Abstract
Background
The emergence of Enterobacteriaceae harboring IMP-4 or IMP-8 carbapenemases is rare. We report an occurrence of Enterobacteriaceae harboring IMP-4 or IMP-8 carbapenemases in a Chinese tertiary care hospital from November 2010 to December 2012.
Methods
The clinical characteristics of 30 patients were described. The genetic relationship of isolates was determined by pulsed-field gel electrophoresis (PFGE). Carbapenemases were detected by modified Hodge test (MHT) and polymerase chain reactions (PCRs). Amplicons were sequenced and blasted to determine the genotype.
Results
Most infected patients were from intensive care unit and had complex and serious underlying illnesses requiring mechanical ventilation. PFGE revealed that Klebsiella pneumoniae showed two major PFGE types. Two Klebsiella oxytoca had an indistinguishable PFGE pattern, while four Enterobacter cloacae were different strains. The sequencing studies showed Enterobacteriaceae harboring IMP-4 or IMP-8 carbapenemase in the 23 infected patients. The majority of patients had infections with the carbapenemase-producing Enterobacteriaceae (CPE) strain, most were successfully treated with a range of antibiotics and discharged.
Conclusion
It is important to maintain a high index of suspicion to screen for carbapenemase-producing Enterobacteriaceae strains. Rapid identification of these strains and implementation of stringent procedures are the key to prevent major outbreaks in a hospital setting.
Keywords: Carbapenemase, Enterobacteriaceae, IMP-4, IMP-8, clinical outcome
Introduction
Since 2000, spread of community-acquired enterobacterial isolates (Escherichia coli) that produce extendedspectrum β-lactamases (ESBLs) capable of hydrolyzing almost all cephalosporins except carbapenems has been reported worldwide1. Carbapenems have become antimicrobial drugs of last resort therefore it is important to maintain the clinical efficacy of these antibiotics. The frequent use of carbapenems, however, combined with the transmissibility of resistance determinants mediated by plasmids, transposons and gene cassettes, has contributed to the increase of carbapenem resistance by Enterobacteriaceae2,3. In countries such as the United States, Israel, Germany and Greece, carbapenemase-producing Enterobacteriaceae (CPE) had emerged and the dissemination of these multidrug-resistant pathogens had become a problem in clinical care of patients and in public health4–7. Invasive infections with these organisms have been associated with high rates of morbidity and mortality, due to their resistance to most available antimicrobial agents8–10. Patients who are hospitalized for prolonged periods and those with severe underlying disease are at high risk of acquiring these types of pathogens11.
The Ambler classification describes various classes of carbapenemases12. Klebsiella pneumoniae carbapenemases (KPC) belong to class A; they are increasingly prevalent in parts of the United States, Israel and Greece and are also an emerging concern in Western Europe13. Class B metallo-beta-lactamases of the types IMP (imipenemase) and VIM (Verona integron-encoded metallo-beta-lactamase) are common in non-fermentative bacteria and have been recently recognized in Enterobacteriaceae worldwide13. Strains carrying the recently reported New Delhi metallobeta-lactamase1 (NDM-1), found throughout India, Pakistan and Bangladesh, have also been rapidly disseminated to Europe and across the globe14. OXA-type (oxacillinhydrolysing) carbapenemases (Class D) are widespread and have been detected primarily in Enterobacteriaceae 15.
CPE have been sporadically reported in China in recent years. In 2009, two cases of pulmonary infection due to multidrug-resistant K. pneumoniae strains producing blaIMP-4 and armA, which contained intervening sequences highly related to those of a Vibrio cholerae plasmid found in Hangzhou, China16. In 2010, a study to investigate the resistance mechanism of 49 Enterobacteriaceae isolates with decreased susceptibility to carbapenems collected at 16 teaching hospitals in China, confirmed the production of KPC-2, IMP-4 and IMP-8 carbapenemase17. In 2011, the first NDM-1 producing E. coli isolate was detected in Hong Kong18.Here we report the emergence of Enterobacteriaceae harboring IMP-4 or IMP-8 carbapenemases in a Chinese tertiary care hospital. This report describes the emergence, the infection control measures implemented, and the results of molecular investigations.
Patients and methods
Setting
Liaocheng People's Hospital is a 3000-bed tertiary care hospital serving about 139,000 inpatients per year. It has a 43-bed general Intensive Care Unit (ICU) including medical ICU and surgical ICU, a 35-bed Neonatal Intensive Care Unit (NICU), a 30-bed Pediatric Intensive Care Unit (PICU) and a single 30-bed ICU in the Brain Hospital. All ICUs are mainly dealing with critically ill patients. Patients who no longer need mechanical ventilation or are in a stable condition will be transferred to general wards from ICU. Before the occurrence described in this report, no screening for carbapenem-non-susceptible Enterobacteriaceae (CNSE) was performed in the ICUs.
Microbiological methods
The non-duplicate Enterobacteriaceae strains were isolated from Liaocheng People's Hospital between November 2010 and December 2012. The strains were sorted and numbered in accordance with the isolates of the time. All isolates were identified using standard conventional microbial methods. The strain identification and antimicrobial susceptibility testing were performed using a Vitek 2 automated system (bioMérieux, France). Susceptibilities were interpreted using the Clinical and Laboratory Standards Institute (CLSI) M100-S20 breakpoints19. If the minimum inhibitory concentration (MIC) to ertapenem was elevated (MIC≥ 2µg/mL), further testing by the modified Hodge test (MHT) was performed as phenotypic confirmatory method in order to investigate the mechanism of resistance.
Detection of carbapenemases
The MHT was performed as previously described19. PCRs were performed for blaKPC, blaIMP, blaVIM, blaOXA-48 and blaNDM-1 genes as previously described20–23. The amplicons were sequenced in Shanghai Sangon company, sequencing instruments and reagents were ABI-PRISM3730 and BigDyeterminator v3.1 respectively. Then the sequencing results blasted to NCBI databases (www.ncbi.nlm.nih.gov/) to identify genetype and registered to GenBank sequence database.
Molecular typing
The genotypes of isolates harboring carbapenemase genes were determined by using PFGE analysis. Salmonella Braenderup H9812 strain DNA was used as standard size markers. The genetic relationship of the test strains was generated by the unweighted pair-group method (UPGMA). Isolates that exhibited a PFGE profile with more than 80% similarity (pulsotype, PT) were considered as closely related strains.
Results
Epidemiological investigations
Thirty CNSE strains were mainly isolated from patients hospitalized in ICUs of Liaocheng People's Hospital, which includes twenty cases in NICU, three cases in Brain Hospital ICU, one case in PICU and one case in surgical ICU. Other five cases were from other clinical departments. Twenty-two patients were neonatal (73.3%) and five patients were over 60 years old (16.7%). All patients suffered from severe underlying diseases and most patients were treated with invasive procedures. Almost all patients had pulmonary infection (n=28), only two patients had orthopedic wound infections. The patient′s characteristics are shown in Table 1.
Table 1.
Characteristics of patients with carbapenem-non-susceptible Enterobacteriaceae
| Patient number/Isolatea |
Sexb/age | Wardc | Underlying diseased | Invasive procedure |
Type of specimen tested |
Infection due to CPE |
Antibiotic therapy | Outcome |
| 1/KPN1 | M/65 years |
Brain Hospital ICU |
TBI | Mechanical ventilation |
Tracheal secretion |
Pneumonia | Levofloxacin | Alive |
| 2/ECL1 | M/24 years |
Orthopedics | Tibia and fibula fracture | Plate fixation | Wound secretion |
Wound infection |
Lomefloxacin | Alive |
| 3/ECL2 | M/78 years |
Department of cardiovascular |
MI, hypertension, diabetes, GCA |
None | Tracheal secretion |
Pneumonia | Ceftizoxime | Alive |
| 4/KPN2 | M/neonatal | NICU | Premature infant, CHD, NRF, RDS, HIE |
Mechanical ventilation |
Tracheal secretion |
Pneumonia | Imipenem | Alive |
| 5/KPN3 | M/neonatal | NICU | Premature infant, MODS, NRF, RDS, HIE |
Mechanical ventilation |
Tracheal secretion |
Pneumonia | Piperacillin/tazobactam | Death unrelated to CPE |
| 6/ECL3 | F/62 years |
Brain Hospital ICU |
ICH | Mechanical ventilation |
Tracheal secretion |
Pneumonia | Ciprofloxacin | Alive |
| 7/KPN4 | F/83 years |
Brain Hospital ICU |
TBI | Mechanical ventilation |
Tracheal secretion |
Pneumonia | Levofloxacin | Alive |
| 8/KOX1 | F/54 years |
Orthopedics | Right knee purulent arthritis |
Arthroscopic Surgery |
Wound secretion |
Wound infection |
Lomefloxacin | Alive |
| 9/KOX2 | F/76 years |
Surgery ICU | NRF, CHD, RA | Mechanical ventilation |
Tracheal secretion |
Pneumonia | Moxifloxacin | Death unrelated to CPE |
| 10/KPN5 | M/neonatal | NICU | Premature infant, NRF, RDS, HIE |
Mechanical ventilation |
Tracheal secretion |
Pneumonia | Meropenem | Alive |
| 11/KPN6 | M/neonatal | NICU | Premature infant, NRF | Mechanical ventilation |
Tracheal secretion |
Pneumonia | Imipenem, cefepime | Alive |
| 12/KPN7 | M/neonatal | NICU | Premature infant, MODS, asphyxia, NRF, RDS, HIE |
Mechanical ventilation |
Tracheal secretion |
Pneumonia | Cefoperazone/sulbactam | Alive |
| 13/KPN8 | F/neonatal | NICU | Premature infant, MODS, NRF, RDS, HIE |
Mechanical ventilation |
Tracheal secretion |
Pneumonia | Cefoperazone/sulbactam, cefepime |
Alive |
| 14/KPN9 | M/neonatal | NICU | Premature infant, MODS, CHD, NRF, RDS, HIE |
Mechanical ventilation |
Tracheal secretion |
Pneumonia | Imipenem, cefepime, cefoperazone/sulbactam |
Alive |
| 15/KPN10 | M/neonatal | NICU | Asphyxia, NRF, RDS, HIE |
Mechanical ventilation |
Tracheal secretion |
Pneumonia | None | Alive |
| 16/KPN11 | M/neonatal | NICU | Premature infant, MODS, RDS, HIE, IVH |
None | Tracheal secretion |
Pneumonia | Piperacillin/tazobactam, ceftriaxone |
Alive |
| 17/KPN12 | M/neonatal | NICU | Premature infant, NRF, RDS |
Mechanical ventilation |
Tracheal secretion |
Pneumonia | Amoxicillin/sulbactam, ceftriaxone |
Alive |
| 18/KPN13 | M/neonatal | NICU | Premature infant, RDS, HIE |
Mechanical ventilation |
Tracheal secretion |
Pneumonia | Amoxicillin/sulbactam | Death unrelated to CPE |
| 19/KPN14 | M/neonatal | NICU | Premature infant, MODS, RDS |
Mechanical ventilation |
Tracheal secretion |
Pneumonia | Amoxicillin/sulbactam | Alive |
| 20/ECL4 | F/2 years |
PICU | CPR, NRF, convulsion | Mechanical ventilation |
Tracheal secretion |
Pneumonia | Amikacin, ceftazidime, cotrimoxazole |
Transferred to other hospitals |
| 21/KPN15 | M/neonatal | Neonatology department(1) |
NRF, MODS | None | Tracheal secretion |
Pneumonia | Meropenem, piperacillin/tazobactam |
Alive |
| 22/KPN16 | F/neonatal | NICU | Fetal macrosomia, HIE, MODS |
Mechanical ventilation |
Tracheal secretion |
Pneumonia | None | Alive |
| 23/KPN17 | F/neonatal | NICU | Premature infant, HIE, NRF, MODS, CHD |
None | Tracheal secretion |
Pneumonia | Piperacillin/tazobactam | Alive |
| 24/KPN18 | M/neonatal | NICU | Premature infant, HIE | None | Tracheal secretion |
Pneumonia | Meropenem, piperacillin/tazobactam |
Alive |
| 25/KPN19 | M/neonatal | NICU | HIE, asphyxia, NRF | Mechanical ventilation |
Tracheal secretion |
Pneumonia | Meropenem | Alive |
| 26/KPN20 | M/neonatal | NICU | Premature infant, HIE | Mechanical ventilation |
Tracheal secretion |
Pneumonia | Ceftriaxone | Alive |
| 27/KPN21 | M/neonatal | NICU | Premature infant, HIE | None | Tracheal secretion |
Pneumonia | Meropenem, Ceftriaxone | Alive |
| 28/KPN22 | F/neonatal | NICU | Premature infant, HIE | Mechanical ventilation |
Tracheal secretion |
Pneumonia | None | Alive |
| 29/KPN23 | M/neonatal | NICU | Premature infant, asphyxia, RDS |
Mechanical ventilation |
Tracheal secretion |
Pneumonia | Imipenem | Alive |
| 30/KPN24 | F/neonatal | Neonatology department(2) |
Premature infant, HIE | None | Tracheal secretion |
Pneumonia | Meropenem | Alive |
KPN, Klebsiella pneumoniae; ECL, Enterobacter cloacae; KOX, Klebsiella oxytoca.
M, male; F, female.
TBI, traumatic brain injury; MI, myocardial infarction; GCA, gastric cardiac adenocarcinoma; CHD, congenital heart disease; NRF, neonatal respiratory failure; RDS, respiratory distress syndrome; HIE, hypoxic-ischemic encephalopathy; ICH, intracerebral hemorrhage; RA, rheumatoid arthritis; IVH, intraventricular hemorrhage; CPR, cardiopulmonary resuscitation.
NICU, neonatal intensive care units; PICU, pediatric intensive care unit.
Infection control measures
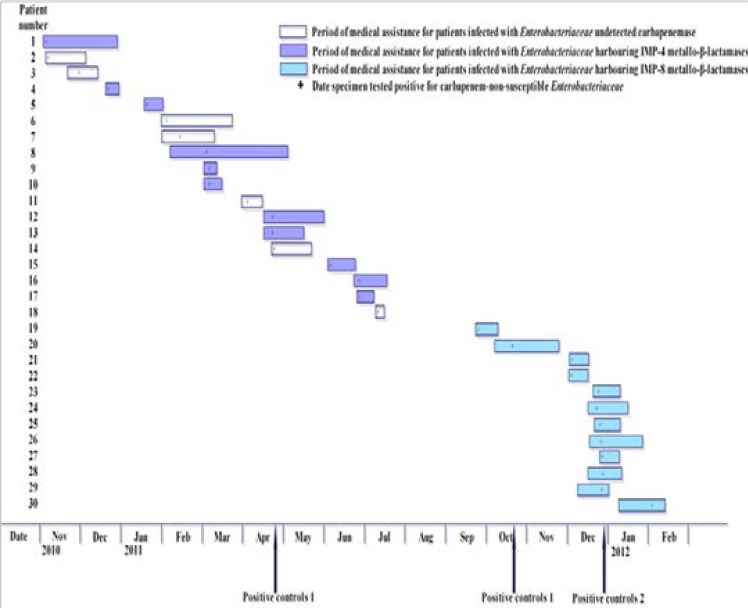
Before April 2011, there had been 10 patients infected with CNSE isolates (patients 1–10, Table 1). Due to the long time interval and lack of experiences, they did not draw the attention of clinical departments and hospital infection administrators. Follow-up medical records showed that these isolates were mainly isolated from the Brain Hospital ICU (patients 1,6,7). In addition, two orthopedic patients (patients 2, 8) were transferred to our hospital after post-operative infection in other hospitals and their information in other hospitals before transferred to our hospital was not retrieved.
Until late April 2011, consecutive cases with invasive infections for CNSE in the NICU (Patients 12–14) were detected, and the strains were all isolated from bronchial aspirate specimen. By PCRs and subsequent sequencing, blaIMP-4 genes were detected from most strains, and the isolated K. pneumoniae and Klebsiella oxytoca were clonally related. Positive control measures were implemented and health care workers were informed about CNSE by leaflets and informative meetings. The outbreak control team was organized and extended infection control measures were implemented in our hospital with the support of hospital administrators. In our hospital, Positive control 1 measures were immediately implemented at the time of admission of CPE-positive patients, to ensure that no patients came into contact with a case. All CPE-positive patients were isolated immediately after detection by contact isolation, and the objective of these measures was to prevent future patient-to-patient transmission and included:
Isolating patients in single rooms or isolating cohorts with the same species and same resistance profile, flagging the presence of cases by displaying a specific poster or logo on the CPE-positive patients beds and in the part of the ward where these patients were cohorted;
All ICU personnel, including cleaning staff and physical therapists, were instructed about standard hygiene procedures such as appropriate hand hygiene, use of gloves and gowns by medical personnel during physical contact, and use of masks by medical personnel when exposure to respiratory secretions was expected;
environmental sites were investigated for CPE contamination. The environmental samples were taken from medical devices, personal computers, telephones, door handles, floors and various other surfaces. Environmental sampling demonstrated no CPE were detected.
Despite the introduction of these measures, during the following three months, four more patients (patients 15–18) were identified in the NICU, who were infected with pneumonia with CNSE. After mid-July, no other cases were detected during the following two months.
However, on September 25, 2011 and October 19, 2011, two cases (patient 19, 20) infected with CNSE were detected, and the resistance was similar to the previous strains. But these strains were confirmed producing IMP-8 carbapenemase, then the same control measures were implemented. From December 5, 2011 to December 29, 2011, nine cases (patient 21–29) infected CNSE led to another larger outbreak. All strains were isolated from tracheal secretions in the NICU, including a case detected in the neonatology department which was transferred from NICU. The outbreak control team drew up more strict control measures, more strict control measures — Positive control 2 was implemented for the key departments, including stopping the transfer of CPE-positive patients to other units or hospitals until the case was discharged and obtaining targeted surveillance samples from all inpatients and contact patients in kinds of ICU for additional measures.
Since then, such strains have been under effective control and have not been detected in the NICU. Only one patient (patient 30) was detected in another neonatology department, and had been transferred from the NICU. The spatio-temporal description of patients infected with CNSE is shown in Figure 1.
Fig 1.
Spatio-temporal description of patients infected with carbapenem-non-susceptible Enterobacteriaceae (n=30)
Clinical outcomes
Although there was no specific antibiotic treatment program, most patients were alive and discharged (n=28). Only two patients died of causes unrelated to CNSE. The clinical outcomes of patients are shown in Table 1.
Antimicrobial susceptibility testing
According to 2010 CLSI criteria19, these recommendations include performing a MHT on isolates resistant to at least one extended-spectrum cephalosporin and with carbapenem MICs at the upper end of the susceptible range (MICs of 2 to 4 µg/ml for imipenem or meropenem or 2 µg/ml for ertapenem), as they may produce carbapenemases. If the MHT is negative, carbapenem MICs are to be reported as indicating susceptibility; if the MHT is positive, carbapenem MICs are to be reported without an interpretation and with the following comment. “This isolate demonstrates carbapenemase production. The clinical efficacy of the carbapenems has not been established for treating infections caused by Enterobacteriaceae that test carbapenem susceptible but demonstrate carbapenemase production in vitro.” The strains exhibited the same multidrug-resistant profile, all strains demonstrated insensitivity to ertapenem and 18 strains showed elevated MIC to imipenem (accounted for 60%).
Most strains exhibited resistance to cephalosporins and sulfonamides, the resistance rates to ceftriaxone, ceftazidime, cefepime and trimethoprim/sulfamethoxazole were 100%, 100%, 80% and 93.3% respectively. However, the isolates showed sensitivity to aminoglycosides, fluoroquinolones and beta-lactamase inhibitor combination, the resistance rates to gentamicin, tobramycin, ciprofloxacin and piperacillin/tazobactam were 56.7%, 16.7%, 13.3% and 20% respectively. No full resistance to tigecycline and polymyxin B was observed among the tested strains (Table 2).
Table 2.
Antimicrobial susceptibility profiles of carbapenem-non-susceptible Enterobacteriaceae
| Antimicrobial agent and MIC(µg/mL) | ||||||||||||
| Isolatea |
MH Tb |
Genoty pe |
Ertapenem | Imipenem | Gentamicin | Tobramycin | Piperacillin/tazobactam | Ceftriaxone | Ceftazidime | Cefepime | Ciprofloxacin | Trimethoprim/sulfamethoxazole |
| KPN1 | + | blaIMP-4 | 4 | 8 | ≤1 | ≤1 | 8 | ≥64 | ≥64 | 32 | ≤0.25 | ≥320 |
| KPN2 | + | blaIMP-4 | ≥8 | 2 | ≥16 | 8 | 64 | ≥64 | ≥64 | ≥64 | 2 | ≥320 |
| KPN3 | + | blaIMP-4 | 4 | ≤1 | ≥16 | 8 | 8 | ≥64 | ≥64 | ≥64 | 1 | ≥320 |
| KPN4 | − | None | 4 | 2 | ≥16 | 8 | 8 | ≥64 | ≥64 | 32 | 1 | ≥320 |
| KPN5 | + | blaIMP-4 | 4 | ≤1 | ≥16 | 8 | 8 | ≥64 | ≥64 | 8 | 0.5 | ≥320 |
| KPN6 | + | None | 4 | 8 | 4 | 8 | 8 | ≥64 | ≥64 | ≥64 | ≤0.25 | ≥320 |
| KPN7 | + | blaIMP-4 | ≥8 | ≤1 | ≥16 | 8 | 64 | ≥64 | ≥64 | 16 | 2 | ≥320 |
| KPN8 | + | blaIMP-4 | 4 | ≤1 | ≥16 | 8 | 32 | ≥64 | ≥64 | ≥64 | 2 | ≥320 |
| KPN9 | + | None | 4 | ≤1 | ≥16 | 8 | 16 | ≥64 | ≥64 | ≥64 | 2 | ≥320 |
| KPN1 0 |
+ | blaIMP-4 | ≥8 | ≤1 | ≥16 | 8 | 8 | ≥64 | ≥64 | 8 | 1 | ≥320 |
| KPN1 1 |
+ | blaIMP-4 | 4 | ≤1 | ≥16 | 8 | 8 | ≥64 | ≥64 | 8 | 2 | ≥320 |
| KPN1 2 |
+ | blaIMP-4 | 4 | 2 | ≥16 | 8 | 8 | ≥64 | ≥64 | ≥64 | 2 | ≥320 |
| KPN1 3 |
+ | None | 4 | ≤1 | ≥16 | 8 | 8 | ≥64 | ≥64 | 8 | 1 | ≥320 |
| KPN1 4 |
+ | blaIMP-8 | 4 | ≤1 | ≥16 | 8 | ≥128 | ≥64 | ≥64 | ≥64 | 1 | ≥320 |
| KPN1 5 |
+ | blaIMP-8 | 4 | 4 | 2 | 8 | 8 | ≥64 | ≥64 | ≥64 | ≤0.25 | ≥320 |
| KPN1 6 |
+ | blaIMP-8 | 4 | 4 | 4 | 8 | 8 | ≥64 | ≥64 | ≥64 | ≤0.25 | ≥320 |
| KPN1 7 |
+ | blaIMP-8 | 4 | 4 | 4 | 8 | 8 | ≥64 | ≥64 | ≥64 | ≤0.25 | ≥320 |
| KPN1 8 |
+ | blaIMP-8 | 4 | 4 | 4 | 8 | 8 | ≥64 | ≥64 | ≥64 | ≤0.25 | ≥320 |
| KPN1 9 |
+ | blaIMP-8 | 4 | 2 | 4 | 8 | 8 | ≥64 | ≥64 | ≥64 | ≤0.25 | ≥320 |
| KPN2 0 |
+ | blaIMP-8 | 4 | 4 | 4 | 8 | 8 | ≥64 | ≥64 | ≥64 | ≤0.25 | ≥320 |
| KPN2 1 |
+ | blaIMP-8 | 4 | 8 | 4 | 8 | 8 | ≥64 | ≥64 | ≥64 | ≤0.25 | ≥320 |
| KPN2 2 |
+ | blaIMP-8 | 4 | 4 | 4 | 8 | 8 | ≥64 | ≥64 | ≥64 | ≤0.25 | ≥320 |
| KPN2 3 |
+ | blaIMP-8 | 4 | 4 | 4 | 8 | 8 | ≥64 | ≥64 | ≥64 | ≤0.25 | ≥320 |
| KPN2 4 |
+ | blaIMP-8 | 4 | 2 | ≤1 | 8 | 8 | ≥64 | ≥64 | 16 | ≤0.25 | ≥320 |
| ECL1 | − | None | 4 | ≤1 | ≥16 | ≥16 | ≥128 | ≥64 | ≥64 | ≥64 | ≥4 | ≥320 |
| ECL2 | − | None | 4 | ≤1 | ≥16 | ≥16 | ≥128 | ≥64 | ≥64 | ≥64 | ≥4 | ≤20 |
| ECL3 | − | None | 4 | ≤1 | ≥16 | ≥16 | ≥128 | ≥64 | ≥64 | ≥64 | 0.5 | ≤20 |
| ECL4 | + | blaIMP-8 | ≥8 | 4 | ≤1 | ≤1 | 8 | ≥64 | ≥64 | 32 | ≤0.25 | ≥320 |
| KOX 1 |
− | blaIMP-4 | ≥8 | 8 | ≥16 | ≥16 | ≥128 | ≥64 | ≥64 | ≥64 | ≥4 | 80 |
| KOX 2 |
+ | blaIMP-4 | ≥8 | ≥16 | ≥16 | ≥16 | ≥128 | ≥64 | ≥64 | ≥64 | ≥4 | ≥320 |
Carbapenemase characterisation
All isolates were screened for carbapenemase production using phenotypic and molecular methods. The MHT yielded positive results in 25 of the 30 isolates, including 23K. pneumoniae (KPN1-KPN3, KPN5-KPN24), one strain of Enterobacter cloacae (ECL4) and one strain of K. oxytoca (KOX2), indicating the presence of a carbapenemase.
By PCRs and subsequent sequencing, blaIMP-4 genes were detected in 11 strains isolated from the patients before May 2011, while isolates from twelve patients had blaIMP-8 genes detected. No blaKPC, blaVIM, blaOXA−48, and blaNDM−1 genes were amplified positive in all strains (Table 2).
Nucleotide sequence accession number
The sequence of the blaIMP-4- containing plasmid in K. pneumoniae was submitted to GenBank under accession no. JQ808503, the blaIMP-4-containing plasmid in K. oxytoca under accession no. JQ820404. And the sequence of the blaIMP-8-containing plasmid in K. pneumoniae was submitted to GenBank under accession no. JQ820405, the blaIMP-8-containing plasmid in E. cloacae under accession no. JQ820406.
Phylogenetic analysis
PFGE of 24 K. pneumoniae isolates identified 6 pulsotypes, pulsotype (PT) PT1 to PT6 using 80% similarity as the cut-off. PT1 and PT2 were the major cluster, included 8isolates (KPN2, KPN3, KPN5, KPN7, KPN8, KPN10-KPN12) and 12 isolates (KPN13-KPN24) respectively.
PT3-6 included the other 4 isolates (KPN1, KPN4, KPN6 KPN9). PFGE revealed that two K. oxytoca had an indistinguishable PFGE pattern, while four E. cloacae were different strains.
Discussion
The emergence of Enterobacteriaceae harboring IMP-4 or IMP-8 carbapenemases described in this report faced two different outbreaks in a tertiary hospital in China. Although cases from other cities in China have been reported, all of these reports were individually IMP-4 or IMP-8 genotype, and the number of cases were small16,24–28. This occurrence included a variety of Enterobacteriaceae, which was mainly K. pneumoniae. Two kinds of PEGF types that presented by K. pneumoniae originated in the NICU, which could have been the source of such strains. Infection control departments were focused on monitoring the departments during the occurrence, and had achieved significant results. Together with the indistinguishable PFGE pattern, this strongly suggests that this mechanical ventilation represented a persistent source of contamination. The source of the IMP-producing K. oxytoca could not be fully elucidated, although the facts suggest that the PFGE pattern was closely related. Because the two patients were transferred to our hospital after post-operative infection in other hospitals. Four E. cloacae had a long separation time interval, and PFGE proved to be a different pattern, the source was not easy to determine.
The main characteristics of this occurrence are that patients were mainly from ICU and had suffered from severe underlying diseases which were treated with invasive procedure. Patients suffering from severe underlying disease are routinely and extensively given broad-spectrum anti-infective agents as prophylaxis or targeted therapy. Broad-spectrum antibiotic therapy exerts a selective pressure towards resistant organisms and affects the normal body flora. Furthermore, intensive care medicine, including use of medical devices, allows pathogens to colonize or infect patients.
In an outbreak described by Yan et al.24, sixteen patients were detected to carry blaIMP-8 and all infections were nosocomially acquired. The organisms caused wound infections in eight patients and bloodstream infections in three patients. They were not directly associated with the death of nine patients24. The outcome of our patients seemed to be better compared to the previously reported data. As the isolates were broadly resistant to most antibiotic classes, various combination therapies were used with little effect according to the in vitro susceptibility results. Fortunately, most patients were successfully treated with quinolones guided by the antimicrobial susceptibility, and the other patients were also successfully treated with broad-spectrum cephalosporins and carbapenem in spite of no reliable in vitro activity. Maybe it could be explained based on principles of PK/PD and clinical data, appropriate drainage in surgical and prompt removal of ventilator in NICU lifted the source of bacterial infection effectively. The favorable clinical outcomes experienced by the patients treated with beta-lactam antibiotics have been previously reported29, but the degree to which the antibiotic treatment contributed to these outcomes is difficult to ascertain. In the United States, three K. pneumoniae harboring IMP-4 carbapenemase were recovered from pediatric patients, and these isolates had a similar resistance with our strains30. Good clinical response to antibiotics with no in vitro activity might have been achieved by the increased antimicrobial concentration.
MHT is CLSI recommended method to detect carbapenem of Enterobacteriaceae, and has a good sensitivity and specificity to detect KPC but still uncertain to Ambler B metals enzyme19. Doyle et al.31 reported that MHT had a good effect to detect KPC and OXA-48 carbapenemase, but was insufficient to the metal enzyme IMP, VIM and NDM-1. Our study demonstrated that MHT had a high sensitivity to detect IMP-4- or IMP-8-type carbapenemase, that confirmed MHT can effectively detect Enterobacteriaceae producing IMP-type carbapenemases. The false-positive results of MHT of the CPE isolates may be attributed to the high level AmpC or ESBLs enzymes producers combined with loss or decreased expression of major porins32. It should be noted that early detection of carbapenem resistant isolates is of utmost importance in allowing adequate antimicrobial therapy to be initiated and in avoiding cross-transmission. Thus, suspicion must be high in multi-resistant enterobacterial isolates with elevated MICs to ertapenem. MICs derived by the Vitek 2 system were usually comparably low for imipenem and meropenem, whereas testing with microbroth dilution revealed higher MICs. This problem has been reported previously33. However, the Vitek 2 system is widely used in Chinese laboratories, and if included in the testing panel such as AST-GN13, ertapenem may serve as an indicator for the detection of IMP carbapenemase. A study recommended using molecular tests for the optimal detection of carbapenemase-producing Enterobacteriaceae31. When obtaining the possibility of CPE strains, they should be promptly examined by PCR to confirm the genotype.
IMP-type metalloenzymes have been reported worldwide, with a higher prevalence in southern Europe and Asia. The first acquired MBL, IMP-1, was reported in Serratia marcescens in Japan in 199134. Since then, IMP-type enzymes has been described worldwide, and endemicity of IMP-type enzymes has been reported in in many countries35. However, Most MBL producers are hospital acquired and multidrug-resistant K. pneumoniae36. The first metallo-β-lactamase reported from China was IMP-4, which was detected on a plasmid in C.youngae and reported in a 2001 publication36, followed by reports of IMP-1 and IMP-9 in a P. aeruginosa isolate37,38.
In an outbreak described by Peleg et al.39, five different gram-negative genera from patients with close epidemiological links were involved. It showed blaIMP-4 appearing highly mobile in different gram-negative strains, which brought greater challenges to infection control. Additional cross-transmission was then prevented by strict patient isolation, intensified hand disinfection and the routine analysis of targeted surveillance cultures. Preventive contact precautions should be carried out for such patients until the negative results of microbiology studies have been obtained. Although we may have terminated the outbreak in our institution, the high number of contact patients that resulted from long hospitalisation times and frequent transferring of the highly care-dependant patients makes it unlikely that the establishment of IMP in China could be prevented.
Conclusion
CPE harboring IMP-4 or IMP-8 carbapenemase strains had emerged in our hospital. Rapid dissemination of CPE was a serious concern in clinical patient care and these pathogens were now also present in China. Therefore, using surveillance cultures and initiating strict hygiene procedures is mandatory for the prevention and early detection of carbapenem-resistant Enterobacteriaceae in units where high-risk patients receive care.
Acknowledgment
This research received no specific grant from any funding agency, commercial or not-for-profit sectors.
Declaration of interest
None.
References
- 1.Pitout JD, Laupland KB. Extended-spectrum beta-lactamase-producing Enterobacteriaceae: an emerging publichealth concern. Lancet Infect Dis. 2008;8:159–166. doi: 10.1016/S1473-3099(08)70041-0. [DOI] [PubMed] [Google Scholar]
- 2.Nordmann P, Dortet L, Poirel L. Carbapenem resistance in Enterobacteriaceae: here is the storm! Trends Mol Med. 2012;18:263–272. doi: 10.1016/j.molmed.2012.03.003. [DOI] [PubMed] [Google Scholar]
- 3.Little ML, Qin X, Zerr DM, Weissman SJ. Molecular diversity in mechanisms of carbapenem resistance in paediatric Enterobacteriaceae. Int J Antimicrob Agents. 2012;39:52–57. doi: 10.1016/j.ijantimicag.2011.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.El-Herte RI, Kanj SS, Matar GM, Araj GF. The threat of carbapenem-resistant Enterobacteriaceae in Lebanon: an update on the regional and local epidemiology. J Infect Public Health. 2012;5:233–243. doi: 10.1016/j.jiph.2012.02.003. [DOI] [PubMed] [Google Scholar]
- 5.Lowe C, Katz K, McGeer A, Muller MP. Toronto ESBL Working Group. Disparity in infection control practices for multidrug-resistant Enterobacteriaceae. Am J Infect Control. 2012;40:836–839. doi: 10.1016/j.ajic.2011.11.008. [DOI] [PubMed] [Google Scholar]
- 6.Borer A, Saidel-Odes L, Eskira S, Nativ R, Riesenberg K, Livshiz-Riven I, Schlaeffer F, Sherf M, Peled N. Risk factors for developing clinical infection with carbapenem-resistant Klebsiella pneumoniae in hospital patients initially only colonized with carbapenem-resistant K pneumoniae. Am J Infect Control. 2012;40:421–425. doi: 10.1016/j.ajic.2011.05.022. [DOI] [PubMed] [Google Scholar]
- 7.Huang SR, Liu MF, Lin CF, Shi ZY. Molecular surveillance and clinical outcomes of carbapenem-resistant Escherichia coli and Klebsiella pneumoniae infections. J Microbiol Immunol Infect. 2012;28 doi: 10.1016/j.jmii.2012.08.029. [DOI] [PubMed] [Google Scholar]
- 8.Patel G, Huprikar S, Factor SH, Jenkins SG, Calfee DP. Outcomes of carbapenem-resistant Klebsiella pneumoniae infection and the impact of antimicrobial and adjunctive therapies. Infect Control Hosp Epidemiol. 2008;29:1099–1106. doi: 10.1086/592412. [DOI] [PubMed] [Google Scholar]
- 9.Livermore DM, Warner M, Mushtaq S, Doumith M, Zhang J, Woodford N. What remains against carbapenem-resistant Enterobacteriaceae? Evaluation of chloramphenicol, ciprofloxacin, colistin, fosfomycin, minocycline, nitrofurantoin, temocillin and tigecycline. Int J Antimicrob Agents. 2011;37:415–419. doi: 10.1016/j.ijantimicag.2011.01.012. [DOI] [PubMed] [Google Scholar]
- 10.Hussein K, Raz-Pasteur A, Finkelstein R, Neuberger A, Shachor-Meyouhas Y, Oren I, Kassis I. Impact of carbapenem resistance on the outcome of patients' hospitalacquired bacteraemia caused by Klebsiella pneumoniae. J Hosp Infect. 2013;10 doi: 10.1016/j.jhin.2012.10.012. [DOI] [PubMed] [Google Scholar]
- 11.Souli M, Galani I, Antoniadou A, Papadomichelakis E, Poulakou G, Panagea T, Vourli S, Zerva L, Armaganidis A, Kanellakopoulou K, Giamarellou H. An outbreak of infection due to beta-Lactamase Klebsiella pneumoniae Carbapenemase 2-producing K. pneumoniae in a Greek University Hospital: molecular characterization, epidemiology, and outcomes. Clin Infect Dis. 2010;50:364–373. doi: 10.1086/649865. [DOI] [PubMed] [Google Scholar]
- 12.Ambler RP. The structure of beta-lactamases. Philos Trans R Soc Lond B Biol Sci. 1980;289:321–331. doi: 10.1098/rstb.1980.0049. [DOI] [PubMed] [Google Scholar]
- 13.Grundmann H, Livermore DM, Giske CG, Canton R, Rossolini GM, Campos J, Vatopoulos A, Gniadkowski M, Toth A, Pfeifer Y, Jarlier V, Carmeli Y. CNSE Working Group. CNSE Working Group. Carbapenem-nonsusceptible Enterobacteriaceae in Europe: conclusions from a meeting of national experts. Euro Surveill. 2010;15:pii. doi: 10.2807/ese.15.46.19711-en. 19711. [DOI] [PubMed] [Google Scholar]
- 14.Moellering RC., Jr NDM-1--a cause for worldwide concern. N Engl J Med. 2010;363:2377–2379. doi: 10.1056/NEJMp1011715. [DOI] [PubMed] [Google Scholar]
- 15.Naas T, Nordmann P. OXA-type beta-lactamases. Curr Pharm Des. 1999;5:865–879. [PubMed] [Google Scholar]
- 16.Liu Y, Zhang B, Cao Q, Huang W, Shen L, Qin X. Two clinical strains of Klebsiella pneumoniae carrying plasmid-borne blaIMP-4, blaSHV-12, and armA isolated at aPediatric Center in Shanghai, China. Antimicrob Agents Chemother. 2009;53:1642–1644. doi: 10.1128/AAC.01325-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang Q, Wang H, Sun H, Chen H, Xu Y, Chen M. Phenotypic and genotypic characterization of Enterobacteriaceae with decreased susceptibility to carbapenems: results from large hospital-based surveillance studies in China. Antimicrob Agents Chemother. 2010;54:573–577. doi: 10.1128/AAC.01099-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ho PL, Lo WU, Yeung MK, Lin CH, Chow KH, Ang I, Tong AH, Bao JY, Lok S, Lo JY. Complete sequencing of pNDM-HK encoding NDM-1 carbapenemase from a multidrug-resistant Escherichia coli strain isolated in Hong Kong. PLoS One. 2011;6:e17989. doi: 10.1371/journal.pone.0017989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.CLSI, author. CLSI document M100-S20. Wayne, PA: Clinical and Laboratory Standards Institute; 2010. Performance standards for antimicrobial susceptibility testing: 20th informational supplement. [Google Scholar]
- 20.Marchaim D, Navon-Venezia S, Schwaber MJ, Carmeli Y. Isolation of imipenem-resistant Enterobacter species: emergence of KPC-2 carbapenemase, molecular characterization, epidemiology, and outcomes. Antimicrob Agents Chemother. 2008;52:1413–1418. doi: 10.1128/AAC.01103-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yan JJ, Hsueh PR, Ko WC, Luh KT, Tsai SH, Wu HM, Wu JJ. Metallo-beta-lactamases in clinical Pseudomonas isolates in Taiwan and identification of VIM-3, a novel variant of the VIM-2 enzyme. Antimicrob Agents Chemother. 2001;45:2224–2228. doi: 10.1128/AAC.45.8.2224-2228.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Poirel L, Héritier C, Tolün V, Nordmann P. Emergence of oxacillinase-mediated resistance to imipenem in Klebsiella pneumoniae. Antimicrob Agents Chemother. 2004;48:15–22. doi: 10.1128/AAC.48.1.15-22.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nordmann P, Poirel L, Carrër A, Toleman MA, Walsh TR. How to detect NDM-1 producers. J Clin Microbiol. 2011;49:718–721. doi: 10.1128/JCM.01773-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yan JJ, Ko WC, Tsai SH, Wu HM, Wu JJ. Outbreak of infection with multidrug-resistant Klebsiella pneumoniae carrying bla(IMP-8) in a university medical center inTaiwan. J Clin Microbiol. 2001;39:4433–4439. doi: 10.1128/JCM.39.12.4433-4439.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee NY, Wu JJ, Lin SH, Ko WC, Tsai LH, Yan JJ. Characterization of carbapenem-nonsusceptible Klebsiella pneumoniae bloodstream isolates at a Taiwanese hospital: clinical impacts of lowered breakpoints for carbapenems. Eur J Clin Microbiol Infect Dis. 2012;31:1941–1950. doi: 10.1007/s10096-011-1525-2. [DOI] [PubMed] [Google Scholar]
- 26.Chen LR, Zhou HW, Cai JC, Zhang R, Chen GX. Combination of IMP-4 metallo-beta-lactamase production and porin deficiency causes carbapenem resistance in aKlebsiella oxytoca clinical isolate. Diagn Microbiol Infect Dis. 2009;65:163–167. doi: 10.1016/j.diagmicrobio.2009.07.002. [DOI] [PubMed] [Google Scholar]
- 27.Yu F, Ying Q, Chen C, Li T, Ding B, Liu Y, Lu Y, Qin Z, Parsons C, Salgado C, Qu D, Pan J, Wang L. Outbreak of pulmonary infection caused by Klebsiella pneumoniae isolates harbouring blaIMP-4 and blaDHA-1 in a neonatal intensive care unit in China. J Med Microbiol. 2012;61:984–989. doi: 10.1099/jmm.0.043000-0. [DOI] [PubMed] [Google Scholar]
- 28.Xia Y, Liang Z, Su X, Xiong Y. Characterization of carbapenemase genes in Enterobacteriaceae species exhibiting decreased susceptibility to carbapenems in a university hospital in Chongqing, China. Ann Lab Med. 2012;32:270–275. doi: 10.3343/alm.2012.32.4.270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Weisenberg SA, Morgan DJ, Espinal-Witter R, Larone DH. Clinical outcomes of patients with Klebsiella pneumoniae carbapenemase-producing K. pneumoniae after treatment with imipenem or meropenem. Diagn Microbiol Infect Dis. 2009;64:233–235. doi: 10.1016/j.diagmicrobio.2009.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Limbago BM, Rasheed JK, Anderson KF, Zhu W, Kitchel B, Watz N, Munro S, Gans H, Banaei N, Kallen AJ. IMP-producing carbapenem-resistant Klebsiella pneumoniae in the United States. J Clin Microbiol. 2011;49:4239–4245. doi: 10.1128/JCM.05297-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Doyle D, Peirano G, Lascols C, Lloyd T, Church DL, Pitout JD. Laboratory detection of enterobacteriaceae that produce carbapenemases. J Clin Microbiol. 2012;50:3877–3880. doi: 10.1128/JCM.02117-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Anderson KF, Lonsway DR, Rasheed JK, et al. Evaluation of methods to identify the Klebsiella pneumoniae carbapenemase in Enterobacteriaceae. J Clin Microbiol. 2007;45:2723–2725. doi: 10.1128/JCM.00015-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tenover FC, Kalsi RK, Williams PP, Carey RB, Stocker S, Lonsway D, Rasheed JK, Biddle JW, McGowan JE, Jr, Hanna B. Carbapenem resistance in Klebsiella pneumoniae not detected by automated susceptibility testing. Emerg Infect Dis. 2006;12:1209–1213. doi: 10.3201/eid1208.060291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ito H, Arakawa Y, Ohsuka S, Wacharotayankun R, Kato N, Ohta M. Plasmid-mediated dissemination of the metallo-beta-lactamase gene blaIMP among clinically isolated strains of Serratia marcescens. Antimicrob Agents Chemother. 1995;39:824–829. doi: 10.1128/aac.39.4.824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Walsh TR, Toleman MA, Poirel L, Nordmann P. Metallo-beta-lactamases: the quiet before the storm? Clin Microbiol Rev. 2005;18:306–325. doi: 10.1128/CMR.18.2.306-325.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hawkey PM, Xiong J, Ye H, Li H, M'Zali FH. Occurrence of a new metallo-beta-lactamase IMP-4 carried on a conjugative plasmid in Citrobacter youngae from the People's Republic of China. FEMS Microbiol Lett. 2001;194:53–57. doi: 10.1111/j.1574-6968.2001.tb09445.x. [DOI] [PubMed] [Google Scholar]
- 37.Wang CX, Mi ZH. IMP-1 metallo-beta-lactamaseproducing Pseudomonas aeruginosa in a university hospital in the People's Republic of China. J Antimicrob Chemother. 2004;54:1159–1160. doi: 10.1093/jac/dkh498. [DOI] [PubMed] [Google Scholar]
- 38.Xiong J, Hynes MF, Ye H, Chen H, Yang Y, M'zali F, Hawkey PM. bla(IMP-9) and its association with large plasmids carried by Pseudomonas aeruginosa isolates from the People's Republic of China. Antimicrob Agents Chemother. 2006;50:355–358. doi: 10.1128/AAC.50.1.355-358.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Peleg AY, Franklin C, Bell JM, Spelman DW. Dissemination of the metallo-beta-lactamase gene blaIMP-4 among gram-negative pathogens in a clinical setting in-Australia. Clin Infect Dis. 2005;41:1549–1556. doi: 10.1086/497831. [DOI] [PubMed] [Google Scholar]