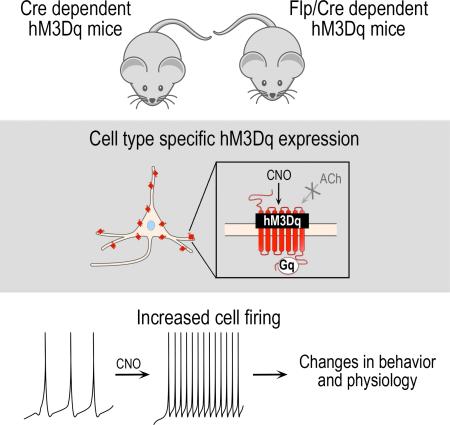
SUMMARY
Chemogenetic technologies, including the mutated human Gq-coupled M3 muscarinic receptor (hM3Dq), have greatly facilitated our ability to directly link changes in cellular activity to altered physiology and behavior. Here, we extend the hM3Dq toolkit with recombinase-responsive mouse lines that permit hM3Dq expression in virtually any cell type. These alleles encode a fusion protein designed to increase effective expression levels by concentrating hM3Dq to the cell body and dendrites. To illustrate their broad utility, we targeted three different genetically defined cell populations: noradrenergic neurons of the compact, bilateral locus coeruleus and two dispersed populations, Camk2a+ neurons and GFAP+ glia. In all three populations, we observed reproducible expression and confirmed that activation of hM3Dq is sufficient to dose-dependently evoke phenotypic changes, without extreme phenotypes associated with hM3Dq overexpression. These alleles offer unprecedented ability to noninvasively control activity of diverse cell types to uncover their function and dysfunction at any developmental stage.
Graphical Abstract
INTRODUCTION
Since their initial description less than ten years ago (Armbruster et al., 2007), the engineered G protein-coupled receptors known as DREADDs (Designer Receptors Exclusively Activated by Designer Drugs) have proven invaluable for linking cellular activity to changes in physiology and behavior. Among the most frequently used DREADDs is hM3Dq, a mutated Gq-coupled human M3 muscarinic receptor that is activated exclusively by the designer drug clozapine N-oxide (CNO), but not its native ligand acetylcholine. By coupling to endogenous signal transduction pathways, hM3Dq has provided insight into the unique functions of cells as diverse as neurons, glia, pancreatic β-cells, and hepatocytes (as reviewed in Urban and Roth, 2015). In neurons, activation of hM3Dq by CNO leads to calcium influx and increased cell firing, thereby promoting neuronal excitability (Alexander et al., 2009). Thus, hM3Dq has been of particular value for identifying the functions encoded by specific neurons in the freely behaving animal.
Previous methods for delivery of hM3Dq to cells in vivo have included viral vectors, transgenes, and targeted knock-in mouse lines (Urban and Roth, 2015; Zhu and Roth, 2014). Each of these strategies drive hM3Dq expression at levels sufficient for behavioral or physiological analyses, but each has limitations. The most commonly used method for delivery of hM3Dq is injection of viral vectors. Although these constructs often generate very high levels of hM3Dq expression, overexpression may result in extreme phenotypes of questionable physiological relevance. Furthermore, the requirement for direct injection limits the use of viruses prenatally and in juvenile/adolescent animals. Multiple injections are required to target dispersed or bilateral cell populations, increasing the possibility of experimentally significant trauma and variable expression among experimental subjects. Some of the experimental limitations of viral vectors can be overcome by the use of transgenes. Given an appropriate promoter, transgene expression can be achieved non-invasively in anatomically dispersed populations at any stage of development. However, each targeted cell population requires generation of a unique transgenic line, and multi-copy transgene arrays can result in unpredictable expression levels. In contrast, single-copy knock-in alleles, particularly recombinase-responsive constructs, offer consistent, reproducible expression and the broadest range of possible cellular targets by partnership with existing cell type-specific recombinase driver lines. However, expression levels of single-copy alleles are likely to be lower than those achieved with viruses or transgenes.
To overcome these experimental challenges, we generated two recombinase responsive knock-in alleles that complement existing hM3Dq constructs by permitting non-invasive targeting of hM3Dq to the cell body (soma) and dendrites of genetically defined cells. This somatodendritic localization is designed to concentrate hM3Dq, thus increasing its effective expression level. Our Cre-responsive allele permits expression of hM3Dq in genetically defined cell populations by taking advantage of the many available cell type-specific Cre driver alleles. Because control by a single recombinase defines relatively broad classes of cells, we have also generated a Flp/Cre-responsive allele which restricts hM3Dq to the narrow intersection of two broader recombinase expression domains. hM3Dq is expressed only in those cells that have expressed Flp and Cre drivers, each controlled by the promoters of different genes, thus allowing functional manipulation of cell populations that cannot be defined by a single gene.
In this manuscript, we confirm that both alleles permit expression of an hM3Dq-mCherry fusion protein detectable at single-cell resolution in live and fixed tissue from embryos or adult mice. Furthermore, we demonstrate the broad utility of the alleles in three different cell populations: noradrenergic neurons of the small, bilateral locus coeruleus and two dispersed cell populations, Camk2a+ neurons and GFAP+ glia. In a proof-of-principle experiment using our Flp/Cre-responsive allele, we demonstrate that CNO increases neuronal firing of hM3Dq-expressing locus coeruleus neurons in vitro. We then demonstrate the in vivo effectiveness of the Flp/Cre-responsive allele by inducing anxiety-like behavior observed following activation of locus coeruleus neurons (McCall et al., 2015). Next, we evaluate the in vivo utility of our Cre-responsive allele by evoking physiological signatures ascribed to activation of Camk2a+ neurons (Alexander et al., 2009) and GFAP+ glia (Agulhon et al., 2013), specifically hippocampal gamma oscillations and profound depression of body temperature, respectively. Collectively, these findings demonstrate that our hM3Dq alleles permit activation of genetically defined cell populations in a reproducible, dose-dependent manner and are valuable additions to the DREADD toolkit.
RESULTS
Generation of Flp/Cre and Cre recombinase-responsive alleles for somatodendritic targeting of hM3Dq
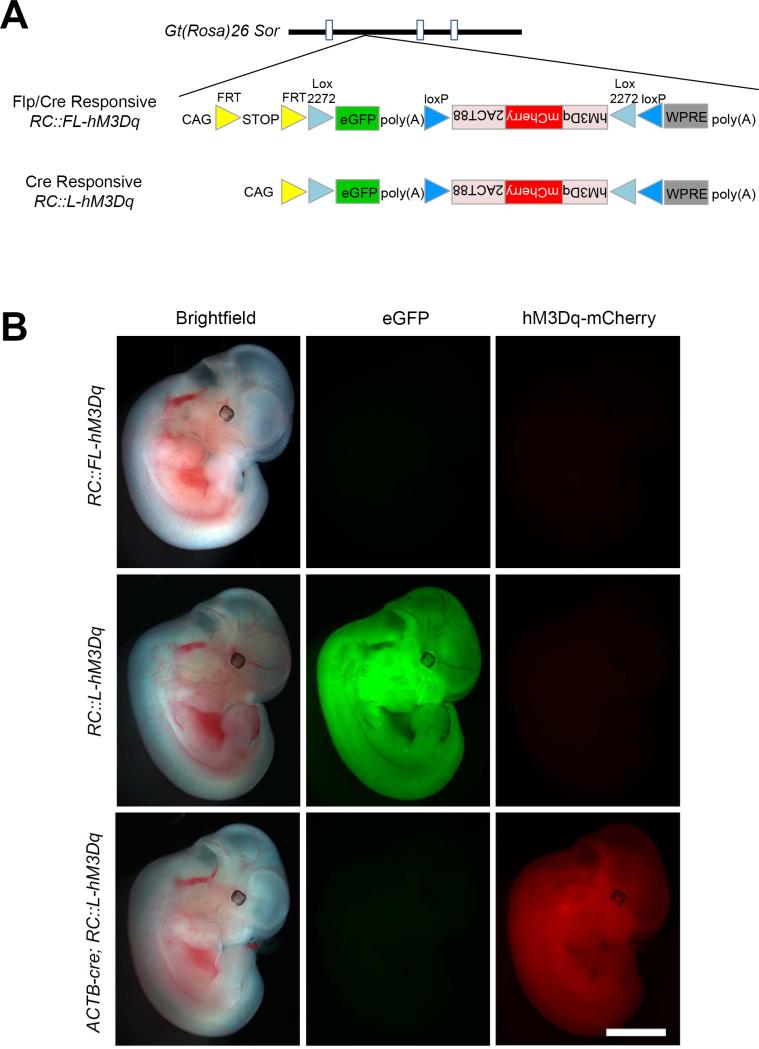
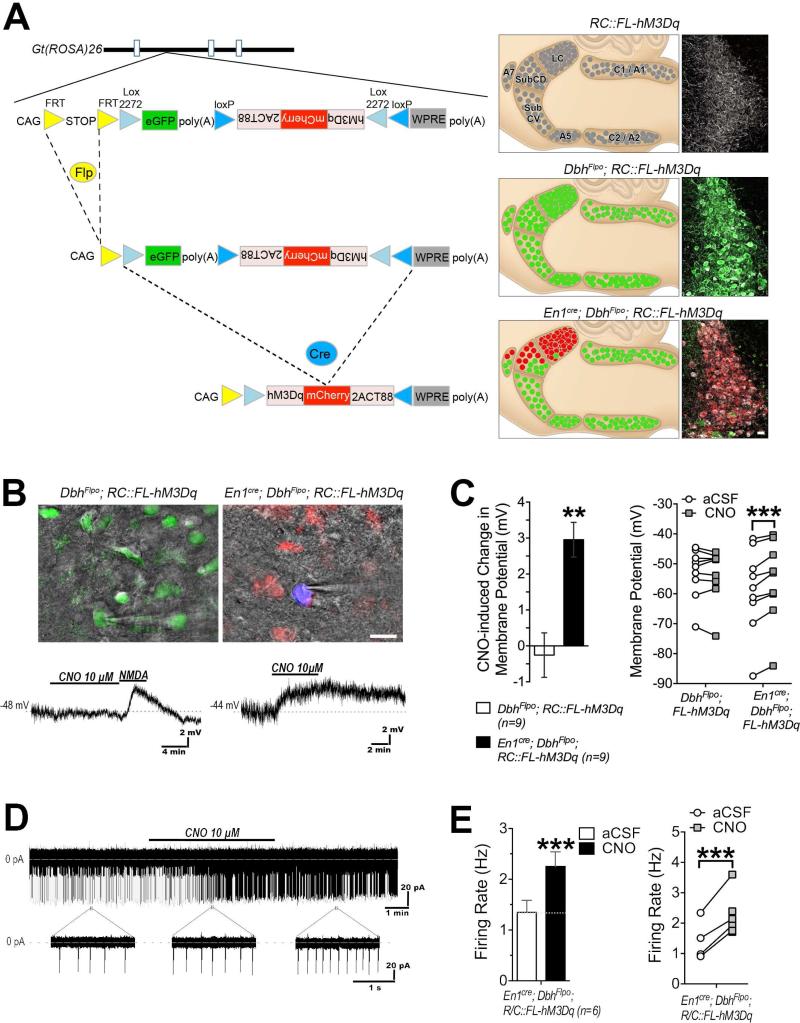
To ensure expression of our Flp/Cre-responsive (RC::FL-hM3Dq) and Cre-responsive (RC::L-hM3Dq) alleles in a wide range of cell types and developmental stages, we used a synthetic CAG promoter (Niwa et al., 1991) and targeted the Gt(ROSA)26Sor locus (Friedrich and Soriano, 1991) in mouse embryonic stem cells (Figure 1A). In RC::FL-hM3Dq (Rosa-CAG-FRT-lox-hM3Dq), a FRT-flanked transcriptional stop cassette (Sauer, 1993) following the CAG promoter prevents transcription of all downstream sequence until it is excised by Flp recombinase. The allele also encodes eGFP and an hM3Dq-mCherry fusion protein with a c-terminal epitope (2ACT88) that mediates localization of the hM3Dq receptor to the cell body (soma) and dendrites (Xia et al., 2003). To prevent expression in the absence of Cre recombination, the hM3Dq-mCherry cDNA is inverted in a Cre-dependent FLEx switch consisting of two pairs of antiparallel lox sites (Atasoy et al., 2008; Schnutgen et al., 2003). Cre activity results in inversion of hM3Dq-mCherry to the proper orientation for transcription. RC::FL-hM3Dq will express eGFP following Flp-mediated recombination and hM3Dq-mCherry after recombination by both Flp and Cre. RC::L-hM3Dq (Rosa-CAG-lox-hM3Dq) lacks the FRT-flanked stop cassette but is otherwise identical to RC::FL-hM3Dq. Therefore, RC::L-hM3Dq will express eGFP in the absence of recombinase activity and hM3Dq-mCherry after Cre-mediated recombination. Because the recombination events are irreversible, cells will express hM3Dq-mCherry continuously, even if recombinase expression is transient. Thus, RC::L-hM3Dq expresses hM3Dq-mCherry in broad populations of cells defined by a single promoter that drives Cre expression. RC::FL-hM3Dq allows restriction of hM3Dq-mCherry to more specific subpopulations defined by shared expression of Flp and Cre driven by different promoters.
Figure 1. Recombinase-responsive alleles for somatodendritic targeting of hM3Dq.
(A) Schematic diagrams of RC::FL-hM3Dq and RC::L-hM3Dq shown in relation to the Gt(ROSA)26Sor locus. In both alleles, a CAG promoter drives gene expression. RC::FL-hM3Dq expresses eGFP after Flp-mediated excision of the FRT-flanked His3-SV40 stop cassette (STOP). hM3Dq-mCherry fusion protein is expressed after excision of the stop cassette and recombination of lox2272 and loxP sites constituting the Cre-dependent FLEx switch. RC::L-hM3Dq expresses eGFP ubiquitously and hM3Dq-mCherry after Cre-mediated recombination of the FLEx switch. 2ACT88, c-terminal fragment of HTR2A; WPRE, woodchuck hepatitis virus posttranscriptional regulatory element; poly(A), bovine growth hormone polyadenylation signal. (B) Native eGFP and mCherry fluorescence in E11.5 embryos. Top: Absence of fluorescence in RC::FL-hM3Dq heterozygote confirms that the FRT-flanked stop cassette blocks expression in the absence of Flp activity. Middle: Ubiquitous eGFP fluorescence, but no mCherry, in RC::L-hM3Dq heterozygote confirms that the FLEx switch blocks hM3Dq expression in the absence of Cre activity. Bottom: Ubiquitous mCherry fluorescence observed after crossing RC:: L-hM3Dq with a ubiquitously expressed ACTB-cre transgene (Lewandoski et al., 2007) confirms recombination of the Cre-dependent FLEx switch. Scale bar: 2 mm.
To confirm that the CAG promoter is broadly active and that hM3Dq-mCherry expression is efficiently controlled by recombinase activity, we examined eGFP and mCherry fluorescence in RC::FL-hM3Dq and RC::L-hM3Dq embryos and adults. In the absence of recombinase activity, we observed neither eGFP nor mCherry expression in RC::FL-hM3Dq embryos (Figure 1B) and adult brain (Figure S1), due to the intact stop cassette and FLEx switch. Following germline excision of the FRT-flanked stop cassette (RC::L-hM3Dq), we observed ubiquitous expression of eGFP but no mCherry. Subsequent germline recombination of the FLEx switch in RC::L-hM3Dq mice resulted in ubiquitous mCherry expression in both embryos and adult brain (Figure 1B and Figure S1). Thus, the CAG promoter is active throughout development, and both the FRT-flanked stop cassette and Cre-dependent FLEx switch are capable of controlling expression of the proteins encoded by our two alleles.
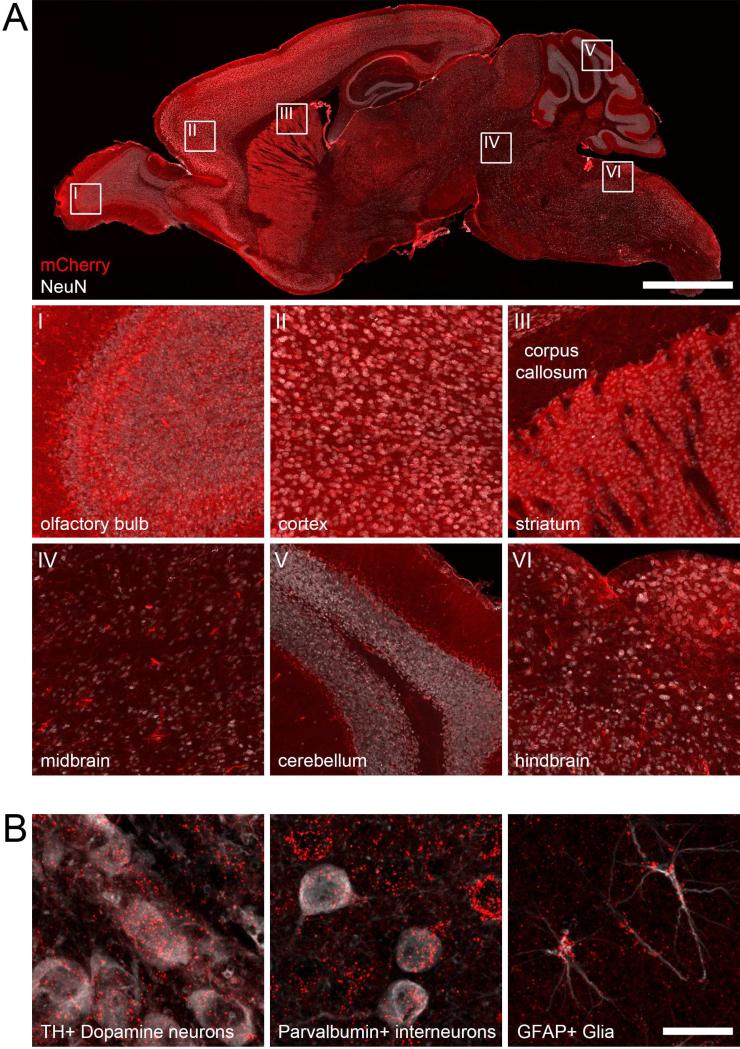
To demonstrate the widest possible expression of hM3Dq-mCherry, we crossed RC::FL-hM3Dq to the ubiquitously expressed ACTB-Flpe (Rodriguez et al., 2000) and ACTB-cre (Lewandoski et al., 1997) recombinase drivers. We observed hM3Dq-mCherry expression in a wide variety of cell types throughout the brain (Figure 2). Next, we used cell type-specific drivers to test whether our alleles can direct somatodendritic localization of hM3Dq-mCherry in restricted cell populations. We used RC::L-hM3Dq in combination with Camk2a-cre (Tsien et al., 1996) or GFAP-creERT2 (Casper et al., 2007) transgenes for expression of hM3Dq-mCherry in neurons or glia, respectively. As expected, mCherry fluorescence was restricted to the soma and dendrites of neurons in the well-documented expression domain of the Camk2a-cre-transgene (Sonner et al., 2005), including cortical and hippocampal neurons (Figure 3A-B, Figure S2). Using the glial-specific GFAP-creERT2 transgene, we observed restricted mCherry fluorescence in the soma and processes of glia defined by Gfap expression (Figure 3C, Figure S3). To demonstrate intersectional Flp/Cre control of RC::FL-hM3Dq, we targeted a population of noradrenergic neurons defined by a shared history of both En1cre and DbhFlpo expression (Robertson et al., 2013). This combination of recombinase drivers restricts hM3Dq-mCherry expression to a subpopulation of noradrenergic neurons (hereafter designated LC complex), which encompasses 99.8% of the compact locus coeruleus (LC) and a portion of the dorsal subcoeruleus and A7 noradrenergic nuclei (Robertson et al., 2013; Plummer et al., 2015). As expected, mCherry fluorescence was restricted to the soma and dendrites of the LC complex (Figure 3D, Figure S4). All remaining noradrenergic neurons, expressing only DbhFlpo, were labeled with eGFP (Figure S4).
Figure 2. Widespread expression of hM3Dq-mCherry after recombination of RC::FL-hM3Dq by ubiquitous Flp and Cre driver alleles.
(A) Sagittal section of ACTB-cre; ACTB-Flpe; RC::FL-hM3Dq brain immunolabeled for mCherry (red) and the neuronal marker NeuN (white). Boxes show location of magnified images (below). Following ubiquitous recombinase expression, mCherry is observed throughout the brain. Fluorescence is attenuated in regions with a high proportion of axons (e.g. corpus callosum), consistent with somatodendritic targeting of hM3Dq-mCherry. Scale bar: 2000 μm (full sagittal section) or 244 μm (magnified images). (B) Sections of ACTB-cre; ACTB-Flpe; RC::FL-hM3Dq brain immunolabeled for mCherry (red) and markers of neuronal and glial subtypes (white). hM3Dq-mcherry is observed in midbrain dopaminergic neurons labeled for tyrosine hydroxylase (TH), cortical interneurons labeled for parvalbumin, and astrocytes labeled for glial fibrillary acidic protein (GFAP). Scale bar: 20 μm.
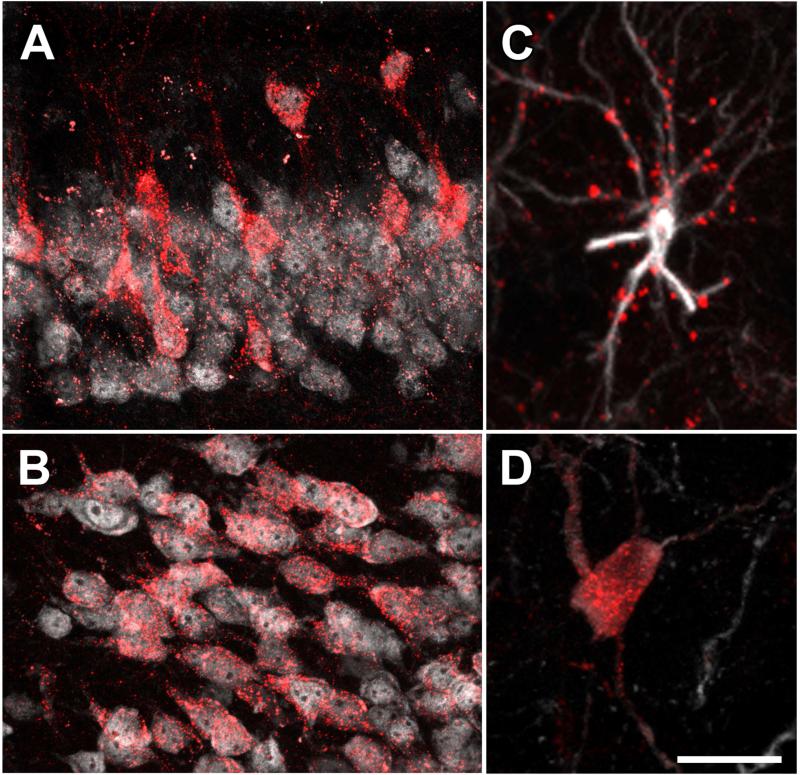
Figure 3. Somatodendritic targeting of hM3Dq-mCherry in neurons and glia.
(A) Hippocampal CA3 neurons in a Camk2a-cre; RC::L-hM3Dq brain immunolabeled for mCherry (red) and nuclear NeuN (white). (B) Cortical neurons from a Camk2a-cre; RC::L-hM3Dq brain immunolabeled for mCherry (red) and NeuN (white). (C) Cortical astrocyte from a GFAP-CreERT2; RC::L-hM3Dq brain immunolabeled for mCherry (red) and GFAP (white). (D) Noradrenergic neuron from an En1Cre; DbhFlpo; RC::FL-hM3Dq brain immunolabeled for mCherry (red) and tyrosine hydroxylase (white). Images taken from coronal sections. Scale bar: 40 μm (A,B) or 20 μm (C,D).
Unexpectedly, we observed faint mCherry fluorescence in cerebellar Purkinje cells that have expressed En1cre but not DbhFlpo (Figure S5A). The absence of ectopic eGFP expression suggested that the ectopic mCherry results from mRNA splicing that occurs after Cre-mediated recombination of the FLEx switch, rather than failure of the FRT-flanked stop cassette. To test this hypothesis, we performed RT-PCR analysis of cerebellum RNA and identified mRNA from RC::FL-hM3Dq in cells that have recombined the FLEx switch (Figure S5B). Due to a cryptic splice acceptor within the hM3Dq cDNA, this mRNA encodes mCherry but not the full hM3Dq-mCherry fusion. These experiments confirm that our alleles faithfully express hM3Dq-mCherry in the defined recombinase expression domains, demonstrating they can deliver hM3Dq to a variety of cell types limited only by the availability of Flp and Cre driver lines.
Selective hM3Dq-mediated activation of noradrenergic neurons in vitro
To evaluate whether the level of hM3Dq expressed from RC::FL-hM3Dq is sufficient to alter cellular activity, we carried out whole-cell patch clamp recordings of noradrenergic neurons in the LC complex that express either hM3Dq-mCherry (En1cre; DbhFlpo; RC::FL-hM3Dq triple heterozygotes) or eGFP (DbhFlpo; RC::FL-hM3Dq controls) (Figure 4A). Fluorescence in the LC complex was readily detected in acute slice preparations (Figure 4B). To evaluate whether CNO would depolarize hM3Dq-expressing neurons, we recorded in the presence of the Na+ channel blocker TTX (500 nM) or TTX plus the L-type Ca++ channel blocker nimodipine (1 μm) to eliminate network effects and oscillations. Average resting membrane potential was −56.0±11.0 mV, consistent with prior observations of locus coeruleus neurons (Williams et al., 1984; de Oliveira et al., 2010). Bath application of 10 μM CNO depolarized hM3Dq-mCherry+ LC complex neurons recorded in TTX (4.04±0.91 mV, n=5 neurons) or TTX+Nimodipine (1.60±0.27 mV, n=4 neurons). The depolarization measured under the two recording conditions was not statistically different (P=0.20), and therefore data were combined, yielding an average depolarization of 2.95±0.48 mV (Figure 4C). In contrast, CNO had no effect on membrane potential of eGFP+ control neurons (Figure 4B-C). Depolarization by 50 μM NMDA, however, confirmed that these cells were healthy and responsive (Figure 4B). In the absence of CNO, hM3Dq-mCherry+ and eGFP+ neurons exhibited similar baseline membrane potential, capacitance, and membrane and input resistance (Figure 4C and data not shown), suggesting that mere expression of hM3Dq-mCherry does not impact membrane properties. To test whether expression of hM3Dq is sufficient to evoke neuronal firing, we next performed cell-attached recordings in the absence of TTX and nimodipine. Bath application of CNO (10 μM) increased the firing rate of hM3Dq-mCherry+ LC neurons by an average of 75.6±30.0% above baseline activity (0.91±0.08 Hz; n=6 cells; p<0.001; Figure 4D-E). To confirm that the ectopically labeled Purkinje cells do not express functional hM3Dq-mCherry fusion protein, we also performed whole-cell recordings of these cells. Consistent with our identification of a nonfunctional mRNA, membrane potential of mCherry+ Purkinje cells was not altered by 10 μM CNO (Figure S5C). Taken together, these results demonstrate that the level of hM3Dq-mCherry expressed from RC::FL-hM3Dq permits chemogenetic activation of an intersectional subpopulation defined by expression of Cre and Flp.
Figure 4. Intersectional genetic strategy for selective, noninvasive expression of hM3Dq and activation of the LC complex.
(A) Schematic diagram of RC::FL-hM3Dq, sagittal diagram of noradrenergic nuclei in the hindbrain, and coronal sections through the LC complex immunolabeled for tyrosine hydroxylase (white), eGFP (green), and mCherry (red). Top: In the absence of recombinase expression, noradrenergic neurons are not labeled by eGFP or mCherry. Middle: Recombination of RC::FL-hM3Dq by DbhFlpo results in eGFP expression in noradrenergic neurons. Bottom: Recombination of RC::FL-hM3Dq by En1cre and DbhFlpo results in hM3Dq-mCherry expression in noradrenergic neurons of the LC complex. (B) Top: Acute hindbrain slices show endogenous expression of eGFP (DbhFlpo; RC::FL-hM3Dq) and hM3Dq-mCherry (En1cre; DbhFlpo; RC::FL-hM3Dq) in LC complex neurons patched using Alexa350-filled electrodes (blue). Scale bar: 20 μm. Bottom: Representative traces from whole-cell recordings taken from LC neurons with TTX (500 nM) and nimodipine (1μM) present. Bath application of CNO (10 μM) depolarizes hM3Dq-mCherry+ neurons from resting membrane potential (dashed line). CNO has no effect on membrane potential of eGFP+ neurons, but NMDA (50 μM) does induce depolarization. (C) Left: Average mV depolarization by CNO (10 μM) from baseline recorded in hM3Dq-mCherry+ (En1cre; DbhFlpo; RC::FL-hM3Dq, n=9 cells from 7 mice) and eGFP+ (DbhFlpo; RC::FL-hM3Dq, n=9 cells from 4 mice) LC complex neurons. **p<0.01 (t-test). Right: Population data show the effect of CNO (10 μM) on membrane potential in individual mCherry+ and eGFP+ LC neurons. ***p<0.001 (Bonferroni). (D) Representative trace of cell-attached recordings from an hM3Dq-mCherry+ neuron. Insets of trace show individual firing events in epochs occurring before (left), during (middle), and after (right) superperfusion of CNO. Dashed line indicates the average holding current. (E) Left: Bath application of CNO (10 μM) increases firing rate of hM3Dq-mCherry+ neurons (En1cre; DbhFlpo; RC::FL-hM3Dq, n=6 cells from 3 mice). Right: Population data show the effect of CNO (10 μM) on firing rate in individual hM3Dq-mCherry+ LC neurons. ***p<0.001 (t-test).
In vivo activation of the LC complex induces anxiety-like behavior and suppresses locomotion
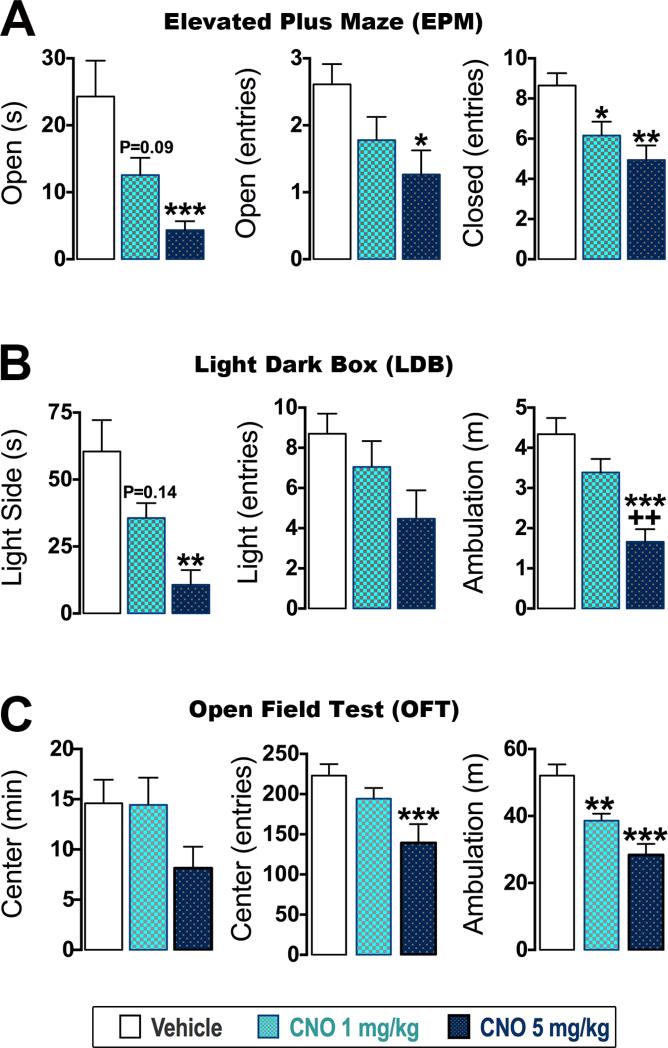
We assessed the in vivo utility of our dual recombinase-responsive allele by testing whether expression of hM3Dq can alter anxiety-like behavior. Optogenetic stimulation of LC neurons at high, tonic frequency (5 Hz) prior to testing evokes anxiety-like behavior and suppresses locomotion (McCall et al., 2015). To test whether hM3Dq would evoke similar responses, we treated mice with CNO (1 mg/kg or 5 mg/kg, IP) or vehicle before testing in three assays: the elevated plus maze (EPM), light dark box (LDB), and open field test (OFT). In all three assays, CNO dose-dependently evoked anxiety-like responses and concomitantly suppressed locomotion of mice expressing hM3Dq-mCherry in the LC complex (Figure 5; Table S1). CNO at 5 mg/kg significantly induced anxiety-like behavior by reducing exploration within the aversive areas of the tests, including open arms of the EPM (Figure 5A), light compartment of the LDB (Figure 5B), and center zone of the OFT (Figure 5C and Figure S6) compared to vehicle. Both doses of CNO also suppressed locomotion, as shown by reduced closed arm entries in the EPM and distance ambulated in LDB and OFT compared to vehicle (Figure 5). CNO had no effect on behavioral performance of all littermate controls (Figure S7). In addition, mice expressing hM3Dq and all littermate controls were indistinguishable when treated with vehicle (Figure S7). Collectively, these data demonstrate that RC::FL-hM3Dq faithfullyexpresses hM3Dq at levels that permit sensitive control of an intersectionally targeted neuronal subpopulation and recapitulates behavioral phenotypes evoked by optogenetic stimulation.
Figure 5. CNO is sufficient to drive anxiety-like behavior and suppress locomotion in mice expressing hM3Dq-mCherry in the LC complex.
(A-C) Behavioral data from elevated plus maze (EPM), light-dark box (LDB), and open field test (OFT). Data are Mean ± SEM for mice given vehicle (n=18-21), CNO 1-mg/kg (n=16-20), and CNO 5-mg/kg (n=13-15) treated En1cre; DbhFlpo; RC::FL-hM3Dq mice. ***p<0.001, **p<0.01, *p<0.05 vs. Vehicle (Bonferroni). ++p<0.01 vs. Vehicle and 1-mg/kg CNO (Bonferroni). Exact p values list trends for a statistical difference versus vehicle.
Dose-dependent activation of hM3Dq drives gamma oscillations in hippocampal neurons in vivo
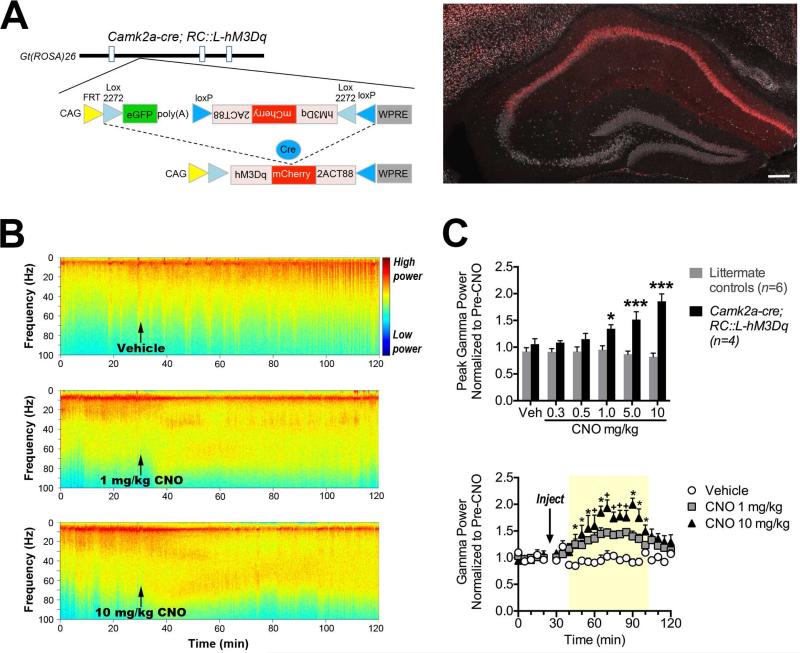
Next, we assessed the efficacy of our single recombinase-responsive allele by testing whether hM3Dq can activate a dispersed neuronal population. In the original description of a tetracycline-controlled hM3Dq transgene, hippocampal gamma power was progressively evoked in Camk2a+ neurons following escalating doses of CNO (Alexander et al., 2009). To determine whether our Cre-responsive allele would evoke similar neural activity, we used the Camk2a-cre transgene (Tsien et al., 1996) to drive hM3Dq-mCherry expression (Figure 6A). We recorded local field potentials in CA3 of the hippocampus of mice expressing hM3Dq-mCherry in Camk2a+ neurons (Camk2a-cre; RC::L-hM3Dq) and littermate controls. Consistent with the original study, we found that CNO dose-dependently increased hippocampal gamma power (50-80 Hz) in mice expressing hM3Dq-mCherry (Figure 6B-C). Doses of CNO as low as 1 mg/kg increased peak gamma power compared to vehicle. The 10-mg/kg dose significantly increased gamma power 20 minutes following CNO injection, and gamma returned to baseline levels an hour thereafter (Figure 6C). In all littermate controls, CNO had no effect on peak gamma power (Figure 6C). These results confirm that RC::L-hM3Dq can be used to efficiently and precisely control the activity of neuronal populations at levels measurable by in vivo electrophysiology.
Figure 6. Dose-dependent induction of hippocampal gamma oscillations by CNO in mice expressing hM3Dq-mCherry in Camk2a+ neurons.
(A) Schematic diagram shows Cre-mediated recombination of the FLEx switch permitting hM3Dq-mCherry expression. Coronal section through the hippocampus of a Camk2a-cre; RC::L-hM3Dq double heterozygote, immunolabeled for mCherry (red) and NeuN (white). Scale bar: 500 μm. (B) Spectrograms show frequency composition of local field potentials (LFP) recorded from Camk2a-cre; RC::L-hM3Dq mice before, during and after administration of vehicle or CNO at 1 or 10 mg/kg i.p. (C) LFP data were divided into five-minute time bins, and power spectral densities were computed. Pre-CNO power was the average gamma power (50-80 Hz) across 0-25 min. The 25-30 min bin was not included for analysis because mice were handled and injected. Gamma power post-CNO injection was normalized to pre-CNO power. Peak gamma power was averaged across 65-80 min bins (i.e., 35-50 min post-CNO) and normalized to pre-CNO. Mice were administered all doses listed. Top: Time course of gamma power for vehicle- and CNO-treated Camk2a-cre; RC::L-hM3Dq double heterozygotes. The yellow window indicates the time period during which CNO increased gamma power. N=4 mice. Bottom: Peak gamma power following CNO injection for Camk2a-cre; RC::L-hM3Dq and littermate control mice demonstrates dose dependence. ***p<0.001, *p<0.05 vs. Vehicle (Bonferroni). +p<0.05 vs. both CNO 1 mg/kg and Vehicle (Bonferroni).
In vivo activation of hM3Dq expressed in glia elicits hypothermia
We further assessed the broad utility of RC::L-hM3Dq by targeting a dispersed population of non-neuronal cells. Activation of hM3Dq expressed in glia by a GFAP-hM3Dq transgene has previously been shown to induce hypothermia (Agulhon et al., 2013). To determine whether RC::L-hM3Dq can recapitulate this effect, we used the tamoxifen-inducible GFAP-creERT2 transgene (Casper et al., 2007) to drive hM3Dq-mCherry expression in glia (Figure 7A). Hypothermia was observed in mice expressing hM3Dq-mCherry in Gfap+ glia starting at 40 minutes after injection of CNO (5 mg/kg) and persisted until the test was stopped at 90 minutes due to dangerously low body temperature (Figure 7B). In contrast, CNO did not alter body temperature in littermate controls (Figure 7C). Furthermore, we observed no effect of vehicle on body temperature (Figure 7B-C). Collectively, these data demonstrate that RC::L-hM3Dq permits activation of dispersed non-neuronal populations at levels sufficient to evoke lasting physiological changes.
Figure 7. Activation of hM3Dq-mCherry in glia is sufficient to elicit hypothermia.
(A) Schematic diagram shows Cre-mediated recombination of the FLEx switch permitting hM3Dq-mCherry expression in glia of mice treated with tamoxifen. (B) Decreased body temperature in GFAP-creErt2; RC::L-hM3Dq double heterozygotes following CNO. Data are Mean ± SEM for mice given vehicle (n=5) or CNO 1 and 5 mg/kg (n=6). *p<0.05 vs. vehicle (Bonferroni). +p<0.05 vs. vehicle and CNO 1 mg/kg (Bonferroni). (C) CNO has no effect on the body temperature of littermate controls. +p<0.05 vs. vehicle and CNO 1 mg/kg (Bonferroni).
DISCUSSION
The experiments described here demonstrate that our Flp/Cre- and Cre-responsive hM3Dq alleles are valuable additions to the DREADD toolkit. The combination of CAG promoter and fusion protein for somatodendritic targeting of hM3Dq drives robust expression that is readily detected in live and fixed tissue across development. As demonstrated by our experiments, this level of expression is sufficient to non-invasively and dose-dependently manipulate cellular activity in diverse cell types, producing behavioral and physiological changes without undesired phenotypes associated with hM3Dq overexpression. A primary strength of DREADD technology is the ability to manipulate cellular activity in freely moving animals, but this advantage is compromised by strategies that require invasive delivery of the DREADD. For instance, viral injection of deep brain structures can result in significant damage to overlying parenchyma and even death of experimental animals. In contrast, our recombinase responsive hM3Dq alleles offer a truly non-invasive means to activate genetically defined cells.
These alleles offer several important advantages not shared by published hM3Dq alleles. RC::FL-hM3Dq offers intersectional genetic control of hM3Dq expression, thus providing increased spatial resolution by precise targeting of hM3Dq to cell populations defined by a shared history of Cre and Flp recombinase expression. Unlike existing Cre-responsive hM3Dq knock-in alleles (Teissier et al., 2015; Jackson Laboratory Stock #026220, unpublished reagent from U. Hochgeschwender and B. Roth), RC::FL-hM3Dq and RC::L-hM3Dq utilize a FLEx switch for efficient control of expression, an mCherry fusion for endogenous fluorescent labeling, and a 2ACT88 epitope for somatodendritic targeting. These features ensure reliable expression of hM3Dq at levels that effectively permit both in vivo visualization and activation of genetically defined cells.
Our characterization of RC::FL-hM3Dq and RC::L-hM3Dq demonstrated that somatodendritic expression of hM3Dq can be obtained in virtually any cell population across tissues and developmental stages. We observed the expected expression of mCherry and eGFP, confirming that the FRT-flanked stop cassette and Cre-dependent FLEx switch provide tight recombinase-responsive control of hM3Dq expression. Unexpectedly, we observed ectopic mCherry fluorescence in Cre+, Flp− cerebellar Purkinje cells of En1cre; DbhFlpo; RC::FL-hM3Dq mice. However, our RT-PCR and in vitro electrophysiology experiments unequivocally demonstrated that functional hM3Dq-mCherry is limited to the intersectional population of noradrenergic neurons that share a history of both Flp and Cre expression. Restriction of ectopic mCherry to a single cell type within the broad En1cre expression domain suggests that any ectopic fluorescence in other intersectional crosses will be similarly limited and will not interfere with experiments.
We validated the in vivo functional capabilities of RC::FL-hM3Dq and RC::L-hM3Dq in three distinct cell types. Activation of hM3Dq expressed bilaterally in the LC complex dose-dependently evoked anxiety-like behavior and suppressed locomotion, consistent with results following unilateral high tonic (5 Hz) optogenetic stimulation of the locus coeruleus (McCall et al., 2015). The subtle changes in anxiety and locomotion we observed were not obscured by the complete behavioral arrest that was reported following prolonged, high frequency optogenetic stimulation (>5 Hz) (Carter et al., 2010). Using RC::L-hM3Dq, we dose-dependently induced gamma oscillations in Camk2a-cre+ hippocampal neurons, consistent with results using the tetracycline-controlled TRE-hM3Dq transgene (Alexander et al., 2009). Notably, the changes in neural activity we observed were not accompanied by limbic seizures, an extreme phenotype observed using the TRE-hM3Dq transgene. Additionally, activation of hM3Dq in GFAP+ glia dose-dependently evoked long-lasting changes in body temperature, consistent with prior results (Agulhon et al., 2013).
While all of these phenotypic changes emerged on the timescale of minutes following activation of hM3Dq, their onset differed across experiments. Given that systemically administered CNO is rapidly observed in the brain (Bender et al. 1994), this varied onset of phenotype is likely dependent on the targeted cell type, phenotypic endpoint, and CNO dose employed. Our findings further suggest that levels of hM3Dq expression also influence the onset of phenotype. For example, the hypothermia we observed was slower to emerge than previously described using a GFAP-hM3Dq transgene (Agulhon et al 2013). This result, together with the absence of extreme phenotypes like seizures or behavioral arrest in the other crosses, is likely due to lower levels of hM3Dq expression from our single-copy knock-in allele compared to transgenic or viral delivery. Taken together, these findings indicate that our hM3Dq alleles modulate cellular activity to produce physiologically relevant changes in behavior without untoward effects that impede or confound interpretation.
In summary, RC::FL-hM3Dq and RC::L-hM3Dq take full advantage of the power of DREADD technology to non-invasively manipulate cellular activity. Our characterization of these alleles demonstrates their capacity to target genetically defined cell populations, regardless of anatomical location or distribution. In addition, the ability to express hM3Dq at any time-point offers the opportunity to uncover the functional consequences of cellular activation throughout development. Together, these capabilities render the hM3Dq alleles broadly useful for controlling the activity of diverse cell types to uncover their function and dysfunction.
EXPERIMENTAL PROCEDURES
Procedures related to the use of mice were conducted according to NIH guidelines and the National Research Council Guide for the Care and Use of Laboratory Animals (Publication No. 85-23, revised 2013). The Institutional Animal Care and Use Committee (IACUC) of the National Institute of Environmental Health Science and University of North Carolina approved all experiments. Detailed methods are provided in the Supplemental Experimental Procedures.
Generation of Mouse Lines
To generate our targeting vector for homologous recombination in embryonic stem cells, we first modified a Cre-dependent FLEx switch (Atasoy et al., 2008; Schnutgen et al., 2003) by insertion of an eGFP cDNA and rabbit β-globin polyadenylation cassette between the 5’ lox2272 and loxP sites. We amplified FRT-flanked and rox-flanked His3-SV40 transcriptional stop cassettes, using as the PCR template a pBS302 plasmid (Sauer, 1993) modified to remove the internal MfeI site. The stop cassettes and FLEx switch containing eGFP were then cloned into the Gt(ROSA)26Sor targeting vector pRC-RFLTG (Plummer et al., 2015) after digestion with MluI and FseI. The digested fragment of pRC-RFLTG provided a CAG promoter (Niwa et al., 1991), woodchuck hepatitis virus posttranscriptional regulatory element (WPRE) (Zufferey et al., 1999), bGH poly(A) cassette, the 5’ and 3’ homology to the Gt(ROSA)26Sor locus, and attB/attP-flanked Neo cassette. A cDNA encoding the hM3Dq-mCherry-2ACT88 fusion protein, which contains the final 264 coding nucleotides of the rat Htr2A gene (Xia et al., 2003), was generously provided by Bryan Roth (University of North Carolina). This cDNA was cloned into the center of the modified FLEx switch, in antisense orientation relative to the CAG promoter and eGFP cDNA, to produce the final targeting vector pRC-RFL-hM3Dq.
Linearized pRC-RFL-hM3Dq was electroporated into G4 embryonic stem cells (George et al., 2007) obtained from the Lunenfeld-Tanenbaum Research Institute (Toronto, Canada), and long range PCR and Southern blotting identified homologous recombinants. A recombinant clone was transiently transfected with pPGKPhiC31obpa (Raymond and Soriano, 2007) to remove the attb-attp-flanked Neo cassette, and two subclones were injected into B6(Cg)-Tyrc-2J/J blastocysts to produce chimeric mice. The RC::FL-hM3Dq strain was established by breeding chimeras with B6;129-Tg(CAG-dre)1Afst mice (Anastassiadis et al., 2009) to excise the rox-flanked stop cassette, and the RC::L-hM3Dq strain was established by crossing RC::FL-hM3Dq with B6.Cg-Tg(ACTFlpe)9205Dym/J mice (ACTB-Flpe) (Rodriguez et al., 2000) to excise the FRT-flanked stop cassette. Both strains were maintained by backcrossing to C57BL/6J mice.
Experimental Crosses
To demonstrate ubiquitous expression of eGFP and hM3Dq-mCherry encoded by our DREADD alleles, we crossed RC::FL-hM3Dq mice with ACTB-Flpe followed by B6;FVB-Tg(ACTB-cre)2Mrt (ACTB-cre) (Lewandoski et al., 1997). Embryos were collected at embryonic day (E) 11.5, and adult organs were collected from 10-week old mice. RC:L-hM3Dq mice were separately crossed with B6(C3)-Tg(GFAP-cre/ERT2)13Kdmc (Casper et al., 2007) and B6.Cg-Tg(Camk2a-cre)T29-1Stl (Tsien et al., 1996) mice. To generate En1cre; DbhFlpo; RC::FL-hM3Dq mice, RC::FL-hM3Dq mice were first crossed to B6;129-Dbhtm1(Flpo)Pjen (DbhFlpo) heterozygotes (Robertson et al., 2013), and offspring were backcrossed to generate animals heterozygous for DbhFlpo and homozygous for RC::FL-hM3Dq. The mice were subsequently crossed with B6.Cg-En1tm2(cre)Wrst (En1cre) heterozygotes (Kimmel et al., 2000) to generate En1cre; DbhFlpo; RC::FL-hM3Dq triple heterozygotes.
Tissue Collection and Immunohistochemistry
Embryos were fixed overnight by immersion in 4% paraformaldehyde (PFA) in 0.01 M phosphate-buffered saline (PBS) at 4°C. Adult mice were anesthetized with sodium pentobarbital (0.1 mL of 50 mg/mL i.p.) and perfused transcardially with PBS followed by 4% PFA in PBS. Brains were postfixed overnight by immersion in 4% PFA at 4°C, and then rinsed in PBS before transfer to 30% sucrose in PBS for 48 hours at 4°C. Tissue for immunohistochemistry was embedded in Tissue Freezing Medium (General Data Company, Cincinnati, OH) and sectioned at 40-μm on a Leica CM3050-S cryostat (Leica Biosystems, Buffalo Grove, IL). Free-floating sections were collected for immunolabeling using antibodies described in Supplemental Experimental Procedures. After staining, sections were mounted on microscope slides and coverslips were attached with Vectashield or Vectashield plus DAPI (Vector Laboratories, Burlingame, CA) before imaging.
Digital Image Processing
Images of native fluorescence in whole embryos and adult brains were collected on a Zeiss SteREO Lumar.V12 stereomicroscope (Carl Zeiss Microscopy, Thornwood, NY). Images of the LC complex from acute slices were acquired using a Zeiss Axio Examiner microscope and camera (Axiocam 503) equipped with Zen 2012 Blue Software (Carl Zeiss). The images of acute slices were processed with ImageJ software (US National Institutes of Health) by merging color channels, adjusting brightness and contrast, and applying a smoothing filter. Images of immunofluorescent-labeled sections were collected on a Zeiss LSM 710, 780, or 880 inverted confocal microscope. Zen 2012 Black Software (Carl Zeiss) was used to convert z-stacks to maximum intensity projections. Images were modified only by brightness and contrast adjustments to optimize the full dynamic range of the fluorescence signal. Anatomical location was confirmed by reference to a mouse brain atlas (Paxinos and Franks, 2013).
Slice Electrophysiology
Following anesthesia of En1cre; DbhFlpo; RC::FL-hM3Dq and littermate mice, coronal hindbrain slices (250 μm) were cut in ice-cold cutting solution and incubated at 28-30°C in oxygenated aCSF for at least 30 min. All solutions are listed in the Supplemental Experimental Procedures. Slices were transferred to a recording chamber and perfused with aCSF (32°C) at 2 mL/min. Patch electrodes (1-5 MΩ) were pulled from borosilicate glass. To determine whether CNO (10 μM) would alter membrane potential, whole-cell current-clamp recordings were made from eGFP+ and hM3Dq-mCherry+ LC neurons in the presence of TTX (500 nM) and nimodipine (1 μM). To confirm the absence of functional hM3Dq, recordings were similarly made from mCherry+ cerebellar Purkinje cells. Cell-attached voltage-clamp recordings were also made from hM3Dq-mCherry+ LC neurons to quantify the increase in firing events elicited by CNO (10 μM). Data were collected at 10 kHz using an EPC 800 amplifier (HEKA) and DigiData 132x digitizer (Molecular Devices). Analyses were performed in ClampFit 10.4 on traces that were filtered at 3 Hz (voltage traces) or 1 Hz (current traces).
Anxiety-like Behavior
En1cre; DbhFlpo; RC::FL-hM3Dq and littermate mice were randomly assigned to receive two i.p. injections of vehicle or CNO (1 or 5 mg/kg). Mice received the first injection immediately before the open field test, and the second injection was 2-3 days later, approximately 15 min before the light dark box. Mice were tested in the elevated plus maze directly after the light dark box. Details of the tests are described below and in Supplemental Experimental Procedures.
Open Field Test
Mice were placed in an open arena (27 × 27 × 20 cm, Med Associates) that was not illuminated (0 lux). Exploration in the center (14.3 × 14.3 cm) and remaining periphery of the arena was measured for 90 min.
Light Dark Box
Mice were allowed to freely explore an arena (27 × 27 × 20 cm, Med Associates) that was divided into two equal compartments. Exploration in the dark (0 lux) and light compartment (~950 lux) of the arena was measured for 10 min.
Elevated Plus Maze
Mice were placed on a ‘+’ shaped maze that was brightly illuminated (300 lux) and elevated 20”. Exploration in the open (11 × 2 inch) and closed arms of the maze (11 × 2 × 5”) was measured for 5 min, as previously described (Sciolino et al., 2015).
In Vivo Electrophysiology
A 10-channel array (44-μm polyimide-coated steel wires, Sandvic), soldered to circuit board (San Francisco Circuits) and connector (Omnetics), was implanted into the hippocampus of anesthetized Camk2a-cre; RC::L-hM3Dq and littermate control mice using the following stereotaxic coordinates (mm): −2.06 anteroposterior, 2.1 mediolateral, 2.1 dorsoventral from bregma (Paxinos and Franks, 2013). Local field potentials were recorded using a 32-channel wireless headstage (Triangle BioSystems International) and Cerebus acquisition system (Blackrock Microsystems). During recordings, mice were placed in a plexiglass chamber (20” × 20”) and given i.p. injection of vehicle or the following escalating doses of CNO: 0.03, 0.1, 0.3, 0.5, 1, 5 and 10 mg/kg, as previously described (Alexander et al. 2009). All mice received each dose of CNO with 2-3 days between treatments. Analyses were performed in Matlab R2014a (MathWorks Inc.) using Chronux software on traces that were band-pass filtered (0.3-500 Hz) and stored at 1 kHz. Details are provided in the Supplemental Experimental Procedures.
Tamoxifen Dosing
Tamoxifen (T5648, Sigma Aldrich, St. Louis, MO) was dissolved in 10:1 corn oil:ethanol at a concentration of 20 mg/ml (w./v.). To induce Cre-mediated recombination, GFAP-creERT2; RC::L-hM3Dq double heterozygous mice and littermate controls received five daily doses of tamoxifen (100 mg/kg i.p., 0.05 ml/10 g body weight) during postnatal week 6 to 12.
Body Temperature
Tamoxifen-treated GFAP-creErt2; RC::L-hM3Dq and littermate mice were briefly restrained for baseline measurement of body temperature using a rectal probe. Following i.p. injection of vehicle or CNO (1 or 5 mg/kg), body temperature was measured in 10 min intervals for 90 min. A heating pad was provided to hypothermic mice after testing. All mice were tested twice with at least 5 days of recovery between the tests.
Data Analysis and Statistics
All data are reported as mean ± SEM. Datasets met normality (Shapiro-Wilk test) and equal variance (Levine test) assumptions. Differences between groups were determined using t-tests or ANOVA. Bonferroni post-hoc tests were performed when necessary to correct for multiple comparisons. Significance for all analyses was set to p<0.05. Analyses were performed using GraphPad Prism 6.0, Matlab R2014a, and IBM SPSS 21.0. Details for the analyses are described in the Supplemental Experimental Procedures.
Supplementary Material
HIGHLIGHTS.
Recombinase-responsive mice express hM3Dq DREADD for induction of cellular activity
Targeting of hM3Dq to cell body and dendrites to increase effective expression levels
mCherry fusion permits visualization of hM3Dq+ cells in vivo
Dose-dependent activation of hM3Dq drives behavioral and physiological changes
ACKNOWLEDGEMENTS
We thank D. D'Agostin, G. Jones, and K. Smith for technical assistance. We thank B. Roth (University of North Carolina) for providing the hM3Dq-mCherry-2ACT88 cDNA. We also thank J. Haam, C. Erxleben, C. Hull (Duke University) and D. Weinshenker (Emory University) for helpful advice. Valuable support was provided by the NIEHS Comparative Medicine Branch, Knockout Mouse Core, Biostatistics and Computational Biology Branch, and Fluorescence Microscopy and Imaging Core. We thank the NIDA Drug Supply Program for providing the CNO. This research was supported by the Intramural Research Program of the US National Institutes of Health, National Institute of Environmental Health Sciences (ES102805 to PJ and ES100221 to SD).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS
NWP and PJ conceived the project and designed the RC::FL-hM3Dq and RC::L-hM3Dq lines. NWP generated the targeting vector. NRS, NWP, YC, SDR, and PJ characterized the mouse lines. NRS and ZAM performed the slice recordings. NRS and PJ conducted the behavioral tests. GMA performed in vivo electrophysiology with assistance from NRS. SD supported GMA. YC and PJ performed the temperature recordings. NRS, YC, GMA, and ZAM performed statistical data analysis. NRS, NWP and PJ wrote the manuscript with input from coauthors.
REFERENCES
- Agulhon C, Boyt KM, Xie AX, Friocourt F, Roth BL, McCarthy KD. Modulation of the autonomic nervous system and behaviour by acute glial cell Gq protein-coupled receptor activation in vivo. J. Physio. 2013;591:5599–5609. doi: 10.1113/jphysiol.2013.261289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander GM, Rogan SC, Abbas AI, Armbruster BN, Pei Y, Allen JA, Nonneman RJ, Hartmann J, Moy SS, Nicolelis MA, et al. Remote control of neuronal activity in transgenic mice expressing evolved G protein-coupled receptors. Neuron. 2009;63:27–39. doi: 10.1016/j.neuron.2009.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anastassiadis K, Fu J, Patsch C, Hu S, Weidlich S, Duerschke K, Buchholz F, Edenhofer F, Stewart AF. Dre recombinase, like Cre, is a highly efficient site-specific recombinase in E. coli, mammalian cells and mice. Dis. Model. Mech. 2009;2:508–515. doi: 10.1242/dmm.003087. [DOI] [PubMed] [Google Scholar]
- Armbruster BN, Li X, Pausch MH, Herlitze S, Roth BL. Evolving the lock to fit the key to create a family of G protein-coupled receptors potently activated by an inert ligand. Proc. Natl. Acad. Sci. USA. 2007;104:5163–5168. doi: 10.1073/pnas.0700293104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atasoy D, Aponte Y, Su HH, Sternson SM. A FLEX switch targets Channelrhodopsin-2 to multiple cell types for imaging and long-range circuit mapping. J. Neurosci. 2008;28:7025–7030. doi: 10.1523/JNEUROSCI.1954-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender D, Holschbach M, Stocklin G. Synthesis of n.c.a. carbon-11 labelled clozapine and its major metabolite clozapine-N-oxide and comparison of their biodistribution in mice. Nucl. Med. Biol. 1994;21:921–925. doi: 10.1016/0969-8051(94)90080-9. [DOI] [PubMed] [Google Scholar]
- Carter ME, Yizhar O, Chikahisa S, Nguyen H, Adamantidis A, Nishino S, Deisseroth K, de Lecea L. Tuning arousal with optogenetic modulation of locus coeruleus neurons. Nat. Neurosci. 2010;13:1526–1533. doi: 10.1038/nn.2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casper KB, Jones K, McCarthy KD. Characterization of astrocyte-specific conditional knockouts. Genesis. 2007;45:292–299. doi: 10.1002/dvg.20287. [DOI] [PubMed] [Google Scholar]
- de Oliveira RB, Howlett MC, Gravina FS, Imtiaz MS, Callister RJ, Brichta AM, van Helden DF. Pacemaker currents in mouse locus coeruleus neurons. Neuroscience. 2010;170:166–177. doi: 10.1016/j.neuroscience.2010.06.028. [DOI] [PubMed] [Google Scholar]
- Friedrich G, Soriano P. Promoter traps in embryonic stem cells: a genetic screen to identify and mutate developmental genes in mice. Genes Dev. 1991;5:1513–1523. doi: 10.1101/gad.5.9.1513. [DOI] [PubMed] [Google Scholar]
- George SH, Gertsenstein M, Vintersten K, Korets-Smith E, Murphy J, Stevens ME, Haigh JJ, Nagy A. Developmental and adult phenotyping directly from mutant embryonic stem cells. Proc. Natl. Acad. Sci. USA. 2007;104:4455–4460. doi: 10.1073/pnas.0609277104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimmel RA, Turnbull DH, Blanquet V, Wurst W, Loomis CA, Joyner AL. Two lineage boundaries coordinate vertebrate apical ectodermal ridge formation. Genes Dev. 2000;14:1377–1389. [PMC free article] [PubMed] [Google Scholar]
- Lewandoski M, Meyers EN, Martin GR. Analysis of Fgf8 gene function in vertebrate development. Cold Spring Harb. Symp. Quant. Biol. 1997;62:159–168. [PubMed] [Google Scholar]
- McCall JG, Al-Hasani R, Siuda ER, Hong DY, Norris AJ, Ford CP, Bruchas MR. CRH Engagement of the Locus Coeruleus Noradrenergic System Mediates Stress-Induced Anxiety. Neuron. 2015;87:605–620. doi: 10.1016/j.neuron.2015.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa H, Yamamura K, Miyazaki J. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene. 1991;108:193–199. doi: 10.1016/0378-1119(91)90434-d. [DOI] [PubMed] [Google Scholar]
- Paxinos G, Franks KBJ. The mouse brain in sterotaxic coordinates. Academic Press; San Diego: 2013. [Google Scholar]
- Plummer NW, Evsyukova IY, Robertson SD, de Marchena J, Tucker CJ, Jensen P. Expanding the power of recombinase-based labeling to uncover cellular diversity. Development. 2015;142:4385–4393. doi: 10.1242/dev.129981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymond CS, Soriano P. High-efficiency FLP and PhiC31 site-specific recombination in mammalian cells. PloS one. 2007;2:e162. doi: 10.1371/journal.pone.0000162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson SD, Plummer NW, de Marchena J, Jensen P. Developmental origins of central norepinephrine neuron diversity. Nat. Neurosci. 2013;16:1016–1023. doi: 10.1038/nn.3458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez CI, Buchholz F, Galloway J, Sequerra R, Kasper J, Ayala R, Stewart AF, Dymecki SM. High-efficiency deleter mice show that FLPe is an alternative to Cre-loxP. Nat. Genet. 2000;25:139–140. doi: 10.1038/75973. [DOI] [PubMed] [Google Scholar]
- Sauer B. Manipulation of transgenes by site-specific recombination: use of Cre recombinase. Methods Enzymol. 1993;225:890–900. doi: 10.1016/0076-6879(93)25056-8. [DOI] [PubMed] [Google Scholar]
- Schnutgen F, Doerflinger N, Calleja C, Wendling O, Chambon P, Ghyselinck NB. A directional strategy for monitoring Cre-mediated recombination at the cellular level in the mouse. Nat. Biotechnol. 2003;21:562–565. doi: 10.1038/nbt811. [DOI] [PubMed] [Google Scholar]
- Sciolino NR, Smith JM, Stranahan AM, Freeman KG, Edwards GL, Weinshenker D, Holmes PV. Galanin mediates features of neural and behavioral stress resilience afforded by exercise. Neuropharmacology. 2015;89:255–264. doi: 10.1016/j.neuropharm.2014.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonner JM, Cascio M, Xing Y, Fanselow MS, Kralic JE, Morrow AL, Korpi ER, Hardy S, Sloat B, Eger EI, 2nd, Homanics GE. Alpha 1 subunit-containing GABA type A receptors in forebrain contribute to the effect of inhaled anesthetics on conditioned fear. Mol. Pharmacol. 2005;68:61–68. doi: 10.1124/mol.104.009936. [DOI] [PubMed] [Google Scholar]
- Teissier A, Chemiakine A, Inbar B, Bagchi S, Ray RS, Palmiter RD, Dymecki SM, Moore H, Ansorge MS. Activity of Raphe Serotonergic Neurons Controls Emotional Behaviors. Cell Rep. 2015;13:1965–1976. doi: 10.1016/j.celrep.2015.10.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsien JZ, Chen DF, Gerber D, Tom C, Mercer EH, Anderson DJ, Mayford M, Kandel ER, Tonegawa S. Subregion- and cell type-restricted gene knockout in mouse brain. Cell. 1996;87:1317–1326. doi: 10.1016/s0092-8674(00)81826-7. [DOI] [PubMed] [Google Scholar]
- Urban DJ, Roth BL. DREADDs (designer receptors exclusively activated by designer drugs): chemogenetic tools with therapeutic utility. Annu. Rev. Pharmacol. Toxicol. 2015;55:399–417. doi: 10.1146/annurev-pharmtox-010814-124803. [DOI] [PubMed] [Google Scholar]
- Williams JT, North RA, Shefner SA, Nishi S, Egan TM. Membrane properties of rat locus coeruleus neurones. Neuroscience. 1984;13:137–156. doi: 10.1016/0306-4522(84)90265-3. [DOI] [PubMed] [Google Scholar]
- Xia Z, Hufeisen SJ, Gray JA, Roth BL. The PDZ-binding domain is essential for the dendritic targeting of 5-HT2A serotonin receptors in cortical pyramidal neurons in vitro. Neuroscience. 2003;122:907–920. doi: 10.1016/s0306-4522(03)00589-x. [DOI] [PubMed] [Google Scholar]
- Zhu H, Roth BL. DREADD: a chemogenetic GPCR signaling platform. Int. J. Neuropsychopharmacol. 2014;18 doi: 10.1093/ijnp/pyu007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zufferey R, Donello JE, Trono D, Hope TJ. Woodchuck hepatitis virus posttranscriptional regulatory element enhances expression of transgenes delivered by retroviral vectors. J. Virol. 1999;73:2886–2892. doi: 10.1128/jvi.73.4.2886-2892.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.