Abstract
tRNAs, a class of non-coding RNAs essential for translation, are unique among cytosolic RNA species in that they shuttle between the nucleus and cytoplasm during their life. Although their export from the nucleus has been studied in detail, limited information on import machinery was available. Our group recently reported that Ssa2p, one of major cytosolic Hsp70s in Saccharomyces cerevisiae, acts as a crucial factor for tRNA import upon nutrient starvation. Ssa2p can bind tRNAs and a nucleoporin directly in an ATP-sensitive manner, suggesting that it acts as a nuclear import carrier for tRNAs, like importin-β proteins. In vitro assays revealed that Ssa2p binds tRNA specifically but has preference for loosely folded tRNAs. In this Extra View, these features of Ssa2p as a new import factor is discussed with other recent findings related to nucleocytoplasmic transport of tRNAs reported from other groups.
Keywords: Hsp70, nuclear import, SSA2, starvation, transport carrier, tRNA, yeast
Introduction
Eukaryotic cells have been enabling complicated biological procedures by assigning whole or parts of them to different membrane-bounded organelles. The most prominent organelle is the nucleus, where the most part of genetic information resides. The nucleus has been thought as a park of RNA factories where RNA synthesis and maturation take place, and many of the product RNAs are then sent out to the cytoplasm for their own jobs. Although some RNAs functioning in the nucleus, such as U snRNAs and the telomerase RNA, have a round-trip ticket to go out to the cytosol for modification and assembly into ribonucleoproteins, all RNAs acting in the cytoplasm were thought to have only a one-way ticket. This idea was disproved about a decade ago: in the yeast Saccharomyces cerevisiae, cytoplasmic splicing of pre-tRNAs was discovered,1,2 and subsequently, nucleocytoplasmic shuttling of tRNAs were reported by 2 groups.3,4 These findings completely changed the view of the tRNAs' life. Indeed, tRNAs are heavily modified during their maturation, and are thought to have many physiological functions more than amino acid-codon adaptors in translation.5 These biological processes are closely related to intracellular dynamics of tRNA species. Thus, the mechanism of nucleocytoplasmic transport of tRNAs and its regulation have been attracting more and more interest.
Export of tRNAs from the nucleus has been studied in detail, and it was already established that the export is conducted by several parallel pathways, which require importin-β family proteins, Los1p/exportin-t and Msn5p/exportin-5, as transport carriers and are driven by the GTPase cycle of Ran (Fig. 1, [1]-[3]).5-11 On the other hand, the molecular entity of import machinery has been obscure though contribution of Mtr10p, an importin-β protein, was reported.4 A recent report has demonstrated that Ssa2p, one of the major cytosolic Hsp70s, is involved in the nuclear import of tRNAs, and has opened the door to detailed understanding of the import mechanism (Fig. 1, [4]).12 In addition, several groups have revealed characteristics of tRNA import, regulation of tRNA transport across the nuclear pore complex (NPC) and the physiological meanings of tRNA import in these years. Here, the recent progresses in analyses of nucleocytoplasmic transport of tRNAs are reviewed comprehensively, focusing on the import mechanism. New questions arisen during these progresses are also summarized for future studies.
Figure 1.
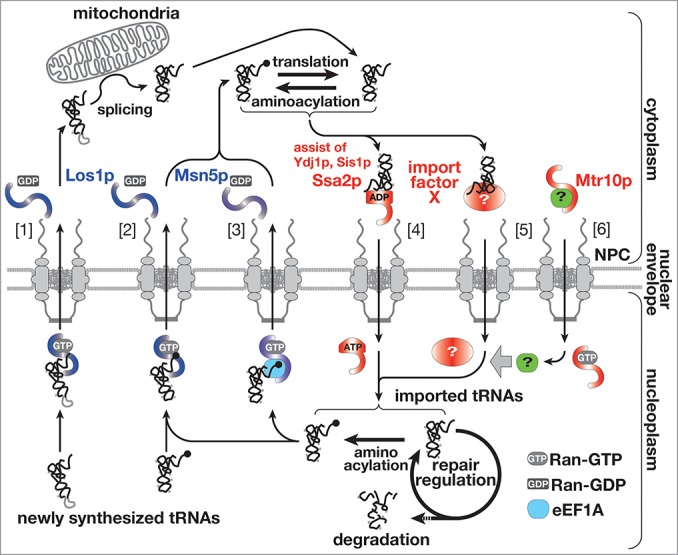
The updated view of tRNA dynamics in S. cerevisiae. Nucleocytoplasmic transport of tRNAs is schematically represented. tRNA export in the yeast is mainly catalyzed by Los1p and Msn5p. Los1p exports newly synthesized and imported mature-types tRNAs [2] in addition to intron-containing precursor tRNAs [1]. Msn5p is responsible for re-export of imported tRNAs complexed with eEF1A [3]. On the other hand, various cytoplasmic tRNAs are imported into the nucleus (here, only a deacylated mature tRNA is shown for simplicity) by Ssa2p [4]. At least, there exists another pathway for nuclear import of tRNAs, which is driven by an unknown import factor X [5] and is assisted by some additional factor imported by Mtr10p (green)[6]. tRNAs imported by such pathways and with healthy appearance are re-exported after aminoacylation. If imported tRNAs have some problems, they are subjected to repair by nuclear processing enzymes or to degradation by nucleases in the nucleus.
How are tRNAs retrograded into the nucleus in S. cerevisiae?
Mtr10p
Mtrp10 was a candidate for an import carrier of tRNAs for years. Anita Hopper's group found that deprivation of various nutrients causes fast and reversible redistribution of cytoplasmic tRNAs to the nucleus in S. cerevisiae and human.4,13,14 They found that the yeast mtr10Δ mutant is defective in nuclear accumulation of tRNAs upon starvation.4 Such a phenotype was also reproduced in MTR10 shut-off strains.12 Mtr10p mediates nuclear import of Npl3p required for mRNA export, and plays a pivotal role in nuclear import of TLC1 RNA, an essential RNA subunit of telomerase, which is assembled into a ribonucleoprotein in the cytoplasm of S. cerevisiae.15,16 Although these characteristics of Mtr10p fit for those of a tRNA import carrier, a recent report by Huang and Hopper demonstrated that there is no obvious interaction between Mtr10p and tRNAs.17 They monitored interaction between several importin-β proteins and tRNAs by formaldehyde-crosslinking and immunoprecipitation followed by RT-PCR. tRNA export carriers, Los1p and Msn5p, clearly bound tRNAs when GTP-locked Gsp1p (a yeast Ran homolog) was overproduced. However, immunoprecipitates of Mtr10p from either GTP-locked or GDP-locked Gsp1p-overexpressing yeast strains did not retain tRNAs above background. These results suggest that Mtr10p is not directly involved in tRNA movement across the NPC as an import carrier, but that Mtr10p regulates localization of some critical protein(s) involved in nuclear import of tRNAs (Fig. 1, [5] [6]).
Ssa2p
Ssa2p was revealed to be a tRNA import factor by a completely different approach; a biochemical search for novel tRNA-binding proteins that affect tRNA localization when their genes are mutated. Indeed, SSA2 deletion compromised tRNA accumulation upon amino acid starvation.12 Ssa2p is one of major cytosolic Hsp70 chaperones and belongs to the Ssa sub-family, which has 4 members, namely Ssa1p–Ssa4p.18 Ssa proteins are involved in various processes, such as protein folding, protein translocation across organellar membranes, assembly and disassembly of protein complexes, protein degradation etc.18-20 Among the 4, Ssa1p and Ssa2p, which share highly homologous sequences, are expressed constitutively while the others are expressed only under stress conditions. Interestingly, Ssa proteins other than Ssa2p do not have major contribution to tRNA import.12
Hsp70s bind proteinaceous substrates when the Hsp70s are in the ADP-binding state and release the substrates when in the ATP-binding state. Their conversion between the 2 states is assisted by so-called co-chaperones. Among the co-chaperones, DnaJ/Hsp40 proteins act as ATPase activating proteins, and help substrate selection of Hsp70s for particular biochemical processes through their variations in eukaryotic cells.20 During mutant analyses of cytosolic major DnaJs, those for Ssa proteins, Sis1p and Ydj1p, were found to be required for nuclear accumulation of tRNAs upon starvation while Zuo1p, a DnaJ homolog for Ssb proteins, another class of Hsp70s associating with ribosomes,21 was not. These results indicate that the ATPase cycle of Ssa2p is required for tRNA import into the nucleus.
Close investigation of Ssa proteins-tRNA interaction unveiled novel features of this type of Hsp70s as RNA interactors.12 First, both Ssa1p and Ssa2p can bind tRNAs directly and specifically in vitro with similar affinity and specificity. This is in marked contrast to the fact that only the ssa2Δ mutant shows apparent defects in tRNA import. Ssa1p and Ssa2p have higher affinities to in vitro-transcribed tRNA-ProUGG than that purified from yeast cells, and both the proteins prefer mutant tRNAs with moderately relaxed acceptor stem variants. These binding characteristics imply that Ssa proteins recognize tRNAs with chaperone-like substrate specificity. For proteinaceous substrates, the substrate-binding domain (SBD) located in the middle of Hsp70s contributes to such binding preference19 while the N-terminal nucleotide-binding domain (NBD) but not the SBD of Ssa proteins recognizes tRNAs, suggesting that the NBD of Ssa proteins has some chaperone-like properties for tRNA binding.12 The NBD of Hsp70s has a cleft where ATP is held to be hydrolyzed, but the cleft is too narrow to accommodate tRNAs. On the other hand, when mutations that have been found around this cleft of various Hsp70s22 were introduced to Ssa proteins, some of them affected tRNA recognition of these Ssa variants, indicating that the adenine nucleotide-binding state of Ssa proteins' NBD alters tRNA binding ability of their putative recognition interface. Interestingly, tRNA binding of Ssa1p and Ssa2p is affected differently by these mutations. For example, E173S and T204 mutations, which compromise ATPase stimulation by DnaJ homologues, cause negative effects only on Ssa2p. Thus, these differences may explain why only Ssa2p, but not Ssa1p, is involved in nuclear import of tRNAs in vivo.
How is Ssa2p involved in nuclear import of tRNAs? An important clue to answer this question was an ability of Ssa proteins to interact with a certain nucleoporin, a component of the NPC. The NPC forms a channel with a selective gate for molecules transported against their concentration gradients across the nuclear envelope, and also acts as a free diffusion channel for molecules with molecular weights less than 40–60 kDa.23 Well-known transport carriers, such as importin-β, bind nucleoporins, through their FG-repeat domains, with moderate affinities. Indeed, Ssa proteins can bind to the FG-repeat domain of Nup116p but not those from Nup100p or Nsp1p in vitro, and mediate binding of tRNAs to Nup116-coated beads.12 Thus, Ssa proteins fulfill characteristics for a transport carrier of tRNAs. In summary, Ssa2p facilitates nuclear import of tRNAs through its direct binding to tRNAs and at least one nucleoporin. Thus, the most straightforward, though not exclusive, interpretation of these results is that Ssa2p serves as an import carrier for tRNAs (Fig. 1, [4]).
Relation between Mtr10p and Ssa2p in nuclear import
There are a couple of possibilities about the relation between Mtr10p and Ssa2p. Takano et al. suggested that the Mtr10p pathway and Ssa2p pathway are parallel from the fact that the mtr10Δ ssa2Δ double mutant shows severer defects in nuclear accumulation of tRNAs than the single mutants.12 It is postulated that the Mtr10p pathway runs constitutively even under the normal growth conditions, and the Ssa2p pathway may account for additional transport capacity under stress conditions (see below). Because the effects of MTR10 mutations on tRNA redistribution upon nutrient starvation are more prominent than those of SSA2 deletion, Mtr10p may be a part of the nuclear import system with the higher capacity.
Regulation of tRNA distribution according to physiological conditions
As described above, various physiological conditions, such as shortage of amino acids, phosphate, glucose etc., alter nucleocytoplasmic distribution of tRNAs.14,24 This means that tRNA export from and/or import into the nucleus is fine-tuned by some signal transduction pathways according to the physiological conditions. There is some controversy on redistribution of tRNAs upon nutrient starvation in detailed points, such as tRNA species affected by nutrient starvation and organisms undergoing tRNA redistribution upon amino acid starvation.4,12,13,25 At least, the extent of tRNA redistribution upon nutrient deprivation varies among tRNA species, strain backgrounds, and even individual yeast cells. However, the regulatory mechanism of tRNA redistribution has become clearer than before. First, the nuclear export systems are controlled by different signal transduction pathways under different stress conditions. Especially, under glucose deprivation, nuclear accumulation of various tRNAs depends on the PKA but not Snf1 protein kinase pathway, and the PKA pathway and another unidentified signaling pathway affect distribution of importin-β family proteins between the nucleus and cytosol.14,25,26 This seems to be achieved not by direct or indirect modification of importin-β proteins, including Los1p and Msn5p, but rather by collapse of the Ran gradient across the nuclear envelope.25-27 On the other hand, the response to amino acid starvation is likely to be achieved by somehow different mechanism. First, the nutrient deprivation is sensed by the TOR and PKA pathways, but not by the Gcn4 pathway. Second, localization of Los1p or Msn5p is not changed under starvation conditions despite the fact that export of tRNAs is suppressed under these conditions.25,26 The finding that Ssa2p is a possible import carrier for tRNAs casts a new light on regulatory mechanism of tRNA redistribution upon nutrient stress. Although it has been postulated that nuclear import of tRNAs is a constitutive process, it is possible that the import capacity is also regulated like the export capacity. This seems to be the case because localization of Ssa2p is moderately affected by nutrient starvation.12 Since Ssa2p is one of major cytosolic chaperones functioning in an array of physiological scenes, it is supposed that only a fraction of Ssa2p is involved in tRNA transport. The abundance of Ssa2p is estimated as about 360,000 molecules/cell, which is 100-fold larger than that of Los1p (3,500 molecules/cell) and Msn5p (3,500 molecules/cell),28 so that such moderate difference of Ssa2p localization may be enough to change distribution of tRNAs. Although further studies are required, tRNA distribution may be regulated at multiple points both in tRNA export and import systems.
Why are tRNAs retrograded?
What are the physiological meanings of retrograde transport of cytoplasmic tRNAs? One clear physiological role of tRNA import was first identified through analysis of tRNA wybutosination by Tsutomu Suzuki's group.29 Wybutosine (yW) is found at the 37th position of tRNA-PheGAA, which is encoded by intron-containing genes, and is made from guanosine 37 (G37) by a series of complex chemical reactions.30 yW formation starts with methylation of G37 to m1G37 by a nuclear enzyme, Trm5p, and Trm5p only recognizes the spliced form of tRNA-PheGAA. Because tRNA splicing is carried out in the yeast cytoplasm,1,2 spliced tRNA-PheGAA with unmodified G37 must be retrograded for yW formation. Indeed, Suzuki's group demonstrated that this is the case. The spliced tRNA-PheGAA with m1G37 is again exported to the cytoplasm for subsequent modifications by Tyw1p-Tyw4p to yield yW37.29 Thus, the maturation of tRNA-PheGAA requires at least one round-trip between the nucleus and cytoplasm in addition to the final export. Probably, nucleocytoplasmic transport acts to organize timing of various maturation processes.
Early studies demonstrated that various tRNA species, including full-length, CCA-less, aminoacylated, and deacylated tRNAs, are subjected to nuclear import, suggesting that the import machinery has wider substrate specificity to tRNA species than the export machinery, which prefers end-matured aminoacylatable tRNAs.3,14,31 The difference in the substrate specificity between the export and import of tRNAs predicted another physiological role that the retrograde transport contributes to quality control of cytosolic tRNAs. Indeed, a recent work from Hopper's group clearly demonstrated that primary tRNA transcripts inadvertently exported from the nucleus and spliced in the cytoplasm are imported back to the nucleus for degradation or repair.32 They also showed that a part of tRNA substrates for Rapid tRNA Decay (RTD), by which cytosolic hypomodified tRNAs are eliminated, is retrograded for nuclear degradation. Another report demonstrated that tRNAs with an unstable acceptor stem receive an extra-CCA sequence to their 3′ terminus by CCA transferase, and such tRNAs are degraded by RTD.33 As mentioned above, Ssa2p prefers tRNAs with the relaxed acceptor stem, so that Ssa2p may also be implicated in sequestration of nonfunctional tRNAs from the cytosolic translational system and in delivering them to the nuclear RTD system if the cytosolic RTD fails to degrade such tRNAs.
Modulation of nucleocytoplasmic concentration of tRNAs may affect availability of certain tRNAs for cytosolic ribosomes. Indeed, los1Δ, msn5Δ, and mtr10Δ mutants were shown to affect the monosome vs. polysome ratio; especially, mtr10Δ cells have more polysomes than the wild type under starvation conditions, suggesting that tRNA redistribution itself regulates overall translation.34 In addition, polysome vs. non-polysome distribution of a group of mRNAs is clearly affected by tRNA redistribution upon amino acid starvation. Especially, expression of genes for biosynthesis of amino acids, such as Met, Arg and Leu, is affected in the translational level when tRNA transport is perturbed. Interestingly, the effect of mtr10Δ and that of msn5Δ on these mRNAs are similar to each other despite the fact that the 2 mutations cause opposite consequences in tRNA localization. This fact suggests that continuing tRNA transport itself, but not tRNA concentration in a certain compartment, is sensed and sends a cue for the translational regulation. Because similar translational regulation was reproduced when the 5′-UTRs of these target mRNAs (ARG3 and MET3) were placed in front of the GFP open reading frame, the cue from nucleocytoplasmic transport is supposed to regulate translational initiation of these mRNAs.
Nucleocytoplasmic shuttling of tRNAs is not only important for eukaryotic cells, but also for their intracellular pathogens, retroviruses. Human immunodeficiency virus-1 (HIV-1) requires certain isodecoders of tRNA-Lys as primers for its reverse transcription and integration into the nuclear genome in non-dividing human T-cells. It was reported that the reverse transcription and pre-integration complexes (RTCs) of HIV-1 are delivered to the nucleus from the cytosol with the help of the tRNA nuclear import system in human.35 Interestingly, this import system hitchhiked by viral RTCs prefers CCA-less tRNAs, which may be sequestered from the cytosol as a part of cytosolic tRNA quality control. In addition, a novel pathway for tRNA export with Transportin 3 as an export carrier is also involved in efficient integration of HIV-1 into the host genome,36 suggesting that tRNA import and export tightly regulating the HIV-1 life cycle may become unique therapeutic targets of AIDS. In summary, tRNA import into the nucleus seems to take pivotal roles in tRNA biosynthesis, maintenance, translational regulation of particular mRNAs, and even the retroviral cell cycle.
Concluding Remarks
Now, we have reached the starting point to investigate detailed mechanism of nuclear import of tRNAs, and total regulation of nucleocytoplasmic shuttling of tRNAs. However, there exist many new and old questions to be solved in front of us. Precise mechanism of Ssa2p action to transport tRNAs across the NPC is still to be investigated although we have a promising working hypothesis that Ssa2p acts as an import carrier for tRNAs. Do tRNAs diffuse into “hydrogel” made of nucleoporins only when complexed with Ssa2p, like nuclear transport cargos bound by importin-β? We do not know how a fraction of Ssa2p is set aside for this unusual task for this cytosolic Hsp70, either. It is also obscure why only Ssa2p, but not highly homologous Ssa1p, is involved in tRNA export. How is the Ssa2p pathway regulated and coordinated with the export pathways according to physiological conditions? In addition, what is the real role of Mtr10p in tRNA import? Are these import systems and their contribution to various physiological procedures conserved in higher eukaryotes? We are stepping into the next stage of tRNA transport analyses to answer these questions.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Funding
This work was supported by JSPS, Grant-in-Aid for Challenging Exploratory Research, Grant Number 26650009, and Grant-in-Aid for Scientific Research on Innovative Areas, Grant Number 15H01542.
References
- 1.Yoshihisa T, Yunoki-Esaki K, Ohshima C, Tanaka N, Endo T. Possibility of cytoplasmic pre-tRNA splicing: the yeast tRNA splicing endonuclease mainly localizes on the mitochondria. Mol Biol Cell 2003; 14:3266-79; PMID:12925762; http://dx.doi.org/ 10.1091/mbc.E02-11-0757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yoshihisa T, Ohshima C, Yunoki-Esaki K, Endo T. Cytoplasmic splicing of tRNA in Saccharomyces cerevisiae. Genes Cells 2007; 12:285-97; PMID:17352735; http://dx.doi.org/ 10.1111/j.1365-2443.2007.01056.x [DOI] [PubMed] [Google Scholar]
- 3.Takano A, Endo T, Yoshihisa T. tRNA actively shuttles between the nucleus and cytosol in yeast. Science 2005; 309:140-2; PMID:15905365; http://dx.doi.org/ 10.1126/science.1113346 [DOI] [PubMed] [Google Scholar]
- 4.Shaheen HH, Hopper AK. Retrograde movement of tRNAs from the cytoplasm to the nucleus in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A 2005; 102:11290-5; PMID:16040803; http://dx.doi.org/ 10.1073/pnas.0503836102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hopper AK. Transfer RNA post-transcriptional processing, turnover, and subcellular dynamics in the yeast Saccharomyces cerevisiae. Genetics 2013; 194:43-67; PMID:23633143; http://dx.doi.org/ 10.1534/genetics.112.147470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sarkar S, Hopper AK. tRNA nuclear export in Saccharomyces cerevisiae: in situ hybridization analysis. Mol Biol Cell 1998; 9:3041-55; PMID:9802895; http://dx.doi.org/ 10.1091/mbc.9.11.3041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hellmuth K, Lau DM, Bischoff FR, Künzler M, Hurt E, Simos G. Yeast Los1p has properties of an exportin-like nucleocytoplasmic transport factor for tRNA. Mol Cell Biol 1998; 18:6374-86; PMID:9774653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kutay U, Lipowsky G, Izaurralde E, Bischoff FR, Schwarzmaier P, Hartmann E, Görlich D. Identification of a tRNA-specific nuclear export receptor. Mol Cell 1998; 1:359-69; PMID:9660920; http://dx.doi.org/ 10.1016/S1097-2765(00)80036-2 [DOI] [PubMed] [Google Scholar]
- 9.Arts G-J, Kuersten S, Romby P, Ehresmann B, Mattaj IW. The role of exportin-t in selective nuclear export of mature tRNAs. EMBO J 1998; 17:7430-41; PMID:9857198; http://dx.doi.org/ 10.1093/emboj/17.24.7430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bohnsack MT, Regener K, Schwappach B, Saffrich R, Paraskeva E, Hartmann E, Görlich D. Exp5 exports eEF1A via tRNA from nuclei and synergizes with other transport pathways to confine translation to the cytoplasm. EMBO J 2002; 21:6205-15; PMID:12426392; http://dx.doi.org/ 10.1093/emboj/cdf613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Calado A, Treichel N, Müller EC, Otto A, Kutay U. Exportin-5-mediated nuclear export of eukaryotic elongation factor 1A and tRNA. EMBO J 2002; 21:6216-24; PMID:12426393; http://dx.doi.org/ 10.1093/emboj/cdf620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Takano A, Kajita T, Mochizuki M, Endo T, Yoshihisa T. Cytosolic Hsp70 and co-chaperones constitute a novel system for tRNA import into the nucleus. eLife 2015; 4:e04659; PMID:25853343; http://dx.doi.org/ 10.7554/eLife.04659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shaheen HH, Horetsky RL, Kimball SR, Murthi A, Jefferson LS, Hopper AK. Retrograde nuclear accumulation of cytoplasmic tRNA in rat hepatoma cells in response to amino acid deprivation. Proc Natl Acad Sci U S A 2007; 104:8845-50; PMID:17502605; http://dx.doi.org/ 10.1073/pnas.0700765104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Whitney ML, Hurto RL, Shaheen HH, Hopper AK. Rapid and reversible nuclear accumulation of cytoplasmic tRNA in response to nutrient availability. Mol Biol Cell 2007; 18:2678-86; PMID:17475781; http://dx.doi.org/ 10.1091/mbc.E07-01-0006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Senger B, Simos G, Bischoff FR, Podtelejnikov A, Mann M, Hurt E. Mtr10p functions as a nuclear import receptor for the mRNA-binding protein Npl3p. EMBO J 1998; 17:2196-207; PMID:9545233; http://dx.doi.org/ 10.1093/emboj/17.8.2196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ferrezuelo F, Steiner B, Aldea M, Futcher B. Biogenesis of yeast telomerase depends on the importin Mtr10. Mol Cell Biol 2002; 22:6046-55; PMID:12167699; http://dx.doi.org/ 10.1128/MCB.22.17.6046-6055.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang HY, Hopper AK. In vivo biochemical analyses reveal distinct roles of β-importins and eEF1A in tRNA subcellular traffic. Genes Dev. 2015; 29:772-83; PMID:25838545; http://dx.doi.org/ 10.1101/gad.258293.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Werner-Washburne M, Stone DE, Craig EA. Complex interactions among members of an essential subfamily of hsp70 genes in Saccharomyces cerevisiae. Mol Cell Biol 1987: 7:2568-77; PMID:3302682; http://dx.doi.org/ 10.1128/MCB.7.7.2568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Young JC, Agashe VR, Siegers K, Hartl F-U. Pathways of chaperone-mediated protein folding in the cytosol. Nat Rev Mol Cell Biol 2004; 5:781-91; PMID:15459659; http://dx.doi.org/ 10.1038/nrm1492 [DOI] [PubMed] [Google Scholar]
- 20.Kampinga HH, Craig EA. The HSP70 chaperone machinery: J proteins as drivers of functional specificity. Nat Rev Mol Cell Biol 2010; 11:579-92; PMID:20651708; http://dx.doi.org/ 10.1038/nrm2941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nelson RJ, Ziegelhoffer T, Nicolet C, Werner-Washburne M, Craig EA. The translation machinery and 70 kd heat shock protein cooperate in protein synthesis. Cell 1992; 71:97-105; PMID:1394434; http://dx.doi.org/ 10.1016/0092-8674(92)90269-I [DOI] [PubMed] [Google Scholar]
- 22.Chang L, Thompson AD, Ung P, Carlson HA, Gestwicki JE. Mutagenesis reveals the complex relationships between ATPase rate and the chaperone activities of Escherichia coli heat shock protein 70 (Hsp70/DnaK). J Biol Chem 2010; 285: 21282-91; PMID:20439464; http://dx.doi.org/ 10.1074/jbc.M110.124149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Adams RL, Wente SR. Uncovering nuclear pore complexity with innovation. Cell 2013; 152:1218-21; PMID:23498931; http://dx.doi.org/ 10.1016/j.cell.2013.02.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hurto RL, Tong AHY, Boone C, Hopper AK. Inorganic phosphate deprivation causes tRNA nuclear accumulation via retrograde transport in Saccharomyces cerevisiae. Genetics 2007; 176:841-52; PMID:17409072; http://dx.doi.org/ 10.1534/genetics.106.069732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chafe SC, Pierce JB, Eswara MBK, McGuire AT, Mangroo D. Nutrient stress does not cause retrograde transport of cytoplasmic tRNA to the nucleus in evolutionarily diverse organisms. Mol Biol Cell 2011; 22:1091-103; PMID:21289100; http://dx.doi.org/ 10.1091/mbc.E09-07-0594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang HY, Hopper AK. Separate responses of karyopherins to glucose and amino acid availability regulate nucleocytoplasmic transport. Mol Biol Cell 2014; 25:2840-52; PMID:25057022; http://dx.doi.org/ 10.1091/mbc.E14-04-0948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chafe SC, Pierce JB, Mangroo D. Nuclear-cytoplasmic trafficking of NTF2, the nuclear import receptor for the RanGTPase, is subjected to regulation. PLoS One 2012; 7:e42501; PMID:22880006; http://dx.doi.org/ 10.1371/journal.pone.0042501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ghaemmaghami S, Huh W-K, Bower K, Howson RW, Belle A, Dephoure N, O'Shea EK, Weissman JS. Global analysis of protein expression in yeast. Nature 2003; 425:737-41; PMID:14562106; http://dx.doi.org/ 10.1038/nature02046 [DOI] [PubMed] [Google Scholar]
- 29.Ohira T, Suzuki T. Retrograde nuclear import of tRNA precursors is required for modified base biogenesis in yeast. Proc Natl Acad Sci U S A 2011; 108:10502-7; PMID:21670254; http://dx.doi.org/ 10.1073/pnas.1105645108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Noma A, Kirino Y, Ikeuchi Y, Suzuki T. Biosynthesis of wybutosine, a hyper-modified nucleoside in eukaryotic phenylalanine tRNA. EMBO J 2006; 25:2142-54; PMID:16642040; http://dx.doi.org/ 10.1038/sj.emboj.7601105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yoshihisa T. tRNA, new aspects in intracellular dynamics. Cell Mol Life Sci 2006; 63:1813-8; PMID:16794784; http://dx.doi.org/ 10.1007/s00018-006-6092-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kramer EB, Hopper AK. Retrograde transfer RNA nuclear import provides a new level of tRNA quality control in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A 2013; 110:21042-7; PMID:24297920; http://dx.doi.org/ 10.1073/pnas.1316579110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wilusz JE, Whipple JM, Phizicky EM, Sharp PA. tRNAs marked with CCACCA are targeted for degradation. Science 2011; 334:817-21; PMID:22076379; http://dx.doi.org/ 10.1126/science.1213671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chu HY, Hopper AK. Genome-wide investigation of the role of the tRNA nuclear-cytoplasmic trafficking pathway in regulation of the yeast Saccharomyces cerevisiae transcriptome and proteome. Mol Cell Biol 2013; 33:4241-54; PMID:23979602; http://dx.doi.org/ 10.1128/MCB.00785-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zaitseva L, Myers R, Fassati A. tRNAs promote nuclear import of HIV-1 intracellular reverse transcription complexes. PLoS Biol 2006; 4:e332; PMID:17020411; http://dx.doi.org/ 10.1371/journal.pbio.0040332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhou L, Sokolskaja E, Jolly C, James W, Cowley SA, Fassati A. Transportin 3 promotes a nuclear maturation step required for efficient HIV-1 integration. PLoS Pathog 2011; 7:e1002194; PMID:21901095; http://dx.doi.org/ 10.1371/journal.ppat.1002194 [DOI] [PMC free article] [PubMed] [Google Scholar]


