Abstract
Previous investigations suggested that lamin A/C gene (LMNA) mutations, which cause a variety of human diseases including muscular dystrophies and cardiomyopathies, alter the nuclear mechanical properties. We hypothesized that biomechanical changes may extend beyond the nucleus.
Keywords: AFM, cardiomyopathy, cardiomyocytes, cell physiology, lamin A/C, relaxation force test, Young Modulus
Combining atomic force microscopy (AFM), molecular and cellular biology, we studied the biomechanical properties of cardiomyocytes expressing the cardiomyopathy LMNA D192G mutation, and attempted rescue through the subsequent introduction of wild-type LMNA.Neonatal rat ventricular myocytes (NRVMs) were infected with adenoviral vectors carrying either human LMNA wild-type or D192G gene. LMNA protein expression was confirmed up to day 6 by western blot. Live-cell AFM force-deformation curves from day 1 through day 6 showed that the nuclei of NRVMs expressing LMNA D192G displayed increased stiffness compared to both uninfected and wild-type expressing cells, with a peak at 48 hours (3-fold increase in nuclear Young modulus, p < 0.0001). Furthermore, mutant NRVMs showed a significant reduction in the adhesion area between AFM probe and cell membrane, impaired cytoskeletal deformation measured by relaxation force test, associated with alteration of the cytoskeletal actin network by confocal microscopy. The altered actin network and mechanical properties of LMNA D192G NRVMs were rescued by the subsequent expression of wild-type LMNA. In conclusion, mutant LMNA deleterious effects appear to extend beyond the increased nuclear stiffness, to include altered cytoskeletal mechanics and defective cell membrane adhesion work, observations that are likely to underpin the changes in cardiac function that characterize this severe cardiomyopathy. Finally, expression of wild-type LMNA restores the mechanical properties of mutant NRVMs.
Introduction
The lamin A/C gene (LMNA) encodes the A-type lamins A and C, which, in differentiated somatic cells, constitute the nuclear lamina.1 A-type lamins are type V intermediate filaments that undergo a series of post-translational modifications and polymerization steps to form a complex filamentous structure that underlies the inner nuclear membrane of the nuclear envelope.1-2 A-type lamins are expressed in a variety of cell types, with high levels in cardiac and skeletal muscle.3 A number of investigations have shown that besides providing structural integrity to the cell nuclear envelope, particularly in heart and skeletal muscle, A-type lamins are involved in a number of other functions including chromatin organization, DNA replication, transcription and repair, nuclear positioning, mechanosensing, and aging.1-3
The variety of tissues involved, and variable functions of A-type lamins, may explain the increasing number of diseases associated with LMNA mutations in humans, first described in Emery-Dreifuss muscular dystrophy.4 Recent reviews list up to 15 different laminopathy disorders, that may occasionally overlap, ranging from striated muscle diseases (cardiomyopathies, muscle dystrophies), to lipodystrophies, nerve and bone disorders, and premature aging.2,5 The full range of functions of lamins and the mechanisms leading to these different phenotypes are complex and still incompletely understood. Overall, genetic mutations of Lamin A gene causing cardiomyopathy are believed to cause ‘loss-of-function’ and, either by haploinsufficiency or by dominant negative effect, lead to altered structural organization and/or altered protein expression (LINC complex, Desmin, Connexin 43) and transcription factors (MAP kinase, AKT/mTOR, Wnt/β-catenin signaling pathways) critical for normal cardiomyocyte function.1-3,6
A-type lamins form a molecular scaffold under the inner nuclear membrane and connect structurally with the cytoplasm by binding integral proteins of the nuclear envelope that act as ‘linkers of the nucleoskeleton and cytoskeleton’ (LINC).7 LINC members include Nesprins 1 and 2, SUN 1 and 2, Emerin and LUMA (encoded by TMEM43 gene). The LINC complex provides support to the nucleus and couples the nucleoskeleton with the cytoskeleton (Desmin, microtubules, actin microfilaments, Titin).1-2,6,8 This network connects the nucleus to the extracellular matrix and provides nuclear anchorage, mechanical stress sensing and resistance to pathologic deformation of the cell.1-2
We were intrigued by the mechanical properties of the nuclear lamina in normal and mutant Lamin A expressing cells. Previous studies had shown that cells expressing LMNA mutations exhibit structural damage of the nuclear envelope: the LMNA D192G mutation, which causes a severe form of dilated cardiomyopathy, is characterized by dramatic ultrastructural changes and disruption of the cardiomyocyte nucleus, as observed in heart tissue from patients and in cellular models.9 Furthermore, a series of investigations based on various techniques and cell models, such as computational imaging of fibroblasts 8,10,11 and micropipette aspiration of nuclei of Xenopus laevis oocytes12 suggested that cardiomyopathy mutations in LMNA lead to increased nuclear fragility resulting in cell death and progressive failure in tissues that are exposed to repetitive mechanical stress, such as seen in normal heart function.8
Atomic Force Microscopy (AFM) is a technique that allows the direct measurement of cellular and subcellular structures, mapping and manipulation of biological surfaces in their native environment at a spatial resolution of few nanometers, with a signal-to-noise ratio superior to that of optical microscopy.13 The AFM force-deformation test, also called single-cell force spectroscopy (SCFS), combines moderately high force and high spatial resolution, as well as the capability of operating under physiological conditions. Using AFM, Kaufmann et al. investigated the mechanical properties of the progeria LMNA mutation E147K in a model of isolated Xenophus oocytes.14 These investigators found an increase in the measured stiffness (Young modulus) in the nuclei expressing this mutant LMNA. While previous investigations indicated defective nuclear mechanics in laminopathies, they relied on model systems (such as amphibian oocytes and fibroblasts) that may not accurately reflect the properties and architecture of striated muscle in particular in cardiomyocytes. Indeed, very little is known about the nuclear and cellular mechanical properties of cardiomyocytes carrying mutant LMNA. Thus, to understand how a mutation of Lamin A protein may impact the mechanical properties of the nucleus and the cell as a whole providing potential novel insights into the underlying mechanisms of laminopathies, we used AFM in a living, single-cardiomyocyte model.
Herein, we report that using AFM single cell force-deformation curve, the nuclear Young modulus, cytoskeletal viscoelastic and cellular work of adhesion can be measured in LMNA D192G neonatal rat ventricular myocytes (NRVMs). The whole cell force-deformation curves derived from AFM investigations carry several pieces of information: (1) the total force required to deform the nucleus, (2) the AFM cantilever deformation at the holding point, (3) the hysteresis area between the loading and unloading cycles, and (4) the area under the deformation curves during the unloading cycle which reflects cell adhesion behavior. Here, we integrated every component of the AFM force-deformation curves during a loading and unloading cycle to provide a more comprehensive insight into whole-cell biomechanical behavior in living cells. Adhesion in this case is meant as the process of detachment of the cardiomyocyte cell membrane (the sarcolemma) to the AFM tip. Upon AFM tip retraction (unloading curve) to the point of maximal cell height, the AFM sphere adheres to the cell membrane and causes an opposite deflection of the probe below the baseline. Both, sequential and/or parallel rupture of bonds established between the tip and the cell and viscous and elastic deformation of the cell body contribute to the AFM retraction curve. Retracting curves are characterized by a peak of force followed by a cascade of rupture events until complete detachment is achieved. The interaction forces involved in these measurements generally range from tens to hundreds of picoNewtons. The work of adhesion (or de-adhesion) in our system was evaluated by integrating the area between the contact point on the surface and the last force interaction, which resulted in the cantilever returning to its null position.
Our findings suggest a complex biomechanical dysfunction in cells expressing the mutant LMNA isoform that alter the nuclear mechanical properties and cell work of adhesion. Moreover our investigations showed that this dysfunction that can be rescued by expression of wild-type (WT) LMNA.
Results
Evaluation of exogenous human LMNA expression
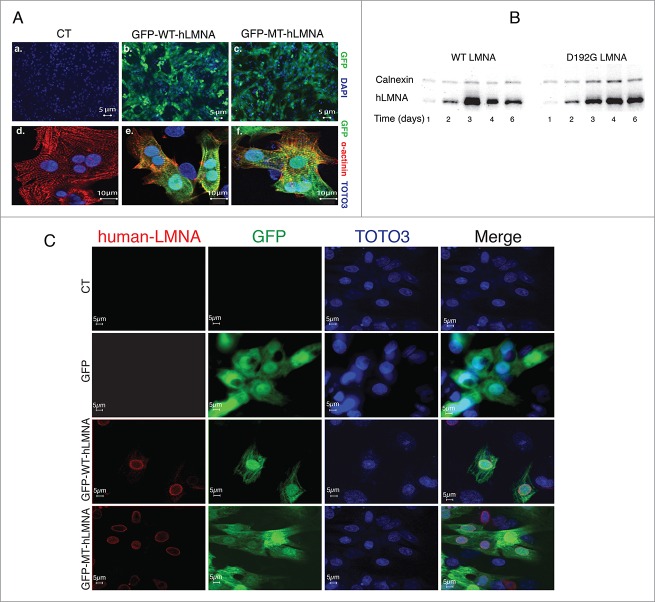
The efficiency of LMNA transduction was determined from the ratio of the GFP fluorescence signal in infected NRVMs (Fig. 1A a–c) over the total DAPI-stained NRVMs nuclei in images collected at 48h post infection; sarcomeric α actinin staining together with GFP expression confirmed the cardiomyocytes infection (Fig. 1A d–f). Increases in cell death were not observed during the course of our studies (data not shown). Expression of exogenous human LMNA was evaluated by reverse transcriptase polymerase chain reaction (RT-PCR) (data not shown), Western Blot and immunofluorescence analyses (Fig. 1B and 1C). Our data indicate that protein expression begins within 12-24 hours post-infection and persists for at least 6 days (Fig. 1B), consistent with the time course of expression of exogenous molecules using the Adenoviral system as previously reported by several other groups.15-18 As shown in Figure 1C, both mutant and wild-type human Lamin A protein are detected by the human specific antibody utilized whereas endogenous rodent LMNA is not recognized.
Figure 1.
Adenoviral human LMNA expression in NRVMs. (A) Indirect immuofluorescence representative images of efficiency of adenoviral transduction in neonatal rat cardiomyocytes showing GFP expression (green) in infected WT (b) and D192G (c) cells. Staining for Sarcomeric alfa-Actinin (red) and GFP fluorescence confirmed human LMNA expression in infected cardiomyocytes (d, e, f). Nuclei were stained with DAPI (a, b, c) or TOTO3 (d, e, f). (B) Western blot showing the time-course of both wild-type and D192G human LMNA expression up to day 6, demonstrating the persistence of human protein transduction. (C) Indirect immunofluorescence indicated co-localization of human LMNA (red, left column) and GFP (green, left-middle column) signals in WT and D192G (MT) NRVMs, transduced by adenoviral bicistronic GFP-LMNA construct. The nuclei are stained in blue (TOTO-3, right-middle column). Uninfected (upper lane) and NVRMs transduced by adenovirus carrying GFP construct only (second lane) displayed no signal for human LMNA protein. The fluorescence is represented in 3 channels.
Cells morphology
Typical NRVMs (>90% of the population) exhibited a characteristic ellipsoidal shape. The long and short axes ranged from 17 to 32 μm and 15 to 20 μm, respectively. The cell height was defined by the distance between the highest point of the cell (consistently at the position of the nucleus) and the substrate and was measured using AFM. The typical height ranged from 2 to 3 μm, corresponding to known dimensions for these cells. We calculated (using the AFM data) the cell height for cells (n = 20) involved in these studies (CT = 2,75 ± 0,43μm, WT = 2,79 ± 0,4μm, MT = 2,37 ± 0,25μm; Anova CT vs. WT: p-value 0.79, CT vs. MT p-value 0.001, WT vs. MT p-value 0.0002).
Elasticity of the nucleus in mutant LMNA D192G NRVM
At day 1 after infection, the Young's modulus, used as an indicator of nuclear elasticity, did not differ within the 3 different conditions as shown in Figure 2A and Table 1 (Anova p-value: 0.089; Tukey HSD mutated vs. control, p-value: 0.627; Tukey HSD wild-type vs. control, p-value: 0.354; Tukey HSD wild-type vs. mutated, p-value: 0.081). Conversely, during the next days of culture, the results seen in D192G LMNA cells significantly differed from uninfected (control, CT) and wild-type (WT) cells.
Figure 2.
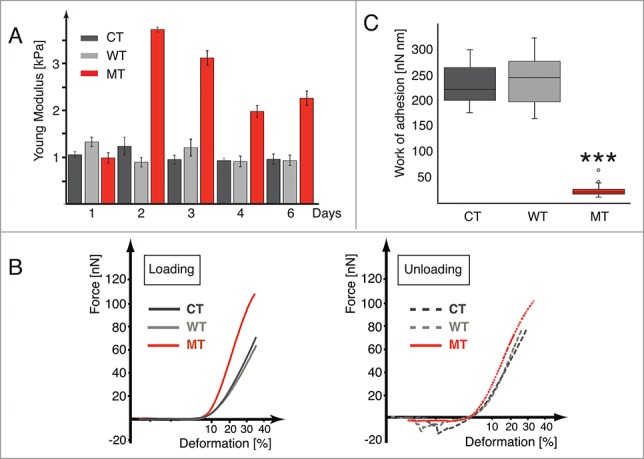
LMNA D192G mutation alters the nuclear and cellular biomechanics. (A) Bar graph indicating the Young modulus values for Control (CT, dark gray), wild-type (WT, light gray) and Mutant (MT, red) cells after 1 to 6 days from the infection (bars indicate SE, corresponding data and statistical significance in Table 1). (B) Examples of loading-unloading AFM curves for CT, WT and MT cells, respectively. (C) Box plot data of the work of adhesion, for the CT, WT and MT NRVMs, respectively, ***p < 0.0005 between MT and both CT and WT.
Table 1.
Young modulus data for CT, WT and MT cells after infection
| DAY | CONTROL Young Modulus [kPa] | WILD-TYPE Young Modulus [kPa] | MUTANT Young Modulus [kPa] | P value |
|---|---|---|---|---|
| 1 | 1.06 SD = 0.31 SE = 0.065 (n = 23) |
1.33 SD = 0.51 SE = 0.102 (n = 25) |
0.99 SD = 0.53 SE = 0.111 (n = 23) |
CT–WT NS WT–MT NS CT–MT NS |
| 2 | 1.24 SD = 0.96 SE = 0.192 (n = 25) |
0.89 SD = 0.49 SE = 0.098 (n = 25) |
3.73 SD = 0.27 SE = 0.057 (n = 23) |
CT–WT NS WT–MT p < 0.0001 CT–MT p < 0.0001 |
| 3 | 0.954 SD = 0.49 SE = 0.098 (n = 25) |
1.208 SD = 0.94 SE = 0.183 (n = 26) |
3.12 SD = 0.75 SE = 0.150 (n = 25) |
CT–WT NS WT–MT p < 0.0001 CT–MT p < 0.0001 |
| 4 | 0.930 SD = 0.32 SE = 0.064 (n = 25) |
0.912 SD = 0.54 SE = 0.117 (n = 20) |
1.98 SD = 0.57 SE = 0.124 (n = 20) |
CT–WT NS WT–MT p < 0.0001 CT–MT p < 0.0001 |
| 6 | 0.962 SD = 0.50 SE = 0.106 (n = 23) |
0.935 SD = 0.51 SE = 0.113 (n = 20) |
2.26 SD = 0.73 SE = 0.156 (n = 22) |
CT–WT NS WT–MT p < 0.0001 CT–MT p < 0.0001 |
| DATA AFTER RESCUE | ||||
|---|---|---|---|---|
| CONTROL Young Modulus [kPa] | WILD TYPE Young Modulus [kPa] | MUTANT Young Modulus [kPa] | MUTANT After rescue Young Modulus [kPa] | P value |
| 0.930 SD = 0.32 SE = 0.064 (n = 25) |
0.912 SD = 0.54 SE = 0.117 (n = 20) |
1.98 SD = 0.57 SE = 0.124 (n = 20) |
1.05 SD = 0.8 SE = 0.212 (n = 14) |
MT–Resc p < 0.0003 |
Data shown in Figure 2A and Table 1 highlight that elasticity of both CT and WT cells remained substantially constant over the 6 days of time-course. In contrast, the nuclear Young modulus for cells expressing the D192G LMNA mutation was significantly higher than that of both CT and WT cells at 48 hours post-infection a point consistent with the time-dependent expression of the mutant protein shown by Western Blot. The Young modulus of MT cells remained higher than those of CT and WT throughout the 6-day time course of the experiments.
Deformation of NRVM nucleus
Figure 2B shows a typical AFM force-deformation curve seen during loading and unloading cycles for CT, WT and MT cells, respectively, taken the second day after adenoviral infection. All curves displayed a smooth and nonlinear deformation profile without irregularities or stress peaks, indicating their elastic nature and suggesting no structural failure of the nuclear envelope. CT and WT cells required a force of 41 ± 8.8 nN and 39 ± 7.1 nN to reach 30% of deformation, respectively while MT required a higher force of 93 ± 5.2 nN. Increasing the deformation to 40% required an even greater force: 72 ± 4.4 and 67 ± 5.3 nN, respectively for CT and WT, while MT required 118 ± 6.1 nN. Remarkably, cells recovered to their original height. In order to verify that cell height do not vary after 5 subsequent loading cycles, we perform an analysis by linear mixed effects models, using 5 cell lines as a random effect. The analysis disclosed that the time effect was not significant (fixed effect slope estimate = −0.000, standard error = 0.003, t = −0.123). This cell height recovery appears to be characteristic of NRVM cells, and differed from that reported in other cells, such as T lymphocytes, which fail beyond 30% deformation and keratinocytes which do not remain viable after the first compression, despite the recovery of cell height.19
AFM loading-unloading curves also contain information about long- and short-range interactions within the cell and represent a basis not only for estimation of Young's modulus but also for assessing the overall cell deformation. As shown in Figure 2B, the area under the deformation axes during the unloading cycle, after the cantilever reached the 100% cell height position, was similar for CT and WT but significantly different for MT cells. We hypothesize that this represents the de-adhesion of the AFM sphere from the cell membrane and that the number of small steps in this curve is related to the disruption of individual adhesion bonds between the AFM sphere and the membrane proteins as described by others.20-21 Specifically, whereas CT and WT have a distinct adhesion area, the MT cells show a much smaller one. Figure 2C shows in box plot form the work of adhesion for NRVMs for the 3 different conditions. Data for CT and WT showed no statistical difference (p > 0.05). In contrast, NRVMs carrying the LMNA D192G mutation showed a much smaller area (p < 0.0005). To confirm the reproducibility of our measurements and the persistence of abnormal work of adhesion behavior, loading/unloading cycles were carried out 10 times on the same MT cell, allowing 2–3 min. of recovery time. The force profile did not statistically change up to 10 cycles and the adhesion area remained extremely small (data not shown).
Cell viscoelasticity
Figure 3A shows the force-relaxation behavior for the 3 NRVMs different conditions: CT, WT and MT. In this test, the cell behavior reflects an initial nuclear compression, followed by a subsequent compression of the cytoskeleton.22 Precisely, the mechanical response of NRVMs to a constant applied deformation appears to involve an initial fast elastic reaction and a subsequent slow viscoelastic process. It is noticeable that while the behavior of CT and WT was almost identical, MT showed a significant downward shift toward a more deformable behavior.
Figure 3.
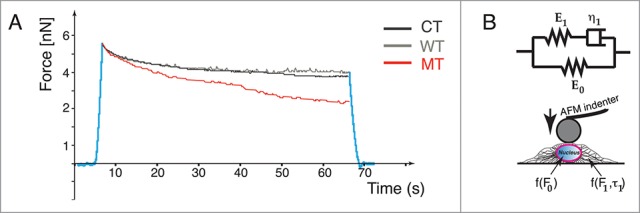
AFM analysis of cell deformation and plasticity. (A) Force-relaxation curves for the 3 NRVMs conditions: Control (CT), wild-type (WT) and Mutant (MT). The MT curve shows altered deformation properties. (B) Standard linear solid model (Zener's model) used to analyze the force-relaxation cell behavior
In our studies the compression force decays with time. As expected, total relaxation force reaches a steady state and is saturated within seconds. Immediately after force application, the cell behaves like an elastic solid follow by a slow viscous period due to the cytoskeletal relaxation. This behavior can be modeled (Fig. 3B) using a standard linear solid model (called also Zener's model) that combines in parallel a pure elastic spring (E0) with one Maxwell element (a purely viscous damper (η1) and a purely elastic spring (E1) connected in series).23 In this case, the force decay with time t is expressed with an exponential equation:
| (1) |
where τ1 = η1/E1, being E0 and E1 the springs Young moduli and η1 the viscosity of the damper, respectively. The first term in Equation 1 can be assumed as the Young modulus of the nucleus. Fitting data for our experiments are shown in Table 2. There were no significant differences in fitting data between CT and WT cells. In contrast, MT data showed that E0 (nuclear Young modulus) was higher than CT and WT while cytoskeletal viscosity was lower.
Table 2.
Fitting data for the constant force experiment
| Exponential decay |
|||
|---|---|---|---|
| CT | WT | MT | |
| E0 | 1.21 ± 0.4 | 1.28 ± 0.2 | 2.27 ± 0.5 |
| E1 | 0.57 ± 0.8 | 0.44 ± 0.3 | 0.36 ± 0.6 |
| τ1 | 3.68 ± 0.3 | 3.51 ± 0.2 | 2.65 ± 0.8 |
| η 1 | 11.3 ± 2.1 | 10.7 ± 1.4 | 3.11 ± 0.8 |
We then examined if changes in the mechanical properties of the mutant NRVMs were associated with altered cytoskeletal structure. D192G mutation of nuclear lamin seems to be associated with an alteration of the length and thickness of actin cytoskeleton elements (Fig. 4). On the contrary, other cytoskeletal compartments such as intermediate filaments (data not shown) and microtubules (Tubulin, Fig. 5) did not appear to be significantly affected by the mutation in our experimental conditions.
Figure 4.
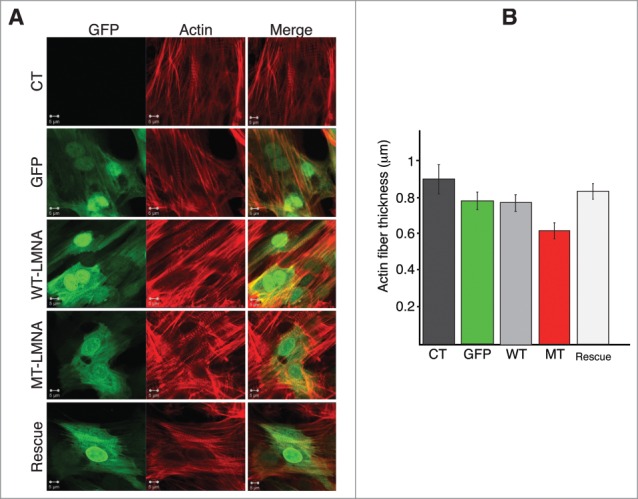
D192G LMNA actin network in NRVMs. (A) Indirect immuofluorescence representative images of NVRMs. Cardiomyocytes expressing mutant LaminA protein display severe decrease of both density and thickness of cytoskeletal actin microfilaments compared to not infected (CT) and NRVMs infected with either WT-LMNA or GFP only. The re-expression of WT Lamin A protein in cardiomyocytes previously infected with MT-LMNA, lead to a restored actin network comparable to controls (Rescue). (B) Quantitative measurement of the actin filaments thickness showed a significant reduction of this cytoskeleton component in MT NRVM compared to both CT and WT cells (p < 0.0001), rescued by the second infection with WT LMNA (p = 0.096 compared to CT).
Figure 5.
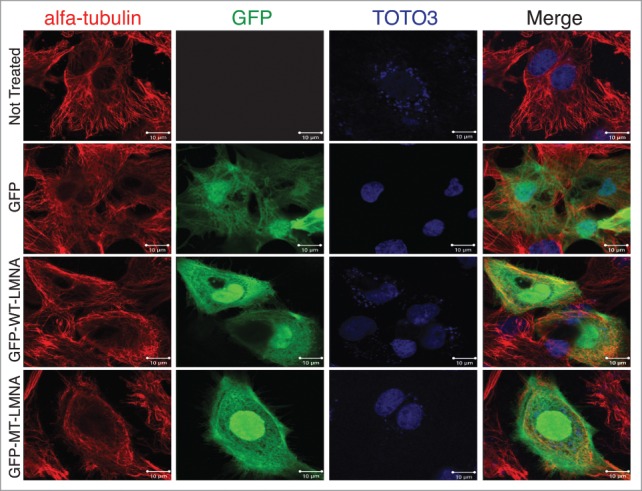
D192G LMNA Tubulin network in NRVMs. From above: uninfected control NRVMs, NRVMs infected with GFP only, WT LMNA, and MT D192G LMNA NRVMs. Mutant cardiomyocytes did not show significantly different Tubulin staining compared to the infected and uninfected controls.
Rescue experiments
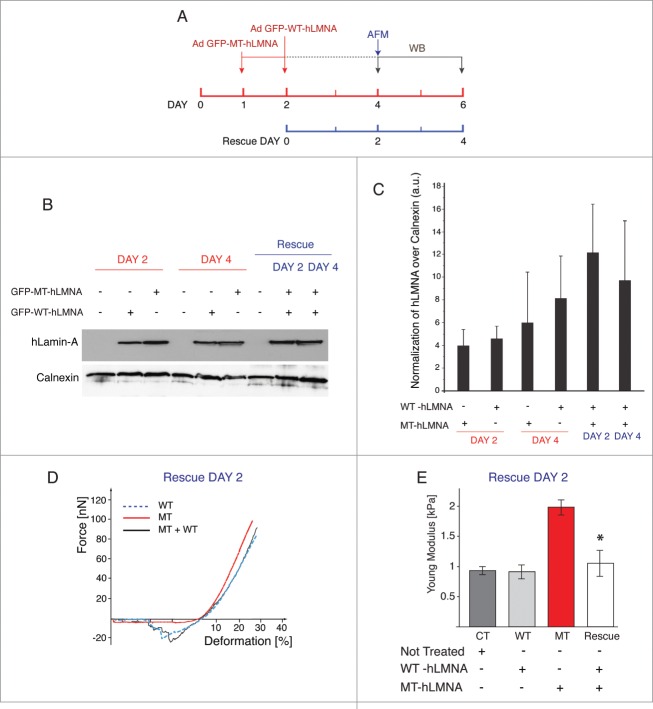
To investigate if the altered biomechanical properties of NRVMs carrying the LMNA D192G mutation could be recovered by the subsequent expression of wild-type Lamin A protein, LMNA D192G cells were subjected to a second round of adenoviral infection with the WT LMNA construct. 48 hours from the second infection, ‘rescued’ NRVMs were tested by AFM (Fig. 6A, D, E), whereas cellular lysates were collected and analyzed by western blot after 48 and 96 h post-infection (Fig. 6A, B, C)
Figure 6.
WT LMNA reverts the effects induced by D192G mutation. (A) Schematic representation of the experimental timeline followed. The day after plating, NRVMs were infected with viral vector carrying human D192G LMNA (red line); the next day the same cells were subjected to a second infection with WT human LMNA. The rescue effect was analyzed by AFM measurement 2 days post second infection whereas western blot were performed at days 2 and 4 (blue line). (B) Western blot analysis of lysates comparing human LMNA expression in single infected (WT or D192G) and double infected (WT and D192G) cells at day 2 and 4. (C) Quantification of the human LMNA (hLMNA) signal over the value of Calnexin used as loading control to assess differences in total LMNA expression (MT together with WT) in the rescue experiments: as expected, there is an increase in total human LMNA protein. (D) Loading-unloading curve showing a de-attachment area in D192G (Mutant) cells after 48 hours from the second infection with WT LMNA. The NRVMs subjected to double infection, displayed a similar work of adhesion properties to that of uninfected (CT) and WT cells with a consistent de-adhesion area during the unloading cycle. (E) Young modulus values highlighted the restoration of the nuclear elasticity in double infected cells. *indicates p < 0.0003 between MT (red bar) and rescued (light gray bar).
As shown in Figure 6D force-deformation for rescued NRVMs show a profile quite similar to that of CT and WT cells with a consistent de-adhesion area during the unloading cycle. Figure 6E and Table 1 show the Young modulus calculated after the rescue experiment indicating that rescued cells showed values similar to those of CT and WT, respectively. Since LMNA D192G is a point-mutation, it was not possible to discriminate between the 2 proteins in the “rescue” experiments, but it was possible to assess differences in total Lamin A expression (MT together with WT) showing the expected increase in total human protein in these cells (Fig. 6B and C). In order to verify if the rescue experiments could have an impact also on the alterations of the actin cytoskeleton induced by the D192G mutant additional immunocytochemistry experiments were performed. Actin has already been suggested to play a role in the regulation of cell growth, differentiation and apoptosis, and disorder of actin-filament integrity relates to the induction of cardiomyocyte apoptosis. This implies that actin loss may cause compromised contractility and also cell death by apoptosis and thus disturb the kinetics of the whole heart.24 Confocal microscopy images showed that in control NRVMs and NRVMs expressing WT LMNA, the actin microfilaments appeared to be highly organized and homogeneously distributed (Fig. 4A). On the contrary, mutants NRVMs were characterized by attenuation and clear disarray of the actin microfilaments (Fig. 4A). A quantitative measurement of actin filaments thickness showed a significant reduction of this cytoskeletal component in MT NRVMs compared to CT and WT cells (p < 0.0001), (Fig. 4B) but remarkably, an almost complete recovery after rescue (p = 0.096 compared to CT) (Fig. 4B).
Discussion
Nuclear stiffness and cell biomechanics are altered in cardiomyocytes expressing LMNA D192G
The main conclusion from our investigations is that the LMNA D192G mutant has a profound effect on nuclear elasticity and cell biomechanics of intact, living NRVMs. These effects include a 3-fold increase in nuclear Young modulus (day 2), an increase in the force required to deform the cell, a more viscous and less elastic properties, and a marked reduction in the adhesion area between AFM probe and cell membrane. Importantly these data indicate that the altered biomechanics in mutant cardiomyocytes extend beyond the nuclear envelope and also involves the mechanical properties of the cell as a whole.
The biomechanical changes found in our study raise some intriguing questions. Clinically, the LMNA mutation D192G (A787G transition) has been reported by Sylvius et al.9 to cause a DCM phenotype characterized by malignant course with severe cardiac failure and premature death. In the myocardial tissue of a D192G carrier, light microscopy revealed non-specific myocyte damage and interstitial fibrosis. However, ultrastructural analysis demonstrated severe alterations of the nuclear envelope with accumulation of mitochondria, glycogen and/or lipofuscin in the nucleoplasm, and chromatin disorganization.9 The D192G mutation affects Coil 1B of the rod domain of LMNA, and suggests an alteration in the coil-coil polymerization of the LMNA dimer.9 In our view, these predicted characteristics made the LMNA D192G mutation an ideal candidate for our biomechanical study. Furthermore, Sylvius et al. reported that the LMNA D192G mutant protein expression in COS7 cell line caused accumulation of SUMO1 in large spots of LMNA at the nuclear envelope, instead of being homogeneously distributed in the nucleus like the wild-type LMNA. SUMO1 is major cell regulator of chromatin organization and gene expression, suggesting that other mechanisms beyond structural integrity may play a role in altering the biomechanical properties of the nuclear envelope.
Since NRVMs expressing wild-type LMNA displayed stiffness comparable to uninfected cells, the higher stiffness in LMNA D192G NRVM appears to be related to the expression of the defective LMNA D192G protein. The time-course of expression of the introduced construct, along with the half-life of the LMNA proteins described by others,25-27 suggests that these mutant proteins compete with endogenous LMNA to cause the abnormal mechanical properties seen. Indeed, consistent with this model, the subsequent introduction of additional wild-type LMNA competes with mutant protein and “rescues” the biomechanical properties of the cells expressing the LMNA D192G mutation. Prior works with the adenoviral expression system have relied on the ability of exogenous protein to “compete” with endogenous systems and this is, in fact, the theory behind the introduction of both “dominant negative” and “constitutively active” constructs.15-17 In support of this being the mechanism of our investigations with the LMNA protein, work by others has shown a very rapid turn-over for LMNA precursor incorporation into the nuclear membrane, on the order of 4 hours: this would mean that our exogenous protein should have the ability to effectively compete for incorporation under both MT and WT (rescue) circumstances over the time-course examined in our work.25-27
The AFM force-deformation curves demonstrate that the alterations in nuclear stiffness of the LMNA D192G cardiomyocytes are accompanied by a series of complex cellular changes ranging from the cell membrane work of adhesion properties to altered whole-cell deformation. By measuring the force curves on cultured NRVMs, we observed that CT and WT cells display similar detachment behavior, with curves showing an initial, large de-adhesion peak followed by smaller peaks, reflecting the energy required to detach the adhesion molecules of the cell membrane from the AFM tip (‘steps’ of the detachment curve). Conversely, MT cells show negligible work of adhesion (Fig. 2B, C). This loss of work of adhesion in MT is intriguing, since it would not necessarily be expected from a mutant nuclear protein. Although our investigations were not designed to assess the specific molecules responsible for the adhesion of the NRVM cell membrane to the AFM tip, we speculate that altered detachment might be explained by the known interaction of lamins with membrane proteins via cytoskeleton. This type of nuclear to cell surface “communication” has also been described in inflammatory and pathologic processes that have been shown to change the cell adhesion properties.28-29 The identification of the molecules involved in the adhesion will be the logical next step of future investigations. Indeed, the link between LMNA, the nucleoskeleton, and cellular adhesion molecules has been previously reported by a number of investigators.1,3,30-31 In the cardiomyocyte, several cell membrane structures could be involved since it is known that lamins binds to a complex series of LMNA binding proteins, the LINC (linkers of the nucleoskeleton to the cytoskeleton) complex, including SUN1/2 and Nesprins, nuclear and cytoskeletal actin, Titin and Desmin. This network forms connections between the lamina nucleoskeleton, and the cytoskeleton 1,3,7,32 that in turn is connected to Vinculin, Integrins and the Dystrophin associated complex at the cell membrane.1,3,17,31,33 An alternative possibility is that the substrate may affect cell mechanics (through Integrins and the cytoskeleton) and cell function in vitro34-35 and in fact increased LMNA expression has been associated with changes in tissue stiffness.36 However, in our study the same substrate was used in all conditions (uninfected control, GFP, wild-type and mutant infected), indicating that the altered stiffness and work of adhesion observed in mutant NRVMs were independent from the characteristics of the substrate.
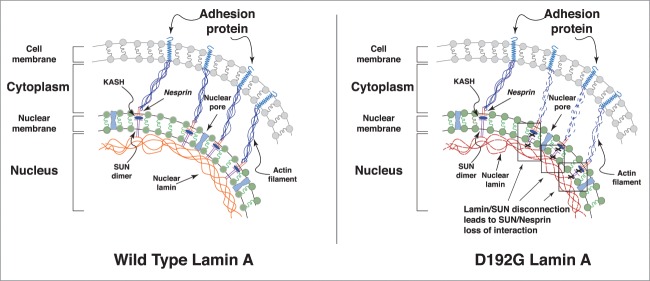
To our knowledge, this is the first evidence that defective cell adhesion between AFM tip and cardiomyocytes cell membrane expressing a mutant LMNA protein has been reported. In an actively contracting organ like the heart forces are transmitted from the extracellular matrix to the cytoskeleton when extracellular forces act on the cardiomyocytes, and from the cytoskeleton to the extracellular environment as cardiomyocytes generate contractile force. It is through cell adhesion molecules that these mechanical forces are transmitted bi-directionally across the cell membrane. We hypothesize that the abnormal mechanical properties of mutant cells could be explained by a decrease in the number or function of adhesion molecules responsible for the adhesion-detachment process at the AFM tip/cell interface. We also speculate that a dysfunction in the interacting members of the LMNA-cytoskeleton-adhesion molecule(s) network could lead to altered mechano-sensing and mechano-transduction in cardiomyocytes expressing mutant D192G LMNA protein (see Fig. 7).
Figure 7.
Hypothesis on the role of defective lamin in altering cell adhesion behavior. Speculative cartoon underlying the potential role of mutated LMNA in cell adhesion properties. The defective nuclear-cytoskeletal connection may lead to adhesion proteins dysfunction, and ultimately to defective adhesion-detachment properties in mutant cells.
Cardiomyocyte viscoelasticity behavior is altered in mutant LMNA expressing myocytes
Under deformation, viscoelastic materials dissipate energy whereas elastic materials store it; in addition, a main feature of viscoelastic materials is the presence of a time-dependent relationship between stress and deformation. In non defective cells, these kinds of adaptive mechanisms are the result of both nuclear elasticity and cytoskeletal reorganization, governed by their component viscoelastic properties. In the past, the viscoelastic properties of cells or isolated nuclei have been mainly investigated by micropipette aspiration that inherently results in a decoupling of the cytoskeletal network of the intact cell.12,37-38 On the contrary, the AFM compression measurements used in our study better reflect the properties of the entire cytoskeleton and the nucleus in intact living cardiomyocytes. Using this methodology, we found a change in viscoelasticity of the MT cytoskeleton compared to CT and WT cells, as shown by the force-relaxation curve reflecting a decreased cytoskeletal viscosity in LMNA D192G NRVMs (Fig. 3A). This change in cytoskeletal viscosity could be related to altered actin polymerization, which has recently been found to be disrupted in LMNA mutant cells models (LMNA−/− and LMNA N195K cells).39-40 Indeed, when the integrity of the actin network is compromised by mutant LMNA, the biomechanical properties of nucleus and the whole cell are disrupted.1,10,32 Our hypothesis that the deleterious effects of LMNA D192G mutation extend beyond the increased nuclear stiffness, involving reduced cell membrane work of adhesion and cytoskeletal modifications is supported in particular by the work of Hale et al.39 by examining the biophysical properties of cells derived from 2 laminopathy mouse models, the authors found softening of the cytoplasm, weakening of the cell and reduction in adhesion properties. Interestingly, compared to our study, the results of Hale et al. were obtained using different methods, such as cell microrheology and static cell adhesion chamber, but they supported our findings. Furthermore, the authors reported that cytoskeletal softening was exclusively a feature of cells derived from laminopathic mouse models (LMNA−/− and LMNAL530P/L530P), but not mutant emerin models.39 Although in their study they were not able to identify overt differences in actin stress fibers in mutant cells, they concluded that the laminopathic defects could stem from dysfunctional connections between the nucleus and cytoskeleton, which would subsequently disrupt cellular functions mediated by the cytoskeleton.39 Our findings and hypothesis are in agreement with published data in which, disruption of the cytoskeletal actin increased damping behavior and decreased cell viscosity, highlighting that actin filaments play the primary role in governing cellular viscoelastic properties.41 Indeed our investigations using confocal microscopy showed structural modifications of the cytoskeletal actin filaments in mutant NRVMs compared to the controls and little influence on other elements of the cytoskeleton (Figs. 4 and 5). The D192G LMNA mutation affects a highly conserved residue of the coil 1B domain. This mutation was first described by Sylvius et al. in a patient with a severe dilated cardiomyopathy.9 In the cardiomyocytes of the carrier, the authors found that the mutation caused severe morphological alterations, including a complete loss of the nuclear envelope, chromatin disorganization and trapping of SUMO1, which may cause altered transcriptional regulation and defect in nucleocytoplasmic transport. Based on our results showing that the mutant cardiomyocytes have a stiffer nucleus, reduced cytoskeletal viscosity due to both decreased actin density and defective actin structure, and lack of cell-cell adhesion, we speculate that the disrupted mechano-transduction could lead to malfunctioning myocardium and heart failure. As proposed in the model in Figure 7, a possible explanation might be related to the loss of interaction between Lamin A and SUN 1 and 2 proteins which, in turns, is reflected to the SUN/Nesprins interactions known to be critical for the mechano-transmission from the nucleus to the cytoskeleton due to their connection with actin intermediate filaments.33,39,42-43
Rescuing the mechanical properties of mutant NRVM
In our study, we have shown that the introduction of wild-type LMNA in mutant NRVMs was able to restore the cell biomechanical properties under investigation. Indeed it is worth to highlight that the re-expression of wild type Lamin A protein lead to a recover of the nuclear stiffness and contributes to normalize the cell membrane work of adhesion. These data suggest that our rescue experiment was able to revert a pathological biomechanical condition induced by D192G LMNA mutation, and indicate that WT LMNA could play a pivotal role, or being at least partially able, to restore the nucleoskeleton and the nucleus-cytoskeletal to cell membrane crosstalk network. These results may provide a hint for new therapeutic strategies and indicate that AFM could represent an innovative biomechanical tool with which to test novel pharmacological approaches to such genetic cardiomyopathies in a preclinical model.
In conclusion, AFM has been shown to be a unique and valuable tool to investigate the mechanical properties of intact, living cardiomyocytes expressing the LMNA D192G mutant protein. AFM has both the spatial and temporal resolution to allow the study of loading/unloading behavior, adhesion and viscoelasticity in these cells. Our results have shown that D192G LMNA mutation causes a profound change in cardiomyocytes biomechanics leading to: (i) increased nuclear stiffness, (ii) nearly completely loss of cell membrane work of adhesion, (iii) altered cellular viscoelasticity associated with modifications of the actin microfilaments network, (iv) all the biomechanical modifications and actin network disarray associated with the D192G LMNA mutation were reverted by the re-expression of wild type LMNA protein. Our findings and recent work form other investigators indicate that lamin and lamina-associated proteins form a unique functional mechanical and mechano-sensing entity that extends from the nucleoskeleton to the cell membrane and which becomes destabilized by changes in any of its component elements.1,7,33,39,42-43
Methods
Cell culture
Neonatal rat ventricular cardiomyocytes (NRVMs) were isolated and cultured from 1-3 days old Wistar rat pups by enzymatic digestion as previously described with minor modifications.44-47 Animal care and treatment were conducted in conformity with institutional guidelines in compliance with national and international laws and policies (European Economic Community Council Directive 86/609, OJL 358, December 12, 1987 and the current Italian law (decree 116/92). Briefly, ventricles were separated from the atria using scissors and then dissociated in CBFHH (calcium and bicarbonate-free Hanks with Hepes) buffer containing 0,5 mg/ml of Collagenase type 2 (Worthington, Biochemical Corporation), and 1 mg/ml of Pancreatine (SIGMA). Cardiomyocytes were enriched (>90% purity) over non-myocytes by a pre-plating step on 100-mm dishes in Dulbecco's modified Eagle Medium (DMEM) 4.5g of glucose, supplemented with 10% FBS and 2 mg/ml vitamin B12 (SIGMA). Myocytes that were either in solution or lightly attached were then separated from the adherent stromal cells by gentle mechanical disaggregation and subsequently plated at a density of 2×105 cells/ml alternatively in primary petri dishes (FALCON) or in multi chamber slides (Lab-Tek II, NUNC) coated with 0.2% gelatin (SIGMA) and cultured as previously described.44-45 After 18 h, the culture medium was changed and cells were subjected to infection with the relevant adenoviral-LMNA constructs as described below.
Adenoviral constructs and infection
Shuttle constructs were generated in Dual CCM plasmid DNA containing GFP gene and human LMNA cDNA. Constructs were bicistronic with the 2 inserts (LMNA and GFP) driven by 2 different CMV promoters to identify cells expressing LMNA protein using GFP as a marker of cellular infection. Constructs contained either human wild-type (WT-LMNA) or the human mutant LMNA D192G (MT-LMNA); the construct carrying only GFP gene was used as a control. NRVMs were infected by adenoviruses at 25 multiplicity of infection (MOI) in serum free medium; 6h post-infection, complete medium was replaced to cardiomyocytes and the cells were incubated at 37°C and 5% CO2. Viability assay was performed to monitor the potentially deleterious effects of adenoviral infection. For these studies, cells were rinsed in PBS 1X and incubated with 2 μM Calcein-AM and 4 μM Ethidium homodimer-1, according to manufacturer instructions. Following incubation, images were acquired with a Leica epifluorescence microscope and quantitation of at least 5 randomly chosen optical fields determined (data not shown). Control experiments were carried out with both uninfected NRVMs and with cells infected with adenovirus carrying GFP only in order to confirm that expression of GFP did not affect the end-points of interest (data not shown). Within each experiment, instruments settings were kept constant.
Analysis of LMNA expression
Expression of human LMNA cDNA was first verified by RT-PCR amplification of mRNA isolated from rat cardiomyocytes 48 h. post infection. Primers specific for either human or rat LMNA cDNA were used and amplified RT-PCR products verified by amplicon sequencing. mRNA was isolated by mirVanaTM miRNA kit (Ambion) and cDNA was synthesized by Thermoscript RT-PCR system kit (Invitrogen) after DNase treatment of total RNA by RNase free DNase (Ambion) according to the manufacturer protocol. DNase treated total RNA was used as a negative template control in reverse transcriptase-PCR to verify the absence of any remaining viral DNA (data not shown).
Expression of human mutant LMNA proteins were verified by western blot using Monoclonal Rabbit anti-Human Lamin A antibody (LS-C137845, Lifespan Biosciences) at dilution 1:1000 for 2h at room temperature. This antibody does not show cross-reactivity with rat LMNA. Secondary anti-Rabbit-HRP conjugate (Sigma cat# A0545) at dilution 1:5000 for 1 h at room temperature was used to detect the antigen-antibody complex. Protein expression was examined at days 1 through 6 after infection by ECL Kit (Bioexpress cat# E-1119-20) and visualized in FluorChem system and normalized against levels of Calnexin (ab75801 1:5000 dilution).
Immunofluorescence
NRVMs were fixed in PBS containing 4% PFA for 20 min at room temperature, aldehydes were quenched with 0.1 M glycine in PBS for 20 min at room temperature. Cells were permeabilized with 1%Triton X-100 for 30 min, blocked with 2% BSA in PBS for 1 h at room temperature and incubated with the proper primary antibodies as follows: Lamin A/C Rabbit anti-Human Monoclonal (EPR4100) antibody, (LS-C137845Lifespan Biosciences) at 1:50 dilution at 4°C and, Sarcomeric α Actinin anti-Mouse Monoclonal (EA-53) antibody (AbCam) at 1:100 dilution prepared in 2% BSA in PBS, all at 4°C overnight. Cells were then washed 3 times for 10 min with PBS and 0.05% Tween 20 and finally incubated with Alexa Fluor 488- and Alexa Fluor 594-conjugated secondary antibodies (Invitrogen) in 2% BSA in PBS for 45 - 60 min at room temperature. F-Actin filaments were counterstained with Alexa Fluor® 594 Phalloidin (Life Technologies) at 1:500 dilution for 45 min in PBS min at room temperature. Nuclei were identified by counter-staining sections with TOTO-3 (Life Technologies) additionally each slides were then mounted in Vectashield with DAPI (Vector Labs).
For image acquisition the following objectives were used: HCX PL Fluotar 100_/1.30 NA, HCX PL apocromatic 63_/1.32_0.6 NA, HCX PL Fluotar 40_/0.75 NA, HCX PL N-Plan 20_/0.40 NA, and HCX PL N-Plan 10_/0.25 NA (all from Leica). Within each experiment, instrument settings were kept constant. A first set of pictures was acquired with a Zeiss LSM 510 Meta confocal microscope and representative images were acquired from at least 3 independent cell preparation. For the purpose of the study a Plan-Apochromat 100X/1.46 objective was used. Results were then confirmed with a Zeiss LSM 710 confocal microscope. Representative images were acquired for all the tested conditions (Controls; GFP; WT; D192G mutant) with a Plan-Apochromat 63X/1.4 objective (zoom 2).
Rescue experiments
Cells were first infected with LMNA D192G adenovirus as already described and 24 hours after the first infection, NRVMs were exposed to a second infection using the LMNA adenoviral construct at the same viral load (25 MOI) and in the same experimental conditions. 48 hours from the second infection, ‘rescued’ NRVMs were tested by AFM. Uninfected NRVM and mutant LMNA D192G NRVMs were used as negative and positive controls, respectively. 48 and 96 hours from the second infection, the ‘rescued’ NRVM, as well as the LMNA D192G and WT were also assessed by western blot using the specific antibodies anti Human LMNA and anti- Rat Calnexin described above.
Atomic force microscopy determination of cellular biomechanics
An AFM Solver Pro-M (NT-MDT, Moscow, Russia) was used to acquire cell morphology as well as force-displacement curves as previously described.48-49 An AFM probe modified with polystyrene microsphere (diameter of ∼10 μm) coated with a layer of gold was used to precisely apply a compression force ‘normal’ to the nucleus. Polystyrene bead, is not a truly rigid body however, the polystyrene Young modulus is about 3×109 Pa much higher than that we measured for our nuclei (around 1×103 Pa).
The AFM sphere probe was chosen instead of a pyramidal sharp tip to increases sensitivity to adhesion forces, prevent local membrane disruption during indentation, and to reduce cell-deformation nonlinearity due to a more homogenous contact between the cells and the probe. The AFM was equipped with a “liquid cell” setup using a standard cantilever holder cell for operating in liquid environment. The scanning measuring head allows a scanning range 100 × 100 × 6 μm, with XY linearity error 0.15% and Z linearity error of 1% and error in determining XY linear dimensions ± 1.2%.
AFM probes were checked every day before the beginning of the experiments. Resonance frequency was verified with the thermal method and was considered acceptable with a variability lower than ± 5% in comparison to the value specified by the supplier (17 kHz). Similarly, also spring constant of the cantilever was verified and considered acceptable between a 20 % variability of the value indicated by the supplier (0,08 N/m). Since AFM tests are sensitive to vibrational noise and thermal drift from the environment, the equipment was placed on a damping isolation table. All SCFS experiments were carried out at constant physiologic temperature 37°C in the Petri dish by equilibrating all components of the AFM setup in advance and through the use of a homemade temperature-controlled sample holder. In AFM tests it is very important to bear in mind that cells can be in different states and thus show distinct properties that can be difficult to compare. Therefore (i) cells have been prepared following a strict protocol for each cell type. Moreover, AFM experiments were always performed on the same day they were collected for biochemical (expressions) analyses, (ii) multiple measurements from different cells have been collected to control for variability and ‘average’ data determined, and (iii) cells were monitored and their morphological details observed (an optical light microscope was used for cell selection throughout the tests). All studies were performed on living, intact cells in cell culture medium. During cell loading and unloading, a force signal was recorded and used to obtain information on nuclear elasticity, whole cell viscoelasticity, as well as cell adhesion/detachment from the AFM probe. Force-deformation curves for single cell compression experiments were plotted as loading force versus relative cell deformation. To avoid hydrodynamic forces (significant at high speed), all cells were deformed at slow speed (0.5 μm/s).50-51 All experiments were performed at the same velocity since these AFM tests are rate-dependent. We assessed the nuclear elasticity using at least 20 cells for each condition (CT, WT and MT) and the same operators performed the experiments.
Measurements performed at the position of the nucleus (associated with maximal cell height) are less affected by artifact due to the substrate stiffness so the tip was moved perpendicular to the cell nucleus. To quantify the cell compression, relative deformation, ε (cell height change/initial cell height) was used. The zero deformation for individual cells was determined from the point of the first detectable force during the loading cycle. Indentation depth was calculated comparing the curve detected on the substrate and the curve recorded on the cell. To calibrate the cantilever deflection signal, curves of force vs. the piezo-displacement were acquired on substrate alone. Cells were selected at random as long as they had a healthy morphology (well spread) and were subjected to single indentation for either force-deformation curves or force-relaxation tests. The total duration of the loading-unloading investigations was never longer than 45-50 min to avoid major variations in the physiological parameters of the cells during measurement.
Nuclear elasticity
Nuclear elasticity was calculated using the Sneddon's modification of the Hertz model contact theory to the AFM force curves.52-53 The model assumes that the indented surface is continuous, frictionless and incompressible at small deformations.54 This is not entirely true for cells; however, this model is satisfactory to yield a general idea of cell elasticity and is conventionally used to assess cell elasticity.53 Only the AFM loading force-deformation data up to nucleus deformation of 10% have been used to assess elasticity since this range has been considered linear elastic.20,51,55-57 The Hertz-Sneddon model for sphere tips has the form,58-59
| (2) |
Where F is the load force, E is the Young modulus, ν the Poisson ratio, R the sphere radius and δ is the probe penetration into the cell. The Poisson's ratio was assumed to be 0.5 because the cell was considered incompressible.60-61 Since the nuclear elasticity depends on the stage of cell division,62 we excluded cells with nuclei showing optical mitosis. Finally, cardiomyocytes that exhibited contractile activity (less than 15% of total cell number) were not included in the analysis due to the difficulty in establishing a reliable baseline.
Cell viscoelasticity
Acquiring force-relaxation responses in the so-called constant height mode assessed the cell viscoelasticity behavior.23 The AFM sphere was brought into contact with the cell membrane for a fixed time, the vertical position of the cantilever was set constant while the cantilever's force varies and it is recorded with time. Stress relaxation measurement was carried out in a central region of the cell to minimize the effect of the solid substrate.
During this test, we assumed an approximately constant strain on the cells while the force was recorded over time. This is because the change in cantilever deflection during relaxation was small enough to be considered negligible when compared to the overall indentation.
Statistical Analysis
Repeated measures data analyses were performed using linear mixed-effects models63 as implemented in lme4 package.64 within open source R65 software. Predictors’ assessment was addressed by the parametric bootstrap model selection technique.66 Baseline multiple comparisons were issued using Anova and Tukey HSD test, as implemented in multicomp67 and sandwich 68 libraries. Normality and homoscedasticity were assessed by Shapiro-Wilk and Levene test respectively. In all instances a significance α = 0.05 level was assumed. Nuclear Young's module data was log-transformed in order to avoid computational concerns. To assess if control and wild-type cells were significantly different from one another, 2 different mixed-effects model were initially examined: the former, an additive model allowing interaction between phenotype and calendar timing; the latter, a pure additive model. Within both models, the replicate measures in the 3-phenotype samples were assumed to be a random effect. As far as the cell height is concerned, repeated measures data have been analyzed by linear mixed-effects models63 as implemented in the lme4 package64 of R.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Itemized List of Contributions
All authors have contributed significantly to the paper, including participation in the conception, design, analysis, interpretation of data, drafting or revising of the manuscript, and final approval.
LM and MRGT conceived the study, TL and OS carried out the AFM tests, LP, VM and GDF prepared the cell cultures and took care of the optical images, MRGT conceived the adenoviral constructs, LP, VM and GDF carried out the analysis of LMNA expression and immunohistochemistry and immunofluorescence, LM and CSL provided animals and helped to draft the manuscript. MB carried out the statistical analysis. OS, LM, and MRGT participated in the design of the study and finalized the paper.
Funding
Financial support from “Foreman-Casali” Foundation, Trieste (Italy) to VM and LM; NIH grants UL1 RR025780 and TR001082, RO1 HL69071, RO1 HL116906 to LM, K23 JL067915 and 1R01HL109209-01A1 to MRGT (USA); supported by MISE-ICE-CRUI 16-06-2010 Project 99, and FVG Region LR 26/2005 Art. 23 to OS (Italy). CSL is supported in part by the Kevin Kauffman Endowed Chair of Cardiology. Financial support from the Foundation Leducq, Transatlantic Network of Excellence (14-CVD 03) to OS, TL, LP, VM, MRGT, LM.
References
- 1.Cattin ME, Muchir A, Bonne G. ‘State-of-the-heart' of cardiac laminopathies. Curr Opin Cardiol 2013; 28:297-304; PMID:23455585; http://dx.doi.org/ 10.1097/HCO.0b013e32835f0c79 [DOI] [PubMed] [Google Scholar]
- 2.Schreiber KH, Kennedy BK. When lamins go bad: Nuclear structure and disease. Cell 2013; 152:1365-75; PMID:23498943; http://dx.doi.org/ 10.1016/j.cell.2013.02.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dahl KN, Ribeiro AJ, Lammerding J. Nuclear shape; mechanics; and mechanotransduction. Circ Res 2008; 102:1307-18; PMID:18535268; http://dx.doi.org/ 10.1161/CIRCRESAHA.108.173989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bonne G, Di Barletta MR, Varnous S, Bécane H, Hammouda E, Merlini L, Muntoni F, Greenberg CR, Gary F, Urtizberea J, et al.. Mutations in the gene encoding lamin a/c cause autosomal dominant emery-dreifuss muscular dystrophy. Nat Genet 1999; 21:285-8; PMID:10080180; http://dx.doi.org/ 10.1038/6799 [DOI] [PubMed] [Google Scholar]
- 5.Burke B, Stewart CL. The nuclear lamins Flexibility in function. Nat Rev Mol Cell Biol 2013; 14:13-24; PMID:23212477; http://dx.doi.org/ 10.1038/nrm3488 [DOI] [PubMed] [Google Scholar]
- 6.Chen SC, Kennedy BK, Lampe PD. Phosphorylation of connexin43 on s279/282 may contribute to laminopathy-associated conduction defects. Exp Cell Res 2013; 319:888-96; PMID:23261543; http://dx.doi.org/ 10.1016/j.yexcr.2012.12.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stroud MJ, Banerjee I, Veevers J, Chen J. Linker of nucleoskeleton and cytoskeleton complex proteins in cardiac structure: function and disease. Circ Res 2014; 114:538-48; PMID:24481844; http://dx.doi.org/ 10.1161/CIRCRESAHA.114.301236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Davidson PM, Lammerding J. Broken nuclei-lamins; nuclear mechanics and disease. Trends Cell Biol 2014; 24:247-56; PMID:24309562; http://dx.doi.org/ 10.1016/j.tcb.2013.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sylvius N, Bilinska ZT, Veinot JP, Fidzianska A, Bolongo PM, Poon S, McKeown P, Davies RA, Chan KL, Tang AS, et al.. In vivo and in vitro examination of the functional significances of novel lamin gene mutations in heart failure patients. J Med Genet 2005; 42:639-47; PMID:16061563; http://dx.doi.org/ 10.1136/jmg.2004.023283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zwerger M, Jaalouk DE, Lombardi ML, Isermann P, Mauermann M, Dialynas G, Herrmann H, Wallrath LL, Lammerding J. Myopathic lamin mutations impair nuclear stability in cells and tissue and disrupt nucleo-cytoskeletal coupling. Hum Mol Genet 2013; 22:2335-49; PMID:23427149; http://dx.doi.org/ 10.1093/hmg/ddt079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gonzalez Avalos P, Reichenzeller M, Eils R, Gladilin E. Probing compressibility of the nuclear interior in wild-type and lamin deficient cells using microscopic imaging and computational modeling. J Biomech 2011; 44:2642-8; PMID:21906741; http://dx.doi.org/ 10.1016/j.jbiomech.2011.08.014 [DOI] [PubMed] [Google Scholar]
- 12.Dahl KN, Kahn SM, Wilson KL, Discher DE. The nuclear envelope lamina network has elasticity and a compressibility limit suggestive of a molecular shock absorber. J Cell Sci 2004; 117:4779-86; PMID:15331638; http://dx.doi.org/ 10.1242/jcs.01357 [DOI] [PubMed] [Google Scholar]
- 13.Muller DJ. AFM A nanotool in membrane biology. Biochem 2008; 47:7986-98; http://dx.doi.org/ 10.1021/bi800753x [DOI] [PubMed] [Google Scholar]
- 14.Kaufmann A, Heinemann F, Radmacher M, Stick R. Amphibian oocyte nuclei expressing lamin a with the progeria mutation e145k exhibit an increased elastic modulus. Nucleus 2011; 2:310-9; PMID:21941106; http://dx.doi.org/ 10.4161/nucl.2.4.16119 [DOI] [PubMed] [Google Scholar]
- 15.Hajjar RJ, Kang JX, Gwathmey JK, Rosenzweig A. Physiological Effects of Adenoviral Gene Transfer of Sarcoplasmic Reticulum Calcium ATPase in Isolated Rat Myocytes. Circulation 1997; 95:423-9; PMID:9008460; http://dx.doi.org/ 10.1161/01.CIR.95.2.423 [DOI] [PubMed] [Google Scholar]
- 16.Kass-Eisler A, Falck-Pederson E, Alvirat M, Rivera J, Buttrick PM, Wittenberg BA, Cipriani L, Leinwand LA. Quantitative determination of adenovirus-mediated gene delivery to rat cardiac myocytes in vitro and in vivo. PNAS 1993; 90:11498-502; PMID:8265580; http://dx.doi.org/ 10.1073/pnas.90.24.11498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kovacic-Milivojević B, Roediger F, Almeida EA, Damsky CH, Gardner DG, Ilić D. Focal adhesion kinase and p130Cas mediate both sarcomeric organization and activation of genes associated with cardiac myocyte hypertrophy. Mol Biol Cell. 2001; 12:2290-307; PMID:11514617; http://dx.doi.org/ 10.1091/mbc.12.8.2290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Östlund C, Bonne G, Schwartz K, Worman HJ. Properties of lamin A mutants found in Emery-Dreifuss muscular dystrophy, cardiomyopathy and Dunnigantype partial lipodystrophy. J. Cell Sci 2001; 114:4435-45; PMID:Can't [DOI] [PubMed] [Google Scholar]
- 19.Lulevich V, Zink T, Chen HY, Liu FT, Liu GY. Cell mechanics using atomic force microscopy-based single-cell compression. Langmuir 2006; 22:8151-5; PMID:16952255; http://dx.doi.org/ 10.1021/la060561p [DOI] [PubMed] [Google Scholar]
- 20.Sen S, Subramanian S, Discher DE. Indentation and adhesive probing of a cell membrane with afm: Theoretical model and experiments. Biophys J 2005; 89:3203-13; PMID:16113121; http://dx.doi.org/ 10.1529/biophysj.105.063826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sunyer R, Trepat X, Fredberg JJ, Farre R, Navajas D. The temperature dependence of cell mechanics measured by atomic force microscopy. Phys Biol 2009; 6:025009; PMID:19571363; http://dx.doi.org/ 10.1088/1478-3975/6/2/025009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stamenović D, Rosenblatt N, Montoya-Zavala M, Matthews BD, Hu S, Suki B, Wang N, Ingber DE. Rheological Behavior of Living Cells Is Timescale-Dependent. Biophys J 2007; 93(8):L39-41; PMID:17693464; http://dx.doi.org/ 10.1529/biophysj.107.116582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moreno-Flores S, Benitez R, Vivanco MD, Toca-Herrera JL. Stress relaxation microscopy Imaging local stress in cells. J Biomech 2010; 43:349-54; PMID:19772964; http://dx.doi.org/ 10.1016/j.jbiomech.2009.07.037 [DOI] [PubMed] [Google Scholar]
- 24.Kumar A, Crawford K, Close L, Madison M, Lorenz J, Doetschman T, Pawlowski S, Duffy J, Neumann J, Robbins J, et al.. Rescue of cardiac alpha-actin-deficient mice by enteric smooth muscle gamma-actin. Proc Natl Acad Sci U S A 1997; 94:4406-11; PMID:9114002; http://dx.doi.org/ 10.1073/pnas.94.9.4406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Qi Y, Li H, Shenoy V, Li Q, Wong F, Zhang L, Raizada MK, Sumners C, Katovich MJ. Moderate cardiac-selective overexpression of angiotensinII type 2 receptor protects cardiac functions from ischaemic injury. Exp Physiol 2011; 97:89-101; PMID:21967903; http://dx.doi.org/ 10.1113/expphysiol.2011.060673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reichart B, Klafke R, Dreger C, Krüger E, Motsch I, Ewald A, Schäfer J, Reichmann H, Müller CR, Dabauvalle MC. Expression and localization of nuclear proteins in autosomal-dominant Emery-Dreifuss muscular dystrophy with LMNA R377H mutation. BMC Cell Biology 2004; 5:12; PMID:15053843; http://dx.doi.org/ 10.1186/1471-2121-5-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sasseville A, Raymond M-J Y. Lamin A precursor is localized to intranuclear foci. J. Cell Sci. 1995; 108:273-85; PMID:7738105 [DOI] [PubMed] [Google Scholar]
- 28.Almqvist N, Bhatia R, Primbs G, Desai N, Banerjee S, Lal R. Elasticity and adhesion force mapping reveals real-time clustering of growth factor receptors and associated changes in local cellular rheological properties. Biophys J 2004; 86:1753-62; PMID:14990502; http://dx.doi.org/ 10.1016/S0006-3495(04)74243-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McNamee CE, Pyo N, Higashitani K. Atomic force microscopy study of the specific adhesion between a colloid particle and a living melanoma cell Effect of the charge and the hydrophobicity of the particle surface. Biophys J 2006; 91:1960-9; PMID:16731555; http://dx.doi.org/ 10.1529/biophysj.106.082420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maniotis AJ, Chen CS, Ingber DE. Demonstration of mechanical connections between integrins; cytoskeletal filaments; and nucleoplasm that stabilize nuclear structure. PNAS 1997; 94:849-54; PMID:9023345; http://dx.doi.org/ 10.1073/pnas.94.3.849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lammerding J, Schulze PC, Takahashi T, Kozlov S, Sullivan T, Kamm RD, Stewart CL, Lee RT. Lamin a/c deficiency causes defective nuclear mechanics and mechanotransduction. J Clin Invest 2004; 113:370-8; PMID:14755334; http://dx.doi.org/ 10.1172/JCI200419670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dahl KN, Kalinowski A. Nucleoskeleton mechanics at a glance. J Cell Sci 2011; 124:675-8; PMID:21321324; http://dx.doi.org/ 10.1242/jcs.069096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ho CY, Jaalouk DE, Vartiainen MK, Lammerding J. Lamin a/c and emerin regulate mkl1-srf activity by modulating actin dynamics. Nature 2013; 497:507-11; PMID:23644458; http://dx.doi.org/ 10.1038/nature12105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wen JH, Vincent LG, Fuhrmann A, Choi YS, Hribar KC, Taylor-Weiner H, Chen S, Engler AJ. Interplay of matrix stiffness and protein tethering in stem cell differentiation. Nature Materials 2014; 13:979-87; PMID:25108614; http://dx.doi.org/ 10.1038/nmat4051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jacot JG, McCulloch AD, Omens JH. Substrate stiffness affects the functional maturation of neonatal rat ventricular myocytes. Biophys J. 2008; 95(7):3479-87; PMID:18586852; http://dx.doi.org/ 10.1529/biophysj.107.124545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Swift J, Ivanovska IL, Buxboim A, Harada T, Dingal PC, Pinter J, Pajerowski JD, Spinler KR, Shin JW, Tewari M, et al.. Nuclear lamin-A scales with tissue stiffness and enhances matrix-directed differentiation. Science 2013; 341:6149; http://dx.doi.org/ 10.1126/science.1240104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hochmuth RM. Micropipette aspiration of living cells. J Biomech 2000; 33:15-22; PMID:10609514; http://dx.doi.org/ 10.1016/S0021-9290(99)00175-X [DOI] [PubMed] [Google Scholar]
- 38.Rowat AC. Physical properties of the nucleus studied by micropipette aspiration. Meth Mol Biol 2009; 464:3-12; http://dx.doi.org/ 10.1007/978-1-60327-461-6_1 [DOI] [PubMed] [Google Scholar]
- 39.Hale CM, Shrestha AL, Khatau SB, Stewart-Hutchinson PJ, Hernandez L, Stewart CL, Hodzic D, Wirtz D. Dysfunctional Connections Between the Nucleus and the Actin and Microtubule Networks in Laminopathic Models. Biophys. J 2008; 95:5462-75; PMID:18790843; http://dx.doi.org/ 10.1529/biophysj.108.139428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Muchir A, Medioni J, Laluc M, Massart C, Arimura T, van der Kooi AJ, Desguerre I, Mayer M, Ferrer X, Briault S, et al.. Nuclear envelope alterations in fibroblasts from patients with muscular dystrophy, cardiomyopathy, and partial lipodystrophy carrying lamin A/C gene mutations. Muscle Nerve 2004; 30:444-50; PMID:15372542; http://dx.doi.org/ 10.1002/mus.20122 [DOI] [PubMed] [Google Scholar]
- 41.Hemmer JD, Nagatomi J, Wood ST, Vertegel AA, Dean D, LaBerge M. Role of Cytoskeletal Componentsin Stress-Relaxation Behavior of Adherent Vascular Smooth Muscle Cells. J Biomech Eng 2009; 131(4):041001 1-9; http://dx.doi.org/ 10.1115/1.3049860 [DOI] [PubMed] [Google Scholar]
- 42.Lombardi ML, Jaalouk DE, Shanahan CM, Burke B, Roux KJ, Lammerding J. The Interaction between Nesprins and Sun Proteins at the Nuclear Envelope Is Critical for Force Transmission between the Nucleus and Cytoskeleton. J Biol Chem 2011; 286:26743-53; PMID:21652697; http://dx.doi.org/ 10.1074/jbc.M111.233700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Banerjee I, Zhang J, Moore-Morris T, Pfeiffer E, Buchholz KS, Liu A, Ouyang K, Stroud MJ, Gerace L, Evans SM, et al.. Targeted Ablation of Nesprin 1 and Nesprin 2 from Murine Myocardium Results in Cardiomyopathy, Altered Nuclear Morphology and Inhibition of the Biomechanical Gene Response. PLoS Genet 2014; 10(2):e1004114; PMID:24586179; http://dx.doi.org/ 10.1371/journal.pgen.1004114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Martinelli V, Cellot G, Toma FM, Long CS, Caldwell JH, Zentilin L, Giacca M, Turco A, Prato M, Ballerini L, et al.. Carbon nanotubes promote growth and spontaneous electrical activity in cultured cardiac myocytes. Nano Lett 2012; 12:1831-8; PMID:22432413; http://dx.doi.org/ 10.1021/nl204064s [DOI] [PubMed] [Google Scholar]
- 45.Martinelli V, Cellot G, Fabbro A, Bosi S, Mestroni L, Ballerini L. Improving cardiac myocytes performance by carbon nanotubes platforms. Frontiers Physiol 2013; 4:239; PMID:24027533; http://dx.doi.org/ 10.3389/fphys.2013.00239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Long CS, Kariya K, Karns L, Simpson PC. Sympathetic modulation of the cardiac myocyte phenotype Studies with a cell-culture model of myocardial hypertrophy. Basic Res Cardiol 1992; 87 Suppl 2:19-31; PMID:1338564 [DOI] [PubMed] [Google Scholar]
- 47.Deng XF, Rokosh DG, Simpson PC. Autonomous and growth factor-induced hypertrophy in cultured neonatal mouse cardiac myocytes Comparison with rat. Circ Res 2000; 87:781-8; PMID:11055982; http://dx.doi.org/ 10.1161/01.RES.87.9.781 [DOI] [PubMed] [Google Scholar]
- 48.Codan B, Del Favero G, Martinelli V, Long CS, Mestroni L, Sbaizero O. Exploring the elasticity and adhesion behavior of cardiac fibroblasts by atomic force microscopy indentation. Mat Sci Eng C 2014; 40:427-34; http://dx.doi.org/ 10.1016/j.msec.2014.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Del Favero G, Florio C, Codan B, Sosa S, Poli M, Sbaizero O, Molgo J, Tubaro A, Lorenzon P. The stretch-activated channel blocker gd(3+) reduces palytoxin toxicity in primary cultures of skeletal muscle cells. Chem Res Toxicol 2012; 25:1912-20; PMID:22900474; http://dx.doi.org/ 10.1021/tx300203x [DOI] [PubMed] [Google Scholar]
- 50.Lu YB, Franze K, Seifert G, Steinhauser C, Kirchhoff F, Wolburg H, Guck J, Janmey P, Wei EQ, Kas J, et al.. Viscoelastic properties of individual glial cells and neurons in the cns. PNAS 2006; 103:17759-64; PMID:17093050; http://dx.doi.org/ 10.1073/pnas.0606150103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mathur AB, Collinsworth AM, Reichert WM, Kraus WE, Truskey GA. Endothelial cardiac muscle and skeletal muscle exhibit different viscous and elastic properties as determined by atomic force microscopy. J Biomech 2001; 34:1545-53; PMID:11716856; http://dx.doi.org/ 10.1016/S0021-9290(01)00149-X [DOI] [PubMed] [Google Scholar]
- 52.Timoshenko SP, Goodier JN. Theory of elasticity; 3rd edition New York McGraw-Hill; 1970 [Google Scholar]
- 53.Kuznetsova TG, Starodubtseva MN, Yegorenkov NI, Chizhik SA, Zhdanov RI. Atomic force microscopy probing of cell elasticity. Micron 2007; 38:824-33; PMID:17709250; http://dx.doi.org/ 10.1016/j.micron.2007.06.011 [DOI] [PubMed] [Google Scholar]
- 54.Carl P, Schillers H. Elasticity measurement of living cells with an atomic force microscope Data acquisition and processing. Pflugers Arch-Eur J Physiol 2008; 457:551-9; http://dx.doi.org/ 10.1007/s00424-008-0524-3 [DOI] [PubMed] [Google Scholar]
- 55.Rico F, Roca-Cusachs P, Gavara N, Farre R, Rotger M, Navajas D. Probing mechanical properties of living cells by atomic force microscopy with blunted pyramidal cantilever tips. Phys Rev E 2005; 72:021914; http://dx.doi.org/ 10.1103/PhysRevE.72.021914 [DOI] [PubMed] [Google Scholar]
- 56.Domke J, Radmacher M. Measuring the elastic properties of thin polymer films with the atomic force microscope. Langmuir 1998; 14:3320-5; http://dx.doi.org/ 10.1021/la9713006 [DOI] [Google Scholar]
- 57.Darling EM, Topel M, Zauscher S, Vail TP, Guilak F. Viscoelastic properties of human mesenchymally-derived stem cells and primary osteoblasts chondrocytes and adipocytes. J Biomech 2008; 41:454-64; PMID:17825308; http://dx.doi.org/ 10.1016/j.jbiomech.2007.06.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hertz H. On the elastic contact of elastic solids. J Reine Angew Math 1881; 92:156-71; [Google Scholar]
- 59.Sneddon IN. The relation between load and penetration in the axisymmetric boussinesq problem for a punch of arbitrary profile. Int J Eng Sci 1965; 3:47-57; http://dx.doi.org/ 10.1016/0020-7225(65)90019-4 [DOI] [Google Scholar]
- 60.Radmacher M. Measuring the elastic properties of living cells by the atomic force microscope. Method Cell Biol 2002; 68:67-90; http://dx.doi.org/ 10.1016/S0091-679X(02)68005-7 [DOI] [PubMed] [Google Scholar]
- 61.Morita Y, Mukai T, Ju Y, Watanabe S. Evaluation of stem cell-to-tenocyte differentiation by atomic force microscopy to measure cellular elastic moduli. Cell Biochem Biophys 2013; 66:73-80; PMID:23090789; http://dx.doi.org/ 10.1007/s12013-012-9455-x [DOI] [PubMed] [Google Scholar]
- 62.Matzke R, Jacobson K, Radmacher M. Direct high-resolution measurement of furrow stiffening during division of adherent cells. Nat Cell Biol 2001; 3:607-10; PMID:11389447; http://dx.doi.org/ 10.1038/35078583 [DOI] [PubMed] [Google Scholar]
- 63.Pinheiro JC, Bates DM. Statistics and computing. Mixed-Effects Models in S and S-PLUS. Springer Verlag, N.Y: 2000 [Google Scholar]
- 64.Bates D, Maechler M, Bolker B, Walker S. lme4: Linear mixed- effects models using Eigen and S4, R package version 2014; 1:1-7 [Google Scholar]
- 65.R Development Core Team R: A Language and Environment for Statistical Computing R Foundation for Statistical Computing, Vienna, Austria: 2014 [Google Scholar]
- 66.Faraway JJ. Chapman and Hall/CRC; 2005. Extending the linear model with R: generalized linear, mixed effects and nonparametric regression models. [Google Scholar]
- 67.Bretz F, Hothorn T, Westfall P. Multiple comparisons using R. CRC Press, 2010 [Google Scholar]
- 68.Zeileis A. Object-oriented computation of sandwich estimators. J Stat Soft 2006; 16(9):1-16 [Google Scholar]