Abstract
In non-plant systems, chromatin association with the nuclear periphery affects gene expression, where interactions with nuclear envelope proteins can repress and interactions with nucleoporins can enhance transcription. In plants, both hetero- and euchromatin can localize at the nuclear periphery, but the effect of proximity to the nuclear periphery on gene expression remains largely unknown. This study explores the putative function of Seh1 and Nup50a nucleoporins on gene expression by using the Lac Operator / Lac Repressor (LacI-LacO) system adapted to Arabidopsis thaliana. We used LacO fused to the luciferase reporter gene (LacO:Luc) to investigate whether binding of the LacO:Luc transgene to nucleoporin:LacI protein fusions alters luciferase expression. Two separate nucleoporin-LacI-YFP fusions were introduced into single insert, homozygous LacO:Luc Arabidopsis plants. Homozygous plants carrying LacO:Luc and a single insert of either Seh1-LacI-YFP or Nup50a-LacI-YFP were tested for luciferase activity and compared to plants containing LacO:Luc only. Seh1-LacI-YFP increased, while Nup50a-LacI-YFP decreased luciferase activity. Seh1-LacI-YFP accumulated at the nuclear periphery as expected, while Nup50a-LacI-YFP was nucleoplasmic and was not selected for further study. Protein and RNA levels of luciferase were quantified by western blotting and RT-qPCR, respectively. Increased luciferase activity in LacO:Luc+Seh1-LacI-YFP plants was correlated with increased luciferase protein and RNA levels. This change of luciferase expression was abolished by disruption of LacI-LacO binding by treating with IPTG in young seedlings, rosette leaves and inflorescences. This study suggests that association with the nuclear periphery is involved in the regulation of gene expression in plants.
Keywords: Arabidopsis thaliana, chromatin organization, gene expression, higher plant, LacO:LacI, Nuclear pore complex, Seh1
Abbreviations
- CCLR
Cell culture lysis reagent
- NPC
Nuclear Pore Complex
- NE
Nuclear Envelope
- SUN
Sad1/Unc84
- CCT
Chromatin Charting
- IPTG
Isopropyl β-D-1-thiogalactopyranoside
- CDS
Coding Sequence.
Introduction
A large body of evidence from metazoan and yeast research shows that the various components of the nuclear periphery - nuclear envelope (NE), lamina and nuclear pore complexes (NPC) - play crucial roles in organizing the genome and in the regulation of gene expression. Chromatin anchorage at the nuclear periphery affects gene expression and nucleic acid metabolism. Generally, it has been found that association of genes with NE proteins or the lamina causes their repression while localization of genes to the nuclear pores is mainly associated with active transcription. In addition to active genes, RNA processing machinery is also associated with nuclear pores, which enhances or accelerates export of mRNA from the nucleus.3,12,13,20 Nucleoporins involved in regulation of gene expression in metazoans and yeast include Nup98/Nup145N, Sec13, Nup62/Nsp1, Nup153/Nup60, Nup50, TPR/Mlp, Nup96, Nup155/Nup107, Nup93 and Nup88/Nup82. These effects are achieved by either directly interacting with chromatin, for instance binding to the promoter region, or by interacting with components of the transcription and RNA processing machinery12,20; thus the effect of nucleoporins on gene expression may occur at the pore or within the nucleus.
While heterochromatic loci such as telomeres and chromocentres are preferentially associated with the nuclear periphery in plants,5,14 it is unknown whether and how the plant NE, lamina and NPC may affect gene expression.17 One reason for this is that until recently only a handful of plant nuclear periphery components were known. Homologues of most of the approximately 60 metazoan and yeast membrane intrinsic NE proteins do not exist in plants,9 with the exception of the Sad1/Unc84 (SUN) domain protein family.9,10 While plants have a structural lamina, BLAST search failed to identify true lamin orthologues of its animal components.2,4,7,8 Plant homologues of nucleoporins were recently identified yet many remain to be functionally characterized.16,22
This study is a first investigation of the effects of plant nucleoporins on gene expression. The nucleoporins AtNup50a and AtSeh1 were chosen because their orthologues in Drosophila melanogaster were previously shown to either enhance (Seh1) or repress (Nup50) gene activity.1 To study changes in gene expression, we chose the marker gene luciferase used in combination with the LacI-LacO gene tethering system pioneered by Rosin et al.19 Rosin et al.19 created Arabidopsis thaliana Col0 chromatin charting (CCT) plants, which contain a transgene consisting of the bacterial lac operator (LacO) fused in frame to the coding sequence of luciferase (Luc; LacO:Luc transgene). In this study, the lac repressor (LacI) was fused to either AtSeh1 or AtNup50a and expressed in the LacO:Luc plants to tether Luc to either AtSeh1 or AtNup50a via LacI-LacO binding interactions. Changes in Luc expression were examined by luciferase assays, western blot and RT-qPCR.
Materials and Methods
Plasmids and cloning
The coding sequence (CDS) for LacI was amplified from pJM71 plasmid19 using primers FLacIr (ATGGTGAAATATGTAACGTTATACGATGTCGC) and RLacIr (TGGAGTACAGGATCCCAGCTGC). cDNA from total RNA extract of Arabidopsis thaliana leaf tissue was used as template to amplify the CDS of Seh1 and Nup50a using the following primers: FSeh1 (ATGGCGAAATCAATGGCG), RSeh1 (GGAGGGAACTGGTTCAAG), LacI-Seh1, FNup50a (ATGGGTGACTCGGAAAACG) and RNup50a (AGTATCTGTAGCTGTTGGAG). LacI-Seh1 and LacI-Nup50a fusions were made by overlapping PCR using primers FNup50aOL (TCTCCAACAGCTACAGATACTAGCAGCCTGATGGTGAAATATGTAACG), RNup50aOL (TAACGTTACATATTTCACCATCAGGCTGCTAGTATCTGTAGCTGTTGGAG), FSeh1OL (GCTTGAACCAGTTCCCTCCTAAGCAGCCTGATGGTGAAATATGTAACG) and RSeh1OL (TAACGTTACATATTTCACCATCAGGCTGCTGGAGGGAACTGGTTCAAG). Gateway cloning was used to insert the constructs in frame with YFP into pCambia1300 vectors. Plasmids were sequenced to ensure mutation-free cloning. Agrobacterium tumefaciens GV3101 cells were heat-shock transformed with pCambia1300:p35S:Seh1-LacI and pCambia1300:p35S:Nup50-LacI and transformed cultures stored at −80°C.
Transgenic plants
All Arabidopsis thaliana lines used here were Col0 ecotype. Arabidopsis plants containing a single insert of the transgene LacO:Luc, where luciferase expression is controlled by a 35S promoter, were obtained from Eric Lam.19 CCT424 and CCT447 lines were selected for the present work due to their moderate level of luciferase activity. Results with the CCT424 line are described in this paper but comparable results were obtained with the CCT447 line (data not shown). Both LacO:Luc lines were floral dipped with agrobacteria containing pCambia1300:p35S:Seh1-LacI-YFP or pCambia1300:p35S:Nup50-LacI-YFP. Transformed CCT lines were selected with hygromycin B and the F3 generation genotyped for homozygous lines and TAIL-PCR was used to identify lines with a single transgene insertion. These lines are referred to as tethered LacO:Luc+Seh1-LacI-YFP and LacO:Luc+Nup50a-LacI-YFP. The homozygous LacO:Luc+Seh1-LacI-YFP line was out-crossed with wild type Col0 to isolate F2 isogenic lines carrying either the LacI or the LacO transgenes and a line with no transgenes as control. Subsequently, F3 seeds were grown to compare the effect of the tethering construct on LacO:Luc expression.
Plant growth
Seeds were surface-sterilised by washing for 10 min in 70% ethanol, 0.05% TritonX-100 followed by 10 min with 96% ethanol and left to dry. Seeds were imbibed at 4°C in the dark in 0.1% agar for 3 to 5 d and were plated in 0.5 x MS agar supplemented with 1% sucrose. Plates were placed in a culture room, at 25°C with 16h light and 8h dark. Seedlings were used at 7 d post germination, then tissue was harvested (approximately 100 mg) and either used immediately or snap frozen in liquid nitrogen and stored at -80°C. Seven-day old seedlings were also moved to soil and grown to adult stage for rosette leaf, stem, inflorescence and seed tissue harvest.
For IPTG treatment, a solution of 100mM IPTG (Sigma) in 0.015% Silwet-l77 (Sigma) was applied with a paintbrush to shoots and roots of seedlings 6 or 7 d after germination growing on a 0.5 x MS, 1% sucrose, agar plate placed vertically. Plates were sealed with micropore tape and placed in a culture room until harvest. Mock controls were treated similarly at the same time but without IPTG. Treatment was applied 20h before tissue harvest.
Luciferase assay
Fresh or frozen tissue was homogenized using a cooled Tissue Lyser (Qiagen) and tungsten beads. Cell culture lysis reagent (CCLR), part of the Luciferase Assay System (Promega), was used as extraction buffer. The samples were spun down at 4°C, 13000 rpm for 10 min and the supernatant removed to fresh tubes and kept on ice. Total protein extract was quantified using the RC/DC assay (BioRad). Samples were diluted with CCLR to the same total protein concentration. For each reaction, 100 µl Luciferase Assay Reagent (LAR, Promega) and 20 µl sample were used and for each sample 4 repeat reactions were prepared. A MicroBetaTriLux luminometer (Perkin Elmer) was used to measure luminescence. In each well, luminescence was counted for 5 s. Luminescence counts per second for each sample were converted to a percentage scale using the average LacO:Luc count as 100%. Experiments were repeated with at least 3 biological and 4 technical replicates and analyzed by Student’s t-test using Microsoft Excel.
SDS-PAGE and immunoblotting
Proteins were extracted as described for the luciferase assay. Ice cold acetone was added and proteins precipitated for 2h at −20°C. Samples were centrifuged for 10min at 13000 rpm at 4°C, the supernatant removed and the pellet dried. The dried pellet was resuspended in Laemmli buffer 11 and stored at −20°C. The protein extract was loaded on a 10% SDS gel and proteins separated by SDS-PAGE. Proteins were blotted onto nitrocellulose membrane and blocked over night at 4°C in 5% milk PBST. Blots were labeled with anti-luciferase antibody (Sigma) as primary antibody and goat anti-rabbit Cy5 (Jackson Immunoresearch) as secondary antibody. Antibody labeling was imaged with a ChemiDoc transluminator (BioRad). Images were analyzed using ImageJ to measure the band intensity. Western blots were repeated more than 3 times and data presented for one blot is representative of these experiments. Band intensities were normalized onto a percentage scale by using the average LacO:Luc band intensity as 100%. Data was analyzed using Microsoft Excel.
RT-qPCR
Tissue was ground in liquid nitrogen with a mortar and pestle and stored at -80C. RNA was extracted using a Nucleospin RNA extraction kit (Macherey-Nagel) following manufacturer’s recommendations. rDNase digestion was performed in solution after elution of RNA and a Nucleospin RNA Clean-up kit (Macherey-Nagel) was used to re-purify the RNA. RNA was quantified with a nanodrop spectrophotometer. The RT reactions were performed with VILO cDNA synthesis kit (Invitrogen) according to the manual (using typically 100ng RNA in 10μl reaction volume). cDNA was diluted fold10- and checked by PCR using GAPC primers to assess contamination with genomic DNA and consistency in samples. For the qPCR, SYBR Green (Applied Biosystems) was used according to manufacturer’s recommendations in an ABI 7500 qPCR cycler, with standard thermal profile. Primer pairs were tested to ensure high and similar amplification efficiencies. LUC expression (primers FLUC1 GGCGTTAATCAGAGAGGCGA and RLUC1 TCGCCTCT`CTGATTAACGCC) was normalized to the expression of OTC (AT1G75330, primers OTC_qRT-F TGAAGGGACAAAGGTTGTGTATGTT and OTC_qRT-R CGCAGACAAAGTGGAATGGA). Seh1-LacI-YFP (no LacO:Luc) and wild-type samples were used to control for unspecific LUC amplification or sample contamination. All samples were run in triplicate. For comparing LUC expression in tethered versus non-tethered lines, data from 5 RT-qPCR experiments were combined yielding a total of 8 biological replicates per line (one extreme outlier in LacO:Luc samples was removed from analysis) . For the IPTG experiment, 3 biological replicates were used. Relative LUC RNA levels were calculated using the ddCt method.21 Data analysis and Student’s t-tests were performed using Microsoft Excel.
Results
Nucleoporin-LacI tethering transgenes alters LacO:luciferase activity
Over 200 CCT lines were created by,19 which contain a single copy of the LacO:Luc transgene randomly inserted in the genome. In some of these lines, luciferase expression, as inferred by luciferase activity assays, was higher than in others indicating the positioning of the transgene was affecting luciferase expression.19 For this study, we chose CCT lines with medium luciferase expression levels to measure a putative increase and/or decrease in luciferase activity. These were transformed with either p35S:Seh1-LacI-YFP or p35S:Nup50a-LacI-YFP. Ten day old homozygous, single insert seedlings were used to measure luciferase activity and therefore detect whether the presence of either Seh1-LacI-YFP or Nup50a-LacI-YFP caused any changes in luciferase levels. Luminescence counts per second were used as measure for luciferase activity and were normalized to a percentage scale, where luminescence in the non-tethered LacO:Luc line was set at 100%. Co-expression of Seh1-LacI-YFP caused a significant increase in luciferase activity (227.7 ± 22.2%; p<0.01), while co-expression of Nup50a-LacI-YFP caused a significant decrease in luciferase activity (17.1 ± 0.5%; p<0.01; Fig. 1).
Figure 1.
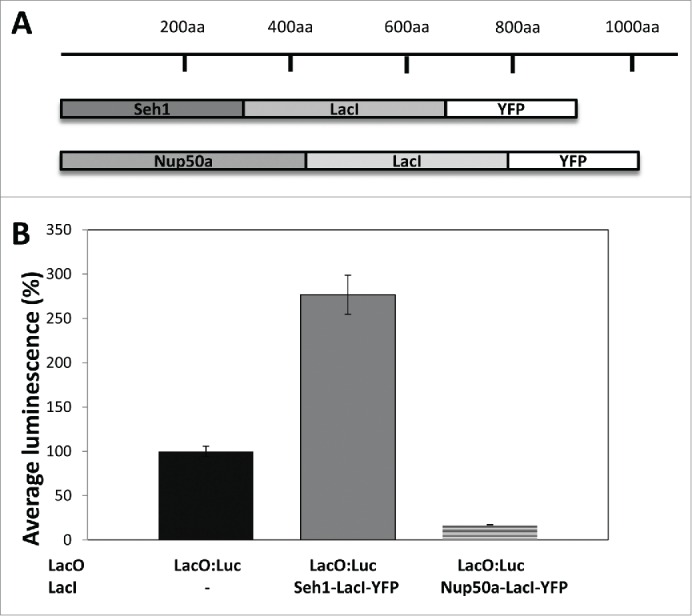
Nucleoporin-LacI tethering transgenes alters LacO:luciferase activity A) Seh1-LacI-YFP and Nup50a-LacI-YFP transgenes. B) Luciferase activity was measured in non-tethered LacO:Luc lines (black) and tethered LacO:Luc lines, where either Seh1-LacI-YFP (gray) or Nup50a-LacI-YFP (dashed line) were present. Seh1 caused an increase (277 ± 22%; p <0.01) and Nup50a a decrease (17.1 ± 0.5%; p<0.01) in luciferase activity. Average ± sem.
Confocal microscopy was used to observe the localization of the nucleoporin-LacI-YFP tethers. Seh1-LacI-YFP was mainly accumulated at the nuclear periphery in root cells, lower epidermal cotyledon cells and trichomes (Figs. 2A–C). On the other hand, Nup50a-LacI-YFP did not localize to the nuclear periphery but instead was nucleoplasmic in all tissues observed (Fig. 2D and E). These subcellular localisations are similar to those reported by 22 and indicate that the LacI moiety does not affect the proper targeting and localization of the 2 nucleoporins. The differing subcellular localisations of the Seh1-LacI-YFP tether and the Nup50a-LacI-YFP tether may partly explain the differing luciferase levels in the 2 lines. In an attempt to investigate further the possible enhancement of gene expression mediated by the NPC, we decided to focus on the Seh1-LacI-YFP tether. The enhanced expression of luciferase in the presence of Seh1-LacI-YFP was further confirmed in F3 isogenic lines derived from initial transgenic plants co-expressing LacO:Luc and Seh1-LacI-YFP (data not shown).
Figure 2.
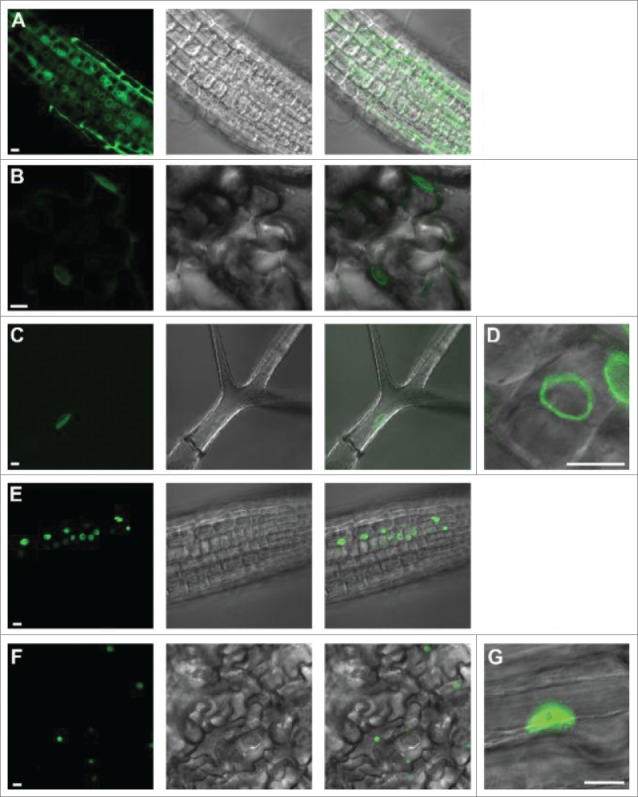
Subcellular localization of LacI-YFP tethers in Arabidopsis thaliana.A-D) Seh1-LacI-YFP is expressed in root tip (A), cotyledon lower epidermal (B) and trichome (C, D) nuclei. In all these tissues, expression is at the nuclear periphery (D). E-G) Nup50a-LacI-YFP is expressed in root tip (E) and cotyledon lower epidermal cells (F, G). In all these tissues expression is nucleoplasmic (G). Size bar = 10 μm (A-F lower left; D, G. right).
Seh1-LacI-YFP increases LacO:Luc transcription
Having found an increase in luciferase activity in the LacO:Luc+Seh1-LacI-YFP line, we wanted to establish whether this was correlated with an increased luc expression at the RNA and protein levels. RT-qPCR was used to compare luc expression levels in the line with no Seh1-LacI-YFP (LacO:Luc only) with the line co-expressing the Seh1-LacI-YFP tether (LacO:Luc+Seh1-LacI-YFP). LUC RNA levels were calculated relative to LacO:Luc. The presence of the Seh1-LacI-YFP tether increased levels of luciferase mRNA by nearly fold2- (1.9 ± 0.18) compared to LacO:Luc only (P<0.001) indicating that luc transcription was upregulated (Fig. 3A). In addition to increased luciferase mRNA, the presence of the Seh1-LacI-YFP tether also significantly increased the level of luciferase protein – 190 ± 7% compared to LacO:Luc luciferase protein levels of 100% (p=0 .001; Fig. 3B). Therefore, our results suggest that increased luciferase mRNA levels lead to increased luciferase protein levels, which in turn result in increased luciferase activity (Fig. 1). Hence, measuring luciferase activity in this system is an appropriate indicator for luc expression levels.
Figure 3.
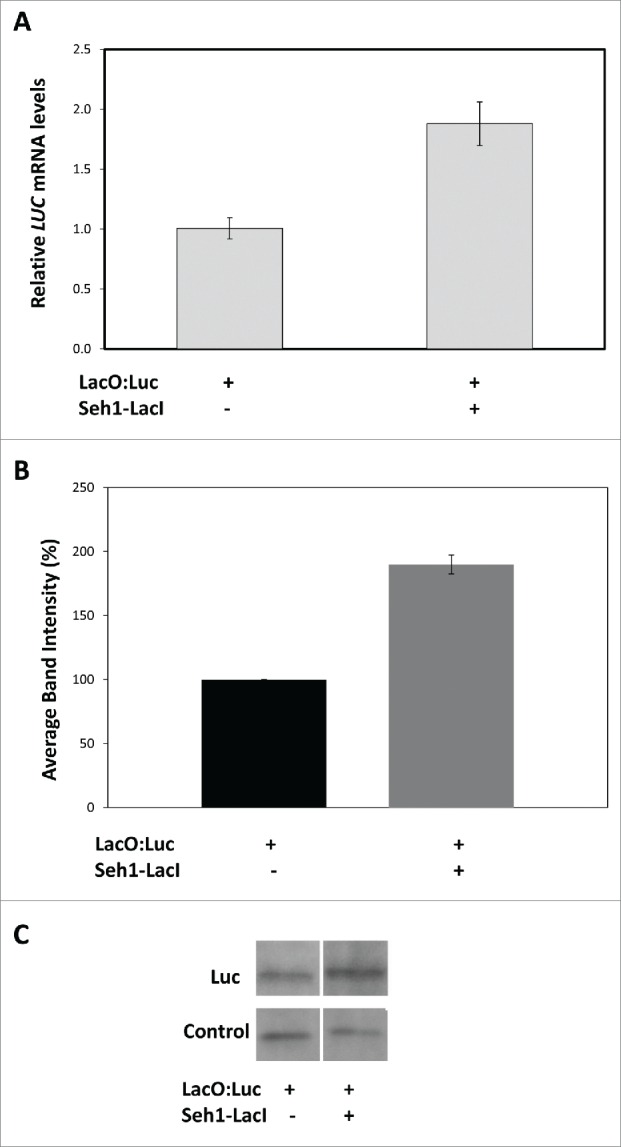
Luciferase mRNA and protein levels in LacO:Luc and LacO:Luc+Seh1-LacI lines. A) Relative expression as measured by RT-qPCR showed a significant increase in luciferase transcription levels in the LacO:Luc+Seh1-LacI-YFP line (1.9 ± 0.18, n=8 ) compared to the LacO:Luc only line (1.0 ± 0.09, n=7 ), p <0.001. Average of biological replicates ± sem ; B and C) Protein levels were inferred from band intensity of the luciferase band and a non-specific control band in western blot analysis. While protein levels remained similar in the control band the intensity of the luciferase increased in LacO:Luc+Seh1-LacI-YFP line (190 ± 7%; p=0 .001) indicating a specific increase in luciferase protein levels upon co-expression with Seh1 transgene. Data from one blot representative of 3 biological repeats.
In order to test whether the enhanced expression of Luc resulted from specific binding of LacI to LacO, we applied isopropyl b-D-1-thiogalactopyranoside (IPTG), which reduces the affinity of LacI for LacO 1000-fold 15,18 used IPTG treatment followed by RT- qPCR to assay the disruption of specific binding of GFP-LacI to LacO in tobacco. Using both RT-qPCR and western blotting, we measured RNA and protein levels 20h post mock-treatment or 100mM IPTG treatment. While IPTG treatment neither affected RNA nor protein levels in the non-tethered LacO:Luc line, it did cause a significant decrease (p=0 .037) in the Seh1-LacI-YFP tethered line (Fig. 4).Thus we conclude that specific interaction was occurring between LacI and LacO in our system.
Figure 4.
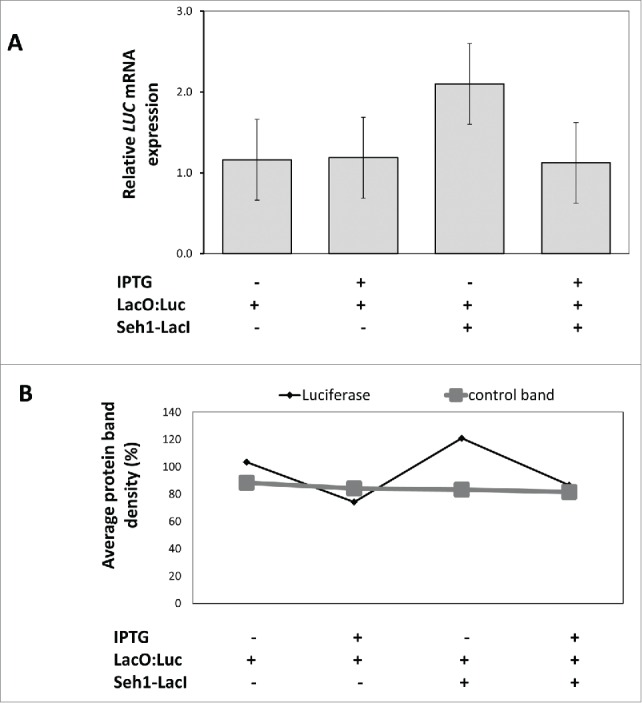
Effect of IPTG on luciferase expression in Seh1-LacI-YFP tethered lines.A) RT-qPCR of total RNA extracted from tethered and non-tethered lines 20h after mock or 100 mM IPTG treatment. RNA levels of luciferase were similar in mock-treated (1.16± 0.47) and IPTG-treated (1.19 ± 0.32) LacO:Luc seedlings (n=3 , p=0 .45). In the LacO:Luc+ Seh1-LacI-YFP tethered line, luciferase RNA levels decreased after IPTG treatment (1.12 ± 0.22) compared to the mock treatment (2.10 ± 0.49) n=3 , p=0 .037. Average of biological replicates ± sem. B) Anti-Luc antibody detected a specific luciferase band and a smaller, non-specific band in total protein extract from tethered and non-tethered lines with mock and IPTG treatment. The average band intensity of the luciferase and the internal band (as control) were measured as an approximation of protein quantity and show that protein levels of luciferase increase significantly in the tethered line and decrease significantly after IPTG treatment. Data from one blot representative of 3 biological repeats.These changes were only seen for the luciferase band and not the internal control bands.
Increased luciferase activity in other tissues
Expression of luciferase in non-tethered LacO:Luc and tethered LacO:Luc+Seh1-LacI-YFP lines was previously established in 7 d old whole seedling extracts. To investigate whether the LacI-LacO tethering effect is present in other plant tissues, we measured luciferase activity in adult rosette leaves, stems, inflorescences and seeds. This time, luminescence of LacO:Luc in leaf tissue was set to 100%. In both leaves and inflorescences, luciferase activity was high and increased significantly when the Seh1-LacI-YFP tether was co-expressed (p<0.001;Fig. 5). In leaf tissue, luciferase levels in LacO:Luc+Seh1-LacI-YFP were 209 ± 3%, higher than in LacO:Luc leaves (100 ± 1%). In inflorescences, in LacO:Luc+Seh1-LacI-YFP luciferase activity levels were 152 ± 1%, higher than in LacO:Luc (72 ± 1%). In stems and seeds, however, luciferase activity was very low – 7.4 ± 0.4% in LacO:Luc and 6.4 ± 0.7% in LacO:Luc+Seh1-LacI-YFP in stems and 5.5 ± 0.4% in LacO:Luc and 7.9 ± 0.5% in LacO:Luc+Seh1-LacI-YFP in seeds. In fact, expression levels in stem and seed samples were similar to the negative control Seh1-LacI-YFP only (9.5 ± 0.5%; data not shown) indicating these are background levels and that luciferase does not appear to be expressed in these tissues, likely as a result of gene silencing or degradation of the product.
Figure 5.
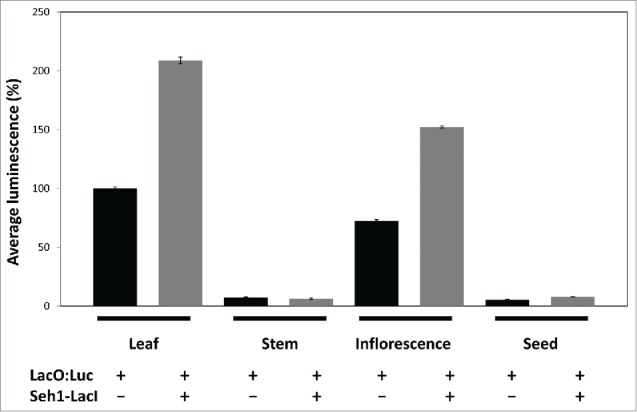
Seh1-LacI-YFP effect on luciferase activity in various tissues. Luciferase activity in rosette leaves, stems, inflorescences and seeds in the absence (black) or presence (gray) of the Seh1-LacI-Luc tether. Luciferase activity increased from 100 ± 1% to 209 ± 3% (p <0.001) in leaf and from 72 ± 1% to 152 ± 1% (p<0.001) in inflorescence tissue, and remained similar in stems (7.4 ± 0.4% and 6.4 ± 0.7%) and seeds (5.5 ± 0.4 and 7.9 ± 0.5%) (p>0.05). Average ± sem.
Discussion
In the chromatin charting lines created by,19 a LacO:Luc transgene is randomly inserted within the chromosomes causing variations in the expression level of the marker gene luciferase. We took the opportunity of this variable level of expression to select lines in which the luciferase is expressed at a moderate level in order to demonstrate that tethering of the LacO:Luc transgene to a nucleoporin via LacI-LacO interactions may change the expression levels of luciferase. While nucleoplasmic Nup50a appears to repress marker gene expression, peripherally localized Seh1 enhances expression. As suggested by,19 sub-nuclear localization of the LacI tether is likely to change the localization of the transgene from random distribution to the site where the tether is localized. Thus, in the case of Seh1-LacI-YFP tether, the LacO:Luc transgene is also likely present at the nuclear periphery (Fig. 2). That the changes in expression levels are due to LacI-LacO mediated tethering of the nucleoporin to the transgene is further suggested by disruption of the tethering with IPTG. Increased expression as a result of interaction with LacI-YFP unbound to Seh1 is unlikely as this is not seen with the NUP50a constructs or in other experiments with both N- and C- constructs including the construct YFP-Seh1-LacI and in other lines (data not shown). Luciferase levels at the mRNA, protein and enzyme activity level increased significantly in the presence of the Seh1-LacI-YFP tether but dropped significantly when the LacI-LacO tethering was disrupted with IPTG. While the data presented clearly indicates an effect of Seh1-LacI-YFP tethering on luciferase expression, we were unable to obtain data demonstrating that this occurs at the NPC and thus other explanations (for instance that Seh1 has transcriptional activator activity) cannot be ruled out. Nucleoporins have been shown to be active in altered gene expression both in the nucleoplasm and at the nuclear periphery.12,20 It proved technologically impossible to establish the location of the tether using 3-D FISH.
Interestingly, drosophila orthologues of Seh1 and Nup50a, DmSec13 and DmNup50, were shown to enhance gene expression.1,20 The enhancing effect observed here for Seh1 may suggest that some similarities in nucleoporin effect on gene expression exist across different kingdoms. The enhancing effects of DmSec13 and DmNup50 were due to direct interactions of the native protein with chromatin.1,20 Whether native Arabidopsis Seh1 and Nup50a interact with chromatin, or other components involved in gene regulation, and how they affect expression of native genes, remains to be identified. In addition, while it is clear that nucleoporin tethering via LacI-LacO affects marker gene expression, the underlying molecular pathways need to be investigated. So far reverse genetics, proteomic approaches,22 yeast 2 hybrid 10 and phenotypic screens on nuclear morphology 4,8,23 were mainly used to identify new components of the nuclear envelope. The characterization of Seh1-LacI-YFP + LacO:Luc line enhancing Luc transcriptional activity would be a new way to screen for mutants affecting Seh1-LacI tethering at the nuclear periphery.
By demonstrating that a nuclear periphery tether enhances the expression of a marker gene in plants, this study is beginning to decipher the role of the plant nuclear periphery in controlling gene expression. Recently, 6 have provided an example of gene repositioning in Arabidopsis regulated by light, suggesting a first biological significance for active localization of regions of chromatin in plants. With more plant NE, NPC and lamina components being identified, this opens an exciting area of future investigations and we continue to explore the biological significance of chromatin tethering in plants.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
The authors are grateful to Eric Lam for donating the CCT lines and pJM71 plasmid DNA and to Camille Bouyer for help with Western blotting. The authors would like to thank Oxford Brookes University for funding this project under the Higher Education Innovation Fund. Katja Graumann is a Leverhulme Early Career Fellow.
References
- 1.Capelson M, Liang Y, Schulte R, Mair W, Wagner U, Hetzer MW. Chromatin-bound nuclear pore components regulate gene expression in higher eukaryotes. Cell 2010; 140:372–383; PMID:20144761; http://dx.doi.org/ 10.1016/j.cell.2009.12.054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ciska M, Masuda K, Moreno Díaz de la Espina S. Lamin-like analogues in plants: the characterization of NMCP1 in Allium cepa. J Exp Bot 2013; 64:1553–64; PMID:23378381; http://dx.doi.org/ 10.1093/jxb/ert020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dieppois G, Stutz F. Connecting the transcription site to the nuclear pore: a multi-tether process that regulates gene expression. J Cell Sci 2010; 123:1989–99; PMID:20519581; http://dx.doi.org/ 10.1242/jcs.053694 [DOI] [PubMed] [Google Scholar]
- 4.Dittmer TA, Stacey NJ, Sugimoto-Shirasu K, Richards EJ. LITTLE NUCLEI genes affecting nuclear morphology in Arabidopsis thaliana. Plant Cell 2007; 19:2793–803; PMID:17873096; http://dx.doi.org/ 10.1105/tpc.107.053231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fang Y, Spector DL. Centromere positioning and dynamics in living Arabidopsis plants. Mol. Biol. Cell 2005; 16:5710–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Feng CM, Qiu Y, Van Buskirk EK, Chen M. Light-regulated gene reporter repositioning in Arabidopsis. Nat Commun 2014; 5:3027; PMID:24390011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fiserova J, Kiseleva E, Goldberg MW. Nuclear envelope and nuclear pore complex structure and organization in tobacco BY-2 cells. Plant J 2009; 59:243–55; http://dx.doi.org/ 10.1111/j.1365-313X.2009.03865.x [DOI] [PubMed] [Google Scholar]
- 8.Goto C, Tamura K, Fukao Y, Shimada T, Hara-Nishimura I. The novel nuclear envelope protein KAKU4 modulates nuclear morphology in Arabidopsis. Plant Cell 2014; 13:122–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Graumann K, Evans DE. Characterisation of SUN-domain proteins at the higher plant nuclear envelope. Plant J 2010; 61:134–44; PMID:19807882; http://dx.doi.org/ 10.1111/j.1365-313X.2009.04038.x [DOI] [PubMed] [Google Scholar]
- 10.Graumann K, Vanrobays E, Tutois S, Probst AV, Evans DE, Tatout C. Characterization of two distinct subfamilies of SUN-domain proteins in Arabidopsis and their interactions with the novel KASH-domain protein AtTIK. J Exp Bot 2014; 65:6499–528; PMID:25217773; http://dx.doi.org/ 10.1093/jxb/eru368 [DOI] [PubMed] [Google Scholar]
- 11.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970; 227:680–5; PMID:5432063; http://dx.doi.org/ 10.1038/227680a0 [DOI] [PubMed] [Google Scholar]
- 12.Liang Y, Hetzer MW. Functional interactions between nucleoporins and chromatin. Curr Opin Cell Biol 2011; 23:65–70; PMID:21030234; http://dx.doi.org/ 10.1016/j.ceb.2010.09.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mekhail K, Moazed D. The nuclear envelope in genome organization, expression and stability. Nat Rev Mol Cell Biol 2010; 11:317–28; PMID:20414256; http://dx.doi.org/ 10.1038/nrm2894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Murphy SP, Gumber HK, Mao Y, Bass HW. A dynamic meiotic SUN belt includes the zygotene-stage telomere bouquet and is disrupted in chromosome segregation mutants of maize (Zea mays L.). Frontiers in Plant Sci 2014; 5:314; http://dx.doi.org/ 10.3389/fpls.2014.00314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Newell and Gray Binding of lac repressor-GFP fusion protein to lac operator sites inserted in the tobacco chloroplast genome examined by chromatin immunoprecipitation. Nucleic Acids Res 2010 Aug; 38(14):e145 Epub 2010 May 19; http://dx.doi.org/ 10.1093/nar/gkq413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Parry G. Components of the Arabidopsis nuclear pore complex play multiple diverse roles in control of plant growth. J Exp Bot 2014; 65:6057–67; PMID:25165147; http://dx.doi.org/ 10.1093/jxb/eru346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Parry G. The plant nuclear envelope and regulation of gene expression. J Exp Bot 2015; 66:1673–85; PMID:25680795; http://dx.doi.org/ 10.1093/jxb/erv023 [DOI] [PubMed] [Google Scholar]
- 18.Riggs AD, Newby RF, Bourgeois S. Lac repressor-operator interaction. II Effect of galactosides and other ligands. J Mol Biol 1970; 51:303–14 [DOI] [PubMed] [Google Scholar]
- 19.Rosin FM, Watanabe N, Cacas J-L, Kato N, Arroyo JM, Fang Y, May B, Vaughn M, Simorowski J, Ramu U, et al.. Genome-wide transposon tagging reveals location-dependent effects on transcription and chromatin organization in Arabidopsis. Plant J 2008; 55:514–25; http://dx.doi.org/ 10.1111/j.1365-313X.2008.03517.x [DOI] [PubMed] [Google Scholar]
- 20.Sarma NJ, Willis K. The new nucleoporin: regulator of transcriptional repression and beyond. Nucleus 2012; 3:508–15; PMID:23047597; http://dx.doi.org/ 10.4161/nucl.22427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc 2008; 3:1101–8; PMID:18546601; http://dx.doi.org/ 10.1038/nprot.2008.73 [DOI] [PubMed] [Google Scholar]
- 22.Tamura K, Fukao Y, Iwamoto M, Haraguchi T, Hara-Nishimura I. Identification and characterization of nuclear pore complex components in Arabidopsis thaliana. Plant Cell 2010; 22:4084–97; PMID:21189294; http://dx.doi.org/ 10.1105/tpc.110.079947 [DOI] [PMC free article] [PubMed] [Google Scholar]


