Highlight
Multiple stress signalling pathways regulate LHCX family gene expression in the diatom Phaeodactylum tricornutum to attune acclimation responses efficiently in highly variable ocean environments.
Key words: Dark, gene expression, iron starvation, LHCX, light, marine diatom, nitrogen starvation, non-photochemical quenching.
Abstract
Diatoms are phytoplanktonic organisms that grow successfully in the ocean where light conditions are highly variable. Studies of the molecular mechanisms of light acclimation in the marine diatom Phaeodactylum tricornutum show that carotenoid de-epoxidation enzymes and LHCX1, a member of the light-harvesting protein family, both contribute to dissipate excess light energy through non-photochemical quenching (NPQ). In this study, we investigate the role of the other members of the LHCX family in diatom stress responses. Our analysis of available genomic data shows that the presence of multiple LHCX genes is a conserved feature of diatom species living in different ecological niches. Moreover, an analysis of the levels of four P. tricornutum LHCX transcripts in relation to protein expression and photosynthetic activity indicates that LHCXs are differentially regulated under different light intensities and nutrient starvation, mostly modulating NPQ capacity. We conclude that multiple abiotic stress signals converge to regulate the LHCX content of cells, providing a way to fine-tune light harvesting and photoprotection. Moreover, our data indicate that the expansion of the LHCX gene family reflects functional diversification of its members which could benefit cells responding to highly variable ocean environments.
Introduction
The perception of environmental signals and the activation of appropriate responses to external stimuli are of major importance in the growth and survival of all organisms. At the cellular level, this requires the presence of complex signal perception and transduction networks, triggering changes in nuclear gene expression (Lee and Yaffe, 2014). External cues such as light, temperature, and nutrient availability strongly affect the physiology and metabolism of photosynthetic organisms, so acclimation mechanisms are needed to cope efficiently with short- and long-term environmental changes to maintain photosynthetic performances (Walters, 2005; Eberhard et al., 2008). In eukaryotic phototrophs, chloroplast biogenesis and activity are integrated in broader regulatory programmes, requiring coordination between the nucleus and chloroplast genomic systems (Rochaix, 2011; Jarvis and Lopez-Juez, 2013). The nucleus responds to stimuli inducing the synthesis of regulatory proteins that modulate chloroplast responses. In turn, molecules originating from the chloroplast activity (e.g. redox state of the photosynthetic electron carriers, reactive oxygen species, plastid gene transcription, tetrapyrroles, and other metabolites) provide a retrograde signal feeding back to the nucleus (Woodson and Chory, 2008).
Marine photosynthesis is dominated by unicellular phytoplanktonic organisms, which are passive drifters in the water column and often experience drastic changes in their surrounding environment (Falkowski et al., 2004; Depauw et al., 2012). Diatoms are among the most abundant and diversified groups of photosynthetic organisms. They are particularly adapted to growing in very dynamic environments such as turbulent coastal waters and upwelling areas, as well as in polar oceans (Margalef, 1978; Field et al., 1998; Kooistra et al., 2007; Arrigo et al., 2012). Several species can survive for long periods at depths where light is limiting for growth, and quickly reactivate their metabolism after returning to the photic zone (Sicko-Goad et al., 1989; Reeves et al., 2011). The adaptive capacity of such algae suggests that they have sophisticated mechanisms to perceive and rapidly respond to environmental variations. Consistent with this notion, genome sequence information of representative diatom model species such as Thalassiosira pseudonana and Phaeodactylum tricornutum (Armbrust et al., 2004; Montsant et al., 2007; Bowler et al., 2008; Rayko et al., 2010), and the availability of transcriptomic and proteomic data in various species exposed to different stimuli and stresses (Nymark et al., 2009; Dyhrman et al., 2012; Thamatrakoln et al., 2012; Ashworth et al., 2013; Nymark et al., 2013; Keeling et al., 2014; Valle et al., 2014; Alipanah et al., 2015; Muhseen et al., 2015) have highlighted the existence of some diatom-specific adaptive strategies, pinpointing molecular regulators of environmental change responses. Several photoreceptors for efficient light colour sensing (Huysman et al., 2013; Schellenberger Costa et al., 2013; Fortunato et al., 2015, 2016) have been identified in diatoms. Peculiar iron acquisition and concentration mechanisms are also known (Allen et al., 2008; Marchetti et al., 2012; Morrissey et al., 2015), which contribute to their survival in iron-limited waters and to their rapid proliferation when iron becomes available (de Baar et al., 2005). Diatoms have peculiar gene sets implicated in nitrogen metabolism, such as a complete urea cycle, that could be used as temporary energy storage or as a sink for photorespiration (Allen et al., 2011). Eventually, diatoms optimize their photosynthesis via extensive energetic exchanges between plastids and mitochondria (Bailleul et al., 2015).
The ecological dominance of diatoms also relies on their capacity to cope with light stresses, thanks to very efficient photoprotective mechanisms. Diatoms possess a high capacity to dissipate excess light energy as heat through high energy quenching (qE) that, together with the photoinhibitory quenching (qI), can be visualized via the non-photochemical quenching (NPQ) of Chl a fluorescence (Lavaud and Goss, 2014; Goss and Lepetit, 2015). The xanthophyll diatoxanthin (Dt) pigment, synthesized from the de-epoxidation of diadinoxanthin (Dd) during illumination (Goss and Jakob, 2010; Lavaud et al., 2012), and the LHCX1 protein, a member of the light-harvesting protein family (Bailleul et al., 2010), have been identified as key components of the qE process in diatoms. P. tricornutum cells with deregulated LHCX1 expression display a significantly reduced NPQ capacity and a decreased fitness, demonstrating a key role for this protein in light acclimation (Bailleul et al., 2010), similarly to the light harvesting complex stress-related (LHCSR) proteins of green algae and mosses (Alboresi et al., 2010; Ballottari et al., 2016).
Multiple nuclear-encoded and plastid-localized LHCX family members have been identified in the genomes of the diatoms P. tricornutum and T. pseudonana. Scattered information derived from independent gene expression analyses indicated that some LHCX isoforms are constitutively expressed while others are expressed in response to stress (Becker and Rhiel, 2006; Allen et al., 2008; Nymark et al., 2009; Zhu and Green, 2010; Bailleul et al., 2010; Beer et al., 2011; Lepetit et al., 2013), similarly to what is observed for the two LHCSR proteins in Physcomitrella patens (Gerotto et al., 2011). In this study, we have extended the characterization of the four P. tricornutum LHCXs, by combining detailed gene expression analysis in cells exposed to different conditions with in vivo analysis of photosynthetic parameters. The result of this analysis revealed a complex regulatory landscape, suggesting that the expansion of the LHCXs reflects a functional diversification of these proteins and may contribute to the regulation of the chloroplast physiology in response to diverse extracellular and intracellular signals.
Materials and methods
Analysis of the LHCXs in the diatom genomes
Phaeodactylum tricornutum and T. pseudonana LHCX gene model identifiers were retrieved from the diatom genomes, respectively, on P. tricornutum Phatr2 and T. pseudonana Thaps3 in the JGI database (http://genome.jgi.doe.gov/). Phaeodactylum tricornutum LHCX proteins were used as query to perform BlastP searches on the Pseudo-nitzschia multiseries (http://genome.jgi.doe.gov/Psemu1/Psemu1.home.html) and Thalassiora oceanica (http://protists.ensembl.org/Thalassiosira_oceanica/Info/Index) genome portals. Best hit sequences were tested on Pfam (http://pfam.xfam.org/) to assess the presence of the Chloroa_b-bind domain (PF00504), characteristic of light-harvesting proteins. Protein alignments were performed with MUSCLE (http://www.ebi.ac.uk/Tools/msa/muscle/).
Diatom growth conditions
The P. tricornutum (Pt1 8.6, CCMP2561) cultures, obtained from the Provasoli-Guillard National Center for Culture of Marine Phytoplankton, were used for the gene expression and photophysiology analyses. Cells were grown in ventilated flasks in f/2 medium (Guillard, 1975) at 18 °C, in a 12h light/12h dark photoperiod using white fluorescence neon lamps (Philips TL-D 90), at 30 μmol m−2 s−1 (low light). High light treatments were performed by irradiating the cells with 500 μmol m−2 s−1 for 5h, 2h after the onset of light, with the same light sources. Dark adaptation treatments were performed for 60h. Blue light (450nm, 1 μmol m−2 s−1) was applied for 10min, 30min, and 1h on dark-adapted cells in the absence and presence of 2 µM DCMU [3-(3,4-dichlorophenyl)-1,1-dimethylurea]. In the iron starvation experiments, P. tricornutum cells at an initial concentration of 2×105 cells ml–1 were grown in f/2 artificial sea water medium (Allen et al., 2008) modified to contain either 11 µM iron (iron-replete) or 5nM iron with the addition of 100 µM of the Fe2+ chelator FerroZine™ (iron-limited) (Stookey, 1970). Cells were harvested after 3 d to perform the analyses. Nitrogen starvation was achieved by diluting P. tricornutum cells to 2×105 cells ml–1 in f/2 medium containing 1mM nitrate (NO3-replete) or 50 µM nitrate (NO3-limited). When cells attained a concentration of 1×106 cells ml–1, they were re-diluted to 2×105 cells ml–1 in their respective media and harvested after 3d, 2h after the onset of light, and then used for experiments.
Generation of transgenic lines overexpressing the LHCX proteins
Vectors for LHCX overexpression were generated by cloning the full-length cDNA sequences of the four LHCX genes in the pKS-FcpBpAt-C-3HA vector (Siaut et al., 2007), using the EcoRI and NotI restriction sites. The LHCX cDNAs were amplified by PCR using the primers described in Supplementary Table S1 at JXB online. Each vector was co-transformed with the pFCPFp-Shble vector for antibiotic selection into P. tricornutum Pt4 cells (DQ085804; De Martino et al., 2007) by microparticle bombardment (Falciatore et al., 1999). Transgenic lines were selected on 100 μg ml–1 phleomycin (Invitrogen) and screened by PCR using primers specific for the four LHCXs (Supplementary Table S1). Transgenic lines overexpressing the LHCX4 isoform in the Pt1 ecotype were also generated, as for the Pt4 ecotype.
RNA extraction and qRT-PCR analysis
Total RNA was isolated from 108 cells with TriPure isolation reagent (Roche Applied Science, IN, USA) according to the manufacturer’s instructions. Quantitative real-time PCR (qRT-PCR) was performed on wild-type cells and on the LHCX-overexpressing clones as described in De Riso et al. (2009). The relative quantification of the different LHCX transcripts was obtained using RPS (ribosomal protein small subunit 30S; ID10847) and H4 (histone H4; ID34971) as reference genes, and by averaging of two reference genes using the geometric mean and the fold changes calculated with the 2−ΔΔCt Livak method (Livak and Schmittgen, 2001). Primer sequences used in qRT-PCR analysis are reported in Supplementary Table S1.
Protein extraction and western blot analysis
Western blot analyses were performed on total cell protein extracts prepared as in Bailleul et al. (2010), and resolved on 14% LDS–PAGE gels. Proteins were detected with different antibodies: anti-LHCSR (gift of G. Peers, University of California, Berkeley, CA, USA) (1:5000); anti-D2 (gift of J.-D. Rochaix, University of Geneva, Switzerland) (1:10 000); anti-PsaF (1:1000) and anti-βCF1 (1:10 000) (gift of F.-A. Wollman, Institut de Biologie Physico-Chimique, Paris, France); and anti-HA primary antibody (Roche) (1:2000). Proteins were revealed with Clarity reagents (Bio-Rad) and an Image Quant LAS4000 camera (GE Healthcare, USA).
Chlorophyll fluorescence measurements
Light-induced fluorescence kinetics were measured using a fluorescence CCD camera recorder (JTS-10, BeamBio, France) as described (Johnson et al., 2009) on cells at 1–2×106 cells ml–1. F v/F m was calculated as (F m–F 0)/F m. NPQ was calculated as (F m–F m')/F m' (Bilger and Bjorkman, 1990), where F m and F m' are the maximum fluorescence emission levels in the dark and light-acclimated cells, measured with a saturating pulse of light. All samples, except the 60h dark-adapted cells, were adapted to dim light (10 μmol m−2 s−1) for 15min at 18 °C before measurements. The maximal NPQ response was measured upon exposure for 10min to saturating green light of 950 μmol m−2 s−1. The relative electron transfer rate (rETRPSII) was measured with a JTS-10 spectrophotometer at different light intensities (20, 170, 260, 320, 520, and 950 µmol m−2 s−1), by changing light every 4min to minimize the photodamage. rETRPSII was calculated as: Y2×light intensity, where Y2 is the efficiency of PSII.
In silico analysis of the LHCX non-coding sequences
Determination of motif occurrence and de novo search of over-represented motifs in the 5'-flanking regions (1000bp or the entire intergenic sequence between the coding gene of interest and the upstream gene) and the introns of the PtLHCX genes were performed by the use of the FIMO (v4.11.1) and MEME Suite (v4.9.1) tools (Bailey et al., 2009), with a p-value cut-off of 0.0001. The Tomtom tool (v4.11.1) on the MEME Suite was used to compare motifs with known transcription binding sites. Microarray data from Alipanah et al. (2015) (accession GSE58946) were downloaded from the GEO database (http://www.ncbi.nlm.nih.gov/pubmed/23193258) using the GEO query package (http://www.ncbi.nlm.nih.gov/pubmed/17496320). Data were loaded and analysed in the R environment using the Limma package (http://www.ncbi.nlm.nih.gov/pubmed/25605792). Selection of transcripts categories: (i) up-regulated, log2 fold change (Fc) >3, adjusted p-value <0.01 (122 genes); (ii) down-regulated, Fc <–3, adjusted p-value <0.01 (200 genes). The P. tricornutum genome sequences and gff mapping of filtered gene models were downloaded from the JGI website and refer to Phatr2 (http://genome.jgi.doe.gov/Phatr2/Phatr2.home.html). The significance of motif enrichment was evaluated using the binomial test with a p-value cut-off of 0.05. All analyses were performed using custom scripts in Perl and R.
Results
LHCX family expansion in the diatom genomes
Several genes belonging to the LHCX family have already been identified in the genome of the pennate diatom P. tricornutum (Bailleul et al., 2010) and the centric diatom T. pseudonana (Zhu and Green, 2010), the most established model species due to the availability of molecular toolkits for genetic manipulations (Apt et al., 1996; Poulsen and Kroger, 2005; De Riso et al., 2009; Trentacoste et al., 2013; Daboussi et al., 2014; Karas et al., 2015). The recently available genome sequences of the pennate diatom P. multiseries, belonging to a widely distributed genus also comprising toxic species (Trainer et al., 2012), and the centric diatom T. oceanica, a species adapted to oligotrophic conditions (Lommer et al., 2012), opened up the possibility to extend this investigation to other ecologically relevant species. As summarized in Table 1, comparative analysis indicates an expansion of the LHCX family in diatoms, compared with the green algae: four members are present in P. tricornutum, five in P. multiseries, T. pseudonana, and T. oceanica, and up to 17 members have been found in the genome of the polar species Fragilariopsis cylindrus (B. Green and T. Mock, personal communication). Analysis of the intron–exon structure of all the available diatom LHCX genes revealed a variable number of introns (from zero to three) as well as variable intron and exon lengths (Table 1).
Table 1.
List of the LHCXs identified in the diatom genomes
| Species | Name | ID | Chromosomal localization | Length (no. of amino acids) | No. of introns |
|---|---|---|---|---|---|
| Thalassiosira pseudonana | LHCX1 | 264921 | chr_23:365603–366232 (–) | 209 | 0 |
| LHCX2 | 38879 | chr_23:368273–368902 (+) | 209 | 0 | |
| LHCX4 | 270228 | chr_5:1446306–1447125 (–) | 231 | 0 | |
| LHCX5 | 31128 | chr_1:2849139–2850176 (–) | 236 | 3 | |
| LHCX6 | 12097 | chr_23:366611–367378 (+) | 255 | 0 | |
| Phaeodactylum tricornutum | LHCX1 | 27278 | chr_7:996379–997300 (+) | 206 | 1 |
| LHCX2 | 56312 | chr_1:2471232–2472170 (+) | 238 | 2 | |
| LHCX3 | 44733 | chr_5:76676–77606 (+) | 206 | 1 | |
| LHCX4 | 38720 | chr_17:53010–53733 (+) | 207 | 1 | |
| Pseudo-nitzschia multiseries | – | 66239 | scaffold_189:181982–182948 (+) | 201 | 1 |
| – | 238335 | scaffold_95:121459–122306 (–) | 202 | 1 | |
| – | 257821 | scaffold_246:124909–125745 (+) | 197 | 1 | |
| – | 264022 | scaffold_1353:8720–9877 (+) | 206 | 1 | |
| – | 283956 | scaffold_38:284133–284828 (–) | 231 | 0 | |
| Thalassiosira oceanica | – | Thaoc_09937 | SuperContig To_g10869: 4.331–5.040 (–) | 210 | 1 |
| – | Thaoc_12733 | SuperContig To_g15184: 10.800–11.435 (–) | 172 | 1 | |
| – | Thaoc_28991 | SuperContig To_g41561: 2.777–3.105 (+) | 81 | 1 | |
| – | Thaoc_31987 | SuperContig To_g45669: 1–1.025 (–) | 205 | 2 | |
| – | Thaoc_32497 | SuperContig To_g46152: 5.664–6.285 (–) | 180 | 1 |
For T. pseudonana (Thaps3), P. tricornutum (Phatr2), and P. multiseries (Psemu1), ID numbers refer to the genome annotation in the JGI database (http://genome.jgi.doe.gov/). For T. oceanica (ThaOc_1.0), ID refers to the Ensembl Protist database (http://protists.ensembl.org/Thalassiosira_oceanica/Info/Index?db=core). (+) and (–) indicate the forward and reverse chromosomal or scaffolds, respectively. The protein length and intron numbers are also indicated.
Light versus dark regulation of expression of the LHCX genes
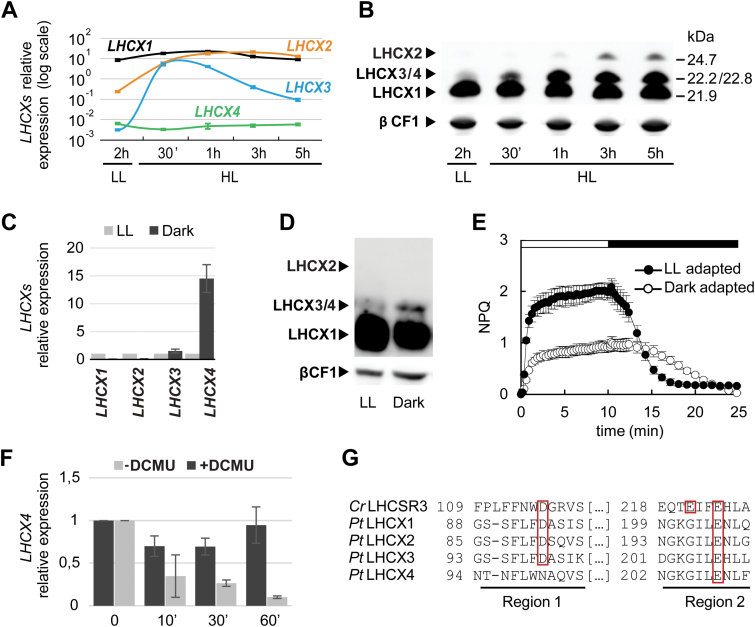
Independent gene expression studies performed in P. tricornutum cells suggest that the LHCX gene family is regulated by light via multiple regulatory pathways. To explore the mechanisms controlling the light responses of the LHCX genes further, we analysed mRNA and protein contents in cells exposed to different light conditions. We first monitored the expression of the LHCX genes in cells grown in low light (LL) and then exposed to high light (HL). In line with previous studies (Bailleul et al., 2010; Lepetit et al., 2013), qRT-PCR and western blot analyses (Fig. 1A and B, respectively) showed that LHCX1 is expressed at very high levels in LL-adapted cells, and that HL treatment slightly increases the LHCX1 content. Conversely, the isoforms 2 and 3 showed different responses to the LL to HL shift. The LHCX2 transcripts, which are significantly less abundant than that of LHCX1 in LL, rapidly increased following HL stress, reaching levels comparable with those of LHCX1 after 1h HL exposure (Fig. 1A). This translated into an increase of the LHCX2 protein observed by western blot (Fig. 1B). However, the increase in the protein content was lower than that of the transcript, possibly because of a low affinity of the LHCSR antibody (Peers et al., 2009) for the LHCX2 isoform. The LHCX3 transcripts that were expressed at very low levels in LL quickly rose upon HL treatment, peaking after 30min and starting to decrease after 1h of light stress. Conversely, a different mRNA expression profile was found in the case of LHCX4, which, unlike the other isoforms, was barely detectable in both LL and HL conditions (Fig. 1A). The LHCX3 and LHCX4 proteins, having very similar molecular weights (22.8kDa and 22.2kDa, respectively), cannot be discriminated by western blot analysis. Based on the different transcriptional regulation of LHCX3 and LHCX4 by light, it is tempting to propose that the light-induced protein of ~22.8kDa reflects the accumulation of the LHCX3 isoform (Fig. 1B). However, in contrast to the transient induction of the LHCX3 mRNAs, this protein is gradually accumulated during the LL to HL shift and it remains stable over the treatment. This discrepancy between transcript and protein expression profiles could be explained assuming that: (i) some post-transcriptional modifications regulate the accumulation of LHCX3 in the light; or (ii) the light-induced protein isoform at 22.8kDa also comprises the LHCX4 protein, which could be present in HL-exposed cells, along with LHCX3.
Fig. 1.
Light and dark regulation of P. tricornutum LHCXs. Analysis of the four LHCX transcripts by qRT-PCR (A) and of LHCX proteins (B) by western blotting in cells adapted to low light (LL) (12L/12D cycles), after exposure to LL for 2h then to high light (HL) for 30min, 1h, 3h, or 5h. mRNA levels were quantified by using RPS as the reference gene (A). Proteins were detected using the anti-LHCSR antibody which recognizes all the PtLHCXs (arrowheads) and the anti-βCF1 antibody as loading control (B). Cells adapted to darkness for 60h were compared with those grown in LL for the analysis of LHCX transcripts (C), proteins (D), and NPQ (E). Relative transcript levels were determined using RPS as a reference, and values were normalized to gene expression levels in LL. LHCX proteins were detected as in (B). The horizontal bar in (E) indicates when the actinic light was on (white) or off (black). (F) LHCX4 mRNAs in 60h dark-adapted cells (Time 0) and in response to 10min, 30min, or 1h of blue light (1 µmol m−2 s−1), in the presence (black) or absence (grey) of the inhibitor DCMU. Transcript levels were quantified by using RPS as the reference, and normalized to gene expression levels in the dark. Error bars represent ±SD of three technical replicates from one representative experiment in (A), and ±SD of three biological replicates in (C), (E), and (F). (G) Alignment of regions 1 and 2 of the Chlamydomonas reinhardtii LHCSR3 and P. tricornutum LHCX1, 2, 3, and 4 protein sequences. The boxes indicate the pH-sensing residues conserved between the LHCXs and LHCSR3. (This figure is available in colour at JXB online).
Previous reports (Lepetit et al., 2013; Nymark et al., 2013) indicate that the LHCX4 transcript is induced in dark-adapted cells. Therefore, we extended the analysis of the expression of the four LHCX genes to cells adapted to prolonged darkness (60h). In these conditions, we observed a significant increase only of the LHCX4 mRNAs (Fig. 1C). For the same reason as described above, we attributed the band of ~22kDa observed in the dark-adapted cells to the LHCX4 protein, although LHCX3 (Fig. 1D) could also be present. In the dark, cells were also showing a decreased NPQ capacity (Fig. 1E) and a slightly reduced PSII maximal quantum yield and overall photosynthetic electron flow capacity (Table 2). Other studies have revealed that blue light photoreceptors (Coesel et al., 2009; Juhas et al., 2014) and the redox state of the chloroplast (Lepetit et al., 2013) could both contribute to the light regulation of LHCX1, 2, and 3 gene expression. Thus, we tested the possible role of these processes in the inhibition of LHCX4 expression upon light exposure. We irradiated dark-adapted cells with low intensity blue light (1 µmol m−2 s−1) during 1h, in the presence or absence of the PSII inhibitor DCMU (Fig. 1F). The analysis revealed that the LHCX4 expression is repressed even at such low light irradiance. Moreover, this repression is lost by poisoning photosynthesis with DCMU, suggesting that this process plays an active role in the light-induced repression of LHCX4.
Table 2.
Photosynthetic parameters of the P. tricornutum wild type and transgenic lines
| Strain | Conditions | F v /F m | rETR PSII | NPQ max |
|---|---|---|---|---|
| Pt1 | LL | 0.66±0.03 | 74.4±1.2 | 2.1±0.1 |
| Pt1 | Dark | 0.60±0.02 | 67.2±4.0 | 1.0±0.1 |
| Pt1 | +Fe | 0.65±0.003 | 72.6±4.8 | 2.2±0.3 |
| Pt1 | –Fe | 0.20±0.004 | 25.7±1.8 | 4.4±0.3 |
| Pt1 | +N | 0.65±0.006 | 74.4±2.4 | 2.1±0.1 |
| Pt1 | –N | 0.40±0.008 | 27.5±3.2 | 3.2±0.4 |
| Pt4 | WT | 0.68±0.01 | 79.3±2.6 | 0.83±0.04 |
| Pt4 | EVL | 0.66±0.01 | 72.5±3.5 | 0.82±0.03 |
| Pt4 | OE1 | 0.67±0.01 | 70.7±2.6 | 1.00±0.1 |
| Pt4 | OE2.5 | 0.67±0.01 | 70.0±1.6 | 1.06±0.05 |
| Pt4 | OE2.20 | 0.66±0.01 | 69.3±1.6 | 1.02±0.08 |
| Pt4 | OE3.12 | 0.66±0.01 | 75.8±4.9 | 1.04±0.07 |
| Pt4 | OE3.33 | 0.68±0.03 | 71.0±3.1 | 1.00±0.1 |
| Pt4 | OE4.11 | 0.59±0.01 | 68.7±1.9 | 1.03±0.01 |
| Pt4 | OE4.13 | 0.58±0.02 | 69.4±1.3 | 1.07±0.04 |
PSII efficiency (F v/F m) and relative electron transport rate (rETRPSII) in different growth conditions are reported. rETRPSII was measured at 260 µmol photons m−2 s−1 light intensity and calculated as: rETRPSII=ɸPSII×actinic light intensity. Non-photochemical quenching (NPQ) was measured with an actinic light intensity of 950 µmol photons m−2 s−1 and calculated as in Maxwell and Johnson (2000). Data are the average of three biological replicates ±SD.
A recent study in Chlamydomonas reinhardtii showed that the activity of the protein LHCSR3 is regulated by the reversible protonation of three specific amino acidic residues following luminal pH acidification in the light (Ballottari et al., 2016). In order to assess if this mechanism is conserved in diatoms, we analysed the P. tricornutum LHCX protein sequences. We found that LHCX1, 2, and 3 possess two of the three amino acids identified in LHCSR3 in conserved positions (Fig. 1G; Supplementary Fig. S2), suggesting that the pH-triggered activation of qE could be conserved in diatoms. On the other hand, only one of these protonatable residues was found in LHCX4.
LHCX expression in iron starvation
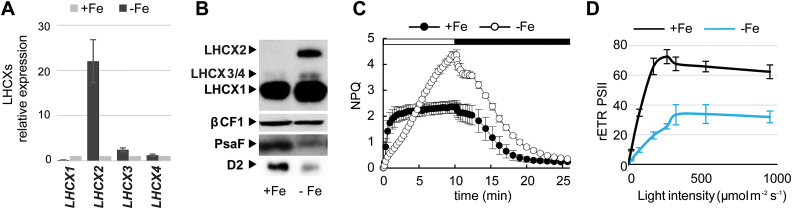
Besides light, nutrient availability also affects chloroplast activity (Wilhelm et al., 2006; Gross, 2012). In many oceanic regions, iron is a major limiting factor for diatom distribution. A general down-regulation of photosynthesis has been reported in iron starvation in several diatom species (Laroche et al., 1995; Allen et al., 2008; Hohner et al., 2013), with a consequent decrease of the carbon fixation reactions, growth rate, and cell size. Since increased NPQ was previously observed in iron-starved P. tricornutum cells (Allen et al., 2008), we compared the expression of the different LHCX isoforms in cells grown under Fe-replete and Fe-limited conditions. We found that while a slight induction of the other LHCX proteins was seen, the LHCX2 transcript was greatly induced in iron-limited cells (Fig. 2A), leading to a significant accumulation of the LHCX2 protein (Fig. 2B). Fe limitation also enhanced NPQ, while slowing down its kinetics (Fig. 2C), possibly because of a slower diadinoxanthin de-epoxidation rate. We also observed a severe impairment of the photosynthetic capacity in iron limitation as indicated by the decrease in F v/F m (Table 2) and in the PSII maximal electron transport rate (rETRPSII) (Fig. 2D; see also Allen et al., 2008). The decreased maximal rETRPSII was probably caused by a diminished capacity for carbon fixation. Moreover, in agreement with previous studies (Allen et al., 2008; Thamatrakoln et al., 2013), we observed a decrease in the amount of PSI (PsaF), which is the complex with the highest Fe content. This complex has been already shown to represent the first target of Fe limitation (Moseley et al., 2002). We also found a significant decrease in PSII (D2 protein), which was probably degraded because of sustained photoinhibition (see also Allen et al., 2008) (Fig. 2B).
Fig. 2.
Effect of iron starvation on P. tricornutum LHCX expression and photophysiology. Experiments were performed on cells grown in iron-replete (11 µM, +Fe) or iron-limited (5nM iron+100 µM FerroZine™, –Fe) conditions: (A) qRT-PCR analysis of LHCX transcripts in –Fe, normalized against the +Fe condition and using RPS and H4 as reference genes. (B) Immunoblot analysis of the LHCX, D2, and PsaF proteins, using βCF1 as loading control. NPQ capacity (C) and relative electron transfer rates (rETRPSII) (D) of cells grown in +Fe or –Fe. The horizontal bar in (C) indicates when the actinic light was on (white) or off (black). rETRPSII was measured at different light intensities (20, 170, 260, 320, 520, and 950 µmol m−2 s−1). In (A), (C), and (D), error bars represent ±SD of three biological replicates. (This figure is available in colour at JXB online).
LHCX expression in nitrogen starvation
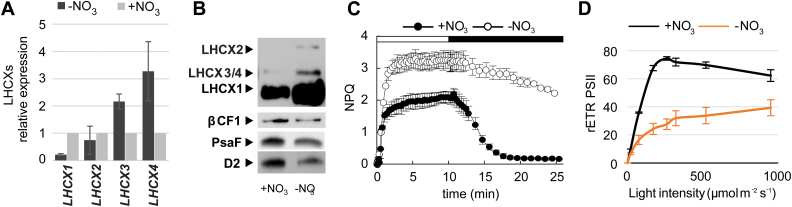
Besides iron, nitrogen (N) is also a limiting resource for diatoms (Mills et al., 2008; Moore et al., 2013; Rogato et al., 2015). Recent transcriptomic and proteomic analysis highlighted important metabolic modifications under N starvation, such as the up-regulation of nitrogen assimilation enzymes, the recycling of intracellular nitrogen-containing compounds from the photosynthetic apparatus and other sources, and the increase in lipid content as a consequence of remodelling of intermediate metabolism (Allen et al., 2011; Palmucci et al., 2011; Hockin et al., 2012; Alipanah et al., 2015; Levitan et al., 2015; Matthijs et al., 2016). We found that N limitation also has a significant effect on the expression of the LHCXs. In particular, N limitation triggered the induction of LHCX3 and LHCX4 mRNAs (Fig. 3A) and of LHCX3/4 proteins (Fig. 3B). The increase of the LHCX1 and 2 isoforms was only visible at the protein level (Fig. 3B). Up-regulation of the LHCX proteins in N limitation correlated with an increase of the NPQ capacity (Fig. 3C). We note that NPQ was slowly relaxing upon dark exposure of N-limited cells, possibly reflecting the repression of the genes encoding the xanthophyll cycle enzymes including the zeaxanthin epoxidase (see Supplementary Fig. S3), when analysing available microarray data from N-depleted cells (Alipanah et al., 2015). The N limitation also led to a drastically reduced F v/F m (Table 2) and a lower maximal rETRPSII (Fig. 3D). We also detected a reduced content of PSII and PSI proteins (Fig. 3B), in line with previous omic studies pointing to a general decrease of the photosynthetic capacity.
Fig. 3.
Effect of nitrogen starvation on P. tricornutum LHCX expression and photophysiology. Experiments were performed on cells grown in nitrogen-replete (1mM, +NO3 –) or nitrogen starvation (50 µM, –NO3 –) conditions: (A) qRT-PCR analysis of LHCX transcripts in –NO3 –, normalized against the values in the +NO3 – condition, and using RPS and H4 as reference genes. (B) Immunoblot analysis of the LHCX, D2, and PsaF proteins, using βCF1 as loading control. NPQ capacity (C) and relative electron transfer rates (rETRPSII) (D) of cells grown in +NO3 – and –NO3 – conditions. The horizontal bar in (C) indicates when the actinic light was on (white) or off (black). rETRPSII was measured at different light intensities (20, 170, 260, 320, 520, and 950 µmol m−2 s−1). In (A), (C), and (D), error bars represent ±SD of three biological replicates. (This figure is available in colour at JXB online).
Analysis of the LHCX non-coding regions
Due to the observed transcriptional responses of LHCX genes in different light and nutrient conditions, we searched for known and potentially novel regulatory motifs in the 5'-flanking regions and the intronic sequences of the four isoforms (see Table 3; Supplementary Fig. S1). Because of their involvement in integrating light signals with CO2/cAMP-induced transcriptional responses, we searched for the three CO2/cAMP-responsive cis-regulatory elements (CCREs) identified in Ohno et al. (2012) and further characterized in Tanaka et al. (2016). Interestingly, we found CCRE-1 in the 5'-flanking sequences of LHCX1 and 4, CCRE-2 in 1, 2, and 4, and CCRE-3 in LHCX4. These cis-regulatory elements may participate in the light-mediated regulation of the four LHCX genes.
Table 3.
The identified regulatory motifs and their occurrence in the P. tricornutum LHCX non-coding sequences
| Sequence | LHCX1 | LHCX2 | LHCX3 | LHCX4 | |
|---|---|---|---|---|---|
| MOTIF 1 | TCA[CT][AT]GTCA | 2 | 2 | 1 | – |
| MOTIF 2 | CGAACCTTGG | – | – | 2 | – |
| MOTIF 3 | CCT[GC]TCCGTA | – | – | 2 | – |
| MOTIF 4 | GAGTCCATCG | – | – | – | 2 |
| MOTIF 5 | CGATCACGGC | – | – | – | 2 |
| MOTIF 6 | [TA]TGACTG | – | 1 | 1 | 1 |
| CCRE-1 | TGACGT | 1 | – | – | 1 |
| CCRE-2 | ACGTCA | 1 | 1 | – | 1 |
| CCRE-3 | TGACGC | – | – | – | 1 |
In contrast, the two P. tricornutum iron-responsive elements identified in Yoshinaga et al. (2014) are not present in the analysed non-coding regions, suggesting that a different transcription factor should be involved in modulating the LHCX2 transcriptional response to iron availability. Similarly, we could not find the two P. tricornutum motifs identified as responsive to short-term nitrogen deprivation (from 4h to 20h) in Matthijs et al. (2016), suggesting that distinct regulatory circuits may act in the short- and long-term acclimation to nitrogen deprivation.
To pinpoint possible novel regulatory motifs, we also scanned the non-coding sequences of the four isoforms using the MEME Suite program (Bailey et al., 2009). The analysis revealed six motifs repeated at least twice in each isoform and/or shared by more than one isoform (Table 3; Supplementary Fig. S1). None of the identified motifs corresponds to a known transcription factor-binding site. This suggests that these motifs could represent novel diatom-specific cis-regulatory elements. In order to examine the potential involvement of the identified motifs in the long-term nitrate deprivation transcriptional responses of LHCX genes, we analysed a published microarray data set performed on 48h and 72h nitrogen-deprived P. tricornutum cells (Alipanah et al., 2015). We compared the frequency of the six identified motifs in the 5'-flanking sequences of responsive and unresponsive transcripts. Interestingly, motif 6 ([T-A]TGACTG) was significantly enriched (p=0.035) in the 5'-flanking sequences of genes up-regulated in response to nitrogen starvation compared with down-regulated genes. The result suggests that motif 6 may be involved in gene transcriptional regulation in cells exposed to prolonged nitrogen starvation.
Modulation of LHCX gene expression in P. tricornutum transgenic lines
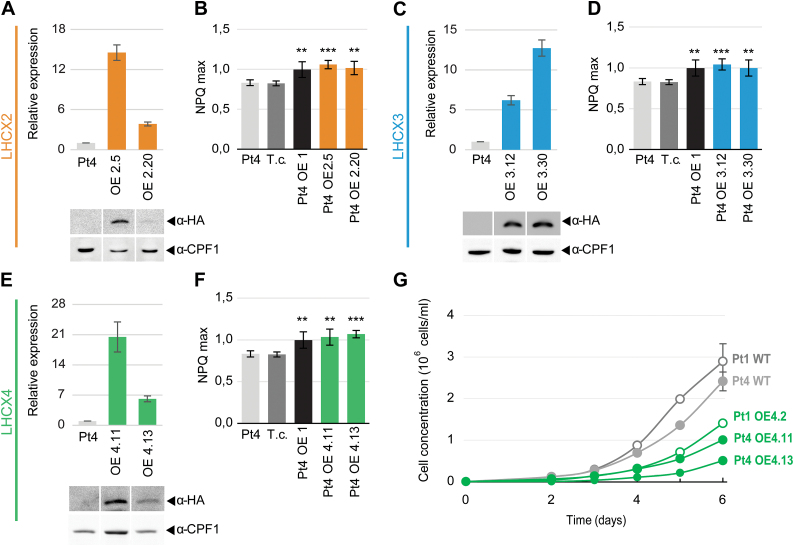
A role in the regulation of the NPQ in P. tricornutum has been proven for the LHCX1 protein by characterizing transgenic lines with a modulated content of LHCX1 by either gene silencing or gene overexpression (Bailleul et al., 2010). Unfortunately, all the attempts to down-regulate the expression of LHCX2, 3, or 4 have been unsuccessful. Therefore, to explore their function, we opted for the strategy used in Bailleul et al. (2010), and tried to rescue the intrinsically lower NPQ capacity of the Pt4 ecotype. To this end, independent transgenic Pt4 lines were generated, bearing a vector in which the LHCX2, 3, or 4 genes were expressed under the control of the P. tricornutum FCPB (LHCF2) promoter. A HA-tag was fused to the C-terminal end of the LHCX transgenes to allow the specific detection of the transgenic proteins. qRT-PCR and western blot analyses (Fig. 4A, C, E) on independent transgenic lines confirmed the expression of the transgenic LHCX isoforms. NPQ analyses (Fig. 4B, D, F) showed that the overexpression of each LHCX isoform generated a modest, but statistically significant, increase in the NPQ capacity compared with the Pt4 wild type as well as compared with a transgenic line transformed only with the antibiotic resistance gene and used as control. Strikingly, we found that all the transgenic lines showed a similar NPQ increase, regardless of which isoform was overexpressed and the different overexpression levels.
Fig. 4.
Phaeodactylum tricornutum Pt4 ecotype lines overexpressing the LHCX genes. (A), (C), (E) LHCX transcript (upper panels) and protein (lower panels) analyses in the Pt4 wild type and transgenic strains overexpressing HA-tagged LHCX2 (A), LHCX3 (C), or LHCX4 (E). Transcript abundance was measured by qRT-PCR using RPS as the reference gene and normalized to the wild type expression value. Tagged proteins were detected by immunoblot using an anti-HA antibody, and an anti-CPF1 antibody as loading control. Bands are taken from the same blots but from non-adjacent lanes. (B), (D), (F) NPQ max capacity in the Pt4 wild type, in a transgenic strain expressing the vector for antibiotic resistance (transformation control, T.c.), and in independent transgenic lines overexpressing LHCX1 (OE1), LHCX2 (OE2), LHCX3 (OE3), and LHCX4 (OE4) genes. Asterisks indicate the results of two-tailed Student t-tests: **p<0.01; ***p<0.001. (G) Growth curves of Pt4 and Pt1 wild-type strains and Pt4 and Pt1 transgenic lines overexpressing the LHCX4 (OE4) gene, grown in 12L/12D cycles (50 µmol m−2 s−1). In all the experiments, n≥3, and bars represent ±SD. (This figure is available in colour at JXB online).
We also checked the possible effect of LHCX overexpression on growth and photosynthetic capacity. For the lines overexpressing the LHCX2 and LHCX3 proteins, we did not observe any altered phenotype (Table 2). In contrast, the Pt4 lines overexpressing LHCX4 showed a reduced PSII efficiency (Table 2). By performing a growth curve analysis, we also observed that these overexpressing lines showed a lag phase lasting 2–3 d (Fig. 4G), which was not the case in wild-type cells. A similar effect on growth was also observed in transgenic lines in which the LHCX4 gene was overexpressed in the Pt1 ecotype (Fig. 4G).
Discussion
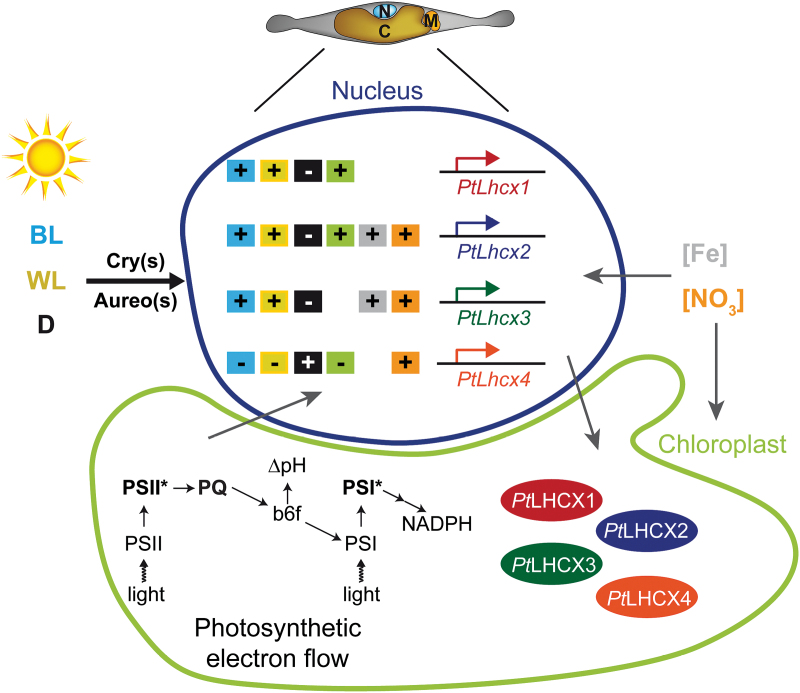
The presence of multiple LHCX genes in all the diatom genomes analysed to date strongly suggests that the expansion of this gene family is a common feature of these algae and may represent an adaptive trait to cope with highly variable environmental conditions. To investigate this scenario, in this work we correlated LHCX expression profiles with the photosynthetic and photoprotective performances in variable experimental conditions, including changes in light irradiance and nutrient availability. These analyses revealed that the four P. tricornutum LHCX genes respond differently to various environmental cues, as summarized in Fig. 5.
Fig. 5.
Model of the P. tricornutum LHCX regulation. Scheme summarizing the multiple external signals and stresses that differentially regulate the expression of the four LHCXs. The LHCX genes are shown in the nucleus and the LHCX proteins in the chloroplast. + and – boxes indicate positive and negative transcriptional regulation, respectively, in response to white light (yellow), blue light (blue, through the cryptochromes, Cry, and aureochromes, Aureo, photoreceptors), darkness (black), chloroplast signals (green), iron starvation (grey), and nitrogen starvation (orange). In the P. tricornutum cell: N, nucleus; C, chloroplast; M, mitochondrion. In the chloroplast: PSI and PSII, photosystem I and II, respectively; PSI* and PSII*, excited photosystems; PQ, plastoquinone pool; b6f, cytochrome b 6 f complex; ΔpH, proton gradient; NADPH, redox potential.
The analyses of the mRNA and protein responses indicate that amounts of the different LHCXs are tightly regulated at the transcriptional, and probably also the post-translational level. As LHCX3 and LHCX4 have a similar size, it was not possible to quantify the amount of these two proteins under the different stresses using one-dimensional electrophoresis. However, considering the transcript and biochemical analyses together (in the case of LHCX2 and LHCX1), it seems that LHCX1 is always expressed at high levels even in non-stress conditions, which is consistent with it having a pivotal role in NPQ regulation and light acclimation as proposed previously (Bailleul et al., 2010).
LHCX2 and 3 are induced following high light stress, where they may contribute to increase the diatom photoprotection capacity. Their induction, as well as the accumulation of LHCX1, may result from the integration of different signals. Two members of the blue light-sensing cryptochrome photolyase family, CPF1 (Coesel et al., 2009) and CRYP (Juhas et al., 2014), modulate the light-dependent expression of LHCX1, LHCX2, and LHCX3. Also, the recently identified Aureochrome 1a blue light photoreceptor, which regulates P. tricornutum photoacclimation (Schellenberger Costa et al., 2013), may affect how much of each LHCX there is in a cell. Moreover, chloroplast activity, through the redox state of the plastoquinone pool, may also regulate LHCX1 and LHCX2 gene expression in HL (Lepetit et al., 2013).
A different regulation pattern is seen in the case of LHCX4, the only isoform which is induced in the absence of light. The amount of LHCX4 mRNA rapidly decreases following a dark to light transition, and this repression is lost when photosynthesis is halted with the PSII inhibitor DCMU. This suggests that chloroplast-derived signals could participate in inhibiting gene expression, even at very low light irradiance, by an as yet unknown process. The peculiar trend observed in the LHCX4 light response suggests a possible role for this protein in P. tricornutum photoacclimation. The increased LHCX4 transcript and possibly protein content is mirrored by a decrease in NPQ capacity and a slightly reduced F v/F m in the dark-adapted cells, compared with cells grown in the light (Fig. 1E; Table 2). Moreover, reduced PSII efficiency and slightly altered growth were observed in cells overexpressing LHCX4 in the light (Fig. 4G), suggesting that LHCX4 could have a negative impact on chloroplast physiology. Indeed, a comparative analysis of the P. tricornutum LHCX protein sequences indicates that LHCX4 lacks key protonatable residues that in Chlamydomonas are involved in NPQ onset when the lumen acidifies (Ballottari et al., 2016). These residues are, however, conserved in the LHCX1, 2, and 3 isoforms. According to the model established in green algae for the protein LHCSR3, these residues diminish their electrostatic repulsion upon protonation, allowing a rearrangement of the protein structure and pigment orientation and enhancement of the quenching capacity (Ballottari et al., 2016). The substitution in LHCX4 of the acidic residues (aspartate and glutamate) with non-protonatable residues (asparagine and glycine) would prevent such regulation. Instead, LHCX4 could contribute to the observed capacity of P. tricornutum to survive long periods in the dark and its repression could be needed for a rapid acclimation following re-illumination (Nymark at al., 2013). Consistent with this, high LHCX gene expression has also been observed in sea-ice algal communities dominated by diatoms that have adapted to the polar night (Pearson et al., 2015).
Besides the light and redox signals discussed above, our study also shows that differences in the availability of iron and nitrogen strongly affect the expression of the different LHCXs. The signalling cascades controlling these responses are still largely unknown, but they probably involve multiple regulatory pathways into the nucleus and chloroplast, considering that these nutrients are essential for diatom photosynthesis and growth (Table 2; Fig. 5). Nitrogen starvation induces a general increase of all the LHCX isoforms, including LHCX4 that is normally repressed in light-grown cells (Fig. 3). We can hypothesize that the general increase of the LHCX content is needed to protect the photosynthetic apparatus, which is strongly affected by nitrogen deprivation, as shown by the drastically reduced F v/F m (Table 2), the lower maximal rETRPSII (Fig. 3D), and the reduction of PSI and PSII protein content. Interestingly, an opposite trend is observed for the main enzymes of the xanthophyll cycle, which are either not induced or are repressed in cells grown in similar nitrogen stress conditions (Supplementary Fig. S3). Thus, in nitrogen starvation, the LHCXs could represent the major contributors to the observed NPQ increase (Fig. 3C).
At variance with nitrogen starvation, iron starvation has a more specific effect on LHCX expression. A strong induction of the LHCX2 mRNA and protein levels (Fig. 2) compared with the other isoforms was observed, pinpointing this isoform as the most likely regulator of the increased NPQ capacity observed in iron stress (Fig. 2C). NPQ in iron-limiting conditions is characterized by a slow induction and a complete relaxation in the dark. These slow induction kinetics might reflect either lower concentrations of the pH-activated de-epoxidase enzyme or its cofactor ascorbate (Grouneva et al., 2006) or slower acidification of the thylakoid lumen due to a reduced photosynthetic activity. Indeed, the photosynthetic capacity is severely impaired when iron is limiting, as demonstrated by the reduction in PSI and PSII subunits (Fig. 2B), but also the lower F v/F m (Table 2) and rETRPSII (Fig. 2D). The decreased electron flow per PSII could also reflect a decrease in the iron-containing cytochrome b 6 f complex, as previously shown for other iron-limited diatoms (Strzepek and Harrison, 2004; Thamatrakoln et al., 2013).
The observations made in this and in previous studies about the complex LHCX regulation in response to different signals prompted us to explore their possible functions in P. tricornutum, by modulating their expression in a natural Pt4 strain characterized by constitutive lower NPQ levels (Fig. 4). We observed that the increased expression of all the tested isoforms generates a small but still consistent increase in the NPQ levels, suggesting a potential involvement of the diverse proteins in NPQ modulation, as previously shown for LHCX1 (Bailleul et al., 2010). However, we also noticed that different overexpressing lines with different transcript and protein levels showed a similar NPQ increase. It is difficult to interpret these first results, especially in the case of lines overexpressing LHCX4, whose endogenous expression is inhibited by light (Fig. 1F). They probably reflect the complexity of NPQ regulation in diatoms, where the presence of multiple players (e.g. several LHCXs and enzymes of the xanthophyll cycle) possibly tend to reduce the consequences on NPQ of genetic modifications of the qE machinery.
Finally, the exploration of the 5'-flanking regions and intronic sequences of the LHCX genes revealed the presence of known and potentially novel cis-regulatory elements that may contribute to the transcriptional regulation of the different isoforms in stress conditions. We revealed an uneven distribution of the CCREs (Ohno et al., 2012; Tanaka et al., 2016) in the four LHCX genes that may be linked to their different light-mediated transcriptional responses. In addition, we identified a 7bp motif in the non-coding sequences of LHCX2, 3, and 4. Using genome-wide transcriptomic data, we found this motif specifically enriched in long-term nitrogen starvation-induced genes, suggesting a possible involvement in the regulation of gene expression in response to nitrogen fluctuations. Although additional studies are required to demonstrate the functionality of these motifs, their discovery may represent a starting point for the identification of the LHCX regulators in the diatom acclimation mechanisms to stress.
Outlook
Here we discovered that the four P. tricornutum LHCXs are regulated in a sophisticated way (Fig. 5). Different and probably interconnected regulatory pathways activated by different signals and stresses tightly control the amount of each LHCX isoform in the cell. By narrowing down the specific growth conditions in which the different LHCXs are required, our results set the basis for future work to define the function of each isoform in the regulation of chloroplast physiology. The generation of new transgenic lines in which the content of each LHCX isoform is specifically modulated will be instrumental in assessing whether they act with the NPQ regulator LHCX1, or play other specific roles. Considering the robustness of LHCX1 expression in all the conditions tested, future studies will probably require the use of new LHCX1 loss-of-function diatom strains. Additional information about the association of LHCXs with photosynthetic complexes and pigments will also be necessary to understand the role played by the expanded LHCX gene family in the efficient acclimation of diatoms to environmental changes.
Supplementary data
Supplementary data are available at JXB online.
Figure S1. Localization of the enriched motifs in non-coding regions of P. tricornutum LHCX genes.
Figure S2. Alignment of the LHCX proteins and three-dimensional model of LHCX1.
Figure S3. Expression of the P. tricornutum xanthophyll cycle genes in nitrogen starvation.
Table S1. List of the oligonucleotides used in this work.
Acknowledgements
This work was supported by Marie-Curie ITNs CALIPSO (ITN 2013 GA 607607) and AccliPhot (ITN 2012 GA 316427) grants and a Gordon and Betty Moore Foundation grant (GBMF 4966) to AF, the French ANR ‘DiaDomOil’ PROGRAMME BIO-MATIERES & ENERGIES to AF and GF, and the Marie Curie Zukunftskolleg Incoming Fellowship and a Zukunftskolleg Interim Grant to BL.
References
- Alboresi A, Gerotto C, Giacometti GM, Bassi R, Morosinotto T. 2010. Physcomitrella patens mutants affected on heat dissipation clarify the evolution of photoprotection mechanisms upon land colonization. Proceedings of the National Academy of Sciences, USA 107, 11128–11133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alipanah L, Rohloff J, Winge P, Bones AM, Brembu T. 2015. Whole-cell response to nitrogen deprivation in the diatom Phaeodactylum tricornutum . Journal of Experimental Botany 66, 6281–6296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen AE, Dupont CL, Obornik M, et al. 2011. Evolution and metabolic significance of the urea cycle in photosynthetic diatoms. Nature 473, 203–207. [DOI] [PubMed] [Google Scholar]
- Allen AE, Laroche J, Maheswari U, Lommer M, Schauer N, Lopez PJ, Finazzi G, Fernie AR, Bowler C. 2008. Whole-cell response of the pennate diatom Phaeodactylum tricornutum to iron starvation. Proceedings of the National Academy of Sciences, USA 105, 10438–10443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apt KE, Kroth-Pancic PG, Grossman AR. 1996. Stable nuclear transformation of the diatom Phaeodactylum tricornutum . Molecular and General Genetics 252, 572–579. [DOI] [PubMed] [Google Scholar]
- Armbrust EV, Berges JA, Bowler C, et al. 2004. The genome of the diatom Thalassiosira pseudonana: ecology, evolution, and metabolism. Science 306, 79–86. [DOI] [PubMed] [Google Scholar]
- Arrigo KR, Perovich DK, Pickart RS, et al. 2012. Massive phytoplankton blooms under arctic sea ice. Science 336, 1408–1408. [DOI] [PubMed] [Google Scholar]
- Ashworth J, Coesel S, Lee A, Armbrust EV, Orellana MV, Baliga NS. 2013. Genome-wide diel growth state transitions in the diatom Thalassiosira pseudonana . Proceedings of the National Academy of Sciences, USA 110, 7518–7523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren JY, Li WW, Noble WS. 2009. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Research 37, W202–W208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailleul B, Berne N, Murik O, et al. 2015. Energetic coupling between plastids and mitochondria drives CO2 assimilation in diatoms. Nature 524, 366–369. [DOI] [PubMed] [Google Scholar]
- Bailleul B, Rogato A, de Martino A, Coesel S, Cardol P, Bowler C, Falciatore A, Finazzi G. 2010. An atypical member of the light-harvesting complex stress-related protein family modulates diatom responses to light. Proceedings of the National Academy of Sciences, USA 107, 18214–18219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballottari M, Truong TB, De Re E, Erickson E, Stella GR, Fleming GR, Bassi R, Niyogi KK. 2016. Identification of pH-sensing sites in the Light Harvesting Complex Stress-Related 3 protein essential for triggering non-photochemical quenching in Chlamydomonas reinhardtii . Journal of Biological Chemistry 291, 7334–7346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker F, Rhiel E. 2006. Immuno-electron microscopic quantification of the fucoxanthin chlorophyll a/c binding polypeptides Fcp2, Fcp4, and Fcp6 of Cyclotella cryptica grown under low- and high-light intensities. International Microbiology 9, 29–36. [PubMed] [Google Scholar]
- Beer A, Juhas M, Buchel C. 2011. Influence of different light intensities and different iron nutrition on the photosynthetic apparatus in the diatom Cyclotella meneghiniana (Bacillariophyceae). Journal of Phycology 47, 1266–1273. [DOI] [PubMed] [Google Scholar]
- Bilger W, Björkman O. 1990. Role of the xanthophyll cycle in photoprotection elucidated by measurements of light-induced absorbance changes, fluorescence and photosynthesis in leaves of Hedera canariensis . Photosynthesis Research 25, 173–185. [DOI] [PubMed] [Google Scholar]
- Bowler C, Allen AE, Badger JH, et al. 2008. The Phaeodactylum genome reveals the evolutionary history of diatom genomes. Nature 456, 239–244. [DOI] [PubMed] [Google Scholar]
- Coesel S, Mangogna M, Ishikawa T, Heijde M, Rogato A, Finazzi G, Todo T, Bowler C, Falciatore A. 2009. Diatom PtCPF1 is a new cryptochrome/photolyase family member with DNA repair and transcription regulation activity. EMBO Reports 10, 655–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daboussi F, Leduc S, Maréchal A, et al. 2014. Genome engineering empowers the diatom Phaeodactylum tricornutum for biotechnology. Nature Communications 5, 3831. [DOI] [PubMed] [Google Scholar]
- de Baar HJW, Boyd PW, Coale KH, et al. 2005. Synthesis of iron fertilization experiments: from the iron age in the age of enlightenment. Journal of Geophysical Research-Oceans 110, C09S16. [Google Scholar]
- De Martino A, Meichenin A, Shi J, Pan K, Bowler C. 2007. Genetic and phenotypic characterization of Phaeodactylum tricornutum (Bacillariophyceae) accessions. Journal of Phycology 43, 992–1009. [Google Scholar]
- Depauw FA, Rogato A, Ribera d’Alcala M, Falciatore A. 2012. Exploring the molecular basis of responses to light in marine diatoms. Journal of Experimental Botany 63, 1575–1591. [DOI] [PubMed] [Google Scholar]
- De Riso V, Raniello R, Maumus F, Rogato A, Bowler C, Falciatore A. 2009. Gene silencing in the marine diatom Phaeodactylum tricornutum . Nucleic Acids Research 37, e96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyhrman ST, Jenkins BD, Rynearson TA, et al. 2012. The transcriptome and proteome of the diatom Thalassiosira pseudonana reveal a diverse phosphorus stress response. PLoS One 7, e33768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberhard S, Finazzi G, Wollman FA. 2008. The dynamics of photosynthesis. Annual Review of Genetics 42, 463–515. [DOI] [PubMed] [Google Scholar]
- Falciatore A, Casotti R, Leblanc C, Abrescia C, Bowler C. 1999. Transformation of nonselectable reporter genes in marine diatoms. Marine Biotechnology 1, 239–251. [DOI] [PubMed] [Google Scholar]
- Falkowski PG, Katz ME, Knoll AH, Quigg A, Raven JA, Schofield O, Taylor FJR. 2004. The evolution of modern eukaryotic phytoplankton. Science 305, 354–360. [DOI] [PubMed] [Google Scholar]
- Field CB, Behrenfeld MJ, Randerson JT, Falkowski P. 1998. Primary production of the biosphere: integrating terrestrial and oceanic components. Science 281, 237–240. [DOI] [PubMed] [Google Scholar]
- Fortunato AE, Annunziata R, Jaubert M, Bouly JP, Falciatore A. 2015. Dealing with light: the widespread and multitasking cryptochrome/photolyase family in photosynthetic organisms. Journal of Plant Physiology 172, 42–54. [DOI] [PubMed] [Google Scholar]
- Fortunato AE, Jaubert M, Enomoto G, et al. 2016. Diatom phytochromes reveal the existence of far-red light based sensing in the ocean. The Plant Cell 28, 616–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerotto C, Alboresi A, Giacometti GM, Bassi R, Morosinotto T. 2011. Role of PSBS and LHCSR in Physcomitrella patens acclimation to high light and low temperature. Plant, Cell and Environment 34, 922–932. [DOI] [PubMed] [Google Scholar]
- Goss R, Jakob T. 2010. Regulation and function of xanthophyll cycle-dependent photoprotection in algae. Photosynthesis Research 106, 103–122. [DOI] [PubMed] [Google Scholar]
- Goss R, Lepetit B. 2015. Biodiversity of NPQ. Journal of Plant Physiology 172, 13–32. [DOI] [PubMed] [Google Scholar]
- Gross M. 2012. The mysteries of the diatoms. Current Biology 22, R581–R585. [DOI] [PubMed] [Google Scholar]
- Grouneva I, Jakob T, Wilhelm C, Goss R. 2006. Influence of ascorbate and pH on the activity of the diatom xanthophyll cycle-enzyme diadinoxanthin de-epoxidase. Physiologia Plantarum 126, 205–211. [Google Scholar]
- Guillard RRL. 1975. Culture of phytoplankton for feeding marine invertebrates. In: Smith WL, Chanley MH, eds. Culture of marine invertebrate animals. New York: Plenum Press, 26–60. [Google Scholar]
- Hockin NL, Mock T, Mulholland F, Kopriva S, Malin G. 2012. The response of diatom central carbon metabolism to nitrogen starvation is different from that of green algae and higher plants. Plant Physiology 158, 299–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohner R, Barth J, Magneschi L, Jaeger D, Niehues A, Bald T, Grossman A, Fufezan C, Hippler M. 2013. The metabolic status drives acclimation of iron deficiency responses in Chlamydomonas reinhardtii as revealed by proteomics based hierarchical clustering and reverse genetics. Molecular and Cellular Proteomics 12, 2774–2790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huysman MJ, Fortunato AE, Matthijs M, et al. 2013. AUREOCHROME1a-mediated induction of the diatom-specific cyclin dsCYC2 controls the onset of cell division in diatoms (Phaeodactylum tricornutum). The Plant Cell 25, 215–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvis P, Lopez-Juez E. 2013. Biogenesis and homeostasis of chloroplasts and other plastids. Nature Reviews Molecular Cell Biology 14, 787–802. [DOI] [PubMed] [Google Scholar]
- Johnson X, Vandystadt G, Bujaldon S, Wollman FA, Dubois R, Roussel P, Alric J, Beal D. 2009. A new setup for in vivo fluorescence imaging of photosynthetic activity. Photosynthesis Research 102, 85–93. [DOI] [PubMed] [Google Scholar]
- Juhas M, von Zadow A, Spexard M, Schmidt M, Kottke T, Buchel C. 2014. A novel cryptochrome in the diatom Phaeodactylum tricornutum influences the regulation of light-harvesting protein levels. FEBS Journal 281, 2299–2311. [DOI] [PubMed] [Google Scholar]
- Karas BJ, Diner RE, Lefebvre SC, et al. 2015. Designer diatom episomes delivered by bacterial conjugation. Nature Commununications 6, 6925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeling PJ, Burki F, Wilcox HM, et al. 2014. The Marine Microbial Eukaryote Transcriptome Sequencing Project (MMETSP): illuminating the functional diversity of eukaryotic life in the oceans through transcriptome sequencing. PLoS Biology 12, e1001889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kooistra WHCF, Gersonde R, Medlin LK, Mann DG. 2007. The origin and evolution of the diatoms: their adaptation to a planktonic existence. In: Falkowski PG, Knoll AH, eds. Evolution of planktonic photoautotrophs. Burlington, MA: Academic Press, 207–249. [Google Scholar]
- Laroche J, Murray H, Orellana M, Newton J. 1995. Flavodoxin expression as an indicator of iron limitation in marine diatoms. Journal of Phycology 31, 520–530. [Google Scholar]
- Lavaud J, Goss R. 2014. The peculiar features of non-photochemical fluorescence quenching in diatoms and brown algae. In: Demmig-Adams B, Garab G, Adams WW, III, Govindjee, eds. Non-photochemical quenching and energy dissipation in plants, algae and cyanobacteria, Dordretch, The Netherlands: Springer, 421–443. [Google Scholar]
- Lavaud J, Materna AC, Sturm S, Vugrinec S, Kroth PG. 2012. Silencing of the violaxanthin de-epoxidase gene in the diatom Phaeodactylum tricornutum reduces diatoxanthin synthesis and non-photochemical quenching. PLoS One 7, e36806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MJ, Yaffe MB. 2014. Protein regulation in signal transduction. In: Cantley LC, Hunter T, Sever R, Thorner J, eds. Signal transduction: principles, pathways, and processes. Cold Spring Harbor, NY: Cold Sprring Harbor Laboratory Press, 31–50. [Google Scholar]
- Lepetit B, Sturm S, Rogato A, Gruber A, Sachse M, Falciatore A, Kroth PG, Lavaud J. 2013. High light acclimation in the secondary plastids containing diatom Phaeodactylum tricornutum is triggered by the redox state of the plastoquinone pool. Plant Physiology 161, 853–865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levitan O, Dinamarca J, Zelzion E, et al. 2015. Remodeling of intermediate metabolism in the diatom Phaeodactylum tricornutum under nitrogen stress. Proceedings of the National Academy of Sciences, USA 112, 412–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2–ΔΔCt method. Methods 25, 402–408. [DOI] [PubMed] [Google Scholar]
- Lommer M, Specht M, Roy AS, et al. 2012. Genome and low-iron response of an oceanic diatom adapted to chronic iron limitation. Genome Biology 13, R66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchetti A, Schruth DM, Durkin CA, Parker MS, Kodner RB, Berthiaume CT, Morales R, Allen AE, Armbrust EV. 2012. Comparative metatranscriptomics identifies molecular bases for the physiological responses of phytoplankton to varying iron availability. Proceedings of the National Academy of Sciences, USA 109, E317–E325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margalef R. 1978. Life-forms of phytoplankton as survival alternatives in an unstable environment. Oceanologica Acta 1, 493–509. [Google Scholar]
- Matthijs M, Fabris M, Broos S, Vyverman W, Goossens A. 2016. Profiling of the early nitrogen stress response in the diatom Phaeodactylum tricornutum reveals a novel family of RING-domain transcription factors. Plant Physiology 170, 489–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxwell K, Johnson GN. 2000. Chlorophyll fluorescence: a practical guide. Journal of Experimental Botany 51, 659–668. [DOI] [PubMed] [Google Scholar]
- Mills MM, Moore CM, Langlois R, Milne A, Achterberg E, Nachtigall K, Lochte K, Geider RJ, La Roche J. 2008. Nitrogen and phosphorus co-limitation of bacterial productivity and growth in the oligotrophic subtropical North Atlantic. Limnology and Oceanography 53, 824–834. [Google Scholar]
- Montsant A, Andrew EA, Coesel S, et al. 2007. Identification and comparative genomic analysis of signaling and regulatory components in the diatom Thalassiosira pseudonana . Journal of Phycology 43, 585–604. [Google Scholar]
- Moore CM, Mills MM, Arrigo KR, et al. 2013. Processes and patterns of oceanic nutrient limitation. Nature Geoscience 6, 701–710. [Google Scholar]
- Morrissey J, Sutak R, Paz-Yepes J, et al. 2015. A novel protein, ubiquitous in marine phytoplankton, concentrates iron at the cell surface and facilitates uptake. Current Biology 25, 364–371. [DOI] [PubMed] [Google Scholar]
- Moseley JL, Allinger T, Herzog S, Hoerth P, Wehinger E, Merchant S, Hippler M. 2002. Adaptation to Fe-deficiency requires remodeling of the photosynthetic apparatus. EMBO Journal 21, 6709–6720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhseen ZT, Xiong Q, Chen Z, Ge F. 2015. Proteomics studies on stress responses in diatoms. Proteomics 15, 3943–3953. [DOI] [PubMed] [Google Scholar]
- Nymark M, Valle KC, Brembu T, Hancke K, Winge P, Andresen K, Johnsen G, Bones AM. 2009. An integrated analysis of molecular acclimation to high light in the marine diatom Phaeodactylum tricornutum . PLoS One 4, e7743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nymark M, Valle KC, Hancke K, Winge P, Andresen K, Johnsen G, Bones AM, Brembu T. 2013. Molecular and photosynthetic responses to prolonged darkness and subsequent acclimation to re-illumination in the diatom Phaeodactylum tricornutum . PLoS One 8, e58722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno N, Inoue T, Yamashiki R, Nakajima K, Kitahara Y, Ishibashi M, Matsuda Y. 2012. CO(2)-cAMP-responsive cis-elements targeted by a transcription factor with CREB/ATF-like basic zipper domain in the marine diatom Phaeodactylum tricornutum . Plant Physiology 158, 499–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmucci M, Ratti S, Giordano M. 2011. Ecological and evolutionary implications of carbon allocation in marine phytoplankton as a function of nitrogen availability: a Fourier transform infrared spectroscopy approach. Journal of Phycology 47, 313–323. [DOI] [PubMed] [Google Scholar]
- Pearson GA, Lago-Leston A, Cánovas F, Cox CJ, Verret F, Lasternas S, Duarte CM, Agusti S, Serrão EA. 2015. Metatranscriptomes reveal functional variation in diatom communities from the Antartic Peninsula. ISME Journal 9, 2275–2289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peers G, Truong TB, Ostendorf E, Busch A, Elrad D, Grossman AR, Hippler M, Niyogi KK. 2009. An ancient light-harvesting protein is critical for the regulation of algal photosynthesis. Nature 462, 518–521. [DOI] [PubMed] [Google Scholar]
- Poulsen N, Kroger N. 2005. A new molecular tool for transgenic diatoms—control of mRNA and protein biosynthesis by an inducible promoter–terminator cassette. FEBS Journal 272, 3413–3423. [DOI] [PubMed] [Google Scholar]
- Rayko E, Maumus F, Maheswari U, Jabbari K, Bowler C. 2010. Transcription factor families inferred from genome sequences of photosynthetic stramenopiles. New Phytologist 188, 52–66. [DOI] [PubMed] [Google Scholar]
- Reeves S, McMinn A, Martin A. 2011. The effect of prolonged darkness on the growth, recovery and survival of Antarctic sea ice diatoms. Polar Biology 34, 1019–1032. [Google Scholar]
- Rochaix JD. 2011. Assembly of the photosynthetic apparatus. Plant Physiology 155, 1493–1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogato A, Amato A, Iudicone D, Chiurazzi M, Ferrante MI, d’Alcala MR. 2015. The diatom molecular toolkit to handle nitrogen uptake. Marine Genomics 24, 95–108. [DOI] [PubMed] [Google Scholar]
- Schellenberger Costa B, Sachse M, Jungandreas A, Bartulos CR, Gruber A, Jakob T, Kroth PG, Wilhelm C. 2013. Aureochrome 1a is involved in the photoacclimation of the diatom Phaeodactylum tricornutum . PLoS One 8, e74451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siaut M, Heijde M, Mangogna M, Montsant A, Coesel S, Allen A, Manfredonia A, Falciatore A, Bowler C. 2007. Molecular toolbox for studying diatom biology in Phaeodactylum tricornutum . Gene 406, 23–35. [DOI] [PubMed] [Google Scholar]
- Sicko-Goad L, Stoermer EF, Kociolek JP. 1989. Diatom resting cell rejuvenation and formation—time course, species records and distribution. Journal of Plankton Research 11, 375–389. [Google Scholar]
- Stookey LL. 1970. Ferrozine—a new spectrophotometric reagent for iron. Analytical Chemistry 42, 779–781. [Google Scholar]
- Strzepek RF, Harrison PJ. 2004. Photosynthetic architecture differs in coastal and oceanic diatoms. Nature 431, 689–692. [DOI] [PubMed] [Google Scholar]
- Tanaka A, Ohno N, Nakajima K, Matsuda Y. 2016. Light and CO2/cAMP signal cross talk on the promoter elements of chloroplastic β-carbonic anhydrase genes in the marine diatom Phaeodactylum tricornutum . Plant Physiology 170, 1105–1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thamatrakoln K, Bailleul B, Brown CM, Gorbunov MY, Kustka AB, Frada M, Joliot PA, Falkowski PG, Bidle KD. 2013. Death-specific protein in a marine diatom regulates photosynthetic responses to iron and light availability. Proceedings of the National Academy of Sciences, USA 110, 20123–20128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thamatrakoln K, Korenovska O, Niheu AK, Bidle KD. 2012. Whole-genome expression analysis reveals a role for death-related genes in stress acclimation of the diatom Thalassiosira pseudonana . Environmental Microbiology 14, 67–81. [DOI] [PubMed] [Google Scholar]
- Trainer VL, Bates SS, Lundholm N, Thessen AE, Cochlan WP, Adams NG, Trick CG. 2012. Pseudo-nitzschia physiological ecology, phylogeny, toxicity, monitoring and impacts on ecosystem health. Harmful Algae 14, 271–300. [Google Scholar]
- Trentacoste EM, Shrestha RP, Smith SR, Glé C, Hartmann AC, Hildebrand M, Gerwick WH. 2013. Metabolic engineering of lipid catabolism increases microalgal lipid accumulation without compromising growth. Proceedings of the National Academy of Sciences, USA 110, 19748–19753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valle KC, Nymark M, Aamot I, Hancke K, Winge P, Andresen K, Johnsen G, Brembu T, Bones AM. 2014. System responses to equal doses of photosynthetically usable radiation of blue, green, and red light in the marine diatom Phaeodactylum tricornutum . PLoS One 9, e114211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walters RG. 2005. Towards an understanding of photosynthetic acclimation. Journal of Experimental Botany 56, 435–447. [DOI] [PubMed] [Google Scholar]
- Wilhelm C, Buchel C, Fisahn J, et al. 2006. The regulation of carbon and nutrient assimilation in diatoms is significantly different from green algae. Protist 157, 91–124. [DOI] [PubMed] [Google Scholar]
- Woodson JD, Chory J. 2008. Coordination of gene expression between organellar and nuclear genomes. Nature Reviews Genetics 9, 383–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshinaga R, Niwa-Kubota M, Matsui H, Matsuda Y. 2014. Characterization of iron-responsive promoters in the marine diatom Phaeodactylum tricornutum . Marine Genomics 16, 55–62. [DOI] [PubMed] [Google Scholar]
- Zhu SH, Green BR. 2010. Photoprotection in the diatom Thalassiosira pseudonana: role of LI818-like proteins in response to high light stress. Biochimica et Biophysica Acta 1797, 1449–1457. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.