Abstract
Background
Aspergillosis has been identified as one of the hospital acquired infections but the contribution of water and inhouse air as possible sources of Aspergillus infection in immunocompromised individuals like HIV-TB patients have not been studied in any hospital setting in Nigeria.
Objective
To identify and investigate genetic relationship between clinical and environmental Aspergillus sp. associated with HIV-TB co infected patients.
Methods
DNA extraction, purification, amplification and sequencing of Internal Transcribed Spacer (ITS) genes were performed using standard protocols. Similarity search using BLAST on NCBI was used for species identification and MEGA 5.0 was used for phylogenetic analysis.
Results
Analyses of sequenced ITS genes of selected fourteen (14) Aspergillus isolates identified in the GenBank database revealed Aspergillus niger (28.57%), A. tubingensis (7.14%), A. flavus (7.14%) and A. fumigatus (57.14%). Aspergillus in sputum of HIV patients were Aspergillus niger, A. fumigatus, A. tubingensis and A. flavus. Also, A. niger and A. fumigatus were identified from water and open-air. Phylogenetic analysis of sequences yielded genetic relatedness between clinical and environmental isolates.
Conclusion
Water and air in health care settings in Nigeria are important sources of Aspergillus sp. for HIV-TB patients.
Keywords: Internal transcribed spacer genes, phylogenetic, genetic relationship, clinical and environmental fungi, HIV-TB
Introduction
Nigeria has the second largest number (3.1 million) of people living with HIV/AIDS (PLWHA) accounting for 10% of the global HIV burden with about 8% of the HIV-people being tuberculosis positive1. Tuberculosis is the most common opportunistic infections affecting HIV individuals. HIV and TB have synergistic interactions that speedily accelerate the weakening and decline of the host immune system, accentuating the progression of each other. People living with HIV/AIDS (PLWHA) are exquisitely vulnerable to TB and are 30–50 times more likely to progress to active TB. The likelihood of progressing to full blown AIDS also increases by 100 folds in HIV-TB co infected patients2. HIV-TB coinfection is on the rise with tuberculosis being the leading cause of death for people infected with HIV3.
Aspergillosis in HIV patients is another opportunistic infection caused by inhalation of Aspergillus spores4–7. Aspergillus spores are numerous in the environment and the organism is the most common ubiquitous opportunistic pathogen affecting the lung8. Spectrum of aspergillosis are allergic bronchopulmonary aspergillosis, chronic necrotizing aspergillosis, aspergilloma and invasive aspergillosis5 with the immune competence of the host largely determining the particular presentation.
Co infection of HIV, TB and aspergillosis9–12 has been documented in the literature unlike reports on the duos of HIV-TB, Aspergillosis-TB and HIV-Aspergillosis. In Nigeria, high prevalence of 51.25% aspergillosis was associated with pulmonary symptoms in patients attending Infectious Diseases Hospital in Kano13. A cytologic assessment of pulmonary aspergillosis in the North-East Nigeria reported 46% in HIV, and 89% in TB patients14. Ogba et al15 reported low incidence of aspergillosis in Calabar South-South Nigeria while Aliyu et al16 reported yeasts and other molds in Benin, Nigeria.
Nosocomial outbreaks of invasive aspergillosis highlight the fact that Aspergillus spores are common in the hospital environment17. Genetic relationship between environmental and clinical strains has been a subject of several studies18–23 with varied conclusion. Molecular characterization of fungi by polymerase chain reaction (PCR) amplification of internal transcribed spacer (ITS) region of the nuclear ribosomal repeat unit of the rRNA gene is a widely used tool. This ITS region has become the primary genetic marker for molecular identification and other species-level pursuits in many groups of fungi24–25. Its usefulness in sequence comparison are based on the PCR successes and high degree variation even between closely related species26,27 and has been proven comparatively with other coding sequence on species resolution28.
Since hospitalized HIV-TB co infected patients are at the risk of exposure to nosocomial infections such as aspergillosis, there is a need to identify the different environmental Aspergillus in hospital settings and to investigate the contribution of water and open-air to Aspergillus dissemination by determining their genetic relationship to those from sputum of HIV-TB patients using PCR-ITS gene sequencing and phylogenetic techniques.
Methods
Study design
This cross-sectional study was conducted between September 2013 and March 2014 in the Tuberculosis (TB) ward of a semi-private hospital in Abeokuta, Ogun State, Nigeria. An Ethical clearance was obtained from the Ethical Committee of the hospital. The hospital has facilities for tuberculosis and HIV/AIDS diagnosis, treatment and monitoring. Patients that were already diagnosed of HIV-TB coinfection were considered eligible for the study. Informed consent (oral) was obtained from the subjects and only consented candidates were enrolled in the study. Demographic characteristics were obtained using a structured form from a total population of thirty (30) HIV-TB patients.
Sampling and sample collection
Details of fungal sampling and isolation from environmental (water and open-air) and clinical (sputum) samples and their morphological classification were as described Shittu et al29. Briefly, water samples were collected from the taps and storage water in the hospital into sterile bottles using standard techniques. Also, different bottled and sachet water sources for drinking water that are sold within and around the hospital premises were purchased for fungal analysis. Also, open-air sampling of the corridor, and indoor (corners and centre) of the wards, nursing section, and bathroom was conducted by exposing duplicate Sabouraud Dextrose Agar (SDA, BIOLAB, Hungary) plates for 2h. Also, sputum samples from HIVTB patients were collected into sterile universal bottles.
Isolation of fungi from environmental and clinical samples
Membrane filter technique was employed in the isolation of fungi from water; by filtering 100 ml in duplicate, using 47 mm membrane filters (Millipore, Bangalore) with a pore size of 0.45µm. The filter papers were placed on both SDA and potato dextrose agar (PDA, LAB M, UK) plates. Also, SDA plates exposed to air were withdrawn after 2h. For fungi isolation from clinical samples, a loopful of sputum was cultured on PDA. All SDA and PDA plates were incubated for 3–5 days at 25°C for growth of fungi.
DNA extraction, purification, amplification and sequencing
DNA extraction, amplification and sequencing were conducted at the International Institute for Tropical Agriculture (IITA), Ibadan, Nigeria. DNA extraction and purification was done with QIAamp DNA Mini Kit (Qiagen, Valencia, CA) with catalogue number 51304 following manufacturer's instruction. The Extracted Nucleic Acid was then treated with RNase and 3µl of the DNA was loaded on 1% agarose and ran at 96–100 volts using 1xTBE for 1h.
DNA amplification primers, ITS-1 (5′-TCCGTAGGTGAACCTGCGG-3′) and ITS-4 (5′-TCCTCCGCTTATTGATATGC-3′) were purchased from Integrated DNA Technologies, Belgium. PCR amplification was performed using GoTaq® Green Master Mix (Promega Corporation, Madison, WI, USA) protocol. PCR Amplification was carried out in a PCR System Thermocycler (Applied Biosystems) according to standard protocol of White et al26. This involved using 5µM of each primer, 2µl genomic DNA and nuclease free water adjusted to a final volume of 50µl. The PCR protocol consisted of initial denaturation of 94°C/5mins, 30 cycles of 72°C/45secs, 56°C/30secs and final extension/elongation of 72°C/5mins. The PCR was purified with absolute ethanol and 70% ethanol respectively. PCR products were separated by electrophoresis on agarose 2.5–3.0% gels. Gels were photographed with a gel documentation system (Gel Doc 1000, BioRad) and the sizes of the PCR products were calculated by comparison with internal mol. wt standards. Finally, 2.5µl of the purified PCR products were sequenced using the Applied Biosystems ABI PRISMTM 3100 DNA sequence Analyzers with the BigDye® Terminator v3.1 Cycle Sequencing kit and protocols.
Molecular identification of sequenced ITS-18S rRNA genes
Fasta formatted sequences obtained from ABI automated sequencer were edited and aligned to generate a consensus sequence using BioEdit Sequence Alignment Editor (version 7.1.9). Consensus sequences were then aligned with sequences deposited in the National Centre for Biotechnological Information (NCBI) genebank by using the Basic Local Alignment search Tool (BLAST) to establish identities of the fungal isolates.
Phylogenetic analysis
To identify homologies, multiple sequence alignment was done using ClustalW alignment algorithm30. Aligned sequences were arranged with MEGA 5.0 for phylogenetic analyses31. The evolutionary history was inferred using the Neighbor-Joining method32. The evolutionary distances were computed using the Maximum Composite Likelihood method33 and were in the units of the number of base substitutions per site. Two thousand bootstrap replicates were performed. All positions containing gaps and missing data were eliminated.
Results
HIV-TB patients' characteristics and fungi isolation
Table 1 shows the sex distribution of HIV-TB patients and positive fungi isolation observed in this study. From a total of thirty patients investigated, there were more female (66.7%) with 95% of culture positive sputum while male (33.3%) had 50% of positive sputum samples.
Table 1.
Sex distribution of HIV-TB patients and fungi isolation
| Sex | Frequency (%) |
Culture Positive n (%) |
Culture Negative n (%) |
| Male | 10 (33.33) | 5(50) | 5(50) |
| Female | 20 (66.67) | 19(95) | 1(5) |
| Total | 30 (100) | 24(80) | 6(20) |
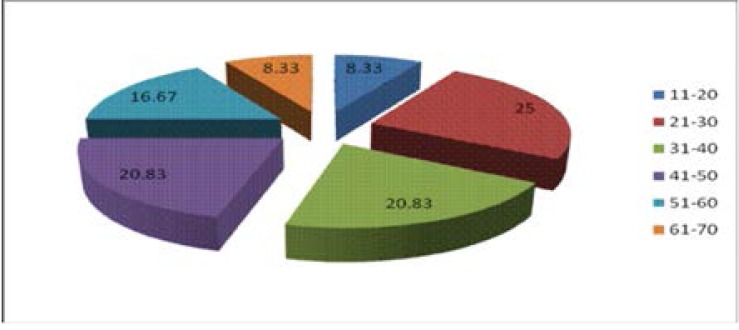
A total of 24 (80%) out of thirty HIV-TB patients were culture positive. Fig. 1 shows the age distribution of HIV-TB co infected patients and fungi isolation with age group 21–30 giving 25% of positive culture.
Fig 1.
Age distribution of HIV-TB co infected patients with percentage fungal infection
PCR amplification and DNA sequences results
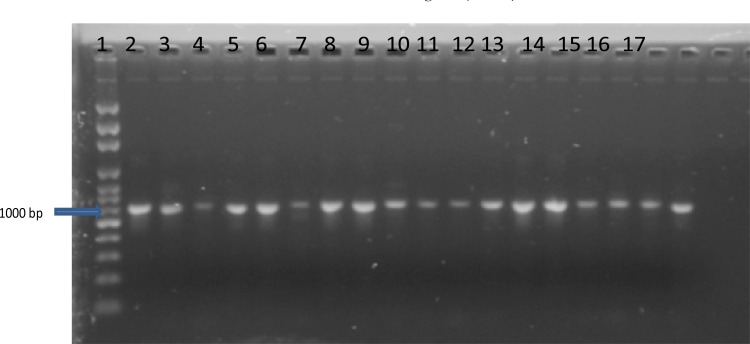
Polymerase Chain Reaction (PCR) amplification of ITS region of rRNA genes with ITS1 and ITS 4 primers yielded distinct DNA bands for all representative isolates investigated (Plate 1).
Plate1.
1%agarose gel electrophoresis of PCR amplicons of the Internal Transcribed Spacer (ITS) region genes of the fungal isolates.
Key: Lane 1- 1kb DNA marker; Lane 2, 3, 14 - Isolate from open air culture; Lane 4, 5, 6, 16, 17 — Isolate from sputum; Lane 7, 8, 13 — Isolates from storage water; Lane 9, 10 — Isolates from tap water; Lane11 — Isolates from bottled water; Lane 12, 15 — Isolates from sachet water.
DNA sequences of Aspergillus sp. identified by similarity searches in the GenBank gave A. fumigatus (57.6%), A. niger (28.6%), A. tubingensis (7.1%), and A. flavus (7.1%) from all sources investigated (Table 2). Aspergillus niger and A. fumigatus were identified from the three environments examined, i.e. water, air, and sputum while A. tubingensis and A. flavus were identified from sputum only.
Table 2.
DNA Sequences and organisms identified by Blast on NCBI
| Isolate codes |
Length of query sequences (bp) |
NCBI Best Match (Identified organisms) |
Sources of isolate |
| A1 | 1037 | A. niger SUMS0061 | Air |
| A2 | 1004 | A. fumigatus HF11 | Air |
| A3 | 568 | A. niger SUMS0061 | Sputum |
| A4 | 1037 | A. fumigatus 871435 | Sputum |
| A5 | 1077 | A. flavus | Sputum |
| A6 | 689 | A. niger SUMS0061 | Storage Water |
| A7 | 1038 | A. fumigatus SUMS0106 | Storage Water |
| A8 | 727 | A. fumigatus SUMS0106 | Tap Water |
| A9 | 720 | A. fumigatus SUMS0106 | Bottled Water |
| A10 | 1032 | A. fumigatus SUMS0106 | Sachet Water |
| A11 | 1046 | A. fumigatus SUMS0106 | Storage Water |
| A12 | 571 | A. niger 20-10 | Air |
| A13 | 559 | A. fumigatusW1-25 | Sachet Water |
| A14 | 391 | A. tubingensis G1-18 | Sputum |
Phylogenetic relationship between isolates
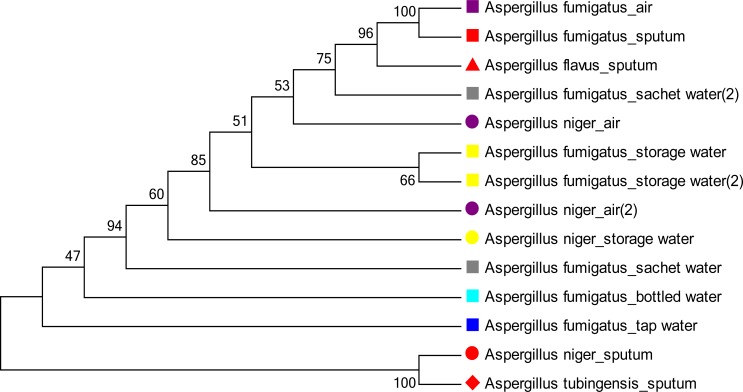
Figure 2 shows an unrooted phylogenetic tree in cladogram format with branch lengths using a neighbor joining method. The optimal tree with the sum of branch length = 3.78942342 is shown. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary relationship between the DNA of the isolates sequenced depicted distinct clades. Analysis of sequences also revealed inter and intra species diversity among A. fumigatus and A. niger from same and different sources. DNA sequences of A. fumigatus from sputum and air showed same branch length and were genetically related with one another.
Fig. 2.
Neighbor joining phylogenetic bootstrap consensus tree of PCR-ITS DNA sequences of environmental and clinical Aspergillus species. The percentage of replicate trees in which the associated taxa related together in the bootstrap test (2000 replicates) is shown next to the branches. Same species are shown in the same shape and sources of isolates in same color.
Discussion
DNA sequences of the ITS regions of fourteen (14) Aspergillus isolates in the GenBank database yielded A. fumigatus, A. niger, A. flavus and A. tubingensis from both clinical and environmental fungal isolates. The same strain of A. fumigatus was isolated from storage water, tap water, sachet water, and bottled water. Notable is the predominance of A. fumigatus in sputum samples. In immunocompromised hosts, A. fumigatus represents a major cause of morbidity and mortality34 and it is the commonest etiologic agent of various clinical forms of bronchopulmonary aspergillosis including allergic, acute invasive and chronic pulmonary aspergillosis8–9,19–22.
In this study, A. tubingensis initially identified as A. niger by microscopic and macroscopic morphology was discovered from sputum samples of patients. This reclassification of A. niger as Aspergillus tubingensis by sequencing agrees with Perrone et al35. This organism was implicated to have caused infectious keratitis36 and osteomyelitis of the maxillary bone of an immunocompromised patient after tooth extraction37.
Phylogenetic analysis in this present study showed that environmental and clinical isolates proceeded from a common ancestor which is similar to that of Lee et al23. Sequences yielded distinct clades revealing inter and intra species similarity and diversity among Aspergillus sp. DNA sequences of A. fumigatus from sputum, air and water were genetically related while Aspergillus tubingensis from sputum was also genetically related with A. niger from air.
This present study has not only revealed the co-existence of Aspergillus sp in 80% of HIV-TB co infected patients, but emphasizes that water and air in health care settings are important sources of Aspergillus portraying serious health risk for HIV-TB co-infected patients.
There is therefore a need to further investigate clinical cases of aspergillosis in HIV-TB patients in Nigeria, a TB endemic region.
To our knowledge, this is the first study on molecular identification of clinical and environmental fungi in Nigeria, a tropical and low incomecountry. This is also the first report on genetic relationship of Aspergillus sp from environmental and clinical sources.
Conclusion
This investigation has shown that both open-air and water in health care settings in Nigeria are important contributors to the transmission of Aspergillus sp. to immunocompromised host.
Conflict of interest
The authors declare that there was no conflict of interest during this study.
References
- 1.Nigeria HIV Fact Sheet 2011. United States Embassy in Nigeria; [28 April 2015]. Available at http://nigeria.usembassy.gov. [Google Scholar]
- 2.Silversides A. HIV/TB Coinfection Rising. Canadian Medical Association Journal. 2006;175(7):725. doi: 10.1503/cmaj.061122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.De Carvalho BM, Monteiro AJ, Neto RJP, Grangeiro TB, Frota CC. Factors related to HIV/ TB coinfection in a Brazilian Reference Hospital. Brazilian Journal ofInfectious Diseases. 2008;12(4):281–286. doi: 10.1590/s1413-86702008000400005. [DOI] [PubMed] [Google Scholar]
- 4.Holding KJ, Dworkin MS, Wan PC, Hanson DL, Klevens RM, Jones JL, Sullivan PS. Aspergillosis among people infected with human immunodeficiency virus: incidence and survival. Adult and Adolescent Spectrum of HIV Disease Project. Clin Infect Dis. 2000;31:1253–1257. doi: 10.1086/317452. [DOI] [PubMed] [Google Scholar]
- 5.Kousha M, Tadi R, Soubani AO. Pulmonary aspergillosis: a clinical review. Eur Respir Rev. 2011;20(121):156–174. doi: 10.1183/09059180.00001011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guazzelli LS, Unis G, Xavierc MO, Severo CB, Picon PD, Severo LC. Fungus ball in HIV-infected patients. Rev Inst Med trop S Paulo. 2009;51(6):345–348. doi: 10.1590/s0036-46652009000600007. [DOI] [PubMed] [Google Scholar]
- 7.Bernhard CD, Vassilios D, Hilmar D, Dimitrios M, Georgios B, Friedrich AS. Surgical treatment of pulmonary aspergillosis mycosis in immunocompromised patients. Interactive Cardio Vascular and Thoracic Surgery. 2008;7:771–776. doi: 10.1510/icvts.2007.171579. [DOI] [PubMed] [Google Scholar]
- 8.Zmeili OS, Soubani AO. Pulmonary aspergillosis: a clinical update. Quarterly Journal of Medicine. 2007;100(6):317–334. doi: 10.1093/qjmed/hcm035. [DOI] [PubMed] [Google Scholar]
- 9.Hasse B, Strebel B, Thurnheer R, Uhlmann F, Krause M. Chronic necrotizing pulmonary aspergillosis after tuberculosis in an HIV-positive woman: an unusual immune reconstitution phenomenon? AIDS. 2005;19(18):2179–2181. doi: 10.1097/01.aids.0000194796.20928.f0. [DOI] [PubMed] [Google Scholar]
- 10.Connolly C, Reid A, Davies G, Sturm W, McAdam KP, Wilkinson D. Relapse and mortality among HIV-infected and uninfected patients with tuberculosis successfully treated with twice weekly directly observed therapy in rural South Africa. AIDS. 1999;13:1543–1547. doi: 10.1097/00002030-199908200-00015. [DOI] [PubMed] [Google Scholar]
- 11.Au-Yeung C, Kanters S, Ding E, Glaziou P, Anema A, Cooper CL, et al. Tuberculosis mortality in HIV-infected individuals: a cross-national systematic assessment. Clin Epidemiol. 2011;3:21–29. doi: 10.2147/CLEP.S15574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Denning DW, Pleuvry A, Cole DC. Global burden of chronic pulmonary aspergillosis as a sequel to pulmonary tuberculosis. Bulletin of the World Health Organization. 2011;89:864–872. doi: 10.2471/BLT.11.089441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Taura DW, Adamu S, Koki YA, Musa MA, Muhammad BB. Mycotic infections associated with pulmonary symptoms in patients attending Infectious Diseases Hospital, Kano. Greener Journal of Microbiology and Antimicrobials. 2014;2(1):015–020. [Google Scholar]
- 14.Adisa JO, Mohammed B, Egbujo EC, Bukar MA. Cytologic Assessment of Pulmonary Aspergillosis in Immunocompromised Subjects in Maiduguri North Eastern, Nigeria. Journal of Medicine and Medical Sciences. 2013;4(6):237–240. [Google Scholar]
- 15.Ogba OM, Abia-Bassey LN, Epoke J. The relationship between opportunistic pulmonary fungal infections and CD4 count levels among HIV-seropositive patients in Calabar, Nigeria. Trans R Soc Trop Med Hyg. 2013;107(3):170–175. doi: 10.1093/trstmh/trs025. [DOI] [PubMed] [Google Scholar]
- 16.Aluyi HS, Otajevivo PA, Iwereibe O. Incidence of pulmonary mycoses in patients with AIDS. Nigerian Journal of Clinical Practice. 2010;3(1):78–83. [Google Scholar]
- 17.Hajjeh RA, Warnock DW. Counterpoint: Invasive Aspergillosis and the Environment - Rethinking Our Approach to Prevention. Clinical Infectious Diseases. 2001;33:1549–1552. doi: 10.1086/322970. [DOI] [PubMed] [Google Scholar]
- 18.Leenders AC, van Belkum A, Behrendt M, Luijendijk A, Verbrugh HA. Density and molecular epidemiology of Aspergillus in air and relationship to outbreaks of Aspergillus infection. Journal of Clinical Microbiology. 1999;37:1752–1757. doi: 10.1128/jcm.37.6.1752-1757.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Anaissie EJ, Stratton SL, Dignani MC, Summerbell RC, Rex JH, Monson TP, et al. Pathogenic Aspergillus species recovered from a hospital water system: A 3-year prospective study. Clinical Infectious Diseases. 2002;34:780–789. doi: 10.1086/338958. [DOI] [PubMed] [Google Scholar]
- 20.Rosehart K, Richards MH, Bidochka MJ. Microsatellite analysis of environmental and clinical isolates of the opportunist fungal pathogen Aspergillus fumigatus. Journal of Medical Microbiology. 2002;51:1128–1134. doi: 10.1099/0022-1317-51-12-1128. [DOI] [PubMed] [Google Scholar]
- 21.Warris A, Klaassen CHW, Meis JFGM, de Ruiter MT, de Valk HA, Abrahamsen TG, Gaustad P, Verweij PE. Molecular epidemiology of Aspergillus fumigatusisolates recovered from water, air and patients shows two related of genetically distinct strains. Journal of Clinical Microbiology. 2003;41:4101–4106. doi: 10.1128/JCM.41.9.4101-4106.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chowdhary A, Kathuria S, Xu J, Sharma C, Sundar G, Singh PK, et al. Clonal expansion and emergence of environmental multiple-triazole-resistant Aspergillus fumigatus strains carrying the tr34/l98h mutations in the cyp51a gene in India. PLoS ONE. 2012;7(12):e52871. doi: 10.1371/journal.pone.0052871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee LD, Hachem RY, Berkheiser M, Hackett B, Jiang Y, Raad IIj. Hospital environment and invasive aspergillosis in patients with hematologic malignancy. American Journal of Infection Control. 2012;40:247–249. doi: 10.1016/j.ajic.2011.03.031. [DOI] [PubMed] [Google Scholar]
- 24.Nilsson RH, Abarenkov K, Larsson K-H, Kõljalg U. Molecular Identification of Fungi: Rationale, Philosophical Concerns, and the UNITE Database. The Open Applied Informatics Journal. 2011;5(Suppl 1-M9):81–86. [Google Scholar]
- 25.Tsui CKM, Woodhall J, Chen W, Lévesque CA, Lau A, Schoen CD, et al. Molecular techniques for pathogen identification and fungus detection in the environment. IMA Fungus. 2011;2(2):177–189. doi: 10.5598/imafungus.2011.02.02.09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.White TJ, Bruns T, Lee S, Taylor J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR Protocols: A Guide to Methods and Applications. 1990;18:315–322. [Google Scholar]
- 27.Peay KG, Kennedy PG, Bruns TD. Fungal community ecology: a hybrid beast with a molecular master. Bio-Science. 2008;58:799–810. [Google Scholar]
- 28.Schocha CL, Seifertb KA, Huhndorfc S, Robertd V, Spougea JL, Levesqueb CA, Chen W Fungal Barcoding Consortiuma, author. Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for Fungi. Proceedings of the National Academy of Sciences. 2012;109(16):1642–1646. doi: 10.1073/pnas.1117018109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shittu OB, Adelaja OM, Obuotor TM, Sam-Wobo SO. Clinical and environmental fungi associated with hospitalized immuno-compromised patients in Abeokuta, south west Nigeria. International Journal of Public Health Research. 2014;2(4):37–40. [Google Scholar]
- 30.Thompson JD, Higgins DG, Gibson TJ, CLUSTAL W. improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Research. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kuma S. MEGA5: Molecular Evolutionary Genetics Analysis Using Maximum Likelihood, Evolutionary Distance, and Maximum Parsimony Methods. Molecular Biology and Evolution. 2011;28(10):2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Saitou N, Nei M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Molecular Biology and Evolution. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 33.Tamura K, Nei M, Kumar S. Prospects for inferring very large phylogenies by using the neighbor-joining method. Proceedings of the National Academy of Sciences (USA) 2004;101:11030–11035. doi: 10.1073/pnas.0404206101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Duarte-Escalante E, Zúñiga G, Ramírez ON, Córdoba S, Refojo N, Arenas R, et al. Mem Inst Oswaldo Cruz. 3. Vol. 104. Rio de Janeiro: 2009. Population structure and diversity of the pathogenic fungus Aspergillus fumigatusisolated from different sources and geographic origins; pp. 427–433. [DOI] [PubMed] [Google Scholar]
- 35.Perrone G, Stea G, Epifani F, Varga J, Frisvad JC, Samson RA. Aspergillus niger contains the cryptic phylogenetic species A. awamori. Fungal Biology. 2011;115:1138–1150. doi: 10.1016/j.funbio.2011.07.008. [DOI] [PubMed] [Google Scholar]
- 36.Kredics L, Varga J, Kocsube S, Rajaraman R, Raghavan A, Do'czi IM, et al. Infectious Keratitis Caused by Aspergillus tubingensis. Cornea. 2009 doi: 10.1097/ICO.0b013e3181967098. [DOI] [PubMed] [Google Scholar]
- 37.Bathoorn E, Salazar NE, Sepehrkhouy S, Meijer M, de Cock H, Haas P-J. Involvement of the opportunistic pathogen Aspergillus tubingensis in osteomyelitis of the maxillary bone: a case report. BMC Infectious Diseases. 2013;13:59. doi: 10.1186/1471-2334-13-59. [DOI] [PMC free article] [PubMed] [Google Scholar]