unmethylated control DNA and methylated control DNA peaks
The low melting point indicates the samples of unmethylated cytosine nucleotides that were converted to uracile nucleotides and the high melting point shows methylated cytosine nucleotides.
The melting temperature for the unmethylated samples was ∼76°C and for the methylated samples, it was ∼81°C. The melting temperature data for the DNA samples and the temperature data for the methylated /positive and unmethylated /negative control samples were evaluated using the “Tm calling” software program. In this algorithm, the melting profiles for the methylated and unmethylated samples showed differences. The methylated DNA had a higher Tm value, due to their CpG sequences in the amplicon. Unmethylated cytosine nucleotides were converted into uracile nucleotides by bisulfite modification and therefore they had a Tm value (Figure 3).
Fig. 3.
The image of the different methylation levels. RARβ peak image of the cases of 27, 36, 41 that are concordant with methylated control DNA according to their degree of methylation (50%, 75%, 100%)
The amplification peak for case 27 was at Tm=81.5°C, which is concordant with the amplification peak of 100% methylated control DNA. Therefore, the case was evaluated as 100% methylated. The amplification peak for case 41 was at Tm= ∼81°C, which corresponds with the amplification peak of 75% methylated control DNA as shown in figure 3. As a result, the case was assessed as 75% methylated. At Tm= ∼80°C, the amplification peak of case 36 was observed, which is concordant with the amplification peak of 50% methylated control DNA. The case was therefore assessed as 50% methylated (Figure 3).
According to the evaluation of the methylation rates for tumor samples from 24 cases, it was determined that 3 were 25% methylated, 5 were 50% methylated, 4 were 75% methylated and 12 cases showed 100% RARβ gene hypermethylation. The mean survival time of the patients with RARβ methylation was calculated to be 19 months and the survival time for unmethylated cases was calculated as 15 months (Table 1).
Table 1.
Mean survival time of our study group according methylation rates of RARβ.
| Survival Time (month) |
|
| Methylated RARβ (n=24) | 19 |
| Unmethylated RARβ (n= 16) | 15 |
In our study group, 28 patients out of 40 received treatment, however, the remaining 12 cases had no specified treatment protocols in their patient files. One of the cases received only chemotherapy treatment, 9 patients received only radiotherapy treatment, and the remaining 17 patients received both radiotherapy and chemotherapy.
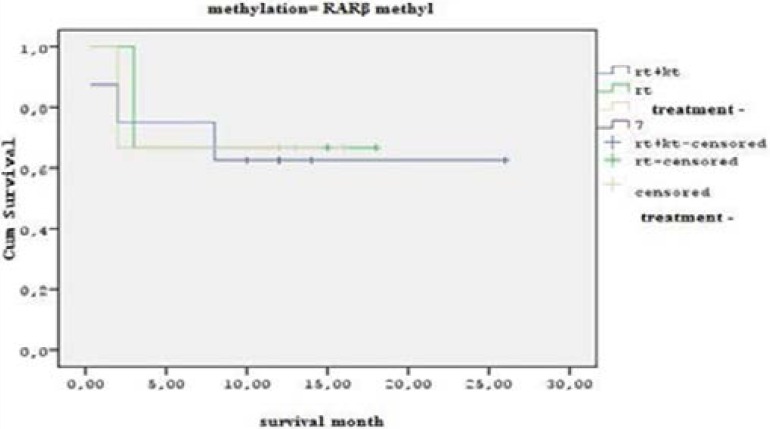
The statistical analysis shows that the RARβ methylated patients who received both chemotherapy and radiotherapy treatment combined had a survival time of 25 months. The patients who received only radiotherapy or had no treatment protocol had a survival time between 15–20 months, which demonstrates a significant difference (P<0.05) (Figure 4).
Fig. 4.
Statistical analysis graphic for the RARβ gene methylation cases, according to their treatment and survival time.
The patients receiving treatment and RARβ genes were also evaluated and it was observed that their survival time was 22 months (P<0.05) (Table 2, Figure 4).
Significant statistical (P<0.05) difference was observed between the survival times of the patients receiving both radiotherapy and chemotherapy treatment and the cases with no applied treatment protocol. Our study group was statistically analyzed by using the Kaplan-Meier test, in order to establish the relationship between RARβ gene quantitative methylation rates (unmethylated, 25%, 50%, 75%, 100% methylated) and patient survival times. Due to the small number of cases, when we divided the samples into 3 groups - unmethylated, methylated <50%, methylated > 50% - there was no statistically significant difference in survival time between the patients with methylation levels higher than 50% and the patients with methylation lower than 50%.
Discussion
DNA methylation can be examined by using MS-PCR, sodium bisulfite sequencing methods or many other similar methods. MS-HRM is a fast, easy and sensitive method that can detect methylated DNA at the rate of 0.1% inside an unmethylated DNA sample9,16.
In our study, successful results have been achieved in all GBM tissue samples with the MS-HRM method. The MS-HRM method was compared in terms of time to the MS-PCR methylation method, which we had been using in our previous studies. We determined that the MS-HRM method was more suitable in routine analysis for screening gene methylation quantitatively.
We aimed to examine the RARβ gene methylation pattern, which has been studied extensively in recent years in different types of cancer, such as in prostate cancer15, carcinoma of cervix, uteri10, lung cancer, gastric cancer, and hematologic neoplasms. However, although a literature review revealed that there was an impact of RARβ gene hypermethylationon gliomagenesis, there was only one study that showed association of the RARβ promoter methylation in GBM samples. Therefore, the research findings obtained in this study were only compared to the data from this particular relevant study. Therefore, our data is open to debate. On the other hand, it is the first study in Turkey that has searched for a relationship between RARβ hypermethylation frequencies in GBM cases.
Piperi and colleagues analyzed the methylation status of four genes (VEGF, COX-2, IL-6, IL-9), which were associated with tumors in 23 glioma samples and they examined the clinical results related to the inflammation and angiogenic agents. They reported that the rate of RARβ methylation was 58.8 % and the rate of MGMT-methylation was 70.58% in the 6 grade II and 17 grade IV GBM samples, respectively. They also reported a positive correlation between tumor stage, necrosis degree and the RARβ and MGMT methylation rates14. According to the researchers' results, the methylation rate in grade IV GBM cases was only 42%. In our study, we detected a 60% hypermethylation sample, which is the same as Piperi and colleagues The researchers in that study used the MS-PCR method and it is known that the MS-HRM method, which we used in our study, is more sensitive for the detection of low rate methylation1,14,17. On the other hand, the limited number of cases and the limited number of studies may have a negative effect on the positive prognostic and predictive impact of the RARβ gene in GBM.
Furthermore, Piperi and colleagues reported RARβ methylation detection in all 6 cases of their early stages glioblastoma patients group. This finding shows thatRARβ gene methylation plays a role in the early stages of the carcinogenesis process. In our study, we only evaluated primary GBM cases and therefore we did not obtain data in this direction.
RARβ is a member of the thyroid-steroid hormone receptor family of nuclear transcriptional regulators. It binds to retinoic acid, which is a biologically active form of vitamin A. RARβ is a tumor suppressor gene, which has a role in many cellular events that include embryonic morphogenesis, cellular growth, cell differentiation and signal transduction14.
Olasz and colleagues showed RARβ gene methylation in head and neck cancer. They reported that the RARβ hypermethylation was correlated with an advanced level of cancer and the gene expression was reduced. However, they also reported that allelic loss of 3p24 RARβ locus (45.2%) did not affect the mRNA level12. Furthermore, it was stated that there was no correlation between gene expression and RARβ allelic loss in breast carcinomas and esophageal cancers. Considering the fact that RARβ is tumor suppressor gene, we can know that wild type tumor suppressor gene was induced by second allele retinoids. However, the increase of RARβ methylation and the significant decrease in gene expression indicates that methylation assumes the main role in RARβ suppression. Olasz and colleagues suggested that methylation pattern analysis of this gene could be a marker for early diagnosis of cancer12. There have been no studies conducted about glial cells in this direction. However, Piperi and colleagues detected 100% methylation in grade II tumor samples in a series of patients14 and this raises the question as to whether it is useful as an early diagnostic marker for glioblastomas. We believe that more studies should be conducted in this direction.
On the other hand, RARβ can be a marker for chemotherapy as it is an agent in response to retinoic acid and together with MGMT methylation also the RARβ methylation may use to chemotherapy planning in GBM. Because up to date the most studied and most known marker for GBM chemotherapy planning was MGMT. These benefits that are already specified for head, neck and breast cancers should also be examined in detail in glioblastomas.
The Kaplan-Meier test was performed to analyze the survival rate of our study group. The results showed no significant statistical difference between the survival time of the cases with or without methylation. It was determined that the mean survival time for the cases without methylation was 15 months and for the cases with RARβ methylation was 19 months. The study by Piperi and colleagues also did not show a statistically significant difference between the cases with methylation and without methylation in terms of survival time12. In line with the fact that methylation increases with age, the patients with and without methylation were compared in terms of their age in our study. The average age of the patients with positive methylation was 52.7, while the average age of patients without methylation was 60.4.
No statistically significant differences were observed between the groups in terms of average age. However, the important aspect of our study is that we compared the survival time of patients who received treatment protocol and the RARβ methylation status. In our study, the patients who received both chemotherapy and radiotherapy treatment combined had a survival time of 25 months. The patients who received only radiotherapy or had no treatment protocol had a survival time between 15–20 months, which demonstrates a statistically significant difference (P<0.05). Therefore, this information is new for glioblastoma treatment planning and it also requires independent confirmation with a series of primary GBMs.
In summary, our results demonstrate the role of the RARβ gene in glioma tumorigenesis, with RARβ promoter methylation being of prognostic value for glioblastoma patients.
Conclusions and recommendations
A high frequency of methylation was determined for the RARβ gene methylation in our study group. The rate of methylation in our group was correlated with previous literature. It is concluded that the MS-HRM application is reliable and applicable for methylation studies of solid tumors. In our study, the mean survival times for the patients with or without methylation were identified, and the longer survival time was observed in the group with the methylated RARβ gene. The results of our study are different from the results of previous study in this regard. Therefore, it was determined to be a new finding in medical literature. Due to the small number of studies on RARβ gene methylation in patients with GBM, we believe that further studies should be carried out to examine larger series of cases and their methylation patterns.
In summary, our results demonstrated the role of RARβ gene in glioma tumorigenesis, with RARβ promoter methylation being a positive prognostic value for glioblastoma patients. Furthermore, a positive correlation between RARβ methylation status and radio/chemotherapy treatment response was observed.
Acknowledgement
This study was supported by grants from Eskişehir Osmangazi University, Eskişehir, Turkey and the project number was 201011034.
List of abbreviations
- DNA
Deoxyribonucleic acid
- GBM
Glioblastoma multiforme
- IDH
Isocitrate dehydrogenase
- MS-HRM
Methylation-sensitive high resolution melting
- NADP
Nicotinamide adenine dinucleotide phosphate
- WT
Wild type
Competing interests section
The authors declare that they have no competing interests.
Ethical approval
The study has been approved by the ethics committee of Osmangazi University Medical School.
Authors' contributions
All authors read and approved the final version of the manuscript.
References
- 1.Burgess R, Jenkins R, Zhang Z. Epigenetic changes in gliomas. Cancer Biol Ther. 2008 Sep;7(9):1326–1334. doi: 10.4161/cbt.7.9.6992. 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cancer Facts and figures. Atlanta: American Cancer Society, Surveillance Research; 2002. [DOI] [Google Scholar]
- 3.CBTRUS (Central Brain Tumor Registry of the United States) 2002-2003-2008, author. Report on Primary Brain Tumors in the United States. Chicago: Central Brain Tumor Registry of the United States; 2003–2008. [DOI] [Google Scholar]
- 4.Costello JF. DNA Methylation in Brain Development and Glioma Genesis. Front Biosci. 2003 Jan 1;8:s175–s184. doi: 10.2741/1027. 2003. [DOI] [PubMed] [Google Scholar]
- 5.Fischer I, Gagner JP, Law M, Newcomb EW, Zagzag D. Angiogenesis in gliomas: biology and molecular pathophysiology. Brain Pathol. 2005;15:297–310. doi: 10.1111/j.1750-3639.2005.tb00115.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kalkan R, Atli Eİ, Özdemir M, Çiftçi E, Aydin HE, Artan S, Arslantaş A. DH1 mutations is prognostic marker for primary glioblastoma multiforme but MGMT hypermethylation is not prognostic for primary glioblastoma multiforme. Gene. 2015 Jan 1;554(1):81–86. doi: 10.1016/j.gene.2014.10.027. [DOI] [PubMed] [Google Scholar]
- 7.Lei Y, Song-tao QI, Zhi-yong LI. Analysis of isocitrate dehydrogenase-1/2 gene mutations in gliomas. Chin Med J. 2010;123(24):3697–3705. doi: 10.3760/cma.j.issn.0366-6999.2010.24.033. PubMed. [DOI] [PubMed] [Google Scholar]
- 8.Losi-guembarovski R, Kuasne H, Guembarovski AL, Rainho CA, Colus IM. DNA methylation patterns of the CDH1, RARβ and SFN genes in choroid plexus tumors. Cancer Genet cytogenet. 2007;179:140–145. doi: 10.2119/molmed.2009.00140. [DOI] [PubMed] [Google Scholar]
- 9.Montgomery J, Wittwer JT, Palais R, ve Zhou R. Simultaneous mutation scanning and genotyping by high-resolution DNA melting analysis. Nature Protocols. 2007;2(1):59–66. doi: 10.1038/nprot.2007.10. [DOI] [PubMed] [Google Scholar]
- 10.Narayan G, Arias-Pulido H, Koul S, Vargas H, Zhang FF, Villella J, Schneider A, Terry MB, Mansukhani M, Murty VV. Frequent promoter methylation of CDH1, DAPK, RARB, and HIC1 genes in carcinoma of cervix uteri: its relationship to clinical outcome. Mol Cancer. 2003 May 13;2:24. doi: 10.1186/1476-4598-2-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ohgaki H, Kleihues P. Population Based Studies on Incidence, Survival Rates, and Genetic Alterations in Astrocytic and Oligodendroglial Gliomas. J Neuropathol Exp Neurol. 2005;64:479–489. doi: 10.1097/01.jnen.0000166799.76946.08. [DOI] [PubMed] [Google Scholar]
- 12.Olasz J, Juhász A, Remenár E, Engi H, Bak M, Csuka O, Kásler M. RAR beta2 suppression in head and neck squamous cell carcinoma correlates with site, histology and age. Oncol Rep. 2007:105–112. doi: 10.3892/or.18.1.105. [DOI] [PubMed] [Google Scholar]
- 13.Pojo M, Costa B M. Molecular hallmarks of gliomas. In: Garami M, editor. Molecular targets of CNS tumors. 2011. pp. 177–200. In Tech. [DOI] [Google Scholar]
- 14.Piperi C, Themistocleous MS, Papavassiliou GA. High Incidence of MGMT and RARβ Promoter Methylation in Primary Glioblastomas: Association with Histopathological Characteristics. Inflammatory Mediators and Clinical Outcome. 2010 doi: 10.2119/molmed.2009.00140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tang D1, Kryvenko ON, Mitrache N, Do KC, Jankowski M, Chitale DA, Trudeau S, Rundle A, Belinsky SA, Rybicki BA. Methylation of the RARB gene increases prostate cancer risk in black Americans. J Urol. 2013 Jul;190(1):317–324. doi: 10.1016/j.juro.2013.01.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wojdacz TK, Dobrovic A, Hansen LL. Methylation-sensitive high-resolution melting. Nature Protocols. 2008;3(1) doi: 10.1038/nprot.2008.191. [DOI] [PubMed] [Google Scholar]
- 17.Wong EM, Dobrovic A. Assessing gene-specific methylation using HRM-based analysis. Methods Mol Biol. 2011;687:207–217. doi: 10.1007/978-1-60761-944-4_14. [DOI] [PubMed] [Google Scholar]