Abstract
Published gene expression studies for radiation-induced thyroid carcinogenesis have used various methodologies. In this study, we identified differential gene expression in a human thyroid epithelial cell line after exposure to high-dose γ-radiation. HTori-3 cells were exposed to 5 or 10 Gy of ionizing radiation using two dose rates (high-dose rate: 4.68 Gy/min, and low-dose rate: 40 mGy/h) and then implanted into the backs of BALB/c nude mice after 4 (10 Gy) or 5 weeks (5 Gy). Decreases in cell viability, increases in giant cell frequency, anchorage-independent growth in vitro, and tumorigenicity in vivo were observed. Particularly, the cells irradiated with 5 Gy at the high-dose rate or 10 Gy at the low-dose rate demonstrated more prominent tumorigenicity. Gene expression profiling was analyzed via microarray. Numerous genes that were significantly altered by a fold-change of >50% following irradiation were identified in each group. Gene expression analysis identified six commonly misregulated genes, including CRYAB, IL-18, ZNF845, CYP24A1, OR4N4 and VN1R4, at all doses. These genes involve apoptosis, the immune response, regulation of transcription, and receptor signaling pathways. Overall, the altered genes in high-dose rate (HDR) 5 Gy and low-dose rate (LDR) 10 Gy were more than those of LDR 5 Gy and HDR 10 Gy. Thus, we investigated genes associated with aggressive tumor development using the two dosage treatments. In this study, the identified gene expression profiles reflect the molecular response following high doses of external radiation exposure and may provide helpful information about radiation-induced thyroid tumors in the high-dose range.
Keywords: thyroid gland, carcinogenesis, cell line, radiation, gene expression
INTRODUCTION
The thyroid gland is sensitive to ionizing radiation, which is a known risk factor for human malignancy. External radiation exposure (such as therapeutic radiation) or internal radionuclide contamination may result in carcinogenesis and mutation of the thyroid gland [1–3]. Ionizing radiation from diagnostic tests and treatments for benign and malignant conditions has been established as a risk factor for thyroid cancer, particularly with exposure early in life [4]. Environmental irradiation experienced by atomic bomb survivors in Japan and by those who endured the fallout of the Chernobyl accident has been indicated as a cause of thyroid cancer, mainly in people who were exposed at younger ages. Numerous reports have documented the increased rate of thyroid cancer incidence in the children of these affected regions [5–9]. More recently, since the Fukushima nuclear accident, public concerns about the long-term health effects of radiation exposure have increased [10].
Exposure to ionizing radiation induces double-stranded DNA breaks, which leads to chromosomal aberrations and cellular transformations. Malignant cell transformation is caused by specific gene alterations, such as the activation of oncogenes or inactivation of tumor suppressor genes [11]. Radiation-induced forms of thyroid cancer are mainly papillary thyroid carcinomas (PTCs), which are also predominantly found in sporadic tumors. As in other cancers, there are several gene alterations that are thought to be involved in thyroid carcinogenesis. Among them, ‘rearranged during transfection’ (RET)/PTC rearrangements have been observed in radiation-associated PTCs, which develop after exposure to radioiodine or external radiation [12–13]. Also, RET/PTC rearrangements are preferentially induced in a dose-dependent manner with high dosages of radiation in vitro and in vivo [14–15]. The exact biological mechanism of radiation-induced thyroid carcinogenesis mediated by genetic alteration has not been demonstrated until now. Additionally, genetic changes related to carcinogenesis are accompanied by other genetic and epigenetic alterations [16–17]. Thus, gene expression profiling has been used to investigate the possible mechanisms of carcinogenesis [18–19]; in most cases, these genetic profiles have been assessed via microarray technology. Gene expression analysis has identified a number of genes associated with radiation-induced thyroid tumors in sporadic tumors. Particularly, studies of post-Chernobyl tumors and post-radiotherapy thyroid tumors using human thyroid tissue have been published [20–24]. Of these reports, there are fewer published studies of post-radiotherapy tumors compared with those of post-Chernobyl tumors.
Additionally, based on an in vitro cellular system, the genes identified in radiation-induced thyroid tumors were found to be misregulated. The HTori-3 cell line is an immortalized and non-tumorigenic human primary thyroid follicular epithelial cell line transfected with the simian virus 40 genome. The resulting immortalized cell line has unique functions, such as iodide trapping and thyroglobulin production, and does not produce tumors in nude mice, despite its growth factor–independent and anchorage-independent growth at low frequency [25].
Taking advantage of these characteristics of the HTori-3 cell line has progressed our understanding of radiation-induced thyroid carcinogenesis. For example, as a result of these features, many studies on chromosomal change, RET/PTC rearrangement, adverse biological effects, and selenomethionine-mediated protection against these adverse biological effects have been performed [14, 26–31]. However, gene expression profiling has only documented the alterations in gene expression caused by space radiation [29–30]; so far only the generation of RET/PTC rearrangements has been studied following exposure to γ-radiation [14].
The purpose of this study was to identify altered gene expression profiles following exposure to high doses of γ-radiation using the HTori-3 cell line.
MATERIALS AND METHODS
Cell culture
HTori-3 cells, which are normal human thyroid cells, were purchased from the European Collection of Cell Cultures (ECACC, UK). The cells were suspended in medium [consisting of equal volumes of Dulbecco's modified Eagle's medium (DMEM, Gibco) and Ham's F12 medium (Sigma) supplemented with 7% fetal bovine serum (FBS), 100 units/ml penicillin, 100 µg/ml streptomycin (Gibco, USA) and 2 mM L-glutamine (Sigma)] and cultured at 37°C in a humidified incubator with a mixture of 95% air and 5% CO2. TPC-1 cells, which are thyroid cancer cells that include the RET/PTC proto-oncogene, were obtained from Marina N. Nikiforova (Ohio State University, USA) and used as the positive control. RAW 264.7 cells, which are murine macrophage cells, were purchased from the American Type Culture Collection (ATCC) and used as the negative control. TPC-1 and RAW 264.7 cells were suspended in DMEM supplemented with 10% FBS and cultured in the same environment.
Irradiation
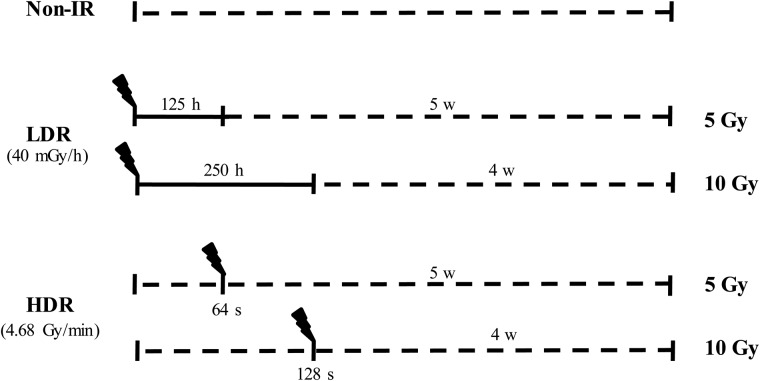
Cells were plated in T75 flasks at one day prior to irradiation. For chronic irradiation, cells were irradiated with 5 Gy (for 125 h) and 10 Gy (for 250 h) using a long-term low-dose-rate irradiation facility (137Cs, 40 mGy/h). In the case of acute exposure, cells were exposed to ionizing radiation (5 Gy for 64 s, or 10 Gy for 128 s) from a 137Cs γ-ray source (4.68 Gy/min, IBL 437C, CIS Bio international, France) at the end of chronic irradiation. After irradiation, both experimental and control cells were subcultured for an additional 4 (10 Gy) or 5 (5 Gy) weeks and were used for the proliferation assay, morphological examination, colony-forming assay, tumorigenicity assay or microarray (Fig. 1).
Fig. 1.
Schematic representation of irradiation protocols. In case of low-dose-rate irradiation, HTori-3 cells were irradiated at 40 mGy/h for 125 h (5 Gy) or 250 h (10 Gy). A high-dose-rate radiation group was irradiated at 4.68 Gy/min for 64 s (5 Gy) or 128 s (10 Gy) (the solid line represents the radiation-irradiated period; the dotted line represents non-irradiated period).
Cell viability assay
The cells were collected in 35-mm dishes (Nunc), irradiated with 0.5, 1, 10 or 30 Gy after 2 d, and incubated for 6–48 h. We confirmed the presence of formazan crystals after 2 h of incubation with the MTT solution (0.1 mg/dish; Sigma). The formazan crystals in each dish were dissolved in 700 µl DMSO (Sigma) and transferred to 96-well plates (200 µl in each of three wells). Spectrophotometry was performed at 540 nm using an iEMS Reader MF (Labsystems, USA).
Morphological examination
The cells were seeded into a 4-well glass chamber slide system (Nunc; 1 × 104 cells/well). After 3 d, the media chamber and the gasket were removed from slides and washed twice with ×1 PBS. The cells on the slides were fixed by adding 3.7% paraformaldehyde (Sigma) at room temperature for 15 min. After washing the cells twice with ×1 PBS, they were mounted in a few drops of crystal/mountTM (Biomeda, USA) with a coverslip and dried at room temperature for 2 h. Imaging was performed with an Apotome microscope (Carl ZEISS, Germany).
Colony-forming assay
One milliliter of medium containing 1% agar was dropped in each well of a 6-well plate (Nunc) and solidified at room temperature. Additionally, a mixture of 1 ml of medium containing 0.3% agar and HTori-3 cells (1 × 103) was seeded in each well. Approximately 200 µl of medium was added every 2–3 d. After incubation for 1 month and staining with iodonitrotetrazolium chloride (Sigma), colonies were photographed with a digital camera and the acquired images were analyzed using the ImageJ software.
Tumorigenicity assay
All mice were housed and maintained at the mouse laboratory at the Radiation Health Institute, in a controlled environment. Four-week-old female BALB/c nude mice (SLC, Japan) were stabilized and managed in a specific pathogen-free (SPF) facility for 1 week. Cell lines were cultured in T-75 flasks. Cells were trypsinized (Gibco) and washed twice with sterilized cold saline at 4°C. The isolated cells (2 × 106) were diluted with 150 µl of saline and injected subcutaneously into the backs of the mice. After the presence of a tumor was noted, the size of the tumor was measured with a caliper once per week.
Histological examination
When the tumor was 1–2 cm in diameter, the tissue was removed from the mouse and the cells were separated for histological examinations. The tissues were stored in 10% buffered formalin and referred to the Department of Pathology, College of Veterinary Medicine, Seoul National University.
RNA isolation
Total RNA for microarrays was isolated using TRIzol reagent (Invitrogen). Cancer cells isolated from the tumors were seeded into 100-mm dishes (Nunc; 7.5 × 105 cells/dish). After 2 d, the medium was discarded, 1 ml of TRIzol reagent was added, and the RNA was harvested: the cells were kept in TRIzol reagent for 5 min at room temperature, 0.2 ml chloroform was added, and the cells were shaken by hand, incubated for 2 min, and centrifuged at 12 000 rpm for 15 min at 4°C.
RT-PCR
cDNA was generated from 2 µg of total RNA per sample using the ImProm-IITM Reverse Transcription System (Promega). A mixture of RNA and 0.5 µg oligo(dT) primer was heated for 5 min at 70°C and incubated for 5 min on ice. Then, the reverse transcription reaction was performed by adding the remnants of the reaction mixture. The reaction was stopped with extension for 1 h at 42°C followed by heating the samples for 15 min at 70°C using the GeneAmp 2400 PCR System (Perkin-Elmer). Two microliters of cDNA was added to the PCR mixture with ×1 PCR buffer, 1.5 mM MgCl2, 20 pmole primer mix, 1 mM dNTP mix, and 2.5 U Top DNA polymerase (Bioneer). Primer sequences were as follows: Human GAPDH sense AGCCTCCCGCTTCGCTCTCT, antisense CCAGGCGCCCAATACGACCA; and mouse GAPDH sense CTGTTCCAGAGACGGCCGCA, antisense CAGGCGCCCAATACGGCCAA.
Microarray
The microarrays on the cells isolated from tumors and the purified RNA isolated from the HTori-3 cell line were performed using the Affymetrix Gene Chip Human Gene 1.0 ST Array (Affymetrix).
Statistical analysis
All data are presented as the mean ± standard deviation. The statistical analysis was performed using Prism 5 (GraphPad, San Diego, CA). All analyses were done using the Student's t-test.
RESULTS
HTori-3 cells exhibit decreased cell survival following irradiation
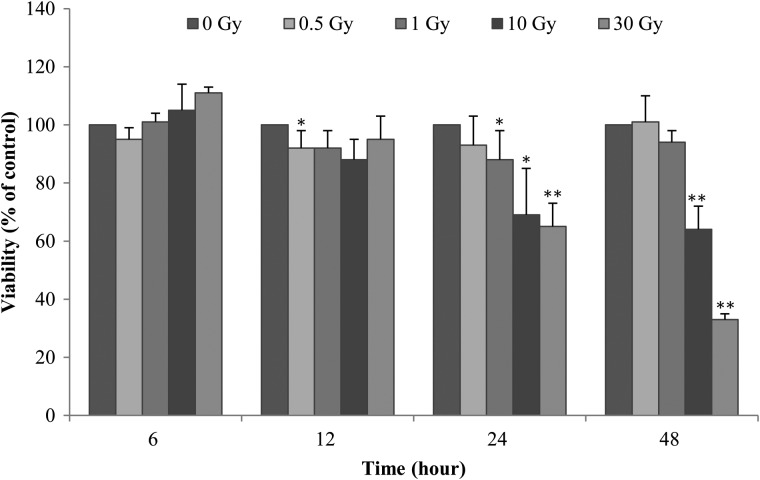
The cell viability assay was performed to investigate cell survival after irradiation. Cell survival after various doses of γ-radiation was evaluated to investigate the characteristics of the HTori-3 cell line and to determine the optimal radiation dose and time for future experiments (Fig. 2). Cell survival decreased in a radiation dose– and time-dependent manner compared with unirradiated cells. Cell survival was similar to that in unirradiated cells 48 h after radiation exposure at a dose of 0.5 Gy. The survival of the cells that were exposed to 1 Gy was increased. In contrast, 24–48 h post-irradiation, the cells exposed to 10 and 30 Gy showed decreased survival in a dose- and time-dependent manner compared with unirradiated cells. Particularly, the survival of the cells exposed to 30 Gy was very low after 48 h. Given these data, exposures of 5 and 10 Gy were used in this study.
Fig. 2.
Time- and dose-dependent effect of γ-radiation on HTori-3 cell viability. HTori-3 cells were exposed to various doses of γ-radiation (at 4.68 Gy/min), and cell viability was determined by MTT assay at the indicated times. The data shown represent three independent experiments, with standard error bars indicated. *P < 0.05 and **P < 0.01.
Tumor development in irradiated HTori-3 cells
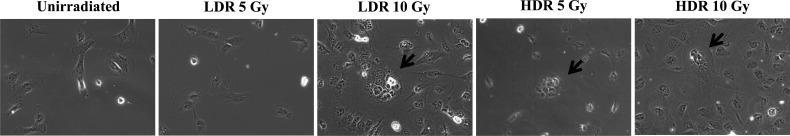
Each group of irradiated cells was evaluated via microscopy to confirm changes in size and shape. Figure 3 highlights cells irradiated with 0, 5 and 10 Gy at either the low- or high-dose rate. HDR 5 Gy (cells irradiated with 5 Gy at the high-dose rate), HDR 10 Gy (cells irradiated with 10 Gy at the high-dose rate), and LDR 10 Gy (cells irradiated with 10 Gy at the low-dose rate) exhibited multinucleated giant cells, which were derived from large plasma membranes. The giant cells associated with LDR 10 Gy were very large and numerous. In contrast, LDR 5 Gy (cells irradiated with 5 Gy at the low-dose rate) results were similar to the control group. When classified by radiation dose, the numbers of giant cells in HDR 5 Gy and LDR 10 Gy were much greater than those of HDR 10 Gy and LDR 5 Gy, respectively.
Fig. 3.
Alterations in the morphology of HTori-3 cells after radiation exposure. HTori-3 cells were exposed to low-dose-rate (40 mGy/h) or high-dose-rate (4.68 Gy/min) radiation. After 5 weeks, the cells were observed and photographed under the inverted microscope. Arrows point to multinucleated giant cells with cytoplasmic vacuoles.
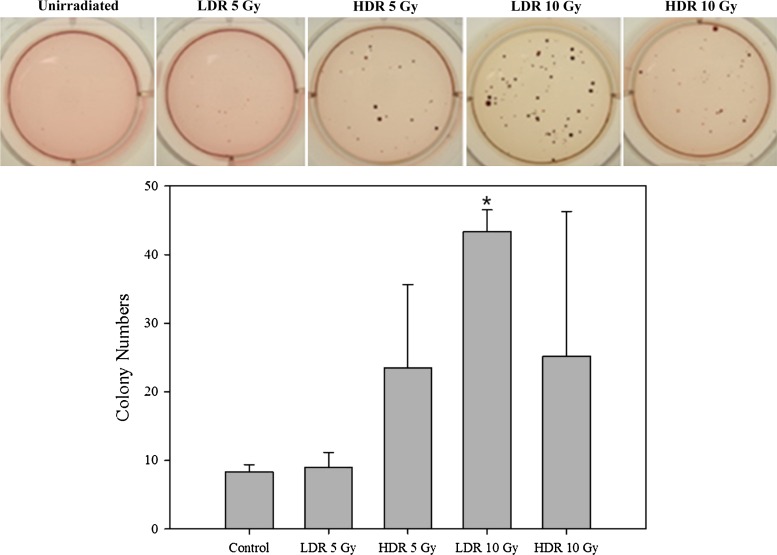
The colony-forming assay was carried out to evaluate anchorage-independent cell growth, which is one of the best in vitro indicators of tumorigenicity. Anchorage-independent cell growth colony formation was verified 2 months after radiation exposure. The size and number of the colonies in the LDR 10 Gy (43.3 ± 3.2, P < 0.0001) were larger and more numerous than those in any other group (HDR 5 Gy: 23.5 ± 12.1, HDR 10 Gy: 25.2 ± 21.2) (Fig. 4). In contrast, the LDR 5 Gy (9.0 ± 2.1) was similar to the control group (8.3 ± 1.0), even though colony formation was suspected.
Fig. 4.
Anchorage-independent cell growth in irradiated HTori-3 cells. A colony-forming assay was used to detect the anchorage-independent growth of transformed cells from the various experimental groups. HTori-3 cells were exposed to low-dose-rate (40 mGy/h) or high-dose-rate (4.68 Gy/min) radiation. After 8 weeks, cells were seeded into 6-well plates in duplicate (1 × 103 cells per well) and incubated for 4 weeks. The resulting colonies were stained with nitroblue tetrazolium. The data shown represent three independent experiments, with standard error bars indicated. *P < 0.0001, using Student's t-test on the differences between control and irradiated groups.
Tumorigenicity in nude mice
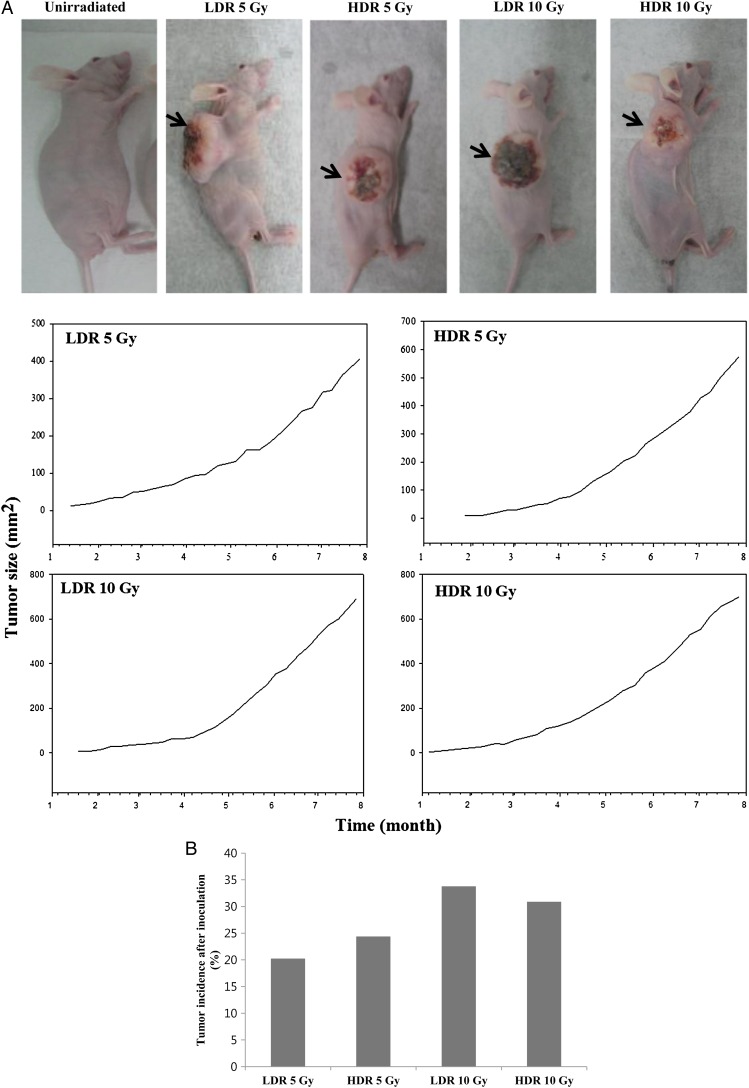
To further investigate the in vivo tumorigenicity of radiation-induced transformed cells, we performed the xenograft transplantation of control cells and of LDR- and HDR-irradiated cells. No tumor growth was seen after injection of control cells, whereas tumors were seen in LDR- and HDR-irradiated cells. Tumors developed after 8–11 weeks; once tumor formation occurred, the size of the tumor was measured with a caliper once per week. Figure 5A illustrates tumor development from representative mice in each group. Of all the groups, LDR 10 Gy exhibited the most rapid changes in tumor development. However, the differences between groups in relative tumor size were not statistically significant. The tumor incidence rate was measured 16 weeks after implantation of the HTori-3 cells into the mice and quantified. When LDR 10 Gy irradiated cells were injected into nude mice, the number of tumors formed, the rate of tumor formation and the size of tumors were higher than in any other groups (Fig. 5B). The tumor incidence rate appeared to follow a pattern similar to that of the changes in cell shape and anchorage-independent growth.
Fig. 5.
Tumorigenicity of HTori-3 cells in nude mice. (A) Representative photographs of tumor formation in nude mice. Arrows indicate location of tumors. Tumor size was monitored once per week. (B) The tumor incidence rate in each group after 16 weeks: low-dose-rate (40 mGy/h) 5 Gy, 20.24%; high-dose-rate (4.68 Gy/min) 5 Gy, 24.39%; low-dose-rate 10 Gy, 33.78%; and high-dose-rate 10 Gy, 30.88%.
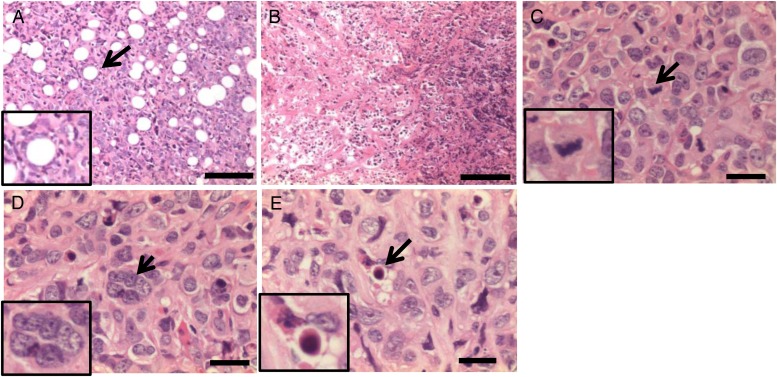
Histological examination suggested that the tumors formed by radiation irradiated cells were remarkable for the phenotype of the malignant tumor. The images highlight adipose tissue invasion, necrosis, giant cells, mitotic structures, and apoptotic bodies, which are frequently visible in tumors (Fig. 6). Similar results were observed in the tumors from all groups. Giant cells are related to undifferentiated thyroid carcinoma; however, the sample size was not sufficient to be statistically significant.
Fig. 6.
Representative histologic image of tumor tissues from xenografts. (A) Adipose tissue invasion, (B) necrosis, (C) mitotic structures, (D) giant cell (multinucleated) and (E) apoptotic body. Scale bars, 300 µm (A, B) and 100 µm (C–E). Insets show malignant tumor-like morphology in enlarged images.
Genetic changes in tumor cells after irradiation
The tumorigenicity of the HTori-3 cells following irradiation in vitro and in vivo was confirmed [32–33]. However, prior to investigating which genes influence these tumorous changes in the cells, the purity of the cells isolated from the tumor tissue was examined to avoid using cultures contaminated with mouse cells, which would confound the results of our experiments. RT-PCR was performed using primers specific to human and mouse GAPDH (data not shown).
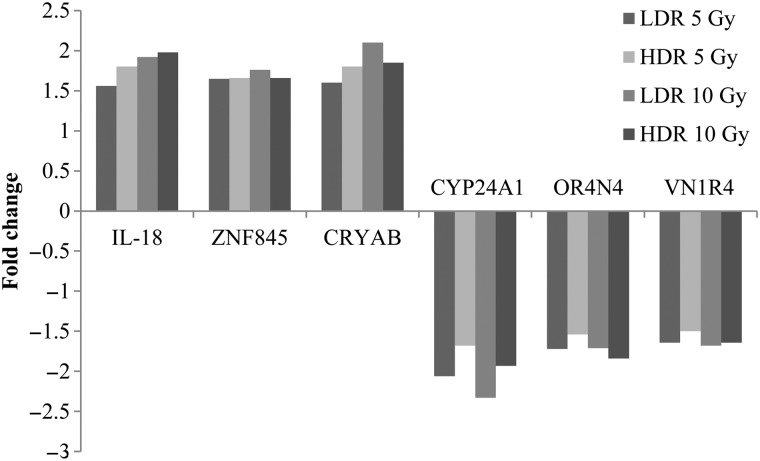
Microarrays were carried out to investigate which genes influence the tumorous changes in normal human cells after irradiation. The gene expression values were determined by comparing irradiated cells with unirradiated controls. Using a threshold of 1.5-fold or more, the expression levels of certain genes were differentially regulated after each dose of radiation exposure (Table 1). We found the highest number of genes affected in the LDR 10 Gy group. In Tables 2 and 3, the key genes indicated to be related to tumor occurrence were listed in order, commencing with the highest association. Genes that were commonly misregulated in all four experimental groups were identified (Fig. 7). The genes in this category were IL-18, ZNF845, CRYAB, CYP24A1, OR4N4 and VN1R4, and the biological processes that these genes affect include apoptosis, the immune response, regulation of transcription, and receptor signaling pathways. We also compared the changes in gene expression between the high-dose rate and the low-dose rate for each radiation dose. By comparing the expression levels of certain genes between HDR 5 Gy and LDR 5 Gy (Table 4) and HDR 10 Gy and LDR 10 Gy (Table 5), we identified genes that are expected to induce aggressive tumor formation. In the HDR 5 Gy group, six genes were misregulated compared with the LDR 5 Gy. Of those, XAGE1D and LOX are genes that are associated with tumor formation. Similarly, in the low-dose 10 Gy group, CD109, SLC7A11 and GDF15 were upregulated, and BMP4 and THBS2 were downregulated.
Table 1.
The number of up- and down-regulated genes in tumor cells in each group
| Irradiation | >2-fold | 1.5–2-fold | <0.5-fold | 0.75–0.5-fold | |
|---|---|---|---|---|---|
| LDR 5 Gy | 25 | 287 | 10 | 371 | |
| HDR 5 Gy | 24 | 296 | 8 | 358 | |
| LDR 10 Gy | 36 | 348 | 7 | 402 | |
| HDR 10 Gy | 23 | 285 | 8 | 340 |
LDR 5 Gy = cells irradiated with 5 Gy at the low-dose rate, HDR 5 Gy = cells irradiated with 5 Gy at the high-dose rate, LDR 10 Gy = cells irradiated with 10 Gy at the low-dose rate, HDR 10 Gy = cells irradiated with 10 Gy at the high-dose rate.
Table 2.
Downregulated gene expression profiles in each group compared with the control group
| Irradiation | Gene symbol | Gene description | Biological process |
|---|---|---|---|
| LDR 5 Gy | CYP24A1 | cytochrome P450, family 24, subfamily A, polypeptide 1 | vitamin D receptor signaling pathway |
| OR4N4 | olfactory receptor, family 4, subfamily N, member 4 | G protein-coupled receptor signaling pathway | |
| SNORD115-32 | small nucleolar RNA, C/D box 115–32 | no data | |
| LOC441233 | hypothetical 441233 | no data | |
| VN1R4 | vomeronasal 1 receptor 4 | G protein-coupled receptor signaling pathway | |
| C6orf58 | chromosome 6 open reading frame 58 | multicellular organismal development | |
| LOC100293539 | similar to ribosomal protein 10 | no data | |
| ZC3H15 | zinc finger CCCH-type containing 15 | cytokine-mediated signaling pathway | |
| HDR 5 Gy | LOC100293539 | similar to ribosomal protein 10 | no data |
| CYP24A1 | cytochrome P450, family 24, subfamily A, polypeptide 1 | vitamin D receptor signaling pathway | |
| LOC441233 | hypothetical 441233 | no data | |
| SNORD115-32 | small nucleolar RNA, C/D box 115–32 | no data | |
| C6orf58 | chromosome 6 open reading frame 58 | multicellular organismal development | |
| OR4N4 | olfactory receptor, family 4, subfamily N, member 4 | G protein-coupled receptor signaling pathway | |
| LDR 10 Gy | CYP24A1 | cytochrome P450, family 24, subfamily A, polypeptide 1 | vitamin D receptor signaling pathway |
| OR4N4 | olfactory receptor, family 4, subfamily N, member 4 | G protein-coupled receptor signaling pathway | |
| VN1R4 | vomeronasal 1 receptor 4 | G protein-coupled receptor signaling pathway | |
| OR5M3 | olfactory receptor, family 5, subfamily M, member 3 | G protein-coupled receptor signaling pathway | |
| LOC100293539 | similar to ribosomal protein 10 | no data | |
| C1orf105 | chromosome 1 open reading frame 105 | no data | |
| HDR 10 Gy | CYP24A1 | cytochrome P450, family 24, subfamily A, polypeptide 1 | vitamin D receptor signaling pathway |
| OR4N4 | olfactory receptor, family 4, subfamily N, member 4 | G protein-coupled receptor signaling pathway | |
| LOC441233 | hypothetical 441233 | no data | |
| VN1R4 | vomeronasal 1 receptor 4 | G protein-coupled receptor signaling pathway | |
| MGST1 | microsomal glutathione S-transferase 1 | glutathione metabolic process | |
| LOC100293539 | similar to ribosomal protein 10 | no data |
LDR 5 Gy = cells irradiated with 5 Gy at the low-dose rate, HDR 5 Gy = cells irradiated with 5 Gy at the high-dose rate, LDR 10 Gy = cells irradiated with 10 Gy at the low-dose rate, HDR 10 Gy = cells irradiated with 10 Gy at the high-dose rate.
Table 3.
Upregulated genes in each group compared with the control group
| Irradiation | Gene symbol | Gene description | Biological process |
|---|---|---|---|
| LDR 5 Gy | ZNF845 | zinc finger protein 845 | regulation of transcription |
| HDR 5 Gy | CRYAB | crystallin, alpha B | apoptotic process involved in morphogenesis |
| IL18 | interleukin 18 | immune response | |
| SNORD113–4 | small nucleolar RNA, C/D box 113–4 | no data | |
| ZNF845 | zinc finger protein 845 | regulation of transcription | |
| SCARNA9 | small Cajal body-specific RNA 9 | no data | |
| LDR 10 Gy | SNORD113–4 | small nucleolar RNA, C/D box 113–4 | no data |
| IL18 | interleukin 18 | immune response | |
| CRYAB | crystallin, alpha B | apoptotic process involved in morphogenesis | |
| AREG | amphiregulin | G protein-coupled receptor signaling pathway | |
| ZNF845 | zinc finger protein 845 | regulation of transcription | |
| HDR 10 Gy | IL18 | interleukin 18 | immune response |
| SNORD113–4 | small nucleolar RNA, C/D box 113–4 | no data | |
| PRAME | preferentially expressed antigen in melanoma | positive regulation of cell proliferation | |
| ZNF845 | zinc finger protein 845 | regulation of transcription |
LDR 5 Gy = cells irradiated with 5 Gy at the low-dose rate, HDR 5 Gy = cells irradiated with 5 Gy at the high-dose rate, LDR 10 Gy = cells irradiated with 10 Gy at the low-dose rate, HDR 10 Gy = cells irradiated with 10 Gy at the high-dose rate.
Fig. 7.
Identification of radiosensitivity genes in γ-radiation–induced tumor cells. Six genes were commonly up- and downexpressed in tumor cells of all groups. IL-18, ZNF845 and CRYAB were upregulated in radiation-induced tumor cells compared with control, whereas CYP24A1, OR4N4 and VN1R4 were downregulated in tumor cells compared with the control.
Table 4.
The list of up- and downregulated genes in the HDR 5 Gy group compared with the LDR 5 Gy group
| Gene expression | Gene symbol | Gene description | Biological process |
|---|---|---|---|
| Upregulated in HDR 5 Gy | XAGE1D | cancer/testis antigen | no data |
| NID2 | nidogen 2 | extracellular matrix organization | |
| LOX | lysyl oxidase | extracellular matrix organization | |
| MAMDC2 | MAM domain-containing 2 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan | |
| LOC100288114 | hypothetical LOC100288114 | no data | |
| Downregulated in HDR 5 Gy | BMP5 | bone morphogenetic protein 5 | negative regulation of cell proliferation |
LDR 5 Gy = cells irradiated with 5 Gy at the low-dose rate, HDR 5 Gy = cells irradiated with 5 Gy at the high-dose rate.
Table 5.
The list of up- and downregulated genes in the LDR 10 Gy group compared with the HDR 10 Gy group
| Gene expression | Gene symbol | Gene description | Biological process |
|---|---|---|---|
| Upregulated in LDR 10 Gy | ACTA2 | actin, α2, smooth muscle | vascular smooth muscle contraction |
| CD109 | GPI-anchored glycoprotein | negative regulation of transforming growth factor β receptor signaling pathway | |
| IL1RAP | IL-1 receptor accessory protein | immune response | |
| SLC7A11 | solute carrier family 7, member 11 | amino acid transport | |
| GDF15 | growth differentiation factor 15 | transforming growth factor β receptor signaling pathway |
|
| Downregulated in LDR 10 Gy | BMP4 | bone morphogenetic protein 4 | angiogenesis |
| THBS2 | thrombospondin 2 | negative regulation of angiogenesis | |
| TANC2 | tetratriopeptide repeat, ankyrin repeat and coiled coil-containing 2 | in utero embryonic development | |
| MKX | mohawk homeobox | muscle organ development | |
| GALNT13 | polypeptide N-acetylgalactosaminyl transferase 13 | integral component of membrane | |
| PRAME | preferentially expressed antigen in melanoma | apoptosis |
LDR 10 Gy = cells irradiated with 10 Gy at the low-dose rate, HDR 10 Gy = cells irradiated with 10 Gy at the high-dose rate.
DISCUSSION
Our study demonstrates that the dose rate is an important factor in the cellular response following exposure to high-dose radiation. For mice irradiated with 5 Gy, the cancer incidence rate of HDR 5 Gy was higher than LDR 5 Gy. The LDR 5 Gy group showed minimal changes in cell features and anchorage-independent growth, even though the total dose of 5 Gy was predicted to generate tumors by the LNT model. Ishizaki et al. identified a very small increase in γ-H2AX-positive foci (which indicate the presence of double-strand breaks) in a human diploid cell line (SuSa/T-n) after exposure to low-dose rate radiation (0.3 mGy/min) compared with high-dose rate radiation [34]. These data support the notion that damaged DNA induced by chronic low-dose-rate irradiation might be repaired during irradiation. After validating cell transformation or confirming anchorage-independent growth of the cells, LDR 10 Gy tended to exhibit the most impressive malignant transformations. Additionally, cancer developed earlier in the LDR 10 Gy group compared with in any other group, which recapitulates our in vitro results. However, it is possible that the mice in the LDR 10 Gy group were injured temporarily by the low-dose-rate irradiation and recovered rapidly. Thus, the main cause of malignant transformation is attributed to the accumulation of genomic instability [26]. In contrast, HDR 10 Gy was expected to demonstrate malignant transformation most frequently, but the degree to which this occurred did not meet our expectations. This may suggest that the cell death is increased by intracellular damage due to high-dose rates of radiation.
We found several specific genes associated with the development of radiation-induced tumors that followed dose- and rate-dependent trends. Several genes were commonly misregulated in all four groups, suggesting that a common molecular mechanism exists that facilitates radiation-induced thyroid carcinogenesis.
CRYAB, which codes for the small heat shock protein, alpha B-crystalin, is one of the genes that is commonly upregulated in all four groups, and it is associated with apoptosis [35].
Ionizing radiation also stimulates the secretion of inflammatory cytokines. Interleukin (IL)-18, a member of the IL-1 family of cytokines, plays vital roles in inflammation and the immune response [36]. Other genes commonly misregulated in all four groups include ZNF845, CYP24A1, ORN4 and VN1R4. ZNF845, which is known to regulate transcription, was upregulated. Furthermore, its function has been associated with the development of various cancers [37]. CYP24A1, which is a key player in 1,25(OH)2 D3 signaling in benign and well-differentiated thyroid tumors, was also increased. Interestingly, however, there was negative staining for CYP24A1 in anaplastic thyroid cancers with high proliferation rates [38]. Considering the histological appearance of the cells and the changes in CYP24A1 gene expression noted herein, we suggest that the thyroid tumors in our study are undifferentiated, anaplastic carcinomas. Finally, OR4N4 has been detected in normal human tissues, such as the testes and the thyroid [39]. VN1R4 has pheromone receptor activity [40] Both OR4N4 and VN1R4 are similarly involved in the G protein-coupled receptor signaling pathways [39–40]. However, the misregulation of these genes in thyroid tumors is still not confirmed.
A number of genome-wide expression analyses using microarray chips have been performed to distinguish between radiation-induced tumors and sporadic tumors [20–24, 41]. Of those, some studies have identified genes that are misregulated in post-Chernobyl or post-radiotherapy tumors [20, 21, 24]. Importantly, the BMP genes, which are also misregulated in post-Chernobyl and post-radiotherapy tumors, were misregulated in the present study. Therefore, the genes that are prominently misregulated in the HDR 5 Gy and LDR 10 Gy groups and the genes commonly misregulated in all four groups suggest that there are possibly biomarkers of thyroid tumors induced by high-dose radiation [42].
We utilized microarray analysis to elucidate gene expression profiles for HTori-3 cells exposed to high-dose γ-radiation. Three previous studies reported that selenomethionine (SeM) protects HTori-3 cells against 1 GeV/neutron iron-ion radiation [29–31]. However, two of these previous studies showed differential gene expression levels at doses of 10 or 20 Gy in vitro, but there were no genes commonly misregulated between our study and these reports [29–30]. Notably, the relative biological effectiveness (RBE) of high linear energy transfer (LET) radiation, such as iron ions, is higher than that of γ-radiation. Therefore, the adverse biological effects are not comparable. One study identified tumorigenicity in athymic nude mice implanted with HTori-3 cells that were irradiated with iron ions at doses of 0.1, 0.2, 0.4, 1 and 2 Gy, but that report did not perform follow-up microarray analysis [31].
One study, using human thyroid tissue, has identified misregulated genes that specify molecular markers in a series of secondary thyroid tumors after radiotherapy [24]. Compared with our study, the human thyroid tissues consisted of follicular thyroid adenomas and papillary thyroid carcinomas, and the dosages used were between 0.1 and 48 Gy [24]. In contrast, the thyroid tumors in our study were induced by exposing human thyroid cells to doses of 5 and 10 Gy, and all of the tumors were undifferentiated carcinomas. Considering the differences in radiation dose, histologic type, and cell nature, it was not surprising that no overlap was found between the misregulated genes identified in the two studies [24, and this study]. We suggest that our results may represent a specific gene signature that can be used to diagnose undifferentiated carcinomas in post-radiotherapy thyroid tumors.
Also, the probability that the thyroid would be exposed to doses of between 5 and 10 Gy by external exposure or internal contamination in large-scale nuclear accidents is unlikely, but it presents high risks. In the Chernobyl accident, the cumulative dose to the thyroid by chronic 131I contamination was generally <1 Gy, but some people experienced doses in excess of 10 Gy [43]. Therefore, different radiation biodosimetry methods have been developed to enhance the ability to correctly estimate exposure doses because appropriate radiological triage approaches are needed following large-scale nuclear accidents. However, the effects of dose rates following large-scale nuclear accidents have not been well studied; thus, biodosimetry methods should be tailored to distinguish dose-rate effects for the estimation of doses in these scenarios [44]. In this study, we identified a gene expression signature that can discriminate between low- and high-dose rates. For example, different effects were observed depending on the dose rate, even when the same overall dose was applied; the HDR 5 Gy and LDR 10 Gy groups showed distinct gene expression profiles compared with the LDR 5 Gy and HDR 10 Gy groups, respectively. However, further validation studies will be needed to confirm this effect.
Undifferentiated thyroid carcinoma is one of the most aggressive tumors and it has poor prognosis. There is still no successful treatment, despite surgery, external beam radiotherapy, and chemotherapy, which may contribute to longer survival in some cases. The efficacy of radiation therapy has been improved by combining it with gene therapy, which requires tightly controlled regulation of transgene expression for clinical success [45]. Ionizing radiation activates inducible promoters via the generation of reactive oxygen species (ROS), and inducible promoters can modulate the expression of therapeutic genes in the irradiated tumor volume both spatially and temporally [46]. TNF-α is a candidate therapeutic gene because it sensitizes tumor cells to radiation, disrupts tumor vasculature, and induces the immune response [47]. IL-2 is an important cytokine with an anti-tumor immune response and anti-angiogenic activity [48–49]. In the present study, IL-18 and CRYAB were associated with the immune response and apoptosis, respectively. Thus, we suggest that these genes could represent potential therapeutic targets for undifferentiated thyroid carcinomas.
We recognize a major caveat of this study. In terms of histologic type, radiation-induced thyroid tumors almost resemble papillary carcinomas. The reason for this phenomenon in our study may be due to the higher proliferation rate of HTori-3 cells compared with human thyroid tissue. This characteristic may have profound implications for the development of undifferentiated carcinomas. After the Chernobyl nuclear accident in Ukraine, 296 people, including children and adolescents, underwent pathological examinations between 1986 and 1997. Of those, undifferentiated thyroid carcinomas were only diagnosed in two cases (0.7%) [50]. Therefore, our results may be less important in terms of nuclear disasters or post-radiotherapy tumors. However, there has been no experimental report about the gene expression profiles of radiation-induced undifferentiated carcinomas using thyroid epithelial cells until now.
In this study, we identified differential gene expression by high-dose irradiation. However, tumor formation itself could induce changes in gene expression. To demonstrate whether these gene changes are solely related to the tumor formation, not to radiation exposure, we investigated only cancer-related genes in thyroid cancer cell lines. We examined differences in gene expression between a normal thyroid cell line (HTori-3) and three different thyroid cancer cell lines [TPC-1 (papillary thyroid carcinoma), SNU-373 (PTC) and SNU-790 (anaplastic thyroid carcinoma)] by using microarray analysis. Eight genes were commonly expressed more in three thyroid cancer cell lines than the normal thyroid cell line (2-fold). One gene (GREM1) was downregulated, and seven genes (CFH, CNTNAP3, IFI44, MCTP1, TM4SF1, TRPS1 and ZNF626) were upregulated (data not shown). In addition, according to microarray analysis in three published studies that have shown thyroid cancer gene expression profiling, genes such as SLC34A2, TM7SF4, COMP, KLK7, KCNJ2, FOXA2, SLC4A4, LYVE-1, TFCP2L1, CDH3, NGEF, PROS1, TGFA, MET and NF-κB showed different expression between the sporadic thyroid carcinoma samples and the normal tissues [51–53]. Our study could not compare the gene expression between the sporadic and the radiation-induced thyroid tumor. However, these tumor-related specific genes didn't contain our target genes. These data imply that our study identified radiation-induced cancer-related genes in human thyroid cells.
In summary, we identified dose-rate-dependent changes in gene expression profiles following high-dose radiation. Particularly, commonly misregulated genes may serve as candidate genes for biomarkers of undifferentiated thyroid carcinomas. Furthermore, these genes could provide helpful information for biodosimetry and targeted gene therapies that can be used in combination with radiation therapy; nevertheless, further study is needed.
CONFLICT OF INTEREST
The authors state that there are no potential conflicts of interest.
FUNDING
This work was supported by a research grant from Korea Hydro & Nuclear Power Co., Ltd [No. E08NS26]. Funding to pay the Open Access publication charges for this article was provided by Korea Hydro & Nuclear Power Co., Ltd [No. E08NS26].
REFERENCES
- 1.Sassolas G, Hafdi-Nejjari Z, Casagranda L et al. Thyroid cancers in children, adolescents, and young adults with and without a history of childhood exposure to therapeutic radiation for other cancers. Thyroid 2013;23:805–10. [DOI] [PubMed] [Google Scholar]
- 2.Hamatani K, Eguchi H, Ito R et al. RET/PTC rearrangements preferentially occurred in papillary thyroid cancer among atomic bomb survivors exposed to high radiation dose. Cancer Res 2008;68:7176–82. [DOI] [PubMed] [Google Scholar]
- 3.Schlumberger M, Le Guen B. Nuclear-power-plant accidents: thyroid cancer incidence and radiation-related health effects from the Chernobyl accident. Med Sci (Paris) 2012;28:746–56. [DOI] [PubMed] [Google Scholar]
- 4.Schonfeld SJ, Lee C, Berrington deGonzález A. Medical exposure to radiation and thyroid cancer. Clin Oncol (R Coll Radiol) 2011;23:244–50. [DOI] [PubMed] [Google Scholar]
- 5.Preston DL, Ron E, Tokuoka S et al. Solid cancer incidence in atomic bomb survivors: 1958–1998. Radiat Res 2007;168:1–64. [DOI] [PubMed] [Google Scholar]
- 6.Tronko MD, Howe GR, Bogdanova TI et al. A cohort study of thyroid cancer and other thyroid diseases after the Chernobyl accident: thyroid cancer in Ukraine detected during first screening. J Natl Cancer Inst 2006;98:897–903. [DOI] [PubMed] [Google Scholar]
- 7.Zablotska LB, Ron E, Rozhko AV et al. Thyroid cancer risk in Belarus among children and adolescents exposed to radioiodine after the Chernobyl accident. Br J Cancer 2011;104:181–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thompson DE, Mabuchi K, Ron E et al. Cancer incidence in atomic bomb survivors. Part II: Solid tumors, 1958–1987. Radiat Res 1994;137 (2 Suppl):S17–67. [PubMed] [Google Scholar]
- 9.Astakhova LN, Anspaugh LR, Beebe GW et al. Chernobyl-related thyroid cancer in children of Belarus: a case–control study. Radiat Res 1998;150:349–56. [PubMed] [Google Scholar]
- 10.Walsh L, Zhang W, Shore RE et al. A framework for estimating radiation-related cancer risks in Japan from the 2011 Fukushima nuclear accident. Radiat Res 2014;182:556–72. [DOI] [PubMed] [Google Scholar]
- 11.Pinto N, Black M, Patel K et al. Genomically driven precision medicine to improve outcomes in anaplastic thyroid cancer. J Oncol 2014;2014:936285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Leeman-Neill RJ, Brenner AV, Little MP et al. RET/PTC and PAX8/PPARγ chromosomal rearrangements in post-Chernobyl thyroid cancer and their association with iodine-131 radiation dose and other characteristics. Cancer 2013;119:1792–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Elisei R, Romei C, Vorontsova T et al. RET/PTC rearrangements in thyroid nodules: studies in irradiated and not irradiated, malignant and benign thyroid lesions in children and adults. J Clin Endocrinol Metab 2001;86:3211–6. [DOI] [PubMed] [Google Scholar]
- 14.Caudill CM, Zhu Z, Ciampi R et al. Dose-dependent generation of RET/PTC in human thyroid cells after in vitro exposure to gamma-radiation: a model of carcinogenic chromosomal rearrangement induced by ionizing radiation. J Clin Endocrinol Metab 2005;90:2364–9. [DOI] [PubMed] [Google Scholar]
- 15.Mizuno T, Iwamoto KS, Kyoizumi S et al. Preferential induction of RET/PTC1 rearrangement by X-ray irradiation. Oncogene 2000;19:438–43. [DOI] [PubMed] [Google Scholar]
- 16.Kikuchi Y, Tsuji E, Yagi K et al. Aberrantly methylated genes in human papillary thyroid cancer and their association with BRAF/RAS mutation. Front Genet 2013;4:271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Catalano MG, Fortunati N, Boccuzzi G. Epigenetics modifications and therapeutic prospects in human thyroid cancer. Front Endocrinol (Lausanne) 2012;3:40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pita JM, Banito A, Cavaco BM et al. Gene expression profiling associated with the progression to poorly differentiated thyroid carcinomas. Br J Cancer 2009;101:1782–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nikolova DN, Zembutsu H, Sechanov T et al. Genome-wide gene expression profiles of thyroid carcinoma: identification of molecular targets for treatment of thyroid carcinoma. Oncol Rep 2008;20:105–21. [PubMed] [Google Scholar]
- 20.Detours V, Delys L, Libert F et al. Genome-wide gene expression profiling suggests distinct radiation susceptibilities in sporadic and post-Chernobyl papillary thyroid cancers. Br J Cancer 2007;97:818–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Port M, Boltze C, Wang Y et al. A radiation-induced gene signature distinguishes post-Chernobyl from sporadic papillary thyroid cancers. Radiat Res 2007;168:639–49. [DOI] [PubMed] [Google Scholar]
- 22.Maenhaut C, Detours V, Dom G et al. Gene expression profiles for radiation-induced thyroid cancer. Clin Oncol (R Coll Radiol) 2011;23:282–8. [DOI] [PubMed] [Google Scholar]
- 23.Stein L, Rothschild J, Luce J et al. Copy number and gene expression alterations in radiation-induced papillary thyroid carcinoma from Chernobyl pediatric patients. Thyroid 2010;20:475–87. [DOI] [PubMed] [Google Scholar]
- 24.Ory C, Ugolin N, Levalois C et al. Gene expression signature discriminates sporadic from post-radiotherapy-induced thyroid tumors. Endocr Relat Cancer 2011;18:193–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lemoine NR, Mayall ES, Jones T et al. Characterisation of human thyroid epithelial cells immortalised in vitro by simian virus 40 DNA transfection. Br J Cancer 1989;60:897–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zitzelsberger H, Bruch J, Smida J et al. Clonal chromosomal aberrations in simian virus 40-transfected human thyroid cells and in derived tumors developed after in vitro irradiation. Int J Cancer (Radiat Oncol Invest) 2001;96:166–77. [DOI] [PubMed] [Google Scholar]
- 27.Evdokimova V, Gandhi M, Rayapureddi J et al. Formation of carcinogenic chromosomal rearrangements in human thyroid cells after induction of double-strand DNA breaks by restriction endonucleases. Endocr Relat Cancer 2012;19:271–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhou Z, Ware JH, Kennedy AR. Carbon and iron radiation-induced cytotoxicity and transformation in vitro . Oncol Lett 2011;2:915–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stewart J, Ware J, Fortina P et al. L-selenomethionine modulates high LFT radiation-induced alterations of gene expression in cultured human thyroid cells. Oncol Rep 2006;16:569–74. [PubMed] [Google Scholar]
- 30.Sanzari JK, Nuth M, Kennedy AR. Induction of cytokine gene expression in human thyroid epithelial cells irradiated with HZE particles (iron ions). Radiat Res 2009;172:437–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ware JH, Zhou Z, Romero-Weaver AL et al. Effects of selenomethionine in irradiated human thyroid epithelial cells and tumorigenicity studies. Nutr Cancer 2011;63:1114–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Riches AC, Herceg Z, Bryant PE et al. Radiation-induced transforamtion of SV40-immortalized human epithelial cells by single and fractionated exposure to gamma-irradiation in vitro. Int J Radiat Biol 1994;66:757–65. [PubMed] [Google Scholar]
- 33.Riches A, Herceg Z, Wang H et al. Radiation-induced carcinogenesis: studies using human epithelial cell lines. Radiat Oncol Investig 1997;5:139–43. [DOI] [PubMed] [Google Scholar]
- 34.Ishizaki K, Hayashi Y, Nakamura H et al. No induction of p53 phosphorylation and few focus formation of phosphorylated H2AX suggest efficient repair of DNA damage during chronic low-dose-rate irradiation in human cells. J Radiat Res 2004;45:521–5. [DOI] [PubMed] [Google Scholar]
- 35.Acunzo J, Katsogiannou M, Rocchi P. Small heat shock proteins HSP27 (HspB1), αB-crystallin (HspB5) and HSP22 (HspB8) as regulators of cell death. Int J Biochem Cell Biol 2012;44:1622–31. [DOI] [PubMed] [Google Scholar]
- 36.Ha CT, Li XH, Fu D et al. Circulating interleukin-18 as a biomarker of total-body radiation exposure in mice, minipigs, and nonhuman primates (NHP). PLoS One 2014;9:e109249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.LifeMap Sciences. The GeneCards Suite: United States of America, Israel and Hong Kong. http://www.genecards.org/cgi-bin/carddisp.pl?gene=ZNF845 (1 August 2010, date last accessed).
- 38.Clinckspoor I, Hauben E, Verlinden L et al. Altered expression of key players in vitamin D metabolism and signaling in malignant and benign thyroid tumors. J Histochem Cytochem 2012;60:502–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.LifeMap Sciences. The GeneCards Suite: United States of America, Israel and Hong Kong. http://www.genecards.org/cgi-bin/carddisp.pl?gene=OR4N4 (1 August 2010, date last accessed).
- 40.LifeMap Sciences. The GeneCards Suite: United States of America, Israel and Hong Kong. http://www.genecards.org/cgi-bin/carddisp.pl?gene=VN1R4 (1 August 2010, date last accessed).
- 41.Detours V, Wattel S, Venet D et al. Absence of a specific radiation signature in post-Chernobyl thyroid cancers. Br J Cancer 2005;92:1545–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lu TP, Hsu YY, Lai LC et al. Identification of gene expression biomarkers for predicting radiation exposure. Sci Rep 2014;4:6293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tronko MD, Howe GR, Bogdanova Tl et al. A cohort study of thyroid cancer and other thyroid diseases after the Chernobyl accident: thyroid cancer in Ukraine detected during first screening. J Natl Cancer Inst 2006;98:897–903. [DOI] [PubMed] [Google Scholar]
- 44.Ghandhi SA, Smilenov LB, Elliston CD et al. Radiation dose-rate effects on gene expression for human biodosimetry. BMC Med Genomics 2015;8:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gridley DS, Slater JM. Combining gene therapy and radiation against cancer. Curr Gene Ther 2004;4:231–48. [DOI] [PubMed] [Google Scholar]
- 46.Weichselbaum RR, Hallahan DE, Sukhatme VP et al. Gene therapy targeted by ionizing radiation. Int J Radiat Oncol Biol Phys 1992;24:565–7. [DOI] [PubMed] [Google Scholar]
- 47.Mauceri HJ, Hanna NN, Wayne JD et al. Tumor necrosis factor alpha (TNF-alpha) gene therapy targeted by ionizing radiation selectively damages tumor vasculature. Cancer Res 1996;56:4311–4. [PubMed] [Google Scholar]
- 48.Trinchieri G. Interleukin-12 and the regulation of innate resistance and adaptive immunity. Nat Rev Immunol 2003;3:133–46. [DOI] [PubMed] [Google Scholar]
- 49.Ogawa M, Yu WG, Umehara K et al. Multiple roles of interferon-gamma in the mediation of interleukin 12-induced tumor regression. Cancer Res 1998;58:2426–32. [PubMed] [Google Scholar]
- 50.Tronko MD, Bogdanova TI, Komissarenko IV et al. Thyroid carcinoma in children and adolescents in Ukraine after the Chernobyl nuclear accident: statistical data and clinicomorphologic characteristics. Cancer 1999;86:149–56. [DOI] [PubMed] [Google Scholar]
- 51.Chung KW, Kim SW, Kim SW. Gene expression profiling of paplillary thyroid carcinomas in Korean patients by oligonucleotide microarrays. J Korean Surg Soc 2012;82:271–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kim HS, Kim do H, Kim JY et al. Microarray analysis of papillary thyroid cancers in Korean. Korean J Intern Med 2010;25:399–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gombos K, Zele E, Kiss I et al. Characterization of microarray gene expression profiles of early stage thyroid tumours. Cancer Genomics Proteomics 2007;4:403–9. [PubMed] [Google Scholar]