Abstract
Paraspeckles are subnuclear structures that assemble on nuclear paraspeckle assembly transcript 1 (NEAT1) long noncoding (lnc)RNA. Paraspeckle formation requires appropriate NEAT1 biogenesis and subsequent assembly with multiple prion-like domain (PLD) containing RNA-binding proteins. We found that SWI/SNF chromatin remodeling complexes function as paraspeckle components that interact with paraspeckle proteins (PSPs) and NEAT1. SWI/SNF complexes play an essential role in paraspeckle formation that does not require their ATP-dependent chromatin remodeling activity. Instead, SWI/SNF complexes facilitate organization of the PSP interaction network required for intact paraspeckle assembly. SWI/SNF complexes may collectively bind multiple PSPs to recruit them onto NEAT1. SWI/SNF complexes are also required for Sat III (Satellite III) lncRNA-dependent formation of nuclear stress bodies under heat shock conditions. Organization of the lncRNA-dependent omega speckle in Drosophila also depends on the chromatin remodeling complex. These findings raise the possibility that a common mechanism controls the formation of lncRNA-dependent nuclear body architecture.
Keywords: long noncoding RNA, nuclear body, paraspeckle, nuclear stress body, omega speckle, chromatin remodeling complex, RNA-binding protein, prion-like domain (PLD)
Abbreviations
- lncRNA
long noncoding RNA
- NEAT1
nuclear paraspeckle assembly transcript 1
- nSB
nuclear stress body
- nt
nucleotide
- PSP
paraspeckle protein
- RNP
ribonucleoprotein
- RRM
RNA recognition motif
- Sat III
Satellite III
- SWI/SNF
SWItch/Sucrose Non-Fermentable.
Introduction
The nucleus is a highly compartmentalized organelle in eukaryotic cells containing genomic DNA that is packaged into chromatin and configured in specific chromosome territories. Numerous large granular structures known as nuclear bodies exist in the interchromatin space.1-3 Nuclear bodies, comprising specific proteins and (usually) RNAs, are involved in specific nuclear events including the biogenesis of macromolecular machinery such as ribosomes and spliceosomes, transcription, RNA processing, protein modification, and protein degradation.1-3 Nuclear bodies are present at a steady state and dynamically respond to various external and/or intrinsic signals. These structures are thought to form through stochastic assembly or ordered assembly of nuclear body components. In addition, a seeding mechanism has also been proposed for nuclear body assembly and maintenance.1
Recent studies have revealed that some nuclear bodies are built on specific long noncoding RNAs (lncRNAs) as their seeds or scaffolds; lncRNAs that function as scaffolds of nuclear bodies are referred to as “architectural RNAs (arcRNAs).”4 To date, 5 types of lncRNA have been classified as arcRNAs 4: nuclear paraspeckle assembly transcript 1 (NEAT1) lncRNA in the paraspeckle,5-8 ribosomal intergenic spacer lncRNAs in the nucleolar detention center,9 human Sat III lncRNAs in the nuclear stress body (nSB),10 Drosophila heat shock RNA (hsr) omega in the omega speckle,11 and fission yeast meiosis-specific RNA species, meiRNA, in the Mei2 dot.12 Thus, arcRNAs are widely present in eukaryotes and function in nuclear body construction in conjunction with multiple proteins. This article mainly describes the building process of the paraspeckle nuclear body that is seeded by and built on architectural NEAT1 lncRNA. In particular, we focus on SWI/SNF chromatin remodeling complexes, which we identify as essential factors for arcRNA-dependent assembly of nuclear bodies.
Paraspeckle Components and their Roles in Paraspeckle Assembly
Paraspeckles were initially defined as interchromatin granule-associated zones, which are frequently located adjacent to interchromatin granules, as observed by electron microscopy (EM).13 Paraspeckles were later defined as nuclear bodies enriched in characteristic RNA-binding proteins, including non-POU domain containing octamer binding (NONO) protein, paraspeckle component 1 (PSPC1), and splicing factor proline/glutamine-rich (SFPQ) protein, all of which belong to the Drosophila behavior and human splicing (DBHS) protein family.14-16 Paraspeckles are RNase sensitive, suggesting that maintenance of the intact paraspeckle structure requires RNA(s).15 In 2009, 4 groups independently discovered that NEAT1 lncRNA is an essential architectural component of paraspeckles.5-8 Paraspeckles average ∼360 nm in diameter,17 indicating that they can be considered to represent huge ribonucleoprotein (RNP) particles, which are more than 1000 times larger than ribosomes.
NEAT1 lncRNA comprises 2 isoforms, 3.7 knt NEAT1_1 and 23 knt NEAT1_2, both of which are transcribed by RNA polymerase II from a common promoter.6,7 The 3′ end of NEAT1_1 is processed by cleavage and polyadenylation specificity factor (CPSF) 5/6 complex to create the canonical 3′ terminus, whereas NEAT1_2 is subjected to non-canonical 3′ end processing by RNase P to create a non-polyadenylated terminus with the characteristic triple helix structure 7,18-20 NEAT1_2, but not NEAT1_1, is essential for paraspeckle formation. EM analysis revealed that the central region of NEAT1_2 is located in the interior part of the paraspeckle and that both the 5′ and 3′ termini of NEAT1_2, as well as NEAT1, are confined to the periphery of the paraspeckle, suggesting that the organization of NEAT1 transcripts constrains the geometry of these bodies.17
More than 40 paraspeckle proteins (PSPs) have been identified, most of which possess characteristic RNA-binding domains including RNA Recognition Motif (RRM), K-Homology motif, and RGG box.18 Further functional analyses revealed that 7 PSPs (NONO, SFPQ, RNA binding motif protein 14 (RBM14), heterogenous nuclear RNP K (HNRNPK), fused in sarcoma (FUS), DAZ-associated protein 1 (DAZAP1), and HNRNPH3) are essential for paraspeckle formation. Functional assignment of the essential PSPs revealed that paraspeckle formation proceeds via 2 distinct steps; first via NEAT1_2 biogenesis and second via further assembly of the intact paraspeckle from the NEAT1_2 subcomplexes (Fig. 1). Direct visualization of NEAT1 and PSPs showed that paraspeckles are formed on the NEAT1 chromosomal locus during transcription of NEAT1 by RNA polymerase II,21 suggesting that paraspeckle formation is initiated co-transcriptionally (Fig. 1). HNRNPK (category 1A protein in Fig. 1) accelerates the synthesis of the essential NEAT1_2 isoform by interfering with the polyadenylation of NEAT1_1 (Fig. 1).18 NONO, SFPQ, and RBM14 (category 1A proteins in Fig. 1) stabilize the synthesized NEAT1_2. FUS, HNRNPH3, and DAZAP1 (category 1B proteins in Fig. 1) scarcely affect NEAT1_2 biogenesis, but are required for assembly of the intact paraspeckle from NEAT1 subcomplexes (Fig. 1).18
Figure 1.
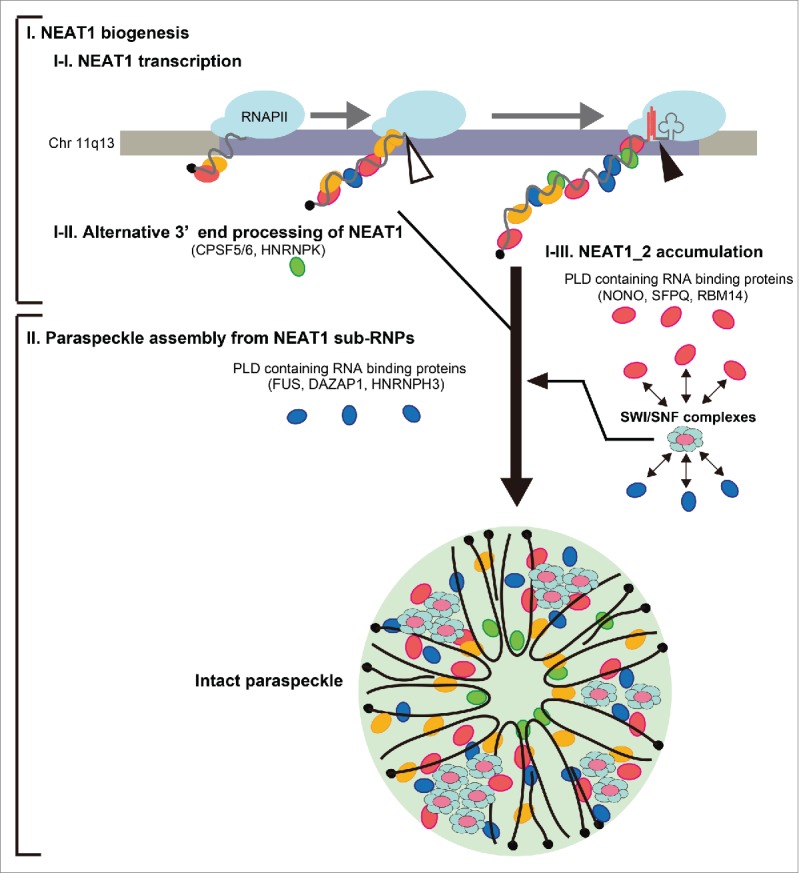
The essential steps in paraspeckle formation. Two essential steps (I and II) in paraspeckle formation are shown. In step I, NEAT1 biogenesis comprises 3 events (I-I, I-II, I-III) that occur continuously. Paraspeckle formation is initiated by NEAT1 transcription by RNA polymerase II (RNAPII) from the human NEAT1 gene locus at Chr 11q13 (I-I). NEAT1 isoforms in the diagram are not drawn to scale. The black, filled circle at the end of NEAT1 represents the cap structure. The red lines and the clover leaf at the 3′ end of NEAT1_2 represent the triple helix structure and tRNA-like structure, respectively, that are required for formation of the non-polyadenylated 3′ end of NEAT1_2. NEAT1_1 and NEAT1_2 are processed by canonical polyadenylation (open triangle) and RNase P-mediated processing (closed triangle), respectively (I-II). HNRNPK (green circle) is an essential category 1A factor in NEAT1_2 synthesis that interferes with CPSF5/6-dependent NEAT1_1 polyadenylation. Three category 1A PLD-containing RNA-binding proteins (NONO, SFPQ, and RBM14) (red circles) stabilize NEAT1_2 (I-III). In step II, SWI/SNF complexes and 3 category 1B PLD-containing RNA-binding proteins (FUS, DAZAP1, and HNRNPH3) (blue circles) act to assemble the intact paraspeckle from the NEAT1 sub-RNPs synthesized in step I. SWI/SNF complexes interact with both category 1A and 1B proteins that may be recruited during paraspeckle assembly. Non-essential paraspeckle proteins are represented by yellow circles.
Novel Function of SWI/SNF Complexes in Paraspeckle Assembly
In a search for factors that interact with the identified PSPs, we found several subunits of SWI/SNF complexes that interact with a number of PSPs.22 A subpopulation of SWI/SNF complexes is diffusely localized to the nucleoplasm, while another subpopulation is enriched in paraspeckles. EM observation revealed that SWI/SNF complexes are distributed in distinct, patchy subdomains in paraspeckles.22
SWI/SNF complexes reposition nucleosomes for transcriptional control, DNA repair, recombination, and chromosome segregation. ATP hydrolysis by the catalytic subunits BRG1 and BRM is required for their nucleosome remodeling activity.23 However, CRISPR/Cas9-mediated mutagenesis of the ATPase motif of BRG1 revealed that the ATP-dependent chromatin remodeling activity of BRG1 is not required for the function of the SWI/SNF complex in paraspeckle assembly.22 This finding suggests that the role of SWI/SNF complexes in paraspeckle assembly is novel and distinct from its well-characterized role in chromatin remodeling. EM observation supports this argument, since BRG1 is concentrated in interior paraspeckle subdomains depleted of chromatin.22
How SWI/SNF complexes facilitate paraspeckle assembly remains largely elusive. Molecular interaction studies have indicated that both BRG1 and BRM directly interact with NEAT1_2 via RNA-protein interactions, and they also interact with a number of essential PSPs via protein-protein interactions.22 Functional depletion of SWI/SNF complexes results in disassembly of paraspeckles, likely due to reduced interactions between the essential PSPs, but not NEAT1-PSP. Thus, SWI/SNF complexes may represent the molecular hub that integrates the assembly of multiple PSPs onto the architectural NEAT1_2 lncRNA (Fig. 2).22 Recently, the prion-like domain (PLD), which resides in several essential PSPs (e.g., FUS and RBM14), was shown to be required for paraspeckle assembly, presumably via the formation of a protein-protein interaction network (Fig. 2).24 An interesting possibility is that SWI/SNF complexes may facilitate interactions between PLDs in PSPs that may trigger liquid-liquid phase separation to drive larger droplet-like granule formation.25 SWI/SNF complexes remain associated with essential PSPs, even when the intact paraspeckle structure is disrupted by depletion of NEAT1, suggesting that SWI/SNF complexes collect PSPs outside the paraspeckle and then recruit them onto NEAT1_2 (Fig. 2).22
Figure 2.
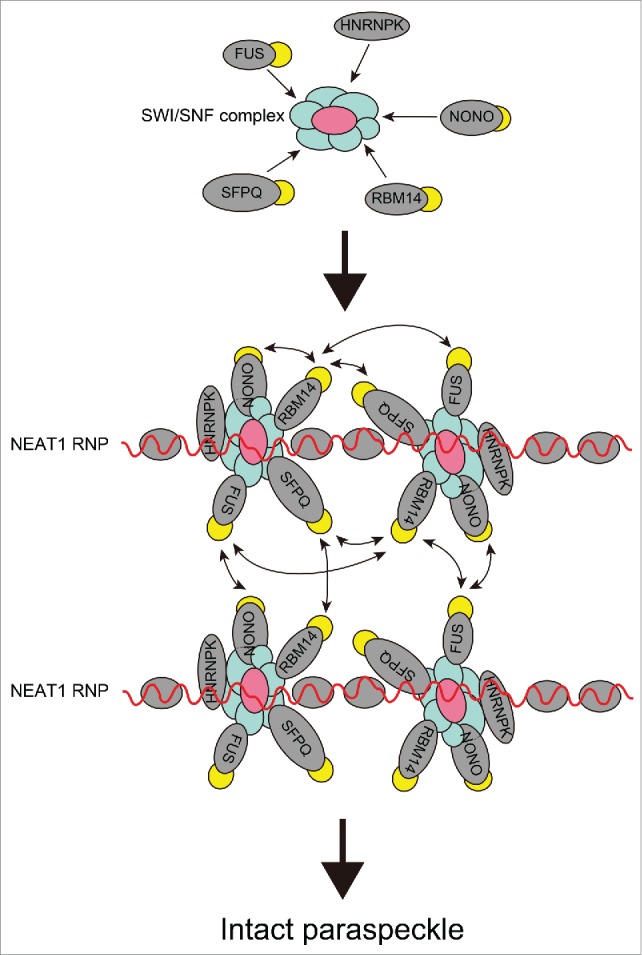
Model of the role of SWI/SNF complexes in paraspeckle assembly. SWI/SNF complexes bind to the essential PSPs outside the paraspeckle and then recruit them onto NEAT1 lncRNA (red wavy lines) to form NEAT1 RNPs. The spatially concentrated NEAT1 RNP may build massive RNP complexes via interactions between PLDs (yellow ovals) and other domains.
BRG1 and BRM lack any canonical RNA-binding motifs, suggesting that novel domain(s) are involved in their interaction with NEAT1_2. Which SWI/SNF subunits directly interact with the essential PSPs remains unknown. In addition to BRG1 and BRM, at least 4 subunits of SWI/SNF complexes (BAF170, BAF155, BAF57, and BAF47) were experimentally shown to contribute to paraspeckle formation; however, the role of each subunit remains uncharacterized. In particular, the subunit composition of paraspeckle-localized SWI/SNF complexes has been poorly investigated. An intriguing possibility is that the specific subunit composition of paraspeckle-localized SWI/SNF complexes enables them to efficiently gather the PSPs onto NEAT1_2 lncRNA.
Chromatin Remodeling Complexes are Required for the Formation of Several Distinct Lncrna-Dependent Nuclear Bodies
SWI/SNF complexes are also essential for the formation of another lncRNA-dependent nuclear body, the nSB. The nSBs form on Sat III lncRNAs that are transcribed from the pericentromeric Sat III repeats located on human chromosomes 9, 12, and 15 in response to heat shock and to various extracellular stresses.10 These nuclear bodies act to sequestrate several splicing-related RNA-binding proteins (e.g., SRSF1, SRSF7, TARDBP, and SAFB) and transcription factors (e.g., HSF1 and NFAT5).10 We showed that SWI/SNF complexes localize to the nSB, where they play an essential role in nSB formation upon heat shock.22 Analogous to its effect on paraspeckle formation, SWI/SNF depletion does not affect Sat III lncRNA expression, suggesting that SWI/SNF complexes function in the assembly of nSBs from individual Sat III RNP complexes. Further study of the interaction between SWI/SNF complexes and nSB components, including Sat III lncRNA, will clarify the commonality of the functions of SWI/SNF complexes in the assembly of these 2 lncRNA-dependent nuclear bodies.
Omega speckles are nuclear bodies in Drosophila that form on hsr-omega lncRNA in response to heat shock stress.11 Omega speckles contain several RNA-binding proteins including NonA, Hrb87F/Hrp36, Hrb57A/Bancal, Hrp59/Rumpelstiltskin, and Hrp40/Squid. NonA, Hrb87F, Hrb57A, and Hrp59 are Drosophila homologs of mammalian DBHS proteins (PSPC1, NONO, and SFPQ), HNRNPA1, HNRNPK, and HNRNPM, respectively, some of which also possess PLDs as well as RRMs. Additionally, Imitation SWI (ISWI) chromatin remodeling complex was identified as the factor that genetically interacts with hsr-omega lncRNA.26 Indeed, ISWI physically interacts with hsr-omega lncRNA and acts as the essential factor for omega speckle formation.26 Thus, paraspeckles, nSBs, and omega speckles share common features, i.e., their subunit compositions and the requirements for their construction. As expected, arcRNAs, PLD-containing RNA-binding proteins, and chromatin remodeling complexes are common factors required for their structural formation (Table 1). The missing pieces are the PLD-containing RNA-binding proteins required for nSB formation. The PLD-containing RNA-binding protein TARDBP was reported to localize to nSBs; however, its role remains to be examined. Further analysis will reveal the PLD-containing RNA-binding protein that functions in nSB formation.
Table 1.
Attributes of lncRNA-dependent nuclear bodies in different animal kingdoms
| Nuclear bodies | ArcRNAs | PLD RNA binding proteins | Chromatin remodeller | Inducing signals | Species detected |
|---|---|---|---|---|---|
| paraspeckle | NEAT1 | NONO, SFPQ, RBM14, FUS, DAZAP1 etc | SWI/SNF | virus infection, proteasome inhibition | mammalian speceis |
| nuclear stress body | Satellite III | TARDBP | SWI/SNF | heat shock, osmotic stress, heavy metal | primate species |
| omega speckle | hsr-omega | Nona, Hrb87F, Hrb57A etc | ISWI | heat shock | Drosophila |
Outlook
LncRNA-dependent nuclear bodies have been discovered, and the architectural roles of lncRNA were revealed as a new, distinct function of lncRNA. As described above, a number of arcRNAs possess similar architectural functions for building specific nuclear bodies. Together with arcRNAs, a number of proteins, including PLD-containing RNA-binding proteins and chromatin remodeling complexes, are involved in nuclear body formation. Our investigation revealed that the reciprocal interactions between arcRNA, PLD-containing RNA-binding proteins, and chromatin remodeler lead to the building of massive RNP complexes comprising nuclear bodies. In the case of paraspeckle formation, the chromatin remodeler may act as the hub to recruit PLD-containing RNA-binding proteins onto arcRNA. It is currently unclear how SWI/SNF complexes specifically target NEAT1 lncRNA. Considering that paraspeckle formation is initiated co-transcriptionally on the NEAT1 chromosome locus in conjunction with NEAT1 transcription, it is possible that SWI/SNF complexes recognize the NEAT1 chromatin region as well as NEAT1 nascent transcripts. Indeed, we have preliminary data suggesting that the association of SWI/SNF complexes on the NEAT1 chromosome locus depends on RNA polymerase II transcription. It would be interesting to pursue the detailed mechanism determining the specific recognition of SWI/SNF complexes to form paraspeckles, as well as other nuclear bodies.
The arcRNA-dependent nuclear bodies act to sequestrate specific RNA-binding proteins with affinity for the respective arcRNAs. The sequestrated proteins are thought to lose their ability to regulate transcription and/or RNA processing; therefore, these nuclear bodies are described as the “molecular sponges” of gene regulation.27,28 Paraspeckles also sequestrate specific mRNA species.16 It would be intriguing to identify the components that are responsible for sequestering respective factors onto paraspeckles and other nuclear bodies. Meanwhile, paraspeckles attach to numerous chromosome regions that are usually transcriptionally active.29 This observation raises the interesting possibility that SWI/SNF complexes localized in paraspeckles affect the activity of genomic loci that are attached to these paraspeckles. Neat1 knockout mice are normally viable, but they have defects in the development of corpus luteum and mammary glands, resulting in reduced female fertility and pup growth.30,31 Additionally, NEAT1 is regulated by estrogen receptors and is the critical modulator of prostate cancer.32 Elucidating the molecular mechanisms underlying these NEAT1-related physiological events and diseases will reveal a great deal about the importance of the interaction of arcRNA with chromatin remodeler in novel types of gene regulatory mechanisms.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Funding
TH was supported by the Funding Program for the Ministry of Education, Culture, Sports, Science, and Technology of Japan (MEXT).
References
- 1.Mao YS, Zhang B, Spector DL. Biogenesis and function of nuclear bodies. Trends Genet 2011; 27:295-306; http://dx.doi.org/ 10.1016/j.tig.2011.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Morimoto M, Boerkoel CF. The role of nuclear bodies in gene expression and disease. Biology (Basel) 2013; 2:976-1033; PMID:24040563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yamazaki T, Hirose T. The building process of the functional paraspeckle with long non-coding RNAs. Front Biosci (Elite Ed) 2015; 7:1-41; PMID:25553361; http://dx.doi.org/ 10.2741/420 [DOI] [PubMed] [Google Scholar]
- 4.Chujo T, Yamazaki T, Hirose T. Architectural RNAs (arcRNAs): A class of long noncoding RNAs that function as the scaffold of nuclear bodies. Biochim Biophys Acta 2016. (in press); http://dx.doi.org/ 10.1016/j.bbagrm.2015.05.007 [DOI] [PubMed] [Google Scholar]
- 5.Clemson CM, Hutchinson JN, Sara SA, Ensminger AW, Fox AH, Chess A, Lawrence JB. An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol Cell 2009; 33:717-26; http://dx.doi.org/ 10.1016/j.molcel.2009.01.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sasaki YT, Ideue T, Sano M, Mituyama T, Hirose T. MENepsilon/β noncoding RNAs are essential for structural integrity of nuclear paraspeckles. Proc Natl Acad Sci U S A 2009; 106:2525-30; http://dx.doi.org/ 10.1073/pnas.0807899106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sunwoo H, Dinger ME, Wilusz JE, Amaral PP, Mattick JS, Spector DL. MEN epsilon/β nuclear-retained non-coding RNAs are up-regulated upon muscle differentiation and are essential components of paraspeckles. Genome Res 2009; 19:347-59; http://dx.doi.org/ 10.1101/gr.087775.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen LL, Carmichael GG: Altered nuclear retention of mRNAs containing inverted repeats in human embryonic stem cells: functional role of a nuclear noncoding RNA. Mol Cell 2009. 35:467-4789; http://dx.doi.org/ 10.1016/j.molcel.2009.06.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Audas TE, Jacob MD, Lee S. Immobilization of proteins in the nucleolus by ribosomal intergenic spacer noncoding RNA. Mol Cell 2012; 45:147-57; http://dx.doi.org/ 10.1016/j.molcel.2011.12.012 [DOI] [PubMed] [Google Scholar]
- 10.Biamonti G, Vourc'h C. Nuclear stress bodies. Cold Spring Harb Perspect Biol 2010; 2:a000695; http://dx.doi.org/ 10.1101/cshperspect.a000695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Prasanth KV, Rajendra TK, Lal AK, Lakhotia SC. Omega speckles - a novel class of nuclear speckles containing hnRNPs associated with noncoding hsr-omega RNA in Drosophila. J Cell Sci 2000; 113 Pt 19:3485-97; PMID:10984439 [DOI] [PubMed] [Google Scholar]
- 12.Watanabe Y, Yamamoto M. S. pombe mei2+ encodes an RNA-binding protein essential for premeiotic DNA synthesis and meiosis I, which cooperates with a novel RNA species meiRNA. Cell 1994; 78:487-98; http://dx.doi.org/ 10.1016/0092-8674(94)90426-X [DOI] [PubMed] [Google Scholar]
- 13.Visa N, Puvion-Dutilleul F, Bachellerie JP, Puvion E. Intranuclear distribution of U1 and U2 snRNAs visualized by high resolution in situ hybridization: revelation of a novel compartment containing U1 but not U2 snRNA in HeLa cells. Eur J Cell Biol 1993; 60:308-21 [PubMed] [Google Scholar]
- 14.Fox AH, Lam YW, Leung AK, Lyon CE, Andersen J, Mann M, Lamond AI. Paraspeckles: a novel nuclear domain. Curr Biol 2002; 12:13-25; http://dx.doi.org/ 10.1016/S0960-9822(01)00632-7 [DOI] [PubMed] [Google Scholar]
- 15.Fox AH, Bond CS, Lamond AI. P54nrb forms a heterodimer with PSP1 that localizes to paraspeckles in an RNA-dependent manner. Mol Biol Cell 2005; 16:5304-15; http://dx.doi.org/ 10.1091/mbc.E05-06-0587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Prasanth KV, Prasanth SG, Xuan Z, Hearn S, Freier SM, Bennett CF, Zhang MQ, Spector DL. Regulating gene expression through RNA nuclear retention. Cell 2005; 123:249-63; http://dx.doi.org/ 10.1016/j.cell.2005.08.033 [DOI] [PubMed] [Google Scholar]
- 17.Souquere S, Beauclair G, Harper F, Fox A, Pierron G. Highly ordered spatial organization of the structural long noncoding NEAT1 RNAs within paraspeckle nuclear bodies. Mol Biol Cell 2010; 21:4020-7; http://dx.doi.org/ 10.1091/mbc.E10-08-0690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Naganuma T, Nakagawa S, Tanigawa A, Sasaki YF, Goshima N, Hirose T. Alternative 3′end processing of long noncoding RNA initiates construction of nuclear paraspeckles. EMBO J 2012; 31:4020-34; http://dx.doi.org/ 10.1038/emboj.2012.251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brown JA, Valenstein ML, Yario TA, Tycowski KT, Steitz JA. Formation of triple-helical structures by the 3′-end sequences of MALAT1 and MENbeta noncoding RNAs. Proc Natl Acad Sci U S A 2012. 109:19202-19207; http://dx.doi.org/ 10.1073/pnas.1217338109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wilusz JE, JnBaptiste CK, Lu LY, Kuhn CD, Joshua-Tor L, Sharp PA. A triple helix stabilizes the 3′ ends of long noncoding RNAs that lack poly(A) tails. Genes Dev 2012. 26: 2392-2407http://dx.doi.org/ 10.1101/gad.204438.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mao YS, Sunwoo H, Zhang B, Spector DL. Direct visualization of the co-transcriptional assembly of a nuclear body by noncoding RNAs. Nat Cell Biol 2011; 13:95-101; http://dx.doi.org/ 10.1038/ncb2140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kawaguchi T, Tanigawa A, Naganuma T, Ohkawa Y, Souquere S, Pierron G, Hirose T. SWI/SNF chromatin-remodeling complexes function in noncoding RNA-dependent assembly of nuclear bodies. Proc Natl Acad Sci U S A 2015; 112:4304-9; http://dx.doi.org/ 10.1073/pnas.1423819112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Clapier CR, Cairns BR. The biology of chromatin remodeling complexes. Annu Rev Biochem 2009; 78:273-304; http://dx.doi.org/ 10.1146/annurev.biochem.77.062706.153223 [DOI] [PubMed] [Google Scholar]
- 24.Hennig S, Kong G, Mannen T, Sadowska A, Kobelke S, Blythe A, Knott GJ, Iyer KS, Ho D, Newcombe EA, et al.. Prion-like domains in RNA binding proteins are essential for building subnuclear paraspeckles. J Cell Biol 2015; 210:529-39; http://dx.doi.org/ 10.1083/jcb.201504117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guo L, Shorter J. It's Raining Liquids: RNA Tunes Viscoelasticity and Dynamics of Membraneless Organelles. Mol Cell 2015; 60:189-19226; http://dx.doi.org/ 10.1016/j.molcel.2015.10.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Onorati MC, Lazzaro S, Mallik M, Ingrassia AM, Carreca AP, Singh AK, Chaturvedi DP, Lakhotia SC, Corona DF. The ISWI chromatin remodeler organizes the hsromega ncRNA-containing omega speckle nuclear compartments. PLoS Genet 2011; 7:e1002096; http://dx.doi.org/ 10.1371/journal.pgen.1002096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hirose T, Virnicchi G, Tanigawa A, Naganuma T, Li R, Kimura H, Yokoi T, Nakagawa S, Benard M, Fox AH, Pierron G. NEAT1 long noncoding RNA regulates transcription via protein sequestration within subnuclear bodies. Mol Biol Cell 2014; 25:169-83; http://dx.doi.org/ 10.1091/mbc.E13-09-0558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Imamura K, Imamachi N, Akizuki G, Kumakura M, Kawaguchi A, Nagata K, Kato A, Kawaguchi Y, Sato H, Yoneda M, et al.. Long Noncoding RNA NEAT1-Dependent SFPQ Relocation from Promoter Region to Paraspeckle Mediates IL8 Expression upon Immune Stimuli. Mol Cell 2014; 53:393-406; http://dx.doi.org/ 10.1016/j.molcel.2014.01.009 [DOI] [PubMed] [Google Scholar]
- 29.West JA, Davis CP, Sunwoo H, Simon MD, Sadreyev RI, Wang PI, Tolstorukov MY, Kingston RE. The long noncoding RNAs NEAT1 and MALAT1 bind active chromatin sites. Mol Cell 2014; 55:791-802; http://dx.doi.org/ 10.1016/j.molcel.2014.07.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nakagawa S, Shimada M, Yanaka K, Mito M, Arai T, Takahashi E, Fujita Y, Fujimori T, Standaert L, Marine JC, et al.. The lncRNA Neat1 is required for corpus luteum formation and the establishment of pregnancy in a subpopulation of mice. Development 2014; 141:4618-27; http://dx.doi.org/ 10.1242/dev.110544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Standaert L, Adriaens C, Radaelli E, Van Keymeulen A, Blanpain C, Hirose T, Nakagawa S, Marine JC. The long noncoding RNA Neat1 is required for mammary gland development and lactation. RNA 2014; 20(12):1844-9; http://dx.doi.org/ 10.1261/rna.047332.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chakravarty D, Sboner A, Nair SS, Giannopoulou E, Li R, Hennig S, Mosquera JM, Pauwels J, Park K, Kossai M, MacDonald TY, et al.. The oestrogen receptor α-regulated lncRNA NEAT1 is a critical modulator of prostate cancer. Nat Commun 2014; 5:5383; http://dx.doi.org/ 10.1038/ncomms6383 [DOI] [PMC free article] [PubMed] [Google Scholar]


