Abstract
Actin and proteins that regulate its dynamics or interactions have well-established roles in the cytoplasm where they function as key components of the cytoskeleton to control diverse processes, including cellular infrastructure, cellular motility, cell signaling, and vesicle transport. Recent work has also uncovered roles for actin and its regulatory proteins in the nucleus, primarily in mechanisms governing gene expression. The Wiskott Aldrich Syndrome (WAS) family of proteins, comprising the WASP/N-WASP, SCAR/WAVE, WHAMM/JMY/WHAMY, and WASH subfamilies, function in the cytoplasm where they activate the Arp2/3 complex to form branched actin filaments. WAS proteins are present in the nucleus and have been implicated as transcriptional regulators. We found that Drosophila Wash, in addition to transcriptional effects, is involved in global nuclear architecture. Here we summarize the regulation and function of nuclear WAS proteins, and highlight how our work with Wash expands the possibilities for the functions of these proteins in the nucleus.
Keywords: actin, JMY, nuclear architecture, nuclear organization, transcription, Wiskott-Aldrich Syndrome, WASH, WASP/N-WASP, WAVE/SCAR, WHAMM
Introduction
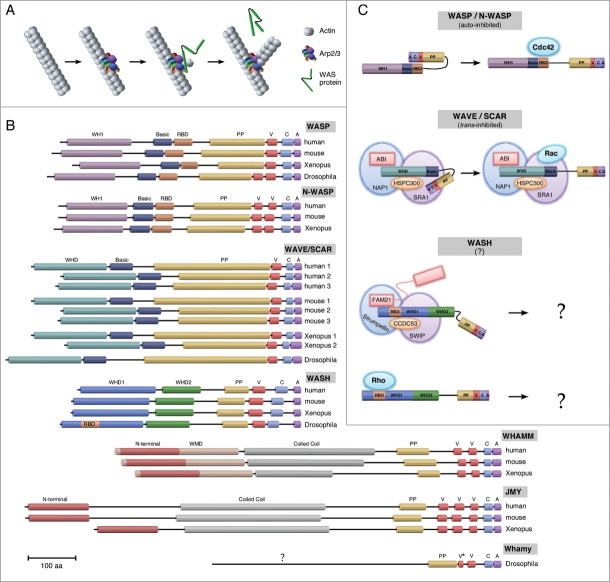
The cytoskeleton undergoes continuous dynamic changes and interactions with other cellular components, such as the plasma membrane, necessary for many essential cellular processes, including those required both inter- and intra-cellularly for communication, attachment/adhesion, migration, and structural integrity (For a review see ref. 1). One family of proteins that mediates membrane-cortical cytoskeleton interactions through their tight spatial and temporal coordination of cytoskeleton dynamics and other cellular machineries, is the Wiskott-Aldrich Syndrome (WAS) family of proteins.2,3 WAS proteins are nucleation-promoting factors (NPFs) that activate the actin-related protein 2/3 (Arp2/3) complex to produce branched actin filaments (Fig. 1A).2-4 WAS family proteins include the well-characterized WASP/N-WASP and WAVE/SCAR founding classes, as well as the more recently recognized WASH and WHAMM/JMY/WHAMY classes (Fig. 1B).3,4 The defining feature of WAS proteins is their conserved Arp2/3-binding catalytic C-terminal VCA domain (verprolin-homology (WH2), cofilin-homology, and acidic domains), with their N-terminal ends conferring their subfamily status (Fig. 1B).5,6
Figure 1.
Schematics of WAS family protein function, domain topology, and activation. (A) The interaction between Arp2/3, actin, and the highly conserved VCA domain of WAS proteins is necessary and sufficient to induce actin polymerization off an existing filament of actin. The WH2 motif binds a G-actin monomer and the CA recruits and changes the conformation of Arp2/3 and interacts with an existing F-actin filament. (B) Topology diagrams of WAS family protein domains for human (H. sapiens), mouse (M. musculus), African clawed frog (X. laevis*), and fruit fly (D. melanogaster). All subfamilies contain a C-terminal VCA domain with class specific N-termini. Drosophila Whamy contains significant differences from both the mammalian WHAMM and JMY subfamilies. Domain Abbreviations: WH1 (WASP homology 1); RBD (Rho binding domain); PP (poly-proline); V/WH2 (Verprolin/WASP homology 2); C (Central/Connector); A (acidic); SHD (Scar homology domain); WHD1 (WASH homology domain 1); WHD2 (WASH homology domain 2); WMD (WHAMM membrane-interacting domain). Scale bar, 100 residues. *X. tropicalis for WHAMM/JMY. (C) Different modes of regulation for WAS family proteins. WASP/N-WASP proteins are auto-inhibited. This inhibition is released upon their direct binding to the Cdc42 small GTPase. WAVE/SCAR proteins interact indirectly with the Rac small GTPase. WAVE/SCAR proteins are trans-inhibited through their constitutive regulation by the WAVE Regulatory Complex (WRC). This inhibition is released upon Rac recruitment by the WRC subunit Sra1. The mechanism of WASH regulation is not yet clear, but appears to be context dependent. WASH proteins are not auto- or trans- inhibited. Nonetheless, WASH has been shown to bind directly to the Rho1 small GTPase, and has also been shown to associate with a WRC-like multiprotein complex (SHRC; Strumpellin, SWIP, CCDC53, FAM21).
WAS family proteins function as downstream effectors of Rho family GTPases, however, they differ in their modes of interaction with these GTPases. WASP/N-WASP proteins are auto-inhibited. This inhibition is released upon their direct binding to the Cdc42 small GTPase (Fig. 1C).2-4,7 Interestingly, one recent study suggests that Rac1 may also be upstream of N-WASP acting in the same capacity as Cdc42.8 WAVE/SCAR proteins are trans-inhibited through their constitutive regulation by the WAVE Regulatory Complex (WRC), comprised of the ABI, SRA1, NAP1 and HSPC300 proteins. GTPase regulation of WAVE/SCAR proteins occurs indirectly through the binding of Rac1 to the WRC component Sra1 (Fig. 1C).2,3,9,10 WASH protein regulation is more complex. The actin nucleation activity of WASH proteins does not appear to be auto- or trans- inhibited. An interaction (direct or indirect) between mammalian WASH and the Rho or Cdc42 small GTPases has not been observed, although a weak interaction with Rac1 has been reported.11 Studies in mammalian cell lines and Dictyostelium suggest that WASH functions as part of a multiprotein complex that regulates endosome trafficking in an Arp2/3-dependent manner.12-15 This multiprotein WASH Regulatory Complex (SHRC; comprised of the SWIP, Strumpellin, CCDC53, and FAM21 proteins) shares some structural features and sequence similarity with WRC proteins, however, the means by which they are activated appear to be different (Fig. 1C).2,11 In contrast, while the SHRC proteins are present in Drosophila and associate with Wash,16 the fly Wash protein also binds directly to the Rho1 small GTPase and this interaction is required for at least a subset of its functions.16,17 It is not yet clear whether Wash's interaction with Rho1 is exclusive to Wash in an SHRC-independent state or if Rho1 is also able to interact with the SHRC. Nonetheless, Drosophila Wash functions as part of the SHRC multiprotein complex (similar to SCAR/WAVE proteins) and functions independently of such a complex (similar to WASP/N-WASP) in a context-dependent manner.16,17 Nothing is yet known about Rho family GTPase regulation of the WHAMM/JMY/WHAMY classes. It will be interesting to determine if Rho family GTPase signaling is a defining feature of their regulation, and if so, which family member is responsible.
In the cytoplasm, the highly conserved WAS family proteins regulate cytoskeletal dynamics important for processes including membrane/vesicle trafficking, morphogenesis, angiogenesis, inflammatory immune response, and when mis-regulated, pathogen infection, bleeding/bruising, immune deficiency and cancer metastasis.18-21 WAS family proteins are also present and necessary in the nucleus, where they have been implicated as significant players primarily in transcriptional roles. Actin and myosin are also present in the nucleus: they are actively imported and exported using the importin/exportin system.22-24 Emerging evidence has shown that nuclear actin is multifunctional, including roles as a transcription factor, as a member of major chromatin remodeling complexes, as part of a system that controls loci movements within the nucleus, and as a component of ribonucleoproteins (RNPs) (For a review see ref. Twenty-five–27). Initially, nuclear actin was thought to be monomeric, however, work in the Xenopus germinal vesicle clearly showed the presence of actin filaments and subsequent studies have demonstrated the formation of actin rod structures in response to various cellular stressors.28,29 Nuclear myosin is likewise multifunctional, including roles in gene expression, nucleoskeleton scaffolds, and in the positioning/organization of chromatin (For a review see ref. 29, 30). As WAS proteins encode various biochemical activities that regulate cytoskeleton dynamics (actin nucleation, actin/microtubule bundling, and cytoskeletal crosslinking), a current challenge is to understand how they regulate these components in the nucleus to carry out their various functions. Here, we discuss the nuclear roles of WAS family proteins, highlighting the mechanisms that regulate their localization to the nucleus and their activities within the nucleus, and when possible, if these activities are actin-dependent.
WASP
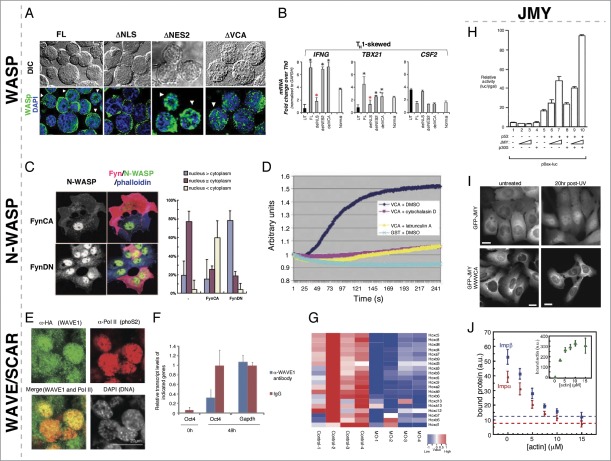
Clinical manifestations of WAS were initially linked to actin-related cytoplasmic functions, however, mutations that have no affect on the actin-nucleation activity of WASP, and even mutations that increase its actin nucleation activity, can still result in severe pathology.31-34 One of the underlying causes of WAS symptoms is a deficiency of CD4+ T-Helper (TH) cells, specifically TH1 cells and, strikingly, WASP proteins were found to accumulate in the nucleus of cells differentiating from TH0 to TH1.35 WASP contains a functional nuclear localization signal (NLS) and nuclear export signal (NES).35 When WASP's NLS is disrupted in human cells, it is unable to localize to the nucleus, and when WASP's NES is disrupted, it localizes primarily in the nucleus (Fig. 2A).36 Currently, the mechanisms, potential binding partners, and signals that regulate WASP distribution between the nucleus and cytoplasm are unknown.
Figure 2.
Nuclear roles for WASP/N-WASP, WAVE/SCAR, and JMY. (A) WASP nuclear localization is regulated by canonical NLS and NES import/export. De-convolved fluorescence micrographs of TH1-skewed WASPNULL cells rescued with the indicated WASP constructs. Expression from the full-length and ΔVCA constructs is found in both the nucleus and cytoplasm, whereas the ΔNLS is only found in the cytoplasm and the ΔNES2 is only found in the nucleus. (B) Nuclear-localized WASP in TH1-skewed cells is responsible for expression of the TH1 regulators IFNG and TBX21. Bar plot graph of gene expression quantitated by RT-qPCR in TH1 WASPNULL (UT) and rescued by full-length (FL), ΔNLS (delNLS), ΔNES2 (delNES2), and ΔVCA (delVCA) constructs compared to TH0 expression levels showing that the TH1 factors IFNG and TBX21 are not-regulated properly in ΔNLS TH1 skewed cells. CSF2 is a non-TH1-specific control. (C) Phosphorylation of N-WASP by the Src kinase, Fyn, leads to enhanced cytoplasmic and decreased nuclear accumulation of N-WASP. Fluorescent micrographs of COS-7 cells expressing ectopic constitutively active Fyn (FynCA) or dominant negative (FynDN) immunostained for anti-Fyn (red), anti-N-WASP (gray/green), and Phalloidin (blue) show that FynCA leads to more cytoplasmic N-WASP, whereas FynDN leads to more nuclear N-WASP. Quantification of N-WASP nuclear versus cytoplasmic N-WASP in COS-7 cells expressing FynCA and FynDN. (D) N-WASP can polymerize actin from nuclear lysates. Pyrene actin assay showing that a GST fusion to the N-WASP VCA domain can polymerize actin, however, it is unable to do so in the presence of the actin polymerization inhibitors cytochalasin D and Latrunculin A. (E) Wave1 is present in mouse C2C12 nuclei after transplantation into the germinal vesicle of Xenopus oocytes. Mouse somatic C2C12 nuclei were transplanted into Xenopus oocytes overexpressing HA-NLS-WAVE1. Immunofluorescence staining for anti-HA (WAVE1) showed WAVE1 co-localizing with active RNA polymerase II in these transplanted nuclei 24 hrs post-nuclear transfer. (F) Nuclear Wave1 is required for transcriptional reprogramming in Xenopus oocytes. Transcriptional activation of the embryonic gene, Oct4, is inhibited in the presence of antibodies against WAVE1 in transplanted somatic nuclei, as measured by QPCR. Expression of the housekeeping gene, Gapdh, was unaffected by the presence of α-WAVE1 antibodies. (G) Nuclear Wave1 is required for hox gene expression. Heat map showing the down-regulation of hox gene expression in WAVE1-morpholino (MO) injected embryos relative to control. (H) JMY and p300 function together to regulate p53-dependent transcription. Bax promoter-luciferase reporter assays were used to measure p53-dependent transcriptional activity. Co-expression of p53 with increasing levels of JMY resulted in a titratable increase in p53 activity that was further enhanced by the expression of p300. (I) The JMY NLS is required for damage-induced nuclear accumulation. Cells expressing GFP-JMY, but not a GFP-JMY truncation mutant removing its actin binding and NLS (GFP-JMYΔWWWCA), show accumulation of GFP signal in the nucleus in response to UV irradiation induced DNA damage. (J) Actin competes with Impα/β for binding to JMY. Actin and importins both bind to a C-terminal fragment of JMY containing tandem WH2 (WWW) motifs and a nested NLS sequence (WWWCA). Quantification of GST pulldowns assaying Impα/β binding to the GST-WWWCA in the presence of increasing concentrations of actin monomers. Permissions. (A–B) Reprinted from Sadhukan et al. (2014).36 The Journal of Immunology 193:150-60. (C) Reprinted with permission from Suetsugu & Takenawa, J. Biol. Chem. 278(43):42515-23.37 © The American Society for Biochemistry and Molecular Biology. Reproduced by permission of The American Society for Biochemistry and Molecular Biology. Permission to reuse must be obtained from the rightsholder. (D) © Macmillan Publishers Ltd: Nature Cell Biology. Reproduced by permission of Macmillan Publishers Ltd: Nature Cell Biology. Permission to reuse must be obtained from the rightsholder. Wu et al., Nat. Cell Biol. 8(7):756–63.40 (E–G) © AAAS. Reproduced by permission of AAAS. Permission to reuse must be obtained from the rightsholder. From Miyamoto et al. (2013) Science 341(6149):1002-5.45 (H) © Elsevier. Reproduced by permission of Elsevier. Permission to reuse must be obtained from the rightsholder. Reprinted from Molecular Cell, Volume 4(3), Shikama et al., A Novel Cofactor for p300 that Regulates the p53 Response, pp. 365-376.46 (I-J) © American Society for Cell Biology. Reproduced by permission of American Society for Cell Biology. Permission to reuse must be obtained from the rightsholder. From Actin binding to WH2 domains regulates nuclear import of the multifunctional actin regulator JMY, Zuchero et al., Mol. Biol. Cell 23:853, 2012.47
Variants of WAS that exhibit a deficiency of TH1 cells, as a result of improper TH0 cell differentiation, are associated with some of the most severe clinical symptoms.31-34 This defect largely correlates to mutations in WASP's N-terminal WH1 domain, which are not expected to alter actin-nucleation activity.34 The result of these mutations is a decrease in mRNA production for 2 key TH0 to TH1 developmental regulators, IFNG and TBX21 (Fig. 2B). Initial ChIP-CHIP experiments revealed the association of nuclear WASP with the cis-regulatory promoter regions of these genes, suggesting a transcriptional role for WASP.35 A series of mass spectrometry and ChIP experiments in human primary or Jurkat TH1-skewed cells suggested that WASp interacts with a number of chromatin remodeling proteins and complexes, which can affect promoter chromatin in multiple ways. First, WASP interacts with epigenetic factors such as the MLL protein, RBBP5, and JMJD6 (a demethylase), which result in increased histone modification H3K4me3 marks on these promoters.35 Second, WASP also affects these promoters through its interaction with the BAF170 and BAF47 subunits of the SWI/SNF chromatin-remodeling complex.34 Significantly, the interacting SWI/SNF complex was shown to contain hBRM as its ATPase, and not Brg1. This suggested that WASP, through this remodeling complex, may act on Notch target genes and, indeed, Notch target genes are affected in WH1 mutant cells.34 Finally, WH1 WASP mutants affect the enrichment of the H2A.z histone variant and subsequent chromatin de-compaction at these cis-regulatory regions through an interaction with EP400.34 Taken together, these findings reflect a role for WASP in the recruitment and/or assembly of multiple chromatin remodeling complexes in a cell-specific and loci-specific manner, independent of any Arp2/3-related activity inside the nucleus, and provide new avenues to understand the biology of this disease.
N-WASP
Similar to WASP, N-WASP contains a predicted and functional NLS and NES.37 Specific deletions of regions containing these signals in COS-7 cells, as well as blocking active nuclear export with Leptomycin B have demonstrated that both the NLS and NES are necessary for canonical nuclear import and export by karyopherins.37 In contrast to WASP, there is growing evidence that the subcellular localization of N-WASP is regulated by activity state and phosphorylation. First, the nuclear fraction of N-WASP was found to be in the open, not auto-inhibited, conformation.38 Secondly, 2 separate studies have indicated that the phosphorylation status of the conserved Tyrosine 256 in humans (T253 in mouse) is indicative of the localization of N-WASp and that blocking the phosphorylation of this residue results in preferential nuclear localization.37,38 Indeed, phosphorylation of this residue may inhibit nuclear importin (NP-1) binding to the NLS: a phosphomimetic point mutant N-WASP was unable to bind in vitro as robustly to NP-1.38 These studies have suggested that 2 different kinases, Fyn and FAK, may be involved in phosphorylation of this residue (Fig. 2C), however, these differences may be cell-type-specific and merit further exploration. In addition to phosphorylation activity state, N-WASP nuclear localization is regulated by FBP11, which interacts with N-WASP and can sequester it inside the nucleus.39
Initial studies demonstrated that localization of un-phosphorylated nuclear N-WASP led to a decrease in HSP90 expression, presumably through a physical interaction with HSTF.37 Of note, HSP90 is essential for Src family kinase activity and as the localization of WASP is dependent on Fyn in some contexts, the down-regulation of HSP90 may act as a possible feedback mechanism to drive N-WASP into the nucleus by decreasing phosphorylation. Subsequent studies in HeLa cells and extracts have highlighted a more global change in transcription when N-WASP is removed or segregated outside of the nucleus. In one of these studies, a proteomics approach identified an interaction between N-WASP and the PSF-NonO complex.40 This multi-functional nuclear complex has proposed roles in many nuclear processes including RNA processing, DNA repair, topoisomerase activity, and transcription (For a review see ref. 41). Fractionation of nuclear lysates demonstrated that PSF-NonO and N-WASP are present in a complex that includes RNA Pol II and actin, with PSF-NonO being necessary for the N-WASP and RNA Pol II association.40 N-WASP was also shown to polymerize actin in nuclear extracts in vitro.40 ChIP experiments showed that this PSF-NonO/N-WASP/actin complex associates with active promoters, and that its disruption by RNAi or antibody interference significantly reduces transcription in in vitro transcription assays.40 Importantly, similar in vitro transcription defects were observed with mutations in N-WASP's VCA domain (disrupting actin polymerization) or with drug inhibition by Latrunculin B and cytochalasin D (preventing actin polymerization) (Fig. 2D).40 BrU incorporation assays in HeLa cells with RNAi and the N-WASPΔVCA protein showed similar global transcriptional defects, however, it is not known if this activity is linked to the formation of de novo F-actin. Interestingly, the Arp2 and Arp3 subunits of the Arp2/3 complex are also found in RNA pol II-containing complexes.42 Removal of these Arp2/3 subunits by RNAi reduces transcription in a similar manner to N-WASP/PSF-NonO. Significantly, mutants that sequester Arp2 or Arp3 in the cytoplasm of HeLa cells have reduced global transcription as well. A recent study demonstrating a role for N-WASP in regulating hair follicle cycling showed N-WASP involvement in gene regulation, however, the number of genes affected was small suggesting that N-WASP may not affect global transcription in all cell types.43 Overall, studies of N-WASP in the nucleus are providing tantalizing hints that WAS family proteins have the capacity to affect actin structures within the nucleus.
WAVE/SCAR
While a nuclear role for WAVE/SCAR family proteins has not yet been described in a specific developmental context, recent experiments implicating WAVE1, Rac1, and nuclear actin in the transcriptional reprogramming of embryonic genes in differentiated nuclei suggests that these contexts do exist. Upon transplantation of differentiated nuclei into the germinal vesicle of Xenopus oocytes, de novo F-actin filaments form in these nuclei followed by a rapid recovery of Oct4 expression (a member of the POU family of homeobox genes), both of which could be abrogated by inhibiting F-actin polymerization.44 Overexpression of Rac1 in Xenopus oocytes resulted in a significant increase in Oct4 transcription leading to a focus on WAVE, the WAS protein whose actin nucleation activity is regulated by this GTPase.44 Similar to Rac1, an accumulation of nuclear WAVE1 coincided with an increase in Oct4 transcription that was lost in the presence of inhibitory antibodies against WAVE1 (Fig. 2E-F).44,45 Overexpression of WAVE1-A and WAVE1-B (but not WAVE1-C) showed a trend toward increasing the expression of embryonic genes, while knockdown of WAVE1 in embryos through MO-injections caused a downregulation of many genes, including hox genes, indicating that nuclear WAVE1 plays a role in embryonic development (Fig. 2G).45 Co-immunoprecipitation experiments with nuclear extracts of an NLS-WAVE1–A or –B fusion protein were conducted to identify nuclear WAVE1 interactors. Both WAVE1–A and –B associated with RNA polymerase II and MLL, a SET domain-containing H3K4 methyltransferase.45 ChIP analysis comparing active RNAPII and H3K4 methylation of hox genes in control versus WAVE1 MO-injected embryos revealed a consistent decrease of RNAPII and H3K4 marks in WAVE1 knockdown embryos. WAVE interaction with both these proteins mapped to its N-terminal Wave Homology Domain (WHD) (Fig. 1B). As such, nuclear WAVE1 functions similarly to N-WASP to regulate global transcription through nucleating F-actin within the nucleus. In the future it will be interesting to uncover in vivo contexts in which WAVE proteins are present in the nucleus, and the mechanisms which regulate its nuclear localization to prevent the ectopic reprogramming of cells.
JMY
Junction-mediating and regulatory protein (JMY) is a special class of WAS family NPFs: while it contains a conserved C-terminal VCA domain, it can nucleate actin with or without the Arp2/3 complex.4 JMY was initially isolated as a transcriptional cofactor of the p53 tumor suppressor protein and exhibits dynamic nuclear/cytoplasmic localization.46,47 In the nucleus, JMY along with CBP/p300 and Strap forms a transcriptional activation complex that regulates p53 transcription (Fig. 2H).46 Interestingly, JMY's nuclear function appears to include an actin component, as nuclear JMY can incorporate actin in an Arp2/3-independent manner and addition of Latrunculin A (inhibits actin polymerization) reduced p53 activity without altering JMY's nuclear localization or levels.48 In the cytoplasm, JMY acts independently from p53 in a manner consistent with classic actin NPF activity. JMY is also a critical target for down-regulating p53 levels. Mdm2 (murine double minute 2), an E3 ubiquitin ligase, can oppose the p53 response by directly ubiquitylating p53, leading to its nuclear export and degradation,49 or by targeting the p53 positive regulator, JMY.50 Co-expression of Mdm2 with JMY in DNA-damaged cells causes a decrease in p53-mediated cell apoptosis, which is restored upon the deletion of the Mdm2 C-terminal region lacking the RING finger domain necessary for JMY binding, as well as ubiquitin ligase activity.49,50 However, under conditions of DNA damage, the Mdm2-JMY interaction is significantly curtailed, leading to an accumulation of JMY in the nucleus to activate p53.50
Exploration of the mechanism of JMY subcellular localization revealed an elegant mechanism for JMY to strategically dictate its cytoplasmic-nuclear distribution (Fig. 2I). JMY has 3 tandem WH2 motifs, each capable of binding to an actin monomer, with an NLS sequence nestled between the first and second of these repeats. Actin monomer binding to these WH2 domains blocks binding of importin-β (Impβ) to JMY's NLS, thereby preventing its nuclear import. Expression of a JMY mutant with a deletion in its C-terminal WWWCA region thereby disabling actin monomer-binding capacity, remained in the cytoplasm upon DNA damage, while wildtype JMY protein localized to the nucleus (Fig. 2I). In addition, Impβ binding to JMY's NLS sequence is out-competed by increasing concentrations of actin (Fig. 2J).47 DNA damage has been shown to induce actin filament assembly in the cytoplasm. When this occurs, a reduced pool of cytoplasmic G-actin unblocks JMY's NLS such that Impβ can transport JMY into the nucleus where it induces p53-dependent transcription for cell-cycle arrest and apoptosis.51 As JMY has only recently been recognized as a WAS subfamily, it is not yet known what the full scope of JMY's cytoplasmic roles are and how JMY's actin-related functions play a role in its activity as a transcription co-factor.
WASH
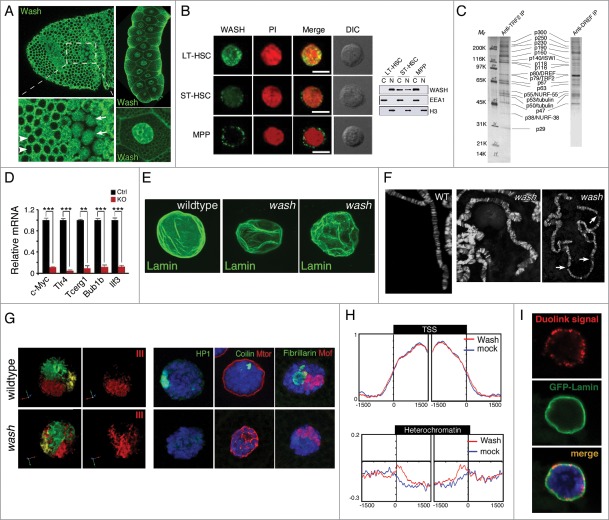
One of the more recently discovered WAS subfamilies, the WASH proteins, are emerging as important cytoplasmic cytoskeleton regulators where they function in endosome sorting and scission, cell migrations, cytoplasmic flows, and other developmental contexts.12,14,16,17,52 In ad-dition to these cytoplasmic roles, WASH family proteins localize to and accumulate within the nucleus in developmental and cell-type specific manners.53-55 In the gastrulating fly embryo, Wash subcellular localization ranges from predominantly cytoplasmic to equally distributed between the cytoplasm and nucleus in spatially restricted regions (Fig. 3A).55 Later in development, there is a striking localization of Wash protein in the nuclei of tissues such as larval salivary glands (Fig. 3A).54 In fly cell lines, Wash is equally distributed in the cytoplasm and nucleus.54,55 Recent work with 3 closely related murine haematopoietic cell lineages show drastically different WASH nuclear vs. cytoplasmic protein distributions: WASH in multipotent progenitor cells (MPPs) is restricted to the cytoplasm; WASH in short-term haematopoietic stem cells (ST-HSCs) is both nuclear and cytoplasmic; and WASH in long-term haematopoietic stem cells (LT-HSCs) is predominantly nuclear (Fig. 3B).53 This spatial regulation may be achieved by the protein's intrinsic properties, as WASH family proteins contain both a conserved bi-partite NLS and an NES.56 Intriguingly, Wash is often part of the stable SHRC, whose subunits (with the exception of CCDC53) have strongly predicted bipartite NLS motifs, or in the case of FAM21, a demonstrated and functional mono-partite NLS.57 Thus, WASH protein nuclear localization may be regulated not only by its own NLS, but also may be shuttled into the nucleus by its binding partners. Indeed, we recently generated transgenic Drosophila Wash protein in which it was not only necessary to disrupt the strongest predicted NLS, but also necessary to attach a strong NES in order to deplete Wash protein from the nucleus.54
Figure 3.
Nuclear localized WASH family proteins function as regulators of nuclear morphology and as transcription factors. (A) Wash accumulates in the nucleus of Drosophila cells in a temporally and spatially specific manner. Confocal projection of stage 7 embryos immunostained with anti-Wash show nuclear localization in specific mitotic domains (arrows) while remaining cytoplasmic in others (arrowheads) (left). Confocal projections of the nuclei of the salivary glands of 3rd instar larvae immunostained with anti-Wash shows nuclear enrichment (right). (B) WASH exhibits different sub-cellular localizations in different sub-populations of haematopoietic stem cells. Micrographs (left) and western blot analysis of lysates (right) of WASH expression in long-term haematopoietic stem cells (LT-HSC; nuclear), short-term haematopoietic stem cells (ST-HSC; nuclear and cytoplasmic) and multipotent progenitor cells (MPP; cytoplasmic). Micrographs are co-stained with PI for nuclear visualization and DIC views are shown (left). (C) Purification of the Drosophila TRF2 complex showing the presence of Wash and its SHRC. Co-immunoprecipitation by TRF2 or DREF monoclonal antibodies purifies a complex containing Wash (p63), SWIP (p116) and Strumpellin (p118), in addition to ISWI, DREF, TRF2, and tubulin. (D) WASH knockout in LT-HSCs leads to reduced c-Myc expression, as well as reduced expression of its transcriptional targets. Bar plot graphs of gene expression levels measured by qPCR showing significantly decreased expression of c-Myc, and its target genes Tlr4, Tcerg1, Bub1b, and Ilf3, in cell-sorted WASH KO LT-HSCs. (E–G) wash mutant salivary glands have altered nuclear morphology compared to wildtype. Confocal projections of wildtype vs. wash mutant nuclei immunostained with anti-Lamin antibody shows crinkled, non-spherical wash nuclei (E). Micrographs of wildtype versus wash mutant salivary gland polytene chromosomes show misalignment and improper banding, as well as extremely fragile chromosomes (F). Three-Dimensional reconstruction of wildtype vs. wash mutant salivary gland nuclei hybridized with chromosome-specific paints (X chromosome: yellow, 2nd chromosome: green, 3rd chromosome: red) showing less compact chromosome territories in wash mutant nuclei (G, left). Micrographs of wildtype verses wash mutant salivary gland nuclei immunostained with anti-HP1 (green; heterochromatin; left), anti-Coilin (green; Cajal bodies; middle), anti-Mtor (red; nuclear envelope protein; middle), anti-Fibrillarin (green; nucleolus; right), anti-MOF (red; X-chromosome; right) showing disrupted nuclear sub-compartments in wash mutant nuclei (G, right). (H) Wash increases chromatin accessibility in heterochromatin regions. Distribution of M.SssI-based chromatin accessibility in control RNAi and wash RNAi treated Drosophila S2 cells, showing increased accessibility in wash knockdown with no affect on transcription start site (TSS) chromatin regions. (I) Wash interacts with B-type Lamin at the nuclear periphery. Duolink proximity ligation assay in Drosophila salivary glands expressing GFP-Lamin using anti-Wash and anti-GFP antibodies shows amplification of Duolink signal at the nuclear periphery. Amplification signal is only observed when the 2 antibodies examined are within 30 nm. Permissions. (A, right) and (E–I) Reprinted from Current Biology, 25(6), Verboon et al., Wash Interacts with Lamin and Affects Global Nuclear Organization, pp. 804-810.54 © Elsevier. Reproduced by permission of Elsevier. Permission to reuse must be obtained from the rightsholder. (B, D) Reprinted with permission from: © Rockefeller University Press. Reproduced by permission of Rockefeller University Press. Permission to reuse must be obtained from the rightsholder. Reprinted from Xia et al. Journal of Experimental Medicine. 211:2119-2134. doi:10.1084/jem.20140169.53 (C) © Macmillan Publishers Ltd: Nature. Reproduced by permission of Macmillan Publishers Ltd: Nature. Permission to reuse must be obtained from the rightsholder. Hochheimer et al., Nature 420:439–45.58
Prior to Wash family proteins being defined as a WAS subfamily, Wash was found as part of the Drosophila TRF2 core promoter transcription complex, made up of known nuclear proteins and remodeling complexes, including TRF2, DREF, ISWI, NURF-38 and −55 (Fig. 3C).58 The TRF2 complex is necessary for the core promoter recognition of the PCNA gene, DNApol 180, as well as a subset of other genes containing a DRE (DNA Replication-related Element) and involved in proliferation. The TRF2 complex was convincingly shown to interact with a DRE containing promoter upstream of the PCNA transcriptional start site, however, the specific biochemical activities of this complex required in this context are not yet known. As the TRF2 complex contains members of the NURF remodeling complex, there is a predicted role in remodeling promoter chromatin. In addition to Wash, 2 isoforms of β-tubulin (mainly thought to be cytoplasmic) and the SHRC members, Strumpellin and SWIP, were found in this remodeling complex. The unexpected inclusion of these usually cytoplasmic factors hint at a potential role for this Wash-containing complex in regulating transcription at a structural level by modulating chromatin and/or the proteins related to the cytoskeleton which function in the nucleoskeleton. Future studies using inhibitors specific to structural proteins such as actin and microtubules, point mutations disrupting Wash's actin/microtubule binding and/or nucleation activities, as well as more modern histone occupancy techniques will be necessary to assess these potential roles.
Recently, Wash was found to be a key transcription factor in murine haematopoietic stem cells (HSC) where it regulates cell differentiation.53 In wild-type LT-HSCs, Wash is primarily localized to the nucleus and upon Wash depletion by RNAi, this population of cells increases due to their inability to differentiate into further lineages as a result of lost c-Myc proto-oncogene transcription (Fig. 3D). Similar to what is observed in flies with the TRF2 complex, murine WASH was shown to interact with the NURF complex (Rbbp4 subunit) and is necessary for this complex to properly associate with the c-Myc promoter, thereby increasing DNA accessibility. Significantly, WASH's function in this nuclear context is actin-dependent as Rbbp4 can only associate with actin when WASH is present, actin inhibition by cytochalasin D decreases DNA accessibility at the promoter, and a WashΔVCA protein does not rescue WASH KO differentiation defects.53 This suggests that WASH's role in the nucleus of HSCs is in the actin-dependent transport of chromatin remodeling factors. Taken together, these studies suggest that nuclear WASH proteins interact with the NURF complex to regulate chromatin accessibility at specific promoters and hint at a larger role for WASH-dependent actin transport within the nucleus.
We recently described a role for Drosophila Wash as a key player in global nuclear architecture. We have found that nuclei from Wash-depleted cells or mutant salivary glands are grossly deformed (Fig. 3E) and that these phenotypes are specific to nuclear Wash.54 wash knockdown or loss-of-function mutants exhibit a number of nuclear defects ranging from mis-localized proteins, which mark nuclear sub-compartments such as coilin (Cajal bodies), fibrillarin (nucleolus), and HP1 (heterochromatin), to fragile DNA and disrupted chromatin territories (Fig. 3F–G).54 We demonstrated that at least a subset of Wash's nuclear phenotypes is due to a novel interaction with the nuclear intermediate filament, Lamin Dm0 (Drosophila B-type Lamin). In particular, we explored the role of Wash and Lamin in modulating heterochromatin and find that Wash and Lamin interact with silent regions of the genome. Interestingly, we found that Wash and Lamin both increased (suppressed PEV) and decreased (enhanced PEV) heterochromatin formation when assaying Wash's ability to modulate chromatin using 2 different in vivo Position Effect Variegation (PEV) assays, suggesting that these reporters are differentially monitoring heterochromatin domains. We also showed a global increase in DNA accessibility at heterochromatic regions in wash and lamin Dm0 mutant backgrounds with no change at transcription start sites (Fig. 3H).
While superficially at odds with the previous transcription studies, our study used global readouts for Wash and Lamin effects on transcriptional start sites, however, this does not preclude the potential for changes in DNA accessibility at specific promoters in the genome similar to those observed for c-Myc and PCNA. Indeed global changes to nuclear organization could manifest as changes in the transcription state of many genes; conversely, changes in the transcription state of many genes could lead to global nuclear organizational changes. Currently, our favored hypothesis is that Wash has more than one role in the nucleus. One line of evidence that supports this hypothesis is that while a subset of Wash interacts with Lamin at the nuclear periphery in salivary gland nuclei (Fig. 3I), Wash is also present throughout the interior of the nucleus both co-localizing with and distinct from the DNA-containing regions (Fig. 3A), suggesting that these different populations of Wash protein may have functionally distinct nuclear roles. It will be interesting in the future to explore Wash's biochemical activities in the nucleus with particular attention to their relation to actin/microtubules, and its interaction with the SHRC, and how these are or are not involved in its role in global nuclear architecture.
Concluding Remarks
Not so long ago, the presence of actin in the nucleus was controversial (For a review see ref. 59). However, definitive experiments by McDonald et al., as well as the advent of new tools for actin visualization in the nucleus (i.e., Actin-Chromobody-TagGFP, LifeAct-GFP fused to an NLS, and Utr230EN) have shown that actin is clearly present.60-62 Recent studies using these new tools have shown nuclear actin present in the form of filaments formed by mDia nucleation, actin monomers in nuclear speckles, and as stable small actin filaments localized to interchromatin spaces.61,62 However, as many of the processes known to involve nuclear actin occur in response to varying stimuli, it will be important to use these tools in a variety of contexts. Seeing as WAS family proteins likely act with, or perhaps act independently of, nuclear actin, it is important to consider the processes in which WAS proteins and nuclear actin are known to overlap, and the structure of the actin within these processes. Thus far, the major described role of WAS family proteins in the nucleus has been altered transcription upon WAS protein perturbation. While WASP seems to act as a canonical bi-partite molecule with a clear separation of its cytoplasmic and nuclear activities, there is some evidence that N-WASP, JMY, and WAVE affect transcription, at least in part, through a mechanism involving nuclear actin.40,42, 45,48 Recently it was also shown that WASH is an important transcription factor for genes regulating T-cell differentiation. These effects occur both through WAS proteins acting as transcription factors and/or as components of chromatin remodeling complexes (CRCs). Nuclear actin has been shown to interact with all 3 RNA polymerases and as an essential component of multiple chromatin remodeling complexes.63-66 Thus far it appears that nuclear actin associated with the transcription machinery is monomeric and inhibiting binding to these actin monomers leads to deficient elongation.67 The question, therefore, is what the role of an F-actin associated protein may be in this process. One interesting proposed model is that actin filaments (non-canonical polymers) associated with WAS proteins and Arp2/3 (N-WASP in this case) and actin-severing proteins may locally increase the concentration of available actin monomers necessary to support elongation.68 It is also possible that non-canonical actin polymers support this process in a structural manner, however, evidence for this does not yet exist and visualization may prove difficult if these polymers are embedded within a locally enriched region of actin monomers. WAS family proteins also exert transcriptional phenotypes through their presence in CRCs. Similar to transcription, convincing evidence also suggests that actin monomers are the functional actin structures in CRCs (For a review see ref. 69), again raising the question of how filament-associated actin regulatory proteins may be involved. In this case, it seems less likely that this discrepancy can be explained by filament severing leading to local actin monomer increases, as the CRC in which actin has been most characterized, INO80, was shown to no longer associate with actin under conditions where actin and cofilin interact in the nucleus.70 As such, the role for WAS proteins and actin in CRCs remains unclear and future experimentation is needed.
Recently, we examined nuclear Wash functions in Drosophila employing both transcriptional/biochemical assays and powerful cell biological techniques and were excited to find gross defects in nuclear architecture, potentially connecting WAS family proteins to nuclear actin functions apart from transcription.54 Wash is essential for nuclear architecture in fly cells (S2R+) and salivary glands, where it interacts with another structural protein besides actin: Lamin B. As such, Wash was found to interact with the same regions of the chromatin as LADs. Although Wash lacks a DNA binding domain, it associates with ∼500 chromatin regions (directly or indirectly) on salivary gland polytene chromosomes. Wash's intrinsic biochemical properties (actin binding, MT binding, Lamin binding, and actin/MT crosslinking) suggest an exciting possibility as a hub for the integration of multiple structural filaments within the nucleus, and potentially as a bridge between these filaments and chromatin. In vitro experiments have shown that Lamin self-assembles into a meshwork under appropriate conditions and that Wash may facilitate this activity and integrate it with other structural proteins to form a higher order scaffold. Indeed, in this same nuclear compartment the Lamin-associated protein Emerin has been show to stabilize actin filament pointed ends suggesting that this region is an important neighborhood for integrating actin and lamin filaments, and perhaps may be involved in the formation of the cortical nucleoskeleton.
Another exciting possibility, which makes sense in lieu of the disrupted organization of the nucleus in wash mutants, is a role for Wash in intra-nuclear movements (loci and chromosomal position), which have been associated with nuclear actin and myosins. It has previously been shown that in response to inducing transcription for specific loci and during the heat shock response, specific loci move to more transcriptionally active regions within the nucleus (or to Cajal bodies for U2 snRNA genes).71,72 These movements, which in some cases also depend on nuclear myosins and encompass movements of up to 5μm, are suggestive of active intra-nuclear transport. No nuclear actin filaments have yet been observed to demonstrate that such movements occur along actin filaments. Thus, it is also possible that these intra-nuclear movements may be driven by a less stable mechanism, such as short actin comet tails. The disorganized nucleus phenotype in the absence of Wash suggests that Wash could regulate these movements such that they are organized and directed or that it is involved in stabilizing against these movements.
While tantalizing, currently it remains unclear if Wash's role in nuclear architecture involves nuclear actin and future studies will aim to use the new actin visualization methods to clarify this. Tied to this is how Wash is regulated in the nucleus. In the cytoplasm, WAS family proteins are generally regulated by Rho family GTPase signaling and, at least in the fly, Wash functions downstream of Rho1.16,17 There is growing evidence suggesting that pools of Rho family GTPases exist inside the nucleus in certain settings (For a review see ref. 26). WAVE1 and N-WASP, whose transcriptional roles are actin-dependent, have been shown to be uninhibited (N-WASP) or correlated with an increase in Rac1 (WAVE1). Interestingly, in contrast to WASP/WAVE proteins that are auto-inhibited or trans-inhibited, respectively (Fig. 1C), WASH family proteins do not appear to be inhibited, suggesting that these proteins are constitutively capable of actin nucleation. Thus, Wash might be able to nucleate actin irrespective of Rho1 signaling. Future experimentation using mutations which specifically disrupt Wash's binding to Rho1, Lamin, actin, its SHRC, or that abolish its actin nucleation or other biochemical activities, will be needed to elucidate the regulation of nuclear Wash, as well as which of its biochemical properties are involved in nuclear architecture.
Another interesting finding is that Wash's nuclear localization is context dependent (Fig. 3A) and thus, likely not required in the same nuclear architecture capacity in all contexts. For instance, in the developing fly embryo nuclear Wash is spatially and temporally regulated in specific developmental domains undergoing synchronized mitosis. One possibility is that Wash is shuttled into the nuclei of cells in these domains function specifically as a transcription factor/cofactor needed for the developmental program of these cells. Another possibility is that these cells are going to divide imminently or have recently divided such that Wash is involved in setting up the machineries required for division or in re-establishing/maintaining nuclear organization after division. Further analysis of the developmental expression of Wash and its expression correlated to specific cell-cycle phases may help address this question. This context specific nature of nuclear Wash function is observed in WAS family proteins as a whole. One theme is that these proteins can accumulate in the nucleus in response to a stimulus or developmental cue, whether it be transplantation into a new cytoplasm for WAVE1, a need to differentiate for WASP and mammalian WASH, heat-shock for N-WASP, or DNA damage for JMY. While WAS family proteins have established transcriptional roles, our Wash data suggests that these transcriptional defects can be secondary to a larger defect like nuclear architecture. Thus, Wash and WAS family proteins in general, provide exciting new entry points to unraveling the roles of classically cytoplasmic protein functions within the nucleus.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank Caitlin Milligan for comments on the manuscript.
Funding
This work was supported by NIH grant GM097083 to SMP.
References
- 1.Bezanilla M, Gladfelter AS, Kovar DR, Lee WL. Cytoskeletal dynamics: A view from the membrane. J Cell Biol 2015; 209:329-37; PMID:25963816; http://dx.doi.org/ 10.1083/jcb.201502062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Burianek LE, Soderling SH. Under lock and key: spatiotemporal regulation of WASP family proteins coordinates separate dynamic cellular processes. Semin Cell Dev Biol 2013; 24:258-66; PMID:23291261; http://dx.doi.org/ 10.1016/j.semcdb.2012.12.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Takenawa T, Suetsugu S. The WASP-WAVE protein network: connecting the membrane to the cytoskeleton. Nat Rev Mol Cell Biol 2007; 8:37-48; PMID:17183359; http://dx.doi.org/ 10.1038/nrm2069 [DOI] [PubMed] [Google Scholar]
- 4.Rottner K, Hanisch J, Campellone KG. WASH, WHAMM and JMY: regulation of Arp2/3 complex and beyond. Trends Cell Biol 2010; 20:650-61; PMID:20888769; http://dx.doi.org/ 10.1016/j.tcb.2010.08.014 [DOI] [PubMed] [Google Scholar]
- 5.Veltman DM, Insall RH. WASP family proteins: their evolution and its physiological implications. Mol Biol Cell 2010; 21:2880-93; PMID:20573979; http://dx.doi.org/ 10.1091/mbc.E10-04-0372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kollmar M, Lbik D, Enge S. Evolution of the eukaryotic ARP2/3 activators of the WASP family: WASP, WAVE, WASH, and WHAMM, and the proposed new family members WAWH and WAML. BMC Res Notes 2012; 5:88; PMID:22316129; http://dx.doi.org/ 10.1186/1756-0500-5-88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim AS, Kakalis LT, Abdul-Manan N, Liu GA, Rosen MK. Autoinhibition and activation mechanisms of the Wiskott-Aldrich syndrome protein. Nature 2000; 404:151-8; PMID:10724160; http://dx.doi.org/ 10.1038/35004513 [DOI] [PubMed] [Google Scholar]
- 8.Tomasevic N, Jia Z, Russell A, Fujii T, Hartman JJ, Clancy S, Wang M, Beraud C, Wood KW, Sakowicz R. Differential regulation of WASP and N-WASP by Cdc42, Rac1, Nck, and PI(4,5)P2. Biochemistry 2007; 46:3494-502; PMID:17302440; http://dx.doi.org/ 10.1021/bi062152y [DOI] [PubMed] [Google Scholar]
- 9.Eden S, Rohatgi R, Podtelejnikov AV, Mann M, Kirschner MW. Mechanism of regulation of WAVE1-induced actin nucleation by Rac1 and Nck. Nature 2002; 418:790-3; PMID:12181570; http://dx.doi.org/ 10.1038/nature00859 [DOI] [PubMed] [Google Scholar]
- 10.Chen Z, Borek D, Padrick SB, Gomez TS, Metlagel Z, Ismail AM, Umetani J, Billadeau DD, Otwinowski Z, Rosen MK. Structure and control of the actin regulatory WAVE complex. Nature 2010; 468:533-8; PMID:21107423; http://dx.doi.org/ 10.1038/nature09623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jia D, Gomez TS, Metlagel Z, Umetani J, Otwinowski Z, Rosen MK, Billadeau DD. WASH and WAVE actin regulators of the Wiskott-Aldrich syndrome protein (WASP) family are controlled by analogous structurally related complexes. Proc Natl Acad Sci U S A 2010; 107:10442-7; PMID:20498093; http://dx.doi.org/ 10.1073/pnas.0913293107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Derivery E, Sousa C, Gautier JJ, Lombard B, Loew D, Gautreau A. The Arp2/3 activator WASH controls the fission of endosomes through a large multiprotein complex. Dev Cell 2009; 17:712-23; PMID:19922875; http://dx.doi.org/ 10.1016/j.devcel.2009.09.010 [DOI] [PubMed] [Google Scholar]
- 13.Duleh SN, Welch MD. WASH and the Arp2/3 complex regulate endosome shape and trafficking. Cytoskeleton (Hoboken) 2010; 67:193-206; PMID:20175130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zech T, Calaminus SD, Caswell P, Spence HJ, Carnell M, Insall RH, Norman J, Machesky LM. The Arp2/3 activator WASH regulates alpha5beta1-integrin-mediated invasive migration. J Cell Sci 2011; 124:3753-9; PMID:22114305; http://dx.doi.org/ 10.1242/jcs.080986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rotty JD, Wu C, Bear JE. New insights into the regulation and cellular functions of the ARP2/3 complex. Nat Rev Mol Cell Biol 2013; 14:7-12; PMID:23212475; http://dx.doi.org/ 10.1038/nrm3492 [DOI] [PubMed] [Google Scholar]
- 16.Verboon JM, Rahe TK, Rodriguez-Mesa E, Parkhurst SM. Wash functions downstream of Rho1 GTPase in a subset of Drosophila immune cell developmental migrations. Mol Biol Cell 2015; 26:1665-74; PMID:25739458; http://dx.doi.org/ 10.1091/mbc.E14-08-1266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu R, Abreu-Blanco MT, Barry KC, Linardopoulou EV, Osborn GE, Parkhurst SM. Wash functions downstream of Rho and links linear and branched actin nucleation factors. Development 2009; 136:2849-60; PMID:19633175; http://dx.doi.org/ 10.1242/dev.035246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Snapper SB, Rosen FS. The Wiskott-Aldrich syndrome protein (WASP): roles in signaling and cytoskeletal organization. Annu Rev Immunol 1999; 17:905-29; PMID:10358777; http://dx.doi.org/ 10.1146/annurev.immunol.17.1.905 [DOI] [PubMed] [Google Scholar]
- 19.Frugtniet B, Jiang WG, Martin TA. Role of the WASP and WAVE family proteins in breast cancer invasion and metastasis. Breast Cancer (Dove Med Press) 2015; 7:99-109; PMID:25941446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thrasher AJ. WASp in immune-system organization and function. Nat Rev Immunol 2002; 2:635-46; PMID:12209132; http://dx.doi.org/ 10.1038/nri884 [DOI] [PubMed] [Google Scholar]
- 21.Villa A, Notarangelo L, Macchi P, Mantuano E, Cavagni G, Brugnoni D, Strina D, Patrosso MC, Ramenghi U, Sacco MG, et al. . X-linked thrombocytopenia and Wiskott-Aldrich syndrome are allelic diseases with mutations in the WASP gene. Nat Genet 1995; 9:414-7; PMID:7795648; http://dx.doi.org/ 10.1038/ng0495-414 [DOI] [PubMed] [Google Scholar]
- 22.Munsie LN, Desmond CR, Truant R. Cofilin nuclear-cytoplasmic shuttling affects cofilin-actin rod formation during stress. J Cell Sci 2012; 125:3977-88; PMID:22623727; http://dx.doi.org/ 10.1242/jcs.097667 [DOI] [PubMed] [Google Scholar]
- 23.Dopie J, Skarp KP, Rajakyla EK, Tanhuanpaa K, Vartiainen MK. Active maintenance of nuclear actin by importin 9 supports transcription. Proc Natl Acad Sci U S A 2012; 109:E544-52; PMID:22323606; http://dx.doi.org/ 10.1073/pnas.1118880109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Skarp KP, Huet G, Vartiainen MK. Steady-state nuclear actin levels are determined by export competent actin pool. Cytoskeleton (Hoboken) 2013; 70:623-34; PMID:23749625; http://dx.doi.org/ 10.1002/cm.21116 [DOI] [PubMed] [Google Scholar]
- 25.Huet G, Skarp KP, Vartiainen MK. Nuclear actin levels as an important transcriptional switch. Transcription 2012; 3:226-30; PMID:22771994; http://dx.doi.org/ 10.4161/trns.21062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rajakyla EK, Vartiainen MK. Rho, nuclear actin, and actin-binding proteins in the regulation of transcription and gene expression. Small GTPases 2014; 5:e27539; PMID:24603113; http://dx.doi.org/ 10.4161/sgtp.27539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Visa N, Percipalle P. Nuclear functions of actin. Cold Spring Harb Perspect Biol 2010; 2:a000620; PMID:20452941; http://dx.doi.org/ 10.1101/cshperspect.a000620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Clark TG, Merriam RW. Diffusible and bound actin nuclei of Xenopus laevis oocytes. Cell 1977; 12:883-91; PMID:563771; http://dx.doi.org/ 10.1016/0092-8674(77)90152-0 [DOI] [PubMed] [Google Scholar]
- 29.de Lanerolle P, Serebryannyy L. Nuclear actin and myosins: life without filaments. Nat Cell Biol 2011; 13:1282-8; PMID:22048410; http://dx.doi.org/ 10.1038/ncb2364 [DOI] [PubMed] [Google Scholar]
- 30.de Lanerolle P. Nuclear actin and myosins at a glance. J Cell Sci 2012; 125:4945-9; PMID:23277533; http://dx.doi.org/ 10.1242/jcs.099754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gulacsy V, Freiberger T, Shcherbina A, Pac M, Chernyshova L, Avcin T, Kondratenko I, Kostyuchenko L, Prokofjeva T, Pasic S, et al. . Genetic characteristics of eighty-seven patients with the Wiskott-Aldrich syndrome. Mol Immunol 2011; 48:788-92; PMID:21185603; http://dx.doi.org/ 10.1016/j.molimm.2010.11.013 [DOI] [PubMed] [Google Scholar]
- 32.Imai K, Morio T, Zhu Y, Jin Y, Itoh S, Kajiwara M, Yata J, Mizutani S, Ochs HD, Nonoyama S. Clinical course of patients with WASP gene mutations. Blood 2004; 103:456-64; PMID:12969986; http://dx.doi.org/ 10.1182/blood-2003-05-1480 [DOI] [PubMed] [Google Scholar]
- 33.Jin Y, Mazza C, Christie JR, Giliani S, Fiorini M, Mella P, Gandellini F, Stewart DM, Zhu Q, Nelson DL, et al. . Mutations of the Wiskott-Aldrich Syndrome Protein (WASP): hotspots, effect on transcription, and translation and phenotype/genotype correlation. Blood 2004; 104:4010-9; PMID:15284122; http://dx.doi.org/ 10.1182/blood-2003-05-1592 [DOI] [PubMed] [Google Scholar]
- 34.Sarkar K, Sadhukhan S, Han SS, Vyas YM. Disruption of hSWI/SNF complexes in T cells by WAS mutations distinguishes X-linked thrombocytopenia from Wiskott-Aldrich syndrome. Blood 2014; 124:3409-19; PMID:25253772; http://dx.doi.org/ 10.1182/blood-2014-07-587642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Taylor MD, Sadhukhan S, Kottangada P, Ramgopal A, Sarkar K, D'Silva S, Selvakumar A, Candotti F, Vyas YM. Nuclear role of WASp in the pathogenesis of dysregulated TH1 immunity in human Wiskott-Aldrich syndrome. Sci Transl Med 2010; 2:37ra44; PMID:20574068; http://dx.doi.org/ 10.1126/scitranslmed.3000813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sadhukhan S, Sarkar K, Taylor M, Candotti F, Vyas YM. Nuclear role of WASp in gene transcription is uncoupled from its ARP2/3-dependent cytoplasmic role in actin polymerization. J Immunol 2014; 193:150-60; PMID:24872192; http://dx.doi.org/ 10.4049/jimmunol.1302923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Suetsugu S, Takenawa T. Translocation of N-WASP by nuclear localization and export signals into the nucleus modulates expression of HSP90. J Biol Chem 2003; 278:42515-23; PMID:12871950; http://dx.doi.org/ 10.1074/jbc.M302177200 [DOI] [PubMed] [Google Scholar]
- 38.Wu X, Suetsugu S, Cooper LA, Takenawa T, Guan JL. Focal adhesion kinase regulation of N-WASP subcellular localization and function. J Biol Chem 2004; 279:9565-76; PMID:14676198; http://dx.doi.org/ 10.1074/jbc.M310739200 [DOI] [PubMed] [Google Scholar]
- 39.Mizutani K, Suetsugu S, Takenawa T. FBP11 regulates nuclear localization of N-WASP and inhibits N-WASP-dependent microspike formation. Biochem Biophys Res Commun 2004; 313:468-74; PMID:14697212; http://dx.doi.org/ 10.1016/j.bbrc.2003.11.139 [DOI] [PubMed] [Google Scholar]
- 40.Wu X, Yoo Y, Okuhama NN, Tucker PW, Liu G, Guan JL. Regulation of RNA-polymerase-II-dependent transcription by N-WASP and its nuclear-binding partners. Nat Cell Biol 2006; 8:756-63; PMID:16767080; http://dx.doi.org/ 10.1038/ncb1433 [DOI] [PubMed] [Google Scholar]
- 41.Shav-Tal Y, Zipori D. PSF and p54(nrb)/NonO–multi-functional nuclear proteins. FEBS Lett 2002; 531:109-14; PMID:12417296; http://dx.doi.org/ 10.1016/S0014-5793(02)03447-6 [DOI] [PubMed] [Google Scholar]
- 42.Yoo Y, Wu X, Guan JL. A novel role of the actin-nucleating Arp2/3 complex in the regulation of RNA polymerase II-dependent transcription. J Biol Chem 2007; 282:7616-23; PMID:17220302; http://dx.doi.org/ 10.1074/jbc.M607596200 [DOI] [PubMed] [Google Scholar]
- 43.Lefever T, Pedersen E, Basse A, Paus R, Quondamatteo F, Stanley AC, Langbein L, Wu X, Wehland J, Lommel S, et al. . N-WASP is a novel regulator of hair-follicle cycling that controls antiproliferative TGF{β} pathways. J Cell Sci 2010; 123:128-40; PMID:20016073; http://dx.doi.org/ 10.1242/jcs.053835 [DOI] [PubMed] [Google Scholar]
- 44.Miyamoto K, Pasque V, Jullien J, Gurdon JB. Nuclear actin polymerization is required for transcriptional reprogramming of Oct4 by oocytes. Genes Dev 2011; 25:946-58; PMID:21536734; http://dx.doi.org/ 10.1101/gad.615211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Miyamoto K, Teperek M, Yusa K, Allen GE, Bradshaw CR, Gurdon JB. Nuclear Wave1 is required for reprogramming transcription in oocytes and for normal development. Science 2013; 341:1002-5; PMID:23990560; http://dx.doi.org/ 10.1126/science.1240376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shikama N, Lee CW, France S, Delavaine L, Lyon J, Krstic-Demonacos M, La Thangue NB. A novel cofactor for p300 that regulates the p53 response. Mol Cell 1999; 4:365-76; PMID:10518217; http://dx.doi.org/ 10.1016/S1097-2765(00)80338-X [DOI] [PubMed] [Google Scholar]
- 47.Zuchero JB, Belin B, Mullins RD. Actin binding to WH2 domains regulates nuclear import of the multifunctional actin regulator JMY. Mol Biol Cell 2012; 23:853-63; PMID:22262458; http://dx.doi.org/ 10.1091/mbc.E11-12-0992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Coutts AS, Weston L, La Thangue NB. A transcription co-factor integrates cell adhesion and motility with the p53 response. Proc Natl Acad Sci U S A 2009; 106:19872-7; PMID:19897726; http://dx.doi.org/ 10.1073/pnas.0906785106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Boyd SD, Tsai KY, Jacks T. An intact HDM2 RING-finger domain is required for nuclear exclusion of p53. Nat Cell Biol 2000; 2:563-8; PMID:10980695; http://dx.doi.org/ 10.1038/35023500 [DOI] [PubMed] [Google Scholar]
- 50.Coutts AS, Boulahbel H, Graham A, La Thangue NB. Mdm2 targets the p53 transcription cofactor JMY for degradation. EMBO Rep 2007; 8:84-90; PMID:17170761; http://dx.doi.org/ 10.1038/sj.embor.7400855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gadea G, de Toledo M, Anguille C, Roux P. Loss of p53 promotes RhoA-ROCK-dependent cell migration and invasion in 3D matrices. J Cell Biol 2007; 178:23-30; PMID:17606864; http://dx.doi.org/ 10.1083/jcb.200701120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gomez TS, Billadeau DD. A FAM21-containing WASH complex regulates retromer-dependent sorting. Dev Cell 2009; 17:699-711; PMID:19922874; http://dx.doi.org/ 10.1016/j.devcel.2009.09.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xia P, Wang S, Huang G, Zhu P, Li M, Ye B, Du Y, Fan Z. WASH is required for the differentiation commitment of hematopoietic stem cells in a c-Myc-dependent manner. J Exp Med 2014; 211:2119-34; PMID:25225459; http://dx.doi.org/ 10.1084/jem.20140169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Verboon JM, Rincon-Arano H, Werwie TR, Delrow JJ, Scalzo D, Nandakumar V, Groudine M, Parkhurst SM. Wash interacts with lamin and affects global nuclear organization. Curr Biol 2015; 25:804-10; PMID:25754639; http://dx.doi.org/ 10.1016/j.cub.2015.01.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rodriguez-Mesa E, Abreu-Blanco MT, Rosales-Nieves AE, Parkhurst SM. Developmental expression of Drosophila Wiskott-Aldrich Syndrome family proteins. Dev Dyn 2012; 241:608-26; PMID:22275148; http://dx.doi.org/ 10.1002/dvdy.23742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Linardopoulou EV, Parghi SS, Friedman C, Osborn GE, Parkhurst SM, Trask BJ. Human subtelomeric WASH genes encode a new subclass of the WASP family. PLoS Genet 2007; 3:e237; PMID:18159949; http://dx.doi.org/ 10.1371/journal.pgen.0030237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Deng ZH, Gomez TS, Osborne DG, Phillips-Krawczak CA, Zhang JS, Billadeau DD. Nuclear FAM21 participates in NF-kappaB-dependent gene regulation in pancreatic cancer cells. J Cell Sci 2015; 128:373-84; PMID:25431135; http://dx.doi.org/ 10.1242/jcs.161513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hochheimer A, Zhou S, Zheng S, Holmes MC, Tjian R. TRF2 associates with DREF and directs promoter-selective gene expression in Drosophila. Nature 2002; 420:439-45; PMID:12459787; http://dx.doi.org/ 10.1038/nature01167 [DOI] [PubMed] [Google Scholar]
- 59.Pederson T, Aebi U. Actin in the nucleus: what form and what for? J Struct Biol 2002; 140:3-9; PMID:12490148; http://dx.doi.org/ 10.1016/S1047-8477(02)00528-2 [DOI] [PubMed] [Google Scholar]
- 60.McDonald D, Carrero G, Andrin C, de Vries G, Hendzel MJ. Nucleoplasmic β-actin exists in a dynamic equilibrium between low-mobility polymeric species and rapidly diffusing populations. J Cell Biol 2006; 172:541-52; PMID:16476775; http://dx.doi.org/ 10.1083/jcb.200507101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Belin BJ, Cimini BA, Blackburn EH, Mullins RD. Visualization of actin filaments and monomers in somatic cell nuclei. Mol Biol Cell 2013; 24:982-94; PMID:23447706; http://dx.doi.org/ 10.1091/mbc.E12-09-0685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Baarlink C, Wang H, Grosse R. Nuclear actin network assembly by formins regulates the SRF coactivator MAL. Science 2013; 340:864-7; PMID:23558171; http://dx.doi.org/ 10.1126/science.1235038 [DOI] [PubMed] [Google Scholar]
- 63.Hu P, Wu S, Hernandez N. A role for β-actin in RNA polymerase III transcription. Genes Dev 2004; 18:3010-5; PMID:15574586; http://dx.doi.org/ 10.1101/gad.1250804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Obrdlik A, Percipalle P. The F-actin severing protein cofilin-1 is required for RNA polymerase II transcription elongation. Nucleus 2011; 2:72-9; PMID:21647301; http://dx.doi.org/ 10.4161/nucl.14508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Philimonenko VV, Zhao J, Iben S, Dingova H, Kysela K, Kahle M, Zentgraf H, Hofmann WA, de Lanerolle P, Hozák P, et al. . Nuclear actin and myosin I are required for RNA polymerase I transcription. Nat Cell Biol 2004; 6:1165-72; PMID:15558034; http://dx.doi.org/ 10.1038/ncb1190 [DOI] [PubMed] [Google Scholar]
- 66.Qi T, Tang W, Wang L, Zhai L, Guo L, Zeng X. G-actin participates in RNA polymerase II-dependent transcription elongation by recruiting positive transcription elongation factor b (P-TEFb). J Biol Chem 2011; 286:15171-81; PMID:21378166; http://dx.doi.org/ 10.1074/jbc.M110.184374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Percipalle P, Fomproix N, Kylberg K, Miralles F, Bjorkroth B, Daneholt B, Visa N. An actin-ribonucleoprotein interaction is involved in transcription by RNA polymerase II. Proc Natl Acad Sci U S A 2003; 100:6475-80; PMID:12743363; http://dx.doi.org/ 10.1073/pnas.1131933100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Miyamoto K, Gurdon JB. Transcriptional regulation and nuclear reprogramming: roles of nuclear actin and actin-binding proteins. Cell Mol Life Sci 2013; 70:3289-302; PMID:23275942; http://dx.doi.org/ 10.1007/s00018-012-1235-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kapoor P, Shen X. Mechanisms of nuclear actin in chromatin-remodeling complexes. Trends Cell Biol 2014; 24:238-46; PMID:24246764; http://dx.doi.org/ 10.1016/j.tcb.2013.10.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kapoor P, Chen M, Winkler DD, Luger K, Shen X. Evidence for monomeric actin function in INO80 chromatin remodeling. Nat Struct Mol Biol 2013; 20:426-32; PMID:23524535; http://dx.doi.org/ 10.1038/nsmb.2529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chuang CH, Carpenter AE, Fuchsova B, Johnson T, de Lanerolle P, Belmont AS. Long-range directional movement of an interphase chromosome site. Curr Biol 2006; 16:825-31; PMID:16631592; http://dx.doi.org/ 10.1016/j.cub.2006.03.059 [DOI] [PubMed] [Google Scholar]
- 72.Dundr M, Ospina JK, Sung MH, John S, Upender M, Ried T, Hager GL, Matera AG. Actin-dependent intranuclear repositioning of an active gene locus in vivo. J Cell Biol 2007; 179:1095-103; PMID:18070915; http://dx.doi.org/ 10.1083/jcb.200710058 [DOI] [PMC free article] [PubMed] [Google Scholar]