Abstract
Localized chromatin organization is now recognized as an important determinant of cell identity and developmental pathways. Recent studies have demonstrated that these epigenetic states are unexpectedly dynamic and malleable. In this Extra view we will highlight the transient nature of stimulus-induced enhancer accessibility and its importance for transcription regulation. Using glucocorticoid receptor (GR) as a model system we will discuss spatiotemporal relationships between receptor/chromatin interactions, lifetimes of the DNase I hypersensitivity sites (DHSs), long-range interactions, and gene regulation. We propose that differential temporal activation and utilization of distal regulatory elements plays a role in directing divergent stimulus-induced transcriptional programs.
Keywords: chromatin accessibility, DNase I, enhancers, gene pulsing, glucocorticoid receptor, long-range interactions, transcription, ultradian
Abbreviations
- ChIP
chromatin immunoprecipitation
- DHSs
DNase I hypersensitivity sites
- eRNA
enhancer RNA
- GR
glucocorticoid receptor
- GREs
GR regulatory elements
- PTMs
posttranslational modifications
- TADs
topologically associating domains
Introduction
Cells in a multicellular organism are subjected to a plethora of signals, including endocrine and paracrine hormones. These temporal changes in cellular environment impact cell physiology and gene transcription. While some of these changes are stochastic, many hormones are released in a complex temporal fashion.1-4 As an example, GR-stimulating glucocorticoids are released from the adrenal glands in a circadian, as well as hourly (ultradian) manner5-9 (Fig. 1A). As a result, GR-expressing cells experience frequent changes in the level of the activating hormone. Intrinsic cellular processes and differences in physiological states of individual cells may further influence hormone signaling and increase cell-to-cell variability of the transcriptional responses. However, the primary driver of the GR-regulated gene expression is pulsatile, cell extrinsic hormonal signaling.
Figure 1.
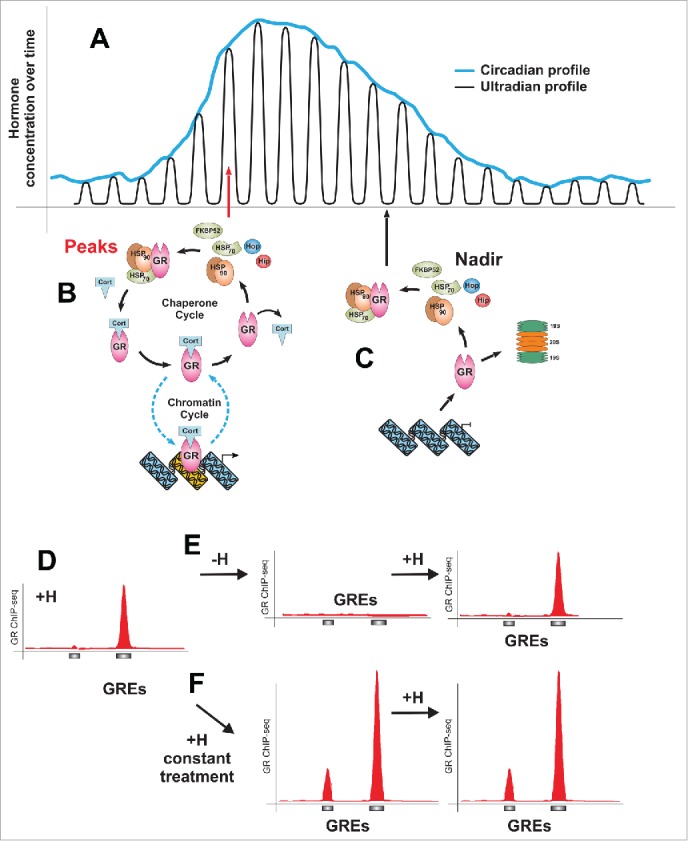
Ultradian mode of glucocorticoid release and its effects on the GR/chromatin interactions. (A) Schematic representation of the glucocorticoid release pattern: circadian profile consists of discrete ultradian (hourly) pulses. (B) At the peak of an ultradian pulse (red arrow) hormone-activated GR interacts with GREs in a transient manner (chromatin cycle). GR molecules undergo a frequent hormone disassociation-reassociation cycles and the hormone-free receptor must interact with the chaperone machinery to regain hormone binding affinity in the nucleus (chaperone cycle). (C) Hormone disassociation-reassociation cycles allow GR to “sense” a decline in the hormone level (nadir, black arrow), leading to an accumulation of hormone-free receptors incapable of productive interactions with GREs. Unbound receptors will either associate with the chaperone machinery or get degraded by the proteasome. (D) GR loading at GREs strictly follows hormone level fluctuations as demonstrated by chromatin immunoprecipitation against GR followed by high throughput sequencing (ChIP-seq). (E) The GR peaks diminish upon hormone withdrawal, while a subsequent hormone pulse reestablishes GR affinity to GREs. (F) Constant stimulation (reminiscent of the hormone release pattern under stress conditions) increases the level of GR binding at the GREs. Considering that ChIP results are representative of the population average, it is unclear whether under this conditions more GR molecules become engaged at a given GRE or whether the GRE becomes occupied in a bigger number of cells.
Considering that endocrine signals govern the function of all tissues and organs in the body, it is surprising how little is known about the genomic effects of the naturally occurring hormone release patterns. The complex and dynamic picture of the GR-mediated transcription regulation10,11 is still not fully appreciated and hormonal fluctuations are frequently dismissed as “noise.” Here we underscore the fact that cells have evolved mechanisms to utilize these naturally occurring transient changes in hormone availability in establishing biologically appropriate patterns of receptor loading, enhancer activation, long-range interactions, and gene activity on the genomic scale.
Dynamic Interactions of TFs with Regulatory Elements
It is becoming apparent that transcription regulation is comprised of many rapid and time-sensitive molecular processes.11-15 To integrate various temporal signals in coherent transcriptional responses, transcription factors (TFs), target chromatin sites, and the transcriptional machinery must detect, adapt, and respond to these changes. It is well established that in vivo the majority of TFs interact with chromatin targets only transiently.12,16-18 Several implications for these dynamic interactions were proposed: (i) regulation of the transcriptional output,19,20 (ii) “sensing” fluctuations in the levels of activating signals,21 and (iii) allowing a noncompetitive transient access for binding of secondary regulatory factors.22 Numerous studies have demonstrated that transient GR association with GR regulatory elements (GREs) in vivo (chromatin cycle) is a key feature of GR signaling.16,19,23-25 In addition, a much slower cycle involving nuclear chaperone machinery has been implicated.21 These two superimposed molecular cycles (Fig. 1B and C) ensure that GR interactions with GREs, detectable by microscopic techniques as well as biochemical methods such as chromatin immunoprecipitation (ChIP), occur in a strictly hormone-dependent manner21,26 (Fig. 1D).
Lifetimes of Chromatin Accessibility
All GR binding takes place at “open” chromatin sites sensitive to DNase I digestion, either at pre-existing (pre-programmed) sites, or sites actively induced by the receptor.27 Once considered just a packaging mechanism for DNA, chromatin is now known to be responsible for the functional organization of the genetic material within the nucleus providing a selective access to genomic regulatory elements.28-30
Chromatin accessibility is regulated by nucleosome remodeling, utilization of histone variants, DNA methylation, and posttranslational modifications (PTMs).15,31,32 Chromatin remodeling complexes can slide nucleosomes, rotate the DNA helix relative to the nucleosome to expose TF binding sites, or evict nucleosomes.31 Multiple remodeling complexes can collaborate or compete at a given site to influence its accessibility as measured by its sensitivity to DNase I digestion.33
Through its ability to interact with remodeling complexes, transient GR binding influences chromatin accessibility in a treatment-specific manner.26 At a number of sites (transient DHSs) hormone withdrawal induced a rapid loss of hypersensitivity (Fig. 2A). This finding is consistent with an early study demonstrating the existence of a transient hypersensitive site near the promoter of a GR-responsive gene.34 However, other hormone-induced DHSs (persistent DHSs) preserve a “memory” of hormone induction. Instead of reverting to their initial level of hypersensitivity, they retain a heightened hypersensitivity state even 40 minutes upon termination of the stimulus (Fig. 2B). Other studies have also described long-lasting hypersensitivity or enhancer activity upon termination of the activating signals.35,36 The basis for different lifetimes of chromatin accessibility is not completely clear, but it appears that the initial chromatin state of the sites plays a role. For example, we find that long-lasting hypersensitivity occurs only at pre-accessible sites, while the de novo and weakly accessible sites always have a short lifetime. Consistent with this finding, persistent DHSs were more likely to be associated with factors implicated in enhancer activation such as p300 and chromatin remodelers (BRG1), even before hormone induction.26
Figure 2.
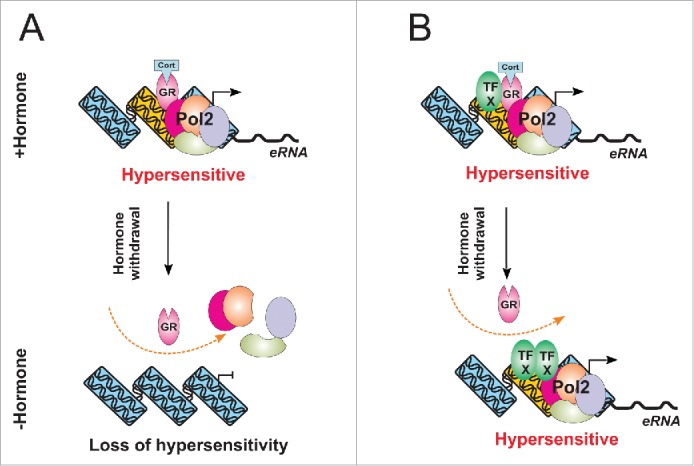
A model for hypersensitivity regulation at transient and persistent DHSs. (A) GR loading at transient sites recruits chromatin remodelers (not shown) leading to an increases accessibility at these sites, as well as Pol II recruitment and release of enhancer RNA (eRNA). Upon hormone withdrawal GR dissociation from GREs leads to dissociation of the remodelers and consequently to a loss of accessibility at these sites. (B) At the persistent DHSs, the GR-created accessibility allows binding of secondary TFs. These factors remain at the sites even after hormone withdrawal and continue to remodel it. This carry-over hypersensitivity may over time return to its initial state or lead to a permanent open state at the site.
While the hypersensitivity of the transient DHSs is clearly GR-dependent, the behavior of the persistent sites cannot be explained by a simple mode of hormone dependent GR binding. Transient GR-mediated remodeling was demonstrated to facilitates the access for binding of secondary regulatory factors, which further influence chromatin state,22,37 and we envision such factors driving accessibility of the persistent sites even after hormone withdrawal (Fig. 2B). In a limited number of cases we found that AP1 was recruited to persistent DHSs in a hormone-dependent manner and remained at the site even after hormone withdrawal (unpublished data). In these instances AP1-mediated chromatin remodeling could be responsible for the sustained level of hypersensitivity at these sites. However, the secondary factors required for the sustenance of hypersensitivity are likely to be site specific as this mode of AP1-mediated remodeling was not apparent at other sites (unpublished data).
Thus, the molecular mechanisms driving diverse DHS dynamics are not completely clear, but the abovementioned differences in the initial state of hypersensitivity, existing variations in DHS-associated motifs targeted by various secondary factors,26 as well as establishment of specific PTMs all could play a role. For example, retention of the H3K4me1 modification at an enhancer upon termination of the activating stimulus was implicated in creation of “memory” of the stimulation as well as in the faster and stronger response upon restimulation.36 It is unclear whether histone modifications at transient and persistent sites differ and additional studies will be required to fully understand the mechanisms behind epigenomic “memory” of hormone induction as well as its implications for transcription regulation.
Enhancer Dynamics, Long-Range Interactions and Transcription Regulation
We previously demonstrated that ultradian hormone release promotes cyclic GR interaction with regulatory elements, leading to cyclic release of nascent RNA from a number of GR regulated genes.21 Our genome-wide studies confirmed that gene pulsing is the predominant transcriptional response of the GR-regulated genes. However, we also uncovered transcription profiles which could not be extrapolated from the strictly hormone-dependent GR binding to GREs.26 Higher order chromatin structure and organization contributes to gene expression regulation.38,39 Thus, it is possible that the diverse lifetimes of accessibility and activation of distal regulatory elements could contribute to the observed divergent patterns of gene regulation through combinatorial long-range interactions. This is especially relevant considering that the majority of the GR binding sites are found away from promoters of GR-responsive genes.27
It was recently discovered that mammalian genomes are hierarchically organized into megabase-sized topologically associating domains (TADs) within the chromosome territories (Fig. 3A and B) and that CTCF and cohesin were both implicated in anchoring the loops formed between TAD boundaries.40-42 TADs are stably maintained during differentiation and development while the organization within the TADs is cell-type specific. Enhancer-promoter looping within the TADs brings distal TF-bound regulatory sites closer to target genes [Reviewed in43,44]. Many of the enhancer-promoter loops were found to be conserved between species while others were found to be tissue-specific and dependent on tissue-specific TFs.42 Interestingly, insertion of artificial zinc fingers to tether a looping factor (or its self-associating domain) to the β-globin promoter resulted in formation of a loop with the LCR as well as activation of the β-globin gene transcription.45 These data suggest that chromatin looping causally underlies gene regulation and that transcription factors through self-association domains could influence the formation of enhancer-promoter loops. GR is frequently found at distal enhancer and can self-associate, suggesting that receptor binding could play a role in initiating or strengthening enhancer-promoter contacts, implicating loops in the regulation of glucocorticoid responsive genes. Indeed, the recent findings26 demonstrate that receptor binding at enhancers and the consequential change in their accessibility correlates with the frequency of interactions of these enhancers with nearby gene targets (Fig 3C-E). It should be noted that nonrandom enhancer-promoter interactions were detectable even before treatment; however receptor loading and alteration of the chromatin structure at enhancers significantly increased the frequency of these contacts.
Figure 3.
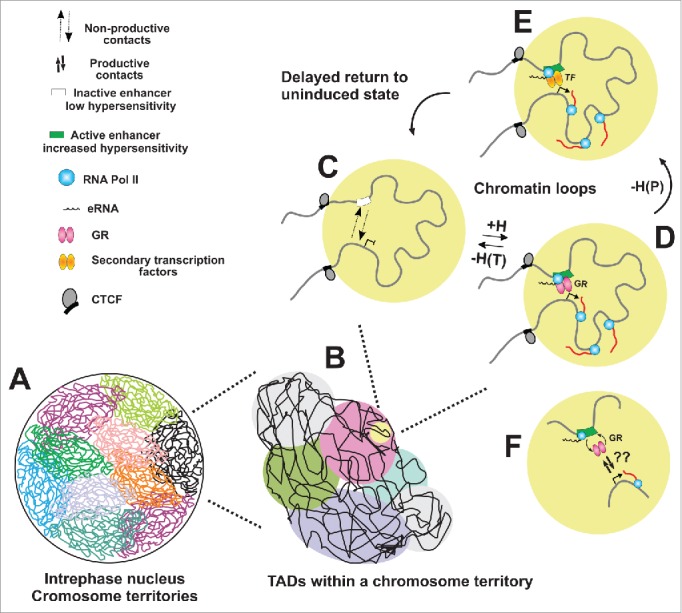
Transient activation of enhancers contributes to gene transcription regulation through dynamic promoter-enhancer contacts. (A) Interphase nucleus is organized in spatially distinct chromosome territories which are further divided into megabase-long topologically associating domains (TADs) (B). TADs are maintained across different cell types while the organization within the TADs is cell-type specific. (C) Local chromatin structure facilitates enhancer-promoter interactions within the TADs brings distal regulatory sites closer to target genes. However, the depicted interactions (D) are non-productive and do not lead to gene activation. In the presence of hormone, GR binds and remodels chromatin at the enhancer element promoting its increased accessibility and activation (D). As a result enhancer-promoter contacts are strengthened, leading to gene activation. Enhancer activation is also characterized by recruitment of Pol II and the mediator, as well as release of eRNA. Upon hormone withdrawal GR dissociation from enhancers at transient DHSs [-H(T)] leads to a loss of hypersensitivity and establishment of the initial state (C). In other cases, [persistent DHSs, -H(P)], secondary factors could continue to remodel the site, keeping it active long after hormone withdrawal (E). Over time the site could either return to its unindicted state (short-term memory) or retain this new remodeled state (long-term memory of hormone induction). A central point of this model is the notion that enhancer-promoter contacts are not static and the hormone-dependent GR binding at enhancers, even though transient, activates these sites leading to stronger and productive enhancer-promoter contacts (F).
A fundamental problem in long range enhancer contacts remains unresolved. As depicted in Fig. 3F, many of the factors acting at these elements, including GR, have residence times in the range of 5-10 sec.12 Domain factors such as CTCF and cohesion are often assumed to have very long residence times, providing potential stability to these interactions. However there is as yet no direct evidence to support this concept. How rapidly exchanging proteins such as GR function within the actual molecular biochemistry at the long-range contacts is a major challenge for the field. Recent advances in live cell imaging, particularly single molecule tracking, hold some promise to address these difficult issues.
In addition to TFs and the well-established architectural proteins CTCF and cohesin, other factors such as mediator, coactivators (p300 and CBP), and eRNAs aree also implicated in establishing promoter-enhancer loops.46-53 In a number of cases we noted hormone-dependent recruitment of Pol II at enhancers and confirmed the release of eRNA in a stimulus-dependent manner (unpublished data). Enhancer transcription could be just a reflection of opportunistic Pol II loading at accessible chromatins. However, previous studies50,54,55 reporting a correlation between the eRNA levels and the expression of nearby genes suggests that eRNA might be important for enhancer function. More systematic studies will be required to determine whether transient release of eRNA in a treatment-specific manner could influence the activity of enhancers and their participation in long-range interactions. Genome editing by CRISPR-Cas9 technology or targeting various factors to specific genomic sites using a variation of the CRISPR-Cas9 system56 will further add to our understanding of the role of the chromatins state of enhancers as well as their function(s).
Overall, our findings suggest that changes in chromatin accessibility may influence the frequency of the enhancer-promoter interactions. However, assuming that transient DHSs are always in the vicinity of transiently induced genes or that persistent DHSs are always in a close proximity to persistently active genes would be an over-simplification. Multiple enhancers likely contribute to the regulation of a gene in a combinatorial manner; furthermore, it is not always apparent which enhancers are functionally involved. Thus, additional studies addressing the role of chromatin structure and accessibility in gene regulation will be required. Individual genes and enhancers should be considered and a combination of high-resolution chromosome conformation capture-based methods as well as super-resolution microscopy methods applied to further elucidate this matter.
In summary, the recent study26 shows that hormone-driven interactions of GR with GREs lead to a complex variety of changes in chromatin accessibility at distal sites. In a number of cases changes in chromatin structure and long-range contacts correlated with gene activity, suggesting that accessibility-driven long-range interactions may have regulatory functions. Under this scenario, differential utilization of distal enhancers could be achieved by temporal alteration of their chromatin structure in a stimulus-dependent manner which may represent a novel principle for gene transcription regulation from a distance.
Relevance to Physiological Hormone Action
Recent advancements in our understanding of the interplay between lifetimes of chromatin states and transcription regulation reinforce the view that complex dynamics on multiple timescales are crucial for the execution of appropriate transcriptional programs. Thus it is not surprising that the pulsed and constant hormone stimulations are associated with divergent transcriptional patterns26 which may provide the molecular basis for the differential physiological outcomes associated with these different modes of hormone exposure.
These findings may have significant implications for glucocorticoid function in vivo and for steroid therapies. Changes in glucocorticoid levels in plasma are paralleled by similar changes in tissues7,57,58 where gene activity is regulated in a dynamic manner.21,59 It is plausible that the disruptions of glucocorticoid release patterns, either as a result of a systemic disease, continuous stress, or treatment with synthetic glucocorticoids, may lead to aberrant patterns of activation of distal regulatory elements and aberrant transcriptional response in the glucocorticoid-target organs that have significant repercussions at the level of their normal physiology. Conversely, restoring the proper hormone delivery pattern may benefit glucocorticoid therapies in various clinical settings.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Funding
This research was supported (in part) by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research.
References
- 1.Rothman MS, Wierman ME. The role of gonadotropin releasing hormone in normal and pathologic endocrine processes. Curr Opin Endocrinol Diabetes Obes 2007; 14:306-10. [DOI] [PubMed] [Google Scholar]
- 2.Chadwick D, Goode JA. Mechanisms and Biological Significance of Pulsatile Hormone Secretion New York, NY: John Wiley & Sons Ltd, 2000. [Google Scholar]
- 3.Belchetz PE, Plant TM, Nakai Y, Keogh EJ, Knobil E. Hypophysial responses to continuous and intermittent delivery of hypopthalamic gonadotropin-releasing hormone. Science 1978; 202:631-3; PMID:100883; http://dx.doi.org/ 10.1126/science.100883 [DOI] [PubMed] [Google Scholar]
- 4.Gore AC. GnRH: The master molecule of reproduction Boston, MA: Kluwer Academic Publishers, 2002. [Google Scholar]
- 5.Atkinson HC, Wood SA, Kershaw YM, Bate E, Lightman SL. Diurnal variation in the responsiveness of the hypothalamic-pituitary-adrenal axis of the male rat to noise stress. J Neuroendocrinol 2006; 18:526-33; PMID:16774501; http://dx.doi.org/ 10.1111/j.1365-2826.2006.01444.x [DOI] [PubMed] [Google Scholar]
- 6.Conway-Campbell BL, Pooley JR, Hager GL, Lightman SL. Molecular dynamics of ultradian glucocorticoid receptor action. Mol Cell Endocrinol 2011; 348:383-93; PMID:21872640; http://dx.doi.org/ 10.1016/j.mce.2011.08.014 [DOI] [PubMed] [Google Scholar]
- 7.Droste SK, de Groote L, Atkinson HC, Lightman SL, Reul JM, Linthorst AC. Corticosterone levels in the brain show a distinct ultradian rhythm but a delayed response to forced swim stress. Endocrinology 2008; 149:3244-53; PMID:18356272; http://dx.doi.org/ 10.1210/en.2008-0103 [DOI] [PubMed] [Google Scholar]
- 8.Lightman SL, Conway-Campbell BL. The crucial role of pulsatile activity of the HPA axis for continuous dynamic equilibration. Nat Rev Neurosci 2010; 11:710-8. [DOI] [PubMed] [Google Scholar]
- 9.Walker JJ, Terry JR, Lightman SL. Origin of ultradian pulsatility in the hypothalamic-pituitary-adrenal axis. ProcBiol Sci 2010; 277:1627-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Miranda TB, Morris SA, Hager GL. Complex genomic interactions in the dynamic regulation of transcription by the glucocorticoid receptor. Mol Cell Endocrinol 2013; 380:16-24; http://dx.doi.org/ 10.1016/j.mce.2013.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stavreva DA, Varticovski L, Hager GL. Complex dynamics of transcription regulation. Biochim Biophys Acta 2012; 1819:657-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hager GL, McNally JG, Misteli T. Transcription dynamics. MolCell 2009; 35:741-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Voss TC, Hager GL. Dynamic regulation of transcriptional states by chromatin and transcription factors. Nat Rev Genet 2014; 15:69-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Coulon A, Chow CC, Singer RH, Larson DR. Eukaryotic transcriptional dynamics: from single molecules to cell populations. Nat Rev Genet 2013; 14:572-84; http://dx.doi.org/ 10.1038/nrg3484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Koster MJ, Snel B, Timmers HT. Genesis of Chromatin and Transcription Dynamics in the Origin of Species. Cell 2015; 161:724-36; PMID:25957681; http://dx.doi.org/ 10.1016/j.cell.2015.04.033 [DOI] [PubMed] [Google Scholar]
- 16.McNally JG, Mueller WG, Walker D, Wolford RG, Hager GL. The glucocorticoid receptor: Rapid exchange with regulatory sites in living cells. Science 2000; 287:1262-5; PMID:10678832; http://dx.doi.org/ 10.1126/science.287.5456.1262 [DOI] [PubMed] [Google Scholar]
- 17.Bosisio D, Marazzi I, Agresti A, Shimizu N, Bianchi ME, Natoli G. A hyper-dynamic equilibrium between promoter-bound and nucleoplasmic dimers controls NF-kB-dependent gene activity. EMBO J 2006; 25:798-810; PMID:16467852; http://dx.doi.org/ 10.1038/sj.emboj.7600977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sharp ZD, Mancini MG, Hinojos CA, Dai F, Berno V, Szafran AT, Smith KP, Lele TP, Ingber DE, Mancini MA. Estrogen-receptor-α exchange and chromatin dynamics are ligand- and domain-dependent. J Cell Sci 2006; 119:4101-16; PMID:16968748; http://dx.doi.org/ 10.1242/jcs.03161 [DOI] [PubMed] [Google Scholar]
- 19.Stavreva DA, Muller WG, Hager GL, Smith CL, McNally JG. Rapid glucocorticoid receptor exchange at a promoter is coupled to transcription and regulated by chaperones and proteasomes. MolCell Biol 2004; 24:2682-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gorski SA, Snyder SK, John S, Grummt I, Misteli T. Modulation of RNA polymerase assembly dynamics in transcriptional regulation. MolCell 2008; 30:486-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stavreva DA, Wiench M, John S, Conway-Campbell BL, McKenna MA, Pooley JR, Johnson TA, Voss TC, Lightman SL, Hager GL. Ultradian hormone stimulation induces glucocorticoid receptor-mediated pulses of gene transcription. NatCell Biol 2009; 11:1093-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Voss TC, Schiltz RL, Sung MH, Yen PM, Stamatoyannopoulos JA, Biddie SC, Johnson TA, Miranda TB, John S, Hager GL. Dynamic exchange at regulatory elements during chromatin remodeling underlies assisted loading mechanism. Cell 2011; 146:544-54; PMID:21835447; http://dx.doi.org/ 10.1016/j.cell.2011.07.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Morisaki T, Muller WG, Golob N, Mazza D, McNally JG. Single-molecule analysis of transcription factor binding at transcription sites in live cells. Nat Commun 2014; 5:4456; PMID:25034201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mueller F, Karpova TS, Mazza D, McNally JG. Monitoring dynamic binding of chromatin proteins in vivo by fluorescence recovery after photobleaching. Methods Mol Biol 2012; 833:153-76; PMID:22183594; http://dx.doi.org/ 10.1007/978-1-61779-477-3_11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mueller F, Mazza D, Stasevich TJ, McNally JG. FRAP and kinetic modeling in the analysis of nuclear protein dynamics: what do we really know? CurrOpinCell Biol 2010; 22:403-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stavreva DA, Coulon A, Sung MH, Baek S, John S, Stixova L, Tesikova M, Hakim O, Miranda T, Hawkins M, et al.. Dynamics of chromatin accessibility and long-range interactions in response to glucocorticoid pulsing. Genome Res 2015; 25:13; http://dx.doi.org/ 10.1101/gr.184168.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.John S, Sabo PJ, Johnson TA, Sung MH, Biddie SC, Lightman SL, Voss TC, Davis SR, Meltzer PS, Stamatoyannopoulos JA, et al.. Interaction of the glucocorticoid receptor with the global chromatin landscape. Mol Cell 2008; 29:611-24. [DOI] [PubMed] [Google Scholar]
- 28.Johnson DG, Dent SY. Chromatin: receiver and quarterback for cellular signals. Cell 2013; 152:685-9; PMID:23375745; http://dx.doi.org/ 10.1016/j.cell.2013.01.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hardison RC, Taylor J. Genomic approaches towards finding cis-regulatory modules in animals. Nat Rev Genet 2012; 13:469-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Spitz F, Furlong EE. Transcription factors: from enhancer binding to developmental control. Nat Rev Genet 2012; 13:613-26. [DOI] [PubMed] [Google Scholar]
- 31.Clapier CR, Cairns BR. The biology of chromatin remodeling complexes. Annu Rev Biochem 2009; 78:273-304. [DOI] [PubMed] [Google Scholar]
- 32.Melters DP, Nye J, Zhao H, Dalal Y. Chromatin Dynamics in Vivo: A Game of Musical Chairs. Genes 2015; 6:751-76; PMID:26262644; http://dx.doi.org/ 10.3390/genes6030751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Morris SA, Baek S, Sung MH, John S, Wiench M, Johnson TA, Schiltz RL, Hager GL. Overlapping chromatin remodeling systems collaborate genome-wide at dynamic chromatin transitions. Nat Struct Mol Biol 2014; 21:73-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reik A, Schutz G, Stewart AF. Glucocorticoids are required for establishment and maintenance of an alteration in chromatin structure: induction leads to a reversible disruption of nucleosomes over an enhancer. EMBO J 1991; 10:2569-76; PMID:1678348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Weintraub H, Beug H, Groudine M, Graf T. Temperature-sensitive changes in the structure of globin chromatin in lines of red cell precursors transformed by ts-AEV. Cell 1982; 28:931-40; PMID:7094019; http://dx.doi.org/ 10.1016/0092-8674(82)90072-1 [DOI] [PubMed] [Google Scholar]
- 36.Ostuni R, Piccolo V, Barozzi I, Polletti S, Termanini A, Bonifacio S, Curina A, Prosperini E, Ghisletti S, Natoli G. Latent enhancers activated by stimulation in differentiated cells. Cell 2013; 152:157-71; PMID:23332752; http://dx.doi.org/ 10.1016/j.cell.2012.12.018 [DOI] [PubMed] [Google Scholar]
- 37.Miranda TB, Voss TC, Sung MH, Baek S, John S, Hawkins M, Grøntved L, Schiltz RL, Hager GL. Reprogramming of the chromatin landscape: interplay of the estrogen and glucocorticoid receptors at the genomic level. Cancer Res 2013; 73:5130-9; PMID:23803465; http://dx.doi.org/ 10.1158/0008-5472.CAN-13-0742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pombo A, Dillon N. Three-dimensional genome architecture: players and mechanisms. Nat Rev Mol Cell Biol 2015; 16:245-57; http://dx.doi.org/ 10.1038/nrm3965 [DOI] [PubMed] [Google Scholar]
- 39.Lupianez DG, Kraft K, Heinrich V, Krawitz P, Brancati F, Klopocki E, Horn D, Kayserili H, Opitz JM, Laxova R, et al.. Disruptions of topological chromatin domains cause pathogenic rewiring of gene-enhancer interactions. Cell 2015; 161:1012-25; PMID:25959774; http://dx.doi.org/ 10.1016/j.cell.2015.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y, Hu M, Liu JS, Ren B. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 2012; 485:376-80; PMID:22495300; http://dx.doi.org/ 10.1038/nature11082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sexton T, Yaffe E, Kenigsberg E, Bantignies F, Leblanc B, Hoichman M, Parrinello H, Tanay A, Cavalli G. Three-dimensional folding and functional organization principles of the Drosophila genome. Cell 2012; 148:458-72; PMID:22265598; http://dx.doi.org/ 10.1016/j.cell.2012.01.010 [DOI] [PubMed] [Google Scholar]
- 42.Rao SS, Huntley MH, Durand NC, Stamenova EK, Bochkov ID, Robinson JT, Sanborn AL, Machol I, Omer AD, Lander ES, et al.. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 2014; 159; 1665-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bouwman BA, de Laat W. Getting the genome in shape: the formation of loops, domains and compartments. Genome Biol 2015; 16:154; PMID:26257189; http://dx.doi.org/ 10.1186/s13059-015-0730-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ea V, Baudement MO, Lesne A, Forne T. Contribution of Topological Domains and Loop Formation to 3D Chromatin Organization. Genes 2015; 6:734-50; PMID:26226004; http://dx.doi.org/ 10.3390/genes6030734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Deng W, Lee J, Wang H, Miller J, Reik A, Gregory PD, Dean A, Blobel GA. Controlling long-range genomic interactions at a native locus by targeted tethering of a looping factor. Cell 2012; 149:1233-44; PMID:22682246; http://dx.doi.org/ 10.1016/j.cell.2012.03.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kagey MH, Newman JJ, Bilodeau S, Zhan Y, Orlando DA, van Berkum NL, Ebmeier CC, Goossens J, Rahl PB, Levine SS, et al.. Mediator and cohesin connect gene expression and chromatin architecture. Nature 2010; 467:430-5; PMID:20720539; http://dx.doi.org/ 10.1038/nature09380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fang F, Xu Y, Chew KK, Chen X, Ng HH, Matsudaira P. Coactivators p300 and CBP maintain the identity of mouse embryonic stem cells by mediating long-range chromatin structure. Stem Cells 2014; 32:1805-16; PMID:24648406; http://dx.doi.org/ 10.1002/stem.1705 [DOI] [PubMed] [Google Scholar]
- 48.De Santa F, Barozzi I, Mietton F, Ghisletti S, Polletti S, Tusi BK, Muller H, Ragoussis J, Wei CL, Natoli G. A large fraction of extragenic RNA pol II transcription sites overlap enhancers. PLoS Biol 2010; 8:e1000384; PMID:20485488; http://dx.doi.org/ 10.1371/journal.pbio.1000384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kim TK, Hemberg M, Gray JM, Costa AM, Bear DM, Wu J, Harmin DA, Laptewicz M, Barbara-Haley K, Kuersten S, et al.. Widespread transcription at neuronal activity-regulated enhancers. Nature 2010; 465:182-7; PMID:20393465; http://dx.doi.org/ 10.1038/nature09033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang D, Garcia-Bassets I, Benner C, Li W, Su X, Zhou Y, Qiu J, Liu W, Kaikkonen MU, Ohgi KA, et al.. Reprogramming transcription by distinct classes of enhancers functionally defined by eRNA. Nature 2011; 474:390-4; PMID:21572438; http://dx.doi.org/ 10.1038/nature10006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lam MT, Cho H, Lesch HP, Gosselin D, Heinz S, Tanaka-Oishi Y, Benner C, Kaikkonen MU, Kim AS, Kosaka M, et al.. Rev-Erbs repress macrophage gene expression by inhibiting enhancer-directed transcription. Nature 2013; 498:511-5; PMID:23728303; http://dx.doi.org/ 10.1038/nature12209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Allen BL, Taatjes DJ. The Mediator complex: a central integrator of transcription. Nat Rev Mol Cell Biol 2015; 16:155-66; PMID:25693131; http://dx.doi.org/ 10.1038/nrm3951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Core LJ, Martins AL, Danko CG, Waters CT, Siepel A, Lis JT. Analysis of nascent RNA identifies a unified architecture of initiation regions at mammalian promoters and enhancers. Nat Genet 2014; 46:1311-20; PMID:25383968; http://dx.doi.org/ 10.1038/ng.3142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kaikkonen MU, Spann NJ, Heinz S, Romanoski CE, Allison KA, Stender JD, Chun HB, Tough DF, Prinjha RK, Benner C, et al.. Remodeling of the Enhancer Landscape during Macrophage Activation Is Coupled to Enhancer Transcription. MolCell 2013; 51:310-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kieffer-Kwon KR, Tang Z, Mathe E, Qian J, Sung MH, Li G, Resch W, Baek S, Pruett N, Grøntved L, et al.. Interactome maps of mouse gene regulatory domains reveal basic principles of transcriptional regulation. Cell 2013; 155:1507-20; PMID:24360274; `http://dx.doi.org/ 10.1016/j.cell.2013.11.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Terns RM, Terns MP. CRISPR-based technologies: prokaryotic defense weapons repurposed. Trends Genet 2014; 30:111-8; PMID:24555991; http://dx.doi.org/ 10.1016/j.tig.2014.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Qian X, Droste SK, Lightman SL, Reul JM, Linthorst AC. Circadian and ultradian rhythms of free glucocorticoid hormone are highly synchronized between the blood, the subcutaneous tissue, and the brain. Endocrinology 2012; 153:4346-53; PMID:22822164; http://dx.doi.org/ 10.1210/en.2012-1484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bhake RC, Leendertz JA, Linthorst AC, Lightman SL. Automated 24-hours sampling of subcutaneous tissue free cortisol in humans. J Med Eng Technol 2013; 37:180-4; PMID:23547774; http://dx.doi.org/ 10.3109/03091902.2013.773096 [DOI] [PubMed] [Google Scholar]
- 59.Conway-Campbell BL, Sarabdjitsingh RA, McKenna MA, Pooley JR, Kershaw YM, Meijer OC, De Kloet ER, Lightman SL. Glucocorticoid ultradian rhythmicity directs cyclical gene pulsing of the clock gene period 1 in rat hippocampus. J Neuroendocrinol 2010; 22:1093-100; PMID:20649850; http://dx.doi.org/ 10.1111/j.1365-2826.2010.02051.x [DOI] [PMC free article] [PubMed] [Google Scholar]


